- 1Transport Biology Section, Department of Plant and Environmental Sciences, University of Copenhagen, Copenhagen, Denmark
- 2Center of Excellence for Membrane Pumps and Disease, PUMPKIN, Aarhus, Denmark
Genetically encoded fluorescent biosensors have long proven to be excellent tools for quantitative live imaging, but sensor applications in plants have been lacking behind those in mammalian systems with respect to the variety of sensors and tissue types used. How can this be improved, and what can be expected for the use of genetically encoded fluorescent biosensors in plants in the future? In this review, we present a table of successful physiological experiments in plant tissue using fluorescent biosensors, and draw some conclusions about the specific challenges plant cell biologists are faced with and some of the ways they have been overcome so far.
Introduction
Genetically encoded fluorescent biosensors are increasingly used as the preferred method to visualize and analyse ion fluxes, signaling components, and metabolites, covering an expanding palette of cellular processes. While fluorescent proteins as such are mainly used for localization and expression studies, genetically encoded fluorescent biosensors in addition allow real time studies of cell metabolism with a similar high spatial and temporal resolution. Cell-specific promoters allow biosensor expression in the target cell of choice in contrast to chemical probes that are inherently dependent on efficient delivery into the cells.
The huge interest and progress in the field is reflected in a large number of recent reviews on fluorescent proteins and genetically encoded sensors, e.g., (Fehr et al., 2004; Lalonde et al., 2005; VanEngelenburg and Palmer, 2008; Frommer et al., 2009; Chudakov et al., 2010; Okumoto, 2010; Mehta and Zhang, 2011; Miyawaki, 2011; Newman et al., 2011; Palmer et al., 2011; Okumoto et al., 2012). Several reviews describe plant specific uses, e.g., (Dixit et al., 2006; Frommer et al., 2009; Swanson et al., 2011; Choi et al., 2012; Ehrhardt and Frommer, 2012; Okumoto, 2012; Okumoto et al., 2012). In addition, http://biosensor.dpb.carnegiescience.edu/ provides a database of selected available biosensors.
In the present context, the term genetically encoded fluorescent biosensors refers to fluorescent proteins coupled with a sensing mechanism that causes a change in fluorescence intensity upon ligand binding. Most sensors can be grouped within two major types of fluorescent biosensors: (1) single fluorescent protein sensors, which can carry the sensing mechanism within the fluorescent protein, such as e.g., pHluorins, or where sensing is coupled to a ligand binding domain. Other options using single fluorescent proteins include protein-protein interactions reported by fluorescent protein reconstitution (biFC) or detection of protein translocation. One notable exception of specific plant relevance is the DII-Venus auxin sensor, where degradation of the fluorescent protein is utilized as sensing mechanism. (2) FRET-based sensors, where ligand binding causes a conformational change of the sensor leading to a change in FRET ratio between two fluorescent proteins, usually CFP/YFP variants. Within these groups many sensor platform designs are possible, which are described in detail elsewhere, see e.g., (Okumoto et al., 2012).
There is general consensus that the field is expanding and far from saturated with respect to sensor targets, the quality and variety of the fluorescent proteins, spatiotemporal resolution, compartmentation, and to imaging techniques. This paper discusses the perspectives for using genetically encoded fluorescent biosensors in plants, summarizing the specific challenges plant cell biologists are faced with and the ways they have been overcome so far. Although, due to space restrictions, this review focuses on fluorescent biosensors, the aspects discussed apply to luminescent biosensors as well. What are the expectations for fluorescent biosensors in plants in the future? Can they help in assigning a function to the many orphan receptor-like kinases or create complete flux maps of metabolite and ions in Arabidopsis and other model organisms as suggested by (Okumoto et al., 2012)? We will point to some challenges that need to be addressed, if biosensors are to be used more widely.
Why are Genetically Encoded Fluorescent Biosensors Less Used in Plants than in Mammalian Cells?
The variety of genetically encoded fluorescent sensors is explored primarily in mammalian systems. Although these sensors offer highly attractive advantages for plant cell imaging, reports on physiological measurements in plants have been comparatively few, which might indicate that using these tools for live physiological experiments in plants is not trivial.
Table 1 below is a compilation of genetically encoded fluorescent sensors used for physiological experiments in plants. Several important insights into plant cell biology have been gained from their use. Notably, applications of the Ca2+-sensing Cameleons have given substantial insight into the role of calcium in stomatal opening (Allen et al., 1999a,b) as well as the role of calcium gradients in growing pollen tubes (Michard et al., 2008), root hairs of Arabidopsis (Monshausen et al., 2007), Nod factor-induced nuclear calcium transients in M. truncatula root hair cells (Capoen et al., 2011) and visualization of Ca2+-dynamics in response to auxin during root gravitropism (Monshausen et al., 2011). Also pH sensors have been useful in plants. The pHluorin sensors have been used to document in detail cytosolic pH gradients and oscillations in growing pollen tubes (Michard et al., 2008), and cell wall pH has been measured by use of pHluorins (Gao et al., 2004) or apo-pHusion (Gjetting et al., 2012) secreted to the apoplast.
