- 1Key Laboratory of Molecular Epigenetics of MOE, Institute of Genetics and Cytology, Northeast Normal University, Changchun, China
- 2Biosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN, USA
MYB transcription factors regulate multiple aspects of plant growth and development. Among the large family of MYB transcription factors, single-repeat R3 MYBs are characterized by their short sequence (<120 amino acids) consisting largely of the single MYB DNA-binding repeat. In the model plant Arabidopsis, R3 MYBs mediate lateral inhibition during epidermal patterning and are best characterized for their regulatory roles in trichome and root hair development. R3 MYBs act as negative regulators for trichome formation but as positive regulators for root hair development. In this article, we provide a comprehensive review on the role of R3 MYBs in the regulation of cell type specification in the model plant Arabidopsis.
Introduction
In plants, MYB transcription factors are encoded by a large family of genes (Stracke et al., 2001; Chen et al., 2006; Dubos et al., 2010; Katiyar et al., 2012). They play important roles in regulating plant growth and development and plant responses to environmental stimuli. There are several sub-families of MYB transcription factors, defined by the number of MYB DNA-binding domain repeats. These include 4R-MYB, 3R-MYB, R2R3-MYB, and 1R-MYB subfamilies that contain four, three, two and one MYB DNA-binding domain repeats, respectively (Dubos et al., 2010).
In the model plant Arabidopsis, there are a total of 64 1R-MYB or MYB-related proteins (Dubos et al., 2010). Among this subfamily, there are a unique set of 1R-MYBs that are characterized by their short sequence (<120 amino acids) consisting largely of the single R3 MYB repeat. These small proteins are referred as single-repeat R3 MYB transcription factors (R3 MYBs) and are subjects of this review article. We provide a comprehensive review about the function and action of R3 MYBs in the model plant Arabidopsis, particularly, in trichome and root hair development.
R3 MYBs in Arabidopsis
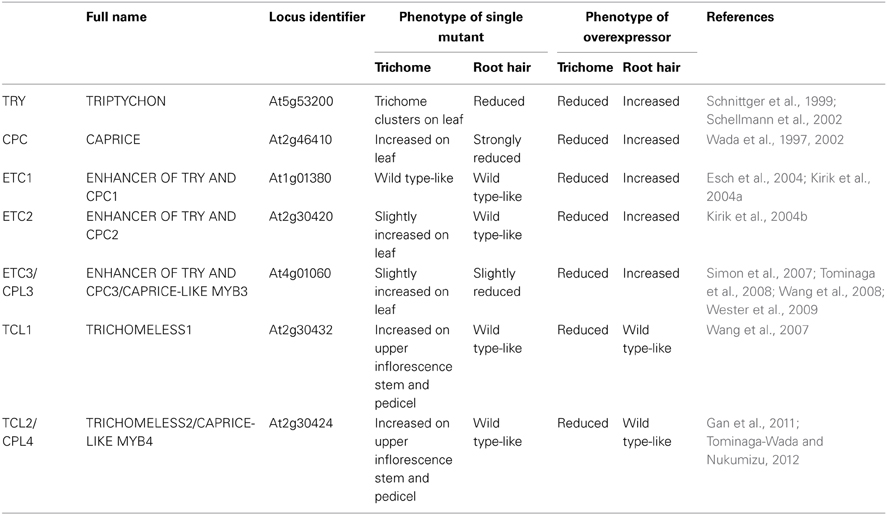
In the completely-sequenced model plant Arabidopsis thaliana (hereafter referred as Arabidopsis), a total of seven genes encoding R3 MYBs have been reported so far. These include TRIPTYCHON (TRY)(Schnittger et al., 1999; Schellmann et al., 2002), CAPRICE (CPC) (Wada et al., 1997, 2002), ENHANCER OF TRY AND CPC1 (ETC1) (Esch et al., 2004; Kirik et al., 2004a), ETC2 (Kirik et al., 2004b), ETC3/CAPRICE-LIKE MYB3 (CPL3) (Simon et al., 2007; Tominaga et al., 2008; Wang et al., 2008), TRICHOMELESS1 (TCL1) (Wang et al., 2007) and TCL2/CPL4 (Gan et al., 2011; Tominaga-Wada and Nukumizu, 2012). These seven R3 MYB proteins share approximately 52–82% similarity, and 37–68% identity to each other at the amino acid level (Figure 1). They consist of largely the single MYB DNA-binding repeat, with the total number of amino acid of each protein ranging from 77 to 112.
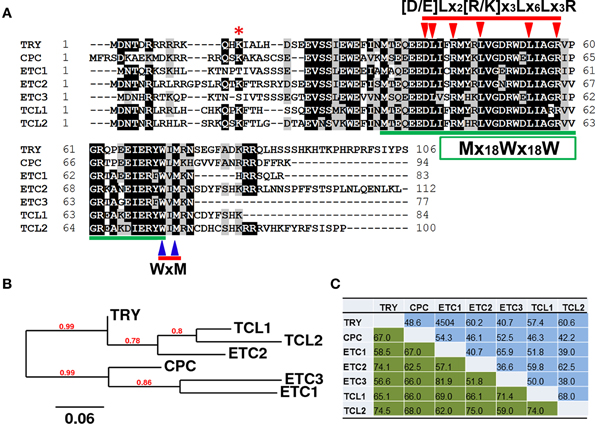
Figure 1. Single-repeat R3 MYB transcription factors in Arabidopsis thaliana. (A) Amino acid sequence alignment of R3 MYBs. Identical and similar amino acids are shaded in black and gray, respectively. The amino acids of [D/E]Lx2[R/K]x3Lx6Lx3R signature that is required for interaction between MYBs and R/B-like bHLH transcription factors are indicated by arrowheads on the top of amino acids. The amino acids of WxM signature that is crucial for cell-to-cell movement are indicated by arrowheads on the bottom of amino acids. The pattern of primary structure of MYB repeat, Mx18Wx18W, is indicated in a box and amino acids are underlined. The Lysine19 (K19) of ETC2 that was proposed to control its stability is indicated by an asterisk. (B) Phylogenetic analysis of R3 MYBs. The full-length amino acid sequences of each R3 MYB were used for the analysis. (C) Similarity and identity of amino acid sequences of R3 MYB proteins.
In order to search for any other potential R3 MYBs encoded by the Arabidopsis genome, the amino acid sequence of TRY was used as a template for searching protein sequence homologs encoded by the fully-sequenced Arabidopsis genome using the “Protein Homologs” tool of Phytozome (www.phytozome.net). In addition to CPC, ETC1, ETC2, ETC3, TCL1 and TCL2, the search identified eight other proteins. These included MYB3, MYB4, MYB5, MYB23, MYB82, GL1, TT2, and WER (Figure S1). However, R3 MYBs were clustered in one distinct branch of the phylogenetic tree (Figure S2). Furthermore, those eight MYBs are mostly R2R3-MYBs, are typically twice the size of R3 MYBs in term of number of amino acids, and share low similarity (17–34%) and identity (10–23%) with R3 MYBs at the amino acid level (Figure S3). Therefore, these MYBs are not considered to be R3 MYB proteins and no additional R3 MYB was identified.
It should be noted that an R3 MYB-related protein, designated as MYBL2, has been shown to be involved in the regulation of flavonoid biosynthesis (Dubos et al., 2008; Matsui et al., 2008). Amino acid alignment and phylogenetic analysis indicated that MYBL2 is more similar to other MYBs than R3 MYBs. For example, the number of amino acid of MYBL2 (195 aa) is almost twice that of R3 MYBs (Figure S1). In addition, the cell-cell movement motif WxM (further discussed in the next section) is not conserved in MYBL2 (Figure S1). Therefore, MYBL2 is not included in our further discussion of R3 MYBs in this review article.
It should also be noted that there are three transcript variants for ETC3, designated as ETC3.1, ETC3.2 and ETC3.3, according to the current annotation by TAIR (http://www.arabidopsis.org) and Phytozome. Proteins encoded by these three ETC3 transcript variants are identical except that compared with ETC.1, ETC3.2 has deletion of three amino acids and ETC3.3 has deletion of two amino acids at a site prior to the conserved bHLH binding sequence signature and cell-to-cell movement motif (described in the next section) (Figure S4). For simplicity, only ETC3.1 was included in our analysis.
