- Institute of Genetics, Ludwig-Maximilians University Munich, Munich, Germany
Plant-specific remorin proteins reside in subdomains of plasma membranes, originally termed membrane rafts. They probably facilitate cellular signal transduction by direct interaction with signaling proteins such as receptor-like kinases and may dynamically modulate their lateral segregation within plasma membranes. Recent evidence suggests such functions of remorins during plant–microbe interactions and innate immune responses, where differential phosphorylation of some of these proteins has been described to be dependent on the perception of the microbe-associated molecular pattern (MAMP) flg22 and the presence of the NBS–LRR resistance protein RPM1. A number of specifically phosphorylated residues in their highly variable and intrinsically disordered N-terminal regions have been identified. Sequence diversity of these evolutionary distinct domains suggests that remorins may serve a wide range of biological functions. Here, we describe patterns and features of intrinsic disorder in remorin protein and discuss possible functional implications of phosphorylation within these rapidly evolving domains.
Introduction
Protein phosphorylation is the most abundant post-translational modification and a key regulatory step in eukaryotic signal transduction (reviewed in Hunter, 2007; Temporini et al., 2008; Gnad et al., 2011). It governs diverse processes such as metabolism, cell cycle, development, hormone signaling, and pathogen-perception (reviewed in Cohen, 1982; Ranjeva and Boudet, 1987; Xing et al., 2002; Cozzone, 2005; Reiland et al., 2009). In plants, most phosphorylation events occur in serine (∼85%) or threonine (∼10%) residues (Sugiyama et al., 2008; Reiland et al., 2009; Nakagami et al., 2010). Among other post-translational modifications phosphorylation of these residues usually occurs in intrinsically disordered regions (Iakoucheva et al., 2004).
Intrinsically disordered regions are flexible and extended protein segments that have no ordered secondary structure under physiological conditions. The discovery of intrinsically disordered proteins led to a revision of the structure–function paradigm, which states that a protein can only fulfill its biological function by folding into a unique and structured state. Their importance is underlined by an exponential growth in the number of intrinsically disordered proteins described in the literature within the last decades (Sickmeier et al., 2007).
Structural pre-requisites as the basis for intrinsic disorder have been described. These regions exhibit large net charges at neutral pH as a consequence of harboring many non-interacting charged groups and few hydrophobic residues. Additionally, their sequences are enriched in disorder-promoting residues (D, K, R, S, Q, P, and E) and devoid of order-promoting residues (W, Y, F, I, V, L, and T). These features concomitantly result in low compactness and globularity but high flexibility of the respective region. Proteins harboring long stretches of intrinsic disorder are frequently involved in essential processes such as transcriptional and translational activation, chromatin remodeling, and signal transduction (Dyson and Wright, 2005; Radivojac et al., 2007; Cortese et al., 2008).
A key feature of signal transduction is the low affinity/high specificity kinase–substrate interaction, where both partners can dissociate rapidly and efficiently. Hypothetically, such combination could be achieved in a coupled binding and folding mechanism consisting of disorder-to-order transitions. In these transitions, the originally disordered region is locked in the binding conformation instead of exploring different conformations free in solution. This results in a reduction of entropy in the free energy of binding, which translates into a weaker binding (low affinity) compared with interactions that involve already ordered proteins with equivalent exposed surface areas (Dyson and Wright, 2005). The specificity is determined by the extension and complementarity of the interaction surface area, which in the case of intrinsically disordered regions, is large even when the interaction domains themselves may be small (Mittag et al., 2009).
Plants, as sessile organisms, require efficient signal transduction cascades to adapt to constantly changing environmental conditions. These cascades are part of complex networks involving hundreds of kinases and thousands of kinase substrates (Stone and Walker, 1995; Dissmeyer and Schnittger, 2011). Accordingly, it has been predicted that 29% of the Arabidopsis thaliana proteome contains intrinsically disordered regions that are 50 residues or longer. Moreover, it has been estimated that around 8% of the total A. thaliana proteome is comprised of completely intrinsically disordered proteins, which mainly serve function as DNA and RNA binding proteins, stress response modulators and signaling components, among others (Dunker et al., 2000).
