- 1Department of Biology and Chemistry, Fitchburg State University, Fitchburg, MA, USA
- 2Plant Sciences, Bond Life Sciences Center, The University of Missouri, Columbia, MO, USA
- 3Department of Biochemistry and Molecular Biology, Department of Chemistry, Michigan State University, East Lansing, MI, USA
Plant responses to insects and wounding involve substantial transcriptional reprogramming that integrates hormonal, metabolic, and physiological events. The ability to respond differentially to various stresses, including wounding, generally involves hormone signaling and trans-acting regulatory factors. Evidence of the importance of transcription factors (TFs) in responses to insects is also accumulating. However, the relationships among hormone signaling, TF activity, and ability to respond specifically to different insects are uncertain. We examined transcriptional and hormonal changes in Arabidopsis thaliana after herbivory by larvae of two lepidopteran species, Spodoptera exigua (Hübner) and Pieris rapae L. over a 24-h time course. Transcriptional responses to the two insects differed and were frequently weaker or absent in response to the specialist P. rapae. Using microarray analysis and qRT-PCR, we found 141 TFs, including many AP2/ERFs (Ethylene Response Factors) and selected defense-related genes, to be differentially regulated in response to the two insect species or wounding. Jasmonic Acid (JA), JA-isoleucine (JA-IL), and ethylene production by Arabidopsis plants increased after attack by both insect species. However, the amounts and timing of ethylene production differed between the two herbivory treatments. Our results support the hypothesis that the different responses to these two insects involve modifications of JA-signaling events and activation of different subsets of ERF TFs, resulting in different degrees of divergence from responses to wounding alone.
Introduction
Plant responses to insects and wounding are complex, involving differential perception, multiple signaling pathways, and extensive transcriptional reprogramming (Delessert et al., 2004; DeVos et al., 2005; Rehrig et al., 2011). Perception of insect attack by plants is thought to occur at the site of herbivory via damage- or herbivore-associated molecular patterns (DAMPS or HAMPs, respectively). DAMPs are plant-derived signals produced after wounding and include ATP, Volatile Organic Compounds or elicitors in cell wall fragments (reviewed by Kaku et al., 2006; Alborn et al., 2007; Heil, 2009; Pieterse et al., 2012). Plants also respond to HAMPs found in insect oral secretions (OS) including fatty-acid/amino acid conjugates and B-glucosidase (Mattiacci et al., 1995; Pare and Tumlinson, 1999; Schmelz et al., 2006; Wu and Baldwin, 2009). Insect feeding patterns may further influence this response (Wittstock et al., 2004; McCartney, 2007). Although the mechanisms are uncertain, it is suggested that plants can identify their attacker and activate species-specific defenses (DeVos et al., 2005; Mewis et al., 2005; Vogel et al., 2007; Galis et al., 2009; Stork et al., 2009). Yet, how hormone concentrations and other important components of plant signaling pathways are interpreted and integrated by the plant to activate appropriate responses after wounding or herbivory by different insects remains unclear.
The plant hormones jasmonic acid (JA), salicylic acid (SA), abscisic acid (ABA), and ethylene (ET) are among the critical players in the events following abiotic and biotic stress, including insect attack. Hormones also appear to modulate the fine-tuning of defenses in response to different insects (Reviewed by Zhu-Salzman et al., 2004; DeVos et al., 2005; Mewis et al., 2005; Thompson and Goggin, 2006; Wasternack and Hause, 2013). The mechanisms of pathway control and integration in plant immunity also involve the hormones ABA (Abe et al., 2003; Bodenhausen and Reymond, 2007), auxin (Grunewald et al., 2009), gibberellic acid (GA) (Hou et al., 2010), and cytokinins (Choi et al., 2011) which interact with stress hormones in a complex network to maximize survivorship of the plants (reviewed by Pieterse et al., 2012). Some of the first signaling events following perception of insect attack are the rapid accumulation and transport of JA and a quick “burst” of ET production (reviewed by Reymond and Farmer, 1998; Winz and Baldwin, 2001; Thaler et al., 2002; Babst et al., 2005; Wu and Baldwin, 2009). The release of ethylene after wounding and herbivory depends on plant species being attacked and the herbivore species involved but is generally greater in response to herbivory than to wounding (Von Dahl and Baldwin, 2007). Von Dahl et al. (2007) hypothesized that ethylene may attenuate or fine-tune responses based on its interaction or cross talk with other phytohormones. Larvae of Pieris rapae grew poorly on mutant plants with compromised ethylene signaling, which was associated with an increase in JA-inducible indolyl glucosinolates (Mewis et al., 2006). Studies conducted by Bodenhausen and Reymond (2007) and Stotz et al. (2000) showed that Arabidopsis plants defective in ethylene signaling had increased resistance to Spodoptera littoralis. On the other hand, DeVos et al. (2006) found that ethylene production after P. rapae feeding primed plants for increased resistance to future viral infection.
