- 1 School of Biological Sciences, Washington State University, Pullman, WA, USA
- 2 Department of Plant Biology, University of Minnesota, St. Paul, MN, USA
Nitrogen is an essential mineral nutrient and it is often transported within living organisms in its reduced form, as amino acids. Transport of amino acids across cellular membranes requires proteins, and here we report the phylogenetic analysis across taxa of two amino acid transporter families, the amino acid permeases (AAPs) and the lysine–histidine-like transporters (LHTs). We found that the two transporter families form two distinct groups in plants supporting the concept that both are essential. AAP transporters seem to be restricted to land plants. They were found in Selaginella moellendorffii and Physcomitrella patens but not in Chlorophyte, Charophyte, or Rhodophyte algae. AAPs were strongly represented in vascular plants, consistent with their major function in phloem (vascular tissue) loading of amino acids for sink nitrogen supply. LHTs on the other hand appeared prior to land plants. LHTs were not found in chlorophyte algae Chlamydomonas reinhardtii and Volvox carterii. However, the characean alga Klebsormidium flaccidum encodes KfLHT13 and phylogenetic analysis indicates that it is basal to land plant LHTs. This is consistent with the hypothesis that characean algae are ancestral to land plants. LHTs were also found in both S. moellendorffii and P. patens as well as in monocots and eudicots. To date, AAPs and LHTs have mainly been characterized in Arabidopsis (eudicots) and these studies provide clues to the functions of the newly identified homologs.
Introduction
Nitrogen is a critical mineral nutrient in all living organisms since it is required for synthesis of a large number of compounds including hormones, nucleotides, and amino acids. As the basic building blocks of proteins, amino acids are needed for metabolism, cellular structure, growth, and development. Amino acid uptake into cells and cellular compartments depends on membrane-integral transporter proteins, and amino acid transporters have been identified in many organisms including bacteria, fungi, animals, and plants (Chang et al., 2004; Boudko, 2010). In plants, amino acid transporters are found in two families within the amino acid–polyamine–choline (APC) transporter superfamily, the amino acid/auxin permease (AAAP), and the APC family. The AAAP family includes transporters from plants, animals, and fungi (Chang et al., 2004), and in plants contains the amino acid permeases (AAPs), lysine–histidine-like transporters (LHTs), proline transporters (ProTs), γ-aminobutyric acid transporters (GATs), ANT1-like aromatic, and neutral amino acid transporters and auxin transporters (AUXs; Wipf et al., 2002; Rentsch et al., 2007). Cationic amino acid transporters (CATs) belong to the APC family and are present in both animals and plants1)
This study addresses the phylogeny of the plant AAP and LHT transporters. These have been characterized in angiosperms (flowering plants), and specifically in eudicots, and detailed overviews on their substrate specificity, localization, and biological functions have recently been presented (see Fischer et al., 2002; Lee and Tegeder, 2004; Rentsch et al., 2007; Tegeder and Rentsch, 2010; Tegeder et al., 2011). In Arabidopsis the AAP family consists of eight members (AtAAP1–8) that generally transport neutral and acidic amino acids with moderate affinity, with the exception of AtAAP3 and AtAAP5 that also transport basic amino acids (Fischer et al., 1995, 2002; Rentsch et al., 2007; Svennerstam et al., 2008). All Arabidopsis AtAAPs analyzed to date have been localized to the plasma membrane and they function as H+-coupled amino acid uptake systems (see Tegeder and Rentsch, 2010). AAPs have been suggested to be involved in a number of physiological processes in plants including amino acid uptake from the soil (Hirner et al., 2006; Lee et al., 2007; Svennerstam et al., 2008), phloem loading or xylem–phloem transfer (Schulze et al., 1999; Okumoto et al., 2002; Koch et al., 2003; Tegeder et al., 2007; Tan et al., 2008; Hunt et al., 2010; Zhang et al., 2010; see also Tegeder and Rentsch, 2010), and seed loading (Schmidt et al., 2007; Tegeder et al., 2007; Tan et al., 2008; Sanders et al., 2009).
Much less is known about the LHTs, a family of 10 members (AtLHT1–10) in Arabidopsis. AtLHT1 was originally described as a lysine and histidine selective transporter (Chen and Bush, 1997), but other studies with AtLHT1 and AtLHT2 suggests that LHTs preferentially transport neutral and acidic amino acids with high affinity (Lee and Tegeder, 2004; Hirner et al., 2006; Svennerstam et al., 2007, 2008). Like the AAPs, AtLHTs are localized to the plasma membrane and transport a broad spectrum of amino acids from the cell wall space into the cell (Hirner et al., 2006; Foster et al., 2008). Based on promoter–GUS studies, LHTs have been suggested to be involved in import of organic nitrogen into root and mesophyll cells (Hirner et al., 2006), as well as into pollen and other cells of reproductive floral tissue (Lee and Tegeder, 2004; Foster et al., 2008).
AAPs and LHTs have not yet been described in any organisms other than angiosperms. With the recent progress in genome sequencing we are however now in the excellent position to determine whether AAP and LHT amino acid transporters are present in ancestors of seed plants and to examine the phylogenetic relationship of AAP and LHT proteins. Three major clades form the large monophyletic plant kingdom. These include the green plants (Viridiplantae), Rhodophytes (red algae), and Glaucophytes (freshwater microscopic algae; Figure 1; Anderberg et al., 2011). The green plants are grouped into the Chlorophytes that contain algae such as Chlamydomonas reinhardtii, and the Streptophytes with algal species (Charophytes) and land plants (Finet et al., 2010; Banks et al., 2011). The land plants are divided in non-vascular plants (Bryophytes; i.e., liverworts, mosses, and hornworts) and vascular plants that split into Lycophytes (non-seed plants) and Euphyllophytes. The Lycophytes contain clubmosses, quilworts, and spikemosses while Euphyllophytes consist of ferns (Monilophytes) and seed-bearing plants (Spermatophytes), which are often grouped into angiosperms (flowering plants) and gymnosperms (i.e., cycads, Ginkgo, conifers, and gnetophytes). However, the evolutionary relationships of Spermatophytes are not clearly resolved (Magallon and Sanderson, 2002; Mathews, 2009).
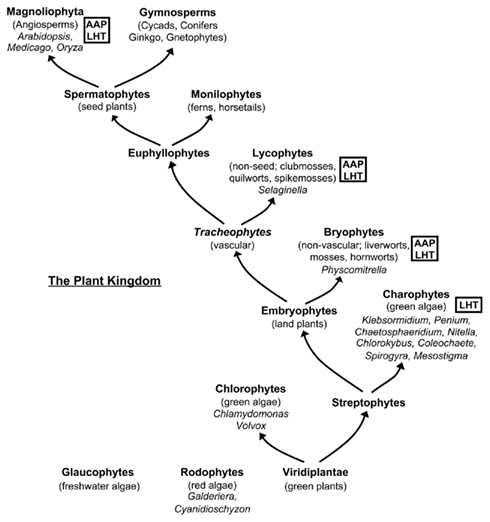
Figure 1. Simplified overview of the plant kingdom and analyzed genera. Taxa containing AAPs and/or LHTs are marked. Note, that genomes of gymnosperms and monilophytes have not been used for the analysis.