Finally, several chimeric transcripts can be formed between ETC2 and TCL2/CPL4, two tandem repeat genes in Chromosome II (ETC2: At2g30420; TCL2/CPL4: At2g30424), resulting from alternative splicing (Tominaga-Wada and Nukumizu, 2012).
In summary, the genome of model plant Arabidopsis encodes a total of seven R3 MYBs, and these seven R3 MYBs are the subjects of this review article.
R3 MYBs Structural Features
In addition to the small size (77–112 aa) and the constitution of a single MYB repeat, there are a number of structural characteristics of R3 MYBs (Figure 1). Firstly, R3 MYBs contain a sequence signature of [D/E]Lx2[R/K]x3Lx6Lx3R that has been shown to be required for the interaction between R3 MYBs and R/B-like bHLH transcription factors (Zimmermann et al., 2004). It should be noted that this bHLH interaction signature is also conserved in many other MYB proteins including those showed sequence homology with R3 MYBs (Figure S1). Secondly, R3 MYBs contain a sequence motif WxM that has been shown to be required for its cell-to-cell movement (Kurata et al., 2005). This cell-to-cell movement motif is not conserved in any other MYBs that showed sequence homology with R3 MYBs (Figure S1). Finally, the primary structure of MYB repeat in R3 MYBs follow a pattern of Mx18Wx18W, instead of [F/I]x18Wx18W that is typically found in the R3 MYB repeat of R2R3-MYB proteins and MYBL2 (Figure S1). Therefore, a combination of these three structural features together with the small size and the constitution of a single MYB repeat define a small family of R3 MYB transcription factors.
Due to its constitution of a single MYB repeat, R3 MYBs lack the activation domain that is typically present in most transcription factors. Therefore, from the domain structural perspective, it is believed that R3 MYBs rely on the interaction with other transcription factors to execute their transcriptional activity. As discussed in details below, this indeed represents a major mechanism of action of R3 MYBs.
Function of R3 MYBs in Trichome Development
Trichomes are hair cells produced by the outward growth of epidermal cells (Marks, 1997; Hülskamp and Schnittger, 1998). They distribute on the surface of aerial organs including leaves, stems and flower organs of most land plants. Trichomes can act as barriers to protect plants from biotic (e.g., insect herbivores) and abiotic stresses, UV light irradiation and excessive transpiration (Mauricio and Rausher, 1997; Eisner et al., 1998; Werker, 2000; Wagner et al., 2004). Some trichomes or trichome-produced compounds are of great commercial value. For example, single-celled seed trichomes of Gossypium hirsutum, cotton fibers, are the most important natural fiber for the textile industry (Kim and Triplett, 2001). Multi-celled glandular trichomes of Artemisia annua accumulates artemisinin, a drug that is widely used for the treatment of malaria (Liu et al., 2006; Lommen et al., 2006). Therefore, study of trichome development may lead to the improvement of agronomical and economical traits in plants such as resistance to insect herbivores, improvement of cotton fiber yield and quality, and increase in yield of oil or other plant products contained in glandular trichomes. Furthermore, trichome has become an excellent system to study cell type specification (Schiefelbein, 2003; Pesch and Hülskamp, 2004; Serna, 2005; Schellmann et al., 2007). In this review article, we specifically focus on discussing the role of R3 MYBs in single-celled trichome development in the model plant Arabidopsis.
The genetic control of trichome development has been studied extensively in the last 20 years. It has been generally recognized that trichome initiation is promoted by an activator complex consisting of a WD40-repeat protein, TRANSPARENT TESTA GLABRA1 (TTG1) (Galway et al., 1994; Walker et al., 1999), an R2R3 MYB-type transcription factor, GLABRA1 (GL1) (Oppenheimer et al., 1991), and a bHLH transcription factor, GLABRA3 (GL3) or ENHANCER OF GLABRA3 (EGL3)(Payne et al., 2000; Zhang et al., 2003). Yeast two hybrid experiments showed that both GL1 and TTG1 bind GL3 but at different regions of the GL3 protein and that TTG1 and GL1 do not interact (Payne et al., 2000). This TTG1-GL3/EGL3-GL1 activator complex induces the expression of a homeodomain protein, GLABRA2 (GL2) (Rerie et al., 1994; Masucci et al., 1996), which is required for trichome initiation. The trichome initiation involves feedback loop controls: in addition to inducing GL2 expression, the TTG1-GL3/EGL3-GL1 activator complex induces the expression of R3 MYB genes. R3 MYBs can move from a trichome precursor cell to its neighboring cell and compete with GL1 for binding GL3 or EGL3 thus disrupting the functionality of the activator complex, resulting in the inhibition of trichome initiation (Hülskamp et al., 1994; Schellmann et al., 2002; Esch et al., 2003; Schiefelbein, 2003; Pesch and Hülskamp, 2004; Ishida et al., 2008; Pesch and Hülskamp, 2009).
According to this model, R3 MYBs act as negative regulators of trichome initiation. Consistent with this view, overexpression of each of seven R3 MYBs resulted in glabrous phenotypes and loss-of-function mutation in R3 MYBs resulted in more trichome formation except etc1 single mutant (Wada et al., 1997; Schnittger et al., 1999; Schellmann et al., 2002; Wada et al., 2002; Esch et al., 2004; Kirik et al., 2004a,b; Simon et al., 2007; Wang et al., 2007; Tominaga et al., 2008; Wang et al., 2008; Wester et al., 2009; Gan et al., 2011; Tominaga-Wada and Nukumizu, 2012). However, differences in severity and pattern of trichome formation have been observed in the single mutants of R3 MYB genes. The trichome phenotypes of R3 MYB single mutants can be categorized into three groups: (i) increased trichome clustering on leaf (try). (ii) increased trichome density on leaf (cpc, etc2, and etc3), and (iii) normal trichome density on leaves but increased trichomes on inflorescence stems and pedicels (tcl1, tcl2). It should be noted that the phenotype of increased trichome number on leaves of etc2 and etc3 single mutants is generally weak, which have resulted in inconsistent results among different studies. For example, the trichome phenotype of etc2 single mutant was not detected in a few studies (Simon et al., 2007; Tominaga et al., 2008; Wang et al., 2008) but was detected in the study by Kirik et al. (2004b). Analysis of Arabidopsis natural variation population supported that ETC2 regulates trichome formation on leaves (Hilscher et al., 2009). As mentioned above, among all single mutants of R3 MYB genes, only etc1 single mutant did not display any detectable trichome phenotypes (Kirik et al., 2004a). Taken together, analysis of single mutants of R3 MYB genes suggested that among seven R3 MYBs, TRY is the predominant member controlling trichome clustering, TCL1 and TCL2 are the predominant members controlling trichome development on inflorescence stem and pedicels, and CPC, ETC2, and ETC3 mainly regulate trichome development on leaves. Table 1 summarizes the trichome and root hair phenotypes of R3 MYB single mutants and overexpression lines.
Analysis of double, triple, quadruple and higher order mutants revealed redundancy among R3 MYB genes in each of these three categories. For example, try cpc double mutants have more trichome clusters than try single mutant and have more trichomes on leaves than cpc single mutant (Schellmann et al., 2002; Kirik et al., 2004a; Wang et al., 2008). Analysis of tcl1 cpc etc1 etc3 quadruple mutants revealed that TCL1 is also involved in the regulation of trichome density on leaves (Wang et al., 2008). Analysis of tcl1 cpc double mutants revealed that CPC is also involved in the regulation of trichome formation on upper inflorescence stem and pedicles (Wang et al., 2007). try cpc etc1 tcl1 quadruple mutants form trichomes in almost all aerial parts of the plant (Wang et al., 2008) (Figure 2). Although etc1 single mutant did not display any detectable trichome phenotypes, analysis of double, triple and quadruple mutants suggested that etc1 mutation can enhance trichome phenotypes in each of those three categories (Kirik et al., 2004a; Wang et al., 2008; Wester et al., 2009).
