- 1Institute of Environmental Biotechnology, Graz University of Technology, Graz, Austria
- 2Austrian Centre of Industrial Biotechnology, Graz, Austria
Stenotrophomonas rhizophila has great potential for applications in biotechnology and biological control due to its ability to both promote plant growth and protect roots against biotic and a-biotic stresses, yet little is known about the mode of interactions in the root-environment system. We studied mechanisms associated with osmotic stress using transcriptomic and microscopic approaches. In response to salt or root extracts, the transcriptome of S. rhizophila DSM14405T changed drastically. We found a notably similar response for several functional gene groups responsible for general stress protection, energy production, and cell motility. However, unique changes in the transcriptome were also observed: the negative regulation of flagella-coding genes together with the up-regulation of the genes responsible for biofilm formation and alginate biosynthesis were identified as a single mechanism of S. rhizophila DSM14405T against salt shock. However, production and excretion of glucosylglycerol (GG) were found as a remarkable mechanism for the stress protection of this Stenotrophomonas strain. For S. rhizophila treated with root exudates, the shift from the planktonic lifestyle to a sessile one was measured as expressed in the down-regulation of flagellar-driven motility. These findings fit well with the observed positive regulation of host colonization genes and microscopic images that show different colonization patterns of oilseed rape roots. Spermidine, described as a plant growth regulator, was also newly identified as a protector against stress. Overall, we identified mechanisms of Stenotrophomonas to protect roots against osmotic stress in the environment. In addition to both the changes in life style and energy metabolism, phytohormons, and osmoprotectants were also found to play a key role in stress protection.
Introduction
Crop cultivation in salinated soils is one of the major challenges facing agriculture today. Salinated areas are increasing world-wide and plants growing under saline or water-imbalance stress are more vulnerable to diseases caused by soil-borne pathogens (FAO, 2005). Biocontrol using salt-tolerant, plant growth-promoting rhizobacteria (PGPR) to protect plant roots against high salinity and pathogens offers sustainable solutions for plant protection, and Stenotrophomonas rhizophila is a model bacterium for a rhizosphere- and phylloplane-competent, salt-tolerant PGPR (Ryan et al., 2009; Berg et al., 2010, 2013). While the species S. maltophilia has become important as a nosocomial human pathogen, no pathogenic potential for humans has ever been observed in the related species S. rhizophila (Wolf et al., 2002). Moreover, both species can be easily distinguished by the production of the osmoprotective substance glucosylglycerol (GG) (only present in S. rhizophila) and the occurrence of specific multidrug-efflux pumps (only present in S. maltophilia) (Ribbeck-Busch et al., 2005).
Plant growth promotion by S. rhizophila strain DSM14405T (syn. strain e-p10) was observed under greenhouse conditions (Schmidt et al., 2012) and in the highly salinated soils of Uzbekistan at levels up to 180% (Egamberdieva et al., 2011). Use of classical physiological and biochemical methods unveiled the mechanisms of plant growth promotion and biocontrol against soil-borne pathogens (Berg and Ballin, 1994; Kobayashi et al., 1995; Jacobi et al., 1996; Dunne et al., 2000; Suckstorff and Berg, 2003) as well as the production of high amounts of osmolytes trehalose and GG in response to salt stress (Roder et al., 2005). Next generation sequencing techniques have allowed for new possibilities to study plant-microbe interaction. For example, genome sequencing has given new insight into the genetic sources that provide beneficial plant-associated bacteria with traits such as plant growth promotion, protection against phytopathogens, and osmoprotection. In general, the described mode of action could be confirmed due to the presence of genes possibly responsible in the genome of S. rhizophila DSM14405T (Berg et al., 2013). For example, S. rhizophila possesses genes responsible for the synthesis and transport of osmoprotective molecules out of the cell. In addition, it contains a number of genes involved in the biocontrol of soil-borne pathogens and important genes that aid in the competition for nutrients and niches as well. Additionally, S. rhizophila is equipped with several genes which may play a role in root colonization, such as those that encode the O-antigen, capsule polysaccharide biosynthesis pathways, hemagglutinin, and outer membrane adhesion proteins. However, despite this knowledge, there is still no evidence that these genes are involved in successful root-microbe interactions under salinated conditions. In addition to the high salinity, the role of root exudates for this interaction was pointed out in other studies (González-Pasayo and Martínez-Romero, 2000; rev. in Bais et al., 2006). The ability of cells to respond appropriately to changing environmental conditions can be investigated using a transcriptomic approach. This technique offers a new and powerful tool to evaluate these hypothetical mechanisms in situ, as shown already by van de Mortel et al. (2012) for the Pseudomonas-Arabidopsis and by López-Guerrero et al. (2012) for the Rhizobium-Phaseolus interaction.
The objective of our study was to investigate the response to changing environmental conditions associated with osmotic stress (1) salt stress and (2) root exudates to understand stress protection against changing osmotic conditions of roots by the endophytic bacterium S. rhizophila DSM14405T in more detail. We hypothesized that there is a general response to changing osmolarities, but also a specific answer to each other of the two parameters which are important for colonizing the root system of plants.
Materials and Methods
Treatment with Oilseed Rape Exudates
Root exudates were collected from oilseed rape cultivar Californium (Kwizda, Austria) and grown for 14 days in gnotobiotic systems of 50 ml of sterilized vermiculite packaged in pots and covered with lids (Metro, Austria). Prior to sowing the seeds, about 50 ml of tap water was amended with 1/10 [v/v] of minimal medium (Gamborgs B5 basal salt mixture; Duchefa), and the seeds were surface-sterilized in sodium hypochlorite (10% wt/wt) for 10 min and washed successively with sterile water under sterile conditions. No seeds were sown in the control system. Plant and control systems were arranged in a replicate randomized block design and maintained at 20°C under 16-h light and 8-h dark conditions. After 14 days, plants were removed and the root exudates and liquid from the control system were collected in sterile bags and squeezed. To corroborate sterility, both root material and exudates were plated on nutrient agar. Root exudates were centrifuged (10 min, 5000 × g), and the supernatant was collected, filter-sterilized (first 0.45 μm, second 0.22 μm filter, Millipore), and stored at −20°C in the dark until use. S. rhizophila DSM14405T was cultivated under agitation in 40 ml CAA (per liter: 5.0 g casamino acids, 1.54 g K2HPO4·3H2O, 0.25 g MgSO4·7H2O) and supplemented with 10 ml of the root exudates and the control liquid, respectively, at 30°C for 48 h. Cells were harvested using centrifugation at 2500 × g for 1 min for RNA extraction.
