- 1Life Science College, Anhui Medical University, Hefei, China
- 2Bio-X Institutes, Key Laboratory for the Genetics of Developmental and Neuropsychiatric Disorders (Ministry of Education), Shanghai Jiao Tong University, Shanghai, China
- 3The Fourth Hospital of Jinan City, Taishan Medical College, Jinan, China
- 4Department of Pharmacy, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai, China
- 5The Third Affiliated Hospital, Guangzhou Medical University, Guangzhou, China
- 6Shanghai Center for Women and Children's Health, Shanghai, China
The efficacy of erlotinib treatment for advanced non-small cell lung cancer (NSCLC) is due to its action as an epidermal growth factor receptor tyrosine kinase inhibitor (EGFR-TKI). Patients treated with erlotinib experience different drug responses. The effect of germline mutations on therapeutic responses and adverse drug responses (ADRs) to erlotinib in Chinese patients requires elucidation. Sixty Han Chinese advanced non-small cell lung cancer patients received erlotinib monotherapy and, for each participant, 76 candidate genes (related to EGFR signaling, drug metabolism and drug transport pathways) were sequenced and analyzed. The single-nucleotide polymorphisms (SNPs) rs1042640 in UGT1A10, rs1060463, and rs1064796 in CYP4F11, and rs2074900 in CYP4F2 were significantly associated with therapeutic responses to erlotinib. Rs1064796 in CYP4F11 and rs10045685 in UGT3A1 were significantly associated with adverse drug reaction. Moreover, analysis of a validation cohort confirmed the significant association between rs10045685 in UGT3A1 and erlotinib adverse drug response(unadjusted p = 0.015). This study provides comprehensive, systematic analyses of genetic variants associated with responses to erlotinib in Chinese advanced non-small cell lung cancer patients. Newly-identified SNPs may serve as promising markers to predict responses and safety in erlotinib-treated advanced non-small cell lung cancer patients after chemotherapy doublet.
Introduction
Lung cancer is the leading cause of cancer-associated death worldwide (Jemal et al., 2011), of which approximately 85% is classified as non-small cell lung cancer (NSCLC). In China, more than 500 thousand deaths each year are attributed to NSCLC, with incidence rising at a rate of 26.9% (Ferlay et al., 2015; Torre et al., 2015). A high proportion of patients are diagnosed with locally advanced disease or distant metastases, making them ineligible for surgery. In these patients, prognosis for radiotherapy or chemotherapy is unsatisfactory with unfavorable side effects, resulting in a 5-year survival rate of less than 17% (Siegel et al., 2013; Canada, 2015). Thus, emergence of effective strategies for the treatment of NSCLC is urgently needed.
The last decade has witnessed significant progress in terms of understanding the biology and genomics of lung cancer. In recent years, novel therapeutic strategies developed in NSCLC that target specific genomic mutations have been firmly established and show promising efficacy. A large body of literature has demonstrated that aberrant epidermal growth factor receptor (EGFR) pathway activity is a backbone of NSCLC cancerogenesis, playing a considerable role in processes decreased in malignancies, such as cell proliferation, differentiation and migration (Bardelli et al., 2003; Sawyers, 2003; Ciardiello and Tortora, 2008). Epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs) were developed to treat patients with EGFR somatic activating mutations; they were first discovered to underpin NSCLC responsiveness to gefitinib in 2004 (Lynch et al., 2004; Mayo et al., 2012).
Erlotinib, a first generation EGFR-TKI, is economical and efficacious for advanced NSCLC patients. Pivotal trials of erlotinib demonstrated meaningful improvements in both progression-free survival (PFS) and overall response rate (ORR) compared with the standard platinum-doublet chemotherapy in EGFR mutant NSCLC patients (Hirsch et al., 2011; La Salvia et al., 2017; Yoshida et al., 2017). The BR21 study confirmed that erlotinib increased overall survival (OS) in patients who were unsuitable for other treatments, with strong efficacies in East Asians, non-smokers and patients with lung adenocarcinomas (Florescu et al., 2008). While EGFR-TKI significantly prolong OS in advanced NSCLC patients, real-world clinical evidence demonstrated considerable individual variability in both therapeutic efficacy and safety signals (Riely et al., 2009; Gainor and Shaw, 2013). Previous studies suggested that patients with deletions in EGFR exon 19 or point mutations in the L858R residue in exon 21 responded well to TKI therapy, whereas patients with the secondary T790M mutation were resistant to first-generation agents (Han et al., 2011). A wealth of additional studies published worldwide, have indicated that mutations in KRAS, BRAF, ALK, RET, ROS1, and PIK3CA influence EGFR-TKI sensitivities in lung adenocarcinomas (Takeuchi et al., 2012; Gainor and Shaw, 2013). Additionally, KRAS mutations may associate with EGFR-TKI resistance in NSCLC patients (Pao et al., 2005). EGFR and KRAS mutations showed mutual gene exclusion in a series of NSCLC patients, which agreed with these previous findings (Shigematsu et al., 2005).
