- 1State Key Laboratory of Biotherapy, Department of Pharmacy, West China Hospital, Sichuan University, Chengdu, China
- 2Department of Cardiology, The Second Hospital of Shandong University, Shandong University, Jinan, China
- 3Laboratory of Clinical Pharmacy and Adverse Drug Reaction, West China Hospital, Sichuan University, Chengdu, China
Sirt6 is one of the sirtuin family members, a kind of NAD+-dependent histone deacetylase and ADP-ribose transferase enzyme. It has an important role in physiological and pathological processes, regulating aging, cancer, obesity, insulin resistance, inflammation, and energy metabolism. Recent studies have suggested that reduced Sirt6 action is related to obesity and diabetes. Aging and overnutrition, two major risk factors for obesity and diabetes, lead to decreased Sirt6 level and function, which results in abnormal glucose and lipid metabolism. Whole-body ablation of Sirt6 in mice results in severe hypoglycemia. Sirt6 deficiency leads to liver steatosis and promotes diet-induced obesity and insulin resistance. Sirt6 has a protective effect on obesity and diabetes. This review surveys evidence for an emerging role of Sirt6 as a regulator of metabolism in mammals and summarizes its major functions in obesity and diabetes.
Introduction
The sirtuins are a highly conserved family of NAD+-dependent deacetylases and ADP-ribosyltransferases that play an important regulatory role in the physiological and pathological processes of the organism. They participate in regulating the life span and aging, cancer, obesity, insulin resistance, inflammatory response and energy metabolism (Michan and Sinclair, 2007). The founding member of the sirtuin family, Sirt2, was originally discovered in Saccharomyces cerevisiae. Currently, seven sirtuins (Sirt1-7) have been found in mammals, each containing the conserved sirtuin core domain that confers NAD+-dependent deacetylase activity (Frye, 1999).
Each member has distinct subcellular localizations, targets and functions. Sirt1 and Sirt2 were found in both the nucleus and cytoplasm; Sirt3, Sirt4, and Sirt5 were found in mitochondria, and Sirt6 and Sirt7 were found in the nucleus (Michishita et al., 2005). Sirt1-3 have strong deacetylase activity, whereas Sirt4 has ADP-ribosyltransferase activity (Michishita et al., 2005). Sirt5 has weak NAD+-dependent deacetylase, desuccinylase and demalonylase activity (Michishita et al., 2005). Sirt7 was recently shown to be a highly specific deacetylase (Michishita et al., 2005). Sirt6 has several enzymatic activities, including NAD+-dependent deacetylase activity and mono-ADP-ribosyl transferase activity of acetyl groups and long-chain acyl groups. The most well-studied sirtuin is Sirt1, and the study of Sirt6 is still in its infancy.
As one of NAD+-dependent deacetylases, Sirt6 was first cloned from a human spleen cDNA library (Liszt et al., 2005). Recent studies have suggested reduced Sirt6 activity related to obesity and diabetes. Aging and overnutrition, two major risk factors for obesity and diabetes, lead to decreased Sirt6 level and function and result in abnormal glucose and lipid metabolism. Whole-body ablation of Sirt6 in mice resulted in severe hypoglycemia (Kanfi et al., 2008, 2010; Kuang et al., 2017; Yao et al., 2017). Hepatic-specific ablation of Sirt6 increased liver steatosis (Kim et al., 2010). Fat-specific deletion of Sirt6 increased blood glucose levels and hepatic steatosis and promoted diet-induced obesity and insulin resistance (Kuang et al., 2017; Xiong et al., 2017; Yao et al., 2017). Neural-specific deletion of Sirt6 in mice promoted diet-induced obesity and insulin resistance (Schwer et al., 2010). In contrast, Sirt6 overexpression protected against diet-induced obesity and insulin resistance (Kanfi et al., 2010).
These recent studies showed that Sirt6 plays an important role in lipid and glucose metabolism. This review summarizes the role of Sirt6 in obesity and diabetes.
Sirt6 and Glucose Metabolism
Glucose is an essential energy source needed by all cells and organs of our bodies. Therefore, blood glucose should be kept at a certain level to maintain tissue and organ energy requirements. The imbalance of glucose homeostasis is an important pathogenic factor of diabetes, obesity and other metabolic diseases. Sirt6 plays an important role in glucose production and metabolism (Figure 1). Mice with whole-body Sirt6 deficiency showed severe hypoglycemia (Mostoslavsky et al., 2006). Subsequent studies found that Sirt6 affects both gluconeogenesis and glycolysis (Zhong et al., 2010; Dominy et al., 2012; Zhang et al., 2014). Moreover, recent reports suggested that Sirt6 regulates pancreatic β-cell function, an important organ for maintaining blood glucose level (Kugel et al., 2016; Demir et al., 2017).
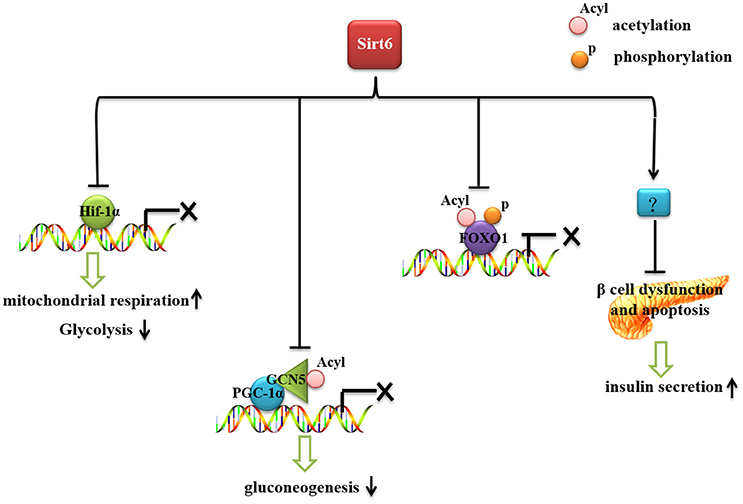
Figure 1. The role of Sirt6 in glucose metabolism. Sirt6 inhibits recruitment of Hif-1α to its target gene promoters, increases mitochondrial respiration and inhibits glycolysis. Sirt6 binds to and activates GCN5, inhibits the acetylation of PGC-1α, and decreases the expression of gluconeogenic genes. Sirt6 can specifically interact with FoxO1, decrease FoxO1 acetylation and phosphorylation level, and inhibit the interaction between FoxO1 and its downstream gene promoters, thereby reducing the expression of gluconeogenic genes. Sirt6 protects against β-cell dysfunction and apoptosis and increases insulin secretion.
Sirt6 and Blood Glucose
The first clue that Sirt6 might play a role in glucose metabolism came from a severe hypoglycemia phenotype observed in Sirt6-deficient mice (Mostoslavsky et al., 2006). Mice with whole-body Sirt6 deficiency showed increased glucose uptake and enhanced insulin signaling (Xiao et al., 2010). Sirt6-deficient mice, although small, appear normal in the first 2 weeks, then show a series of acute degenerative phenotypes, dying at about age 1 month (Mostoslavsky et al., 2006). The phenotypes include lymphopenia, osteopenia, lordokyphosis and loss of subcutaneous fat (Mostoslavsky et al., 2006). The most striking phenotype is severe hypoglycemia, which could be the main factor killing the mice before age 1 month. However, the severe hypoglycemia phenotype may be not triggered by the abnormal insulin level, because blood insulin levels are even lower than in wild-type mice (Xiao et al., 2010). When the mice were fed water containing 10% glucose, blood glucose was increased and about 83% of the mice with whole-body Sirt6 deficiency survived, indicating the hypoglycemia was a main factor in the postnatal lethality (Xiao et al., 2010).