Salt Shock
S. rhizophila DSM14405T was cultivated in 50 ml CAA under agitation at 30°C for 13 h (per liter: 5.0 g casamino acids, 1.54 g K2HPO4·3H2O, 0.25 g MgSO4·7H2O) until an optical density of OD600 0.8 was reached. A final salt (NaCl) concentration of 3% in the medium was reached by using a sterile concentrated sodium chloride stock solution (0.3 g l−1). After 2.7 h cultivation in the medium containing 3% salt, the S. rhizophila DSM14405T culture (OD600 = 0.9) was used for RNA extraction. Two independent replicates were performed as described above.
RNA Extraction and Transcriptomic Analyses
RNA was extracted using the RNAprotect® Bacteria Reagent (Qiagen, Hilden, Germany). Total rRNA was removed and mRNA was enriched using the MICROBExpress kit, according to the manufacturer's protocol (Invitrogen, Carlsbad, USA). The mRNA was sequenced using LGC Genomics (Berlin, Germany), and data collection was performed using MicroDiscovery (Berlin, Germany). The data used for assessing the changes in gene transcription correspond to normalized values for the number of reads that uniquely mapped to each CDS. Transcription fold change for each CDS was assessed by dividing the corresponding value from the cells that were either treated with root exudates or exposed to salt shock by those from the control group. Of the total genes either up or down-regulated, only those showing fold changes greater than or equal to 1.5 and less than or equal to 0.6 were considered significantly impacted.
Germination Pouch Colonization Assay
A batch of 200 oilseed rape seeds were surface-sterilized with 40 ml of 3% NaOCl for 1 min and subsequently washed twice with 40 ml of water for 1 min each time. Surface-sterilized seeds were inoculated with S. rhizophila DSM14405T by incubating in a 2 ml cell suspension containing 107 CFU ml−1. The control included seeds treated with 0.85% NaCl. Twelve seeds per treatment were placed into 2 (6 seeds per pouch) sterile Cyg™ germination pouches (Mega International, West St. Paul, MN, USA) wetted with 10 ml of sterilized deionized water or 1.25% NaCl solution. Germination pouches were then placed in sterile, aseptically sealed containers and placed in a growing chamber for 9 days with controlled day and night settings (12 h of light at 25°C and 12 h of dark at 20°C). After 9 days of growth, roots of 3 seedlings were combined for determination of cell counts resulting in 4 replicates per individual treatment. All root material was cut and transferred to Whirlpak® bags (Carl Roth, Karlsruhe, Germany) containing 2 ml of 0.85% NaCl solution. The roots in the bags were then crushed using a pestle to form a homogenous suspension, which was subsequently serially diluted and drop-streaked onto LB Petri dishes. The plates were then incubated at 30°C for 24 h.
Fluorescent in situ Hybridization (FISH)
To study the oilseed rape colonization ability of S. rhizophila DSM14405T using confocal microscopy, the oilseed rape roots grown in seed germination pouches were fixed with 4% paraformaldehyde/phosphate buffered saline (PBS) (3:1 vol/vol). The control group contained roots without bacterial treatment. The fixed samples were then stored in PBS/ 96% ethanol (1:1) at −20°C. The FISH probes were purchased from genXpress® (Wiener Neudorf, Austria), and the in-tube FISH was performed as described by Cardinale et al. (2008). The FISH probes used for the hybridization step were labeled with the fluorescent dye Cy3 and included EUB338 (Amman et al., 1990), EUB338 II, and EUB338 III (Daims et al., 1999), all directing eubacteria. An equimolar ratio of the FISH probes was used for the hybridization step to detect S. rhizophila DSM14405T. In this step, 30% formamide was added to the samples which were then subsequently incubated in a water bath (43°C) for 90 min. After hybridization, the samples were washed at 44°C for 15 min. Microscopy and image capturing were performed using a Leica TCS SPE confocal microscope (Leica Microsystems, Wetzlar, Germany) with the Leica ACS APO 63X OIL CS objective (NA: 1.30). A z-step of 0.4–0.8 μm was applied to acquire confocal stacks.