It is well-established that JA and its amino acid conjugate, JA-isoleucine (JA-IL) are critical players in plant responses to insects as well as wounding (Chini et al., 2007; Thines et al., 2007). Plants with mutations in the JA signaling pathway such as coi1 and jar1 have increased susceptibility to insect attack (Thaler et al., 2002; Mewis et al., 2005; Bodenhausen and Reymond, 2007; Verhage et al., 2011). ET modulates JA-signaled defenses (Lorenzo et al., 2004) and SA often inhibits them (Beckers and Spoel, 2006). One goal of this study was to elucidate how the interactions among JA, SA, and ET production may influence transcriptional responses to attack by different insects.
Transcription factors (TFs) provide a likely mechanism for translating hormone signaling into the subsequent activation of defense genes differentially expressed in response to attack by different insect species. For example, the activation of TFs in the WRKY and APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) families has been shown to be required for differential responses in planta to JA treatment, pathogenesis, herbivory, and wounding (Delessert et al., 2004; Reymond et al., 2004; Lu et al., 2011). Additional transcriptional responses to wounding, JA treatment, or JA and ET often include the up-regulation of genes involved in plant defense such as PDF1.2, VSP2, LOX2, and chitinases (reviewed by Boter et al., 2004; Bodenhausen and Reymond, 2007; Pieterse et al., 2012; Wasternack and Hause, 2013), which are largely under transcriptional control of ERFs and the ABA/JA responsive-TF, MYC2 (reviewed by Brown et al., 2003; Lorenzo and Solano, 2005; Dombrecht et al., 2007; reviewed by Wasternack and Hause, 2013). Because ERFs, members of the MYC family and other TFs serve as points of cross talk between the JA, ET, and SA hormone pathways (Li et al., 1999; Abe et al., 2003; Lorenzo et al., 2003; Yadav et al., 2005; Robert-Seilaniantz et al., 2011; Verhage et al., 2011; Schweizer et al., 2013), they are likely to be critical players in responses to different biotic stresses, including insect herbivory.
Using microarray analysis and qRT-PCR, we identified and quantified transcripts whose expression in Arabidopsis thaliana (Columbia) was altered in attacked (local) and unattacked (systemic) tissues after feeding by S. exigua and P. rapae. We hypothesized that the two insects elicit different patterns of hormone production, differential expression of TF genes, and differential expression of selected defense-related genes. We identified several unique transcriptional patterns within the AP2-ERF gene family and associated this with both increased JA and ET elicitation and differential regulation of defense-related gene expression at different times after feeding by the 2 different insects.
Materials and Methods
Plants and Insect Care
Eggs of the caterpillar Spodoptera exigua Hübner (Noctuidae) were obtained from Benzon Research (Carlisle, PA, USA) and larvae were reared on artificial diet (Bioserv, Frenchtown, NJ, USA). The caterpillar Pieris rapae L. (Pieridae) was maintained as a culture on pak-choi and originated from the Carolina Biological Supply Company (North Carolina). Both caterpillar species were transferred to Arabidopsis thaliana (L.) ecotype Columbia (Col-0) plants 1 day before the experiments to acclimate to the new host. Col-0 seeds were vernalized in 2% agar and sown into 6 × 5 cm pots containing sterile Metromix 200 soil (Sun Gro Horticulture) and Osmocote. Plants were chamber grown at 22 ± 1°C, 65 ± 5% relative humidity, and 200 μmol m−2s−1 light intensity on a short-day (8:16 (L:D)) photoperiod to delay flowering and keep plants in the rosette stage. Plants were watered as needed.