Xiao et al. found that Sirt6 negatively regulates Akt phosphorylation at Thr 308 and Ser 473 by inhibiting multiple upstream molecules, insulin receptors, and insulin receptor substrate 1 and 2 (Xiao et al., 2010). With Sirt6 deficiency, insulin signaling is activated and Akt phosphorylation is increased (Xiao et al., 2010). Then, glucose uptake is increased, which may cause hypoglycemia. In contrast, Sirt6 deficiency does not affect intestinal glucose absorption and does not lead to higher glucose secretion in the kidney (Zhong et al., 2010). In vivo study by 18F-fluorodeoxyglucose-positron emission tomography (FDG-PET) in Sirt6-deficient mice showed increased glucose uptake in both brown adipose tissue and muscle but not liver, brain or heart, which could further explain the hypoglycemic phenotype (Zhong et al., 2010). The increased glucose uptake in these tissues could be explained by higher expression of glucose transporter 1 (Glut1), one of main glucose transporters that modulates basal uptake of glucose, independent of insulin or growth factor (Zhong et al., 2010). The central nervous system also plays an important role in regulating glucose metabolism. Growth hormone and insulin-like growth factor 1 (IGF-1) levels were lower in mice with brain-specific Sirt6 knockout than control mice, similar to mice with whole-body Sirt6 knockout (Schwer et al., 2010). Thus, under certain physiological conditions, Sirt6 may affect glucose metabolism and insulin sensitivity via growth hormone/IGF-1 signaling.
Sirt6 and Glycolysis
With sufficient oxygen, glucose is metabolized to pyruvate, which is further converted to ATP in mitochondria. However, in the absence of nutrients or during hypoxia, cells undergo anaerobic respiration and pyruvate is converted to lactic acid (Aragonés et al., 2009; Vander Heiden et al., 2009). In understanding the hypoglycemia seen in Sirt6-deficient mice, Zhong et al. demonstrated that Sirt6 regulates glucose homoeostasis by suppressing the expression of multiple glycolytic genes (Zhong et al., 2010). This suppression results in efficient ATP production via mitochondrial oxidative phosphorylation instead of glycolysis. Loss of Sirt6 increases glycolysis and diminishes mitochondrial respiration (Aragonés et al., 2009). The role of Sirt6 in glycolysis is mediated by hypoxia-inducible factor 1α (Hif-1α), known to regulate glycolysis and mitochondrial respiration in a coordinated manner (Zhong et al., 2010). Sirt6 deficiency induced Hif-1α activity and then increased the expression of glycolysis-related genes such as Glut1, lactate dehydrogenase (LDH), phosphoglycerate kinase (PGK1), glucose-6-phosphate isomerase (GPI), and phosphofructokinase 1 (PFK-1), and promoted glycolysis (Hu et al., 2006; Zhong et al., 2010). Simultaneously, activated Hif-1α directly inhibited mitochondrial respiration by increasing the expression of dehydrogenase kinase (PDK) (Kim et al., 2006; Papandreou et al., 2006). Moreover, when mice with Sirt6 deficiency were treated with an Hif-1α inhibitor, the hypoglycemia phenotype was rescued, which suggests that increased activity of Hif-1α contributes to the impaired glucose metabolism in these mice (Zhong et al., 2010).
Further study revealed that Sirt6 could regulate Hif-1α via two plausible scenarios: (1) Sirt6 inhibits recruitment of Hif-1α (accelerating its degradation) to its target gene promoters, or (2) Hif-1α could already localize to the promoters under normoglycemia, but the presence of Sirt6 would inhibit its transcriptional activity (Zhong et al., 2010).
Sirt6 and Gluconeogenesis
In addition to regulating glycolysis, Sirt6 affects gluconeogenesis. In the absence of Sirt6, hepatic gluconeogenesis was significantly elevated, which suggests a compensatory response to hypoglycemia (Dominy et al., 2012). Gluconeogenesis is tightly controlled by various cellular signaling pathways and transcription factors (Magnusson et al., 1992). Peroxisome proliferator-activated receptor γ coactivator 1-α (PGC-1α) is a key transcriptional regulator for gluconeogenesis. PGC-1α increases the expression of gluconeogenic enzymes such as glucose-6-phosphatase (G6p) and phosphoenolpyruvate carboxykinase (Pepck) (Puigserver et al., 2003). The transcriptional activity of PGC-1α is negatively regulated by its acetylation level. General control non-repressed protein 5 (GCN5) increased the acetylation level of PGC-1α and decreased PGC-1α transcriptional activity (Lerin et al., 2006). Sirt6 could directly bind to and activate GCN5 (Dominy et al., 2012). With knockout of Sirt6, GCN5 activity is decreased, the acetylation level of PGC-1α is reduced and PGC-1α controls the expression of gluconeogenic genes (Dominy et al., 2012).
Forkhead box protein O1 (FoxO1) also plays an important role in regulating gluconeogenesis. FoxO1 activates gluconeogenesis by directly binding the promoter regions of G6p and Pepck (Schilling et al., 2006). With mutation of the FoxO1 transcriptional activation domain and activity abolished, gluconeogenesis was significantly diminished (Nakae et al., 2001). FoxO1 deficiency significantly impaired the fasting-induced expression of G6p and Pepck (Matsumoto et al., 2007). The transcriptional activity of FoxO1 is mainly regulated by its phosphorylation and acetylation (Brunet et al., 2004; Yamagata et al., 2008; Zhao et al., 2010). In Sirt6-deleted cardiomyocytes, FoxO1 phosphorylation was increased (Sundaresan et al., 2012). The phosphorylation of FoxO1 promotes the translocation of FoxO1 from the nucleus to the cytoplasm, thereby reducing its transcriptional activity. Subsequent studies found that Sirt6 can specifically interact with FoxO1, thereby inhibiting the interaction between FoxO1 and its downstream genes G6p and Pepck, to reduce the expression of gluconeogenic genes (Zhang et al., 2014).
