- Laboratory of Health Science, Graduate School of Life and Environmental Sciences, Kyoto Prefectural University, Kyoto, Japan
MicroRNAs (miRNAs) are non-coding RNAs that can regulate the expression of mRNAs and proteins by degrading mRNA molecules or by inhibiting their translation. It has been predicted that miRNAs regulate approximately 60% of protein-coding genes that could be involved in a wide range of biological processes. Research over the last 5 years suggests that miRNAs play important roles in skeletal muscle function and several miRNAs have been identified as modulators of myogenesis, muscle mass, and nutrient metabolism in physiological and pathological states. In addition, some miRNAs can be incorporated into intracellular vesicles, released into the circulation, transported to other cells, and possibly function in other organs in an endocrine manner. This phenomenon might explain the interactions between skeletal muscles and other organs. Thus, far, several muscle-secreted miRNAs have been identified and their involvement in muscle biology has been debated. Based on the recent understanding, this perspective article describes the potential valuable role of miRNAs in skeletal muscle function, delineates its limitations, and outlines its future perspectives.
Introduction
In recent years, there has been a significant advancement in our understanding of the importance and role of non-coding RNAs in several biological processes. Several non-coding RNAs, besides transfer and ribosomal RNAs, are involved in the transcriptional and translational regulation of other RNAs; these are referred to as functional RNAs. Among non-coding RNAs, microRNAs (miRNAs), which are small non-coding RNAs (approximately 19–22 nucleotides in length), are diverse and regulate over 1500 target mRNAs (Kozomara and Griffiths-Jones, 2011) by perfectly or partially hybridizing to complementary binding sites located in the 3′ untranslated regions of the mRNAs and by inhibiting translation via mRNA cleavage or steric hindrance. miRNAs hold promise for presenting new findings in cellular and molecular biological systems and contribute to the biomedical field by acting as biomarkers for clinical diagnosis and as drug targets for the treatment of diseases. The role and regulation of miRNAs in skeletal muscle function have also been evaluated; however, multiple aspects remain to be elucidated. The identification of an association between miRNAs and skeletal muscle function after several years of research has been a turning point in skeletal muscle research. Here, I discuss the advancements in skeletal muscle miRNA-based research and provide a perspective on its potential future implications.
MicroRNA in Muscular Physiology and Pathology
Research has suggested that miRNA-mediated gene regulation is a fundamental mechanism of post-transcriptional regulation and that it may have diverse functional effects. It has previously been shown that the levels of several proteins do not frequently correspond to the levels of their mRNA; this phenomenon could be partly attributed to translational regulation by miRNAs. For instance, the mRNA expression level of peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC1-α) was markedly increased following exercise although the protein amount was not changed (Watt et al., 2004; Gibala et al., 2009), could be attributed to translational regulation by miRNAs. Indeed, earlier studies on muscular miRNA have shown that the levels of both miR-23 and miR-696, which hybridize with PGC1-α mRNA and reduce the protein amount, decrease in response to acute or chronic exercise (Safdar et al., 2009; Aoi et al., 2010). Approximately 60% of protein-coding genes may be regulated by miRNAs (Friedman et al., 2009). Therefore, miRNAs can influence the biological phenotype with regard to overall development, maintenance of homeostasis, cell death, carcinogenesis, and age-related changes by regulating protein content. Studies on skeletal muscles have suggested that several miRNAs could also function as modulators of myogenesis, muscle mass, and nutrient metabolism in skeletal muscle (Chen et al., 2006; Small et al., 2010; Dey et al., 2011; Gagan et al., 2011; Zhang et al., 2012; Hitachi et al., 2014), thus raising questions regarding the association of miRNAs with exercise-induced physiological changes, muscular pathogenesis, and age-related muscle dysfunction (Figure 1).