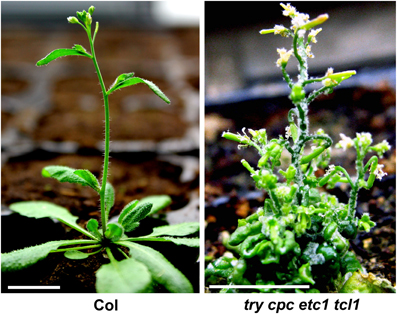
Figure 2. R3 MYB transcription factors function redundantly to control trichome formation in Arabidopsis. In Col wild type plant (left), trichomes were observed on leaves, lower part of stems and flower organs. In try cpc etc1 tcl1 quadruple mutant (right), trichomes were observed in almost all aerial parts of the plants.
Taken together, all seven R3 MYBs negatively regulate trichome formation in a largely redundant manner. Specificities have also been observed, which will be further discussed in a later section.
Function of R3 MYBs in Root Hair Development
Root hair is a tubular outgrowth of a trichoblast, a hair-forming cell on the epidermis of a plant root (Gilroy and Jones, 2000). Root hairs are formed in the region of maturation zone of the root. They are lateral extensions of a single cell and rarely branched. Root hairs help plant collect water and mineral nutrients (Gilroy and Jones, 2000). Similar to the trichome system, root hair has become an excellent system to study cell type specification (Ishida et al., 2008; Schiefelbein et al., 2009; Tominaga-Wada et al., 2011; Grebe, 2012; Ryu et al., 2013). Root hair development follows a position-dependent pattern. Epidermal cells in contact with two underlying cortical cells differentiate into hair cells (H cells; trichoblasts) whereas cells that contact only a single cortical cell differentiate into mature hairless cells (N cells; atrichoblasts) (Schiefelbein, 2000; Dolan and Costa, 2001).
The genetic control of root hair formation is remarkably similar to that of trichome initiation with all same components in the activator complex except that GL1 is replaced by another R2R3-MYB transcription factor, WEREWOLF (WER) (Lee and Schiefelbein, 1999). Recently, it was found that WER positively regulates the expression of MYB23, an R2R3 MYB, during root epidermis development (Kang et al., 2009). Similar to WER, MYB23 specifies non-hair cell type and can substitute for the function of WER. Unlike WER, MYB23 regulates its own expression, providing a positive feedback loop to reinforce cell fate decisions and root epidermal patterning (Kang et al., 2009). The TTG1-GL3/EGL3-WER activator complex induces the expression of GL2 and R3 MYBs. R3 MYBs can move from an N cell to a neighboring H cell to compete with WER for binding GL3 or EGL3, thus limiting the activity of the TTG1-GL3/EGL3-WER activator complex (Ishida et al., 2008; Schiefelbein et al., 2009; Tominaga-Wada et al., 2011; Grebe, 2012; Ryu et al., 2013). Opposing to that in the trichome initiation system, positive regulators of trichome initiation function as negative regulators of root hair formation and vice versa. Therefore, R3 MYBs are positive regulators of root hair formation.
According to this model, overexpression of R3 MYBs is expected to result in increased root hair formation whereas loss-of-function mutation results in decreased root hair formation. This is indeed the general trend. However, differences have been observed. TCL1 and TCL2 appeared to have diverged the most among R3 MYB subfamily because overexpression of TRY, CPC, ETC1, ETC2, and ETC3 rendered more root hair formation (Tominaga et al., 2008) whereas overexpression of TCL1 and TCL2 had no effect on root hair formation (Wang et al., 2007; Gan et al., 2011). Among all single mutants of R3 MYBs, only try and cpc single mutants have been consistently shown to have decreased root hair formation and cpc single mutant had stronger root hair phenotype than try mutant (Wada et al., 1997, 2002; Schellmann et al., 2002). Although etc3 single mutant was shown to display wild-type root hair phenotypes in two studies (Simon et al., 2007; Wang et al., 2008), it was shown to produce less root hairs in two other studies (Tominaga et al., 2008; Wester et al., 2009), indicating that the root hair phenotype of etc3 mutant is generally weak. Other single mutants including etc1, etc2, tcl1, and tcl2 displayed little or no root hair phenotypes. These results suggested that CPC is the predominant member of R3 MYB family controlling root hair formation.
Analysis of double, triple and quadruple mutants revealed that other R3 MYB genes may function redundantly with CPC to positively regulate root hair formation. Analysis of etc1 cpc double mutant and etc1 try cpc triple mutant revealed that ETC1 functioned redundantly with TRY and CPC to control root hair formation (Kirik et al., 2004a). Analysis of cpc etc1 etc3 tcl1 quadruple mutants indicated that TCL1 positively regulate root hair formation (Wang et al., 2008). Analysis of etc3 cpc double mutant revealed that ETC3 functioned redundantly with CPC to control root hair formation (Tominaga et al., 2008). Taken together, five of the seven R3 MYB genes have been shown to be involved in the regulation of root hair formation. So far, a role of ETC2 and TCL2 in root hair formation has not been reported.
Other Functions of R3 MYBs
In addition to regulating trichome and root hair development, R3 MYBs have been shown to be involved in the regulation of other processes including flowering, anthocyanin accumulation and stomatal formation. This is consistent with the observation that the expression of R3 MYB genes is not restricted to trichomes and root hairs. For example, TRY is also expressed in inflorescence and siliques (Schellmann et al., 2002).
etc3/cpl3 mutant was shown to flower earlier with fewer leaves than the wild-type (Tominaga et al., 2008; Tominaga-Wada et al., 2013b) although the early flowering phenotype was not reproduced by another study (Wester et al., 2009). Subsequently, it was shown that mutations in TRY or CPC delayed flowering of etc3/cpl3 mutant and a mutation in ETC1 did not further delay flowering but reduced plant size (Tominaga-Wada et al., 2013b). Mutations in ETC3/CPL3 were also shown to affect endoreduplication (Tominaga et al., 2008).
CPC was found to be a positive regulator of stomatal formation in the hypocotyl (Serna, 2008). CPC was localized in the nucleus and peripheral cytoplasm of fully differentiated epidermal cells in hypocotyl. CPC expression in differentiating stomaless-forming cells was shown to be positively regulated by TOO MANY MOUTHS (TMM), a leucine-rich repeat-containing receptor-like protein expressed in proliferative postprotodermal cells (Nadeau and Sack, 2002). Furthermore, CPC acts redundantly with TRY to promote stomata formation.
Because GL3, EGL3, and TTG1 of the activator complex are also involved in the regulation of seed coat mucilage and anthocyanin production, in addition to the regulation of trichome and root hair formation (Zhang et al., 2003), and that TTG1-GL3/EGL3-GLl and TTG1-GL3/EGL3-WER complexes activate the expression of R3 MYB genes, it raises a question of whether R3 MYBs also regulate anthocyanin and seed coat mucilage production. Molecular and genetic studies suggested that R3 MYBs regulate anthocyanin biosynthesis but not seed coat mucilage production.
CPC was shown to be a negative regulator of anthocyanin biosynthesis (Zhu et al., 2009). Overexpression of CPC repressed a total of 85 genes at the whole genome level. Of these 85 genes, seven are later anthocyanin biosynthesis genes. As discussed above, the action of R3 MYBs in the regulation of trichome and root hair development involves the TTG1-GL3/EGL3-GL1 and TTG1-GL3/EGL3-WER activator complex, respectively. The regulation of anthocyanin biosynthesis involves a similar TTG1-bHLH-MYB activator complex (Walker et al., 1999; Zhang et al., 2003; Zimmermann et al., 2004; Gonzalez et al., 2008). Unlike the trichome and root hair activator complexes, the MYB function is executed by PAP1 and PAP2. GL3/EGL3 interacts with both TTG1 and PAP1/PAP2, acting as positive regulators of anthocyanin biosynthesis (Zhang et al., 2003; Zimmermann et al., 2004). Transient expression analysis indicated that CPC competes with PAP1/PAP2 for binding with GL3/EGL3. Similarly, a tomato ortholog of TRY was also shown to negatively regulate anthocyanin accumulation (Nukumizu et al., 2013). Among all R3 MYBs, only CPC appears to act in the negative feedback on anthocyanin accumulation in response to nitrogen starvation conditions (Nemie-Feyissa et al., 2014).