Sirt6 and Pancreatic β-Cell Function
The connection between Sirt6 and glucose metabolism is strengthened by the critical role of Sirt6 in promoting glucose-stimulated insulin secretion and ATP production in pancreatic β-cells (Xiong et al., 2016). These effects might be related to evidence of mitochondria damage (mitochondrial function and Ca2+ dynamic regulation in β cells impaired in Sirt6-deficient mice) and lower rate of oxygen consumption seen in Sirt6-deficient pancreatic β cells (Xiong et al., 2016). Sirt6 ablation also increases cell apoptosis and impairs insulin secretion in response to glucose in MIN6 cells (β-cell lines). Conversely, Sirt6 overexpression protects against palmitate-induced β-cell dysfunction and apoptosis (Song et al., 2016; Xiong et al., 2016). Recently, a new study found that Sirt6 is critical for pancreatic β-cell function and survival in mice (Qin et al., 2018). Sirt6 deficiency does not affect endocrine morphology, β-cell mass or insulin production but did result in glucose intolerance and defective glucose-stimulated insulin secretion in mice (Qin et al., 2018). β-cell specific deletion of Sirt6 reproduced the defect in insulin secretion. Loss of Sirt6 increases acetylation of histone H3K9Ac, H3K56Ac, and activates RNA polymerase II at the promoter region of thioredoxin-interacting protein (TXNIP) (Qin et al., 2018). TXNIP expression is negatively associated with glucose-stimulated insulin secretion in β-cells and that overexpression of TXNIP inhibits insulin secretion (Rani et al., 2010; Yoshihara et al., 2010; Luo et al., 2014). Sirt6-deficiency in β-cells exhibited a time-dependent increase in H3K9Ac, H3K56Ac, and TXNIP levels. Finally, β-cell specific Sirt6-deficient mice showed increased sensitivity to streptozotocin induced β-cell apoptosis (Qin et al., 2018). Together, this report indicates that Sirt6 has a key role in pancreatic function.
Sirt6 and Calorie Restriction
Calorie restriction (CR) reduces cellular NADH concentration, thereby increasing the NAD+/NADH ratio and promoting Sirt2 activity (Tasselli et al., 2016a). Sirt6 activity is significantly modulated by CR. It is increased by nutrient depletion or long-term CR in the brain, white adipose tissue (WAT), muscle, liver and kidney in mice (Kanfi et al., 2008; Kuang et al., 2017). Sirt6 also mediates the effects of CR, which is known to delay the onset of age-associated diseases, including diabetes and cardiovascular diseases. Sirt6 ablation abolished CR-induced life extension. Moreover, CR-activated Sirt6 suppressed NF-κB signaling and delayed aging (Zhang et al., 2013). Transgenic mice overexpressing Sirt6 showed multiple phenotypes resembling CR, including reduced body weight, enhanced metabolic activity, and reduced serum levels of cholesterol, adipokines, insulin, and glucose, which further demonstrates the regulatory role of Sirt6 in energy metabolism (Kanfi et al., 2010, 2012).
Sirt6 and Lipid Metabolism
Recent reports have shown that Sirt6 is an important regulator of lipid metabolism (Figure 2). Sirt6 regulates the hepatic accumulation of triglycerides (TG), which is associated with fatty liver disease (Kugel and Mostoslavsky, 2014; Vitiello et al., 2017). Sirt6 deficiency promotes lipogenesis and fatty acid uptake but inhibits β-oxidation (Kim et al., 2010). Cholesterol synthesis is also negatively regulated by Sirt6 (Elhanati et al., 2013). Sirt6 positively regulates lipid mobilization and thermogenesis in adipose tissue (Chen et al., 2017; Kuang et al., 2017; Xiong et al., 2017; Yao et al., 2017). Moreover, Sirt6 regulates circadian metabolic programs, accompanied by changes in lipid metabolism (Masri et al., 2014).
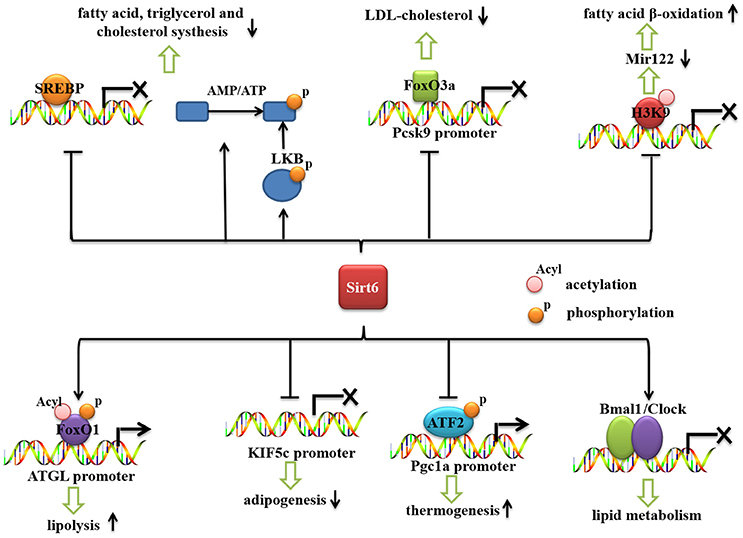
Figure 2. The role of Sirt6 in lipid metabolism. Sirt6 represses the mRNA expression of SREBP1/2 and inhibits the cleavage of SREBPs into their active forms. Sirt6 activates AMPK (by increasing the AMP/ATP ratio and activating LKB), which phosphorylates and inactivates SREBP. Sirt6 can be recruited by FoxO3a to the Pcsk9 promoter, suppress its gene expression and decrease the level of LDL-cholesterol. Sirt6 downregulates miR-122 by deacetylating H3K56, reduces miR-122 expression, and increases fatty acid β-oxidation. Sirt6 also specifically interacts with FoxO1, decreases FoxO1 acetylation and phosphorylation, and promotes the interaction between FoxO1 and the promoter of its downstream gene, ATGL, thereby increasing ATGL expression and inducing lipolysis in adipose tissue. Sirt6 inhibits KIF5C gene expression and impairs adipogenesis. Sirt6 regulates phospho-ATF2 (p-ATF2) binding to the PGC-1α promoter and subsequently affects the thermogenic program. Sirt6 regulates the circadian rhythm in the liver by maintaining a proper cyclic recruitment of CLOCK/BMAL1.
Sirt6 and Hepatic Lipid Metabolism
The liver is a key metabolic organ controlling aspects of lipid metabolism in response to hormonal or nutritional signals. Under starvation, the liver converts lipid stores into available energy via fatty acid oxidation. In the fed condition, metabolic programs in the liver are switched on to store energy in the form of lipid droplets via the process of lipogenesis (Puigserver et al., 2003).
In addition to its critical regulatory role in glucose metabolism, Sirt6 plays an important regulatory role in hepatic lipid metabolism. Hepatic-specific disruption of Sirt6 in mice resulted in fatty liver formation (Kim et al., 2010). Hepatic TG accumulation in liver is mainly regulated by fatty acid uptake, β-oxidation, and TG synthesis and secretion. Deletion of Sirt6 increased the expression of hepatic fatty acid transport genes and reduced that of fatty acid β-oxidation genes (carnitine palmitoyltransferase 1 [CPT1α], acyl-coenzyme A oxidase 1 [ACOX1]). Genetic deletion of Sirt6 in mice increased the mRNA levels of lipogenic genes such as acetyl CoA carboxylase 1 (ACC1), fatty acid synthase (FAS), and stearoyl-CoA desaturase-1 (SCD1) (Kim et al., 2010).