Based on the current literature, only few plant signaling components have been experimentally characterized with respect to their intrinsic disorder. According to the DisProt database1, chaperones, molecular assembly proteins, and molecular recognition effectors are over-represented classes of intrinsically disordered proteins in plants. Examples of chaperones are the A. thaliana ERD10 and ERD14 proteins belonging to the dehydrin family that are induced by abiotic stress (Mouillon et al., 2006; Kovacs et al., 2008). Furthermore, the N-terminal region of the A. thaliana Hy5 bZIP transcription factor involved in photomorphogenesis (Yoon et al., 2006) and the Hordeum vulgare (barley) senescence-associated NAC transcription factor (Kjaersgaard et al., 2011) are examples of intrinsically disordered proteins involved in molecular assembly. The group of intrinsically disordered molecular recognition effectors is represented by a number of A. thaliana chloroplast translocases (Richardson et al., 2009), the C-terminal region of the Alb3 membrane insertase (Falk et al., 2010) and the N-terminal domain of DELLA proteins that are involved in gibberellic acid signaling (Sun et al., 2010).
Remorin Proteins-Phosphorylated Residents of the Plasma Membrane
Recently, remorin proteins have emerged as new components of signal transduction cascades (Jarsch and Ott, 2011). Remorins are plant-specific proteins present in all land plants including ferns and mosses and constitute a multi-gene family that comprises six subgroups. All described members tightly associate to the plasma membrane in specialized compartments known as membrane rafts and/or can be enriched in detergent insoluble membranes (Raffaele et al., 2009; Lefebvre et al., 2010). In mammals and plants, these rafts have been suggested to constitute platforms for signal transduction, pathogen infection, and apoptosis, among other processes (Simons and Toomre, 2000; Jarsch and Ott, 2011; Simon-Plas et al., 2011).
Remorin proteins do not show any significant sequence similarity to other described proteins or domains (Raffaele et al., 2007). They are characterized by the presence of a conserved C-terminal region (Pfam domain Remorin_C; PF03763) that encodes a predicted coiled-coil motif and a putative membrane-anchoring motif. On the other hand, the N-terminal region is extremely diverse and, for example, in the case of members belonging to the phylogenetic group 3 even absent. The current annotation of this region (Pfam domain Remorin_N; PF03766) can thereby not be applied to the whole remorin family but is restricted to subgroup 1b. Among the few motifs that can be predicted in these N-terminal regions are putative phosphorylation sites for serine/threonine protein kinases (i.e., PKA, PIKK, GSK3, MAPK; Raffaele et al., 2007).
Stimulus-dependent phosphorylation has been shown for some remorins. The potato remorin StREM1.3 was originally discovered as a plasma membrane associated protein, which is strongly phosphorylated upon treatment with polygalacturonic acid (Farmer et al., 1989). In A. thaliana differential phosphorylation of the group 1 remorins AtREM1.2 (At3g61260) and AtREM1.3 (At2g45820) is dependent on the presence of the NBS–LRR resistance protein RPM1 (Widjaja et al., 2009) and triggered upon perception of microbe-associated molecular pattern (MAMPs; Benschop et al., 2007), respectively. The predominant detection of phosphopeptides from group 1b remorins in proteomic screens certainly reflects the high abundance of these members in plant cells. However, the identification of phosphopeptides from several other remorin proteins in a number of independent studies suggests phosphorylation to be common feature of this protein family.