For seed coat mucilage, in addition to TTG1, it requires bHLH transcription factor EGL3 and TT8, and MYB transcription factor MYB61 (Zhang et al., 2003). Analysis of single, double and higher order R3 MYB mutants did not reveal defects in seed mucilage production (Zhu et al., 2009; Wang et al., 2010), suggesting that R3 MYBs do not regulate seed mucilage biosynthesis.
Redundancy and Specificity of R3 MYBs
As discussed above, R3 MYBs can function redundantly to regulate trichome and root hair development. However, although R3 MYBs are small in size (77–112 aa) and are similar (52–82%) to each other at the amino acid level, some members exhibit distinct function than others in trichome and root hair development. For example, among all R3 MYBs, TRY is characteristic of its regulatory role in trichome clustering, CPC is characteristic of its role in root hair development, and TCL1 and TCL2 are characteristic of their roles in trichome development on the inflorescence stem and pedicels. What determines the functional specificity of each R3 MYB? In general, there are a number of attributes of functional specificity of any given genes including the expression level, spatiotemporal expression of the gene and the biochemical property of the protein (e.g., transcriptional activity, protein stability and protein subcellular localization). The expression level and pattern of a gene are largely determined by the promoter activity whereas the biochemical properties of the protein are determined by the amino acid constitution of the protein. R3 MYBs display differences in both categories.
Promoter Activity
At the transcript level, differences in tissue/organ expression of R3 MYB genes have been observed. Among all seven R3 MYB genes, ETC2 and TCL1 were not expressed in the root (Kirik et al., 2004b; Wang et al., 2007). The transcript of TCL2/CPL4 was also not detected or at very low level in the root as determined by RT-PCR (Gan et al., 2011; Tominaga-Wada and Nukumizu, 2012). These results implied that these three R3 MYB genes do not play a major role in regulating root hair formation. This view is consistent with the observation that none of etc2, tcl1, and tcl2 single mutants displayed root hair phenotypes. Therefore, the differences in tissue/organ expression patterns of R3 MYBs determine the first level of specificities. Nonetheless, all of these three genes tested were able to partially rescue the root hair phenotype of cpc mutant when their expression was driven by CPC promoter (Simon et al., 2007). These results reinforce the concept that the normal functions of these genes are restricted by their promoter activities.
Results from promoter-swap experiments supported that transcriptional regulation is important for the functional diversity of R3 MYB genes (Wester et al., 2009). When ETC3 was expressed under the promoter of ETC3, TRY or CPC, it could equally rescue the trichome phenotype of etc3 mutant (Wester et al., 2009), suggesting that the promoters of TRY, CPC, and ETC3 are interchangeable with respect to etc3 single mutant rescue. However, when these constructs (pETC3:ETC3, pTRY:ETC3, and pCPC:ETC3) were used to complement triple mutants containing etc3, qualitative differences were observed. pETC3:ETC3 construct rescued the cpc try etc3 mutant to the same extent as the try cpc mutant whereas pTRY:ETC3 and pCPC:ETC3 displayed an over-rescued phenotype resembling that of try. Differences were also observed between pTRY:ETC3 and pCPC:ETC3 in terms of trichome cluster phenotypes. These results suggest that the regulation of expression of R3 MYB genes is important for their functioning.
Protein Properties
TRY and CPC displayed indistinguishable expression patterns in leaves (Schellmann et al., 2002), yet the trichome phenotypes of try and cpc single mutants are very different. These suggested that the amino acid sequences of R3 MYBs play important roles in determining its functionality. The importance of protein properties in determining its functionality has been supported by genetic complementation studies, in particularly promoter swapping assays. For example, when TCL1 was expressed under the control of TRY or CPC promoter in try and cpc mutant background, respectively, TCL1 was only able to partially rescue the trichome and root hair phenotypes of try and cpc mutants (Wang et al., 2007), indicating that TCL1 does not function equivalently with TRY or CPC. On the other hand, the expression of TRY under the control of TRY or CPC promoter could completely rescue the trichome clustering phenotype of try mutant whereas the expression of CPC under TRY promoter could not (Pesch and Hülskamp, 2011), suggesting that the specific role of TRY in regulating trichome cluster formation is not based on its transcriptional regulation but on specific protein properties. Furthermore, it has been suggested that TRY protein has specific properties relevant in the context of both cluster formation and trichome density (Pesch and Hülskamp, 2011).
Differences in R3 MYB protein properties have also been observed in other studies. For example, by using promoter-swap experiments, Simon et al. (2007) compared the ability of several R3 MYBs to rescue the root hair phenotype of cpc mutant under CPC promoter. It was found that ETC1 possessed the best ability, followed by ETC3, TRY, and ETC2, revealing differences in protein properties.
Taken together, the functional specificity of each R3 MYB was determined both by the promoter activity of each gene and the biochemical property of each protein. Which aspect of protein properties do R3 MYBs differ? The precise answer for this question is unclear. Recent studies, however, have revealed some possibilities.
Binding strength to GL3/EGL3
During trichome patterning, two important actions of R3 MYBs at the protein level are the competition with GL1 for bindings GL3 and the movement from trichome cell to its neighboring cells. The transcriptional activity and their binding strength with bHLH transcription factors (GL3/EGL3) of R3 MYBs have been studied in the yeast system and the bi-molecular fluorescence complementation (BiFC) system (Wester et al., 2009), and in the Arabidopsis mesophyll protoplast transient expression system (Wang et al., 2008). Although these assays could not distinguish differences in protein stability, these studies indicate that differences in biochemical property do exist among members of R3 MYBs. It was found that R3 MYBs including TRY, CPC, ETC1, ETC2, and ETC3 differ in their binding strength to GL3 (Wester et al., 2009). Furthermore, by using the yeast three-hybrid system, it was found that R3 MYBs' capacity to compete with GL1 for binding to GL3 also differs with CPC being the most potent inhibitor followed by ETC1, TRY, ETC3, and ETC2 (Wester et al., 2009). Such differences in competing GL1 for binding GL3 may contribute to the functional specificities of R3 MYBs.
Cell-to-cell movement
The cell-to-cell movement motif is conserved in all R3 MYB proteins, and the cell-to-cell movement has been experimentally demonstrated for CPC and ETC3 (Kurata et al., 2005; Wester et al., 2009). It has been shown that CPC is able to move readily within the root epidermis when its expression level is high (Kang et al., 2013). CPC is capable of moving from the stele tissue in the center of the root to the outermost epidermal layer, where it can induce the hair cell fate (Kang et al., 2013). The accumulation and localization of CPC in the nuclei of H-position cells require EGL3. During trichome patterning, it was found that ETC3 protein was localized in the nucleus as well as in the cytoplasm in trichome initials whereas ETC3 protein was restricted to the nucleus in surrounding cells (Wester et al., 2009), indicating that ETC3 protein likely moves from the trichome initial into the neighboring epidermal cells. The cell-cell movement of ETC3 was further confirmed using the particle bombardment in single leaf epidermal cells (Wester et al., 2009). Mathematical analysis suggests that the mobility of the inhibitors depends on their affinity for GL3 and predicted that ETC3 moves faster than CPC (Wester et al., 2009). This prediction was experimentally validated in particle bombardment experiments (Wester et al., 2009). Therefore, the difference in cell-to-cell movement ability may also contribute to the function specificities of R3 MYB proteins.
Protein subcellular localization
At the protein level, all R3 MYBs are localized in the nucleus as expected for transcription factors but several R3MYB proteins such as TCL1 (Wang et al., 2007), CPC (Serna, 2008) and ETC3 (Wester et al., 2009) are also found to be localized to a site around plasma membrane. Recently, it has been found that subcellular localization of R3 MYBs and GL1 affects their functionality (Pesch et al., 2013). AtMYC1, a homolog of GL3 and EGL3, can regulate the intracellular localization of GL1, TRY and CPC. AtMYC1 can relocate GL1 from the nucleus into the cytoplasm and can be recruited into the nucleus by TRY and CPC. It was suggested that AtMYC1 represses the activity of TRY and CPC (Pesch et al., 2013). It remains unclear to what extent, differences in protein subcellular location of R3 MYBs may affect their functionalities.
