- 1 School of Life Sciences, Warwick University, Coventry, UK
- 2 Research Centre for Plant RNA Signalling, College of Life and Environmental Sciences, Hangzhou Normal University, Hangzhou, China
FLOWERING LOCUS T (FT) protein is known to be part of the mobile flowering inducing “florigen” signal in plants, but it may not be acting alone. This article reviews the data that FT mRNA can also move systemically throughout the plant and into the shoot apical meristem (SAM) independently of the FT protein. There is a promotion of flowering when increased levels of virally expressed FT mRNA are present together with endogenously produced FT protein in inducing conditions, even if the additional FT mRNA is non-translatable and thus not increasing the overall levels of FT protein. A specific sequence, or “zip code” of the FT mRNA is required for systemic movement and this sequence binds a specific protein(s) in plant extracts. This raises the possibility the FT mRNA may be moving systemically through the plant and into the SAM as an RNA–protein complex, whether FT protein is also a component of this mobile complex remains to be determined.
In Arabidopsis the FLOWERING LOCUS T (FT) gene plays a key role in the induction of flowering (Abe et al., 2005; Wigge et al., 2005). The Arabidopsis FT protein (Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007) and its orthologs from rice (Tamaki et al., 2007), cucurbit (Lin et al., 2007), and tomato (Lifschitz et al., 2006), have been shown to be a component of a systemic flowering signal that moves through the phloem from the leaves to the shoot apical meristem (SAM) to induce the switch from vegetative growth to flower formation. However more recently it has been demonstrated that, in addition to FT protein, FT RNA is also able to move systemically through the plant from leaves to the SAM (Li et al., 2009, 2011) and here we review the evidence that systemic movement of FT mRNA may have a role to play in floral induction.
To analyze the movement of FT RNA an RNA movement assay was developed based upon a virus expression system. In this assay a PVX virus which had its coat protein gene deleted from its viral genome, and which because of the lack of coat protein was therefore was unable to move from cell to cell, was used to express Arabidopsis FT RNA fused downstream of a GFP coding sequence. As a control the same viral vector was used that expressed GFP RNA alone. It was found that both the viral RNA and GFP RNA from this control virus vector was only detectable in those leaves of the tobacco plants that had been inoculated with the virus construct, and not in any other leaves of the plants. This is because the lack of viral coat protein prevented intercellular and systemic movement of the viral RNA and associated GFP RNA. In plants inoculated with the virus vector expressing the Arabidopsis FT RNA, however, both virus RNA and GFP-FT RNA were detected in inoculated leaves, and in systemic young leaves of the tobacco plant which had grown after the viral inoculation had taken place (and thus the presence of viral derived RNA in these samples could not be due to accidental cross-contamination during inoculation; Li et al., 2009). In addition to the full genomic RNA, the PVX virus expresses two sub-genomic RNAs from internal promoters in its genome, which because the GFP-FT sequence replaced the coat protein gene at the 3′ end of the viral genome, both contain the GFP-FT RNA sequence and thus were mobile and detectable in systemic leaves. Thus the addition of the FT RNA sequence to the GFP RNA sequence in the viral RNA genome/sub-genomes enabled it to overcome the movement deficiency caused by the absence of the coat protein gene.