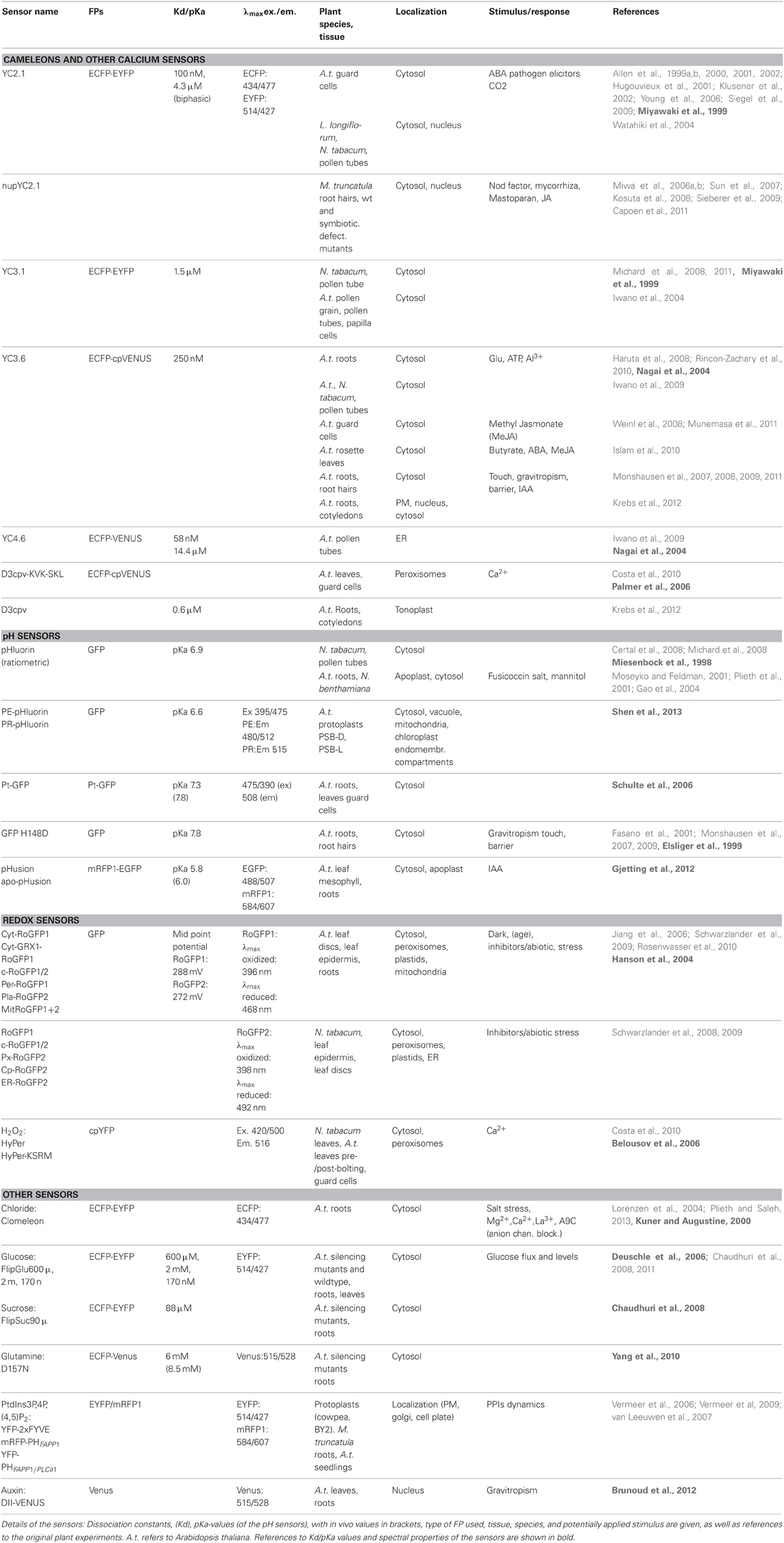
Table 1. Genetically encoded fluorescent sensors used for physiological experiments in plants, subdivided by sensor function.
Looking closer at these experiments, some obvious similarities are seen. It can be argued that successful experiments are often carried out in single cell systems, such as guard cells, root hairs and pollen tubes, where complex cell-to-cell communication is limited. These experiments are all studies of ion signaling, that can be directly correlated with a growth or turgor response, making them attractive experimental setups. Although this is a trend, indeed, several experiments have been successfully carried out in intact tissue, very often in roots, (Fasano et al., 2001; Rincon-Zachary et al., 2010; Monshausen et al., 2011; Gjetting et al., 2012) where autofluorescence is negligible and access is not hindered by the waxy cuticle. Sensors that were successfully used for physiological measurements in intact tissue were often developed specifically for use in plants (e.g., the auxin sensor, DII-Venus, and the apoplastic pH-sensor apo-pHusion). Secondly, it is noted that overall only few sensor platforms and targets were used, again reflecting the fact that many sensors are originally designed for mammalian purposes. Thirdly, sensors were most often expressed in the cytosol, which is the default expression if not specifically targeted to other compartments, and finally most experiments were carried out in Arabidopsis or tobacco, which may not be surprising, since these are easy to manipulate. These observations emphasize some specific challenges that have to be addressed in order to broaden the palette of successful sensor applications in plants.
Plant-Specific Features That Limit the Applicability of Genetically Encoded Fluorescent Sensors
Autofluorescence from Chloroplasts and Cell Walls
Plants are complex, multicellular organisms to work with, and fluorescent probes do not always penetrate multiple cell layers, largely due to the barrier formed by the waxy cuticle and the cell wall. Therefore, genetically encoded sensors are ideally suited for plant cell imaging. However, in plants autofluorescence is a major challenge, particularly in photosynthesizing tissue (chlorophyll ex. 420–460 nm/em. 600–700 nm) and from the cell wall (various components are excited by UV to blue wavelengths, emitting mainly blue light), which can be addressed by the choice of fluorophores in sensor design, or may be circumvented, when lower photon counts/densities can be tolerated, by the use of bioluminescent proteins, such as Aequorin (Mehlmer et al., 2012), where excitation is caused by a chemical reaction instead of light, thus avoiding excitation of autofluorescence.