In addition to EGFR gene mutations, expression levels of genes involved in enzymatic drug metabolism, drug transport, and intracellular signaling have correlated with erlotinib efficacy and ADR. Both erlotinib and gefitinib are transported by the ATP-binding cassette family proteins, ABCB1, and ABCG2, and then metabolized in the liver by cytochrome P450 (CYP450) and the UDP glucuronosyltransferase (UGT) family (Han et al., 2007; Kim et al., 2010). Activities of these gene products determine pharmacokinetics of the agents and therefore influence ADR and therapeutic response rates. Harmsen et al. reported that gefitinib- and erlotinib-induced ABCB1 increase activated accumulations of ABCB1 substrates in vitro, suggesting that ABCB1 influenced drug resistance in these models (Harmsen et al., 2013). Another study showed that reduced CYP2D6 activity associated with high frequencies of skin rash in patients undergoing gefitinib therapy (Suzumura et al., 2012). UGT is one of vital roles in constitutive cellular metabolic pathways. Inter-individual variability in UGT pathway activities have been associated with drug metabolism and diseases (Suzuki and Sugiyama, 2002; Na et al., 2017).
Previous studies have focused on analyzing associations between candidate genes and drug responses in NSCLC; most studies emphasized somatic variants and failed to systematically and comprehensively identify and validate germline candidate biomarkers (Schmid et al., 2016). There is a pressing need to carry out non-biased studies identifying genetic variants in NSCLC that associate with responses to erlotinib and changes in safety signals. These data would guide clinical decision making when implementing personalized medicine. Here, single-nucleotide polymorphisms (SNP) in the EGFR signal pathway, as well as the drug metabolism and transport pathways, were investigated with next-generation sequencing (NGS) technologies to identify SNPs associated with drug responses to erlotinib in Chinese advanced NSCLC patients. Application of multiple genomic alteration screens will allow for improved treatment planning. Novel molecular biomarkers identified here may be useful for future research of therapeutic strategies in NSCLC patients.
Materials and Methods
Study Design
The objective of this study was to discover the genetic variants associated with drug reactions of erlotinib of lung cancer patients in Chinese population. The whole study was designed to be conducted in two steps. In the first step, using NGS method, we systematically investigated the germ line genetic variants of 76 genes contributions to EGFR signaling, EGFR antagonist drug metabolism, or transport in discovery cohort (60 samples). In the second step, the candidate genetic variant identified in the first step were verified invalidation cohort (134 samples).
Patient Recruitment
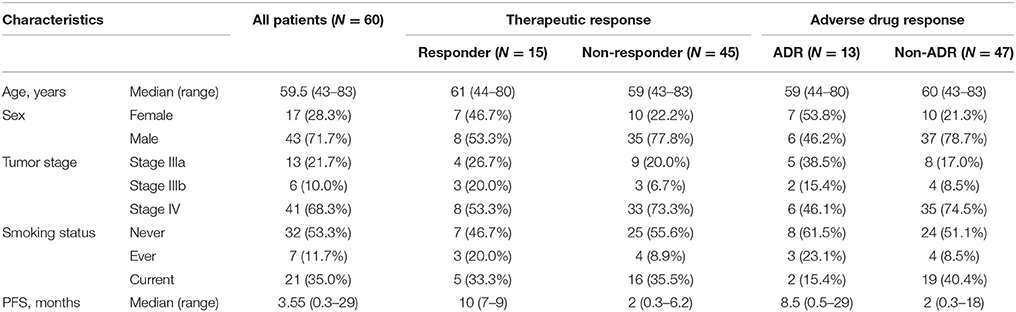
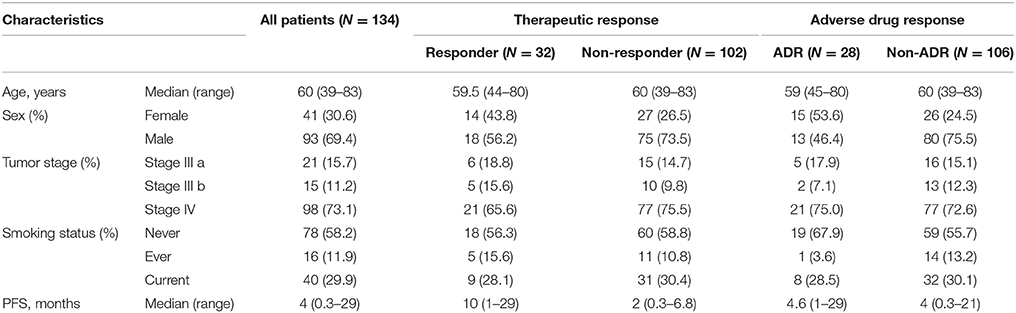
Blood samples were collected from 60 Chinese Han patients who underwent erlotinib monotherapy after prior standard chemotherapy for advanced NSCLC at the Shanghai Pulmonary Hospital. The patients with complete clinical information, accurate pathological diagnosis and classification criteria and without other diseases were recruited. Patient clinical records were obtained, including: gender, age at presentation, smoking status, cancer diagnosis, pathologic stage, PFS, and ADR (e.g., skin rash and/or digestive tract injury). The EGFR of each patients have been sequenced by NGS and validated by SNapShot. All cases in study carry EGFR mutation and none of the genetic variants has been reported associated with drug response of advanced NSCLC, so the patients recruited in our study received erlotinib as second-line treatment. In the validation study, 134 Chinese erlotinib monotherapy-treated patients with advanced NSCLC following prior chemotherapy were enrolled. Clinical demographics are given in Table 5; allelic type and frequency are given in Tables 1, 2.