Rosiglitazone (RGZ), an agonist of peroxisome proliferator-activated receptor gamma (PPARγ), plays an important role in protecting against fatty liver disease. Activation of PPARγ by RGZ increased Sirt6 expression (Yang et al., 2011). RGZ treatment ameliorated hepatic lipid accumulation and increased the expression of Sirt6, PGC-1α, and FoxO1 in rat liver (Yang et al., 2011). In addition, AMP-activated protein kinase (AMPK) phosphorylation was increased by RGZ. Sirt6 knockdown increased hepatocyte lipid accumulation and abolished the effect of RGZ on hepatic steatosis (Yang et al., 2011). Sirt6 knockdown also abolished the effect of RGZ on the mRNA and protein expression of PGC1-α and FoxO1 and phosphorylation levels of AMPK, which suggests that Sirt6 is involved in the RGZ-mediated metabolic effects (Yang et al., 2011). However, this result contradicts a report that Sirt6 inhibits the activity of PGC-1a by increasing GCN5 expression (Dominy et al., 2012), so Sirt6 may have different regulatory roles in different models. MicroRNA-122 (miR-122), a microRNA (miRNA) highly expressed in liver, constitutes 70% of the total miRNA pool in liver (Chang et al., 2004; Bhattacharyya et al., 2006; Jopling, 2012). MiR-122 plays an important regulatory role in many metabolic processes, including cholesterol synthesis and fatty acid oxidation (Krützfeldt et al., 2005; Esau et al., 2006). Sirt6 and miR-122 are reciprocally regulated to control the gene expression of fatty acid oxidation (Elhanati et al., 2016). Sirt6 downregulates miR-122 by deacetylating H3K56. MiR-122 binds to three sites on the Sirt6 3' untranslated region and reduces its levels. Overexpression of Sirt6 in mouse liver reduced miR-122 expression and increased that of fatty acid β-oxidation genes (Elhanati et al., 2016).
Sirt6 also regulates low-density lipoprotein (LDL)-cholesterol levels (Kanfi et al., 2010). Sirt6 regulates cholesterol level via the lipogenic transcription factors sterol regulatory element binding proteins 1/2 (SREBP1/2) and AMPK (Elhanati et al., 2013). It represses the expression of SREBP1/2 and by three mechanisms: it represses the transcription levels of SREBP1/2 and inhibits their cleavage into their active forms (Elhanati et al., 2013) and it activates AMPK (by increasing the AMP/ATP ratio and activating LKB), which phosphorylates and inactivates SREBP1 (Elhanati et al., 2013). Proprotein convertase subtilisin/kexin type 9 (PCSK9) is a crucial gene regulating LDL-cholesterol (Tao et al., 2013). Sirt6 plays a critical role in regulating Pcsk9 gene expression. Sirt6 can be recruited by FoxO3a to the Pcsk9 gene promoter and deacetylate histone H3 at Lys 9 and 56, thereby suppressing the gene expression (Tao et al., 2013). Conversely, overexpression of Sirt6 decreased LDL-cholesterol level in high-fat diet (HFD)-fed mice (Kanfi et al., 2010).
Sirt6 and Adipose Tissue Lipolysis
Sirt6 regulates the lipid metabolism of adipose tissues. Sirt6 ablation mediated by ap2-CRE or adiponectin-CRE in adipose tissue increased HFD-induced obesity and insulin resistance (Kuang et al., 2017; Yao et al., 2017), and Sirt6 overexpression inhibited HFD-induced obesity and insulin resistance (Kanfi et al., 2010). Sirt6 ablation increased diet-induced obesity via adipocyte hypertrophy rather than abnormal adipocyte differentiation (Kuang et al., 2017). Adipocyte hypertrophy in Sirt6-deficient mice might be attributed to impaired lipolytic activity, which causes fat storage synthesis exceeding lipolysis and results in obesity. Adipose TG lipase (ATGL) is the key lipase that hydrolyzes TG into diglycerides (Zimmermann et al., 2004; Kuang et al., 2017); Sirt6 ablation suppressed ATGL expression. Sirt6 physically interacts with FoxO1, and Sirt6 deficiency increased the acetylation and phosphorylation of FoxO1, thereby promoting its nuclear exclusion and decreasing its transcriptional activity, which downregulated ATGL expression (Kuang et al., 2017). The role of Sirt6 in regulating FoxO1 is contradictory in liver and adipose tissue (Zhang et al., 2014; Kuang et al., 2017), which indicates that the regulatory function of Sirt6 is tissue-specific. Sirt6 overexpression protects against HFD-induced physiological damage by blocking the lipotoxicity of obesity and restoring glucose homeostasis via specific reduction of PPARγ signaling and level of diacylglycerol acyltransferase 1 (DGAT1), a key regulator of TG synthesis (Kanfi et al., 2010).
Sirt6 and Adipogenesis
Adipose tissue, differentiated from pre-adipocytes, is regulated by multiple transcriptional factors, including PPARγ and CCAAT/enhancer-binding protein α, β, and δ (C/EBPα/β/δ). Recently, Chen et al found that Sirt6 deficiency in preadipocytes blocks their adipogenesis (Chen et al., 2017). Sirt6 deficiency impairs adipogenesis, and Sirt6 is an essential factor for mitotic clonal expansion during adipogenesis by inhibiting the expression of kinesin family member 5C (KIF5C) and enhancing casein kinase 2 (CK2) activity. KIF5C is negatively regulated by Sirt6. Moreover, this study showed that KIF5C is a negative factor for adipogenesis by interacting with CK2a, a catalytic subunit of CK2 (Chen et al., 2017).
Sirt6 and Thermogenesis
Obesity is due to a chronic imbalance between energy intake and energy expenditure. WAT is essential for TG storage and insulin resistance, whereas brown adipose tissue (BAT) generates heat by dissipating energy via uncoupled respiration mediated by uncoupling protein 1 (UCP1) (Lowell and Spiegelman, 2000; Nedergaard and Cannon, 2010). WAT could be converted into brown-like adipocytes (beige cells). Cells undergoing a browning process have been suggested to have strong anti-diabetic or anti-obesity benefit (Cypess et al., 2009; Barbatelli et al., 2010; Petrovic et al., 2010; Wu et al., 2012). Recent studies show that Sirt6 has a critical role in regulating the thermogenesis of fat (Yao et al., 2017). Cold exposure and a β-adrenergic agonist markedly induced Sirt6 expression in fat. Fat-specific ablation of Sirt6 mediated by Ap2-CRE impairs the thermogenic function of brown adipocytes, thereby causing a morphological “whitening” of brown fat, and decreased oxygen consumption, core body temperature and cold sensitivity (Yao et al., 2017). PGC-1α is highly expressed in BAT and is a central regulator of brown fat thermogenesis (Puigserver et al., 1998). White fat cells overexpressing PGC-1α show mitochondrial oxidation phosphorylation and expression of thermogenesis genes (Puigserver et al., 1998). Sirt6 depletion markedly decreased the expression of PGC-1α and other thermogenic genes (Yao et al., 2017). Sirt6-depleted adipocytes also decreased basal mitochondrial respiration and maximal mitochondrial respiratory capacity. Mitochondrial oxidative phosphorylation and the expression of biogenesis genes are decreased significantly in primary brown adipocytes with Sirt6 deletion (Yao et al., 2017). Decreased PGC-1α expression in brown fat was not attributed to changes in acetylation levels of H3K9 or H3K56 in its promoter region in Sirt6-deficient mice (Yao et al., 2017). Activating transcription factor 2 (ATF2) is recruited to the PGC-1α promoter after β-adrenergic receptor activation in BAT (Herzig et al., 2001; Cao et al., 2004). Sirt6 depletion reduced phosphorylated ATF2 binding to the PGC-1α promoter and subsequently decreased the thermogenic program of brown fat and led to obesity (Yao et al., 2017).