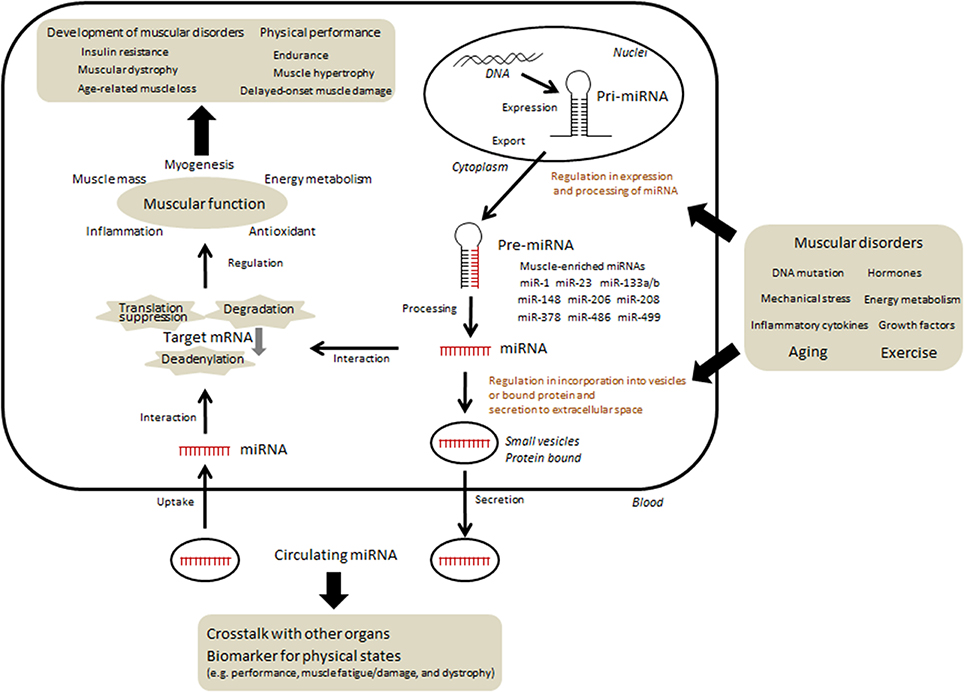
Figure 1. Current understanding and hypothesis regarding the regulation and function of miRNAs in skeletal muscle. The expression of several miRNAs changed according to various conditions such as physical activity, nutrients, diseases, and aging. In contrast, several miRNAs could function as modulators of myogenesis, muscle mass, and nutrient metabolism. Some of miRNAs can be taken up into intracellular vesicles and secreted into extracellular fluid. Circulating miRNAs migrate to skeletal muscle as well as other organs and may regulate certain functions. In addition, specific c-miRNAs may be useful in determining precise muscle-related events.
A tissue-specific miRNA is defined as a miRNA that is expressed in a specific tissue at levels that are >20-fold higher than its mean level in all other tissues (Lee et al., 2008). Several miRNAs are highly enriched in muscle tissue and are often referred to as myomiRs. Four myomiRs, namely, miR-1, miR-133a, miR-133b, and miR-206, together account for nearly 25% of miRNA expression in skeletal muscles in both humans and mice (Sempere et al., 2004; McCarthy, 2008). In addition, miR-208, miR-486, and miR-499 are encoded by muscle-specific genes such as ankyrin and myosin heavy chain (McCarthy et al., 2009; Small et al., 2010). The presence of miR-378 at high levels in muscle has been validated in several studies (Davidsen et al., 2011; Gagan et al., 2011). Therefore, skeletal muscle research is focused on the regulation of miRNAs and their association with muscle functions. A growing amount of evidences has suggested that these muscle-specific miRNAs, along with other miRNAs, affect various phenotypic changes in skeletal muscles, including exercise adaptation, immobilization, and muscular diseases (Eisenberg et al., 2007; Allen et al., 2009; Safdar et al., 2009; Aoi et al., 2010; Nielsen et al., 2010; Davidsen et al., 2011; Roberts et al., 2012; Russell et al., 2013; Alexander et al., 2014). In addition, expression of several miRNAs has been shown to be altered during aging in animals and humans (Hamrick et al., 2010; Mercken et al., 2013; Nielsen et al., 2014b; Rivas et al., 2014). Changes in the expression of some age-regulated miRNAs are reversed by calorie restriction, which is often adopted as a daily habit to prevent the development of age-related events and diseases (Mercken et al., 2013); this finding supports the significance of miRNA in aging. Recently, additional miRNAs that possibly play critical roles in muscle disorders related to diabetes, cancer, and inflammation have also been identified (Jiang et al., 2013; Chen et al., 2014; Georgantas et al., 2014; Rowlands et al., 2014; Sato et al., 2014). These findings provide novel information regarding the regulatory system of miRNAs, as described by several authors on this “research topics” (Zacharewicz et al., 2013; Aoi and Sakuma, 2014; Hitachi and Tsuchida, 2014; Sharma et al., 2014).