Results
Transcriptional Response of S. rhizophila DSM14405T to Salt Stress
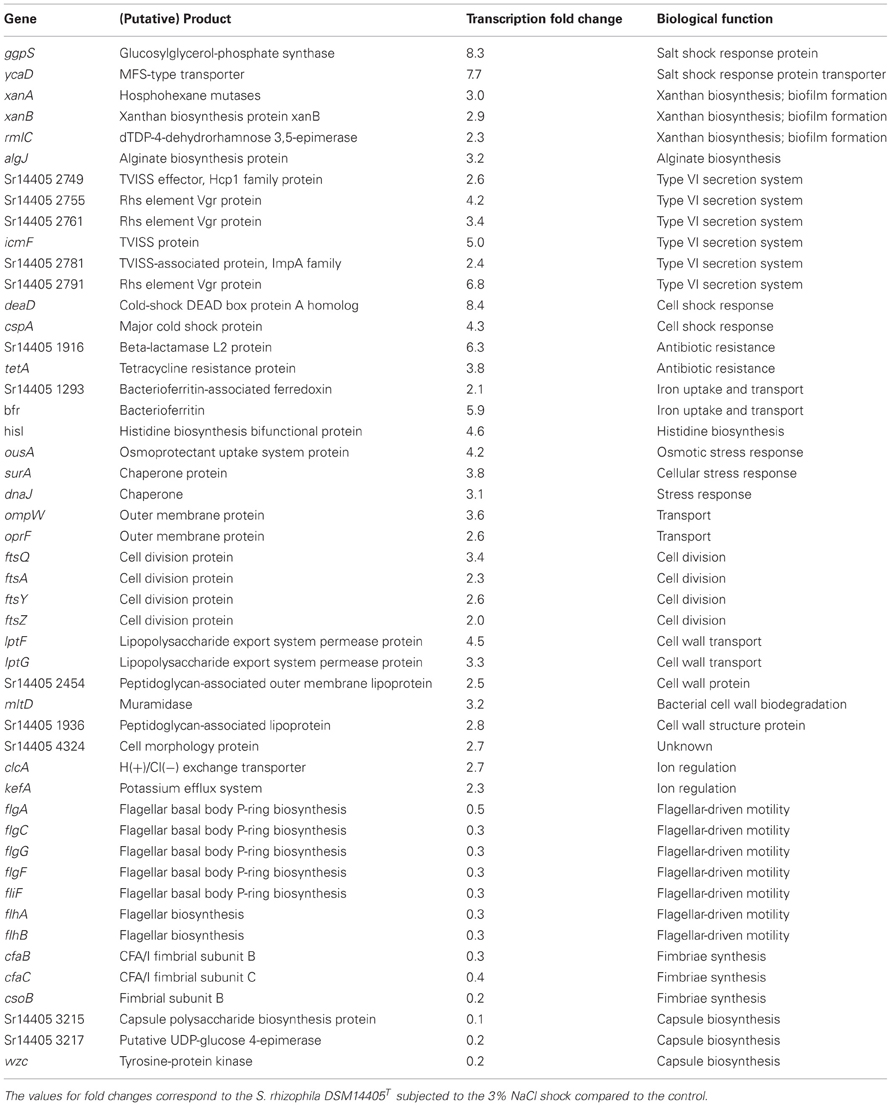
Under salt stress of 3% NaCl, a total number of 912 and 1521 genes of S. rhizophila DSM14405T were significantly up and down-regulated, respectively. The impact of salt shock on the transcription of S. rhizophila DSM14405T with respect to various functional gene groups is shown in Figure 1. The majority of functional groups were strongly affected, such as up-regulated genes involved in translation, synthesis of the cell wall, outer or cytoplasm membrane, nucleotide and amino acid transport and metabolism, and the production and conversion of energy. In contrast, genes involved in cell motility, secretion, intracellular trafficking, defense mechanisms, and the transport and metabolism of carbohydrates and inorganic ions are down-regulated. Moreover, genes responsible for lipid metabolism and hypothetical genes are somewhat ambiguously affected by salt stress as some are up while others down-regulated.
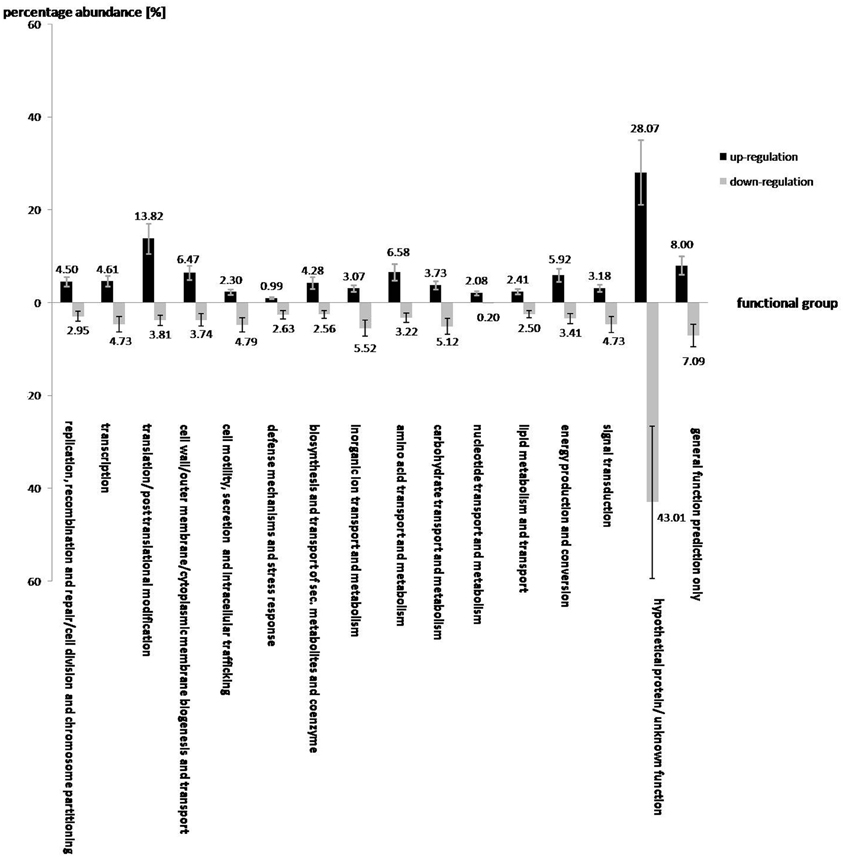
Figure 1. The effect of salt shock on the gene expression of various functional gene groups in S. rhizophila DSM14405T. A total of 912 and 1521 genes were significantly up and down-regulated, respectively. The impact of salt stress on most functional gene groups is clearly pronounced, as a given functional group shows either an increase or decrease in the transcription of genes belonging to that group. Genes involved in translation, synthesis of the cell wall, outer or cytoplasm membrane, nucleotide and amino acid transport and metabolism, energy production and conversion are up-regulated. In contrast, genes involved in cell motility, secretion, and intracellular trafficking, defense mechanisms, and transport and metabolism of carbohydrates and inorganic ions are down-regulated. Genes involved in lipid metabolism, and the hypothetical genes are rather ambiguously affected by salt stress, as some of these are up and others down-regulated. The values above each column correspond to the percentage abundance of the corresponding functional group relative to the total count of the up and down-regulated genes. The transcription fold change for each CDS corresponds to the ratio calculated for S. rhizophila under salt shock compared with the control. Data are presented as the mean value of two independent replicates. The error bar shown on each functional group corresponds to the mean value of errors for all genes belonging to that functional group.