Sirt6 and Circadian Rhythm
Circadian rhythm refers to changes in the life cycle during 24 h. Circadian rhythms play an important role in regulating body metabolism (Kohsaka et al., 2007; Barnea et al., 2009). In mice with circadian rhythm disruption, energy metabolism, especially glucose metabolism, was disturbed. Both behavioral and molecular circadian rhythms were changed in mice with metabolic disorders caused by an HFD (Kohsaka et al., 2007; Barnea et al., 2009). The clock has a central role in the circadian rhythm (Feng and Lazar, 2012). Bmal1 gene (also called aryl hydrocarbon receptor nuclear translocator-like [ARNTL]) is the central part of the biological clock transcription and translation feedback loop (Kiyohara et al., 2006). These two genes, encoding the protein together to form a Clock/Bmal1 isomer, play an important role in the feedback loop. Sirt6 is the only constitutive chromatin-associated sirtuin and is prominently present at transcriptionally active genomic loci (Masri et al., 2014). Sirt6 interacts with CLOCK/BMAL1 and, differently from SIRT1, governs their chromatin recruitment to gene promoters (Masri et al., 2014). Sirt6 contributes to chromatin recruitment of both the circadian machinery as well as SREBP-1 (Masri et al., 2014). Liver-specific deletion of Sirt6 downregulates hepatic rhythmic transcription, accompanied by changes in lipid metabolism (Masri et al., 2014). Deletion of Sirt6 leads to decreased binding of Clock/Bmal to the clock control gene (CCG) promoter and the binding of SREBP1 with its downstream gene promoters, thereby affecting lipid metabolism in the organism (Masri et al., 2014). The relationship between Sirt6 and fatty acid metabolism remains unclear. Sirt6 regulation of the circadian metabolic programs sheds new light on how the enzyme couples chromatin dynamics to metabolism (Masri et al., 2014; Tasselli et al., 2016b).
Sirt6 and Inflammation
Sirt6 deficiency increases the inflammatory response in many tissues. In adipose tissue, Sirt6 deficiency increases macrophage infiltration and adipose tissue inflammation and promotes HFD-induced insulin resistance (Kuang et al., 2017; Xiong et al., 2017). Sirt6 deficiency in mouse immune cells leads to liver inflammation and fibrosis (Xiao et al., 2012). In particular, in pancreatic cancer cells, Sirt6 induces the expression of proinflammatory cyto-/chemokines [interleukin 8 and tumor necrosis factor α (TNFα)] (Lappas, 2012). In endothelial cells, Sirt6 deficiency increased the expression of proinflammatory cytokines, extracellular matrix remodeling enzymes and adhesion molecules (Lappas, 2012). Loss of Sirt6 increased the expression of NF-κB, whereas overexpression of Sirt6 decreased NF-κB transcriptional activity (Lappas, 2012). Sirt6 interacts with the NF-κB subunit, deacetylates histone H3 lysine 9 (H3K9) at NF-kB target gene promoters, and inhibits the expression of downstream target genes, thus inhibiting the inflammatory reaction (Kawahara et al., 2009). In Sirt6-deficient cells, RELA promoter occupancy was increased; it enhanced hyperacetylation of NF-κB target gene promoters and induced NF-κB–dependent gene expression, cellular senescence and apoptosis (Kawahara et al., 2009). Sirt6 also binds to c-Jun and decreases the expression of its downstream target genes IL-6, monocyte chemoattractant protein 1, TNFα and H3K9, thereby inhibiting the expression of these genes (Xiao et al., 2012; Kuang et al., 2017).
Sirt6 and DNA Damage and Diabetes
DNA damage is a permanent change of nucleotide sequence during DNA replication, resulting in the change of corresponding genetic characteristics (Shimizu et al., 2014). It is also related to the occurrence and development of chronic diseases, such as cancer and diabetes (Blasiak et al., 2004; Grindel et al., 2016). Sirt6 has been characterized as a histone deacetylase (HDAC) that targets specific sites (Michishita et al., 2008, 2009; Yang et al., 2009; Tasselli et al., 2016a). Sirt6 deacetylates the histone H3 on acetylated K9, K56 (Michishita et al., 2008, 2009) and the more recently identified K18 residue (Yang et al., 2009), causing the repression of many genes involved in inflammation, aging, genome stability, metabolic pathways and telomere integrity (Vitiello et al., 2017). Loss of Sirt6 affects the pathway of ATM/CHK2 and recruitment of repair factors to sites of DNA damage (Cagnetta et al., 2018). Sirt6 interacts, deacetylates and affects telomere repeat binding factor 2 (TRF2) stability, which may be part of a higher order complex with functional impacts on DNA damage response (DDR), cancer and aging (Rizzo et al., 2017). Moreover, following DNA damage, Sirt6 is recruited to double-strand breaks ensuring the proper activation of downstream DDR factors leading to an efficient DNA repair. Studies in diabetic patients showed greater oxidative damage to DNA. This indicates that the role of Sirt6 in DNA damage may as a new therapeutic pathway for cancer and diabetes related disorders.
Conclusion
Excessive intake of carbohydrates or fat can lead to a range of metabolic syndromes, such as obesity, fatty liver and diabetes. With evidence that plays an important regulatory role in energy metabolism, it may be a potential therapeutic target for obesity and diabetes mellitus. Clinical trials investigating the use of sirtuin activators for treating diabetes are under way; such activators show promise as alternatives to current diabetes therapies. Thus, further research of sirtuin activators may result in a new class of safe, effective diabetes treatments.
Author Contributions
JK designed and wrote the manuscript. LC, QT, JZ, and YL contributed to the discussion and review of the manuscript. JH obtained funding and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81471068 and 81270926), Distinguished Young Scientists of Sichuan Province (2014JQ0034) and Young Scientist Fellowship of Sichuan University (2013SCU04A17).
Abbreviations
ACC1, acetyl CoA carboxylase 1; ACOX1, acyl-coenzyme A oxidase 1; AMPK, AMP-activated protein kinase; ATF2, activating transcription factor 2; Bmal1, aryl Hydrocarbon Receptor Nuclear Translocator Like; C/EBPα/β/δ, CCAAT/enhancer-binding protein α, β, and δ; CCGs, clock control gene; CK2, enhancing casein kinase 2; CPT1α, carnitine palmitoyltransferase 1; DGAT1, diacylglycerol acyltransferase 1; FAS, fatty acid synthase; FoxO1, Forkhead box protein O1; G6p, glucose-6-phosphatase; GCN5, general control non-repressed protein 5; Glut1, glucose transporter 1; GPI, glucose-6-phosphate isomerase; HFD, high-fat diet; Hif-1α, hypoxia-inducible factor-1α; IGF-1, insulin-like growth factor 1; KIF5C, kinesin family member 5C; LDH, lactate dehydrogenase; PCSK9, proprotein convertase subtilisin/kexin type 9; PDK, dehydrogenase kinase; Pepck, phosphoenolpyruvate carboxykinase; PFK-1, phosphofructokinase-1; PGC-1α, peroxisome proliferator activated receptor γ coactivator 1-α; PGK1, phosphoglycerate kinase; PPARγ, peroxisome proliferator-activated receptor gamma; SCD1, stearoyl-CoA desaturase-1; SREBP 1/2, sterol regulatory element binding proteins 1/2; UCP1, uncoupling protein 1; ATGL, adipose triglyceride lipase; CR, calorie restriction; WAT, white adipose tissue; TG, triglycerides; LDL, low-density lipoprotein; TNFα, tumor necrosis factor α; IL-8, interleukin 8.