Protein stability
It is known that the degradation of GL3 and EGL3 proteins is ubiquitin/26S proteasome-dependent (Patra et al., 2013). However, no study has been conducted to directly compare the protein stability of R3 MYBs though it was suggested that a mutation at Lysine19 (K19) of ETC2 may affect its protein stability (Hilscher et al., 2009). This amino acid is also conserved in TRY, CPC, TCL1, and TCL2, but not in ETC1 and ETC3 (Figure 1). It is unclear whether such a difference may contribute to the function specificities of R3 MYB proteins
Regulation of Expression of R3 MYB Genes
As discussed above, the trichome and root hair initiation involves feedback loop controls. In addition to inducing GL2 expression, the TTG1-GL3/EGL3-GL1 (in trichome initiation) and TTG1-GL3/EGLs-WER (in root hair formation) activator complexes also induce the expression of R3 MYB genes. Consistent with these modes, CPC has been identified as a direct target gene for WER (Koshino-Kimura et al., 2005; Ryu et al., 2005). WER protein binds three sites in the CPC promoter region, designated WBSI, WBSII/CPCMBSI and CPCMBSII, and regulates its transcription. Furthermore, both GL1 and GL3 have been shown to be recruited to the promoter region of CPC and ETC1 (Morohashi et al., 2007; Zhao et al., 2008).
Recent studies suggested the expression of R3 MYB gene may also be controlled by other mechanisms. By using an Arabidopsis protoplast transient expression system, it was found that co-transfection of GL1 or WER, with GL3 or EGL3, is required and sufficient to activate TRY, CPC, ETC1, and ETC3, but not TCL1, TCL2 or ETC2 (Wang et al., 2008; Gan et al., 2011). Furthermore, MIR156-regulated SPLs (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE), which are known to play important roles in regulating phase transition and flowering, have been shown to be able to directly activate the expression of TCL1 and TRY through binding to their promoters and that this activation is independent of GL1 (Yu et al., 2010). Reduced expression of TCL2 in 35S:MIR156 transgenic plants suggested that MIR156-targeted SPLs may also regulate the expression of TCL2 (Gan et al., 2011).
Finally, the proposed regulatory feedback loop between the activators and the inhibitors can also lead to an auto-repression of the inhibitors. Specifically, binding of R3 MYB to GL3 can displace GL1 thereby inactivating the activator complex by forming a different protein complex through the replacement of GL1 with R3 MYBs. Consequently, this creates a shortcut of the regulatory feedback and results in the repression of R3 MYB own expression.
Modes of Action of R3 MYBs
As discussed above, the mode of action of R3 MYBs in trichome and root hair development is very similar. The action of R3 MYBs in root hair patterning has been discussed in several recent review articles (Ishida et al., 2008; Schiefelbein et al., 2009; Tominaga-Wada et al., 2011; Grebe, 2012; Ryu et al., 2013; Schiefelbein et al., 2014). To avoid redundancy, here we only focus on describing the action of R3 MYBs in trichome development. Genetic and molecular studies suggested that R3 MYBs move from a trichome precursor cell to its neighboring cell to compete with GL1 for binding GL3 or EGL3, thus limiting the activity of the TTG1-GL3/EGL3-GL1 activator complex and resulting in decreased expression of GL2; this results in the inhibition of trichome formation (Figure 3). During trichome patterning, regulatory feedback loops include several important events: (1) the activation of the inhibitors by the activators, (2) the movement of inhibitors between cells, (3) the repression of the activators by the inhibitors, and (4) the auto-repression of the inhibitors. These events created differences between cells and ultimately result in a pattern of trichome and non-trichome cells. This model of action of R3 MYBs is supported both by the binding between R3 MYBs and GL3 and the cell-to-cell movement of MYBs. For example, all seven R3 MYBs have been shown to interact with GL3 in Arabidopsis protoplasts (Wang et al., 2008; Gan et al., 2011). CPC and ETC3 have been shown to be able to move from cell to cell (Kurata et al., 2005; Wester et al., 2009). Although the cell-to-cell movement of other R3 MYBs has not been experimentally demonstrated, the cell-to-cell movement motif is conserved in all seven R3 MYBs (Figure 1).
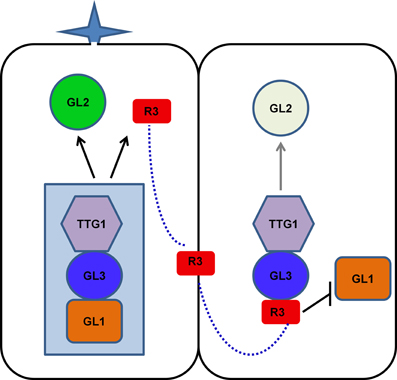
Figure 3. Model of action of R3 MYB transcription factors in controlling trichome formation. TTG1, GL3/EGL3, and GL1 form an activator complex to regulate the transcription of GL2 and R3 MYB genes. R3 MYBs can move from a trichome precursor cell (left) to its neighboring cell (right) to compete with GL1 for binding GL3 or EGL3, thus limiting the activity of the TTG1-GL3/EGL3-GL1 activator complex and resulting in decreased expression of GL2. In addition, some R3 MYBs, such as TCL1 and TCL2, can suppress the transcription of GL1. These events create differences between cells and ultimately result in a pattern of trichome and non-trichome cells. For simplicity, EGL3 is not shown in the model. R3, R3 MYBs.
Recent studies revealed that R3 MYBs also use other distinctive mechanisms to regulate trichome development. Both TCL1 and TCL2 can directly suppress the transcription of GL1 (Wang et al., 2007; Gan et al., 2011), a member of the TTG1-GL3/EGL3-GL1 activator complex. Chromatin immunoprecipitation assay indicated that TCL1 or its protein complex can directly bind the cis-acting elements that are required for the proper expression of GL1 (Wang et al., 2007). Overexpression of TCL1 resulted in suppression of GL1 and loss-of-function of TCL1 resulted in elevated expression of GL1 (Wang et al., 2007). Thus, some R3 MYBs, such as TCL1 and TCL2, have dual roles in disrupting the functionality of the activator complex: competing with GL1 for binding GL3 and suppressing the expression of GL1. The suppression of GL1 expression by R3 MYBs provides an additional regulation loop to control the activity of activator complex. However, it is unknown if this represent a general mechanism for all members of R3 MYBs. It has been shown that overexpression of CPC could also drastically suppress the expression of GL1 (Wang et al., 2007), suggesting other members of R3 MYB family could also function in a similar manner as TCL1 to directly suppress the transcription of GL1. However, it remains elusive whether such a suppression mechanism may also operate similarly during root hair patterning because overexpression of CPC was not able to change either the expression pattern or expression level of WER in roots (Song et al., 2011).
Cell fate in both trichome and root hair development is determined by competition between positive regulators and negative regulators. For both systems, models explaining pattern formation are similar. These models are largely based on a feedback loop in which the positive regulators activate the negative regulators and the negative regulators inhibit the activators, with the negative regulators being able to move between cells. Although the same machinery is central to the spatial regulation of root hair and trichome patterning, the context is different. Trichome formation can occur without a recognizable reference to other structures except for other trichomes whereas root hair patterning is position-dependent with root hairs normally form only in epidermal cells overlying a cleft between two underlying cortex cells. Such a position-dependent pattern is dependent on a leucine-rich repeat receptor-like kinase SCRAMBLED (SCM) which inhibits WER expression in the H cell position (Kwak et al., 2005; Kwak and Schiefelbein, 2007).
Since both GL2 and R3 MYB genes are activated by the same TTG1-GL3/EGL3-GL1 activator complex but having opposite roles in trichome formation, one might wonder the relationship between GL2 and R3 MYBs in regulating trichome formation. One genetic approach answering this question is to analyze double mutants and higher order mutants between gl2 and R3 MYB mutants. By doing that, it was found that R3 MYBs could still negatively regulate trichome formation in a redundant manner in gl2 mutant background (Wang et al., 2010). These studies suggested that R3 MYBs could control trichome formation in a GL2-independent manner. The implication of these studies is that trichome formation may require the expression of other genes in addition to GL2. These studies called for additional models for the regulation of trichome development.