Generally, phosphorylation of remorins appears to be restricted to serine and threonine residues despite putative phosphorylation of few tyrosine residues can be predicted. Examples are phosphopeptides of two A. thaliana (AtREM1.2 and AtREM1.3) and two Medicago truncatula [MtREM1.1 (MtC60319) and MtREM1.2 (MtC00278)] 1b remorins that were detected in independent studies (Benschop et al., 2007; Niittyla et al., 2007; Nuhse et al., 2007; Sugiyama et al., 2008; Whiteman et al., 2008; Li et al., 2009; Reiland et al., 2009). Interestingly, phosphorylation of a serine residue that is conserved across group 1b, was reported in all of these four proteins (S88, S66, S85, and S76 in AtREM1.2, AtREM1.3, MtREM1.1, and MtREM1.2, respectively), which suggests functional conservation in this amino acid position. An additional feature is that all sites are located in the lowly conserved N-terminal region. In case of groups with longer N-terminal regions, e.g., group 5 and group 6, several other phosphopeptides have been detected. In group 5, phosphorylation of one A. thaliana (AtREM5.1; At1g45207) and two Oryza sativa (rice) remorins (Os08g36760 and Os02g52810.1) have been reported (Benschop et al., 2007; Nuhse et al., 2007; Sugiyama et al., 2008; Nakagami et al., 2010). However, no patterns of evolutionary conservation in the phosphorylated residues were found. A similar situation is present in group 6, where phosphorylation of four A. thaliana [AtREM6.1 (At2g02170), AtREM6.2 (At1g30320), AtREM6.4 (At4g36970), and AtREM6.5 (At1g67590)] and two M. truncatula (Medtr4g115770, Medtr5g046550) remorins was described in non-conserved residues (Benschop et al., 2007; Nuhse et al., 2007; Sugiyama et al., 2008; Reiland et al., 2009; Grimsrud et al., 2010; Nakagami et al., 2010). Furthermore, a serine residue (S48) in the N-terminal region of the symbiotic remorin LjSYMREM1 (remorin group 2) of Lotus japonicus is phosphorylated by the kinase domains of the receptor proteins NFR1 and SYMRK in vitro (Toth et al., 2012).
Remorins and Intrinsic Disorder
It was described earlier that a combination of low mean hydrophobicity and relatively high net charge results in the absence of a compact structure in proteins under physiological conditions (Uversky et al., 2000). Thus, both parameters can be used to discriminate disordered from folded proteins. Investigating a subset of 84 remorin proteins distributed throughout the six phylogenetic groups revealed a common pattern that suggests a tendency toward intrinsic disorder with respect to their net charge and hydrophobicity (Figure 1A). Only 15 remorins did not fulfill the disorder criteria according to the charge-hydrophobicity phase space. However, there is no apparent clustering among members of the same subgroup. This supports an overall clear tendency toward intrinsic disorder (Figure 1A) but also highlights the diversity among remorin proteins.
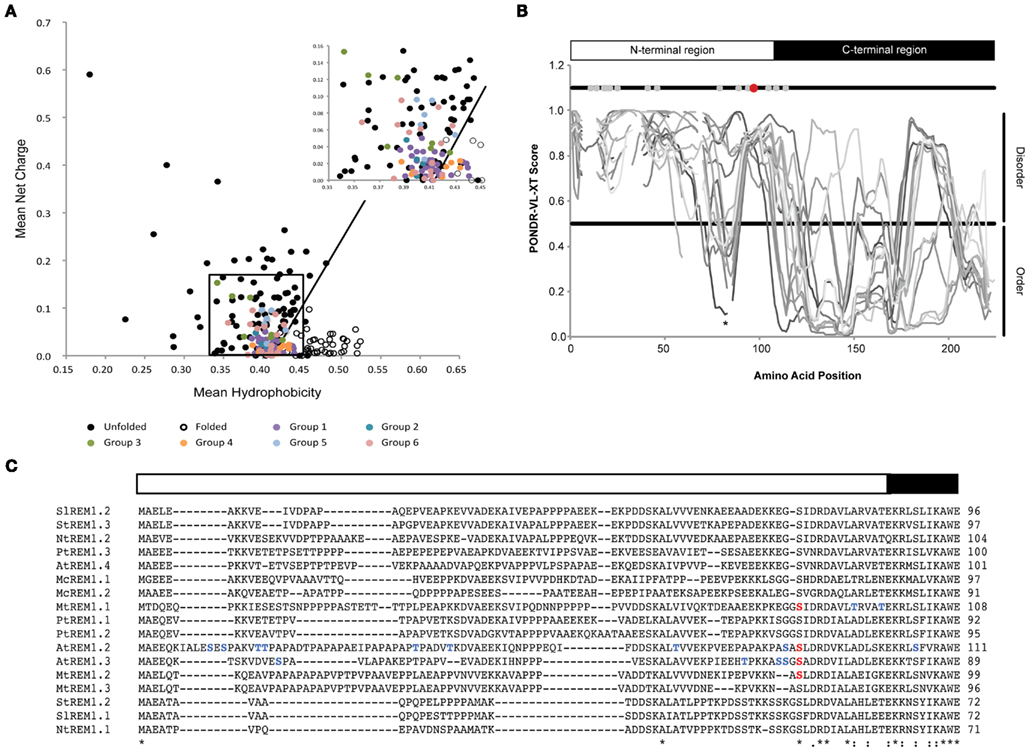
Figure 1. Intrinsic disorder in remorin proteins. (A) Charge-hydrophobicity phase space for the remorin protein family. Black circles depict intrinsically disordered proteins described in the literature (data taken partially from Uversky et al., 2000). White circles depict natively folded proteins obtained from PDB database. Colored circles depict remorin proteins belonging to the six phylogenetic groups. The black box highlights zoomed region on the right corner. The solid black line represents border between intrinsically disorder proteins and folded proteins and is described by the formula: 〈R〉 = 2.785 〈H〉 −1.151, where 〈R〉 and 〈H〉 are the mean net charge and the mean net hydrophobicity, respectively. (B) Intrinsic