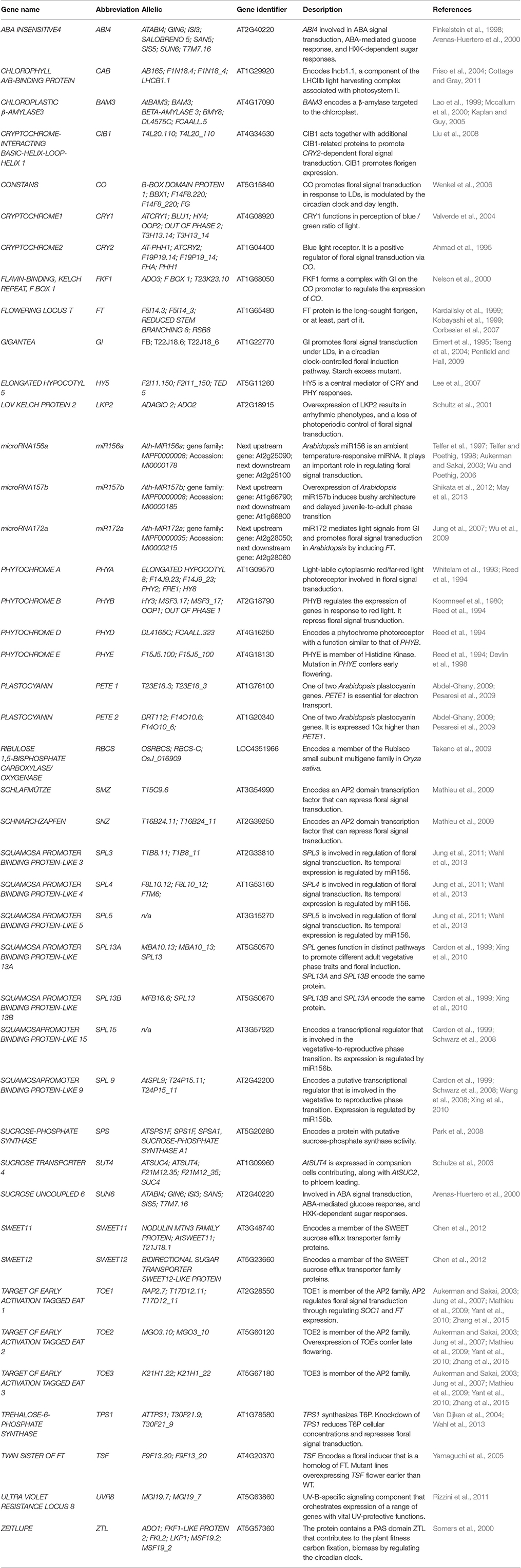
- School of Life Sciences, University of Warwick, Coventry, United Kingdom
Over the past decade, integrated genetic, cellular, proteomic and genomic approaches have begun to unravel the surprisingly crosstalk between photoreceptors and sugar signaling in regulation of floral signal transduction. Although a number of physiological factors in the pathway have been identified, the molecular genetic interactions of some components are less well understood. The further elucidation of the crosstalk mechanisms between photoreceptors and sugar signaling will certainly contribute to our better understanding of the developmental circuitry that controls floral signal transduction. This article summarizes our current knowledge of this crosstalk, which has not received much attention, and suggests possible directions for future research.
Introduction: Light, Sugars and Floral Signal Transduction
Post-embryonic development progresses through distinct developmental phase transitions. It has been proposed (Matsoukas, 2014a) that the prolonged juvenile-to-adult and vegetative-to-reproductive phase transitions might be due to several antiflorigenic signals, which affect the transcription levels of florigen FLOWERING LOCUS T (FT; Corbesier et al., 2007), and SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL; Shikata et al., 2009) genes.
Juvenility can be defined as the early period of development during which the abundance of antiflorigenic signals such as miR156/miR157 (Lauter et al., 2005; Martin et al., 2009; Lee et al., 2010; Varkonyi-Gasic et al., 2010) is sufficiently high to suppress the expression of FT and SPLs (Shikata et al., 2009, 2012; Wang et al., 2009; Jung et al., 2011). On the other hand, expression of miR172 in leaves activates FT (Aukerman and Sakai, 2003; Jung et al., 2007), through repression of AP2-like transcripts SCHLAFMÜTZE (SMZ), SCHNARCHZAPFEN (SNZ) and TARGET OF EAT 1–3 (TOE1-3; Jung et al., 2007; Mathieu et al., 2009), whereas the increase in SPLs at the shoot apical meristem (SAM), leads to the activation of floral meristem identity genes (Wang et al., 2009; Yamaguchi et al., 2009), which result in vegetative-to-reproductive phase transition.
Light is a key regulator of the juvenile-to-adult and vegetative-to-reproductive phase transitions (Turck et al., 2008; Matsoukas et al., 2012; Lifschitz et al., 2014; Matsoukas, 2015). It constitutes a critical environmental growth indicator, which is estimated by the duration, quality, direction and intensity, as well as the essential energy source for the synthesis of carbohydrates by the photosynthetic apparatus. Light perception is mediated through the action of photoreceptors, namely PHYTOCHROMES (PHYs; derives from Greek phyto- “relating to plants” and khrōma “color”; Chen and Chory, 2011), CRYPTOCHROMES (CRYs; derives from Greek kruptós “hidden” and khrōma “color”; Chaves et al., 2011), the ultraviolet B photoreceptor ULTRA VIOLET RESISTANCE LOCUS 8 (Jenkins, 2014), phototropins (Christie, 2007) and the ZEITLUPE (ZTL) family members ZTL, FLAVIN-BINDING, KELCH-REPEAT F-BOX (FKF1), and LOV KELCH PROTEIN 2 (LKP2; Kim et al., 2007; Suetsugu and Wada, 2013). Members of each of these photoreceptor families have direct interactions with circadian clock genes and proteins.
Several molecular mechanisms that mediate sugar responses have been identified in plants (reviewed in Rolland et al., 2006; Smeekens et al., 2010; Dobrenel et al., 2013; Lastdrager et al., 2014; Smeekens and Hellmann, 2014; Van den Ende, 2014; Li and Sheen, 2016). Sugar signals can be generated either by carbohydrate concentration and relative ratios to other metabolites, such as hormones and carbon-nitrogen ratio, or by flux through sugar-specific transporters and/or sensors (Matsoukas, 2014b). Glucose, sucrose and trehalose-6-phosphate (T6P) have been recognized as pivotal integrating regulatory molecules that control the expression of genes involved in floral signal transduction (reviewed in Ponnu et al., 2011; Bolouri Moghaddam and Van den Ende, 2013; Matsoukas, 2014b).
Glucose-mediated signal transduction is largely dependent on HEXOKINASE1 (HXK1)-dependent pathway, HXK1-independent pathway, and glycolysis-dependent pathway, which utilizes the SUCROSE NONFERMENTING RELATED KINASE1 (SnRK1)/TARGET OF RAPAMYCIN (TOR) pathway (Moore et al., 2003; Baena-Gonzalez et al., 2007; Ren et al., 2012). SnRK1 has a role when sugars are in extremely limited supply, whereas HXK and Tre6P play a role in the presence of excess sugar. Sucrose plays an essential role in the regulation of important metabolic processes (reviewed in Tognetti et al., 2013). Its concentration tends to be directly related to light intensity (LI), and inversely related to temperature. It has been shown that sucrose, together with T6P act as proxies for the carbohydrate status in plant tissues (Lunn et al., 2006; Wahl et al., 2013; Xing et al., 2015). It is notable that T6P inhibits the activity of the SnRK1 in sugar metabolic control of floral signal transduction (Zhang et al., 2009). In particular, mutations in SNRK1 confer early flowering, whereas SnRK1 overexpression delays flowering (Baena-Gonzalez et al., 2007; Tsai and Gazzarrini, 2012). Several lines of evidence suggest that Tre6P inhibits SnRK1 when sucrose is above a threshold level (Polge and Thomas, 2007; Zhang et al., 2009). When the sucrose concentration decreases, with Tre6P decreasing as well, SnRK1 is released from repression, promoting the expression of genes involved in photosynthesis-related events, so that more carbon is made available (Delatte et al., 2011). Mutations in T6P signaling pathway confer late flowering. This late flowering phenotype was found to be due to reduced expression levels of FT, the elevated levels of miR156, and reduced levels of at least three miR156-regulated transcripts: SPL3, 4, 5 (Wahl et al., 2013). However, T6P not only signals sucrose availability (Lunn et al., 2006), but it also negatively regulates sucrose levels by restricting sucrose synthesis and/or promoting sucrose catabolism (Yadav et al., 2014). Interestingly, the regulatory effects of T6P on growth and development would be an effective means for manipulating carbon partitioning and plant yield (Smeekens, 2015).