Written informed consent was obtained from the participants in our study prior to enrollment. The Ethics Committee of Shanghai and the Ethical Committee of Human Genetic Resources approved the protocol used in this study. Patient recruiting, blood sample collection, and clinical data collection and usage were all performed according to the guidelines and regulations of the committees.
Next Generation Sequencing
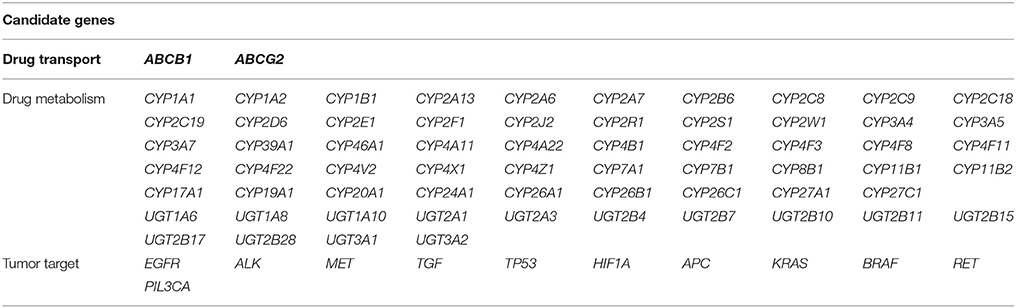
Seventy-six candidate genes were assessed. These genes were chosen due to their known contributions to EGFR signaling, EGFR antagonist drug metabolism, or transport in NSCLC. Candidate genes were submitted for HaloPlex probe design (Agilent Technologies, Santa Clara, CA, USA) using the Sure-Design service (Table 3) (https://earray.chem.agilent.com/suredesign/index.htm). Genomic DNA specimens were extracted from whole peripheral blood using AxyPrep Blood Genomic DNA Miniprep Kits (AxyGen, Shanghai, China) in strict accordance with standard protocols. The qualities and quantities of resultant DNA specimens obtained were measured with a NanoDrop 2000 spectrophotometer (Thermo Scientific, Commonwealth of Massachusetts, USA). DNA concentrations were adjusted to 20 ng/μl and stored at −20°C until use. Target regions were captured and enriched using HaloPlex Target Enrichment Kits (Agilent Technologies, Santa Clara, USA) according to the manufacturer's instructions. Libraries were pooled in equimolar aliquots. Amplicon sequencing reactions were performed at a 3 nM concentration per sample to counterbalance differences in sample quality. Sequencing was performed on an Illumina MiSeq benchtop sequencer (Illumina, San Diego, CA, USA). The variant call cutoff was 5%, and results were only interpreted if the coverage was >40x.
Statistical Analyses
Raw data from successful sequencing runs were analyzed using Sure-Call software (version 2.0). SNP sites with success rates <90%, Minor allele frequency (MAF) <1%, or those that were homogeneous across all samples were excluded. The Response Evaluation Criteria in Solid Tumors system (RECIST guidelines version 1.1) was used to classify responses as CR, PR, SD, or PD. Associations were identified between patient genotypes and objective responses to erlotinib within the first month of medication. To analyze ADR, patients were subcategorized according to clinical incidence of treatment-related adverse events.
Hardy-Weinberg genetic equilibriums, allelic and genotypic frequencies, odds ratios (ORs) and 95% confidence intervals (CIs), SNP case-control association analyses and linkage disequilibrium analyses were performed using SHEsis software (http://analysis.bio-x.cn/myAnalysis.php.). P-values < 0.05 were considered statistically significant. Correlations between objective responses to EGFR-TKI and ADR, as well as multiple logistic regression analyses were calculated using SPSS software (version 18.0) (http://www-01.ibm.com/software/analytics/spss/). In validation trials, genotypic data were analyzed using SHEsis software and statistical analyses were performed using SPSS. P-values of two-tailed tests < 0.05 were considered statistically significant.
Results
Clinical Characteristics
The clinical characteristics of the first discovery cohort are described in Table 1. NSCLC patients who showed complete response or partial response (PR) were considered erlotinib sensitive-responders and patients who showed progressive disease (PD) or stable disease (SD) were considered erlotinib-resistant non-responders (RECIST guidelines version 1.1).
The cohort consisted of 43 males and 17 females, 32 never-smokers, seven ever-smokers and 21 currently-smoking patients. Among the 60 patient-cohort, the median PFS was 3.55 months, ranging from 0.3 to 29 months. Fifteen patients were responders with a median PFS of 12.57 months. Forty-five patients were non-responders, with a median PFS of 2.7 months. Thirteen patients presented with adverse drug reactions. The most common adverse event was skin rash, which correlated with therapeutic response (Table 1). No significant differences were observed between responders and non-responders in characteristics including: sex, age and smoking status; indicating that the populations were adequately matched.