Insect and Wounding Treatments
Insect treatments for the microarray analysis were conducted as described by Appel et al. (2014), (this edition). Control plants were grown in the same conditions as insect-treated plants except cages without insects were placed on them. Experiments were designed to minimize circadian influences and capture gene expression in full-rosette leaves. Two to three second- and third-instar S. exigua and P. rapae caterpillars were allowed to feed on 5–6 week old plants until 20–30% of the leaf area of 4 leaves was removed. Caterpillars were kept in custom-made soft cages, which were checked periodically for plant damage. To maximize damage in minimal time (<30–45 min), caterpillars that were not eating were replaced with ones that were more cooperative. Furthermore, caterpillars were withheld food for 24 h to encourage eating. This technique was used so that we could observe early responses to caterpillars, which would be missed if an aggressive time line were not imposed. For example, plants with insects imposing 20–30% damage quickest were used for the 15-min and subsequent samples, respectively. Control and treatment plants were kept in the same experimental area. Experiments with each insect were conducted on separate days and were repeated 3 times. Pictures of damaged and control plants can be found in the Supplementary Material. Once sufficient damage was achieved, caterpillars were removed and the plants were returned to the growth chamber until processed for RNA or hormone analysis. For wounding treatments, insect damage was simulated using a sterile damage wheel across both sides of the mid-rib on 6 leaves. Leaves were harvested for gene expression or hormone analysis beginning at 15 min up until 48 h after cage removal or wounding. Four plants were used for each bioreplicate and four bioreplicates were collected per treatment. We harvested the four treated leaves on each plant for different assays: two for RNA tissue, one for JA/SA measurement, and one for an initial ethylene analysis. RNA and JA/SA sample leaves were weighed, flash frozen in grinding tubes immersed in liquid nitrogen, and then stored at −80°C. Ethylene samples were processed immediately.
Microarray Analysis and Transcription Factor Identification
Plants were harvested and RNA was isolated for microarray analysis as described by Appel et al. (2014). Briefly, RNA was isolated using the TRIZOL method (Invitrogen, Carlsbad, CA, USA) and its quality was determined by an Agilent 2100 Bioanalyzer. RNA was reverse transcribed into labeled cDNA using a T17 primer, dNTPs and Cyanidin-3 and 5 dUTPs. cDNA was purified and hybridized to an Operon v1 microarray chip with 26,090 Arabidopsis gene specific 70-mer oligonucleotides. Four replicate chips were used for each treatment. Analysis of the data, including statistics and identification of false positives was done using the methods described by Ehlting et al. (2005, 2008), Storey and Tibshirani (2003), and Pylatuik and Fobert (2005). To analyze the microarray data for putative TFs, we conducted a literature search and used the online databases, GenBank, Gene Annotation tool (GO) from the TAIR website (www.Arabidopsis.org) and DATF (Database of Arabidopsis TFs; Guo et al., 2005). A complete list of all Transcription Factor gene names and abbreviations can be found in Table S3.
Gene Expression via Real Time qRT-PCR
The expression of ERFs, MYC2 (JIN1), Housekeeping genes, and 6 defense-related marker genes was measured by semi-quantitative Real-Time PCR. A list of genes and primers can be found in Table S1. Total RNA from insect-attacked and control tissue samples was extracted using Sigma Total Plant RNA kits (STRN50, St. Louis, MO, USA) or the TRIZOL method (microarray experiment only). The same RNA used for the microarray was used for qPCR for the initial ERF analysis to confirm the array results. Otherwise, RNA was isolated from insect-treated plant material from subsequent experiments. RNA quality was confirmed using a Bio-Rad Experion automated electrophoresis system (Hercules, CA, USA) and a Bio-Rad RNA standard sensitivity kit which adequately detects and quantifies nanogram levels of RNA.
Primers were designed and tested using methods described in Rehrig et al., 2011. We used Primer 3 Software (Rozen and Skaletsky, 2000) and Invitrogen's Vector NTI Software (Carlsbad, CA, USA) as well as IDT's on-line tool, OligoAnalyzer for further prediction of primer dimers. All primers were BLASTed in NCBI to ensure specificity of amplification. We performed gel electrophoresis of PCR products and detected single bands of expected size. Additionally, melting curve analysis of all PCR products was done via real-time PCR. All PCR products were sequenced to ensure that only gene products of interest were being amplified.