The identification of downstream components of photoreceptor signaling that involved in floral signal transduction has revealed a crosstalk between pathways of different light qualities as well as with other seemingly unrelated signaling pathways. One such crosstalk that has not received much attention and involves carbohydrates, forms the focus of this article.
Light Perception and Circadian Clock
The circadian [derived from the Latin roots “circa” (around) and “diem” (day)] system is a complex regulatory network. It is consists of a set of proteins that forms an interconnected feedback network with multiple loops. This system provides temporal information to organisms to coordinate developmental and metabolic responses in coincidence with the environmental conditions. One of the main functions of light in regulation of floral signal transduction is in the initiation of cues that interact with the circadian oscillator and entrain the circadian rhythm. Several reviews have been published on the circadian clock system recently (Romanowski and Yanovsky, 2015; Endo, 2016; Sanchez and Kay, 2016), so the circadian clock will not be described in great detail here. The circadian clock system has three primary components. First is the central oscillator/pacemaker that generates the 24 h oscillators. A model for the Arabidopsis circadian oscillator described a series of multiple interlocked transcriptional–translational feedback loops referred to as the morning, core, and evening loops (Huang et al., 2012; Pokhilko et al., 2012). The “morning complex” comprises the genes encoding the proteins CIRCADIAN CLOCK ASSOCIATED 1 (CCA1; Wang and Tobin, 1998) and LATE ELONGATED HYPOCOTYL (LHY). Both genes increase their expression prior to dawn (Schaffer et al., 1998). The “morning complex” genes encoding PSEUDO-RESPONSE REGULATOR (PRR) 5, 7, and 9 increase their expression after dawn (Matsushika et al., 2000; Farre et al., 2005). The “evening loop” comprises genes encoding GI (Fowler et al., 1999; Park et al., 1999) and TIME OF CAB EXPRESSION 1 (TOC1; Strayer et al., 2000) as well as the evening complex genes encoding EARLY FLOWERING (ELF) 3, 4 (Herrero et al., 2012), and LUX ARRHYTHMO (LUX; Hazen et al., 2005; Nusinow et al., 2011). The “evening complex” genes increase their expression prior to, and after dusk. The “morning” and “evening” complex proteins regulate each other through a series of promoter cis-acting elements (Harmer et al., 2000; Alabadi et al., 2001; Covington et al., 2008), and protein–protein interactions (Kim et al., 2007; Nusinow et al., 2011; Chow and Kay, 2013). These type of interactions create a robust and tunable oscillator that modulate gene expression in a coordinated 24 h rhythm.
The second component is the input pathway that synchronizes or entrains the oscillator with environmental cues. The best-characterized signal is light (reviewed in Kami et al., 2010). In Arabidopsis, red/far-red light perception is mediated by PHYs. Blue light perception is mediated by CRYs and the blue-light sensing proteins ZTL, FKF1, and LKP2. The third component is the output pathway that links the oscillator to processes under circadian rhythm such as photoperiodic induction and floral signal transduction.
The plant circadian oscillator is also entrained by daily temperature rhythms (Wenden et al., 2011) and sugars (Blasing et al., 2005; Dodd et al., 2005; Knight et al., 2008; Dalchau et al., 2011; Haydon et al., 2013). However, the perception and transduction of such signals are not fully understood. Considering that photosynthates can contribute to the fine-tuning of the circadian clock (reviewed in Sanchez and Kay, 2016) and that floral signal transduction in LDs is also controlled by the circadian clock (Matsoukas et al., 2012; Song et al., 2013), it has been hypothesized that photosynthates might have a role in modulating the photoperiodic timing mechanism, which includes the PHYs and CRYs (Dodd et al., 2015).
PRRs have been identified as components of the circadian clock (Nakamichi et al., 2007; Ito et al., 2008). Generally, it has been proposed that PRRs contribute to photoperiod measurement through regulation of the time-keeping mechanism associated with CO transcription (Strayer et al., 2000; Yanovsky and Kay, 2002; Nakamichi et al., 2007, 2010). Recently, it was shown that PRRs form a light-signaling mechanism dedicated to photoperiodic flowering through their accumulation during the day, transferring information on light exposure to CO protein (Hayama et al., 2017), which acts upstream of FT and TSF. Interestingly, PRR7 expression is coordinately modulated not only by light but also by photosynthesis, permitting PRR7 to act as a transcriptional repressor in circadian sugar signaling (Haydon et al., 2013). Therefore, specific circadian-clock components not only transfer temporal information to a photoperiodic time-keeping mechanism but also convey qualitative and quantitative information on light exposure to the time-keeping mechanism, establishing measurement of day length.
Interplay Between Sugar and Phytochrome Signaling Modulates Floral Signal Transduction
In Arabidopsis, the PHY family consists of PHYA, PHYB, PHYD, and PHYE (Table 1; Clack et al., 1994). PHYA is predominately involved in physiological responses to continuous far-red light, whereas PHYB is involved in responses to red light. The phyA mutant flowers significantly later than wild type (WT) in long days (LDs), which indicates that PHYA acts to promote flowering (Johnson et al., 1994). In antithesis, the early flowering phenotype of phyB mutant under short day (SD) and LD conditions demonstrates the repressive role of PHYB in floral signal transduction (Guo et al., 1998). Interestingly, the identification of downstream components of photoreceptor-signaling that involved in floral induction has revealed a crosstalk between pathways of different light qualities as well as with other seemingly unrelated pathways such as phytohormones (Matsoukas, 2014b) and carbohydrate metabolism-related events (Dijkwel et al., 1997; Short, 1999; Kozuka et al., 2005; Ghassemian et al., 2006).
Carbohydrates modulate development through PHY-mediated responses (Tsukaya et al., 1991; Barnes et al., 1996; Dijkwel et al., 1997; Short, 1999). PHYA is involved in activation of several photosynthetic genes, such as RIBULOSE 1,5-BISPHOSPHATE CARBOXYLASE/OXYGENASE (RBCS), CHLOROPHYLL A/B-BINDING PROTEIN (CAB), and PLASTOCYANIN (PC). CAB, RBCS, and PC are repressed by sucrose or glucose (Dijkwel et al., 1997; Takano et al., 2009; Cottage and Gray, 2011). Exogenous sucrose application or high light intensity (LI) reverses the late-flowering phenotype of the Arabidopsis phyA mutant. It has been proposed that the late-flowering phenotype of phyA might be due to a reduced photosynthetic input to FT (King et al., 2008). This is supported by the fact that high LI reverses its late flowering phenotype, the mutant has half the WT leaf area and, in addition, a reduced photosynthetic pigment content (Walters et al., 1999; Bagnall and King, 2001; King et al., 2008).
Overexpression of PHYs in Nicotiana tabacum (Sharkey et al., 1991) and Solanum tuberosum (Sharkey et al., 1991; Yanovsky et al., 1998) increase the transcription of SUCROSE-PHOSPHATE SYNTHASE (SPS). Interestingly, ectopic expression of SPS has been shown to promote flowering in several plant species (Micallef et al., 1995; Baxter et al., 2003). On the other hand, loss of PHYs in Oryza sativa phyA phyB phyC triple mutant affect sugar metabolism, carbon partitioning and sugar transport (Jumtee et al., 2009). In Arabidopsis, the circadian regulated sugar-induced β-AMYLASE3 (BAM3) gene is induced by PHYA transcription (reviewed in Kaplan et al., 2006). BAM3 is essential for maltose production (Niittyla et al., 2004), whereas it regulates the juvenile-to-adult and vegetative-to-reproductive phase transitions via starch catabolism-related events (Matsoukas et al., 2013).