Of the genes that are significantly impacted by salt stress in S. rhizophila DSM14405T (Table 1, Tables S1A, S1B), a number of those responsible for general and specific stress responses are up-regulated. While surA and dnaJ code for general stress chaperones, ggpS and ycaD build a well-known salt stress response mechanism in S. rhizophila DSM14405T through the synthesis of the osmolyte GG and show a fold change of 8.3, and 7.7, respectively (Hagemann et al., 2008). Moreover, two genes responsible for cold shock, deaD and cspA, are also strongly up-regulated under salt stress. In addition, the transcription of ousA that codes for an osmotic stress protein is positively affected as well. Cellular ion exchange mechanisms and some iron uptake genes are also up-regulated in S. rhizophila DSM14405T as the result of salt stress.
Although unable to synthesize xanthan, S. rhizophila DSM14405T possesses some of the up-regulated xanthan-coding genes including xanA, xanB, and rmlAC. These genes are involved in biofilm formation in addition to their role in xanthan biosynthesis (Huang et al., 2006). Likewise, the alginate coding gene algJ shows a fold change of 3.2 as a result of salt shock. Alginate is an exopolysaccharide involved in the development and architecture of biofilms that protect bacteria from antibiotics and other harmful environmental factors (Monday and Schiller, 1996; Stapper et al., 2004). Furthermore, specific secretion and transport systems such as those that code for the type VI secretion system (TVISS) are strongly up-regulated in S. rhizophila DSM14405T under salt shock, however, it should be noted that closely related plant-associated Stenotrophomonas strains such as S. maltophilia R551-3 lack the TVISS. Moreover, genes involved in the conversion and transport of substances through the cell wall and those responsible for cell division are also up-regulated.
Genes responsible for flagellar apparatus and fimbriae-biosynthesis genes are comparatively down-regulated in S. rhizophila DSM14405T under salt shock. Similarly, salt shock also negatively impacted the predicted capsule biosynthesis genes.
Transcriptional Response of S. rhizophila DSM14405T to Root Exudates
A total of 763 and 246 genes were significantly up and down-regulated, respectively, as a result of the addition of oilseed-rape root exudates (Figure 2). In general, the effect of root exudates on the functional groups is indeterminate, as some genes of a particular group are up-regulated while others are transcribed in lesser numbers. However, some functional groups are equally affected by plant root exudates, as the transcription of almost all corresponding gene sequences is either up or down-regulated. For instance, as shown in Figure 2, root exudates have only a positive effect on the transcription of genes responsible for amino acid, nucleotide, and carbohydrate transport and metabolism, as well as biogenesis of cell membranes, transport of substances through the cell, and the genes responsible for the transport of secondary metabolites and coenzymes. Conversely, genes involved in the secretion, transport, and metabolism of inorganic ions as well as in cell motility are mainly down regulated in response to root exudate stress.
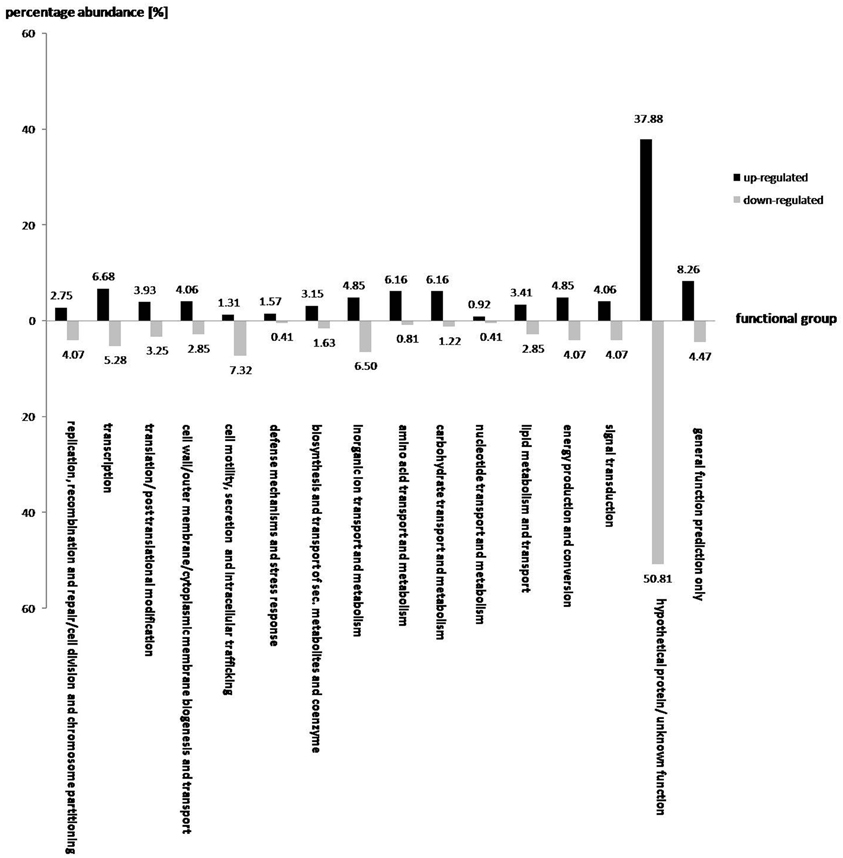
Figure 2. The effect of oilseed rape seedling exudates on gene expression of various functional gene groups in S. rhizophila DSM14405T. A total of 763 and 246 genes were significantly up and down-regulated, respectively. While some functional groups are both positively and negatively regulated by root exudates, others show a clear and pronounced alteration, as the majority of the corresponding genes are either up or down-regulated. For example, genes responsible for amino acid, nucleotide, and carbohydrate transport and metabolism, and those involved in cell wall, outer-membrane or cytoplasmic membrane biogenesis and transport as well as genes responsible for the transport of secondary metabolites and coenzymes are mainly up-regulated. In contrast, genes involved in cell motility and secretion, and those responsible for the transport and metabolism of inorganic ions are mainly down-regulated. The value above each column corresponds to percentage abundance of the corresponding functional group in the total count of the up or down-regulated genes.