SNP Associated With Objective Responses to Erlotinib
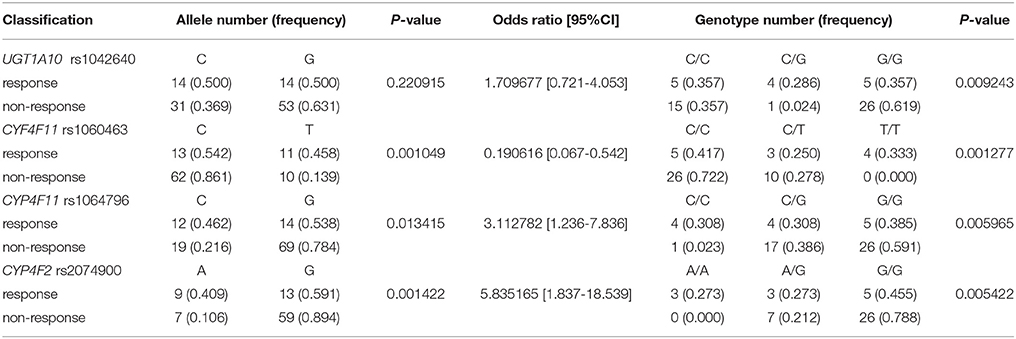
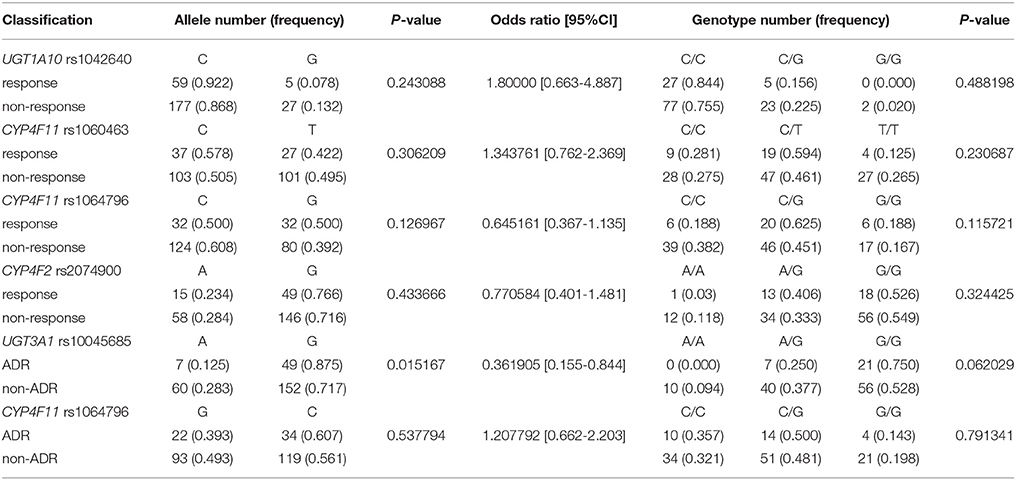
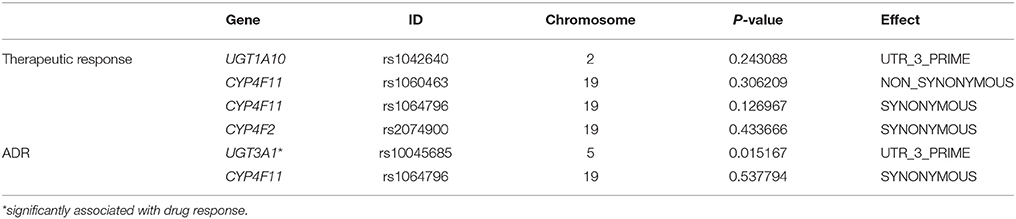
Four SNP sites, all from drug metabolism genes, associated with objective therapeutic responses to erlotinib (Table 4). Namely, rs1042640 (located in UGT1A10; p = 0.009), rs1060463 and rs1064796 (located in CYP4F11; p = 0.001 and 0.013, respectively), and rs2074900 (located in CYP4F2; p = 0.001) associated with therapeutic responses to erlotinib. Interestingly, the rs1060463 SNP in CYP4F11 (a C>T point mutation) had the weakest association with objective therapeutic response. However, none of the associations between these SNP and response rates remain statistically meaningful after multiple-testing corrections.
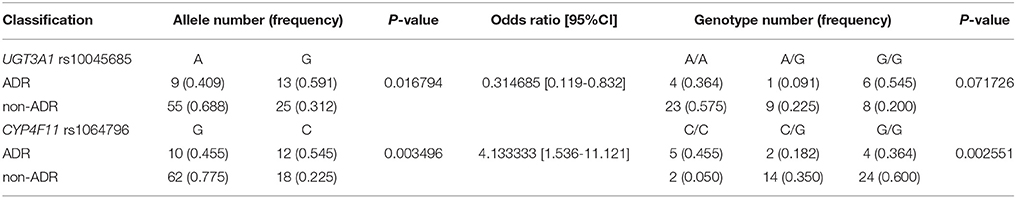
SNP Associated With Erlotinib-Induced Adverse Events
Two SNP associated with adverse drug reactions (Table 5). Specifically, rs1064796 in CYP4F11 (a G>C point mutation; p = 0.003) and rs10045685 in UGT3A1 (an A>G point mutation; p = 0.017) associated with decreased risk of ADR. Interestingly, one SNP was associated with both response rates to erlotinib and improved safety signals. The rs10045685 mutation in the UGT3A1 3′untranslated region (UTR) significantly associated with both therapeutic responses and reduced ADR after erlotinib treatment. However, the associations between the A>G mutant allele and lowered risk of ADR were not found after either Bonferroni or false discovery rate (FDR) corrections.