The SUCROSE UNCOUPLED6 (SUN6) gene of Arabidopsis is involved in hexose kinase-mediated sugar sensing (Huijser et al., 2000). Gene expression analysis in the sugar insensitive sun6 mutant has shown that PHYA signaling is not repressed by sugars (Dijkwel et al., 1997). SUN6 was shown to be allelic to ABA INSENSITIVE 4 (ABI4). Functional analysis of the abi4 mutant has shown that it is defective in ABA metabolism or response (Dijkwel et al., 1997; Huijser et al., 2000). Therefore, the early flowering phenotype of sun6, at least in LDs, demonstrates a tight interplay between light quality, sugar and phytohormone pathways in regulation of floral induction in Arabidopsis.
Further evidence on interaction between carbohydrate-metabolism repression and light signaling is provided by the inhibitory activity of PHYB in the control of hypocotyl elongation by PHYA, in presence of exogenous sucrose or glucose (Short, 1999). Down-regulation or over-expression of SUT4 in Solanum tuberosum delays or promotes floral induction, respectively (Chincinska et al., 2008). Besides floral induction, in the same work evidence was provided on SUT4 involvement in the shade avoidance response. This suggest that PHY-dependent and photoperiod-dependent developmental responses, such as floral signal transduction and shade avoidance share a common downstream mechanism in which sucrose accumulation levels are actively involved.
Interplay Between Sugar and Cryptochrome Signaling Modulates Floral Induction
CRYPTOCHROMES (CRYs) comprise flavoproteins that are able to detect blue light (Guo et al., 1998). The role of CRY1 in promoting floral induction in Arabidopsis has been demonstrated by the late flowering phenotype of cry1 mutants compared to WT in various light conditions (Mozley and Thomas, 1995). Similarly, the cry2/fha1 (fha-1 is a mutant allele of CRY2 in Landsberg erecta background) mutant flowers later than the WT in LDs but not in SDs, whereas transgenic plants overexpressing CRY2 flowered slightly early in SDs but not in LDs (Koornneef et al., 1991). It has been shown that CRY2 interacts with bHLH proteins CRYPTOCHROME-INTERACTING BASIC-HELIX-LOOP-HELIX (CIB) proteins to regulate the FT expression and floral signal transduction (Liu et al., 2008; Liu H. et al., 2013; Liu Y. et al., 2013).
Further evidence for the interaction between photosynthetic assimilates and CRYs is provided by a microarray analysis revealing regulation of CRY1 and CRY2 transcription levels by glucose (Li et al., 2006). It has been reported that PHYA interacts with CRY1, and PHYB binds CRY2 (Ahmad et al., 1998; Mas et al., 2000), so red and blue light may crosstalk at multiple layers to co-ordinately regulate developmental transitions. PHYB, CONSTANS (CO) and, indirectly, PHYA are under the regulation of CRYs (Valverde et al., 2004; Thomas, 2006). Therefore, any modification on CRYs transcription levels would also affect the other photoreceptors and CO, which act directly upstream of FT and TWIN SISTER OF FT (TSF) with catalytic effects on the juvenile-to-adult and vegetative-to-reproductive phase transitions.
Mutants lacking CRYs or having defects in their signaling pathway show changes in chloroplast composition and disturbance of normal acclimation (Smith et al., 1993; Walters et al., 1999). The fact that CRY1 and CRY2 can also act as sensors of irradiance (Guo et al., 1998) could provide a further link between light quality and carbohydrate metabolism in regulation of floral signal transduction.
The Arabidopsis ELONGATED HYPOCOTYL 5 protein (HY5) is a central mediator of CRY and PHY responses (Lee et al., 2007). It integrates multiple environmental and phytohormonal signaling inputs (Catala et al., 2011; Xu et al., 2014) by mediating homeostatic coordination of sugars (Chen et al., 2016), and maintaining chlorophyll levels and CO2 uptake. It appears that HY5 might operate in conjunction with the circadian oscillator to adjust levels of rhythmic photosynthetic gene expression (Toledo-Ortiz et al., 2014). Interestingly, HY5 regulates both sucrose metabolism and subsequent movement of sucrose into phloem cells for shoot-root translocation by promoting the expression levels of SWEET11 and SWEET12 (Chen et al., 2016), genes encoding sucrose efflux transporters (Chen et al., 2012), and TPS1 (Chen et al., 2016), a gene encoding T6P. The T6P pathway controls the expression of SPLs, partially via miR156, and partly independently of the miR156-dependent pathway via the florigen FT (Wahl et al., 2013). Evidence have been provided that miR156, and possibly miR172, are directly regulated by HY5 (Zhang et al., 2011). Taken together, these data could provide a potential mechanistic link, at the molecular level, on how the photoreceptor-sugar crosstalk might be involved in regulation of floral signal transduction via the HY5 and TPS1-miR156-SPL module.
Light Intensity and Floral Signal Transduction
LI seems to be particularly important during the juvenile-to-adult and vegetative-to-reproductive phase transition (Figure 1). It has been proposed that the inability to flower during the juvenile period is because of a foliar inability to produce floral signals, the presence of antiflorigens, and/or of the incompetence of the SAM to respond (Zeevaart, 1985; Matsoukas et al., 2012, 2013; Matsoukas, 2015). The length of the juvenile vegetative phase in daylenth-sensitive plants can be revealed by reciprocal transfers between inductive and non-inductive photoperiods (Adams et al., 2003; Matsoukas et al., 2013; Matsoukas, 2014a).
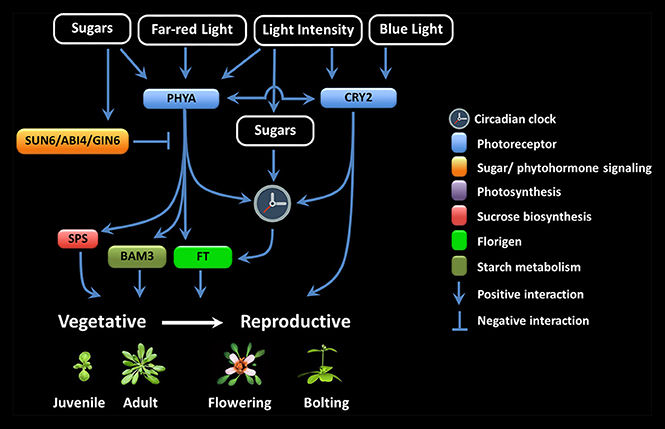
Figure 1. Multiple interactions among the components involved in floral signal transduction in response to photoreceptor and sugar signaling crosstalk.
Exposure to low or high LI levels can delay or hasten time to flowering, respectively. For instance, Achillea millefolium grown under a 16 h d−1 photoperiod in controlled environment conditions flowered after 57, 45, and 37 d when grown under 100, 200, or 300 μmol m−2 s−1, respectively (Zhang et al., 1996). Similarly, Adams et al. (1999) demonstrated that Petunia flowering was hastened by LDs, but that decreased LI prolonged time to flowering. Arabidopsis plants flower rapidly under non-inductive SDs after exposure to 8–12 d at a high LI. It has been shown that this “photosynthetic” response is FT-independent. In contrast, the IDD8 locus of Arabidopsis was reported to have a role in FT-dependent induction of flowering by modulating sugar transport and metabolism by regulating SUCROSE SYNTHASE4 activity (Seo et al., 2011).
However, the effect of LI on time to flowering can be unpredictable in several species. Hence, the term “facultative irradiance response” (FI) has been coined to describe a developmental hastening of flowering by addition of supplemental light (Erwin and Warner, 2000). Species such as Antirrhinum [LD plant (LDP)], Nicotiana [LDP or SD plant (SDP)], and Petunia (LDP) that exhibit a FI response, show a decrease in leaf numbers and days to flower as irradiance increases. In contrast, the term “irradiance indifferent” (II) refers to species such as Salvia (SDP or facultative LDP) and Zinnia (day neutral plant or facultative SDP) that do not show any response to increased irradiance (Thomas and Vince-Prue, 1997; Erwin and Warner, 2000; Mattson and Erwin, 2005; Thomas, 2006).