Precautions for Mounting Procedures and Studying Externally Applied Chemical Stimuli
Genetically encoded sensors as such are non-invasive, but their application to study cellular responses to chemical stimuli requires a perfusion setup and immobilization for microscopy, which can potentially harm the cells. The cuticle covering aerial tissues is an entrance barrier for many compounds, and sometimes even for the ligand itself, making in vivo calibrations difficult when using such sensors. Efficient immobilization methods ensure that no movement of the specimen takes place, while at the same time allowing for perfusion of the chemical stimulus and plant growth. It was, however, recently shown that the commonly used method to immobilize Arabidopsis tissue with a medical adhesive severely impairs cell viability of root cells (Gjetting et al., 2012), making alternative methods necessary. An alternative could be the newly developed root chip (Grossmann et al., 2011) or more simply mounting roots on agarose (Gjetting et al., 2012). Another common method used e.g., for cross-fixing pollen tubes on polylysine slides (Michard et al., 2008) was also shown to disrupt Arabidopsis root cells. In general, the act of handling living tissue under a microscope will inevitably cause disturbance of the tissue and induce various stress responses and tropisms. This of course affects live imaging methods of genetically encoded fluorescent sensors as well as other methods.
Gene Silencing May be Caused by Choice of Promoter or Tandem Fluorescent Proteins
Gene silencing has often been mentioned as a particular problem for plant expression of genetically encoded fluorescent biosensors, particularly when used in tandem repeats, or driven by the 35S promoter (Miyawaki et al., 1997; Deuschle et al., 2006; Krebs et al., 2012). This problem was solved in one case by replacing the 35S-promoter of viral origin with the plant-derived UBQ10 promoter in Arabidopsis (Krebs et al., 2012), or by expressing the sensor in transgene silencing mutants (Deuschle et al., 2006). The use of silencing mutants however, is not optimal, since their general growth pattern is changed, and may influence the measurements in unpredictable ways. In our lab, the 35S-promoter did not provoke inhibitory gene silencing when driving the expression of either FRET-based sensors or ratiometric pH sensors (Gjetting et al., 2012 and unpublished results). Transgene silencing in root tips and seedlings was reported to cause a reduction in fluorescence intensity and thus undetectable FRET changes after 10–15 days of growth (Deuschle et al., 2006; Chaudhuri et al., 2011). In contrast, we were able to monitor pH changes in leaves of 1–2 months old plants which were not subject to silencing (Gjetting et al., 2012).
The Apoplast
This plant-specific extracellular compartment plays a major role in transport regulation, but obviously only plant scientists are interested in developing tools to study its dynamics. Sensors for apoplastic measurements must deal with the low pH values, which are disruptive to many fluorescent proteins, and also be able to measure large differences in ion or solute concentration in the much less buffered apoplast. Apoplastic pH sensors have been used to measure salt stress (Schulte et al., 2006) and the effect of externally applied auxin (Gjetting et al., 2012), but the targeting of sensor protein to the apoplast results in accumulation in the ER, which should be taken into account when measuring the ratio. However, this accumulated protein could potentially be used as an internal pH reference or even as a tool to study pH in the endomembrane system as well. Another issue with apoplastic measurements relate to the structure of GFP in that an oxidizing environment, such as the cell wall and ER can impair proper folding of the fluorescent protein. The use of superfolder GFP (sfGFP) variants may in time be helpful in plants for solving this problem (Aronson et al., 2011).
Improvement of Sensor Applications in Plants
Increasing Sensor Target Range to Include, e.g., Hormones and Kinases
There are many possibilities to expand the range of sensor targets in plants. Developing sensors for central, plant-specific signaling events, like hormone action or activity of plant-specific receptor-like kinases would be major landmarks. An example is a recently developed auxin sensor, which is a fusion of the YFP variant Venus to the Aux/IAA auxin-interaction domain DII (Brunoud et al., 2012), targeted to the nucleus. Using this sensor, auxin distribution was mapped during gravity sensing and lateral shoot formation in Arabidopsis. For mammalian cells, e.g., a variety of GFP-based biosensors exist for kinases, GTPases, phosphatidylinositols (PtdIns) (Kimber et al., 2002; Yoshizaki et al., 2006; Zhang and Allen, 2007). Such sensors (PtdIns) have only recently emerged in the plant community (Munnik and Nielsen, 2011), probably because plants use different signaling components that cannot be targeted by sensors developed for mammalian systems.
pH Measurements in Acidic Compartments
Sensors in plants have so far mainly been expressed in the cytosol, although several other compartments have also been explored (see Table 1). Indeed, targeted sensors are desirable, e.g., to study cell wall pH-dynamics (Gao et al., 2004; Gjetting et al., 2012). Sensor secretion to the apoplast involves accumulation of protein in transit in the endomembrane system, which is a problem to be considered carefully. This may be the reason that some researchers prefer pH-sensitive, small molecular weight fluorescent probes for surface pH measurements in Arabidopsis (Bibikova et al., 1998; Monshausen et al., 2011; Geilfus and Muhling, 2012). However, an apoplastic sensor, stably expressed in cells throughout the tissue, and not just the surface is preferable e.g., in roots to study details of the extracellular pH signature of gravitropic responses and auxin signaling (Swarup et al., 2005). The localization of pH sensors in the acidic compartment of the apoplast or vacuole is also hindered by the sensitivity of GFP to acidity (Tsien, 1998). The pH sensor ptGFP, derived from the Orange Seapen, Ptilosarcus gurneyi showed increased acid stability compared with avGFP derived pHluorins. PtGFP fluorescence could be fully restored after exposure to pH 3.5, and partially restored down to pH 2.5 and may therefore be more suitable for acidic measurements. In contrast, pHluorins were completely denatured at pH 3.5 (Schulte et al., 2006). Recently, a pHluorin-derived sensor, based on a solubility-modified GFP (sm-GFP) was targeted to the vacuole, and to other endomembrane compartments and used to determine pH of the different compartments (Shen et al., 2013).