Expression of FT RNA from a PVX viral vector with a functional coat protein resulted in the production of Arabidopsis FT protein throughout the plant, and the presence of this FT protein was able to induce flowering in the short-day (SD) requiring Maryland Mammoth tobacco plants even in non-inducing long day (LD) photoperiods. To show that the movement of the FT RNA was a property of the RNA itself and not due to the phloem-mobile FT protein that was being produced from the viral construct, another viral construct was tested that expressed a mutated FT gene. The mutation converted the ATG start codon to a stop codon thus preventing translation of FT protein from this mutant FT (mFT) RNA. Plants inoculated with virus constructs expressing mFT were not induced to flower in non-inducing LD photoperiods and remained vegetative the same as mock inoculated control plants. Interestingly, movement of the mFT associated RNAs was unaffected and they were still detectable in systemic leaves even without FT protein production (Li et al., 2009). To show that this movement ability was not due to any associated PVX viral sequences, a different viral vector (Turnip Crinkle Virus, TCV) was used to express GFP, GFP-FT, or GFP-mFT in Arabidopsis. Confirming previous results, only viral RNAs associated with the FT and mFT sequences were able to move systemically though the Arabidopsis plants from inoculated leaves to non-inoculated newly formed systemic leaves. Furthermore, FT mRNA was shown to be able to move in the complete absence of any viral RNA. This was demonstrated in a transient assay where tobacco leaves were infiltrated with Agrobacterium that were expressing GFP-FT or GFP-mFT fusion constructs. Both RNAs were subsequently detected in both infiltrated, and non-infiltrated systemic leaves showing that the FT and mFT RNA could move through the plant independently of any viral RNA sequences (Li et al., 2009).
If FT mRNA movement is involved in the induction of flowering then the FT mRNA must be able to move into the SAM. Evidence that this is the case was obtained from in situ immune-detection and RNA silencing assays which showed that viruses expressing FT or mFT were able to enter the SAM, unlike viruses expressing GFP alone which were prevented from entering the SAM by the normal meristem exclusion process (Li et al., 2011). Thus FT mRNA is able to overcome the selective meristem exclusion system that plants have evolved to prevent entry of viral and other endogenous RNAs into the SAM. As both mFT RNA as well as FT RNA are able to direct entry into the SAM it shows that the FT protein is not involved in overcoming meristem exclusion. Indeed, whilst the mechanism by which FT and mFT RNA overcome meristem exclusion is not yet understood, it may be that this function of the FT mRNA is necessary in order for the FT protein to enter the SAM to induce flowering and thus that FT mRNA may be a necessary component of the florigen signal. This is a hypothesis that deserves further examination and we will consider the arguments both for and against in more detail.
In rice, both the Hd3a protein and low levels of Hd3a mRNA have been detected in the SAM, although in Arabidopsis only FT protein and no FT mRNA was detected in the SAM (Corbesier et al., 2007; Tamaki et al., 2007), this difference may be explained by differences in the sensitivity of the methods of detection used in these analyses. There have been elegant experiments designed to try to uncouple the effects of FT mRNA and protein on floral induction (Jaeger and Wigge, 2007; Mathieu et al., 2007; Notaguchi et al., 2008). In these cases movement of FT protein, but not FT mRNA, was inhibited by either attachment of a nuclear localization signal and/or fusion of multiple YFP proteins. As these prevented movement of the FT protein out of phloem companion cells into the SAM then flowering was inhibited demonstrating that movement of the FT protein into the SAM is required to induce flowering. In those cases where movement of FT protein out of the phloem is completely prevented then any effect of FT mRNA in facilitating entry into the SAM would not be observed. In other experiments the levels of FT mRNA in different parts of the plant where reduced through the expression of artificial microRNAs against FT mRNA (amiR-FT). Expression of amiR-FT throughout the plant, or in phloem companion cells, inhibited flowering presumably because it prevented production of FT protein, whereas expression of amiR-FT in just the SAM did not inhibit flowering indicating that FT mRNA does not play any role within the SAM itself (Mathieu et al., 2007). However, this is not inconsistent with the hypothesis that FT mRNA may be involved in facilitating movement of the FT protein into the SAM as FT mRNA does not have to be acting inside the SAM to have this effect.
Evidence that FT mRNA itself has an effect in promoting flowering is difficult to obtain because active FT protein needs to be present in order for flowering to occur and it is hard to distinguish what contribution to the induction of flowering is made by the FT mRNA alone. However it has been successfully demonstrated that FT mRNA does have a promotive effect on floral induction using tobacco plants that are induced to flower. These plants were Maryland Mammoth tobacco plants that were grown in SD inducing photoperiods, and were thus producing endogenous FT protein which would move to the SAM to induce flowering. Some of these plants were inoculated with a virus expressing the mFT RNA (hence no further FT protein was produced), and this promoted earlier flowering over the mock inoculated controls which flowered at the normal time in SD (Li et al., 2011). This demonstrates that FT mRNA (or mFT RNA) itself is able to enhance the induction of flowering by the FT protein which was being produced endogenously in the plant in SD conditions.