Concluding Remarks and Future Directions
R3 MYBs are best characterized for their regulatory roles in trichome and root hair development. In the last two decades, great progress has been made to elucidate the molecular mechanism of action of R3 MYBs in cell type specification. However, several questions remained to be answered. Firstly, the regulation of expression of R3 MYB genes needs further exploration. Not all members of R3 MYB genes are regulated by the activator complex. Only the expression of TRY, CPC, ETC1, and ETC3 were found to be regulated by the TTG1-GL3/EGL3-GL3 activator complex. The expression of TCL1, TCL2, and TRY can be regulated by MIR156-mediated SPLs. Little is known about how the expression of ETC2 is regulated. When tested in protoplasts, none of the SPLs tested could activate Gal4:GUS reporter gene when recruited to the Gal4 promoter by a Gal4 DNA binding domain, suggesting that SPLs may need co-activator(s) to activate the transcription of R3 MYB genes (Gan et al., 2011). Secondly, the molecular mechanism of action of R3 MYBs in regulating trichome formation needs to be investigated further. It has been previously thought that R3 MYBs inhibit trichome formation via their competition with GL1 for binding GL3/EGL3, thus limiting the activity of the TTG1-GL3/EGL3-GL1 activator complex and resulting in the down regulation of GL2. Now it is known some R3 MYBs, such as TCL1 and TCL2, also directly suppress the expression of GL1, which also results in the down regulation of GL2 that is required for trichome formation. Adding another layer of complexity, recent results suggested R3 MYBs may regulate trichome formation in a GL2-independent manner (Wang et al., 2010). Therefore, the relationship between GL2 and R3 MYB genes in the regulation of trichome formation deserves further investigation. A comprehensive systems approach integrating genetic, genomic and computational analyses would be essential for dissecting complex regulatory networks (Bruex et al., 2012). Thirdly, the movement of R3 MYBs and components of activator complex deserves further investigation. Mathematical model suggested that the movement of both R3 MYBs and GL3 are important for root epidermal patterning (Savage et al., 2008). However, discrepancies in the movement of components of activator complex have been observed. In roots, it was reported that GL3 protein moves from the hair cells to the non-hair cells (Bernhardt et al., 2005). However, in a particle bombardment experiment using GFP fusion proteins, it was found that TRY and CPC, but not GL1 or GL3, can move between cells (Digiuni et al., 2008). In a similar particle bombardment experiment, it was shown that none of TTG1, GL3, GL1 or GL2 moves between adjacent epidermal cells whereas CPC moves to neighboring cells (Zhao et al., 2008). Furthermore, it is unclear whether the cell-to-cell movement of R3 MYB proteins is directional. If so, what controls the directional movement of R3 MYB proteins? A recent study has shed lights on the mechanism of possible directional cell-to-cell movement of R3 MYB proteins (Kang et al., 2013). Specifically, CPC is preferentially expressed in non-hair cells but acts in the root-hair cells. This directional cell-to-cell movement of CPC involves EGL3 which traps CPC protein in the root hair cells (Kang et al., 2013). Fourthly, trichome and root hair patterning involve both negative and positive feedback within and between cells. Much is known about the negative feedback mechanism in which R3 MYBs disrupt the functionality of TTG1-bHLH-MYB activator complex but less is known about the positive feedback. In root hair patterning, MYB23 functions in the positive feedback loop which can substitute the function of WER and bind to its own promoter (Kang et al., 2009). Such a positive feedback can reinforce cell fate decision and ensure robust establishment of the cell type pattern. Finally, R3 MYBs are wildly distributed in plant kingdom. It would be of great interest to examine if they regulate trichome formation in other plant species in a similar manner as that in Arabidopsis. Emerging evidence supports that this is likely the case (e.g., Tominaga-Wada et al., 2013a). Trichomes in many plant species (e.g., cotton fiber) are of great economic values. Manipulation of R3 MYBs expression can potentially contribute to the increase of productivity and quality in economic crops.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant no. 31170262), the Program for Introducing Talents to Universities (B07017) and a startup fund from Northeast Normal University to Shucai Wang, and by the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory to Jin-Gui Chen. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. Department of Energy under Contract No. DE-AC05-00OR22725. This manuscript has been authored by UT-Battelle, LLC, under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00133/abstract
Figure S1. Amino acid sequence alignment of R3 MYBs with other MYBs. Identical and similar amino acids are shaded in black and gray, respectively. The amino acids of [D/E]Lx2[R/K]x3Lx6Lx3R signature that is required for interaction between MYBs and R/B-like bHLH transcription factors are indicated by red arrowheads on the top of amino acids. The amino acids of WxM motif that is crucial for cell-to-cell movement are indicated by blue arrowhead on the top of amino acids. The pattern of primary structure of MYB repeat, Mx18Wx18W, is indicated by arrows on the bottom of amino acids.
Figure S2. Phylogenetic analysis of R3 MYBs with other MYBs. The entire amino acid sequences of single repeat R3 MYB transcription factors were used for phylogenetic analysis using tool on www.phylogeny.fr with default settings.
Figure S3. Amino acid similarity and identity of R3 MYBs and other MYBs.
Figure S4. Amino acid sequence alignment of ETC3 variants. Identical and similar amino acids are shaded in black and gray, respectively. The amino acids of [D/E]Lx2[R/K]x3Lx6Lx3R signature that is required for interaction between MYBs and R/B-like bHLH transcription factors are indicated by arrowheads on the top of amino acids. The amino acids of WxM motif that is crucial for cell-to-cell movement are indicated by arrowheads on the bottom of amino acids.
Abbreviations
CPC, CAPRICE; CPL3, CAPRICE-LIKE MYB3; CPL4, CAPRICE-LIKE MYB4; EGL3, ENHANCER OF GLABRA3; ETC1, ENHANCER OF TRY AND CPC1; ETC2, ENHANCER OF TRY AND CPC2; ETC3, ENHANCER OF TRY AND CPC3; GL1, GLABRA1; GL2, GLABRA2; GL3, GLABRA3; TCL1, TRICHOMELESS1; TCL2, TRICHOMELESS2; TRY, TRIPTYCHON; TTG1, TRANSPARENT TESTA GLABRA1.