Here, sequences from red algae (Galdieria sulfuraria and Cyanidioschyzon merolae), green algae (Chlorophytes: Chlamydomonas reinhardtii and Volvox carterii; Charophytes: Penium marinum, Spirogyra praetensis, Coleochaete sp., Chaetosphaeridium globosum, Mesostigma viride, Nitella hyalina, Klebsormidium flaccidum, Chlorokybus atmosphyticus), and basal non-vascular (Physcomitrella patens), non-seed vascular (Selaginella moellendorffii), and vascular land plants (eudicots: Arabidopsis thaliana, Medicago sativa; monocots: Oryza sativa) were analyzed for AAP and LHT proteins (Figure 1). Phylogenetic reconstruction was performed to determine diversification of the AAP and LHT amino acid transporters as well as their lineage association.
Results and Discussion
AAP and LHT Transporters Form Two Distinct Groups
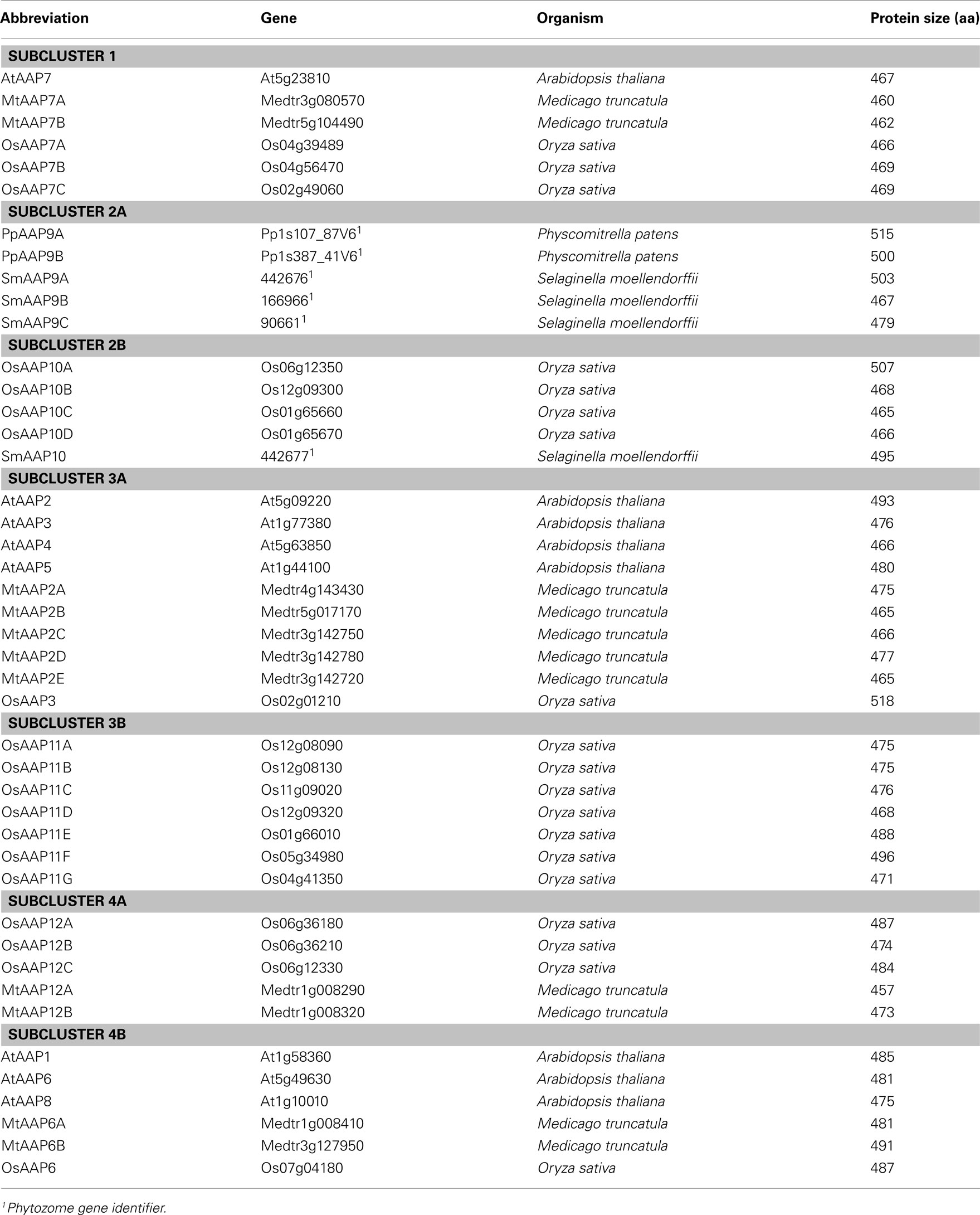
Database searches for AAP and LHT proteins in red algae, green algae, basal non-vascular and vascular land plants, and seed plants resulted in 44 AAP and 39 LHT protein sequences (Figure 2; Tables 1 and 2). Predicted protein sequences for the AAPs averaged 478 ± 14 amino acids (mean ± SD). The length of LHT sequences was similar (463 ± 31 amino acids). The LHT sequence from Klebsormidium flaccidum (KfLHT13) is an incomplete cDNA and contains the C-terminal 388 amino acids. A maximum-likelihood tree was constructed using PhyML 3.0 (Guindon et al., 2010) based on the alignment of full-length AAP and LHT sequences and the truncated KfLHT13 (Figure 2). In addition, trees were made using alignments in which the variable-length N- and C-terminal regions of the alignment were removed (data not shown). These trees did not differ from those based on full-length AAPs and LHTs, and KfLHT13 (Figures 2–4). Both AAPs and LHTs were found in eudicots, monocots, Selaginella and Physcomitrella, but AAPs and LHTs form two distinct groups supporting functional differences between the two transporter families in the analyzed organisms (see also Figure 1). The absence of AAP or LHT genes in Chlorophytes is consistent with the hypothesis that chlorophyte algae are not ancestors of land plants (Turmel et al., 1999; Karol et al., 2001; Kapraun, 2007).
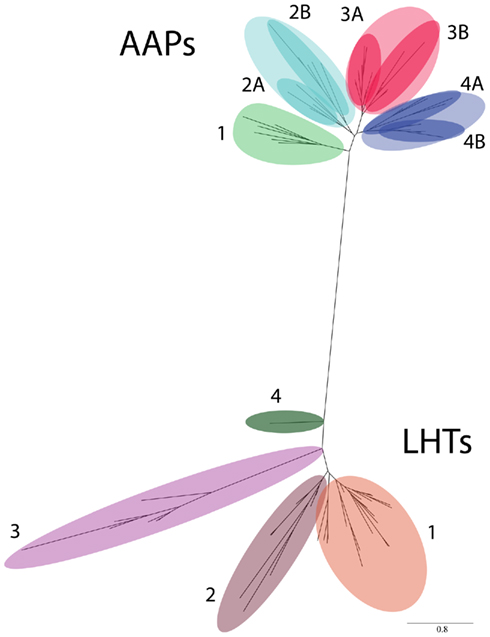
Figure 2. Phylogenetic tree of AAP and LHT proteins found across taxa. A total of 44 AAP and 39 LHT proteins were identified. AAPs and LHTs form two distinct groups, each sub-divided in clusters or subclusters. The clusters (numbers) and subclusters (letters) were labeled and color-coded.