References
Aragonés, J., Fraisl, P., Baes, M., and Carmeliet, P. (2009). Oxygen sensors at the crossroad of metabolism. Cell Metab. 9, 11–22. doi: 10.1016/j.cmet.2008.10.001
Barbatelli, G., Murano, I., Madsen, L., Hao, Q., Jimenez, M., Kristiansen, K., et al. (2010). The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte transdifferentiation. Am. J. Physiol. Endocrinol. Metab. 298, E1244–E1253. doi: 10.1152/ajpendo.00600.2009
Barnea, M., Madar, Z., and Froy, O. (2009). High-fat diet delays and fasting advances the circadian expression of adiponectin signaling components in mouse liver. Endocrinology 150, 161–168. doi: 10.1210/en.2008-0944
Bhattacharyya, S. N., Habermacher, R., Martine, U., Closs, E. I., and Filipowicz, W. (2006). Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125, 1111–1124. doi: 10.1016/j.cell.2006.04.031
Blasiak, J., Arabski, M., Krupa, R., Wozniak, K., Zadrozny, M., Kasznicki, J., et al. (2004). DNA damage and repair in type 2 diabetes mellitus. Mutat. Res. 554, 297–304. doi: 10.1016/j.mrfmmm.2004.05.011
Brunet, A., Sweeney, L. B., Sturgill, J. F., Chua, K. F., Greer, P. L., Lin, Y., et al. (2004). Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303, 2011–2015. doi: 10.1126/science.1094637
Cagnetta, A., Soncini, D., Orecchioni, S., Talarico, G., Minetto, P., Guolo, F., et al. (2018). Depletion of SIRT6 enzymatic activity increases acute myeloid leukemia cells' vulnerability to DNA-damaging agents. Haematologica 103, 80–90. doi: 10.3324/haematol.2017.176248
Cao, W., Daniel, K. W., Robidoux, J., Puigserver, P., Medvedev, A. V., Bai, X., et al. (2004). p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol. Cell. Biol. 24, 3057–3067. doi: 10.1128/MCB.24.7.3057-3067.2004
Chang, J., Nicolas, E., Marks, D., Sander, C., Lerro, A., Buendia, M. A., et al. (2004). miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 1, 106–113. doi: 10.4161/rna.1.2.1066
Chen, Q., Hao, W., Xiao, C., Wang, R., Xu, X., Lu, H., et al. (2017). SIRT6 is essential for adipocyte differentiation by regulating mitotic clonal expansion. Cell Rep. 18, 3155–3166. doi: 10.1016/j.celrep.2017.03.006
Cypess, A. M., Lehman, S., Williams, G., Tal, I., Rodman, D., Goldfine, A. B., et al. (2009). Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 360, 1509–1517. doi: 10.1056/NEJMoa0810780
Demir, I. E., Ceyhan, G. O., and Friess, H. (2017). Epigenomic therapies: the potential of targeting SIRT6 for the treatment of pancreatic cancer. Expert Opin. Ther. Targets 21, 1–3. doi: 10.1080/14728222.2017.1265507
Dominy, J. E. Jr., Lee, Y., Jedrychowski, M. P., Chim, H., Jurczak, M. J., Camporez, J. P., et al. (2012). The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Mol. Cell 48, 900–913. doi: 10.1016/j.molcel.2012.09.030
Elhanati, S., Ben-Hamo, R., Kanfi, Y., Varvak, A., Glazz, R., Lerrer, B., et al. (2016). Reciprocal regulation between SIRT6 and miR-122 controls liver metabolism and predicts hepatocarcinoma prognosis. Cell Rep. 14, 234–242. doi: 10.1016/j.celrep.2015.12.023
Elhanati, S., Kanfi, Y., Varvak, A., Roichman, A., Carmel-Gross, I., Barth, S., et al. (2013). Multiple regulatory layers of SREBP1/2 by SIRT6. Cell Rep. 4, 905–912. doi: 10.1016/j.celrep.2013.08.006
Esau, C., Davis, S., Murray, S. F., Yu, X. X., Pandey, S. K., Pear, M., et al. (2006). miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3, 87–98. doi: 10.1016/j.cmet.2006.01.005
Feng, D., and Lazar, M. A. (2012). Clocks, metabolism, and the epigenome. Mol. Cell 47, 158–167. doi: 10.1016/j.molcel.2012.06.026
Frye, R. A. (1999). Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 260, 273–279. doi: 10.1006/bbrc.1999.0897
Grindel, A., Guggenberger, B., Eichberger, L., Poppelmeyer, C., Gschaider, M., Tosevska, A., et al. (2016). Oxidative stress, DNA damage and DNA repair in female patients with diabetes mellitus type 2. PLoS ONE 11:e0162082. doi: 10.1371/journal.pone.0162082
Herzig, S., Long, F., Jhala, U. S., Hedrick, S., Quinn, R., Bauer, A., et al. (2001). CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413, 179–183. doi: 10.1038/35093131
Hu, C. J., Iyer, S., Sataur, A., Covello, K. L., Chodosh, L. A., and Simon, M. C. (2006). Differential regulation of the transcriptional activities of hypoxia-inducible factor 1 alpha (HIF-1alpha) and HIF-2alpha in stem cells. Mol. Cell. Biol. 26, 3514–3526. doi: 10.1128/MCB.26.9.3514-3526.2006
Jopling, C. (2012). Liver-specific microRNA-122: biogenesis and function. RNA Biol. 9, 137–142. doi: 10.4161/rna.18827
Kanfi, Y., Naiman, S., Amir, G., Peshti, V., Zinman, G., Nahum, L., et al. (2012). The sirtuin SIRT6 regulates lifespan in male mice. Nature 483, 218–221. doi: 10.1038/nature10815
Kanfi, Y., Peshti, V., Gil, R., Naiman, S., Nahum, L., Levin, E., et al. (2010). SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell 9, 162–173. doi: 10.1111/j.1474-9726.2009.00544.x
Kanfi, Y., Shalman, R., Peshti, V., Pilosof, S. N., Gozlan, Y. M., Pearson, K. J., et al. (2008). Regulation of SIRT6 protein levels by nutrient availability. FEBS Lett. 582, 543–548. doi: 10.1016/j.febslet.2008.01.019
Kawahara, T. L., Michishita, E., Adler, A. S., Damian, M., Berber, E., Lin, M., et al. (2009). SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell 136, 62–74. doi: 10.1016/j.cell.2008.10.052
Kim, H. S., Xiao, C., Wang, R. H., Lahusen, T., Xu, X., Vassilopoulos, A., et al. (2010). Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 12, 224–236. doi: 10.1016/j.cmet.2010.06.009
Kim, J. W., Tchernyshyov, I., Semenza, G. L., and Dang, C. V. (2006). HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3, 177–185. doi: 10.1016/j.cmet.2006.02.002