Despite the high sensitivity of FI species to elevated levels of LI, the majority does not show a hastened flowering phenotype with increasing irradiance. It has been shown for Pelargonium x hortorum that a linear relationship between LI and days to flower, for an increased irradiance developmental response, exists until a threshold level between 6.89 and 9.01 μmol m−2 d−1 (Erickson et al., 1980). However, some species require greater threshold levels. For instance, absolute flowering of Digitalis was reached with LI > 11 μmol m d (Fausey et al., 2001). Furthermore, giving supplemental irradiance (at 30, 60, and 90 μmol m−2 s−1) to Gerbera hastened flowering by up to 23 d in the winter, but only up to 11 d during the Spring (Gagnon and Dansereau, 1989). This suggests that the impact of supplemental irradiance on floral signal transduction can be dependent on season's ambient light conditions and species' threshold requirement.
What is not clear is the precise molecular genetic mechanisms by which LI, if acting through photosynthates can regulate the floral signal transduction. It may well be that assimilates themselves act as part of the florigen (Périlleux and Bernier, 2002; Bernier and Perilleux, 2005). Interestingly, long-distance floral signal transport is now accepted as more complex than the movement of a single type of signal molecule (Matsoukas et al., 2012; Matsoukas, 2015). It is possible that total carbohydrate, or a particular carbohydrate level may be required to reach a specific threshold in order to sustain a steady supply of sufficient bulk flow through the phloem from the leaves to the SAM to enable delivery of florigen. This would be necessary to render the SAM competent to flower.
Concluding Remarks
Floral signal transduction has been the focus of a great deal of attention during the last few decades. The molecular mechanisms underlying light perception and the downstream signaling pathways that regulate the floral signal transduction have been intensively challenged. The fact that some photoreceptors can also act as sensors of irradiance provides a promising link between light qualities and assimilate partitioning and resource utilization in regulation of floral signal transduction.
Numerous reports highlight the role of several molecules that integrate light, clock, temperature, and hormone signaling pathways in orchestration of floral signal transduction. However, further investigation is vital for the elucidation of the molecular mechanism underlying photoreceptor-mediated signal integration at the subcellular, tissue-specific and temporal level in response to sugar signaling. This research field is prosperous and technical advances in “-OMICS” tools might shed light on the underlying molecular genetic mechanisms.
Author Contributions
The author confirms being the sole contributor of this work and approved it for publication.
Funding
Research in the laboratory of IM is supported by a Jenkinson TIRI Award and the University of Bolton, UK.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author would like to thank Professor Brian Thomas (University of Warwick, UK) and Dr. Andrea Massiah (University of Warwick, UK) for valuable discussions, and the three independent reviewers for their insightful comments on the article.
References
Abdel-Ghany, S. E. (2009). Contribution of plastocyanin isoforms to photosynthesis and copper homeostasis in Arabidopsis thaliana grown at different copper regimes. Planta 229, 767–779. doi: 10.1007/s00425-008-0869-z
Adams, S. R., Munir, M., Valdes, V. M., Langton, F. A., and Jackson, S. D. (2003). Using flowering times and leaf numbers to model the phases of photoperiod sensitivity in Antirrhinum majus L. Ann. Bot. 92, 689–696. doi: 10.1093/aob/mcg194
Adams, S. R., Pearson, S., Hadley, P., and Patefield, W. (1999). The effects of temperature and light integral on the phases of photoperiod sensitivity in Petunia x hybrida. Ann. Bot. 83, 263–269. doi: 10.1006/anbo.1998.0817
Ahmad, M., Jarillo, J. A., Smirnova, O., and Cashmore, A. R. (1998). The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol. Cell 1, 939–948. doi: 10.1016/S1097-2765(00)80094-5
Ahmad, M., Lin, C., and Cashmore, A. (1995). Mutations throughout an Arabidopsis blue-light photoreceptor impair blue-light-responsive anthocyanin accumulation and inhibition of hypocotyl elongation. Plant J. 8, 653–658. doi: 10.1046/j.1365-313X.1995.08050653.x
Alabadi, D., Oyama, T., Yanovsky, M. J., Harmon, F. G., Mas, P., and Kay, S. A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883. doi: 10.1126/science.1061320
Arenas-Huertero, F., Arroyo, A., Zhou, L., Sheen, J., and Leon, P. (2000). Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 14, 2085–2096. doi: 10.1101/gad.14.16.2085
Aukerman, M. J., and Sakai, H. (2003). Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15, 2730–2741. doi: 10.1105/tpc.016238
Baena-Gonzalez, E., Rolland, F., Thevelein, J. M., and Sheen, J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942. doi: 10.1038/nature06069
Bagnall, D. J., and King, R. W. (2001). Phytochrome, photosynthesis and flowering of Arabidopsis thaliana: photophysiological studies using mutants and transgenic lines. Funct. Plant Biol. 28, 401–408. doi: 10.1071/PP99123
Barnes, S. A., Nishizawa, N. K., Quaggio, R. B., Whitelam, G. C., and Chua, N. H. (1996). Far-red light blocks greening of Arabidopsis seedlings via a phytochrome A-mediated change in plastid development. Plant Cell 8, 601–615. doi: 10.1105/tpc.8.4.601
Baxter, C. J., Foyer, C. H., Turner, J., Rolfe, S. A., and Quick, W. P. (2003). Elevated sucrose-phosphate synthase activity in transgenic tobacco sustains photosynthesis in older leaves and alters development. J. Exp. Bot. 54, 1813–1820. doi: 10.1093/jxb/erg196
Bernier, G., and Perilleux, C. (2005). A physiological overview of the genetics of flowering time control. Plant Biotechnol. J. 3, 3–16. doi: 10.1111/j.1467-7652.2004.00114.x
Blasing, O. E., Gibon, Y., Gunther, M., Hohne, M., Morcuende, R., Osuna, D., et al. (2005). Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17, 3257–3281. doi: 10.1105/tpc.105.035261
Bolouri Moghaddam, M. R., and Van den Ende, W. (2013). Sugars, the clock and transition to flowering. Front. Plant Sci. 4:22. doi: 10.3389/fpls.2013.00022
Cardon, G., Hohmann, S., Klein, J., Nettesheim, K., Saedler, H., and Huijser, P. (1999). Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237, 91–104. doi: 10.1016/S0378-1119(99)00308-X
Catala, R., Medina, J., and Salinas, J. (2011). Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 16475–16480. doi: 10.1073/pnas.1107161108
Chaves, I., Pokorny, R., Byrdin, M., Hoang, N., Ritz, T., Brettel, K., et al. (2011). The cryptochromes: blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 62, 335–364. doi: 10.1146/annurev-arplant-042110-103759
Chen, L. Q., Qu, X. Q., Hou, B. H., Sosso, D., Osorio, S., Fernie, A. R., et al. (2012). Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335, 207–211. doi: 10.1126/science.1213351
Chen, M., and Chory, J. (2011). Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 21, 664–671. doi: 10.1016/j.tcb.2011.07.002
Chen, X., Yao, Q., Gao, X., Jiang, C., Harberd, N. P., and Fu, X. (2016). Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 26, 640–646. doi: 10.1016/j.cub.2015.12.066
Chincinska, I., Liesche, J., Krugel, U., Michalska, J., Geigenberger, P., Grimm, B., et al. (2008). Sucrose transporter StSUT4 from potato affects flowering, tuberization, and shade avoidance response. Plant Physiol. 146, 515–528. doi: 10.1104/pp.107.112334
Chow, B. Y., and Kay, S. A. (2013). Global approaches for telling time: omics and the Arabidopsis circadian clock. Semin. Cell Dev. Biol. 24, 383–392. doi: 10.1016/j.semcdb.2013.02.005
Christie, J. M. (2007). Phototropin blue-light receptors. Annu. Rev. Plant Biol. 58, 21–45. doi: 10.1146/annurev.arplant.58.032806.103951
Clack, T., Mathews, S., and Sharrock, R. A. (1994). The phytochrome apoprotein family in Arabidopsis is encoded by 5 genes: the sequences and expression of PHYD and PHYE. Plant Mol. Biol. 25, 413–427. doi: 10.1007/BF00043870