AAPs Evolved at the Same Time as Land Plants
When searching the databases, AAP proteins were found in non-vascular land plants (Physcomitrella patens; 2 proteins), non-seed vascular plants (Selaginella moellendorffii, 4 proteins), and seed plants including Arabidopsis thaliana (8 proteins), Oryza sativa (19 proteins), and Medicago truncatula (11 proteins). No AAPs were found in algal sequences of Rhodophytes (Galdieria and Cyanidioschyzon), Chlorophytes (Chlamydomonas and Volvox), or Charophytes (Penium, Spirogyra, Coleochaete, Chaetosphaeridium, Nitella, Klebsormidium, and Chlorokybus). However, the Charophyte search was based on EST sequences, and until the whole genome sequences are available we cannot rule out that AAPs are present in Charophytes. The identified AAP proteins are grouped into four main clusters (1, 2, 3, and 4), with cluster 2–4 being sub-divided into two subclusters A and B (Table 1; Figure 3). Cluster 1, 3, and 4 only contain AAPs of seed plants while cluster 2 contains non-vascular and non-seed vascular plant, and angiosperm proteins.
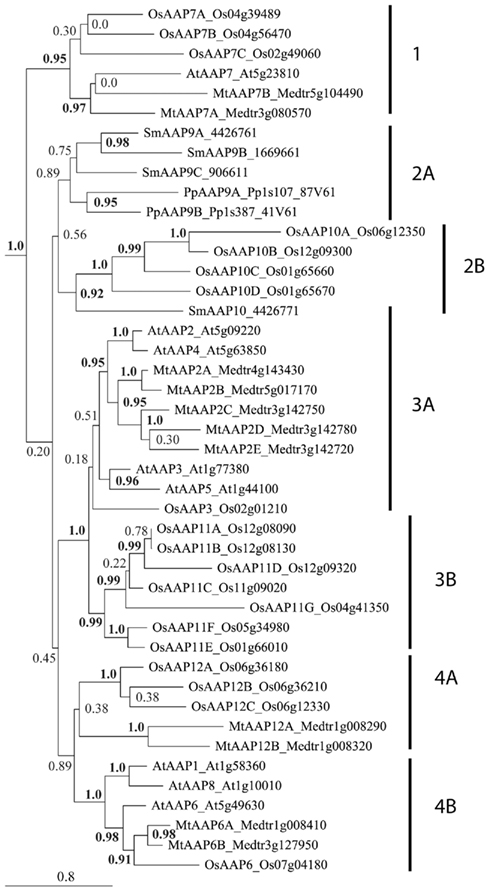
Figure 3. Phylogenetic analysis of AAP proteins found in Arabidopsis thaliana (At), Oryza sativa (Os), Medicago truncatula (Mt), Selaginella moellendorffii (Sm), and Physcomitrella patens (Pp). Multiple protein sequence alignment was done using CLUSTAL X (Thompson et al., 2007). A maximum-likelihood tree was constructed using PhyML 3.0 (Guindon et al., 2010). Numbers at the nodes indicate SH-like branch support (Shimodaira and Hasegawa, 1999; Buckley et al., 2001). Values above 0.9 (in bold) show significant phylogenetic support. Accession numbers of sequences (see Table 1) are provided after the species transporter names. The clusters (numbers) and subclusters (letters) are labeled.
Cluster 1 contains proteins from monocots and eudicots that are related to Arabidopsis AtAAP7. It holds AtAAP7 and two Medicago proteins (MtAAP7A and 7B) consistent with a genome duplication in legumes relative to Arabidopsis (Cannon et al., 2006). In addition, it includes three rice AAPs (OsAAP7A–7C) that likely represent an amplification of AAP7 genes in monocots. While the specific function of AtAAP7 and related proteins is still unknown, the phylogenetic analysis supports that they are important for seed plants since they are maintained in both monocot and eudicot lineages.
Cluster 2 contains AAP proteins from non-vascular and non-seed plants, and monocots, but lacks eudicot proteins. Subcluster 2A includes proteins only from the moss Physcomitrella patens (PpAAP9A and 9B) and spikemoss Selaginella moellendorffii (SmAAP9A–9C), but no proteins from seed plants, suggesting differences in amino acid transporter function between early and higher land plants. Differences in function might be based on (i) differences in phloem loading or source-sink transport between Spermatophytes that have complex leaf venation and the Bryophytes and Lycophytes with no vasculature or microphylls with only a single vascular strands (Reinhart and Thomas, 1981; Aldous, 2002; Beerling, 2005), or on (ii) differences in reproduction (flower versus spores and spore-bearing structures; Prigge and Bezanilla, 2010). For example, specific transporters might be needed for uptake of amino acids into moss sporophytes (Caussin et al., 1983).
In contrast, subcluster 2B contains four proteins from rice (OsAAP10A–10D) and one from S. moellendorffii. It is interesting that cluster 2 lacks eudicot representation, which indicates that AAPs of this type were lost from eudicot genomes after divergence from monocots. This also suggests that AAPs in cluster 2 represent the earliest AAP sequences. No information is available concerning the function of AAPs in cluster 2. However, the presence of monocot AAPs indicates that an essential amino acid transport activity, required for non-vascular and non-seed vascular plants, was maintained in monocots and likely replaced in eudicots by other AAPs or amino acid transporters in other families.
Cluster 3 contains only monocot and eudicot sequences. Subcluster 3A includes Arabidopsis AtAAP3 and AtAAP5 and one related rice protein (OsAAP3), as well as Arabidopsis AtAAP2 and AtAAP4 and five legume/Medicago AAPs (MtAAP2A–2E). All of the Arabidopsis AtAAPs in subcluster 3A appear to be involved in loading of amino acids into the phloem. With the evolution of vascular plants, two vascular tissues were established, the xylem and the phloem. While the xylem functions in water and nutrient transport from the root to the shoot, phloem is important for long-distance transport of nutrients from source (e.g., mature leaves or roots) to sinks such as developing roots, flowers and seeds. In most herbaceous plants such as Arabidopsis, rice, or Medicago, phloem loading follows the apoplasmic route, where nutrients are loaded from the apoplast into the sieve element-companion cell complex of the collection phloem (Rennie and Turgeon, 2009). In addition, xylem to phloem amino acid transfer might occur along the transport pathway from source to sink (Pate et al., 1975, 1977). These loading steps into the collection or transport phloem require the activity of plasma membrane transporters.
In Arabidopsis, AtAAP3 function in the phloem seems to be restricted to the root (Okumoto et al., 2004), while AtAAP5 probably functions in import of amino acids into the companion cells (Brady et al., 2007; Zhang et al., 2008) of different organs including roots and leaves (Fischer et al., 1995; Cartwright et al., 2009; see also Tegeder and Rentsch, 2010). Preliminary results from the Tegeder lab indicate that AtAAP4 also plays a role in leaf phloem loading (Garneau and Tegeder, unpublished). AtAAP2 on the other hand is essential for phloem loading along the transport path (Hirner et al., 1998; Zhang et al., 2010). One rice and five Medicago proteins are related to the Arabidopsis phloem loaders and we cautiously speculate that they are involved in amino acid import into the sieve element/companion cells complex in legumes and monocot species. This prediction however requires proof through cellular and subcellular localization studies, and functional analysis in plants in future. Nevertheless, it receives some support from the fact that other legume AAPs, specifically Phaseolus vulgaris PvAAP1 and Pisum sativum PsAAP1, have also been localized to the phloem and group within cluster 3A (Tegeder et al., 2007; Tan et al., 2008).
Cluster 3B only contains seven rice proteins (OAAP11A–11G); eudicots are not represented. It is possible that this large group of monocot AAPs all function in phloem loading of amino acids in different tissues considering the similarity of subcluster 3B to 3A and the presence of only one rice AAP (and multiple eudicot sequences) in 3A. Note that the placement of OsAAP3 in cluster 3A only has weak phylogenetic support. Further, the lack of Selaginella and Physcomitrella sequences in cluster 3B suggests that AAPs developed independently in monocots, rather than the alternative, that AAPs of this group were lost in eudicots. None of the currently known eudicot amino acid transporters including AAPs from Arabidopsis (Figure 3), tomato (LeAAPs), potato (StAAPs), pea (PsAAPs), faba bean (VfAAPs), canola (BnAAPs), and Ricinus (RcAAPs) falls into cluster 3B (see Tan et al., 2008), providing further support for this hypothesis. Future research needs to determine if the monocot AAP proteins of cluster 3B differ in function from eudicot AAP proteins.