References
Bernhardt, C., Zhao, M., Gonzalez, A., Lloyd, A., and Schiefelbein, J. (2005). The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132, 291–298. doi: 10.1242/dev.01565
Bruex, A., Kainkaryam, R. M., Wieckowski, Y., Kang, Y. H., Bernhardt, C., Xia, Y., et al. (2012). A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 8:e1002446. doi: 10.1371/journal.pgen.1002446
Chen, Y., Yang, X., He, K., Liu, M., Li, J., Gao, Z., et al. (2006). The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60, 107–124. doi: 10.1007/s11103-005-2910-y
Digiuni, S., Schellmann, S., Geier, F., Greese, B., Pesch, M., Wester, K., et al. (2008). A competitive complex formation mechanism underlies trichome patterning on Arabidopsis leaves. Mol. Syst. Biol. 4, 217. doi: 10.1038/msb.2008.54
Dolan, L., and Costa, S. (2001). Evolution and genetics of root hair stripes in the root epidermis. J. Exp. Bot. 52, 413–417. doi: 10.1093/jexbot/52.suppl_1.413
Dubos, C., Le Gourrierec, J., Baudry, A., Huep, G., Lanet, E., Debeaujon, I., et al. (2008). MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 55, 940–953. doi: 10.1111/j.1365-313X.2008.03564.x
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Eisner, T., Eisner, M., and Hoebeke, E. R. (1998). When defence backfires: detrimental effect of a plant's protective trichomes on an insect beneficial to the plant. Proc. Natl. Acad. Sci. U.S.A. 95, 4410–4414. doi: 10.1073/pnas.95.8.4410
Esch, J. J., Chen, M. A., Hillestad, M., and Marks, M. D. (2004). Comparison of TRY and the closely related At1g01380 gene in controlling Arabidopsis trichome patterning. Plant J. 40, 860–869. doi: 10.1111/j.1365-313X.2004.02259.x
Esch, J. J., Chen, M., Sanders, M., Hillestad, M., Ndkium, S., Idelkope, B., et al. (2003). A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130, 5885–5894 doi: 10.1242/dev.00812
Galway, M. E., Masucci, J. D., Lloyd, A. M., Walbot, V., Davis, R. W., and Schiefelbein, J. W. (1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166, 740–754. doi: 10.1006/dbio.1994.1352
Gan, L., Xia, K., Chen, J. G., and Wang, S. (2011). Functional characterization of TRICHOMELESS2, a new single repeat R3 MYB transcription factor in the regulation of trichome patterning in Arabidopsis. BMC Plant Biol. 11:176. doi: 10.1186/1471-2229-11-176
Gonzalez, A., Zhao, M., Leavitt, J. M., and Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Grebe, M. (2012). The patterning of epidermal hairs in Arabidopsis–updated. Curr. Opin. Plant Biol. 15, 31–37. doi: 10.1016/j.pbi.2011.10.010
Hilscher, J., Schlötterer, C., and Hauser, M. T. (2009). A single amino acid replacement in ETC2 shapes trichome patterning in natural Arabidopsis populations. Curr. Biol. 19, 1747–1751. doi: 10.1016/j.cub.2009.08.057
Hülskamp, M., Misra, S., and Jürgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–566. doi: 10.1016/0092-8674(94)90118-X
Hülskamp, M., and Schnittger, A. (1998). Spatial regulation of trichome formation in Arabidopsis thaliana. Semin. Cell Dev. Biol. 9, 213–220. doi: 10.1006/scdb.1997.0209
Ishida, T., Kurata, T., Okada, K., and Wada, T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59, 364–386. doi: 10.1146/annurev.arplant.59.032607.092949
Gilroy, S1., and Jones, D. L. (2000). Through form to function: root hair development and nutrient uptake. Trends Plant Sci. 5, 56–60. doi: 10.1016/S1360-1385(99)01551-4
Kang, Y. H., Kirik, V., Hulskamp, M., Nam, K. H., Hagely, K., Lee, M. M., et al. (2009). The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell 21, 1080–1094. doi: 10.1105/tpc.108.063180
Kang, Y. H., Song, S. K., Schiefelbein, J., and Lee, M. M. (2013). Nuclear trapping controls the position-dependent localization of CAPRICE in the root epidermis of Arabidopsis. Plant Physiol. 163, 193–204. doi: 10.1104/pp.113.221028
Katiyar, A., Smita, S., Lenka, S. K., Rajwanshi, R., Chinnusamy, V., and Bansal, K. C. (2012). Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13:544. doi: 10.1186/1471-2164-13-544
Kim, H. J., and Triplett, B. A. (2001). Cotton fibre growth in planta and in vitro: models for plant cell elongation and cell wall biogenesis. Plant Physiol. 127, 1361–1366. doi: 10.1104/pp.010724
Kirik, V., Simon, M., Hülskamp, M., and Schiefelbein, J. (2004a). The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268, 506–513. doi: 10.1016/j.ydbio.2003.12.037
Kirik, V., Simon, M., Wester, K., Schiefelbein, J., and Hülskamp, M. (2004b). ENHANCER of TRY and CPC 2(ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol. Biol. 55, 389–398. doi: 10.1007/s11103-004-0893-8
Koshino-Kimura, Y., Wada, T., Tachibana, T., Tsugeki, R., Ishiguro, S., and Okada, K. (2005). Regulation of CAPRICE transcription by MYB proteins for root epidermis differentiation in Arabidopsis. Plant Cell Physiol. 46, 817–826. doi: 10.1093/pcp/pci096
Kurata, T., Ishida, T., Kawabata-Awai, C., Noguchi, M., Hattori, S., Sano, R., et al. (2005). Cell-to-cell movement of the CAPRICEprotein in Arabidopsis root epidermal cell differentiation. Development 132, 5387–5398. doi: 10.1242/dev.02139
Kwak, S. H., and Schiefelbein, J. (2007). The role of the SCRAMBLED receptor-like kniase in patterning the Arabidopsis root epidermis. Dev. Biol. 302, 118–131. doi: 10.1016/j.ydbio.2006.09.009
Kwak, S. H., Shen, R., and Schiefelbein, J. (2005). Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307, 1111–1113. doi: 10.1126/science.1105373
Lee, M. M., and Schiefelbein, J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99, 473–483. doi: 10.1016/S0092-8674(00)81536-6
Liu, C., Zhao, Y., and Wang, Y. (2006). Artemisinin: current state and perspectives for biotechnological production of an anti malarial drug. Appl. Microbiol. Biotechnol. 72, 11–20. doi: 10.1007/s00253-006-0452-0
Lommen, W. J., Schenk, E., Bouwmeester, H. J., and Verstappen, F. W. (2006). Trichome dynamics and artemisinin accumulation during development and senescence of Artemisia annua leaves. Planta Med. 72, 336–345. doi: 10.1055/s-2005-916202
Marks, M. D. (1997). Molecular genetic analysis of trichome development in Arabidopsis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 137–163. doi: 10.1146/annurev.arplant.48.1.137
Masucci, J. D., Rerie, W. G., Foreman, D. R., Zhang, M., Galway, M. E., Marks, M. D., et al. (1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122, 1253–1260.
Matsui, K., Umemura, Y., and Ohme-Takagi, M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55, 954–967. doi: 10.1111/j.1365-313X.2008.03565.x
Mauricio, R., and Rausher, M. D. (1997). Experimental manipulation of putative selective agents provides evidence for the role of natural enemies in the evolution of plant defense. Evolution 51, 1435–1444. doi: 10.2307/2411196
Morohashi, K., Zhao, M., Yang, M., Read, B., Lloyd, A., Lamb, R., et al. (2007). Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol. 145, 736–746. doi: 10.1104/pp.107.104521
Nadeau, J. A., and Sack, F. D. (2002). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296, 1697–1700. doi: 10.1126/science.1069596
Nemie-Feyissa, D., Olafsdottir, S. M., Heidari, B., and Lillo, C. (2014). Nitrogen depletion and small R3-MYB transcription factors affecting anthocyanin accumulation in Arabidopsis leaves. Phytochemistry 98, 34–40. doi: 10.1016/j.phytochem.2013.12.006
Nukumizu, Y., Wada, T., and Tominaga-Wada, R. (2013). Tomato (Solanum lycopersicum) homologs of TRIPTYCHON (SlTRY) and GLABRA3 (SlGL3) are involved in anthocyanin accumulation. Plant Signal. Behav. 8, e24575. doi: 10.4161/psb.24575
Oppenheimer, D. G., Herman, P. L., Sivakumaran, S., Esch, J., and Marks, M. D. (1991). A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67, 483–493. doi: 10.1016/0092-8674(91)90523-2
Patra, B., Pattanaik, S., and Yuan, L. (2013). Ubiquitin protein ligase 3 mediates the proteasomal degradation of GLABROUS 3 and ENHANCER OF GLABROUS 3, regulators of trichome development and flavonoid biosynthesis in Arabidopsis. Plant J. 74, 435–447. doi: 10.1111/tpj.12132
Payne, C. T., Zhang, F., and Lloyd, A. M. (2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156, 1349–1362.