Another important characteristic of miRNAs is that a single miRNA regulates the expression of approximately hundreds of mRNAs and proteins by degrading mRNA molecules or by inhibiting their translation (Bartel, 2004; Djuranovic et al., 2012; Pasquinelli, 2012). This feature suggests that some miRNA's are functionally redundant and that the loss of functional regulation of a single miRNA does not always result in alterations in the expression of its target protein. Indeed, it has been observed that certain phenotypes are preserved despite impairments in specific regulatory miRNAs (Jin et al., 2009; Concepcion et al., 2012; Heyer et al., 2012), which is indicative of a cross-talk within complicated network of miRNAs involved in the modulation of skeletal muscle function. Thus, various miRNAs complement and cooperate with each other, making them essential molecular systems that maintain cellular homeostasis.
Circulating microRNA and Skeletal Muscle
Several miRNAs are secreted from cells into the circulation or are taken up from circulation into cells, suggesting that minimal miRNA degradation occurs due to RNases present in body fluids (Mitchell et al., 2008). This may be attributed to the protection of miRNAs from RNases by intracellular small vesicles such as exosomes, microvesicles, and apoptotic bodies, or by their binding with non-vesicle-associated proteins such as lipoprotein particles (Vickers et al., 2011; Raposo and Stoorvogel, 2013). Modulation of the function of recipient cells by circulating miRNAs (c-miRNAs) could explain the communication between skeletal muscles and other organs in physiological and pathological conditions (Figure 1). It has been suggested that exercise transiently or adaptively changes the level of c-miRNAs in animals and humans (Baggish et al., 2011; Aoi et al., 2013; Bye et al., 2013; Sawada et al., 2013; Nielsen et al., 2014a), leading to post-transcriptional regulation of proteins associated with energy metabolism and angiogenesis in adipocytes, hepatocytes, and endothelial cells. The circulating levels of several muscle-enriched miRNAs are also altered in muscle disorders (Miyachi et al., 2010; Mizuno et al., 2011; Roberts et al., 2013) and may be involved in such pathologies. In addition, such c-miRNAs have a potential role as useful biomarkers owing to their stability in body fluids, which could determine the various interactions between tissues and reflect their physiological and pathological states. c-miRNAs have been evaluated as biomarkers for clinical diagnosis, particularly in cancer studies. As previously mentioned, since several specific miRNAs are associated with skeletal muscles, determining their c-miRNA levels may be useful for the estimation of muscle-related events.
It has previously been reported that several muscle-specific miRNAs can be detected in plasma and serum; the levels of these miRNAs are altered in some muscle disorders. Serum levels of several muscle-enriched miRNAs such as miR-1, miR-133, and miR-206 are elevated in Duchenne muscular dystrophy in humans and animals (Cacchiarelli et al., 2011; Mizuno et al., 2011; Hu et al., 2014). In addition to these miRNAs, other candidate c-miRNAs, such as miR-378 and miR-31 have been suggested to be associated with dystrophy in a recent human study (Vignier et al., 2013). Furthermore, circulating levels of muscle-enriched miRNAs such as miR-1, miR-206, and miR-499 are higher in patients with chronic obstructive pulmonary disease, who often exhibit reduced muscle fiber size and proportions compared with those of normal healthy subjects (Donaldson et al., 2013). Roberts et al. (2013) showed that the dystrophy-associated miRNAs dystromiRs exist in a protein-bound form but not in the extracellular vesicles. Levels of muscle-specific miRNAs (miR-1, miR-133a, miR-133b, and miR-206) are also higher in the serum of patients with rhabdomyosarcoma tumors than in the serum of control subjects (Miyachi et al., 2010). In muscular atrophy, the levels of miR-23a are decreased and miR-23a is secreted into the extracellular space after being taken up by exosomes (Hudson et al., 2014). In contrast, the majority of the miRNAs that are highly expressed in skeletal muscle are difficult to detect in circulation under normal physiological condition. Further, these miRNAs exhibit no changes in response to exercise (Baggish et al., 2011; Nielsen et al., 2014a), except for miR-486, the levels of which is decreased in response to acute and chronic moderate exercise associated with lower aerobic performance (Aoi et al., 2013). However, other researchers reported that the levels of several c-miRNAs are elevated after strenuous/prolonged exercise such as running a marathon and that, in particular, alterations in the levels of c-miR-1, c-miR-133a, c-miR-206, and c-miR-208 are closely associated with performance and muscle damage parameters (Baggish et al., 2014; Gomes et al., 2014; Mooren et al., 2014). These conflicting results could be attributed to variations in the kind, intensity, and duration of exercises evaluated in the different studies. It has been shown that levels of common miRNAs are influenced by both exercise and myopathy, rendering it difficult to establish the use of certain miRNAs as reliable biomarkers for muscle disease. This warrants the development of further parameters upon consideration of miRNAs as diagnostic tools.