Amino acid permeases in cluster 4 are also divided into two subclusters. Cluster 4A holds three rice (OsAAP12A–12C) and two Medicago AAPs (MtAAP12A and 12B) with unknown function. Subcluster 4B contains Arabidopsis AtAAP6 related proteins and is branched into a group with AtAAP6 and two (duplicated) Medicago proteins, a related single rice protein (OsAAP6), and a group harboring Arabidopsis amino acid transporters AtAAP1 and AtAAP8. AtAAP6 is localized to the leaf xylem parenchyma (Okumoto et al., 2002). Although not directly involved in phloem loading, it is predicted to be important for xylem to phloem transfer of amino acids in Arabidopsis (Okumoto et al., 2002; Hunt et al., 2010), and the AAP6 relatives in legumes and monocots might have similar functions. It is interesting that one group within cluster 4B only contains two Arabidopsis transporters. It seems that Arabidopsis has gained two extra copies of AtAAP6: AtAAP1 and AtAAP8. Both AtAAP1 and AtAAP8 proteins are involved in seed loading, rather than phloem loading of amino acids (Schmidt et al., 2007; Sanders et al., 2009), supporting that they are AAP6 paralogs. It is tempting to hypothesize that monocot AAPs in group 4A function in seed loading as nothing is known to date concerning the function of OsAAP12A, B, or C.
Nevertheless, it is important to point out that AAP expression is generally not phloem or seed specific (see Ortiz-Lopez et al., 2000; Rentsch et al., 2007; Tegeder and Rentsch, 2010). For example, Arabidopsis AAPs (i.e., AtAAP1 and AtAAP5) are also expressed in root epidermal and cortex cells suggesting that they fulfill additional functions in plants including amino acid uptake from the soil (Lee et al., 2007; Svennerstam et al., 2008; Cartwright et al., 2009). Further, the function of AAPs and other amino acid transporters including LHTs seems to be influenced by the physiology of the plant as nitrogen starvation and nitrate re-feeding affects their expression patterns (Liu and Bush, 2006).
In angiosperm, analysis of the relatedness of the rice and legume AAPs with Arabidopsis proteins might help with prediction of their function. For example a placement of rice and/or Medicago proteins with Arabidopsis AAP2, 3, 4, 5, and 6 might suggest a function in phloem loading. Interestingly, rice lacks close relatives of AAP2 and AAP4, and Medicago has no AAP3 and AAP5 phloem loaders. Some of the duplicated Arabidopsis AAPs may be functionally redundant. At least for AtAAP3 this appears to be the case, since mutant analysis did not result in a functional phenotype (Okumoto et al., 2004). On the other hand, some of the evolved AAPs in legumes (Medicago) and monocots seem not to be present in Arabidopsis/non-legume dicots (see Figure 3, subcluster 2B, 3B, and 4A) further supporting differences in AAP function among angiosperms. This is also in agreement with the large variation in the number of AAP proteins between Arabidopsis (8 proteins), Medicago (11 proteins), and rice (19 proteins). For example monocots might require additional or different amino acid transporters than eudicots due to differences in morphology and physiology between these distinct groups of seed plants. In legumes, additional AAP proteins might be needed for amino acid transport processes related to N2 fixation and nodule function.
Taken together, AAPs are mainly found in euphyllophytes, including monocots and eudicots/legumes, which is in agreement with the main functions of AAPs in phloem and seed loading in support of amino acid translocation from source to sink (seeds). Non-vascular and non-seed vascular plants only have AAPs that are more closely related to AAP7, a transporter that remains to be characterized and might differ in function from the other AAPs.
LHTs Evolved Prior to Land Plants
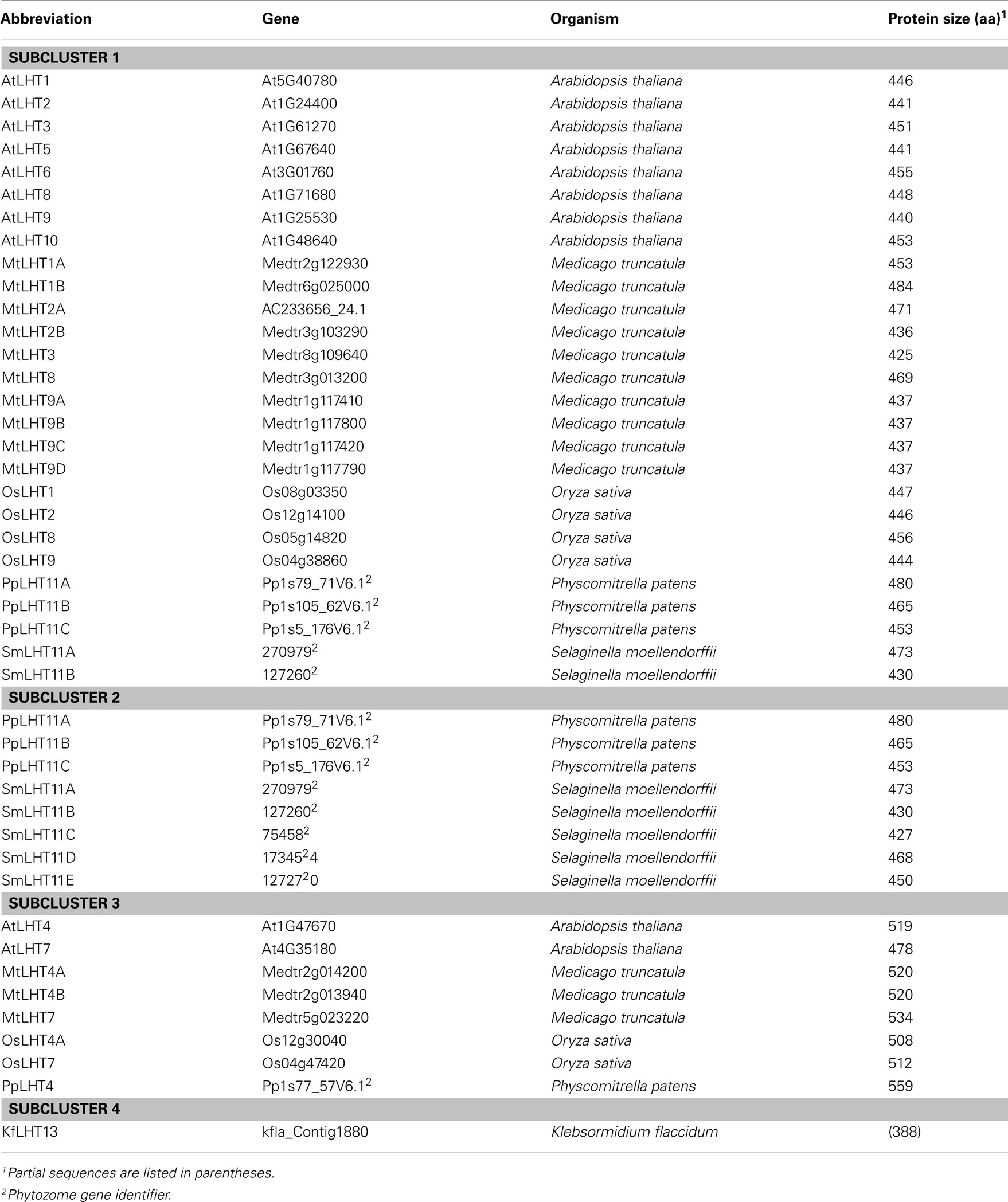
Phylogenetic analysis revealed that LHTs are present in Charophytes (Klebsormidium flaccidum, 1 protein), non-vascular land plants (P. patens, 4 proteins), non-seed vascular plants (S. moellendorffii, 5 proteins), and seed plants (A. thaliana, 10 proteins; O. sativa, 6 proteins; M. truncatula, 13 proteins), demonstrating that LHTs evolved before the occurrence of early land plants (Table 2). The identified LHT proteins group into four clusters (Table 2; Figure 4). Cluster 1 includes LHT proteins of euphyllophytes, cluster 2 has five Selaginella and three Physcomitrella LHT proteins, cluster 3 contains euphyllophyte sequences and one Physcomitrella LHT and cluster 4 only contains one Klebsormidium protein.