disorder prediction of group 1b remorin proteins. Predictions were performed using the PONDR VL-XT program on a set of 17 group 1b remorins, which belong to Arabidopsis thaliana, Mesembryanthemum crystallinum, Medicago truncatula, Nicotiana tabaccum, Populus trichocarpa, Solanum lycopersicum, and Solanum tuberosum. Gaps in the plot represent gaps in the sequence alignment. Gray and black bars depict N- and C-terminal regions, respectively. Gray dots indicate residues that have been found to the phosphorylated in AtREM1.2, AtREM1.3, MtREM1.1, and MtREM1.2. The red dot indicates the residue that was described to be phosphorylated and conserved among all group 1b remorin proteins. The asterisk marks a conserved region of order within the disordered N-terminal region. (C) Protein alignment of group 1b remorin proteins. The alignment was performed using the ClustalW algorithm on the full-length protein sequences. For simplicity only the N-terminal region and the first residues of the C-terminal region are shown. Non-conserved phosphorylation sites are depicted in blue, whereas the conserved phospho-serine residue is indicated in red. Asterisks (*) indicate positions that have a single, fully conserved residue. A colon (:) indicates conservation between amino acids with highly similar chemical properties. A period (.) indicates conservation between amino acids with weakly similar chemical properties.
As already mentioned above, most available data regarding remorin phosphorylation cover group 1b. Deeper insights into the intrinsic disorder profiles of remorins belonging this subgroup were gained by using the PONDR VL-XT algorithm2. Alignment of the PONDR VL-XT disorder prediction scores that were obtained for 17 group 1b remorin proteins distributed over seven different species showed that the N-terminal region is intrinsically disordered in all selected sequences (Figure 1B). Additionally, a segment of predicted order was located around positions 70–90 within this predicted unstructured region in 6 out of 17 group 1b remorins (Figure 1B). Conservation of this segment anticipates its functional importance. Furthermore, molecular recognition motives involved in protein–protein interactions may coincide with segments of local order within disordered regions (Oldfield et al., 2005; Mohan et al., 2006). However, on protein sequence level, remorin N-terminal regions show little conservation even within the same phylogenetic group. In contrast, the C-terminal region is conserved throughout the remorin protein family. Structurally, the latter region in group 1b remorins is mostly ordered (Figure 1B). Interestingly, the transition between disordered and ordered segments coincides well with the borders between the phylogenetically defined N- and C-terminal regions that were described previously (Raffaele et al., 2007).
An overlay of experimentally determined phosphorylation sites on the disorder prediction plot of group 1b remorins showed that they cluster in the disordered N-terminal region (Figures 1B,C). Expansion of such analyses to group 5 and 6 remorins with experimentally determined phosphoserines and phosphothreonines revealed that these residues predominantly accumulate within long stretches of predicted disorder. In contrast, phosphorylation sites predicted with the NetPhos 2.0 server3 are randomly distributed along remorin protein sequences (Figure 2). This reinforces the idea that serine and threonine phosphorylation occurs preferentially in flexible regions (Iakoucheva et al., 2004). The DISPHOS 1.3 predictor4 incorporates intrinsic disorder prediction to improve the discrimination between phosphorylation sites.