We treated samples with Turbo DNAse (Ambion, Austin, TX, USA) according to the manufacturer's specifications. RNA quantity after DNAse treatment was measured using a NanoDrop (ThermoScientific, Wilmington, DE, USA) in triplicate for each sample immediately before the reverse transcription reaction. We followed the protocol for Invitrogen's Superscript III 2-step qRT-PCR kit with Platinum SYBR Green qPCR Super-Mix UDG (Carlsbad, CA, USA) with minor modifications. To acquire sufficient amounts of cDNA for all of the subsequent real time PCR reactions, 4 reverse transcription reactions were performed for each RNA sample. These were done in 96-well plates and the volumes of 4 technical replicates for each sample were pooled, sub-sampled for a standard curve mix, and diluted 5×.
All PCR reactions were run in 96-well plates. Each bioreplicate was run in triplicate. Five mL of cDNA template, 5 mM primer pair mixes, molecular-grade water, and Platinum SYBR Green for a total of 20 mL was used for PCR. Amplification was then conducted under the following conditions on a MJ Research Opticon 2 DNA Engine: 50°C UDG treatment for 2 min, 95°C denaturation for 2 min, followed by 40 cycles of 95°C denaturation for 15 s, 56°C annealing for 30 s and 72°C extension for 30 s. After extension, but prior to fluorescence measurement reads, the temperature was ramped to approximately 1.5–2.0°C below the gene product melting curve start (Tm, –dl/dT min). A final 5 min extension at 72°C followed by a complete melting curve analysis from 72 to 95°C were then conducted.
Data Analysis of qRT-PCR Data
qRT-PCR data were acquired using the standard curve method (Larionov et al., 2005). All data were initially analyzed using Opticon 3 Monitor Software. We used LinReg PCR (Ramakers et al., 2003) to identify a value for the threshold of fluorescence. We entered this value into the Opticon Software Program, which automatically calculated expression values from the Ct values based on the regression equation of the standard curve. Expression values for 24-h data from RNA from the microarray analysis were normalized against the geometric mean of 18S and G6PD5. Because a suitable housekeeping (HK) gene could not be found for the 6-h data, all 6-h expression levels were normalized to the total amount of cDNA in the PCR reaction using a correction factor. Because we had experimentally demonstrated that HK genes were inappropriate normalization factors for measuring gene expression after insect attack, later experiments with defense-related gene expression were normalized to the expression of an exogenous Luciferase RNA spike added prior to reverse transcription (Rehrig et al., 2011). Outliers for qRT-PCR and hormone measurements were identified using a one-pass Extreme Studentized Deviate (ESD) test (Pillai and Tienzo, 1959) and eliminated from the analyses. Statistically significant differences in final gene expression ratios between treatments and controls for both the P. rapae and the S. exigua experiment were identified using the PROC NPAR1WAY command in SAS and Kruskal-Wallis analyses (SAS Institute, Cary, NC, USA). Gene expression data displayed in Figure 6 were transformed using the Log2 values of fold changes. A Hierarchal cluster analysis was done using the Spearman Rank Correlation feature in the software Cluster 3.0 (Eisen et al., 1998) and monitored with Java TreeView 1.1.3 (Saldanha, 2004).
Ethylene Measurements
For ethylene analysis, additional experiments were conducted using similar methods as described except the time course was extended to include the original time points (15 min, 30 min, 1 h, 2 h, 6 h, 24 h) as well as 12, 36, 48, and 72 h treatments. Four leaves from 1 plant (1 bioreplicate) from either insect-attacked or control plants were placed in sealed 10cc glass vials and allowed to incubate for 30–90 min. Four bioreplicates were taken for each treatment. Air was then drawn off using a 5cc syringe and manually injected into an HP Gas Chromatograph. ET levels were calculated using a regression equation of a standard curve and corrected for fresh weight and incubation time. To identify differences in ethylene, JA, JA-IL, and SA levels among treatments, we conducted an ANOVA in SAS (Cary, NC, USA). Statistically significant differences between treatments at a p-value of 0.05 or lower were determined using the PROC GLM command and post-hoc Tukey values.