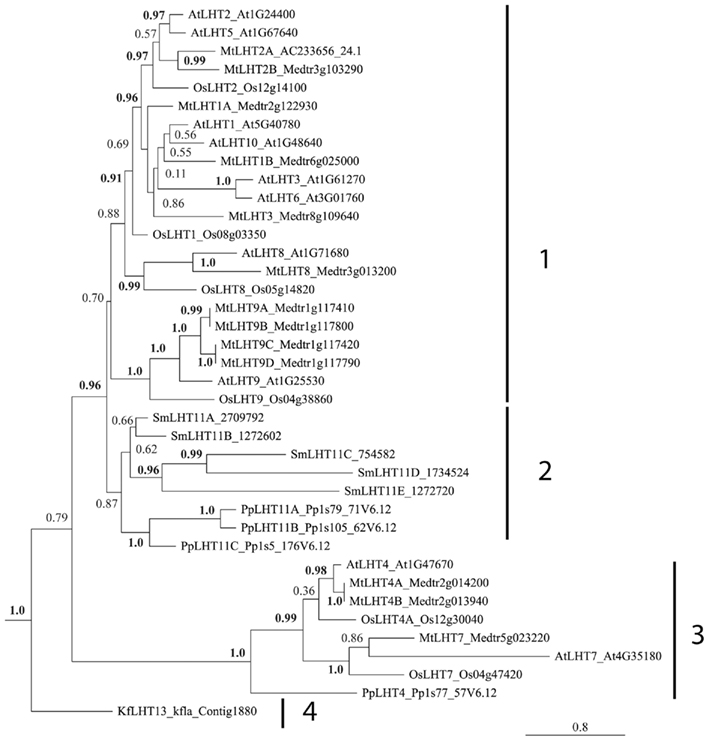
Figure 4. Phylogenetic analysis of LHT proteins found in Arabidopsis thaliana (At), Oryza sativa (Os), Medicago truncatula (Mt), Selaginella moellendorffii (Sm), Physcomitrella patens (Pp), and Klebsormidium flaccidum (Kf). Multiple protein sequence alignment was done using CLUSTAL X (Thompson et al., 2007). A maximum-likelihood tree was constructed using PhyML 3.0 (Guindon et al., 2010). Numbers at the nodes indicate SH-like branch support (Shimodaira and Hasegawa, 1999; Buckley et al., 2001). Values above 0.9 (in bold) show significant phylogenetic support. Accession numbers of sequences (see Table 2) are provided after the species transporter names. The clusters are numbered.
Cluster 1 contains 8 of the 10 Arabidopsis AtLHTs, including AtLHT1, 2, 3, 5, 6, 8, 9, and 10. It appears that a duplication event has occurred in Arabidopsis and placement of the LHT proteins suggests that AtLHT2 and AtLHT5, AtLHT1 and AtLHT10, and AtLHT3 and AtLHT6, respectively are the result of such duplication. Relatives of AtLHTs are present in Medicago and 10 of the 13 MtLHTs are present in cluster 1. The presence of only four rice LHT sequences in group 1 indicates that LHTs were not as extensively duplicated in monocots as in eudicots. Recent studies have shown that Arabidopsis LHT transporters of cluster 1 including AtLHT1, 2, 4, 5, and 6 are expressed in male and female floral tissue, such as anther tissue, tapetum, mature pollen, pollen tubes, and pistil transmitting tissue (Hirner et al., 2006; Foster et al., 2008; see also Tegeder and Rentsch, 2010), and it was suggested that they might be essential for successful sexual plant reproduction. This is also in agreement with the observation that LHTs of cluster 1 are only present in flowering plant species. However, experimental proof for LHT function in reproduction is still missing, and AtLHTs of cluster 1 seem to have additional functions in plants as they are expressed in other organs besides flowers (Hirner et al., 2006; see Arabidopsis eFP Browser, Winter et al., 2007). As for example recently demonstrated for AtLHT1, the transporter is important for amino acid uptake into root and mesophyll cells (Hirner et al., 2006; Svennerstam et al., 2008). LHT8 and LHT9 proteins of cluster 1 form separate subgroups. One group includes Arabidopsis AtLHT8 and an ortholog each in rice and Medicago, and the second group contains Arabidopsis AtLHT9, rice OsLHT9, and four closely related Medicago MtLHT9 transporters. Localization of these transporters has not been resolved and similar to most other LHTs, their physiological functions remain to be elucidated.
Cluster 2 includes three P. patens (PpLHT11A–C) and five S. moellendorffii LHT11 proteins (SmLHT11A–E) but none from Spermatophytes. LHT11 genes may have evolved independently in Physcomitrella and Selaginella suggesting that in early land plants these LHTs serve functions in cellular amino acid transport processes that are not required in higher plants (see above). However, as in seed plants, Bryophytes and Lycophytes seem to need both LHTs and AAPs for growth and development (Figures 1–4).
Cluster 3 contains LHT4 and LHT7 proteins from angiosperms and Physcomitrella PpLHT4. Recent expression studies suggest that Arabidopsis AtLHT4 and AtLHT7 might be involved in reproduction, specifically in anther and pollen development (Bock et al., 2006; Foster et al., 2008). However, at least AtLHT4 has most certainly additional functions since it is also expressed in root and stem (Winter et al., 2007). This might explain its phylogenetic divergence from other LHTs and its placement with PpLHT4. In early land plants, transporters may be critical for amino acid movement over relatively short distances. As plants colonized dry land, translocation of amino acids from source to sink cells occurred probably by cell to cell transport (symplasmic) and between cells (apoplasmic), especially in non-vascular mosses (Trachtenberg and Zamski, 1978; Reinhart and Thomas, 1981). Uptake of the apoplasmic amino acids required membrane proteins including H+-coupled, high affinity LHT symporters, as indicated by the phylogenetic analysis.
Cluster 4 only contains a LHT protein from green algae Klebsormidium called KfLHT13, suggesting its evolutionary divergence from LHTs of land plants and differences in function. While we are not aware of amino acid transport studies in Charophytes, research with Chlorophytes such a Chlamydomonas and Chlorella spp. and marine microalgae demonstrate that in algae different transport systems are present (Kirk and Kirk, 1978a; Cho et al., 1981; Cho and Komor, 1985; Shehawy and Kleiner, 2001; Kato et al., 2006; see also Flynn and Butler, 1986 and references within). Although an LHT transporter was only found in Klebsormidium, the screened charophyte sequences were obtained from EST projects and we predict that LHTs are also present in other charophytes besides Klebsormidium. Placement of KfLHT13 further supports that LHTs have evolved before land plants and that their function is important to green algae as well.