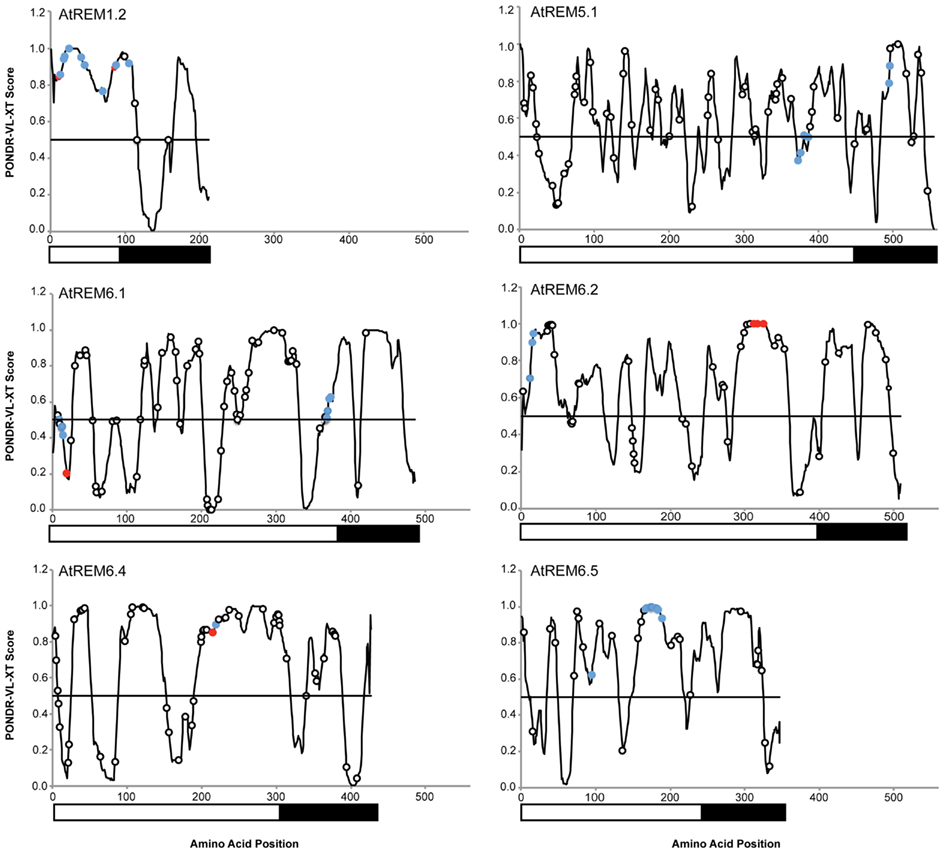
Figure 2. Phosphorylation of serine and threonine residues clusters in intrinsic disordered regions. Blue circles represent sites mapped under control (non-stimulated) conditions. Red circles represent sites mapped upon application of MAMPs. Black and white boxes under the plots indicate the N- and C-terminal regions, respectively. Disorder prediction for A. thaliana remorin proteins was performed using the PONDR VL-XT algorithm. Predicted phosphorylation sites were obtained using the NetPhos 2.0 server.
Intrinsic Disorder and Evolution of Remorin Proteins
Disordered regions exhibit different patterns with respect to the distribution of gained point mutations and generally higher rates of insertions and deletions compared to ordered proteins. This is probably due to their sequence composition and the fact that, despite high rates of sequence evolution, functionality is maintained by retaining domain flexibility under physiological conditions (Brown et al., 2011).