Variety of FPs and Technology
Expanding the variety of sensor fluorescent proteins, e.g., by the development of different FRET donor/acceptors would facilitate the study of several ions/metabolites simultaneously, e.g., the commonly linked signaling cascade of intracellular calcium/apoplastic pH, as well as same ion fluxes in several compartments or complex protein-protein interactions. Multiplexed FRET (Piljic and Schultz, 2008) and fluorescence lifetime imaging (FLIM)-FRET (Grant et al., 2008) are becoming more feasible as the variety of spectral variants increases. In N. benthamiana leaves a FRET-FLIM assay was used to detect known protein interactions using a FRET pair of the GFP variant TSapphire as donor, and mOrange as acceptor (Bayle et al., 2008). A similar approach was used to detect a flavonoid metabolon in Arabidopsis protoplasts (Crosby et al., 2011). These examples are not using genetically encoded sensors as such, and are based on transient expression in single cell systems, but further illustrate the possibilities of using fluorescent protein technology in plants and may be useful for sensor construction at a later stage.
Identification of New Genes and Gene Function
Genetically encoded sensors may also be used to identify the role of genes of unknown function. A new class of glucose efflux transporters, SWEETs was identified by FRET-based glucose sensors (Chen et al., 2010), and repeated with a sucrose sensor, identifying a subclade of SWEET efflux transporters involved in sucrose transport, indicating a role in phloem loading (Chen et al., 2011). Another promising sensor application known from animal systems, addressed the functional identification of unknown signaling components. This idea was elegantly adapted to Arabidopsis, where the luminescent calcium sensor Aequorin was used to identify an extracellular signaling peptide, AtRALF1, (rapid alkalinization factor) by its ability to induce a cytosolic Ca2+-increase (Haruta et al., 2008). The effect of this peptide was subsequently analysed in detail in Arabidopsis roots expressing the Cameleon sensor YC3.6.
Concluding Remarks
The use of genetically encoded sensors in plants faces some specific challenges not shared with the mammalian world, which need to be addressed by plant scientists. Nevertheless, the continuous development and refinement of fluorescent proteins, sensor design and bioimaging techniques make genetically encoded sensors very promising tools for elucidating metabolic networks and signaling events in plant cells in the future.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Allen, G. J., Chu, S. P., Harrington, C. L., Schumacher, K., Hoffmann, T., Tang, Y. Y., and et al. (2001). A defined range of guard cell calcium oscillation parameters encodes stomatal movements. Nature 411, 1053–1057. doi: 10.1038/35082575
Allen, G. J., Chu, S. P., Schumacher, K., Shimazaki, C. T., Vafeados, D., Kemper, A., and et al. (2000). Alteration of stimulus-specific guard cell calcium oscillations and stomatal closing in Arabidopsis det3 mutant. Science 289, 2338–2342. doi: 10.1126/science.289.5488.2338
Allen, G. J., Kuchitsu, K., Chu, S. P., Murata, Y., and Schroeder, J. I. (1999a). Arabidopsis abi1-1 and abi2-1 phosphatase mutations reduce abscisic acid-induced cytoplasmic calcium rises in guard cells. Plant Cell 11, 1785–1798.
Allen, G. J., Kwak, J. M., Chu, S. P., Llopis, J., Tsien, R. Y., Harper, J. F., and et al. (1999b). Cameleon calcium indicator reports cytoplasmic calcium dynamics in Arabidopsis guard cells. Plant J. 19, 735–747. doi: 10.1046/j.1365-313x.1999.00574.x
Allen, G. J., Murata, Y., Chu, S. P., Nafisi, M., and Schroeder, J. I. (2002). Hypersensitivity of abscisic acid-induced cytosolic calcium increases in the Arabidopsis farnesyltransferase mutant era1-2. Plant Cell 14, 1649–1662. doi: 10.1105/tpc.010448
Aronson, D. E., Costantini, L. M., and Snapp, E. L. (2011). Superfolder GFP is fluorescent in oxidizing environments when targeted via the Sec translocon. Traffic 12, 543–548. doi: 10.1111/j.1600-0854.2011.01168.x
Bayle, V., Nussaume, L., and Bhat, R. A. (2008). Combination of novel green fluorescent protein mutant TSapphire and DsRed variant mOrange to set up a versatile in planta FRET-FLIM assay. Plant Physiol. 148, 51–60. doi: 10.1104/pp.108.117358
Belousov, V. V., Fradkov, A. F., Lukyanov, K. A., Staroverov, D. B., Shakhbazov, K. S., Terskikh, A. V., and et al. (2006). Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods 3, 281–286. doi: 10.1038/nmeth866
Bibikova, T. N., Jacob, T., Dahse, I., and Gilroy, S. (1998). Localized changes in apoplastic and cytoplasmic pH are associated with root hair development in Arabidopsis thaliana. Development 125, 2925–2934.