Limitations and Prospects for Future Studies
The understanding of the role of microRNA in the field of skeletal muscle research has developed rapidly by examining muscle tissue and blood samples obtained not only from animal models but also from healthy human volunteers and patients. However, despite the rapid progress, several aspects regarding miRNAs are still unclear. Typical technical issues like the small size (length of approximately 20 nucleotides) and the sequence similarity between different miRNAs make it difficult to accurately quantify the levels of miRNAs and design specific probes and primers. miRNAs can exert their function even if they do not completely hybridize with their targets (Bartel, 2009; Fabian et al., 2010), which may be one of the reasons why several miRNAs show changes in expression that are common among exercised, dystrophied, and aging muscles. In addition, the development of protocols for quantification of c-miRNA in circulation is underway. It is important to avoid hemolysis during sampling not only because it is difficult to extract miRNAs from serum or plasma, but also because blood cells contain various miRNAs and can significantly affect the c-miRNA level (Pritchard et al., 2012). Strenuous exercise frequently induces hemolysis by mechanical, osmotic, and oxidative stress (Telford et al., 2003; Sentürk et al., 2005; Peeling et al., 2009), which could affect the profile of c-miRNAs.
Although a limited number of experimental systems have evaluated c-miRNAs (Kosaka et al., 2010; Mittelbrunn et al., 2011; Vickers et al., 2011; Hudson et al., 2014), mechanisms underlying the uptake of c-miRNAs into certain cells, their secretion, and their release into the circulation are still unclear. c-miRNA-binding proteins in exosomes and other extracellular vesicles need to be studied further. It should also be considered that miRNAs can bind to lipoproteins in circulation, in case of physiological and pathological conditions that affect the level of lipoproteins which could easily change with exercise and age. Furthermore, an approach involving the identification of miRNAs from other body fluids such as saliva, urine, and sweat to establish them as biomarkers is necessary for relatively simpler diagnosis, without the need for muscle biopsy or blood collection in clinical setting and in the field of athletics. Further research involving the characterization of detailed mechanisms and the physiological and pathological changes in miRNAs based on appropriate evaluation/assessment protocols is warranted. Nevertheless, the field of miRNA research is attractive and is expected to present more novel findings for researchers.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alexander, M. S., Casar, J. C., Motohashi, N., Vieira, N. M., Eisenberg, I., Marshall, J. L., et al. (2014). MicroRNA-486-dependent modulation of DOCK3/PTEN/AKT signaling pathways improves muscular dystrophy-associated symptoms. J. Clin. Invest. 124, 2651–2667. doi: 10.1172/JCI73579
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Allen, D. L., Bandstra, E. R., Harrison, B. C., Thorng, S., Stodieck, L. S., Kostenuik, P. J., et al. (2009). Effects of spaceflight on murine skeletal muscle gene expression. J. Appl. Physiol. 106, 582–595. doi: 10.1152/japplphysiol.90780.2008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Aoi, W., Ichikawa, H., Mune, K., Tanimura, Y., Mizushima, K., Naito, Y., et al. (2013). Muscle-enriched microRNA miR-486 decreases in circulation in response to exercise in young men. Front. Physiol. 4:80. doi: 10.3389/fphys.2013.00080
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Aoi, W., Naito, Y., Mizushima, K., Takanami, Y., Kawai, Y., Ichikawa, H., et al. (2010). The microRNA miR-696 regulates PGC-1alpha in mouse skeletal muscle in response to physical activity. Am. J. Physiol. Endocrinol. Metab. 298, E799–E806. doi: 10.1152/ajpendo.00448.2009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Aoi, W., and Sakuma, K. (2014). Does regulation of skeletal muscle function involve circulating microRNAs? Front. Physiol. 5:39. doi: 10.3389/fphys.2014.00039
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baggish, A. L., Hale, A., Weiner, R. B., Lewis, G. D., Systrom, D., Wang, F., et al. (2011). Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J. Physiol. 589, 3983–3994. doi: 10.1113/jphysiol.2011.213363