Of those genes with a significant transcription fold change discussed above, some code for products with a known physiological function and are presented in Table 2. The complete list of S. rhizophila DSM14405T genes with a significant transcription fold change is presented in Tables S2A, S2B. Cell wall breakdown and cell adherence are early and crucial steps in host-plant colonization. As presented in Table 2, the treatment of S. rhizophila DSM14405T cells with oilseed rape seedling exudates resulted in enhanced expression of cbg-1 and xynB that code for beta-glucosidase and xylanase B, respectively, and are involved in cell wall breakdown. Furthermore, both these genes are conserved among plant-associated Stenotrophomonas strains as they are present in both S. rhizophila DSM14405T and the plant-benefiting S. maltophilia R551-3, but absent from the human-pathogenic S. maltophilia K279a. In addition, Sr14405 2818, which is also up-regulated by 2.4 folds, codes for an adhesin protein and is homologous to the haemagglutinin-like protein coding gene from the human-pathogenic S. maltophilia K279a (Table 2). Other up-regulated genes include two adjacent genes, mdtI and mdtJ that both code for spermidine export proteins. Spermidine is a plant growth regulator and has been recently shown to strongly promote the growth of arugula plants (Al-Whaibi et al., 2012). Moreover, several genes that code for multidrug resistance pumps, efflux transporters, heavy metal transport systems, and resistance against antibiotics are positively affected by seedling exudates.
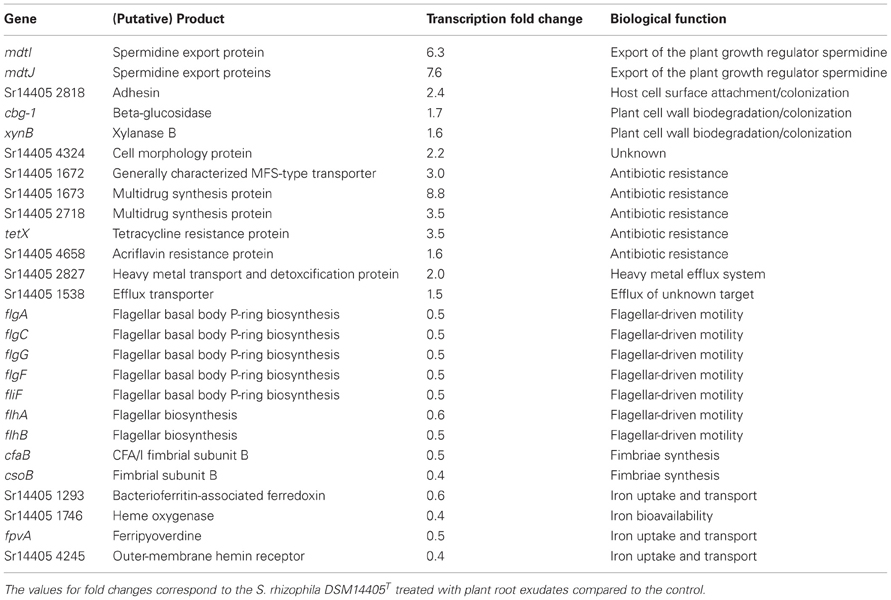
Table 2. Selected S. rhizophila DSM14405T genes with known biological roles impacted by plant root exudates.
S. rhizophila DSM14405T contains two flagella-encoding gene blocks that are almost entirely negatively affected by the addition of oilseed rape seedling exudates. The complete list of the flagellar apparatus-coding genes that are down-regulated is not confined to those noted in Table 2, and is presented in Tables S2A, S2B. Likewise, the expression of the genes responsible for fimbriae-driven cell motility, such as cfaB and csoB is negatively impacted by seedling exudates. Moreover, genes involved in the uptake, transport, and bioavailability of iron are also down-regulated.
Similarities in the Transcriptional Response of S. rhizophila DSM14405T to Salt and Root Exudates
In response to both oilseed rape root exudates and salt shock, S. rhizophila DSM14405T copes with osmotic stress in a surprisingly similar way through the alteration of gene expression. Numerous functional gene groups are up-regulated in response to osmotic stress factors and include those involved in energy production, as well as those involved in the synthesis and transport of cell wall, outer membrane, and cytoplasmic membrane, and those responsible for the metabolism and transport of amino acids, nucelotides, and secondary metabolites (Figure 3). Conversely, genes responsible for cell motility, secretion, intracellular trafficking, and the transport and metabolism of inorganic ions are down-regulated under both salt stress and treatment with root exudates.