Validation of Associations Between SNP and Erlotinib Responses
Validation experiments were conducted in 134 NSCLC patients who received erlotinib monotherapy; clinical information of the cohort is given in Table 2; allelic type and frequency are given in Table 6. Multiplex SNapShot assays were performed to confirm whether the five above-identified SNP significantly associated with erlotinib responses in this second, larger independent population. Results showed that rs10045685 (an A>G point mutation in the UGT3A1 3′UTR) associated with erlotinib-induced adverse responses (unadjusted p = 0.015; Table 7).
UGT3A1-miRNA Functional Predictions
To predict the function of UGT3A1 in drug metabolism, an additional analysis was performed using the miRTarBase(http://mirtarbase.mbc.nctu.edu.tw/). UGT3A1 is a target gene of the microRNA hsa-miR-181a-5p. This suggested that activity of this microRNA may related to responses to erlotinib treatment in EGFR-activated NSCLC. Moreover from The Cancer Genome Atlas (TCGA) database, hsa-miR-181a-5p was identified as an oncogene that targeted UGT3A1 in lung adenocarcinomas.
Discussion
EGFR-TKI treatment for NSCLC have an impressive profile of therapeutic responses and ADR compared with standard chemotherapies. Previous studies showed relationships between both somatic activating mutations in EGFR coding sequences and SNP of drug metabolism genes with therapeutic responses to EGFR-TKI (Ganzinelli et al., 2015; Lópezayllón et al., 2015; Ruan et al., 2016). For example, rs884225 in EGFR is associated with erlotinib-induced ADR. Given that drug responses have highly correlated with clinical outcomes, a systematic multi-gene approach was used to investigate genetic variations in therapeutic responses and ADR in erlotinib-treated advanced NSCLC Chinese Han patients. A significant association was identified between the SNP rs10045685 A>G in the 3′UTR of UGT3A1 and reduced risk of erlotinib-induced ADR. Validation studies confirmed that rs10045685 associated with erlotinib-induced ADR, which might making this mutation a promising biomarker for responses to erlotinib in NSCLC patients.
Few previous studies have investigated UGT3A1 variants in NSCLC. However, a recent study found that the UGT3A gene family catalyze biotransformation of endogenous and environmental carcinogens including drugs (Lu et al., 2015). Additionally, UGT3A1 differentiated primate species by participation in sugar conjugation during metabolism and elimination of lipophilic metabolites and exogenous chemicals (Meech and Mackenzie, 2010). Also, increased presence of a UGT3A1 inactivating mutations may have significant therapeutic and/or toxicological implications (Mackenzie et al., 2008). Therefore, the UGT3A1 SNP rs10045685 may be a predictive molecular marker of improved erlotinib-induced ADR in NSCLC patients.
Additionally, the current study identified four SNP associated with objective responses to erlotinib: rs1042640 in UGT1A10, rs1060463, and rs1064796 in CYP4F11 and rs2074900 in CYP4F2. Previous studies have shown that CYP450 family members function in cancer drug metabolism, including in NSCLC. Among CYPs, CYP4F subfamily is thought to be primarily involved in the metabolism of fatty acids and arachidonic acid metabolites (Dhar et al., 2008). To these authors' knowledge, associations between CYP4F expression and cancer have not been reported in any tumor type. Previous studies showed that increase of TNF-α-induced CYP4F11 was in response to jun N-terminal protein kinase (JNK) pathway activity (Sechler et al., 2013). Another finding suggested that the CYP ω-hydroxylase, consisting of CYP4A and CYP4F molecules, promoted angiogenesis and metastatic potential in human NSCLC cells through up-regulation of Vascular endothelial growth factor (VEGF) and Matrix metallopeptidase 9 (MMP-9) (Wang et al., 2010; Bell and Strobel, 2012). Previously, CYP2D6 was found to be another important CYP450 enzyme involved in drug responses, accounting for 30% of metabolism of all medications (Gurley et al., 2008). However, no significant associations were found, and the exact roles of CYP2D6 in clinical settings were unclear (Guillemette et al., 2000; Mckillop et al., 2006). Additionally, UGT1A7 contributed to detoxification by catalyzing the binding and subsequent deactivation of glucuronic acid with tobacco carcinogens, including: nitrosamines, benzopyrenes (Xiao et al., 2015; Rajappa et al., 2017). In contrast, mutations in UGT1A10 associated with therapeutic responses to erlotinib in this study.
There were limitations to the current study. First, the blood samples tested were collected from a small cohort of patients that received erlotinib as second- or third-line therapy following chemotherapy (Rosell et al., 2012; Greenhalgh et al., 2015). First-line chemotherapy in EGFR-mutant NSCLC may decrease numbers of cancer cells, and diminish the overall clinical benefit of subsequent EGFR-TKI therapies. Second, most of the SNP associations identified in this study were not strong enough to persist after Bonferroni corrections. Given the strictness of multiple-testing corrections, type II error may have occurred when multiple sites were tested in these relatively small samples. Third, subcategorizing the patients into responder/non-responder, ADR/non-ADR is complicated clinically, which might have an effect on discovering genetic variants associated with TKI response. Also, false positive results could occur in small-samples studies, and further validation studies in larger samples should be conducted to verify whether erlotinib-induced ADR and therapeutic response had specific sets of predictive biomarkers and share common biomarkers.