Brunoud, G., Wells, D. M., Oliva, M., Larrieu, A., Mirabet, V., Burrow, A. H., and et al. (2012). A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482, 103–106. doi: 10.1038/nature10791
Capoen, W., Sun, J., Wysham, D., Otegui, M. S., Venkateshwaran, M., Hirsch, S., and et al. (2011). Nuclear membranes control symbiotic calcium signaling of legumes. Proc. Natl. Acad. Sci. U.S.A. 108, 14348–14353. doi: 10.1073/pnas.1107912108
Certal, A. C., Almeida, R. B., Carvalho, L. M., Wong, E., Moreno, N., Michard, E., and et al. (2008). Exclusion of a proton ATPase from the apical membrane is associated with cell polarity and tip growth in Nicotiana tabacum pollen tubes. Plant Cell 20, 614–634. doi: 10.1105/tpc.106.047423
Chaudhuri, B., Hormann, F., and Frommer, W. B. (2011). Dynamic imaging of glucose flux impedance using FRET sensors in wild-type Arabidopsis plants. J. Exp. Bot. 62, 2411–2417. doi: 10.1093/jxb/erq444
Chaudhuri, B., Hormann, F., Lalonde, S., Brady, S. M., Orlando, D. A., Benfey, P., and et al. (2008). Protonophore- and pH-insensitive glucose and sucrose accumulation detected by FRET nanosensors in Arabidopsis root tips. Plant J. 56, 948–962. doi: 10.1111/j.1365-313X.2008.03652.x
Chen, L. Q., Hou, B. H., Lalonde, S., Takanaga, H., Hartung, M. L., Qu, X. Q., and et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532. doi: 10.1038/nature09606
Chen, L. Q., Qu, X. Q., Hou, B. H., Sosso, D., Osorio, S., Fernie, A. R., and et al. (2011). Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335, 207–211. doi: 10.1126/science.1213351
Choi, W. G., Swanson, S. J., and Gilroy, S. (2012). High-resolution imaging of Ca2+, redox status, ROS and pH using GFP biosensors. Plant J. 70, 118–128. doi: 10.1111/j.1365-313X.2012.04917.x
Chudakov, D. M., Matz, M. V., Lukyanov, S., and Lukyanov, K. A. (2010). Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 90, 1103–1163. doi: 10.1152/physrev.00038.2009
Costa, A., Drago, I., Behera, S., Zottini, M., Pizzo, P., Schroeder, J. I., and et al. (2010). H2O2 in plant peroxisomes: an in vivo analysis uncovers a Ca2+-dependent scavenging system. Plant J. 62, 760–772. doi: 10.1111/j.1365-313X.2010.04190.x
Crosby, K. C., Pietraszewska-Bogiel, A., Gadella, T. W. Jr., and Winkel, B. S. (2011). Forster resonance energy transfer demonstrates a flavonoid metabolon in living plant cells that displays competitive interactions between enzymes. FEBS Lett. 585, 2193–2198. doi: 10.1016/j.febslet.2011.05.066
Deuschle, K., Chaudhuri, B., Okumoto, S., Lager, I., Lalonde, S., and Frommer, W. B. (2006). Rapid metabolism of glucose detected with FRET glucose nanosensors in epidermal cells and intact roots of Arabidopsis RNA-silencing mutants. Plant Cell 18, 2314–2325. doi: 10.1105/tpc.106.044073
Dixit, R., Cyr, R., and Gilroy, S. (2006). Using intrinsically fluorescent proteins for plant cell imaging. Plant J. 45, 599–615. doi: 10.1111/j.1365-313X.2006.02658.x
Ehrhardt, D. W., and Frommer, W. B. (2012). New technologies for 21st century plant science. Plant Cell 24, 374–394. doi: 10.1105/tpc.111.093302
Elsliger, M. A., Wachter, R. M., Hanson, G. T., Kallio, K., and Remington, S. J. (1999). Structural and spectral response of green fluorescent protein variants to changes in pH. Biochemistry 38, 5296–5301. doi: 10.1021/bi9902182
Fasano, J. M., Swanson, S. J., Blancaflor, E. B., Dowd, P. E., Kao, T. H., and Gilroy, S. (2001). Changes in root cap pH are required for the gravity response of the Arabidopsis root. Plant Cell 13, 907–921.
Fehr, M., Ehrhardt, D. W., Lalonde, S., and Frommer, W. B. (2004). Minimally invasive dynamic imaging of ions and metabolites in living cells. Curr. Opin. Plant Biol. 7, 345–351. doi: 10.1016/j.pbi.2004.03.015
Frommer, W. B., Davidson, M. W., and Campbell, R. E. (2009). Genetically encoded biosensors based on engineered fluorescent proteins. Chem. Soc. Rev. 38, 2833–2841. doi: 10.1039/b907749a
Gao, D., Knight, M. R., Trewavas, A. J., Sattelmacher, B., and Plieth, C. (2004). Self-reporting Arabidopsis expressing pH and Ca2+ indicators unveil ion dynamics in the cytoplasm and in the apoplast under abiotic stress. Plant Physiol. 134, 898–908. doi: 10.1104/pp.103.032508
Geilfus, C. M., and Muhling, K. H. (2012). Transient alkalinization in the leaf apoplast of Vicia faba L. depends on NaCl stress intensity: an in situ ratio imaging study. Plant Cell Environ. 35, 578–587. doi: 10.1111/j.1365-3040.2011.02437.x
Gjetting, K. S., Ytting, C. K., Schulz, A., and Fuglsang, A. T. (2012). Live imaging of intra- and extracellular pH in plants using pHusion, a novel genetically encoded biosensor. J. Exp. Bot. 63, 3207–3218. doi: 10.1093/jxb/ers040
Grant, D. M., Zhang, W., McGhee, E. J., Bunney, T. D., Talbot, C. B., Kumar, S., and et al. (2008). Multiplexed FRET to image multiple signaling events in live cells. Biophys. J. 95, L69–L71. doi: 10.1529/biophysj.108.139204
Grossmann, G., Guo, W. J., Ehrhardt, D. W., Frommer, W. B., Sit, R. V., Quake, S. R., and et al. (2011). The RootChip: an integrated microfluidic chip for plant science. Plant Cell 23, 4234–4240. doi: 10.1105/tpc.111.092577
Hanson, G. T., Aggeler, R., Oglesbee, D., Cannon, M., Capaldi, R. A., Tsien, R. Y., and et al. (2004). Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 279, 13044–13053. doi: 10.1074/jbc.M312846200