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baggish, A. L., Park, J., Min, P. K., Isaacs, S., Parker, B. A., Thompson, P. D., et al. (2014). Rapid upregulation and clearance of distinct circulating microRNAs after prolonged aerobic exercise. J. Appl. Physiol. (1985) 116, 522–531. doi: 10.1152/japplphysiol.01141.2013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bye, A., Røsjø, H., Aspenes, S. T., Condorelli, G., Omland, T., and Wisløff, U. (2013). Circulating microRNAs and aerobic fitness–the HUNT-Study. PLoS ONE 8:e57496. doi: 10.1371/journal.pone.0057496
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cacchiarelli, D., Legnini, I., Martone, J., Cazzella, V., D'Amico, A., Bertini, E., et al. (2011). miRNAs as serum biomarkers for Duchenne muscular dystrophy. EMBO Mol. Med. 3, 258–265. doi: 10.1002/emmm.201100133
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, D., Goswami, C. P., Burnett, R. M., Anjanappa, M., Bhat-Nakshatri, P., Muller, W., et al. (2014). Cancer affects microRNA expression, release, and function in cardiac and skeletal muscle. Cancer Res. 74, 4270–4281. doi: 10.1158/0008-5472.CAN-13-2817
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, J. F., Mandel, E. M., Thomson, J. M., Wu, Q., Callis, T. E., Hammond, S. M., et al. (2006). The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233. doi: 10.1038/ng1725
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Concepcion, C. P., Han, Y. C., Mu, P., Bonetti, C., Yao, E., D'Andrea, A., et al. (2012). Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 8:e1002797. doi: 10.1371/journal.pgen.1002797
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Davidsen, P. K., Gallagher, I. J., Hartman, J. W., Tarnopolsky, M. A., Dela, F., Helge, J. W., et al. (2011). High responders to resistance exercise training demonstrate differential regulation of skeletal muscle microRNA expression. J. Appl. Physiol. 110, 309–317. doi: 10.1152/japplphysiol.00901.2010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dey, B. K., Gagan, J., and Dutta, A. (2011). miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol. Cell Biol. 31, 203–214. doi: 10.1128/MCB.01009-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Djuranovic, S., Nahvi, A., and Green, R. (2012). miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336, 237–240. doi: 10.1126/science.1215691
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Donaldson, A., Natanek, S. A., Lewis, A., Man, W. D., Hopkinson, N. S., Polkey, M. I., et al. (2013). Increased skeletal muscle-specific microRNA in the blood of patients with COPD. Thorax. 68, 1140–1149. doi: 10.1136/thoraxjnl-2012-203129
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eisenberg, I., Eran, A., Nishino, I., Moggio, M., Lamperti, C., Amato, A. A., et al. (2007). Distinctive patterns ofmicroRNA expression in primary muscular disorders. Proc. Natl. Acad. Sci. U.S.A. 104, 17016–17021. doi: 10.1073/pnas.0708115104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fabian, M. R., Sonenberg, N., and Filipowicz, W. (2010). Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379. doi: 10.1146/annurev-biochem-060308-103103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Friedman, R. C., Farh, K. K., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. doi: 10.1101/gr.082701.108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gagan, J., Dey, B. K., Layer, R., Yan, Z., and Dutta, A. (2011). MicroRNA-378 targets the myogenic repressor MyoR during myoblast differentiation. J. Biol. Chem. 286, 19431–19438. doi: 10.1074/jbc.M111.219006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Georgantas, R. W., Streicher, K., Greenberg, S. A., Greenlees, L. M., Zhu, W., Brohawn, P. Z., et al. (2014). Inhibition of myogenic microRNAs 1, 133, and 206 by inflammatory cytokines links inflammation and muscle degeneration in adult inflammatory myopathies. Arthritis Rheumatol. 66, 1022–1033. doi: 10.1002/art.38292
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gibala, M. J., McGee, S. L., Garnham, A. P., Howlett, K. F., Snow, R. J., and Hargreaves, M. (2009). Brief intense interval exercise activates AMPK and p38 MAPK signaling and increases the expression of PGC-1{alpha} in human skeletal muscle. J. Appl. Physiol. 106, 929–934. doi: 10.1152/japplphysiol.90880.2008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gomes, C. P., Oliveira, G. P. Jr., Madrid, B., Almeida, J. A., Franco, O. L., and Pereira, R. W. (2014). Circulating miR-1, miR-133a, and miR-206 levels are increased after a half-marathon run. Biomarkers 19, 585–589. doi: 10.3109/1354750X.2014.952663