Kiyohara, Y. B., Tagao, S., Tamanini, F., Morita, A., Sugisawa, Y., Yasuda, M., et al. (2006). The BMAL1 C terminus regulates the circadian transcription feedback loop. Proc. Natl. Acad. Sci. U.S.A. 103, 10074–10079. doi: 10.1073/pnas.0601416103
Kohsaka, A., Laposky, A. D., Ramsey, K. M., Estrada, C., Joshu, C., Kobayashi, Y., et al. (2007). High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 6, 414–421. doi: 10.1016/j.cmet.2007.09.006
Krützfeldt, J., Rajewsky, N., Braich, R., Rajeev, K. G., Tuschl, T., Manoharan, M., et al. (2005). Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438, 685–689. doi: 10.1038/nature04303
Kuang, J., Zhang, Y., Liu, Q., Shen, J., Pu, S., Cheng, S., et al. (2017). Fat-specific Sirt6 ablation sensitizes mice to high-fat diet-induced obesity and insulin resistance by inhibiting lipolysis. Diabetes 66, 1159–1171. doi: 10.2337/db16-1225
Kugel, S., and Mostoslavsky, R. (2014). Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem. Sci. 39, 72–81. doi: 10.1016/j.tibs.2013.12.002
Kugel, S., Sebastian, C., Fitamant, J., Ross, K. N., Saha, S. K., Jain, E., et al. (2016). SIRT6 suppresses pancreatic cancer through control of Lin28b. Cell 165, 1401–1415. doi: 10.1016/j.cell.2016.04.033
Lappas, M. (2012). Anti-inflammatory properties of sirtuin 6 in human umbilical vein endothelial cells. Mediators Inflamm. 2012:597514. doi: 10.1155/2012/597514
Lerin, C., Rodgers, J. T., Kalume, D. E., Kim, S. H., Pandey, A., and Puigserver, P. (2006). GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 3, 429–438. doi: 10.1016/j.cmet.2006.04.013
Liszt, G., Ford, E., Kurtev, M., and Guarente, L. (2005). Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 280, 21313–21320. doi: 10.1074/jbc.M413296200
Lowell, B. B., and Spiegelman, B. M. (2000). Towards a molecular understanding of adaptive thermogenesis. Nature 404, 652–660. doi: 10.1038/35007527
Luo, Y., He, F., Hu, L., Hai, L., Huang, M., Xu, Z., et al. (2014). Transcription factor Ets1 regulates expression of thioredoxin-interacting protein and inhibits insulin secretion in pancreatic beta-cells. PLoS ONE 9:e99049. doi: 10.1371/journal.pone.0099049
Magnusson, I., Rothman, D. L., Katz, L. D., Shulman, R. G., and Shulman, G. I. (1992). Increased rate of gluconeogenesis in type II diabetes mellitus. A 13C nuclear magnetic resonance study. J. Clin. Invest. 90, 1323–1327. doi: 10.1172/JCI115997
Masri, S., Rigor, P., Cervantes, M., Ceglia, N., Sebastian, C., Xiao, C., et al. (2014). Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 158, 659–672. doi: 10.1016/j.cell.2014.06.050
Matsumoto, M., Pocai, A., Rossetti, L., Depinho, R. A., and Accili, D. (2007). Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 6, 208–216. doi: 10.1016/j.cmet.2007.08.006
Michan, S., and Sinclair, D. (2007). Sirtuins in mammals: insights into their biological function. Biochem. J. 404, 1–13. doi: 10.1042/BJ20070140
Michishita, E., McCord, R. A., Berber, E., Kioi, M., Padilla-Nash, H., Damian, M., et al. (2008). SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452, 492–496. doi: 10.1038/nature06736
Michishita, E., McCord, R. A., Boxer, L. D., Barber, M. F., Hong, T., Gozani, O., et al. (2009). Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 8, 2664–2666. doi: 10.4161/cc.8.16.9367
Michishita, E., Park, J. Y., Burneskis, J. M., Barrett, J. C., and Horikawa, I. (2005). Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 16, 4623–4635. doi: 10.1091/mbc.E05-01-0033
Mostoslavsky, R., Chua, K. F., Lombard, D. B., Pang, W. W., Fischer, M. R., Gellon, L., et al. (2006). Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124, 315–329. doi: 10.1016/j.cell.2005.11.044
Nakae, J., Kitamura, T., Silver, D. L., and Accili, D. (2001). The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J. Clin. Invest. 108, 1359–1367. doi: 10.1172/JCI200112876
Nedergaard, J., and Cannon, B. (2010). The changed metabolic world with human brown adipose tissue: therapeutic visions. Cell Metab. 11, 268–272. doi: 10.1016/j.cmet.2010.03.007
Papandreou, I., Cairns, R. A., Fontana, L., Lim, A. L., and Denko, N. C. (2006). HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 3, 187–197. doi: 10.1016/j.cmet.2006.01.012
Petrovic, N., Walden, T. B., Shabalina, I. G., Timmons, J. A., Cannon, B., and Nedergaard, J. (2010). Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J. Biol. Chem. 285, 7153–7164. doi: 10.1074/jbc.M109.053942
Puigserver, P., Rhee, J., Donovan, J., Walkey, C. J., Yoon, J. C., Oriente, F., et al. (2003). Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature 423, 550–555. doi: 10.1038/nature01667
Puigserver, P., Wu, Z., Park, C. W., Graves, R., Wright, M., and Spiegelman, B. M. (1998). A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92, 829–839. doi: 10.1016/S0092-8674(00)81410-5
Qin, K., Zhang, N., Zhang, Z., Nipper, M., Zhu, Z., Leighton, J., et al. (2018). SIRT6-mediated transcriptional suppression of Txnip is critical for pancreatic beta cell function and survival in mice. Diabetologia doi: 10.1007/s00125-017-4542-6. [Epub ahead of print]
Rani, S., Mehta, J. P., Barron, N., Doolan, P., Jeppesen, P. B., Clynes, M., et al. (2010). Decreasing Txnip mRNA and protein levels in pancreatic MIN6 cells reduces reactive oxygen species and restores glucose regulated insulin secretion. Cell. Physiol. Biochem. 25, 667–674. doi: 10.1159/000315086
Rizzo, A., Iachettini, S., Salvati, E., Zizza, P., Maresca, C., D'angelo, C., et al. (2017). SIRT6 interacts with TRF2 and promotes its degradation in response to DNA damage. Nucleic Acids Res. 45, 1820–1834. doi: 10.1093/nar/gkw1202
Schilling, M. M., Oeser, J. K., Boustead, J. N., Flemming, B. P., and O'brien, R. M. (2006). Gluconeogenesis: re-evaluating the FOXO1-PGC-1alpha connection. Nature 443, E10–E11. doi: 10.1038/nature05288