Gene function of lysine–histidine-like transporters was likely important in ancestors of plants, as a gene is detected in a charophyte, contributing to its high affinity and substrate selectivity for neutral and acid amino acids. Localization and expression studies of Arabidopsis AtLHTs suggest that, in addition to other functions, LHTs have a major role in sexual plant reproduction in seed plants. This also indicates a difference in LHT function between angiosperms and non-seed/non-land plants. While LHT functions still need to be demonstrated in planta, this is in agreement with the phylogenetic analysis showing a grouping of angiosperm LHTs while LHT proteins from non-vascular and non-seed plants, and green algae are present in separate groups.
Based on the phylogeny, genes in the LHT family of land plants likely arose from an ancestral gene similar to the charophyte LHT. The ancestral gene diversified as plants colonized dry land, as seen by the presence of multiple LHT in moss, a non-vascular plant. In contrast, no algal genes encoding AAP transporters were detected in our analysis of the Charophytes, but it may be too early to conclude whether genes belonging to the AAP family are present, as the charophyte genome has not been sequenced completely. Algae generally acquire amino acids from the environment for growth, and some variation with respect of the kind and amount of amino acids that are taken up has been observed between and within species (Cho et al., 1981; Cho and Komor 1983, 1985; Flynn and Butler, 1986; Kato et al., 2006). In addition, leakage of amino acids from the cells into the apoplast might occur and requires transporters for retrieval. Physiological studies have demonstrated that in algae active transport systems with varying specificity and affinities (high and low) are present (Kirk and Kirk, 1978a,b; Sauer et al., 1983; Cho and Komor, 1985; Shehawy and Kleiner, 2001; Kato et al., 2006; see also Flynn and Butler, 1986), which might point to the presence of both, LHT and AAP transporters, or additional amino acid transporters in Charophyte algae.
Conclusion
Recent functional studies support that AAP and LHT proteins have essential roles in transport of a broad range of amino acids in eudicots (see Tegeder and Rentsch, 2010). Here, phylogenetic analysis supports that AAPs, which generally present moderate and low affinity systems for neutral and acidic amino acids, are important to land plants with a main function in phloem loading and that they are not required in red algae or green algae (Charophytes or Chlorophytes). In contrast, LHTs are found in green algae, non-seed plants and angiosperms suggesting the need for high affinity amino acid transporters across the different organisms.
Both AAPs and LHTs were found in all land plants analyzed consistent with essential and distinct functions for both transporter families. To date, information on the role of AAP and LHT transporters is almost exclusively based on studies in Arabidopsis and in some cases in legumes, and suggests differences between AAPs and LHTs in substrate selectivity, transport affinity and cellular function (see Rentsch et al., 2007; Tegeder and Rentsch, 2010; see also above). However, phylogenetic analyses indicates that function of some AAP and LHT transporters diverged in monocots, non-seed vascular plants, non-vascular plants and green algae, and future studies need to address the role of the amino acid transporters across land plants and in green algae.
Materials and Methods
Identification and Annotation of AAP and LHT Proteins
Genome sequences are available for Arabidopsis thaliana, rice (Oryza sativa), Medicago truncatula, Selaginella moellendorffii, Physcomitrella patens, Chlamydomonas reinhardtii, and Volvox carterii. AAP and LHT sequences were selected from rice, M. truncatula, S. moellendorffii, and P. patens predicted protein sequences using BLAST searches with known Arabidopsis AAP and LHT transporters (see Tables 1 and 2) on the Phytozome website2. The same database was searched for AAP and LHT protein sequences from the Chlorophytes C. reinhardtii and V. carterii. Dr. Charles F. Delwiche and Dr. James Thierer, University of Maryland provided support by screening their EST (Expressed Sequence Tag) databases for AAP and LHT relatives in charophytes, specifically in Penium marinum, Spirogyra praetensis, Coleochaete sp., and Chaetosphaeridium globosum, Mesostigma viride, Nitella hyalina, Klebsormidium flaccidum, Chlorokybus atmosphyticus3. In addition, the genome (protein) sequences of the red algae Galdieria sulfuraria available through http://genomics.msu.edu/cgi-bin/galdieria/blast.cgi (Barbier et al., 2005) and Cyanidioschyzon merolae at http://merolae.biol.s.u-tokyo.ac.jp/and were searched for the presence of AAPs and LHTs.
Sequence Alignments and Phylogenetic Analysis
Multiple protein sequence alignments were generated with Clustal X (Thompson et al., 2007) and, for comparison, with MUSCLE (Edgar, 2004). Phylogenetic analysis was performed through the iPlant Collaborative website4. Maximum-likelihood analysis was done using PhyML 3.0 (Guindon and Gascuel, 2003; Guindon et al., 2010) and statistical analysis of phylogenetic trees was performed using a Shimodaira–Hasegawa-like test (SH-like test; Shimodaira and Hasegawa, 1999; Buckley et al., 2001). The values for SH-like branch support are presented at the nodes on the trees. Values above 0.9 show significant phylogenetic support. Trees were visualized using the FigTree program5
Naming of AAP and LHT Transporters
The identified AAP and LHT sequences were named based on clustering with Arabidopsis AtAAP and AtLHT protein sequences. In Arabidopsis 8 AAP (AtAAP1–8) and 10 LHT (AtLHT1–10) transporters have been previously identified (see Rentsch et al., 2007). Phylogenetic grouping was used to name the transporters from other species. In cases where more than one Arabidopsis relative was found from a given species, letter labeling was chosen in addition. For example, three rice relatives of AtAAP7 were named OsAAP7A, 7B, and 7C. Transporters that did not group with Arabidopsis proteins were given numbers not found for the Arabidopsis transporters such as AAP9 or LHT11.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Charles Delwiche and Mr. James Thierer, Cell Biology and Molecular Genetics, University of Maryland for providing sequences from Charophyte algae prior to publication. Mechthild Tegeder appreciates the financial support by the National Science Foundation Grant IOS 1021286 and the Agricultural and Food Research Initiative Competitive Grant no. 2010-65115-20382 from the USDA National Institute of Food and Agriculture.
Footnotes
References
Aldous, A. R. (2002). Nitrogen translocation in Sphagnum mosses: effects of atmospheric nitrogen deposition. New Phytol. 156, 241–253.
Anderberg, H. I., Danielson, J. Å. H., and Johanson, U. (2011). Algal MIPs, high diversity and conserved motifs. BMC Evol. Biol. 11, 110. doi:10.1186/1471-2148-11–110
Banks, J. A., Nishiyama, T., Hasebe, M., Bowman, J. L., Gribskov, M., dePamphilis, C., Albert, V. A., Aono, N., Aoyama, T., Ambrose, B. A., Ashton, N. W., Axtell, M. J., Barker, E., Barker, M. S., Bennetzen, J. L., Bonawitz, N. D., Chapple, C., Cheng, C., Correa, L. G., Dacre, M., DeBarry, J., Dreyer, I., Elias, M., Engstrom, E. M., Estelle, M., Feng, L., Finet, C., Floyd, S. K., Frommer, W. B., Fujita, T., Gramzow, L., Gutensohn, M., Harholt, J., Hattori, M., Heyl, A., Hirai, T., Hiwatashi, Y., Ishikawa, M., Iwata, M., Karol, K. G., Koehler, B., Kolukisaoglu, U., Kubo, M., Kurata, T., Lalonde, S., Li, K., Li, Y., Litt, A., Lyons, E., Manning, G., Maruyama, T., Michael, T. P., Mikami, K., Miyazaki, S., Morinaga, S., Murata, T., Mueller-Roeber, B., Nelson, D. R., Obara, M., Oguri, Y., Olmstead, R. G., Onodera, N., Petersen, B. L., Pils, B., Prigge, M., Rensing, S. A., Riaño-Pachón, D. M., Roberts, A. W., Sato, Y., Scheller, H. V., Schulz, B., Schulz, C., Shakirov, E. V., Shibagaki, N., Shinohara, N., Shippen, D. E., Sørensen, I., Sotooka, R., Sugimoto, N., Sugita, M., Sumikawa, N., Tanurdzic, M., Theissen, G., Ulvskov, P., Wakazuki, S., Weng, J. K., Willats, W. W., Wipf, D., Wolf, P. G., Yang, L., Zimmer, A. D., Zhu, Q., Mitros, T., Hellsten, U., Loqué, D., Otillar, R., Salamov, A., Schmutz, J., Shapiro, H., Lindquist, E., Lucas, S., Rokhsar, D., and Grigoriev, I. V. (2011). The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963.