Haruta, M., Monshausen, G., Gilroy, S., and Sussman, M. R. (2008). A cytoplasmic Ca2+ functional assay for identifying and purifying endogenous cell signaling peptides in Arabidopsis seedlings: identification of AtRALF1 peptide. Biochemistry 47, 6311–6321. doi: 10.1021/bi8001488
Hugouvieux, V., Kwak, J. M., and Schroeder, J. I. (2001). An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106, 477–487. doi: 10.1016/S0092-8674(01)00460-3
Islam, M. M., Hossain, M. A., Jannat, R., Munemasa, S., Nakamura, Y., Mori, I. C., and et al. (2010). Cytosolic alkalization and cytosolic calcium oscillation in arabidopsis guard cells response to ABA and MeJA. Plant Cell Physiol. 51, 1721–1730. doi: 10.1093/pcp/pcq131
Iwano, M., Entani, T., Shiba, H., Kakita, M., Nagai, T., Mizuno, H., and et al. (2009). Fine-tuning of the cytoplasmic Ca2+ concentration is essential for pollen tube growth. Plant Physiol. 150, 1322–1334. doi: 10.1104/pp.109.139329
Iwano, M., Shiba, H., Miwa, T., Che, F. S., Takayama, S., Nagai, T., and et al. (2004). Ca2+ dynamics in a pollen grain and papilla cell during pollination of Arabidopsis. Plant Physiol. 136, 3562–3571. doi: 10.1104/pp.104.046961
Jiang, K., Schwarzer, C., Lally, E., Zhang, S., Ruzin, S., Machen, T., and et al. (2006). Expression and characterization of a redox-sensing green fluorescent protein (reduction-oxidation-sensitive green fluorescent protein) in Arabidopsis. Plant Physiol. 141, 397–403. doi: 10.1104/pp.106.078246
Kimber, W. A., Trinkle-Mulcahy, L., Cheung, P. C., Deak, M., Marsden, L. J., Kieloch, A., and et al. (2002). Evidence that the tandem-pleckstrin-homology-domain-containing protein TAPP1 interacts with Ptd(3, 4)P2 and the multi-PDZ-domain-containing protein MUPP1 in vivo. Biochem. J. 361, 525–536. doi: 10.1042/0264-6021:3610525
Klusener, B., Young, J. J., Murata, Y., Allen, G. J., Mori, I. C., Hugouvieux, V., and et al. (2002). Convergence of calcium signaling pathways of pathogenic elicitors and abscisic acid in Arabidopsis guard cells. Plant Physiol. 130, 2152–2163. doi: 10.1104/pp.012187
Kosuta, S., Hazledine, S., Sun, J., Miwa, H., Morris, R. J., Downie, J. A., and et al. (2008). Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc. Natl. Acad. Sci. U.S.A. 105, 9823–9828. doi: 10.1073/pnas.0803499105
Krebs, M., Held, K., Binder, A., Hashimoto, K., Den Herder, G., Parniske, M., and et al. (2012). FRET-based genetically encoded sensors allow high-resolution live cell imaging of Ca2+ dynamics. Plant J. 69, 181–192. doi: 10.1111/j.1365-313X.2011.04780.x
Kuner, T., and Augustine, G. J. (2000). A genetically encoded ratiometric indicator for chloride: capturing chloride transients in cultured hippocampal neurons. Neuron 27, 447–459. doi: 10.1016/S0896-6273(00)00056-8
Lalonde, S., Ehrhardt, D. W., and Frommer, W. B. (2005). Shining light on signaling and metabolic networks by genetically encoded biosensors. Curr. Opin. Plant Biol. 8, 574–581. doi: 10.1016/j.pbi.2005.09.015
Lorenzen, I., Aberle, T., and Plieth, C. (2004). Salt stress-induced chloride flux: a study using transgenic Arabidopsis expressing a fluorescent anion probe. Plant J. 38, 539–544. doi: 10.1111/j.0960-7412.2004.02053.x
Mehlmer, N., Parvin, N., Hurst, C. H., Knight, M. R., Teige, M., and Vothknecht, U. C. (2012). A toolset of aequorin expression vectors for in planta studies of subcellular calcium concentrations in Arabidopsis thaliana. J. Exp. Bot. 63, 1751–1761. doi: 10.1093/jxb/err406
Mehta, S., and Zhang, J. (2011). Reporting from the field: genetically encoded fluorescent reporters uncover signaling dynamics in living biological systems. Annu. Rev. Biochem. 80, 375–401. doi: 10.1146/annurev-biochem-060409-093259
Michard, E., Dias, P., and Feijo, J. (2008). Tobacco pollen tubes as cellular models for ion dynamics: improved spatial and temporal resolution of extracellular flux and free cytosolic concentration of calcium and protons using pHluorin and YC3.1 Cameleon. Sex Plant Reprod. 21, 169–181. doi: 10.1007/s00497-008-0076-x
Michard, E., Lima, P. T., Borges, F., Silva, A. C., Portes, M. T., Carvalho, J. E., and et al. (2011). Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science 332, 434–437. doi: 10.1126/science.1201101
Miesenbock, G., De Angelis, D. A., and Rothman, J. E. (1998). Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394, 192–195. doi: 10.1038/28190
Miwa, H., Sun, J., Oldroyd, G. E., and Downie, J. A. (2006a). Analysis of Nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol. Plant Microbe Interact. 19, 914–923. doi: 10.1094/MPMI-19-0914
Miwa, H., Sun, J., Oldroyd, G. E., and Downie, J. A. (2006b). Analysis of calcium spiking using a Cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. Plant J. 48, 883–894. doi: 10.1111/j.1365-313X.2006.02926.x
Miyawaki, A. (2011). Proteins on the move: insights gained from fluorescent protein technologies. Nat. Rev. Mol. Cell Biol. 12, 656–668. doi: 10.1038/nrm3199
Miyawaki, A., Griesbeck, O., Heim, R., and Tsien, R. Y. (1999). Dynamic and quantitative Ca2+ measurements using improved Cameleons. Proc. Natl. Acad. Sci. U.S.A. 96, 2135–2140. doi: 10.1073/pnas.96.5.2135