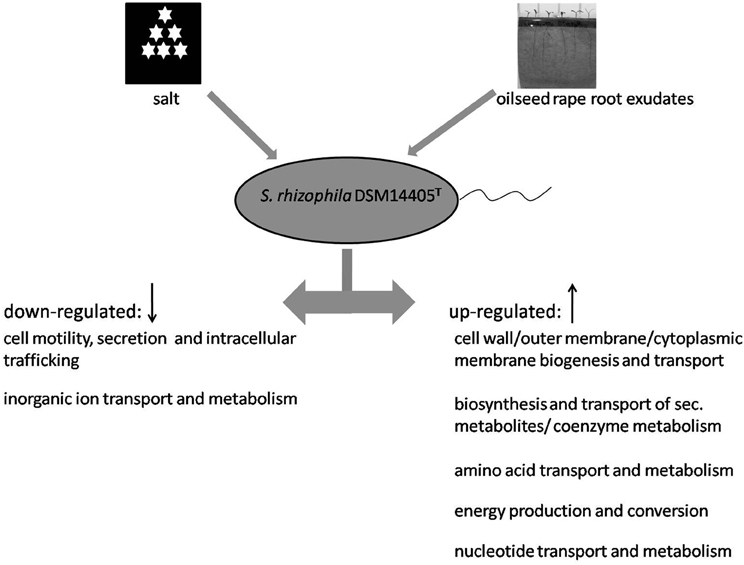
Figure 3. Model showing the response of S. rhizophila DSM14405T to osmotic stress: salt shock and root exudates. Functional gene groups shared in the response to oilseed rape root exudates and salt shock are presented. Several functional gene groups are up-regulated as a result of both oilseed rape root exudates and salt shock including those responsible for the synthesis and transport of cell wall, outer membrane, and cytoplasmic membrane, the metabolism and transport of amino acids, nucleotide, and secondary metabolites, and energy production. In contrast, genes responsible for cell motility, secretion and intracellular trafficking, and the transport and metabolism of inorganic ions are down-regulated.
Colonization Patterns of S. rhizophila DSM14405T on Roots Under Stress
S. rhizophila DSM14405T intensely colonizes oilseed rape plants, as revealed by the cell count of log10 9.47 CFU g−1 root fresh weight (±0.08) for seeds treated with deionized water. The treatment of seeds with 1.25% NaCl, however, decreased the colonization ability by nearly half resulting in a cell count of log10 9.09 CFU g−1 root fresh weight (±0.18). Furthermore, microscopic images captured using FISH combined with confocal laser scanning microscopy (CLSM) also revealed a significant decrease in the colonization of oilseed rape roots treated with 1.25% NaCl (Figure 4).
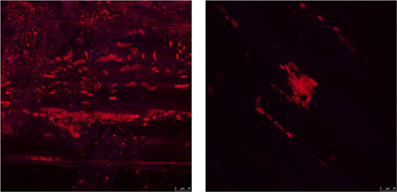
Figure 4. The impact of salt stress on the capability of S. rhizophila DSM14405T to colonize the oilseed rape rhizosphere visualized using FISH-CLSM. S. rhizophila DSM14405T intensely colonizes the oilseed rape rhizosphere (left) while the treatment of seeds with 1.25% NaCl (right) severely decreases the colonization capability. An equimolar ratio of the FISH probes EUB338, EUB338 II, and EUB338 III labeled with the fluorescent dye Cy3 was used in the hybridization step. Microscopic images were captured using a Leica TCS SPE confocal microscope. The Leica ACS APO 63X OIL CS objective (NA: 1.30) was used to acquire confocal stacks by applying a z-step of 0.4–0.8 μm.
Discussion
We studied the response of S. rhizophila DSM14405T to osmotic changes in the form of plant root exudates and salt shock at both the physiological and molecular level. Even though we found a notable similarity in how the cell copes with these stressors, the individual responses included a great deal of specificity at the gene level thus. The response of S. rhizophila DSM14405T to both oilseed rape root exudates and salt corresponds with several functional gene groups including those responsible for the synthesis and transport of cell wall, outer membrane, and cytoplasmic membrane, the metabolism and transport of amino acids, nucleotide, and secondary metabolites, energy production, cell motility, secretion and intracellular trafficking, and the transport and metabolism of inorganic ions. For S. rhizophila treated with root exudates, however, the shift from the planktonic lifestyle to a sessile one as expressed in the down-regulation of flagellar-driven motility is targeted to colonize the plant host, and is well in accordance with the observed positive regulation of host colonization genes. In addition to the changes in behavior and lifestyle of the bacterium, several bioactive substances were identified as key factors in stress protection. The first among them is the plant growth regulator, spermidine. Although this substance is known to strongly promote growth, this is the first evidence to show its involvement in stress protection of roots. The second group includes osmoprotective substances which were both produced and excreted in high volumes as described earlier (Roder et al., 2005).
Spermidine is a well-known plant growth regulator and has been revealed to play a critical role in plant embryo development (Imai et al., 2004). Moreover, it has been recently shown to strongly promote the growth of arugula plants (Al-Whaibi et al., 2012). In addition, spermidine affects biofilm formation in various bacterial species via multiple pathways that involve both transport and signaling networks (McGinnis et al., 2009). As a result, enhanced biofilm formation or possible plant growth regulation resulting from the up-regulation of S. rhizophila spermidine export genes would well-serve the lifestyle shift that ultimately leads to efficient colonization of the plant host in the presence of oilseed rape exudates. Spermidine was found to prolong the life span of several eukaryotic model organisms including yeasts, nematodes, flies, and plants as well as significantly reduce age-related oxidative protein damage in mice which could indicate a potential universal anti-aging drug for eukaryotes (Imai et al., 2004; Eisenberg et al., 2009).
GG and trehalose are well-studied general osmoprotective substances that protect cells from high salt concentrations (Ferjani et al., 2003; Hincha and Hagemann, 2004). While both species produce trehalose, GG is synthesized exclusively in S. rhizophila thus distinguishing itself from the pathogenic S. maltophilia (Ribbeck-Busch et al., 2005; Roder et al., 2005). In S. rhizophila DSM14405T, ggpS and ycaD are both strongly up-regulated under 3% salt and are essential for the synthesis and transport of GG. This finding corresponds completely with both the general role of GG as a cell protector and previous findings that the amount of GG excreted into the medium increases substantially in comparison with intracellular GG content resulting from a shift of lower (less than 2%) to higher salt concentrations (Roder et al., 2005). Thus, GG production is the specific mechanism of S. rhizophila DSM14405T to cope with salt stress.