Barbier, G., Oesterhelt, C., Larson, M. D., Halgren, R. G., Wilkerson, C., Garavito, R. M., Benning, C., and Weber, A. P. (2005). Comparative genomics of two closely related unicellular thermo-acidophilic red algae, Galdieria sulphuraria and Cyanidioschyzon merolae, reveals the molecular basis of the metabolic flexibility of Galdieria sulphuraria and significant differences in carbohydrate metabolism of both algae. Plant Physiol. 137, 460–474.
Bock, K. W., Honys, D., Ward, J. M., Padmanaban, S., Nawrocki, E. P., Hirschi, K. D., Twell, D., and Sze, H. (2006). Integrating membrane transport with male gametophyte development and function through transcriptomics. Plant Physiol. 140, 1151–1168.
Boudko, D. Y. (2010). “Molecular ontology of amino acid transport,” in Epithelial Transport Physiology, ed. G. A. Gerencser (New York, NY: Springer), 379–472.
Brady, S. M., Orlando, D. A., Lee, J. Y., Wang, J. Y., Koch, J., Dinneny, J. R., Mace, D., Ohler, U., and Benfey, P. N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806.
Buckley, T. R., Simon, C., Shimodaira, H., and Chambers, G. K. (2001). Evaluating hypotheses on the origin and evolution of the New Zealand alpine cicadas (Maoricicada) using multiple-comparison tests of tree topology. Mol. Biol. Evol. 18, 223–234.
Cannon, S. B., Sterck, L., Rombauts, S., Sato, S., Cheung, F., Gouzy, J., Wang, X., Mudge, J., Vasdewani, J., Schiex, T., Spannagl, M., Monaghan, E., Nicholson, C., Humphray, S. J., Schoof, H., Mayer, K. F., Rogers, J., Quétier, F., Oldroyd, G. E., Debellé, F., Cook, D. R., Retzel, E. F., Roe, B. A., Town, C. D., Tabata, S., Van de Peer, Y., and Young, N. D. (2006). Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proc. Natl. Acad. Sci. U.S.A. 103, 14959–14964.
Cartwright, D. A., Brady, S. M., Orlando, D. A., Strumfels, B., and Benfey, P. N. (2009). Reconstructing spatiotemporal gene expression data from partial observations. Bioinformatics 25, 2581–2587.
Caussin, C., Fleurat-Lessard, P., and Bonnemain, J. L. (1983). Absorption of some amino acids by sporophytes isolated from Polytrichum formosum and ultrastructural characteristics of the haustorium transfer cells. Ann. Bot. 51, 167–173.
Chang, A. B., Lin, R., Studley, W. K., Tran, C. V., and Saier, M. H. Jr. (2004). Phylogeny as a guide to structure and function of membrane transport proteins. Mol. Membr. Biol. 21, 171–181.
Chen, L., and Bush, D. R. (1997). LHT1, a lysine- and histidine-specific amino acid transporter in Arabidopsis. Plant Physiol. 115, 1127–1134.
Cho, B. H., and Komor, E. (1983). Mechanism of proline uptake by Chlorella vulgaris. Biochim. Biophys. Acta 735, 361–366.
Cho, B. H., and Komor, E. (1985). The amino acid transport systems of the autotrophically grown green alga Chlorella. Biochim. Biophys. Acta 821, 384–392.
Cho, B. H., Sauer, N., Komor, E., and Tanner, W. (1981). Glucose induces two amino acid transport systems in Chlorella. Proc. Natl. Acad. Sci. U.S.A. 78, 3591–3594.
Edgar, R. C. (2004). MUSCLE multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797.
Finet, C., Timme, R. E., Delwiche, C. F., and Marlétaz, F. (2010). Multigene phylogeny of the green lineage reveals the origin and diversification of land plants. Curr. Biol. 20, 2217–2222.
Fischer, W. N., Kwart, M., Hummel, S., and Frommer, W. B. (1995). Substrate specificity and expression profile of amino acid transporters (AAPs) in Arabidopsis. J. Biol. Chem. 270, 16315–16320.
Fischer, W. N., Loo, D. D. F., Koch, W., Ludewig, U., Boorer, K. J., Tegeder, M., Rentsch, D., Wright, E. M., and Frommer, W. B. (2002). Low and high affinity amino acid H+-cotransporters for cellular import of neutral and charged amino acids. Plant J. 29, 717–731.
Flynn, K. J., and Butler, I. (1986). Nitrogen sources for the growth of marine microalgae: role of dissolved free amino acids. Mar. Ecol. Prog. Ser. 34, 281–304.
Foster, J., Lee, Y. H., and Tegeder, M. (2008). Distinct expression of members of the LHT amino acid transporter family in flowers indicates specific roles in plant reproduction. Sex. Plant Reprod. 21, 143–152.
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321.
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704.
Hirner, A., Ladwig, F., Stransky, H., Okumoto, S., Keinath, M., Harms, A., Frommer, W. B., and Koch, W. (2006). Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. Plant Cell 18, 1931–1946.
Hirner, B., Fischer, W. N., Rentsch, D., Kwart, M., and Frommer, W. B. (1998). Developmental control of H+/amino acid permease gene expression during seed development of Arabidopsis. Plant J. 14, 535–544.
Hunt, E., Gattolin, S., Newbury, H. J., Bale, J. S., Tseng, H. M., Barrett, D. A., and Pritchard, J. (2010). A mutation in amino acid permease AAP6 reduces the amino acid content of the Arabidopsis sieve elements but leaves aphid herbivores unaffected. J. Exp. Bot. 61, 55–64.
Kapraun, D. F. (2007). Nuclear DNA content estimates in green algal lineages: Chlorophyta and Streptophyta. Ann. Bot. 99, 677–701.
Karol, K. G., McCourt, R. M., Cimino, M. T., and Delwiche, C. F. (2001). The closest living relatives of land plants. Science 294, 2351–2353.
Kato, Y., Ueno, S., and Imamura, N. (2006). Studies on the nitrogen utilization of endosymbiotic algae isolated from Japanese Paramecium bursaria. Plant Sci. 170, 481–486.
Kirk, D. L., and Kirk, M. M. (1978a). Carrier-mediated uptake of arginine and urea by Chlamydomonas reinhardtii. Plant Physiol. 61, 556–560.