What enables FT mRNA to move and have its effect on flowering time? It is apparent that some feature of the FT mRNA not only enables this RNA to move, and confers this on associated RNA molecules (e.g., FT-GFP RNA moves but GFP RNA alone does not), but it may also enhance the movement (or activity) of the FT protein. A deletion analysis of the FT mRNA showed that a short 102 nucleotide sequence at the 5′ end of the FT mRNA was all that was necessary to direct systemic RNA movement, all FT RNA sequences containing this 102 nucleotide domain were able to move within the plant whereas FT RNAs lacking this domain were unable to move (Li et al., 2009). Further deletion analysis of this 102 nucleotide domain indicated that there may be more than one sequence in this domain involved in promoting FT RNA movement (Li, unpublished results), these cis-acting sequences may be “zip code” sequences that Lucas et al. (2001) proposed may interact with zip code binding proteins to form RNA–protein complexes that are able to be transported through plasmodesmata. It has been well established that such zip codes in the 3′ UTR are important for intracellular trafficking of mRNA molecules such as β-actin (St Johnston, 2005), it is now apparent that similar sequence motifs are also involved in the intercellular movement of mRNA molecules. Viruses have specific sequences that promote viral RNA movement (Wang and Ding, 2010), and this has also been shown to be the case for endogenous plant mRNAs such as GIBBERELLIC ACID INSENSITIVE (GAI) mRNA which has specific motifs that enable it to move systemically throughout the Arabidopsis plant (Huang and Yu, 2009). In pumpkin (Cucurbita maxima), the GAI mRNA forms an RNA–protein complex with about 17 proteins present in the phloem sap one of which is RBP50, a phloem-mobile polypyrimidine tract binding (PTB) protein that selectively binds the GAI mRNA (Ham et al., 2009). It is not known if FT mRNA binds to PTB proteins, however both the FT mRNA and the 102 nucleotide movement sequence were found to bind specifically to a protein(s) of around 20 kDa in Arabidopsis and 24 kDa in tobacco in protein extracts made from vegetative tobacco plants (Li et al., unpublished). No binding was detected to proteins extracted from leaves of flowering Arabidopsis or tobacco plants indicating that the expression of these FT mRNA-binding proteins might be developmentally regulated, being switched off once floral induction has been initiated. This could potentially add another layer ofcontrol in the regulation of flowering.
Notaguchi et al. (2008) argued that the FT mRNA sequence is not important for the long distance promotion of flowering by the FT protein. This is based on experiments where the sequence of the FT mRNA but not the protein was altered by synonymous substitutions in 171 of the 175 codons, and replacement of the 5′ and 3′ untranslated regions, to create a synonymous FT (synFT) mRNA which shared only 63% identity to the ORFs in the original FT sequence. All these changes did not affect the long distance and graft-transmissible ability of the FT protein to induce flowering. As the specific protein binding sites in the FT mRNA sequence (specifically within the 102 nucleotide domain) have not yet been identified, it is not yet known which nucleotides are essential for binding and which ones can be varied without affecting binding. What has been demonstrated is that a non-translatable FT mRNA (mFT) is capable of enhancing the promotion of flowering by an endogenous FT protein, that a 102 nucleotide sequence at the 5′ end of the FT mRNA binds a specific protein(s), and that deletion of this 102 nucleotide sequence prevents movement of the FT mRNA. This raises the possibility that protein binding to the 102 nucleotide region of the FT mRNA may be necessary for FT mRNA movement and the resulting enhancement of floral induction by the FT protein (Figure 1).