An interesting example is SYMREM1, a group 2 remorin protein, which has been shown to be involved in bacterial infection during root nodule symbiosis and was proposed as a novel signaling component in this mutualistic interaction (Lefebvre et al., 2010; Toth et al., 2012). Two proteins, encoded by orthologous genes, have been described in the phylogenetically closely related model legumes M. truncatula and L. japonicus. In line with their evolutionary proximity essential signaling proteins are highly conserved on amino acid level: e.g., the LRR receptor-like kinases (RLKs) NFP/NFR5 (82.8% similarity), the LysM-RLKs DMI2/SYMRK (87.6% similarity), the calcium-/calmodulin-dependent kinases DMI3/CCAMK (92.2% similarity) and the putative transcription factor NSP2 (83.4% similarity). In contrast the orthologous remorins MtSYMREM1 and LjSYMREM1 exhibit an overall similarity of only 67.1%. This is mainly due to low sequence conservation in their predicted intrinsically disordered N-terminal regions (38.3%), while their ordered C-terminal regions with a similarity of 85.3% reflect the evolutionary distance between these two legumes (Toth et al., 2012).
Conclusion and Perspective
To understand signal transduction processes and regulatory functions on a structural and mechanistic level, detailed analysis of intrinsically disordered proteins is inevitable. Remorins that have been proposed to be components of plant innate immunity signal transduction, represent prominent examples for intrinsically disordered proteins. Analysis of their intrinsically disordered regions may help to unravel their so far unknown physiological roles in the future. However, accumulating evidence suggests that their molecular functions may be as diverse as their protein sequences. The two common features of remorins, phosphorylation proclivity within their N-terminal regions and intrinsic disorder, make them ideal candidates to elicit signaling functions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We kindly thank the Collaborative Research Centre (Sonderforschungsbereich) SFB924, the Emmy Noether programme (grant OT423/2-1) both funded by the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG) and the Centre of Advanced Studies (CAS) of the Ludwig-Maximilians-Universität München (LMU) for financial support.
Footnotes
References
Benschop, J. J., Mohammed, S., O’Flaherty, M., Heck, A. J., Slijper, M., and Menke, F. L. (2007). Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell Proteomics 6, 1198–1214.
Brown, C. J., Johnson, A. K., Dunker, A. K., and Daughdrill, G. W. (2011). Evolution and disorder. Curr. Opin. Struct. Biol. 21, 441–446.
Cohen, P. (1982). The role of protein phosphorylation in neural and hormonal control of cellular activity. Nature 296, 613–620.
Cortese, M. S., Uversky, V. N., and Dunker, A. K. (2008). Intrinsic disorder in scaffold proteins: getting more from less. Prog. Biophys. Mol. Biol. 98, 85–106.
Cozzone, A. J. (2005). Role of protein phosphorylation on serine/threonine and tyrosine in the virulence of bacterial pathogens. J. Mol. Microbiol. Biotechnol. 9, 198–213.
Dunker, A. K., Obradovic, Z., Romero, P., Garner, E. C., and Brown, C. J. (2000). Intrinsic protein disorder in complete genomes. Genome. Inform. Ser. Workshop Genome Inform. 11, 161–171.
Dyson, H. J., and Wright, P. E. (2005). Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208.
Falk, S., Ravaud, S., Koch, J., and Sinning, I. (2010). The C terminus of the Alb3 membrane insertase recruits cpSRP43 to the thylakoid membrane. J. Biol. Chem. 285, 5954–5962.
Farmer, E. E., Pearce, G., and Ryan, C. A. (1989). In vitro phosphorylation of plant plasma membrane proteins in response to the proteinase inhibitor inducing factor. Proc. Natl. Acad. Sci. U.S.A. 86, 1539–1542.
Gnad, F., Gunawardena, J., and Mann, M. (2011). PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res. 39, D253–D260.
Grimsrud, P. A., Den Os, D., Wenger, C. D., Swaney, D. L., Schwartz, D., Sussman, M. R., Ane, J. M., and Coon, J. J. (2010). Large-scale phosphoprotein analysis in Medicago truncatula roots provides insight into in vivo kinase activity in legumes. Plant Physiol. 152, 19–28.
Hunter, T. (2007). The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738.
Iakoucheva, L. M., Radivojac, P., Brown, C. J., O’Connor, T. R., Sikes, J. G., Obradovic, Z., and Dunker, A. K. (2004). The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 32, 1037–1049.
Jarsch, I. K., and Ott, T. (2011). Perspectives on remorin proteins, membrane rafts, and their role during plant-microbe interactions. Mol. Plant Microbe Interact. 24, 7–12.