Kirk, M. M., and Kirk, D. L. (1978b). Carrier-mediated uptake of arginine and urea by Volvox carteri f. nagariensis. Plant Physiol. 61, 549–555
Koch, W., Kwart, M., Laubner, M., Heineke, D., Stransky, H., Frommer, W. B., and Tegeder, M. (2003). Reduced amino acid content in transgenic potato tubers due to antisense inhibition of the leaf H+/amino acid symporter StAAP1. Plant J. 33, 211–220.
Lee, Y. H., Foster, J., Chen, J., Voll, L. M., Weber, A. P. M., and Tegeder, M. (2007). AAP1 transports uncharged amino acids into roots of Arabidopsis. Plant J. 50, 305–319.
Lee, Y. H., and Tegeder, M. (2004). Selective expression of a novel high-affinity transport system for acidic and neutral amino acids in the tapetum cells of Arabidopsis flowers. Plant J. 40, 60–74.
Liu, X., and Bush, D. R. (2006). Expression and transcriptional regulation of amino acid transporters in plants. Amino Acids 30, 113–120.
Magallon, S., and Sanderson, M. J. (2002). Relationships among seed plants inferred from highly conserved genes: sorting conflicting phylogenetic signals among ancient lineage. Am. J. Bot. 89, 1991–2006.
Mathews, S. (2009). Phylogenetic relationships among seed plants: persistent questions and the limits of DNA sequence data. Am. J. Bot. 96, 228–236.
Okumoto, S., Koch, W., Tegeder, M., Fischer, W. N., Biehl, A., Leister, D., Stierhof, Y. D., and Frommer, W. B. (2004). Root phloem-specific expression of the plasma membrane amino acid proton co-transporter AAP3. J. Exp. Bot. 55, 2155–2168.
Okumoto, S., Schmidt, R., Tegeder, M., Fischer, W. N., Rentsch, D., Frommer, W. B., and Koch, W. (2002). High affinity amino acid transporters specifically expressed in xylem parenchyma and developing seeds of Arabidopsis. J. Biol. Chem. 277, 45338–45346.
Ortiz-Lopez, A., Chang, H. C., and Bush, D. R. (2000). Amino acid transporters in plants. Biochim. Biophys. Acta 1465, 275–280.
Pate, J. S., Sharkey, P. J., and Atkins, C. A. (1977). Nutrition of a developing legume fruit: functional economy in terms of carbon, nitrogen, water. Plant Physiol. 59, 506–510.
Pate, J. S., Sharkey, P. J., and Lewis, O. A. M. (1975). Xylem to phloem transfer of solutes in fruiting shoots of legumes, studied by a phloem bleeding technique. Planta 122, 11–26.
Prigge, M. J., and Bezanilla, M. (2010). Evolutionary crossroads in developmental biology: Physcomitrella patens. Development 137, 3535–3543.
Reinhart, D. A., and Thomas, R. J. (1981). Sucrose uptake and transport in conducting cells of Polytrichum commune. Bryologist 84, 59–64.
Rennie, E. A., and Turgeon, R. (2009). A comprehensive picture of phloem loading strategies. Proc. Natl. Acad. Sci. U.S.A. 106, 14162–14167
Rentsch, D., Schmidt, S., and Tegeder, M. (2007). Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Lett. 581, 2281–2289.
Sanders, A., Collier, R., Trethewy, A., Gould, G., Sieker, R., and Tegeder, M. (2009). AAP1 regulates import of amino acids into developing Arabidopsis embryos. Plant J. 59, 540–552.
Sauer, N., Komor, E., and Tanner, W. (1983). Regulation and characterization of two inducible amino-acid transport systems in Chlorella vulgaris. Planta 159, 404–410.
Schmidt, R., Stransky, H., and Koch, W. (2007). The amino acid permease AAP8 is important for early seed development in Arabidopsis thaliana. Planta 226, 805–813.
Schulze, W., Frommer, W. B., and Ward, J. M. (1999). Transporters for ammonium, amino acids and peptides are expressed in pitchers of the carnivorous plant Nepenthes. Plant J. 17, 637–646.
Shehawy, R. M., and Kleiner, D. (2001). “Nitrogen limitation,” in Algal Adaptation to Environmental Stresses-Physiological, Biochemical and Molecular Mechanisms, eds L. C. Rai and J. P. Gaur (Berlin: Springer), 45–64.
Shimodaira, H., and Hasegawa, M. (1999). Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16, 1114–1116.
Svennerstam, H., Ganeteg, U., Bellini, C., and Näsholm, T. (2007). Comprehensive screening of Arabidopsis mutants suggests the lysine histidine transporter 1 to be involved in plant uptake of amino acids. Plant Physiol. 143, 1853–1860.
Svennerstam, H., Ganeteg, U., and Näsholm, T. (2008). Root uptake of cationic amino acids by Arabidopsis depends on functional expression of amino acid permease. New Phytol. 180, 620–630.
Tan, Q. M., Grennan, A. K., Pelissier, H. C., Rentsch, D., and Tegeder, M. (2008). Characterization and expression of French bean amino acid transporter PvAAP1. Plant Sci. 174, 348–356.
Tegeder, M., and Rentsch, D. (2010). Uptake and partitioning of amino acids and peptides. Mol. Plant 3, 997–1011.
Tegeder, M., Rentsch, D., and Patrick, J. W. (2011). “Organic carbon and nitrogen transporters,” in Plant Plasma Membrane: Plant Cell Monographs, eds A. Murphy, W. Peer, and B. Schulz (Berlin: Springer), 331–352.
Tegeder, M., Tan, Q., Grennan, A. K., and Patrick, J. W. (2007). Amino acid transporter expression and localisation studies in pea (Pisum sativum). Funct. Plant Biol. 34, 1019–1028.
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (2007). The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882.
Trachtenberg, S., and Zamski, E. (1978). Conduction of ionic solutes and assimilates in the leptom of Polytrichum juniperinum Willd. J. Exp. Bot. 29, 719–727.
Turmel, M., Lemieux, C., Burger, G., Lang, B. F., Otis, C., Plante, I., and Gray, M. W. (1999). The complete mitochondrial DNA sequences of Nephoroselmis olivacea and Pedinomonas minor: two radically different evolutionary patterns within green algae. Plant Cell 11, 1717–1730.
Winter, D., Vinegar, B., Nahal, H., Ammar, R., Wilson, G., and Provart, N. (2007). An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2, e718. doi:10.1371/journal.pone.0000718
Wipf, D., Ludewig, U., Tegeder, M., Rentsch, D., Koch, W., and Frommer, W. B. (2002). Conservation of amino acid transporters in fungi, plants and animals. Trends Biochem. Sci. 27, 139–147.
Zhang, C. Q., Barthelson, R. A., Lambert, G. M., and Galbraith, D. W. (2008). Global characterization of cell-specific gene expression through fluorescence-activated sorting of nuclei. Plant Physiol. 147, 300–340.
Keywords: amino acid, nitrogen, transporter, AAP, LHT, membrane, evolution
Citation: Tegeder M and Ward JM (2012) Molecular evolution of plant AAP and LHT amino acid transporters. Front. Plant Sci. 3:21. doi: 10.3389/fpls.2012.00021
Received: 15 October 2011;
Accepted: 19 January 2012;
Published online: 13 February 2012.
Edited by:
Heven Sze, University of Maryland, USAReviewed by:
Daniel R. Bush, Colorado State University, USAGuillaume Pilot, Virginia Tech, USA
Torgny Näsholm, Swedish University of Agricultural Sciences, Sweden
Copyright: © 2012 Tegeder and Ward. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Mechthild Tegeder, School of Biological Sciences, Washington State University, Abelson Hall 401, Pullman, WA 99164-4236, USA. e-mail: tegeder@wsu.edu