Corbesier, L., Vincent, C., Jang, S. H., Fornara, F., Fan, Q. Z., Searle, I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. doi: 10.1126/science.1141752
Cottage, A., and Gray, J. C. (2011). Timing the switch to phototrophic growth: a possible role of GUN1. Plant Signal. Behav. 6, 578–582. doi: 10.4161/psb.6.4.14900
Covington, M. F., Maloof, J. N., Straume, M., Kay, S. A., and Harmer, S. L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9:R130. doi: 10.1186/gb-2008-9-8-r130
Dalchau, N., Baek, S. J., Briggs, H. M., Robertson, F. C., Dodd, A. N., Gardner, M. J., et al. (2011). The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc. Natl. Acad. Sci. U.S.A. 108, 5104–5109. doi: 10.1073/pnas.1015452108
Delatte, T. L., Sedijani, P., Kondou, Y., Matsui, M., De Jong, G. J., Somsen, G. W., et al. (2011). Growth arrest by trehalose-6-phosphate: an astonishing case of primary metabolite control over growth by way of the SnRK1 signaling pathway. Plant Physiol. 157, 160–174. doi: 10.1104/pp.111.180422
Devlin, P. F., Patel, S. R., and Whitelam, G. C. (1998). Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10, 1479–1487. doi: 10.1105/tpc.10.9.1479
Dijkwel, P. P., Huijser, C., Weisbeek, P. J., Chua, N. H., and Smeekens, S. C. (1997). Sucrose control of phytochrome A signaling in Arabidopsis. Plant Cell 9, 583–595. doi: 10.1105/tpc.9.4.583
Dobrenel, T., Marchive, C., Azzopardi, M., Clement, G., Moreau, M., Sormani, R., et al. (2013). Sugar metabolism and the plant target of rapamycin kinase: a sweet operaTOR? Front. Plant Sci. 4:93. doi: 10.3389/fpls.2013.00093
Dodd, A. N., Belbin, E., Frank, A., and Webb, A. A. (2015). Interactions between circadian clocks and photosynthesis for the temporal and spatial coordination of metabolism. Front. Plant Sci. 6:245. doi: 10.3389/fpls.2015.00245
Dodd, A. N., Salathia, N., Hall, A., Kevei, E., Toth, R., Nagy, F., et al. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309, 630–633. doi: 10.1126/science.1115581
Eimert, K., Wang, S. M., Lue, W. I., and Chen, J. (1995). Monogenic recessive mutations causing both late floral initiation and excess starch accumulation in Arabidopsis. Plant Cell 7, 1703–1712. doi: 10.1105/tpc.7.10.1703
Endo, M. (2016). Tissue-specific circadian clocks in plants. Curr. Opin. Plant Biol. 29, 44–49. doi: 10.1016/j.pbi.2015.11.003
Erickson, V., Armitage, A., Carlson, W., and Miranda, R. (1980). The effect of cumulative photosynthetically active radiation on the growth and flowering of the seedling geranium, Pelargonium x hortorum. HortScience 15, 815–817.
Erwin, J., and Warner, R. (2000). Determination of photoperiodic response group and effect of supplemental irradiance on flowering of several bedding plant species. Acta Hortic. 580, 95–99. doi: 10.17660/ActaHortic.2002.580.11
Farre, E. M., Harmer, S. L., Harmon, F. G., Yanovsky, M. J., and Kay, S. A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15, 47–54. doi: 10.1016/j.cub.2004.12.067
Fausey, B., Cameron, A., and Heins, R. (2001). Daily light integral, photoperiod, and vernalization affect flowering of Digitalis purpurea L.‘Foxy’. HortScience 36:565.
Finkelstein, R. R., Wang, M. L., Lynch, T. J., Rao, S., and Goodman, H. M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10, 1043–1054. doi: 10.1105/tpc.10.6.1043
Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., et al. (1999). GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18, 4679–4688. doi: 10.1093/emboj/18.17.4679
Friso, G., Giacomelli, L., Ytterberg, A. J., Peltier, J. B., Rudella, A., Sun, Q., et al. (2004). In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16, 478–499. doi: 10.1105/tpc.017814
Gagnon, S., and Dansereau, B. (1989). Influence of light and photoperiod on growth and development of gerbera. Acta Hortic. 272, 145–152.
Ghassemian, M., Lutes, J., Tepperman, J. M., Chang, H. S., Zhu, T., Wang, X., et al. (2006). Integrative analysis of transcript and metabolite profiling data sets to evaluate the regulation of biochemical pathways during photomorphogenesis. Arch. Biochem. Biophys. 448, 45–59. doi: 10.1016/j.abb.2005.11.020
Guo, H., Yang, H., Mockler, T. C., and Lin, C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279, 1360–1363. doi: 10.1126/science.279.5355.1360
Harmer, S. L., Hogenesch, J. B., Straume, M., Chang, H. S., Han, B., Zhu, T., et al. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113. doi: 10.1126/science.290.5499.2110
Hayama, R., Sarid-Krebs, L., Richter, R., Fernandez, V., Jang, S., and Coupland, G. (2017). PSEUDO RESPONSE REGULATORs stabilize CONSTANS protein to promote flowering in response to day length. EMBO J. 36, 904–918. doi: 10.15252/embj.201693907
Haydon, M. J., Mielczarek, O., Robertson, F. C., Hubbard, K. E., and Webb, A. A. (2013). Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 502, 689–692. doi: 10.1038/nature12603
Hazen, S. P., Schultz, T. F., Pruneda-Paz, J. L., Borevitz, J. O., Ecker, J. R., and Kay, S. A. (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. U.S.A. 102, 10387–10392. doi: 10.1073/pnas.0503029102
Herrero, E., Kolmos, E., Bujdoso, N., Yuan, Y., Wang, M., Berns, M. C., et al. (2012). EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24, 428–443. doi: 10.1105/tpc.111.093807
Huang, W., Perez-Garcia, P., Pokhilko, A., Millar, A. J., Antoshechkin, I., Riechmann, J. L., et al. (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336, 75–79. doi: 10.1126/science.1219075
Huijser, C., Kortstee, A., Pego, J., Weisbeek, P., Wisman, E., and Smeekens, S. (2000). The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE4: involvement of abscisic acid in sugar responses. Plant J. 23, 577–585. doi: 10.1046/j.1365-313x.2000.00822.x
Ito, S., Niwa, Y., Nakamichi, N., Kawamura, H., Yamashino, T., and Mizuno, T. (2008). Insight into missing genetic links between two evening-expressed pseudo-response regulator genes TOC1 and PRR5 in the circadian clock-controlled circuitry in Arabidopsis thaliana. Plant Cell Physiol. 49, 201–213. doi: 10.1093/pcp/pcm178
Jenkins, G. I. (2014). The UV-B photoreceptor UVR8: from structure to physiology. Plant Cell 26, 21–37. doi: 10.1105/tpc.113.119446
Johnson, E., Bradley, M., Harberd, N. P., and Whitelam, G. C. (1994). Photoresponses of light-grown phyA mutants of Arabidopsis (phytochrome A is required for the perception of daylength extensions). Plant Physiol. 105, 141–149. doi: 10.1104/pp.105.1.141
Jumtee, K., Okazawa, A., Harada, K., Fukusaki, E., Takano, M., and Kobayashi, A. (2009). Comprehensive metabolite profiling of phyA phyB phyC triple mutants to reveal their associated metabolic phenotype in rice leaves. J. Biosci. Bioeng. 108, 151–159. doi: 10.1016/j.jbiosc.2009.03.010
Jung, J. H., Seo, P. J., Kang, S. K., and Park, C. M. (2011). miR172 signals are incorporated into the miR156 signaling pathway at the SPL3/4/5 genes in Arabidopsis developmental transitions. Plant Mol. Biol. 76, 35–45. doi: 10.1007/s11103-011-9759-z
Jung, J. H., Seo, Y. H., Seo, P. J., Reyes, J. L., Yun, J., Chua, N. H., et al. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19, 2736–2748. doi: 10.1105/tpc.107.054528
Kami, C., Lorrain, S., Hornitschek, P., and Fankhauser, C. (2010). Light-regulated plant growth and development. Curr. Top. Dev. Biol. 91, 29–66. doi: 10.1016/S0070-2153(10)91002-8
Kaplan, F., and Guy, C. L. (2005). RNA interference of Arabidopsis beta-amylase8 prevents maltose accumulation upon cold shock and increases sensitivity of PSII photochemical efficiency to freezing stress. Plant J. 44, 730–743. doi: 10.1111/j.1365-313X.2005.02565.x
Kaplan, F., Sung, D. Y., and Guy, C. L. (2006). Roles of β-amylase and starch breakdown during temperatures stress. Physiol. Plant. 126, 120–128. doi: 10.1111/j.1399-3054.2006.00604.x
Kardailsky, I., Shukla, V. K., Ahn, J. H., Dagenais, N., Christensen, S. K., Nguyen, J. T., et al. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965. doi: 10.1126/science.286.5446.1962
Kim, W. Y., Fujiwara, S., Suh, S. S., Kim, J., Kim, Y., Han, L., et al. (2007). ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449, 356–360. doi: 10.1038/nature06132