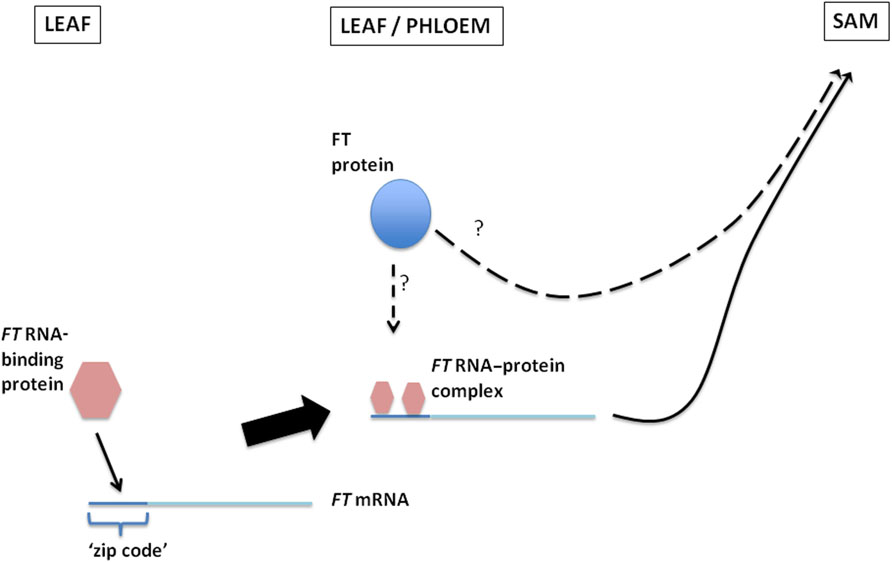
FIGURE 1. A proposed model illustrating the binding of an FT RNA-binding protein(s) to “zip codes” located within 102 nt of the 5′end of the FT mRNA to form an RNA–protein complex which then moves from the leaf through the phloem and into the SAM. FT protein may either associate, and be transported, with this RNA–protein complex, or it may move independently of the FT mRNA with its movement through the plasmodesmata and into the SAM being facilitated by the FT RNA–protein complex.
The systemic movement of RNA molecules is tightly regulated, not all RNA molecules can move (e.g., GFP mRNA) although many do as thousands of mRNA transcripts have been detected in the phloem of Arabidopsis (Deeken et al., 2008). Of those viral and cellular-derived RNA molecules that are present in the phloem only a few are able to enter into the meristem, these include the mRNAs of FT, GAI, CmNACP, and LeT6 a tomato KNOX gene (Ruiz-Medrano et al., 1999; Kim et al., 2001; Haywood et al., 2005; Li et al., 2011). Viral RNAs are prevented from entering the meristem by a selective surveillance mechanism (Foster et al., 2002) which involves RNA silencing as well as other mechanisms such as selective transport through plasmodesmata mediated by proteins with trafficking signal domains (Kim et al., 2005; Qu et al., 2005; Schwach et al., 2005). It is known that FT mRNA is able to enter the meristem. It remains to be determined whether the protein(s) that bind to the first 102 nucleotides of the FT mRNA have trafficking signals that enable it to do this and whether they also facilitate the entry of FT protein, perhaps as part of a RNA–protein complex, into the meristem to promote flowering at the same time.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., Ichinoki, H., Notaguchi, M., Goto, K., and Araki, T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056.
Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., Giakountis, A., Farrona, S., Gissot, L., Turnbull, C., and Coupland, G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033.
Deeken, R., Ache, P., Kajahn, I., Klinkenberg, J., Bringmann, G., and Hedrich, R. (2008). Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. Plant J. 55, 746–759.
Foster, T. M., Lough, T. J., Emerson, S. J., Lee, R. H., Bowman, J. L., Forster, R. L., and Lucas, W. J. (2002). A surveillance system regulates selective entry of RNA into the shoot apex. Plant Cell 14, 1497–1508.
Ham, B. K., Brandom, J. L., Xoconostle-Cázares, B., Ringgold, V., Lough, T. J., and Lucas, W. J. (2009). A polypyrimidine tract binding protein, pumpkin RBP50, forms the basis of a phloem-mobile ribonucleoprotein complex. Plant Cell 21, 197–215.