Miyawaki, A., Llopis, J., Heim, R., McCaffery, J. M., Adams, J. A., Ikura, M., and et al. (1997). Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388, 882–887. doi: 10.1038/42264
Monshausen, G. B., Bibikova, T. N., Messerli, M. A., Shi, C, and Gilroy, S. (2007). Oscillations in extracellular pH and reactive oxygen species modulate tip growth of Arabidopsis root hairs. Proc. Natl. Acad. Sci. U.S.A. 104, 20996–21001. doi: 10.1073/pnas.0708586104
Monshausen, G. B., Bibikova, T. N., Weisenseel, M. H., and Gilroy, S. (2009). Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots. Plant Cell 21, 2341–2356. doi: 10.1105/tpc.109.068395
Monshausen, G. B., Messerli, M. A., and Gilroy, S. (2008). Imaging of the Yellow Cameleon 3.6 indicator reveals that elevations in cytosolic Ca2+ follow oscillating increases in growth in root hairs of Arabidopsis. Plant Physiol. 147, 1690–1698. doi: 10.1104/pp.108.123638
Monshausen, G. B., Miller, N. D., Murphy, A. S., and Gilroy, S. (2011). Dynamics of auxin-dependent Ca2+ and pH signaling in root growth revealed by integrating high-resolution imaging with automated computer vision-based analysis. Plant J. 65, 309–318. doi: 10.1111/j.1365-313X.2010.04423.x
Moseyko, N., and Feldman, L. J. (2001). Expression of pH-sensitive green fluorescent protein in Arabidopsis thaliana. Plant Cell Environ. 24, 557–563. doi: 10.1046/j.1365-3040.2001.00703.x
Munemasa, S., Hossain, M. A., Nakamura, Y., Mori, I. C., and Murata, Y. (2011). The Arabidopsis calcium-dependent protein kinase, CPK6, functions as a positive regulator of methyl jasmonate signaling in guard cells. Plant Physiol. 155, 553–561. doi: 10.1104/pp.110.162750
Munnik, T., and Nielsen, E. (2011). Green light for polyphosphoinositide signals in plants. Curr. Opin. Plant Biol. 14, 489–497. doi: 10.1016/j.pbi.2011.06.007
Nagai, T., Yamada, S., Tominaga, T., Ichikawa, M., and Miyawaki, A. (2004). Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 10554–10559. doi: 10.1073/pnas.0400417101
Newman, R. H., Fosbrink, M. D., and Zhang, J. (2011). Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem. Rev. 111, 3614–3666. doi: 10.1021/cr100002u
Okumoto, S. (2010). Imaging approach for monitoring cellular metabolites and ions using genetically encoded biosensors. Curr. Opin. Biotechnol. 21, 45–54. doi: 10.1016/j.copbio.2010.01.009
Okumoto, S. (2012). Quantitative imaging using genetically encoded sensors for small molecules in plants. Plant J. 70, 108–117. doi: 10.1111/j.1365-313X.2012.04910.x
Okumoto, S., Jones, A., and Frommer, W. B. (2012). Quantitative imaging with fluorescent biosensors: advanced tools for spatiotemporal analysis of biodynamics in cells. Annu. Rev. Plant Biol. 63, 663–706. doi: 10.1146/annurev-arplant-042110-103745
Palmer, A. E., Giacomello, M., Kortemme, T., Hires, S. A., Lev-Ram, V., Baker, D., and et al. (2006). Ca2+ indicators based on computationally redesigned calmodulin-peptide pairs. Chem. Biol. 13, 521–530. doi: 10.1016/j.chembiol.2006.03.007
Palmer, A. E., Qin, Y., Park, J. G., and McCombs, J. E. (2011). Design and application of genetically encoded biosensors. Trends Biotechnol. 29, 144–152. doi: 10.1016/j.tibtech.2010.12.004
Piljic, A., and Schultz, C. (2008). Simultaneous recording of multiple cellular events by FRET. ACS Chem. Biol. 3, 156–160. doi: 10.1021/cb700247q
Plieth, C., and Saleh, L. (2013). A9C sensitive Cl(-)-accumulation in A. thaliana root cells during salt stress is controlled by internal and external calcium. Plant Signal. Behav. [Epub ahead of print].
Plieth, C., Sattelmacher, B., Trewavas, A., Hansen, U., and Knight, M. (2001). Engineering plants expressiong calcium and pH indicators in the cytoplasm and the apoplast. Plant Nutri. 92, 252–253. doi: 10.1007/0-306-47624-X_121
Rincon-Zachary, M., Teaster, N. D., Sparks, J. A., Valster, A. H., Motes, C. M., and Blancaflor, E. B. (2010). Fluorescence resonance energy transfer-sensitized emission of yellow Cameleon 3.60 reveals root zone-specific calcium signatures in Arabidopsis in response to aluminum and other trivalent cations. Plant Physiol. 152, 1442–1458. doi: 10.1104/pp.109.147256
Rosenwasser, S., Rot, I., Meyer, A. J., Feldman, L., Jiang, K., and Friedman, H. (2010). A fluorometer-based method for monitoring oxidation of redox-sensitive GFP (roGFP) during development and extended dark stress. Physiol. Plant 138, 493–502. doi: 10.1111/j.1399-3054.2009.01334.x
Schulte, A., Lorenzen, I., Bottcher, M., and Plieth, C. (2006). A novel fluorescent pH probe for expression in plants. Plant Methods 2, 7. doi: 10.1186/1746-4811-2-7
Schwarzlander, M., Fricker, M. D., Muller, C., Marty, L., Brach, T., Novak, J., and et al. (2008). Confocal imaging of glutathione redox potential in living plant cells. J. Microsc. 231, 299–316. doi: 10.1111/j.1365-2818.2008.02030.x
Schwarzlander, M., Fricker, M. D., and Sweetlove, L. J. (2009). Monitoring the in vivo redox state of plant mitochondria: effect of respiratory inhibitors, abiotic stress and assessment of recovery from oxidative challenge. Biochim. Biophys. Acta 1787, 468–475. doi: 10.1016/j.bbabio.2009.01.020
Shen, J., Zeng, Y., Zhuang, X., Sun, L., Yao, X., Pimpl, P., and et al. (2013). Organelle pH in the Arabidopsis endomembrane system. Mol. Plant. doi: 10.1093/mp/sst079. [Epub ahead of print].