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hamrick, M. W., Herberg, S., Arounleut, P., He, H. Z., Shiver, A., Qi, R. Q., et al. (2010). The adipokine leptin increases skeletal muscle mass and significantly alters skeletal muscle miRNA expression profile in aged mice. Biochem. Biophys. Res. Commun. 400, 379–383. doi: 10.1016/j.bbrc.2010.08.079
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heyer, M. P., Pani, A. K., Smeyne, R. J., Kenny, P. J., and Feng, G. (2012). Normal midbrain dopaminergic neuron development and function in miR-133b mutant mice. J. Neurosci. 32, 10887–10894. doi: 10.1523/JNEUROSCI.1732-12.2012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hitachi, K., Nakatani, M., and Tsuchida, K. (2014). Myostatin signaling regulates Akt activity via the regulation of miR-486 expression. Int. J. Biochem. Cell. Biol. 47, 93–103. doi: 10.1016/j.biocel.2013.12.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hitachi, K., and Tsuchida, K. (2014). Role of microRNAs in skeletal muscle hypertrophy. Front. Physiol. 4:408. doi: 10.3389/fphys.2013.00408
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hu, J., Kong, M., Ye, Y., Hong, S., Cheng, L., and Jiang, L. (2014). Serum miR-206 and other muscle-specific microRNAs as non-invasive biomarkers for Duchenne muscular dystrophy. J. Neurochem. 129, 877–883. doi: 10.1111/jnc.12662
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hudson, M. B., Woodworth-Hobbs, M. E., Zheng, B., Rahnert, J. A., Blount, M. A., Gooch, J. L., et al. (2014). miR-23a is decreased during muscle atrophy by a mechanism that includes calcineurin signaling and exosome-mediated export. Am. J. Physiol. Cell Physiol. 306, C551–C558. doi: 10.1152/ajpcell.00266.2013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jiang, L. Q., Franck, N., Egan, B., Sjögren, R. J., Katayama, M., Duque-Guimaraes, D., et al. (2013). Autocrine role of interleukin-13 on skeletal muscle glucose metabolism in type 2 diabetic patients involves microRNA let-7. Am. J. Physiol. Endocrinol. Metab. 305, E1359–E1366. doi: 10.1152/ajpendo.00236.2013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jin, Z. B., Hirokawa, G., Gui, L., Takahashi, R., Osakada, F., Hiura, Y., et al. (2009). Targeted deletion of miR-182, an abundant retinal microRNA. Mol. Vis. 15, 523–533. Available online at: http://www.molvis.org/molvis/v15/a53/
Kosaka, N., Iguchi, H., Yoshioka, Y., Takeshita, F., Matsuki, Y., and Ochiya, T. (2010). Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 285, 17442–17445. doi: 10.1074/jbc.M110.107821
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kozomara, A., and Griffiths-Jones, S. (2011). miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157. doi: 10.1093/nar/gkq1027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, E. J., Baek, M., Gusev, Y., Brackett, D. J., Nuovo, G. J., Chen, C., et al. (2008). Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA 14, 35–42. doi: 10.1261/rna.804508
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McCarthy, J. J. (2008). MicroRNA-206: the skeletal muscle-specific myomiR. Biochim. Biophys. Acta 1779, 682–691. doi: 10.1016/j.bbagrm.2008.03.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McCarthy, J. J., Esser, K. A., Peterson, C. A., and Dupont-Versteegden, E. E. (2009). Evidence of MyomiR network regulation of beta-myosin heavy chain gene expression during skeletal muscle atrophy. Physiol. Genomics 39, 219–226. doi: 10.1152/physiolgenomics.00042.2009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mercken, E. M., Majounie, E., Ding, J., Guo, R., Kim, J., Bernier, M., et al. (2013). Age-associated miRNA alterations in skeletal muscle from rhesus monkeys reversed by caloric restric-tion. Aging (Albany, NY) 5, 692–703. Available online at: http://www.impactaging.com/papers/v5/n9/full/100598.html
Mitchell, P. S., Parkin, R. K., Kroh, E. M., Fritz, B. R., Wyman, S. K., Pogosova-Agadjanyan, E. L., et al. (2008). Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 105, 10513–10518. doi: 10.1073/pnas.0804549105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mittelbrunn, M., Gutiérrez-Vázquez, C., Villarroya-Beltri, C., González, S., Sánchez-Cabo, F., González, M. Á., et al. (2011). Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2, 282. doi: 10.1038/ncomms1285