Haywood, V., Yu, T. S., Huang, N. C., and Lucas, W. J. (2005). Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J. 42, 49–68.
Huang, N. C., and Yu, T. S. (2009). The sequences of Arabidopsis GA-INSENSITIVE RNA constitute the motifs that are necessary and sufficient for RNA long-distance trafficking. Plant J. 59, 921–929.
Jaeger, K. E., and Wigge, P. A. (2007). FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 17, 1050–1054.
Kim, J. Y., Rim, Y., Wang, J., and Jackson, D. (2005). A novel cell-to-cell trafficking assay indicates that the KNOX homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev. 19, 788–793.
Kim, M., Canio, W., Kessler, S., and Sinha, N. (2001). Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293, 287–289.
Li, C., Gu, M., Shi, N., Zhang, H., Ynag, X., Osman, T., Liu, Y., Wang, H., Vatish, M., Jackson, S., and Hong, Y. (2011). Mobile FT mRNA contributes to the systemic florigen signaling in floral induction. Sci. Rep. 1, 73.
Li, C., Zhang, K., Zeng, X., Jackson, S., Zhou, Y., and Hong, Y. (2009). A cis element within flowering locus T mRNA determines its mobility and facilitates trafficking of heterologous viral RNA. J. Virol. 83, 3540–3548.
Lifschitz, E., Eviatar, T., Rozman, A., Shalit, A., Goldshmidt, A., Amsellem, Z., Alvarez, J. P., and Eshed, Y. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. U.S.A. 103, 6398–6403.
Lin, M., Belanger, H., Lee, Y., VarkonyiGasic, E., Taoka, K.-C., Miura, E., XoconostleCazares, B., Gendler, K., Jorgensen, R., Phinney, B., Lough, T., and Lucas, W. (2007). FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell 19, 1488–1506.
Lucas, W. J., Yoo, B. C., and Kragler, F. (2001). RNA as a long-distance information macromolecule in plants. Nat. Rev. Mol. Cell Biol. 2, 849–857.
Mathieu, J., Warthmann, N., Kuttner, F., and Schmid, M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17, 1055–1060.
Notaguchi, M., Abe, M., Kimura, T., Daimon, Y., Kobayashi, T., Yamaguchi, A., Tomita, Y., Dohi, K., Mori, M., and Araki, T. (2008). Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol. 49, 1645–1658.
Qu, F., Ye, X., Hou, G., Sato, S., Clemente, T. E., and Morris, T. J. (2005). RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J. Virol. 79, 15209–15217.
Ruiz-Medrano, R., Xoconostle-Cázares, B., and Lucas, W. J. (1999). Phloem long-distance transport of CmNACP mRNA: implications for supracellular regulation in plants. Development 126, 4405–4419.
Schwach, F., Vaistij, F. E., Jones, L., and Baulcombe, D. C. (2005). An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138, 1842–1852.
St Johnston, D. (2005). Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 6, 363–375.
Tamaki, S., Matsuo, S., Wong, H. L., Yokoi, S., and Shimamoto, K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036.
Wang, Y., and Ding, B. (2010). Viroids: small probes for exploring the vast universe of RNA trafficking in plants. J. Integr. Plant Biol. 52, 28–39.
Keywords: flowering locus T, FT, mRNA, flowering, tobacco
Citation: Jackson SD and Hong Y (2012) Systemic movement of FT mRNA and a possible role in floral induction. Front. Plant Sci. 3:127. doi: 10.3389/fpls.2012.00127
Received: 10 February 2012; Accepted: 28 May 2012;
Published online: 12 June 2012.
Edited by:
Steven Huber, United States Department of Agriculture – Agricultural Research Service, USAReviewed by:
Uener Kolukisaoglu, University of Tübingen, GermanyFabio Fornara, University of Milan, Italy
Copyright: © 2012 Jackson and Hong. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Stephen D. Jackson, School of Life Sciences, Warwick University, Gibbet Hill, Coventry CV4 7AL, UK. e-mail: stephen.jackson@warwick.ac.uk