In conclusion, in the present study we conducted a pilot study for high-throughput germline genetic variant analysis of drug response of NSCLC using second generation sequencing, which might provide potential candidate bio-targets for personalized medicine of erlotinib in clinic.
Author Contributions
Conception and design: SQ; Sample process: XZ, YL, FC, HD, and YS; Analysis: FC, YL, CW, QX, ML, LH, and YR; Interpretation of data: CW, FC, LG, LS, XS, HD, and YL; Drafting the article: CW, FC, YL, JM, and SQ. All authors contributed critical revision and final approval.
Funding
This work was supported by grants from the 863 Program (2012AA02A515,2012AA021802), the National Nature Science Foundation of China (81421061, 81273596, J1210047,30900799, 81361120389, 30972823), National key research and development program (2016YFC0905000, 2016YFC1200200,2016YFC0906400). The 4th Three-year Action Plan for Public Health of Shanghai (The Project No.: 15GWZK0101), Shanghai Key Laboratory of Psychotic Disorders (13dz2260500), Public Science and Technology Research Funds (201210056), The Fourth Round of Shanghai Three-year Action Plan on Public Health Discipline and Talent Program: Women and Children's Health (No. 15GWZK0401), The Research Fund for LH's Academician Workstation of New Medicine and Clinical Translation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely thank all the patients who agreed to participate in this study and all the staff who helped with sample collection and loads of work.
References
Bardelli, A., Parsons, D. W., Silliman, N., Ptak, J., Szabo, S., Saha, S., et al. (2003). Mutational analysis of the tyrosine kinome in colorectal cancers. Science 300, 949–949. doi: 10.1126/science.1082596
Bell, J. C., and Strobel, H. W. (2012). Regulation of cytochrome P450 4F11 by nuclear transcription factor-κB. Drug Metab. Disposition Biol. Fate Chem. 40, 205–211. doi: 10.1124/dmd.111.041178
Ciardiello, F., and Tortora, G. (2008). Drug therapy: EGFR antagonists in cancer treatment. N. Engl. J. Med. 358, 1160–1174. doi: 10.1056/NEJMra0707704
Dhar, M., Sepkovic, D. W., Hirani, V., Magnusson, R. P., and Lasker, J. M. (2008). Omega oxidation of 3-hydroxy fatty acids by the human CYP4F gene subfamily enzyme CYP4F11. J. Lipid Res. 49, 612–624. doi: 10.1194/jlr.M700450-JLR200
Ferlay, J., Soerjomataram, I., Dikshit, R., Eser, S., Mathers, C., Rebelo, M., et al. (2015). Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136:E359. doi: 10.1002/ijc.29210
Florescu, M., Hasan, B., Seymour, L., Ding, K., and Shepherd, F. A. (2008). A clinical prognostic index for patients treated with erlotinib in National Cancer Institute of Canada clinical trials group study BR.21. J. Thorac. Oncol. 3, 590–598. doi: 10.1097/JTO.0b013e3181729299
Gainor, J. F., and Shaw, A. T. (2013). Novel targets in non-small cell lung cancer: ROS1 and RET fusions. Oncologist 18, 865–875. doi: 10.1634/theoncologist.2013-0095
Ganzinelli, M., Rulli, E., Caiola, E., Garassino, M. C., Broggini, M., Copreni, E., et al. (2015). Role of KRAS-LCS6 polymorphism in advanced NSCLC patients treated with erlotinib or docetaxel in second line treatment (TAILOR). Sci. Rep. 5:16331. doi: 10.1038/srep16331
Greenhalgh, J., Bagust, A., Boland, A., Dwan, K., Beale, S., Hockenhull, J., et al. (2015). Erlotinib and gefitinib for treating non-small cell lung cancer that has progressed following prior chemotherapy (review of NICE technology appraisals 162 and 175): a systematic review and economic evaluation. Health Technol. Assess. 19, 1–134. doi: 10.3310/hta19470
Guillemette, C., Ritter, J. K., Auyeung, D. J., Kessler, F. K., and Housman, D. E. (2000). Structural heterogeneity at the UDP-glucuronosyltransferase 1 locus: functional consequences of three novel missense mutations in the human UGT1A7 gene. Pharmacogenetics 10:629. doi: 10.1097/00008571-200010000-00006
Gurley, B. J., Swain, A., Hubbard, M. A., Williams, D. K., Barone, G., Hartsfield, F., et al. (2008). Clinical assessment of CYP2D6-mediated herb–drug interactions in humans: effects of milk thistle, black cohosh, goldenseal, kava kava, St. John's wort, and Echinacea. Mol. Nutr. Food Res. 52, 755–763. doi: 10.1002/mnfr.200600300
Han, B., Zhou, X., Zhang, R., Zang, W., Chen, Z., Song, H., et al. (2011). Mutations of the epidermal growth factor receptor gene in NSCLC patients. Oncol. Lett. 2, 1233–1237. doi: 10.3892/ol.2011.366