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miyachi, M., Tsuchiya, K., Yoshida, H., Yagyu, S., Kikuchi, K., Misawa, A., et al. (2010). Circulating muscle-specific microRNA, miR-206, as a potential diagnostic marker for rhabdomyosarcoma. Biochem. Biophys. Res. Commun. 400, 89–93. doi: 10.1016/j.bbrc.2010.08.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mizuno, H., Nakamura, A., Aoki, Y., Ito, N., Kishi, S., Yamamoto, K., et al. (2011). Identification of muscle-specific microRNAs in serum of muscular dystrophy animal models: promising novel blood-based markers for muscular dystrophy. PLoS ONE 6:e18388. doi: 10.1371/journal.pone.0018388
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mooren, F. C., Viereck, J., Krüger, K., and Thum, T. (2014). Circulating microRNAs as potential biomarkers of aerobic exercise capacity. Am. J. Physiol. Heart Circ. Physiol. 306, H557–H563. doi: 10.1152/ajpheart.00711.2013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nielsen, S., Åkerström, T., Rinnov, A., Yfanti, C., Scheele, C., Pedersen, B. K., et al. (2014a). The miRNA plasma signature in response to acute aerobic exercise and endurance training. PLoS ONE 9:e87308. doi: 10.1371/journal.pone.0087308
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nielsen, S., Hvid, T., Kelly, M., Lindegaard, B., Dethlefsen, C., Winding, K., et al. (2014b). Muscle specific miRNAs are induced by testosterone and independently upregulated by age. Front. Physiol. 4:394. doi: 10.3389/fphys.2013.00394
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nielsen, S., Scheele, C., Yfanti, C., Akerström, T., Nielsen, A. R., Pedersen, B. K., et al. (2010). Muscle specific microRNAs are regulated by endurance exercise in human skeletal muscle. J. Physiol. (Lond.) 588, 4029–4037. doi: 10.1113/jphysiol.2010.189860
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pasquinelli, A. E. (2012). MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 13, 271–282. doi: 10.1038/nrg3162
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Peeling, P., Dawson, B., Goodman, C., Landers, G., Wiegerinck, E. T., Swinkels, D. W., et al. (2009). Training surface and intensity: inflammation, hemolysis, and hepcidin expression. Med. Sci. Sports Exerc. 41, 1138–1145. doi: 10.1249/MSS.0b013e318192ce58
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pritchard, C. C., Kroh, E., Wood, B., Arroyo, J. D., Dougherty, K. J., Miyaji, M. M., et al. (2012). Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev. Res. (Phila.) 5, 492–497. doi: 10.1158/1940-6207.CAPR-11-0370
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Raposo, G., and Stoorvogel, W. (2013). Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383. doi: 10.1083/jcb.201211138
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rivas, D. A., Lessard, S. J., Rice, N. P., Lustgarten, M. S., So, K., Goodyear, L. J., et al. (2014). Diminished skeletal muscle microRNA expression with aging is associated with attenuated muscle plasticity and inhibition of IGF-1 signaling. FASEB J. 28, 4133–4147. doi: 10.1096/fj.14-254490
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roberts, T. C., Blomberg, K. E. M., McClorey, G., El Andaloussi, S., Godfrey, C., Betts, C., et al. (2012). Expression analysis in multiple muscle groups and serum reveals complexity in the microRNA tran-scriptome of the mdx mouse with implications for therapy. Mol. Ther. Nucleic Acids 1, e39. doi: 10.1038/mtna.2012.26
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roberts, T. C., Godfrey, C., McClorey, G., Vader, P., Briggs, D., Gardiner, C., et al. (2013). Extracellular microRNAs are dynamic non-vesicular biomarkers of muscle turnover. Nucleic Acids Res. 41, 9500–9513. doi: 10.1093/nar/gkt724
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rowlands, D. S., Page, R. A., Sukala, W. R., Giri, M., Ghimbovschi, S. D., Hayat, I., et al. (2014). Multi-Omic integrated networks connect DNA methylation and microRNA with skeletal muscle plasticity to chronic exercise in type 2 diabetic obesity. Physiol. Genomics 46, 747–765. doi: 10.1152/physiolgenomics.00024.2014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Russell, A. P., Lamon, S., Boon, H., Wada, S., Güller, I., Brown, E. L., et al. (2013). Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short-term endurance training. J. Physiol. 591, 4637–4653. doi: 10.1113/jphysiol.2013.255695