Pesch, M., and Hülskamp, M. (2004). Creating a two-dimensional pattern de novo during Arabidopsis trichome and root hair initiation. Curr. Opin. Genet. Dev. 14, 422–427. doi: 10.1016/j.gde.2004.06.007
Pesch, M., and Hülskamp, M. (2009). One, two, three...models for trichome patterning in Arabidopsis? Curr. Opin. Plant Biol. 12, 587–592. doi: 10.1016/j.pbi.2009.07.015
Pesch, M., and Hülskamp, M. (2011). Role of TRIPTYCHON in trichome patterning in Arabidopsis. BMC Plant Biol. 11:130. doi: 10.1186/1471-2229-11-130
Pesch, M., Schultheiß, I., Digiuni, S., Uhrig, J. F., and Hülskamp, M. (2013). Mutual control of intracellular localisation of the patterning proteins AtMYC1, GL1 and TRY/CPC in Arabidopsis. Development 140, 3456–3467. doi: 10.1242/dev.094698
Rerie, W. G., Feldmann, K. A., and Marks, M. D. (1994). The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8, 1388–1399. doi: 10.1101/gad.8.12.1388
Ryu, K. H., Kang, Y. H., Park, Y. H., Hwang, I., Schiefelbein, J., and Lee, M. M. (2005). The WEREWOLF MYB protein directly regulates CAPRICE transcription during cell fate specification in the Arabidopsis root epidermis. Development 132, 4765–4775. doi: 10.1242/dev.02055
Ryu, K. H., Zheng, X., Huang, L., and Schiefelbein, J. (2013). Computational modeling of epidermal cell fate determination systems. Curr. Opin. Plant Biol. 16, 5–10. doi: 10.1016/j.pbi.2012.12.003
Savage, N. S., Walker, T., Wieckowski, Y., Schiefelbein, J., Dolan, L., and Monk, N. A. (2008). A mutual support mechanism through intercellular movement of CAPRICE and GLABRA3 can pattern the Arabidopsis root epidermis. PLoS Biol. 6:e235. doi: 10.1371/journal.pbio.0060235
Schellmann, S., Hülskamp, M., and Uhrig, J. (2007). Epidermal pattern formation in the root and shoot of Arabidopsis. Biochem. Soc. Trans. 35, 146–148. doi: 10.1042/BST0350146
Schellmann, S., Schnittger, A., Kirik, V., Wada, T., Okada, K., Beermann, A., et al. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21, 5036–5046. doi: 10.1093/emboj/cdf524
Schiefelbein, J. (2003). Cell-fate specification in the epidermis: a common patterning mechanism in the root and shoot. Curr. Opin. Plant Biol. 6, 74–78. doi: 10.1016/S136952660200002X
Schiefelbein, J., Huang, L., and Zheng, X. (2014). Regulation of epidermal cell fate in Arabidopsis roots: the importance of multiple feedback loops. Front. Plant Sci. 5:47. doi: 10.3389/fpls.2014.00047
Schiefelbein, J., Kwak, S. H., Wieckowski, Y., Barron, C., and Bruex, A. (2009). The gene regulatory network for root epidermal cell-type pattern formation in Arabidopsis. J. Exp. Bot. 60, 1515–1521. doi: 10.1093/jxb/ern339
Schiefelbein, J. W. (2000). Constructing a plant cell. The genetic control of root hair development. Plant Physiol. 124, 1525–1531. doi: 10.1104/pp.124.4.1525
Schnittger, A., Folkers, U., Schwab, B., Jürgens, G., and Hülskamp, M. (1999). Generation of a spacing pattern: the role of TRIPTYCHON in trichome patterning in Arabidopsis. Plant Cell 11, 1105–1116.
Serna, L. (2005). Epidermal cell patterning and differentiation throughout the apical-basal axis of the seedling. J. Exp. Bot. 56, 1983–1989. doi: 10.1093/jxb/eri213
Serna, L. (2008). CAPRICE positively regulates stomatal formation in the Arabidopsis hypocotyl. Plant Signal. Behav. 3, 1077–1082. doi: 10.4161/psb.3.12.6254
Simon, M., Lee, M. M., Lin, Y., Gish, L., and Schiefelbein, J. (2007). Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev. Biol. 311, 566–578. doi: 10.1016/j.ydbio.2007.09.001
Song, S. K., Ryu, K. H., Kang, Y. H., Song, J. H., Cho, Y. H., Yoo, S. D., et al. (2011). Cell fate in the Arabidopsis root epidermis is determined by competition between WEREWOLF and CAPRICE. Plant Physiol. 157, 1196–1208. doi: 10.1104/pp.111.185785
Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/S1369-5266(00)00199-0
Tominaga, R., Iwata, M., Sano, R., Inoue, K., Okada, K., and Wada, T. (2008). Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development 135, 1335–1345. doi: 10.1242/dev.017947
Tominaga-Wada, R., Ishida, T., and Wada, T. (2011). New insights into the mechanism of development of Arabidopsis root hairs and trichomes. Int. Rev. Cell Mol. Biol. 286, 67–106. doi: 10.1016/B978-0-12-385859-7.00002-1
Tominaga-Wada, R., and Nukumizu, Y. (2012). Expression analysis of an R3-Type MYB transcription factor CPC-LIKE MYB4 (TRICHOMELESS2) and CPL4-related transcripts in Arabidopsis. Int. J. Mol. Sci. 13, 3478–3491. doi: 10.3390/ijms13033478
Tominaga-Wada, R., Nukumizu, Y., Sato, S., and Wada, T. (2013a). Control of plant trichome and root-hair development by a tomato (Solanum lycopersicum) R3 MYB transcription factor. PLoS ONE 8:e54019. doi: 10.1371/journal.pone.0054019
Tominaga-Wada, R., Nukumizu, Y., and Wada, T. (2013b). Flowering is delayed by mutations in homologous genes CAPRICE and TRYPTICHON in the early flowering Arabidopsis cpl3 mutant. J. Plant Physiol. 170, 1466–1468. doi: 10.1016/j.jplph.2013.05.013
Wada, T., Kurata, T., Tominaga, R., Koshino-Kimura, Y., Tachibana, T., Goto, K., et al. (2002). Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129, 5409–5419. doi: 10.1242/dev.00111
Wada, T., Tachibana, T., Shimura, Y., and Okada, K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277, 1113–1116. doi: 10.1126/science.277.5329.1113
Wagner, G. J., Wang, E., and Shepherd, R. W. (2004). New approaches for studying and exploiting an old protuberance, the plant trichome. Ann. Bot. 93, 3–11. doi: 10.1093/aob/mch011
Walker, A. R., Davison, P. A., Bolognesi-Winfield, A. C., James, C. M., Srinivasan, N., Blundell, T. L., et al. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11, 1337–1350.
Wang, S., Barron, C., Schiefelbein, J., and Chen, J. G. (2010). Distinct relationships between GLABRA2 and single-repeat R3 MYB transcription factors in the regulation of trichome and root hair patterning in Arabidopsis. New Phytol. 185, 387–400. doi: 10.1111/j.1469-8137.2009.03067.x
Wang, S., Hubbard, L., Chang, Y., Guo, J., Schiefelbein, J., and Chen, J. G. (2008). Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol. 8:81. doi: 10.1186/1471-2229-8-81
Wang, S., Kwak, S. H., Zeng, Q., Ellis, B. E., Chen, X. Y., Schiefelbein, J., et al. (2007). TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 134, 3873–3882. doi: 10.1242/dev.009597
Werker, E. (2000). Trichome diversity and development. Adv. Bot. Res. 31, 1–35. doi: 10.1016/S0065-2296(00)31005-9
Wester, K., Digiuni, S., Geier, F., Timmer, J., Fleck, C., and Hülskamp, M. (2009). Functional diversity of R3 single-repeat genes in trichome development. Development 136, 1487–1496. doi: 10.1242/dev.021733
Yu, N., Cai, W. J., Wang, S., Shan, C. M., Wang, L. J., and Chen, X. Y. (2010). Temporal control of trichome distribution by MicroRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 22, 2322–2335. doi: 10.1105/tpc.109.072579
Zhang, F., Gonzalez, A., Zhao, M., Payne, C. T., and Lloyd, A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130, 4859–4869. doi: 10.1242/dev.00681
Zhao, M., Morohashi, K., Hatlestad, G., Grotewold, E., and Lloyd, A. (2008). The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135, 1991–1999. doi: 10.1242/dev.016873
Zhu, H. F., Fitzsimmons, K., Khandelwal, A., and Kranz, R. G. (2009). CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol. Plant 2, 790–802. doi: 10.1093/mp/ssp030
Keywords: CPC, GL1, GL2, GL3, R3 MYB, root hair, trichome, TRY
Citation: Wang S and Chen J-G (2014) Regulation of cell fate determination by single-repeat R3 MYB transcription factors in Arabidopsis. Front. Plant Sci. 5:133. doi: 10.3389/fpls.2014.00133
Received: 19 February 2014; Paper pending published: 10 March 2014;
Accepted: 20 March 2014; Published online: 08 April 2014.
Edited by:
John Schiefelbein, University of Michigan, USAReviewed by:
Hyung-Taeg Cho, Seoul National University, South KoreaMyeong Min Lee, Yonsei University, South Korea
Copyright © 2014 Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin-Gui Chen, Biosciences Division, Oak Ridge National Laboratory, 1 Bethel Valley Rd., Oak Ridge, TN 37831, USA e-mail: chenj@ornl.gov
 Shucai Wang
Shucai Wang Jin-Gui Chen
Jin-Gui Chen