TVISS genes represent a novel key virulence system used by many important pathogenic bacteria in eukaryotic host infection (Bingle et al., 2008; Pieper et al., 2009) and are intensely up-regulated under salt shock. In addition, plant growth promotion increased up to 180% in the highly salinated soils of Uzbekistan in the presence of S. rhizophila DSM14405T (Egamberdieva et al., 2011). Similarly, Schmidt et al. (2012) reported that this plant growth promotion effect was more pronounced in soil than under gnotobiotic conditions suggesting it is due to the control of diseases and deleterious microorganisms. Together with the absence of TVISS genes from the other known plant-beneficial Stenotrophomonas strains with no plant growth promoting effect under saline conditions, these findings imply that the salt-stimulated S. rhizophila DSM14405T TVISS is indirectly harnessed to promote plant growth by eliminating harmful and deleterious microorganisms in soil.
The treatment of S. rhizophila DSM14405T with oilseed rape exudates resulted in the down-regulation of iron uptake and transport genes. This change is possibly due to the fact that once treated with oilseed rape exudates, S. rhizophila is provided with biologically available iron bound with plant siderophores resulting in less demand for the synthesis of bacterial siderophores to bind and uptake biologically unavailable iron ions present in the medium. Moreover, treatment with root exudates resulted in the up-regulation of several multidrug resistance pumps thus demonstrating that the role of the multidrug pumps is not confined to the export of antibiotics out of the cell, but also includes a more general function with the transport of other substances once in a hyperosmotic environment.
Several questions still remain, such as the reason for positive regulation of several genes responsible for iron uptake and transport, as well as the reason for cell division under salt shock or the role of other remaining genes that are significantly up or down-regulated by osmotic stress factors. However, this work has shed light on the so far unknown mode of action of S. rhizophila DSM14405T to a-biotic changes by unveiling the mechanisms that are harnessed to establish in highly salinated plant root ecosystems.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the Austrian Science Foundation FWF (P 20542-B16) by a grant to Gabriele Berg. The genome sequence was funded by a project in the Austrian Centre of Industrial Biotechnology, which has been supported by the Austrian BMWFJ, BMVIT, SFG, Standortagentur Tirol, and ZIT through the Austrian FFG-COMET-Funding Program.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/Functional_Plant_Ecology/10.3389/fpls.2013.00141/abstract
Table S1A | Significantly up-regulated genes in S. rhizophila DSM14405T under salt shock.
Table S1B | Significantly down-regulated genes in S. rhizophila DSM14405T under salt shock.
Table S2A | Significantly up-regulated genes in S. rhizophila DSM14405T under treatment with root exudates.
Table S2B | Significantly down-regulated genes in S. rhizophila DSM14405T under treatment with root exudates.
References
Al-Whaibi, M. H., Siddiqui, M. H., Al-Munqadhi, B. M. A., Sakran, A. M., Ali, H. M., and Basalah, M. O. (2012). Influence of plant growth regulators on growth performance and photosynthetic pigments status of Eruca sativa Mill. J. Med. Plants Res. 6, 1948–1954.
Amman, R. I., Binder, B. J., Olson, R. J., Chisholm, S. W., Devereux, R., and Stahl, D. A. (1990). Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56, 1919–1925.
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., and Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266.
Berg, G., Alavi, M., Schmidt, C. S., Egamberdieva, D., Kamilova, F., and Lugtenberg, B. (2013). “Biocontrol and osmoprotection for plants under saline conditions,” in Molecular Microbial Ecology of the Rhizosphere, ed F. J. de Bruijn (John Wiley and Sons, Inc.).
Berg, G., and Ballin, G. (1994). Bacterial antagonists to Verticillium dahliae. J. Phytopathol. 141, 99–110.
Berg, G., Egamberdieva, D., Lugtenberg, B., and Hagemann, M. (2010). “Symbiotic plant-microbe interactions: stress protection, plant growth promotion and biocontrol by Stenotrophomonas,” in Symbiosis and Stress, eds J. M. G. Seckbach and M. Grube (Dordrecht; Heidelberg; London; New York: Springer), 445–460.
Bingle, L. E. H., Bailey, C. M., and Pallen, M. J. (2008). Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 11, 3–8.
Cardinale, M., De Castro, J. V. Jr. Müller, H., Berg, G., and Grube, M. (2008). In situ analysis of the bacterial community associated with the reindeer lichen Cladonia arbuscula reveals predominance of Alphaproteobacteria. FEMS Microbiol. Ecol. 66, 1–9.
Daims, H., Brühl, A., Amann, R., Schleifer, K. H., and Wagner, M. (1999). The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22, 434–444.
Dunne, C., Moenne-Loccoz, Y., de Bruijn, F. J., and O'Gara, F. (2000). Overproduction of an inducible extracellular serine protease improves biological control of Pythium ultimum by Stenotrophomonas maltophilia strain W81. Microbiology 146, 2069–2078.
Egamberdieva, D., Kucharova, Z., Davranov, K., Berg, G., Makarova, N., Azarova, T., et al. (2011). Bacteria able to control foot and root rot and to promote growth of cucumber in salinated soils. Biol. Fertil. Soils 47, 197–205.
Eisenberg, T., Knauer, H., Schauer, A., Büttner, S., and Ruckenstuhl, C. (2009). Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 11, 1305–1314.
FAO, Rome (Italy), Land, Water Development Div.; International Programme for Technology and Research in Irrigation and Drainage, Rome (Italy). Secretariat; Centre Virtuel de l'Eau Agricole et ses Usages, Rome (Italy). Centre d'Information sur l'Eau Agricole et ses Usage. (2005). Management of Irrigation-Induced Salt-Affected Soils. Rome: CISEAU/FAO/IPTRID.
Ferjani, A., Mustardy, L., Sulpice, R., Marin, K., Suzuki, I., Hagemann, M., et al. (2003). Glucosylglycerol, a compatible solute, sustains cell division under salt stress. Plant Physiol. 131, 1628–1637.