Kjaersgaard, T., Jensen, M. K., Christiansen, M. W., Gregersen, P., Kragelund, B. B., and Skriver, K. (2011). Senescence-associated barley NAC (NAM, ATAF1,2, CUC) transcription factor interacts with radical-induced cell death 1 through a disordered regulatory domain. J. Biol. Chem. 286, 35418–35429.
Kovacs, D., Kalmar, E., Torok, Z., and Tompa, P. (2008). Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol. 147, 381–390.
Lefebvre, B., Timmers, T., Mbengue, M., Moreau, S., Herve, C., Toth, K., Bittencourt-Silvestre, J., Klaus, D., Deslandes, L., Godiard, L., Murray, J. D., Udvardi, M. K., Raffaele, S., Mongrand, S., Cullimore, J., Gamas, P., Niebel, A., and Ott, T. (2010). A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 107, 2343–2348.
Li, H., Wong, W. S., Zhu, L., Guo, H. W., Ecker, J., and Li, N. (2009). Phosphoproteomic analysis of ethylene-regulated protein phosphorylation in etiolated seedlings of Arabidopsis mutant ein2 using two-dimensional separations coupled with a hybrid quadrupole time-of-flight mass spectrometer. Proteomics 9, 1646–1661.
Mittag, T., Kay, L. E., and Forman-Kay, J. D. (2009). Protein dynamics and conformational disorder in molecular recognition. J. Mol. Recognit. 105–116.
Mohan, A., Oldfield, C. J., Radivojac, P., Vacic, V., Cortese, M. S., Dunker, A. K., and Uversky, V. N. (2006). Analysis of molecular recognition features (MoRFs). J. Mol. Biol. 362, 1043–1059.
Mouillon, J. M., Gustafsson, P., and Harryson, P. (2006). Structural investigation of disordered stress proteins. Comparison of full-length dehydrins with isolated peptides of their conserved segments. Plant Physiol. 141, 638–650.
Nakagami, H., Sugiyama, N., Mochida, K., Daudi, A., Yoshida, Y., Toyoda, T., Tomita, M., Ishihama, Y., and Shirasu, K. (2010). Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 153, 1161–1174.
Niittyla, T., Fuglsang, A. T., Palmgren, M. G., Frommer, W. B., and Schulze, W. X. (2007). Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Mol. Cell Proteomics 6, 1711–1726.
Nuhse, T. S., Bottrill, A. R., Jones, A. M. E., and Peck, S. C. (2007). Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 51, 931–940.
Oldfield, C. J., Cheng, Y., Cortese, M. S., Romero, P., Uversky, V. N., and Dunker, A. K. (2005). Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry 44, 12454–12470.
Radivojac, P., Iakoucheva, L. M., Oldfield, C. J., Obradovic, Z., Uversky, V. N., and Dunker, A. K. (2007). Intrinsic disorder and functional proteomics. Biophys. J. 92, 1439–1456.
Raffaele, S., Bayer, E., Lafarge, D., Cluzet, S., German Retana, S., Boubekeur, T., Leborgne-Castel, N., Carde, J. P., Lherminier, J., Noirot, E., Satiat-Jeunemaitre, B., Laroche-Traineau, J., Moreau, P., Ott, T., Maule, A. J., Reymond, P., Simon-Plas, F., Farmer, E. E., Bessoule, J. J., and Mongrand, S. (2009). Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21, 1541–1555.
Raffaele, S., Mongrand, S., Gamas, P., Niebel, A., and Ott, T. (2007). Genome-wide annotation of remorins, a plant-specific protein family: evolutionary and functional perspectives. Plant Physiol. 145, 593–600.
Ranjeva, R., and Boudet, A. M. (1987). Phosphorylation of proteins in plants – regulatory effects and potential involvement in stimulus-response coupling. Annu. Rev. Plant Physiol. 38, 73–93.
Reiland, S., Messerli, G., Baerenfaller, K., Gerrits, B., Endler, A., Grossmann, J., Gruissem, W., and Baginsky, S. (2009). Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 150, 889–903.