Sieberer, B. J., Chabaud, M., Timmers, A. C., Monin, A., Fournier, J., and Barker, D. G. (2009). A nuclear-targeted Cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiol. 151, 1197–1206. doi: 10.1104/pp.109.142851
Siegel, R. S., Xue, S., Murata, Y., Yang, Y., Nishimura, N., Wang, A., and et al. (2009). Calcium elevation-dependent and attenuated resting calcium-dependent abscisic acid induction of stomatal closure and abscisic acid-induced enhancement of calcium sensitivities of S-type anion and inward-rectifying K channels in Arabidopsis guard cells. Plant J. 59, 207–220. doi: 10.1111/j.1365-313X.2009.03872.x
Sun, J., Miwa, H., Downie, J. A., and Oldroyd, G. E. (2007). Mastoparan activates calcium spiking analogous to Nod factor-induced responses in Medicago truncatula root hair cells. Plant Physiol. 144, 695–702. doi: 10.1104/pp.106.093294
Swanson, S. J., Choi, W. G., Chanoca, A., and Gilroy, S. (2011). In vivo imaging of Ca2+, pH, and reactive oxygen species using fluorescent probes in plants. Annu. Rev. Plant Biol. 62, 273–297. doi: 10.1146/annurev-arplant-042110-103832
Swarup, R., Kramer, E. M., Perry, P., Knox, K., Leyser, H. M., Haseloff, J., and et al. (2005). Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat. Cell Biol. 7, 1057–1065. doi: 10.1038/ncb1316
Tsien, R. Y. (1998). The green fluorescent protein. Annu. Rev. Biochem. 67, 509–544. doi: 10.1146/annurev.biochem.67.1.509
VanEngelenburg, S. B., and Palmer, A. E. (2008). Fluorescent biosensors of protein function. Curr. Opin. Chem. Biol. 12, 60–65. doi: 10.1016/j.cbpa.2008.01.020
van Leeuwen, W., Vermeer, J. E., Gadella, T. W. Jr., and Munnik, T. (2007). Visualization of phosphatidylinositol 4, 5-bisphosphate in the plasma membrane of suspension-cultured tobacco BY-2 cells and whole Arabidopsis seedlings. Plant J. 52, 1014–1026. doi: 10.1111/j.1365-313X.2007.03292.x
Vermeer, J. E., Thole, J. M., Goedhart, J., Nielsen, E., Munnik, T., and Gadella, T. W. Jr. (2009). Imaging phosphatidylinositol2 4-phosphate dynamics in living plant cells. Plant J. 57, 356–372. doi: 10.1111/j.1365-313X.2008.03679.x
Vermeer, J. E., van Leeuwen, W., Tobena-Santamaria, R., Laxalt, A. M., Jones, D. R., Divecha, N., and et al. (2006). Visualization of PtdIns3P dynamics in living plant cells. Plant J. 47, 687–700. doi: 10.1111/j.1365-313X.2006.02830.x
Watahiki, M. K., Trewavas, A. J., and Parton, R. M. (2004). Fluctuations in the pollen tube tip-focused calcium gradient are not reflected in nuclear calcium level: a comparative analysis using recombinant yellow Cameleon calcium reporter. Sex. Plant Reprod. 17, 125–130. doi: 10.1007/s00497-004-0224-x
Weinl, S., Held, K., Schlucking, K., Steinhorst, L., Kuhlgert, S., Hippler, M., and et al. (2008). A plastid protein crucial for Ca2+-regulated stomatal responses. New Phytol. 179, 675–686. doi: 10.1111/j.1469-8137.2008.02492.x
Yang, H., Bogner, M., Stierhof, Y. D., and Ludewig, U. (2010). H-independent glutamine transport in plant root tips. PLoS ONE 5:e8917. doi: 10.1371/journal.pone.0008917
Yoshizaki, H., Aoki, K., Nakamura, T., and Matsuda, M. (2006). Regulation of RalA GTPase by phosphatidylinositol 3-kinase as visualized by FRET probes. Biochem. Soc. Trans. 34, 851–854. doi: 10.1042/BST0340851
Young, J. J., Mehta, S., Israelsson, M., Godoski, J., Grill, E., and Schroeder, J. I. (2006). CO2 signaling in guard cells: calcium sensitivity response modulation, a Ca2+-independent phase, and CO(2) insensitivity of the gca2 mutant. Proc. Natl. Acad. Sci. U.S.A. 103, 7506–7511. doi: 10.1073/pnas.0602225103
Keywords: plant bioimaging, genetically encoded biosensors, fluorescence microscopy, calcium sensor, phosphatidylinositol sensor, pH sensors
Citation: Gjetting SK, Schulz A and Fuglsang AT (2013) Perspectives for using genetically encoded fluorescent biosensors in plants. Front. Plant Sci. 4:234. doi: 10.3389/fpls.2013.00234
Received: 29 April 2013; Accepted: 13 June 2013;
Published online: 12 July 2013.
Edited by:
Elison B. Blancaflor, The Samuel Roberts Noble Foundation, USAReviewed by:
Elison B. Blancaflor, The Samuel Roberts Noble Foundation, USAGabriele B. Monshausen, Pennsylvania State University, USA
Copyright © 2013 Gjetting, Schulz and Fuglsang. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Anja T. Fuglsang, Transport Biology Section, Department of Plant and Environmental Sciences, University of Copenhagen, Thorvaldsensvej 40, Copenhagen, DK-1871, Denmark e-mail: atf@life.ku.dk