González-Pasayo, R., and Martínez-Romero, E. (2000). Multiresistance genes of Rhizobium etli CFN42. Mol. Plant Microbe Interact. 13, 572–577.
Hagemann, M., Ribbeck-Busch, K., Klähn, S., Hasse, D., Steinbruch, R., and Berg, G. (2008). The plant-associated bacterium Stenotrophomonas rhizophila expresses a new enzyme for the synthesis of the compatible solute glucosylglycerol. J. Bacteriol. 190, 5898–5906.
Hincha, D. K., and Hagemann, M. (2004). Stabilization of model membranes during drying by compatible solutes involved in the stress tolerance of plants and microorganisms. Biochem. J. 383, 277–283.
Huang, T. P., Somers, E. B., and Wong, A. C. (2006). Differential biofilm formation and motility associated with lipopolysaccharide/exopolysaccharide-coupled biosynthetic genes in Stenotrophomonas maltophilia. J. Bacteriol. 188, 3116–3120.
Imai, A., Matsuyama, T., Hanzawa, Y., Akiyama, T., Tamaoki, M., and Saji, H. (2004). Spermidine synthase genes are essential for survival of Arabidopsis. Plant Physiol. 135, 1565–1573.
Jacobi, M., Kaiser, D., Berg, G., Jung, G., Winkelmann, G., and Bahl, H. (1996). Maltophilin – a new antifungal compound produced by Stenotrophomomas maltophilia R3089. J. Antib. 49, 1101–1104.
Kobayashi, D. Y., Gugliemoni, M., and Clarke, B. B. (1995). Isolation of chitinolytic bacteria Xanthomonas maltophilia and Serratia marcescens as biological control agents for summer patch disease of turf grass. Soil Biol. Biochem. 27, 1479–1487.
López-Guerrero, M. G., Ormeño-Orrillo, E., Acosta, J. L., Mendoza-Vargas, A., Rogel, M. A., Ramírez, M. A., et al. (2012). Rhizobial extrachromosomal replicon variability, stability and expression in natural niches. Plasmid 68, 149–158.
McGinnis, M. W., Parker, Z. M., Walter, N. E., Rutkovsky, A. C., Cartaya-Marin, C., and Karatan, E. (2009). Spermidine regulates Vibrio cholerae biofilm formation via transport and signaling pathways. FEMS Microbiol. Lett. 299, 166–174.
Monday, S. R., and Schiller, N. L. (1996). Alginate synthesis in Pseudomonas aeruginosa: the role of algL (alginate lyase) and algX. J. Bacteriol. 178, 625–632.
Pieper, R., Huang, S. T., Robinson, J. M., Clark, D. J., Alami, H., Parmar, P. P., et al. (2009). Temperature and growth phase influence the outer-membrane proteome and the expression of a type VI secretion system in Yersinia pestis. Microbiology 155, 498–512.
Ribbeck-Busch, K., Roder, A., Hasse, D., de Boer, W., Martínez, J. L., Hagemann, M., et al. (2005). A molecular biological protocol to distinguish potentially human-pathogenic strains of Stenotrophomonas maltophilia from non-pathogenic S. rhizophila strains. Environ. Microbiol. 7, 1853–1858.
Roder, A., Hoffmann, E., Hagemann, M., and Berg, G. (2005). Synthesis of the compatible solutes glucosylglycerol and trehalose by salt-stressed cells of Stenotrophomonas strains. FEMS Microbiol. Lett. 243, 219–226.
Ryan, R. P., Monchy, S., Cardinale, M., Taghavi, S., Crossman, L., Avison, M. B., et al. (2009). Versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Microbiol. Rev. 7, 514–525.
Schmidt, C. S., Alavi, M., Cardinale, M., Müller, H., and Berg, G. (2012). Stenotrophomonas rhizophila DSM14405T promotes plant growth probably by altering fungal communities in the rhizosphere. Biol. Fertil. Soils 48, 947–960.
Stapper, A. P., Narasimhan, G., Ohman, D. E., Barakat, J., Hentzer, M., Molin, S., et al. (2004). Alginate production affects Pseudomonas aeruginosa biofilm development and architecture, but is not essential for biofilm formation. J. Med. Microbiol. 53, 679–690.
Suckstorff, I., and Berg, G. (2003). Evidence for dose-dependent effects on plant growth by Stenotrophomonas strains from different origins. J. Appl. Microbiol. 95, 656–663.
van de Mortel, J. E., de Vos, R. C., Dekkers, E., Pineda, A., Guillod, L., Bouwmeester, K., et al. (2012). Metabolic and transcriptomic changes induced in Arabidopsis by the rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol. 160, 2173–8218.
Keywords: plant-microbe interaction, oilseed rape, PGPR, SPA, transcriptomics, root exudates, FISH–CLSM
Citation: Alavi P, Starcher MR, Zachow C, Müller H and Berg G (2013) Root-microbe systems: the effect and mode of interaction of Stress Protecting Agent (SPA) Stenotrophomonas rhizophila DSM14405T. Front. Plant Sci. 4:141. doi: 10.3389/fpls.2013.00141
Received: 28 March 2013; Paper pending published: 12 April 2013;
Accepted: 25 April 2013; Published online: 14 May 2013.
Edited by:
Boris Rewald, University of Natural Resources and Life Sciences (BOKU), AustriaReviewed by:
Robert P. Ryan, University College Cork, IrelandEsperanza Martinez-Romero, Universidad Nacional Autonoma de Mexico, Mexico
Copyright © 2013 Alavi, Starcher, Zachow, Müller and Berg. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Gabriele Berg, Institute of Environmental Biotechnology, Graz University of Technology, Petersgasse 12, 8010 Graz, Austria. e-mail: gabriele.berg@tugraz.at
 Peyman Alavi1
Peyman Alavi1