Richardson, L. G., Jelokhani-Niaraki, M., and Smith, M. D. (2009). The acidic domains of the Toc159 chloroplast preprotein receptor family are intrinsically disordered protein domains. BMC Biochem. 10, 35. doi:10.1186/1471-2091-10-35
Sickmeier, M., Hamilton, J. A., Legall, T., Vacic, V., Cortese, M. S., Tantos, A., Szabo, B., Tompa, P., Chen, J., Uversky, V. N., Obradovic, Z., and Dunker, A. K. (2007). DisProt: the database of disordered proteins. Nucleic Acids Res. 35, D786–D793.
Simon-Plas, F., Perraki, A., Bayer, E., Gerbeau-Pissot, P., and Mongrand, S. (2011). An update on plant membrane rafts. Curr. Opin. Plant Biol. 14, 642–649.
Simons, K., and Toomre, D. (2000). Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1, 31–39.
Stone, J. M., and Walker, J. C. (1995). Plant protein kinase families and signal transduction. Plant Physiol. 108, 451–457.
Sugiyama, N., Nakagami, H., Mochida, K., Daudi, A., Tomita, M., Shirasu, K., and Ishihama, Y. (2008). Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol. Syst. Biol. 4, 193.
Sun, X., Jones, W. T., Harvey, D., Edwards, P. J., Pascal, S. M., Kirk, C., Considine, T., Sheerin, D. J., Rakonjac, J., Oldfield, C. J., Xue, B., Dunker, A. K., and Uversky, V. N. (2010). N-terminal domains of DELLA proteins are intrinsically unstructured in the absence of interaction with GID1/gibberellic acid receptors. J. Biol. Chem. 285, 11557–11571.
Temporini, C., Calleri, E., Massolini, G., and Caccialanza, G. (2008). Integrated analytical strategies for the study of phosphorylation and glycosylation in proteins. Mass Spectrom. Rev. 27, 207–236.
Toth, K., Stratil, T. F., Madsen, E. B., Ye, J., Popp, C., Antolin-Llovera, M., Grossmann, C., Jensen, O. N., Schussler, A., Parniske, M., and Ott, T. (2012). Functional domain analysis of the remorin protein LjSYMREM1 in Lotus japonicus. PLoS ONE 7, e30817. doi:10.1371/journal.pone.0030817
Uversky, V. N., Gillespie, J. R., and Fink, A. L. (2000). Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 41, 415–427.
Whiteman, S. A., Serazetdinova, L., Jones, A. M., Sanders, D., Rathjen, J., Peck, S. C., and Maathuis, F. J. (2008). Identification of novel proteins and phosphorylation sites in a tonoplast enriched membrane fraction of Arabidopsis thaliana. Proteomics 8, 3536–3547.
Widjaja, I., Naumann, K., Roth, U., Wolf, N., Mackey, D., Dangl, J. L., Scheel, D., and Lee, J. (2009). Combining subproteome enrichment and rubisco depletion enables identification of low abundance proteins differentially regulated during plant defense. Proteomics 9, 138–147.
Xing, T., Ouellet, T., and Miki, B. L. (2002). Towards genomic and proteomic studies of protein phosphorylation in plant-pathogen interactions. Trends Plant Sci. 7, 224–230.
Keywords: remorin, phosphorylation, intrinsic disorder, signaling
Citation: Marín M and Ott T (2012) Phosphorylation of intrinsically disordered regions in remorin proteins. Front. Plant Sci. 3:86. doi: 10.3389/fpls.2012.00086
Received: 27 February 2012; Paper pending published: 28 March 2012;
Accepted: 18 April 2012; Published online: 07 May 2012.
Edited by:
Gabriel Schaaf, ZMBP Tübingen, GermanyReviewed by:
Dominique Loqué, Lawrence Berkeley National Laboratory, USAThomas Sebastian Nuhse, The University of Manchester, UK
Copyright: © 2012 Marín and Ott. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Thomas Ott, Institute of Genetics, Ludwig-Maximilians University Munich, Grosshaderner Strasse 2-4, Munich, 82152 Martinsried, Germany. e-mail: thomas.ott@biologie.uni-muenchen.de
 Macarena Marín
Macarena Marín