Han, J., Lim, H., Yoo, Y., Shin, E., Park, Y., Lee, S., et al. (2007). Associations of ABCB1, ABCC2, and ABCG2 polymorphisms with irinotecan-pharmacokinetics and clinical outcome in patients with advanced non-small cell lung cancer. Cancer 110, 138–147. doi: 10.1002/cncr.22760
Harmsen, S., Meijerman, I., Maasbakker, R. F., Beijnen, J. H., and Schellens, J. H. (2013). PXR-mediated P-glycoprotein induction by small molecule tyrosine kinase inhibitors. Eur. J. Pharmaceut. Sci. 48, 644–649. doi: 10.1016/j.ejps.2012.12.019
Hirsch, F. R., Kabbinavar, F., Eisen, T., Martins, R., Schnell, F. M., Dziadziuszko, R., et al. (2011). A randomized, phase ii, biomarker-selected study comparing erlotinib to erlotinib intercalated with chemotherapy in first-line therapy for advanced non–small-cell lung cancer. J. Clin. Oncol. 29, 3567–3573. doi: 10.1200/JCO.2010.34.4929
Jemal, A., Bray, F., Center, M., Ferlay, J., Ward, E., and Forman, D. (2011). Global cancer statistics. CA Cancer J. Clin. 61, 69–90. doi: 10.3322/caac.20107
Kim, W. J., Lee, J. H., Yi, J., Cho, Y. J., Heo, K., Lee, S. H., et al. (2010). A nonsynonymous variation in MRP2/ABCC2 is associated with neurological adverse drug reactions of carbamazepine in patients with epilepsy. Pharmacogenet. Genom. 20, 249–256. doi: 10.1097/FPC.0b013e328338073a
La Salvia, A., Rossi, A., Galetta, D., Gobbini, E., De Luca, E., Novello, S., et al. (2017). Intercalated chemotherapy and epidermal growth factor receptor inhibitors for patients with advanced non–small-cell lung cancer: a systematic review and meta-analysis. Clin. Lung Cancer 18, 23.e21–33.e21. doi: 10.1016/j.cllc.2016.08.006
Lópezayllón, B. D., De, C. J., Rodriguez, C., Pernía, O., Iba-ezd, C. I., Beldainiesta, C., et al. (2015). Biomarkers of erlotinib response in non-small cell lung cancer tumors that do not harbor the more common epidermal growth factor receptor mutations. Int. J. Clin. Exp. Pathol. 8:2888.
Lu, L., Zhou, J., Shi, J., Peng, X., Qi, X., Wang, Y., et al. (2015). Drug-metabolizing activity, protein and gene expression of UDP-Glucuronosyltransferases are significantly altered in hepatocellular carcinoma patients. PLoS ONE 10:e0127524. doi: 10.1371/journal.pone.0127524
Lynch, T. J., Bell, D. W., Sordella, R., Gurubhagavatula, S., Okimoto, R. A., Brannigan, B. W., et al. (2004). Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N. Engl. J. Med. 350, 2129–2139. doi: 10.1056/NEJMoa040938
Mackenzie, P., Rogers, A., Treloar, J., Jorgensen, B., Miners, J., and Meech, R. (2008). Identification of UDP Glycosyltransferase 3A1 as a UDP N- Acetylglucosaminyltransferase. J. Biol. Chem. 283, 36205–36210. doi: 10.1074/jbc.M807961200
Mayo, C., Bertran-Alamillo, J., Molina-Vila, M., Giménez-Capitán, A., Costa, C., and Rosell, R. (2012). Pharmacogenetics of EGFR in lung cancer: perspectives and clinical applications. Pharmacogenomics 13, 789–802. doi: 10.2217/pgs.12.54
Mckillop, D., Guy, S. P., Spence, M. P., Kendrew, J., Kemp, J. V., Bushby, N., et al. (2006). Minimal contribution of desmethyl-gefitinib, the major human plasma metabolite of gefitinib, to epidermal growth factor receptor (EGFR)-mediated tumour growth inhibition. Xenobiotica 36, 29–39. doi: 10.1080/00498250500523253
Meech, R., and Mackenzie, P. I. (2010). UGT3A: novel UDP-glycosyltransferases of the UGT superfamily. Drug Metab. Rev. 42:45. doi: 10.3109/03602530903205823
Na, Y., Sun, R., Liao, X., Aa, J., and Wang, G. (2017). UDP-glucuronosyltransferases (UGTs) and their related metabolic cross-talk with internal homeostasis: a systematic review of UGT isoforms for precision medicine. Pharmacol. Res. 121, 169–183. doi: 10.1016/j.phrs.2017.05.001
Pao, W., Wang, T., Riely, G., Miller, V., Pan, Q., Ladanyi, M., et al. (2005). KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2:e17. doi: 10.1371/journal.pmed.0020017
Rajappa, S., Doval, D. C., Biswas, J., Patil, S., Somani, N., Srinivasan, S., et al. (2017). Efficacy of erlotinib as first-line maintenance therapy in patients with locally advanced or metastatic nonsmall cell lung cancer who have not experienced disease progression or unacceptable toxicity during chemotherapy. South Asian J. Cancer 6, 1–5. doi: 10.4103/2278-330X.202573