King, R., Hisamatsu, T., Goldschmidt, E. E., and Blundell, C. (2008). The nature of floral signals in Arabidopsis. I. Photosynthesis and a far-red photoresponse independently regulate flowering by increasing expression of FLOWERING LOCUS T (FT). J. Exp. Bot. 59, 3811–3820. doi: 10.1093/jxb/ern231
Knight, H., Thomson, A. J., and Mcwatters, H. G. (2008). Sensitive to freezing6 integrates cellular and environmental inputs to the plant circadian clock. Plant Physiol. 148, 293–303. doi: 10.1104/pp.108.123901
Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. doi: 10.1126/science.286.5446.1960
Koornneef, M., Hanhart, C. J., and Vanderveen, J. H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66. doi: 10.1007/BF00264213
Koornneef, M., Rolff, E., and Spruit, C. (1980). Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Get the document, find related information or use other SFX services. Z. Pflanzenphysiol. 100, 147–160. doi: 10.1016/S0044-328X(80)80208-X
Kozuka, T., Horiguchi, G., Kim, G. T., Ohgishi, M., Sakai, T., and Tsukaya, H. (2005). The different growth responses of the Arabidopsis thaliana leaf blade and the petiole during shade avoidance are regulated by Photoreceptors and sugar. Plant Cell Physiol. 46, 213–223. doi: 10.1093/pcp/pci016
Lao, N. T., Schoneveld, O., Mould, R. M., Hibberd, J. M., Gray, J. C., and Kavanagh, T. A. (1999). An Arabidopsis gene encoding a chloroplast-targeted beta-amylase. Plant J. 20, 519–527. doi: 10.1046/j.1365-313X.1999.00625.x
Lastdrager, J., Hanson, J., and Smeekens, S. (2014). Sugar signals and the control of plant growth and development. J. Exp. Bot. 65, 799–807. doi: 10.1093/jxb/ert474
Lauter, N., Kampani, A., Carlson, S., Goebel, M., and Moose, S. P. (2005). microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. U.S.A. 102, 9412–9417. doi: 10.1073/pnas.0503927102
Lee, H., Yoo, S. J., Lee, J. H., Kim, W., Yoo, S. K., Fitzgerald, H., et al. (2010). Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res. 38, 3081–3093. doi: 10.1093/nar/gkp1240
Lee, J., He, K., Stolc, V., Lee, H., Figueroa, P., Gao, Y., et al. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749. doi: 10.1105/tpc.106.047688
Li, L., and Sheen, J. (2016). Dynamic and diverse sugar signaling. Curr. Opin. Plant Biol. 33, 116–125. doi: 10.1016/j.pbi.2016.06.018
Li, Y., Lee, K. K., Walsh, S., Smith, C., Hadingham, S., Sorefan, K., et al. (2006). Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Res. 16, 414–427. doi: 10.1101/gr.4237406
Lifschitz, E., Ayre, B. G., and Eshed, Y. (2014). Florigen and anti-florigen-a systemic mechanism for coordinating growth and termination in flowering plants. Front. Plant Sci. 5:465. doi: 10.3389/fpls.2014.00465
Liu, H., Wang, Q., Liu, Y., Zhao, X., Imaizumi, T., Somers, D. E., et al. (2013). Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 110, 17582–17587. doi: 10.1073/pnas.1308987110
Liu, H., Yu, X., Li, K., Klejnot, J., Yang, H., Lisiero, D., et al. (2008). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535–1539. doi: 10.1126/science.1163927
Liu, Y., Li, X., Li, K., Liu, H., and Lin, C. (2013). Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet. 9:e1003861. doi: 10.1371/journal.pgen.1003861
Lunn, J. E., Feil, R., Hendriks, J. H., Gibon, Y., Morcuende, R., Osuna, D., et al. (2006). Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem. J. 397, 139–148. doi: 10.1042/BJ20060083
Martin, A., Adam, H., Diaz-Mendoza, M., Zurczak, M., Gonzalez-Schain, N. D., and Suarez-Lopez, P. (2009). Graft-transmissible induction of potato tuberization by the microRNA miR172. Development 136, 2873–2881. doi: 10.1242/dev.031658
Mas, P., Devlin, P. F., Panda, S., and Kay, S. A. (2000). Functional interaction of phytochrome B and cryptochrome 2. Nature 408, 207–211. doi: 10.1038/35041583
Mathieu, J., Yant, L. J., Murdter, F., Kuttner, F., and Schmid, M. (2009). Repression of flowering by the miR172 target SMZ. PLoS Biol. 7:e1000148. doi: 10.1371/journal.pbio.1000148
Matsoukas, I. G. (2014a). Attainment of reproductive competence, phase transition, and quantification of juvenility in mutant genetic screens. Front. Plant Sci. 5:32. doi: 10.3389/fpls.2014.00032
Matsoukas, I. G. (2014b). Interplay between sugar and hormone signaling pathways modulate floral signal transduction. Front. Genet. 5:218. doi: 10.3389/fgene.2014.00218
Matsoukas, I. G. (2015). Florigens and antiflorigens: a molecular genetic understanding. Essays Biochem. 58, 133–149. doi: 10.1042/bse0580133
Matsoukas, I. G., Massiah, A. J., and Thomas, B. (2012). Florigenic and antiflorigenic signalling in plants. Plant Cell Physiol. 53, 1827–1842. doi: 10.1093/pcp/pcs130
Matsoukas, I. G., Massiah, A. J., and Thomas, B. (2013). Starch metabolism and antiflorigenic signals modulate the juvenile-to-adult phase transition in Arabidopsis. Plant Cell Environ. 36, 1802–1811. doi: 10.1111/pce.12088
Matsushika, A., Makino, S., Kojima, M., and Mizuno, T. (2000). Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol. 41, 1002–1012. doi: 10.1093/pcp/pcd043
Mattson, N., and Erwin, J. (2005). The impact of photoperiod and irradiance on flowering of several herbaceous ornamentals. Sci. Horticult. 104, 275–292. doi: 10.1016/j.scienta.2004.08.018
May, P., Liao, W., Wu, Y., Shuai, B., Mccombie, W. R., Zhang, M. Q., et al. (2013). The effects of carbon dioxide and temperature on microRNA expression in Arabidopsis development. Nat. Commun. 4:2145. doi: 10.1038/ncomms3145
Mccallum, C. M., Comai, L., Greene, E. A., and Henikoff, S. (2000). Targeted screening for induced mutations. Nat. Biotechnol. 18, 455–457. doi: 10.1038/74542
Micallef, B. J., Haskins, K. A., Vanderveer, P. J., Roh, K. S., Shewmaker, C. K., and Sharkey, T. D. (1995). Altered photosynthesis, flowering, and fruiting in transgenic tomato plants that have an increased capacity for sucrose synthesis. Planta 196, 327–334. doi: 10.1007/BF00201392
Moore, B., Zhou, L., Rolland, F., Hall, Q., Cheng, W. H., Liu, Y. X., et al. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300, 332–336. doi: 10.1126/science.1080585
Mozley, D., and Thomas, B. (1995). Developmental and photobiological factors affecting photoperiodic induction in Arabidopsis thaliana Heynh Landsberg erecta. J. Exp. Bot. 46, 173–179. doi: 10.1093/jxb/46.2.173
Nakamichi, N., Kiba, T., Henriques, R., Mizuno, T., Chua, N. H., and Sakakibara, H. (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22, 594–605. doi: 10.1105/tpc.109.072892
Nakamichi, N., Kita, M., Niinuma, K., Ito, S., Yamashino, T., Mizoguchi, T., et al. (2007). Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 48, 822–832. doi: 10.1093/pcp/pcm056
Nelson, D., Lasswell, J., Rogg, L., Cohen, M., and Bartel, B. (2000). FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101, 331–340. doi: 10.1016/S0092-8674(00)80842-9
Niittyla, T., Messerli, G., Trevisan, M., Chen, J., Smith, A. M., and Zeeman, S. C. (2004). A previously unknown maltose transporter essential for starch degradation in leaves. Science 303, 87–89. doi: 10.1126/science.1091811
Nusinow, D. A., Helfer, A., Hamilton, E. E., King, J. J., Imaizumi, T., Schultz, T. F., et al. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475, 398–402. doi: 10.1038/nature10182
Park, D. H., Somers, D. E., Kim, Y. S., Choy, Y. H., Lim, H. K., Soh, M. S., et al. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285, 1579–1582. doi: 10.1126/science.285.5433.1579
Park, J. Y., Canam, T., Kang, K. Y., Ellis, D. D., and Mansfield, S. D. (2008). Over-expression of an Arabidopsis family A sucrose phosphate synthase (SPS) gene alters plant growth and fibre development. Transgenic Res. 17, 181–192. doi: 10.1007/s11248-007-9090-2
Penfield, S., and Hall, A. (2009). A role for multiple circadian clock genes in the response to signals that break seed dormancy in Arabidopsis. Plant Cell 21, 1722–1732. doi: 10.1105/tpc.108.064022
Périlleux, C., and Bernier, G. (2002). “The control of flowering: do genetical and physiological approaches converge?,” in Plant Reproduction, Annual Plant Reviews, eds S. D. O'neill and J. A. Roberts (Sheffield: Sheffield Academic Press), 1–32.