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Safdar, A., Abadi, A., Akhtar, M., Hettinga, B. P., and Tarnopolsky, M. A. (2009). miRNA in the regulation of skeletal muscle adaptation to acute endurance exercise in C57Bl/6J male mice. PLoS ONE 4:e5610. doi: 10.1371/journal.pone.0005610
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sato, T., Yamamoto, T., and Sehara-Fujisawa, A. (2014). miR-195/497 induce postnatal quiescence of skeletal muscle stem cells. Nat. Commun. 5, 4597. doi: 10.1038/ncomms5597
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sawada, S., Kon, M., Wada, S., Ushida, T., Suzuki, K., and Akimoto, T. (2013). Profiling of circulating microRNAs after a bout of acute resistance exercise in humans. PLoS ONE 8:e70823. doi: 10.1371/journal.pone.0070823
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sempere, L. F., Freemantle, S., Pitha-Rowe, I., Moss, E., Dmitrovsky, E., and Ambros, V. (2004). Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5:R13. doi: 10.1186/gb-2004-5-3-r13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sentürk, U. K., Gündüz, F., Kuru, O., Koçer, G., Ozkaya, Y. G., Yesilkaya, A., et al. (2005). Exercise-induced oxidative stress leads hemolysis in sedentary but not trained humans. J. Appl. Physiol. (1985) 99, 1434–1441. doi: 10.1152/japplphysiol.01392.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sharma, M. J., Juvvuna, P. K., Kukreti, H., and McFarlane, C. (2014). Mega roles of microRNAs in regulation of skeletal muscle health and disease. Front. Physiol. 5:239. doi: 10.3389/fphys.2014.00239
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Small, E. M., O'Rourke, J. R., Moresi, V., Sutherland, L. B., McAnally, J., Gerard, R. D., et al. (2010). Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc. Natl. Acad. Sci. U.S.A. 107, 4218–4223. doi: 10.1073/pnas.1000300107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Telford, R. D., Sly, G. J., Hahn, A. G., Cunningham, R. B., Bryant, C., and Smith, J. A. (2003). Footstrike is the major cause of hemolysis during running. J. Appl. Physiol. (1985) 94, 38–42. doi: 10.1152/japplphysiol.00631.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vickers, K. C., Palmisano, B. T., Shoucri, B. M., Shamburek, R. D., and Remaley, A. T. (2011). MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 13, 423–433. doi: 10.1038/ncb2210
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vignier, N., Amor, F., Fogel, P., Duvallet, A., Poupiot, J., Charrier, S., et al. (2013). Distinctive serum miRNA profile in mouse models of striated muscular pathologies. PLoS ONE 8:e55281. doi: 10.1371/journal.pone.0055281
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Watt, M. J., Southgate, R. J., Holmes, A. G., and Febbraio, M. A. (2004). Suppression of plasma free fatty acids upregulates peroxisome proliferator-activated receptor (PPAR) alpha and delta and PPAR coactivator 1alpha in human skeletal muscle, but not lipid regulatory genes. J. Mol. Endocrinol. 33, 533–544. doi: 10.1677/jme.1.01499
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zacharewicz, E., Lamon, S., and Russell, A. P. (2013). MicroRNAs in skeletal muscle and their regulation with exercise, ageing, and disease. Front. Physiol. 4:266. doi: 10.3389/fphys.2013.00266
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, J., Ying, Z. Z., Tang, Z. L., Long, L. Q., and Li, K. (2012). MicroRNA-148a promotes myogenic differentiation by targeting the ROCK1 gene. J. Biol. Chem. 287, 21093–21101. doi: 10.1074/jbc.M111.330381
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: microRNAs, skeletal muscle, exercise, aging and longetivity, exosomes
Citation: Aoi W (2015) Frontier impact of microRNAs in skeletal muscle research: a future perspective. Front. Physiol. 5:495. doi: 10.3389/fphys.2014.00495
Received: 08 October 2014; Accepted: 01 December 2014;
Published online: 05 January 2015.
Edited by:
Kenneth S. Campbell, University of Kentucky, USAReviewed by:
Esther Dupont-Versteegden, University of Kentucky, USAAshok Kumar, University of Louisville, USA
Copyright © 2015 Aoi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wataru Aoi, Laboratory of Health Science, Graduate School of Life and Environmental Sciences, Kyoto Prefectural University, 1-5 Hangi-cho Shimogamo, Sakyo-ku, Kyoto 606-8522, Japan e-mail: waoi@kpu.ac.jp
 Wataru Aoi
Wataru Aoi