Schwer, B., Schumacher, B., Lombard, D. B., Xiao, C., Kurtev, M. V., Gao, J., et al. (2010). Neural sirtuin 6 (Sirt6) ablation attenuates somatic growth and causes obesity. Proc. Natl. Acad. Sci. U.S.A. 107, 21790–21794. doi: 10.1073/pnas.1016306107
Shimizu, I., Yoshida, Y., Suda, M., and Minamino, T. (2014). DNA damage response and metabolic disease. Cell Metab. 20, 967–977. doi: 10.1016/j.cmet.2014.10.008
Song, M. Y., Wang, J., Ka, S. O., Bae, E. J., and Park, B. H. (2016). Insulin secretion impairment in Sirt6 knockout pancreatic beta cells is mediated by suppression of the FoxO1-Pdx1-Glut2 pathway. Sci. Rep. 6:30321. doi: 10.1038/srep30321
Sundaresan, N. R., Vasudevan, P., Zhong, L., Kim, G., Samant, S., Parekh, V., et al. (2012). The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat. Med. 18, 1643–1650. doi: 10.1038/nm.2961
Tao, R., Xiong, X., Depinho, R. A., Deng, C. X., and Dong, X. C. (2013). FoxO3 transcription factor and Sirt6 deacetylase regulate low density lipoprotein (LDL)-cholesterol homeostasis via control of the proprotein convertase subtilisin/kexin type 9 (Pcsk9) gene expression. J. Biol. Chem. 288, 29252–29259. doi: 10.1074/jbc.M113.481473
Tasselli, L., Xi, Y., Zheng, W., Tennen, R. I., Odrowaz, Z., Simeoni, F., et al. (2016a). SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat. Struct. Mol. Biol. 23, 434–440. doi: 10.1038/nsmb.3202
Tasselli, L., Zheng, W., and Chua, K. F. (2016b). SIRT6: novel mechanisms and links to aging and disease. Trends Endocrinol. Metab. 28, 168–185. doi: 10.1016/j.tem.2016.10.002
Vander Heiden, M. G., Cantley, L. C., and Thompson, C. B. (2009). Understanding the warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033. doi: 10.1126/science.1160809
Vitiello, M., Zullo, A., Servillo, L., Mancini, F. P., Borriello, A., Giovane, A., et al. (2017). Multiple pathways of SIRT6 at the crossroads in the control of longevity, cancer, and cardiovascular diseases. Ageing Res. Rev. 35, 301–311. doi: 10.1016/j.arr.2016.10.008
Wu, J., Boström, P., Sparks, L. M., Ye, L., Choi, J. H., Giang, A. H., et al. (2012). Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150, 366–376. doi: 10.1016/j.cell.2012.05.016
Xiao, C., Kim, H. S., Lahusen, T., Wang, R. H., Xu, X., Gavrilova, O., et al. (2010). SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J. Biol. Chem. 285, 36776–36784. doi: 10.1074/jbc.M110.168039
Xiao, C., Wang, R. H., Lahusen, T. J., Park, O., Bertola, A., Maruyama, T., et al. (2012). Progression of chronic liver inflammation and fibrosis driven by activation of c-JUN signaling in Sirt6 mutant mice. J. Biol. Chem. 287, 41903–41913. doi: 10.1074/jbc.M112.415182
Xiong, X., Sun, X., Wang, Q., Qian, X., Zhang, Y., Pan, X., et al. (2016). SIRT6 protects against palmitate-induced pancreatic beta-cell dysfunction and apoptosis. J. Endocrinol. 231, 159–165. doi: 10.1530/JOE-16-0317
Xiong, X., Zhang, C., Zhang, Y., Fan, R., Qian, X., and Dong, X. C. (2017). Fabp4-Cre-mediated Sirt6 deletion impairs adipose tissue function and metabolic homeostasis in mice. J. Endocrinol. 233, 307–314. doi: 10.1530/JOE-17-0033
Yamagata, K., Daitoku, H., Takahashi, Y., Namiki, K., Hisatake, K., Kako, K., et al. (2008). Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol. Cell 32, 221–231. doi: 10.1016/j.molcel.2008.09.013
Yang, B., Zwaans, B. M., Eckersdorff, M., and Lombard, D. B. (2009). The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle 8, 2662–2663. doi: 10.4161/cc.8.16.9329
Yang, S. J., Choi, J. M., Chae, S. W., Kim, W. J., Park, S. E., Rhee, E. J., et al. (2011). Activation of peroxisome proliferator-activated receptor gamma by rosiglitazone increases sirt6 expression and ameliorates hepatic steatosis in rats. PLoS ONE 6:e17057. doi: 10.1371/journal.pone.0017057
Yao, L., Cui, X., Chen, Q., Yang, X., Fang, F., Zhang, J., et al. (2017). Cold-inducible SIRT6 Regulates thermogenesis of brown and beige fat. Cell Rep. 20, 641–654. doi: 10.1016/j.celrep.2017.06.069
Yoshihara, E., Fujimoto, S., Inagaki, N., Okawa, K., Masaki, S., Yodoi, J., et al. (2010). Disruption of TBP-2 ameliorates insulin sensitivity and secretion without affecting obesity. Nat. Commun. 1:127. doi: 10.1038/ncomms1127
Zhang, H. J., Zhang, X. F., Ma, Z. M., Pan, L. L., Chen, Z., Han, H. W., et al. (2013). Irisin is inversely associated with intrahepatic triglyceride contents in obese adults. J. Hepatol. 59, 557–562. doi: 10.1016/j.jhep.2013.04.030
Zhang, P., Tu, B., Wang, H., Cao, Z., Tang, M., Zhang, C., et al. (2014). Tumor suppressor p53 cooperates with SIRT6 to regulate gluconeogenesis by promoting FoxO1 nuclear exclusion. Proc. Natl. Acad. Sci. U.S.A. 111, 10684–10689. doi: 10.1073/pnas.1411026111
Zhao, Y., Yang, J., Liao, W., Liu, X., Zhang, H., Wang, S., et al. (2010). Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat. Cell Biol. 12, 665–675. doi: 10.1038/ncb2069
Zhong, L., D'urso, A., Toiber, D., Sebastian, C., Henry, R. E., Vadysirisack, D. D., et al. (2010). The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell 140, 280–293. doi: 10.1016/j.cell.2009.12.041
Keywords: Sirt6, obesity, diabetes mellitus, type 2, LiPo, gluconeogenesis
Citation: Kuang J, Chen L, Tang Q, Zhang J, Li Y and He J (2018) The Role of Sirt6 in Obesity and Diabetes. Front. Physiol. 9:135. doi: 10.3389/fphys.2018.00135
Received: 07 January 2018; Accepted: 12 February 2018;
Published: 27 February 2018.
Edited by:
Dechun Feng, National Institute on Alcohol Abuse and Alcoholism, United StatesReviewed by:
Hua Wang, Anhui Medical University, ChinaZIyi Liu, National Institutes of Health (NIH), United States
Copyright © 2018 Kuang, Chen, Tang, Zhang, Li and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinhan He, jinhanhe@scu.edu.cn
 Jiangying Kuang1,2
Jiangying Kuang1,2 Jinhan He
Jinhan He