Riely, G. J., Marks, J., and Pao, W. (2009). KRAS mutations in non-small cell lung cancer. Proc. Am. Thorac. Soc. 6, 201–205. doi: 10.1513/pats.200809-107LC
Rosell, R., Carcereny, E., Gervais, R., Vergnenegre, A., Massuti, B., Felip, E., et al. (2012). Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 13:239. doi: 10.1016/S1470-2045(11)70393-X
Ruan, Y., Jiang, J., Guo, L., Li, Y., Huang, H., Shen, L., et al. (2016). Genetic association of curative and adverse reactions to tyrosine kinase inhibitors in Chinese advanced non-small cell lung cancer patients. Sci. Rep. 6:23368. doi: 10.1038/srep23368
Sawyers, C. L. (2003). Opportunities and challenges in the development of kinase inhibitor therapy for cancer. Genes Dev. 24, 2998–3010. doi: 10.1101/gad.1152403
Schmid, S., Gautschi, O., Rothschild, S., Mark, M., Froesch, P., Klingbiel, D., et al. (2016). Clinical outcome of ALK -positive non–small cell lung cancer (NSCLC) patients with De Novo EGFR or KRAS co-mutations receiving tyrosine kinase inhibitors (TKIs). J. Thorac. Oncol. 12, 681–688. doi: 10.1016/j.jtho.2016.12.003
Sechler, M., Cizmic, A. D., Avasarala, S., Scoyk, M. V., Brzezinski, C., Kelley, N., et al. (2013). Non-small-cell lung cancer: molecular targeted therapy and personalized medicine drug resistance, mechanisms, and strategies. Pharmacogenom. Personal. Med. 6, 25–36. doi: 10.2147/PGPM.S26058
Shigematsu, H., Lin, L., Takahashi, T., Nomura, M., Suzuki, M., Wistuba, I. I., et al. (2005). Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J. Natl. Cancer Inst. 97:339. doi: 10.1093/jnci/dji055
Siegel, R., Naishadham, D., and Jemal, A. (2013). Cancer statistics, 2013. Cancer J. Clin. 63, 11–30. doi: 10.3322/caac.21166
Suzumura, T., Kimura, T., Kudoh, S., Umekawa, K., Nagata, M., Matsuura, K., et al. (2012). Reduced CYP2D6 function is associated with gefitinib-induced rash in patients with non-small cell lung cancer. BMC Cancer 12:568. doi: 10.1186/1471-2407-12-568
Suzuki, H., and Sugiyama, Y. (2002). Single nucleotide polymorphisms in multidrug resistance associated protein 2 (MRP2/ABCC2): its impact on drug disposition. Adv. Drug Deliv. Rev. 54, 1311–1331. doi: 10.1016/S0169-409X(02)00075-3
Takeuchi, K., Soda, M., Togashi, Y., Suzuki, R., Sakata, S., Hatano, S., et al. (2012). RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 18, 378–81. doi: 10.1038/nm.2658
Torre, L. A., Bray, F., Siegel, R. L., Ferlay, J., Lortet-Tieulent, J., and Jemal, A. (2015). Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108. doi: 10.3322/caac.21262
Wang, Y., Bell, J. C., Keeney, D. S., and Strobel, H. W. (2010). Gene Regulation of CYP4F11 in human keratinocyte HaCaT Cells. Drug Metabol. Disposition 38, 100–107. doi: 10.1124/dmd.109.029025
Xiao, X. G., Xia, S., Zou, M., Mei, Q., Zhou, L., Wang, S. J., et al. (2015). The relationship between UGT1A1 gene polymorphism and irinotecan effect on extensive-stage small-cell lung cancer. Oncotargets Ther. 8, 3575–3583. doi: 10.2147/OTT.S95149
Yoshida, T., Kuroda, H., Oya, Y., Shimizu, J., Horio, Y., Sakao, Y., et al. (2017). Clinical outcomes of platinum-based chemotherapy according to T790M mutation status in EGFR-positive non-small cell lung cancer patients after initial EGFR-TKI failure. Lung Cancer 109, 89–91. doi: 10.1016/j.lungcan.2017.05.001
Keywords: non-small cell lung cancer, erlotinib, single-nucleotide polymorphism (SNP), therapeutic responses, adverse drug responses
Citation: Wang C, Chen F, Liu Y, Xu Q, Guo L, Zhang X, Ruan Y, Shi Y, Shen L, Li M, Du H, Sun X, Ma J, He L and Qin S (2018) Genetic Association of Drug Response to Erlotinib in Chinese Advanced Non-small Cell Lung Cancer Patients. Front. Pharmacol. 9:360. doi: 10.3389/fphar.2018.00360
Received: 06 February 2018; Accepted: 27 March 2018;
Published: 11 April 2018.
Edited by:
Hervé Emonard, Université de Reims Champagne Ardenne, FranceReviewed by:
Natasa Djordjevic, University of Kragujevac, SerbiaPavankumar Tandra, University of Nebraska Medical Center, United States
Copyright © 2018 Wang, Chen, Liu, Xu, Guo, Zhang, Ruan, Shi, Shen, Li, Du, Sun, Ma, He and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin He, helin@sjtu.edu.cn
Shengying Qin, chinsir@sjtu.edu.cn
†These authors have contributed equally to this work.
 Cong Wang1,2†
Cong Wang1,2† Qingqing Xu
Qingqing Xu Shengying Qin
Shengying Qin