Pesaresi, P., Scharfenberg, M., Weigel, M., Granlund, I., Schroder, W. P., Finazzi, G., et al. (2009). Mutants, overexpressors, and interactors of Arabidopsis plastocyanin isoforms: revised roles of plastocyanin in photosynthetic electron flow and thylakoid redox state. Mol. Plant 2, 236–248. doi: 10.1093/mp/ssn041
Pokhilko, A., Fernandez, A. P., Edwards, K. D., Southern, M. M., Halliday, K. J., and Millar, A. J. (2012). The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 8:574. doi: 10.1038/msb.2012.6
Polge, C., and Thomas, M. (2007). SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 12, 20–28. doi: 10.1016/j.tplants.2006.11.005
Ponnu, J., Wahl, V., and Schmid, M. (2011). Trehalose-6-phosphate: connecting plant metabolism and development. Front. Plant Sci. 2:70. doi: 10.3389/fpls.2011.00070
Reed, J., Nagatani, A., Elich, T., Fagan, M., and Chory, J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 1139–1149. doi: 10.1104/pp.104.4.1139
Ren, M., Venglat, P., Qiu, S., Feng, L., Cao, Y., Wang, E., et al. (2012). Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell 24, 4850–4874. doi: 10.1105/tpc.112.107144
Rizzini, L., Favory, J. J., Cloix, C., Faggionato, D., O'hara, A., Kaiserli, E., et al. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332, 103–106. doi: 10.1126/science.1200660
Rolland, F., Baena-Gonzalez, E., and Sheen, J. (2006). Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol. 57, 675–709. doi: 10.1146/annurev.arplant.57.032905.105441
Romanowski, A., and Yanovsky, M. J. (2015). Circadian rhythms and post-transcriptional regulation in higher plants. Front. Plant Sci. 6:437. doi: 10.3389/fpls.2015.00437
Sanchez, S. E., and Kay, S. A. (2016). The plant circadian clock: from a simple timekeeper to a complex developmental manager. Cold Spring Harb. Perspect. Biol. 8:a027748. doi: 10.1101/cshperspect.a027748
Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carre, I. A., et al. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93, 1219–1229. doi: 10.1016/S0092-8674(00)81465-8
Schultz, T., Kiyosue, T., Yanovsky, M., Wada, M., and Kay, S. (2001). A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell 13, 2659–2670. doi: 10.1105/tpc.13.12.2659
Schulze, W. X., Reinders, A., Ward, J. M., Lalonde, S., and Frommer, W. B. (2003). Interactions between co-expressed Arabidopsis sucrose transporters in the split-ubiquitin system. BMC Biochem. 4:3. doi: 10.1186/1471-2091-4-3
Schwarz, S., Grande, A. V., Bujdoso, N., Saedler, H., and Huijser, P. (2008). The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 67, 183–195. doi: 10.1007/s11103-008-9310-z
Seo, P. J., Ryu, J., Kang, S. K., and Park, C. M. (2011). Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis. Plant J. 65, 418–429. doi: 10.1111/j.1365-313X.2010.04432.x
Sharkey, T. D., Vassey, T. L., Vanderveer, P. J., and Vierstra, R. D. (1991). Carbon metabolism enzymes and photosynthesis in transgenic tobacco (Nicotiana tabacum L.) having excess phytochrome. Planta 185, 287–296. doi: 10.1007/BF00201046
Shikata, M., Koyama, T., Mitsuda, N., and Ohme-Takagi, M. (2009). Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 50, 2133–2145. doi: 10.1093/pcp/pcp148
Shikata, M., Yamaguchi, H., Sasaki, K., and Ohtsubo, N. (2012). Overexpression of Arabidopsis miR157b induces bushy architecture and delayed phase transition in Torenia fournieri. Planta 236, 1027–1035. doi: 10.1007/s00425-012-1649-3
Short, T. W. (1999). Overexpression of Arabidopsis phytochrome B inhibits phytochrome A function in the presence of sucrose. Plant Physiol. 119, 1497–1506. doi: 10.1104/pp.119.4.1497
Smeekens, S. (2015). From leaf to kernel: trehalose-6-Phosphate signaling moves carbon in the field. Plant Physiol. 169, 912–913. doi: 10.1104/pp.15.01177
Smeekens, S., and Hellmann, H. A. (2014). Sugar sensing and signaling in plants. Front. Plant Sci. 5:113. doi: 10.3389/fpls.2014.00113
Smeekens, S., Ma, J., Hanson, J., and Rolland, F. (2010). Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 13, 274–279. doi: 10.1016/j.pbi.2009.12.002
Smith, H., Samson, G., and Fork, D. (1993). Photosynthetic acclimation to shade: probing the role of phytochromes using photomorphogenic mutants of tomato. Plant Cell Environ. 16, 929–937. doi: 10.1111/j.1365-3040.1993.tb00516.x
Somers, D., Schultz, T., Milnamow, M., and Kay, S. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101, 319–329. doi: 10.1016/S0092-8674(00)80841-7
Song, Y. H., Ito, S., and Imaizumi, T. (2013). Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 18, 575–583. doi: 10.1016/j.tplants.2013.05.003
Strayer, C., Oyama, T., Schultz, T. F., Raman, R., Somers, D. E., Mas, P., et al. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771. doi: 10.1126/science.289.5480.768
Suetsugu, N., and Wada, M. (2013). Evolution of three LOV blue light receptor families in green plants and photosynthetic stramenopiles: phototropin, ZTL/FKF1/LKP2 and aureochrome. Plant Cell Physiol. 54, 8–23. doi: 10.1093/pcp/pcs165
Takano, M., Inagaki, N., Xie, X., Kiyota, S., Baba-Kasai, A., Tanabata, T., et al. (2009). Phytochromes are the sole photoreceptors for perceiving red/far-red light in rice. Proc. Natl. Acad. Sci. U.S.A. 106, 14705–14710. doi: 10.1073/pnas.0907378106
Telfer, A., Bollman, K. M., and Poethig, R. S. (1997). Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124, 645–654.
Telfer, A., and Poethig, R. S. (1998). HASTY: a gene that regulates the timing of shoot maturation in Arabidopsis thaliana. Development 125, 1889–1898.
Tognetti, J. A., Pontis, H. G., and Martinez-Noel, G. M. (2013). Sucrose signaling in plants: a world yet to be explored. Plant Signal. Behav. 8:e23316. doi: 10.4161/psb.23316
Toledo-Ortiz, G., Johansson, H., Lee, K. P., Bou-Torrent, J., Stewart, K., Steel, G., et al. (2014). The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 10:e1004416. doi: 10.1371/journal.pgen.1004416
Tsai, A. Y., and Gazzarrini, S. (2012). AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 69, 809–821. doi: 10.1111/j.1365-313X.2011.04832.x
Tseng, T. S., Salome, P. A., Mcclung, C. R., and Olszewski, N. E. (2004). SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 16, 1550–1563. doi: 10.1105/tpc.019224
Tsukaya, H., Ohshima, T., Naito, S., Chino, M., and Komeda, Y. (1991). Sugar-dependent expression of the CHS-A gene for chalcone synthase from petunia in transgenic Arabidopsis. Plant Physiol. 97, 1414–1421. doi: 10.1104/pp.97.4.1414
Turck, F., Fornara, F., and Coupland, G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594. doi: 10.1146/annurev.arplant.59.032607.092755
Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., and Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006. doi: 10.1126/science.1091761
Van den Ende, W. (2014). Sugars take a central position in plant growth, development and, stress responses. A focus on apical dominance. Front. Plant Sci. 5:313. doi: 10.3389/fpls.2014.00313
Van Dijken, A. J., Schluepmann, H., and Smeekens, S. C. (2004). Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiol. 135, 969–977. doi: 10.1104/pp.104.039743
Varkonyi-Gasic, E., Gould, N., Sandanayaka, M., Sutherland, P., and Macdiarmid, R. M. (2010). Characterisation of microRNAs from apple (Malus domestica 'Royal Gala') vascular tissue and phloem sap. BMC Plant Biol. 10:159. doi: 10.1186/1471-2229-10-159
Wahl, V., Ponnu, J., Schlereth, A., Arrivault, S., Langenecker, T., Franke, A., et al. (2013). Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339, 704–707. doi: 10.1126/science.1230406
Walters, R. G., Rogers, J. J., Shephard, F., and Horton, P. (1999). Acclimation of Arabidopsis thaliana to the light environment: the role of photoreceptors. Planta 209, 517–527. doi: 10.1007/s004250050756
Wang, J.-W., Czech, B., and Weigel, D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749. doi: 10.1016/j.cell.2009.06.014
Wang, J. W., Schwab, R., Czech, B., Mica, E., and Weigel, D. (2008). Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20, 1231–1243. doi: 10.1105/tpc.108.058180
Wang, Z. Y., and Tobin, E. M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217. doi: 10.1016/S0092-8674(00)81464-6
Wenden, B., Kozma-Bognar, L., Edwards, K. D., Hall, A. J. W., Locke, J. C. W., and Millar, A. J. (2011). Light inputs shape the Arabidopsis circadian system. Plant J. 66, 480–491. doi: 10.1111/j.1365-313X.2011.04505.x
Wenkel, S., Turck, F., Singer, K., Gissot, L., Le Gourrierec, J., Samach, A., et al. (2006). CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18, 2971–2984. doi: 10.1105/tpc.106.043299
Whitelam, G., Johnson, E., Peng, J., Carol, P., Anderson, M., and Cowl, J. E. A. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5, 757–768. doi: 10.1105/tpc.5.7.757
Wu, G., Park, M. Y., Conway, S. R., Wang, J. W., Weigel, D., and Poethig, R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. doi: 10.1016/j.cell.2009.06.031
Wu, G., and Poethig, R. S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133, 3539–3547. doi: 10.1242/dev.02521
Xing, L. B., Zhang, D., Li, Y. M., Shen, Y. W., Zhao, C. P., Ma, J. J., et al. (2015). Transcription profiles reveal sugar and hormone signaling pathways mediating flower induction in Apple (Malus domestica Borkh.). Plant Cell Physiol. 56, 2052–2068. doi: 10.1093/pcp/pcv124
Xing, S., Salinas, M., Hohmann, S., Berndtgen, R., and Huijser, P. (2010). miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22, 3935–3950. doi: 10.1105/tpc.110.079343
Xu, D., Li, J., Gangappa, S. N., Hettiarachchi, C., Lin, F., Andersson, M. X., et al. (2014). Convergence of light and ABA signaling on the ABI5 promoter. PLoS Genet. 10:e1004197. doi: 10.1371/journal.pgen.1004197
Yadav, U. P., Ivakov, A., Feil, R., Duan, G. Y., Walther, D., Giavalisco, P., et al. (2014). The sucrose-trehalose 6-phosphate (Tre6P) nexus: specificity and mechanisms of sucrose signalling by Tre6P. J. Exp. Bot. 65, 1051–1068. doi: 10.1093/jxb/ert457
Yamaguchi, A., Kobayashi, Y., Goto, K., Abe, M., and Araki, T. (2005). TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 46, 1175–1189. doi: 10.1093/pcp/pci151
Yamaguchi, A., Wu, M. F., Yang, L., Wu, G., Poethig, R. S., and Wagner, D. (2009). The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 17, 268–278. doi: 10.1016/j.devcel.2009.06.007
Yanovsky, M. J., Alconada-Magliano, T. M., Mazzella, M. A., Gatz, C., Thomas, B., and Casal, J. J. (1998). Phytochrome A affects stem growth, anthocyanin synthesis, sucrose-phosphate-synthase activity and neighbour detection in sunlight-grown potato. Planta 205, 235–241. doi: 10.1007/s004250050316
Yanovsky, M. J., and Kay, S. A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419, 308–312. doi: 10.1038/nature00996
Yant, L., Mathieu, J., Dinh, T. T., Ott, F., Lanz, C., Wollmann, H., et al. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22, 2156–2170. doi: 10.1105/tpc.110.075606
Zeevaart, J. A. D. (1985). “Bryophyllum,” in Handbook of Flowering, ed A. H. Halevy (Boca Raton, FL: CRC Press Inc), 89–100.
Zhang, B., Wang, L., Zeng, L., Zhang, C., and Ma, H. (2015). Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev. 29, 975–987. doi: 10.1101/gad.251520.114
Zhang, D., Armitage, A., Affolter, J., and Dirr, M. (1996). Environmental control of flowering and growth of Achillea millefolium L. ‘summer pastels’. HortScience 31, 364–365.
Zhang, H., He, H., Wang, X., Wang, X., Yang, X., Li, L., et al. (2011). Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65, 346–358. doi: 10.1111/j.1365-313X.2010.04426.x
Zhang, Y., Primavesi, L. F., Jhurreea, D., Andralojc, P. J., Mitchell, R. A., Powers, S. J., et al. (2009). Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol. 149, 1860–1871. doi: 10.1104/pp.108.133934
Nomenclature
The following nomenclature will be used in this article:
• Names of genes are written in italicized upper-case letters, e.g., CRY2.
• Names of proteins are written in non-italicized upper-case letters, e.g., CRY2.
• Names of mutants are written in italicized lower-case letters, e.g., cry2.
Keywords: cryptochromes, developmental phase transitions, floral signal transduction, florigen, juvenile-to-adult phase transition, photoreceptors, phytochromes, sugar signaling
Citation: Matsoukas IG (2017) Crosstalk between Photoreceptor and Sugar Signaling Modulates Floral Signal Transduction. Front. Physiol. 8:382. doi: 10.3389/fphys.2017.00382
Received: 15 December 2016; Accepted: 22 May 2017;
Published: 12 June 2017.
Edited by:
Wim Van den Ende, KU Leuven, BelgiumReviewed by:
Matthew Paul, Rothamsted Research (BBSRC), United KingdomGaurav Zinta, Shanghai Center for Plant Stress Biology (PSC), CAS, China
Soulaiman Sakr, Agrocampus Ouest, France
Copyright © 2017 Matsoukas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ianis G. Matsoukas, i.matsoukas@bolton.ac.uk
†Present Address: Ianis G. Matsoukas, School of Sport and Biological Sciences, University of Bolton, Bolton, United Kingdom
 Ianis G. Matsoukas
Ianis G. Matsoukas