JA and JA Conjugate Measurements
JA and JA-IL levels were quantified using an ethyl acetate extraction method in conjunction with HPLC/MS similar to that described in Chung et al. (2008). Briefly, samples (approximately 150 mg tissue) were frozen in liquid nitrogen and hormones were extracted using 1 mL of extraction solvent (80:20 methanol:water + 0.1% formic acid) for 18 h at −20 C. Extracts were then centrifuged (10,000 × g for 10 min at 4°C) and the supernatant was transferred to autosampler vials. Five μL of each supernatant were injected into a Waters UPLC BEH C18 column (2.1 × 50 mm; 1.7 μm particles) held at 50°C on a Waters (Milford, MA, USA) Acquity ultraperformance liquid chromatography (UPLC) system that was coupled to a Waters Quattro Premier XE tandem quadrupole mass spectrometer. Separation was performed using a linear gradient based upon 0.15% aqueous formic acid (A) and methanol (B) over a 3-min program using a total flow rate of 0.4 mL/min. Quantification of JA and SA was performed using electrospray ionization in negative-ion mode using multiple reaction monitoring (MRM), using m/z 209 ≥ 59 for JA, m/z 322 ≥ 130 for JA-IL (Chung et al., 2008), and m/z 137 ≥ 93 for SA (Zeng et al., 2011). Peak areas were integrated, and calibration curves generated, using Waters QuanLynx software.
Insect Feeding and Glucosinolate Assays
Insect feeding bioassays with P. rapae and S. exigua were conducted separately. Insects were weighed before and after feeding. One insect was placed on one plant (N = 12−31) of either WT or erf104, erf105, erf5, erf6 mutant plants (Shuqun Zhang, personal communication) and enclosed using customized plastic cages with mesh lids. Control plants from each genotype were also enclosed in cages, but received no insect treatments. Plants were placed under growing lights under 12 h days and insects were allowed to feed for 24–48 h, leaving some tissue for GS analysis. Growth rates were calculated according to actual time spent feeding on the plants. The performance of the insects was determined by a suite of nutritional indices that describe the consumption, growth, and efficiency with which food is converted to growth (Slansky and Scriber, 1985). Relative Growth Rates (RGR), Relative Consumption Rates (RCR), and Efficiency of Conversion Indices (ECI) were calculated (see Supplementary Material). Any insects that died, pupated or molted during the experiment were eliminated from the analysis. The amount of tissue eaten was determined using a digital phenotyping protocol described by Green et al. (2012). Total indolyl and aliphatic glucosinolate content in the remaining tissue was measured using a method described by Mewis et al. (2006). Insect-attacked plants were used to determine induced GS levels, while control plants served as baselines for constitutive levels.
Results
Overall Patterns of Transcription Factor Expression in Response to Insects or Wounding
Of the approximately 1500 putative TFs in the Arabidopsis genome (Riechmann et al., 2000), we identified 141 genes encoding TFs whose expression was statistically significantly altered by wounding, P. rapae or S. exigua attack (Table S2). The TFs differentially expressed in response to insects or wounding represented 25 of the 50 families characterized in the AgrisTF Database (Davuluri et al., 2003). Families with the most members represented were the AP2-ERF/RAV (18), MYB (18) Homeobox (11), bHLH (10), and NAC (9) as well as ZIM-Related Proteins/JAZ (7). The two caterpillars and wounding elicited different expression patterns of TFs (Table S4). This is most prominently seen in the AP2/ERF Family where only RAV2 was affected by both insects, and its expression was increased by S. exigua feeding and decreased by P. rapae feeding. Similarly, ZAT10, ZAT5, ZAT12, WIP4, and AZF3, members of the C2H2 transcription factor family, were up-regulated by S. exigua or wounding, but not by P. rapae in any tissue or treatment. Only wounding caused a change in the expression of LOB genes, which are involved in organ development (Husbands et al., 2007). Genes in JAZ family, which are well-documented to be JA-responsive (Chung et al., 2008) were widely elicited by both insects and wounding. Although JAZ proteins are not true TFs, they strongly interact with MYC2 and other important transcriptional regulators such as NINJA and TPL within the nucleus to control the expression of JA-induced genes (Pauwels et al., 2010). Therefore, they were included in our analysis.
We confirmed the contrasts for AP2/ERF TF genes with qRT-PCR on 17 of the 24 affected AP2/ERF TFs (including 2 RAV genes). We were able to statistically validate 20/26 of the microarray expression values for the ERFs; and, if we include all instances where qRT-PCR values were in the same direction as those of the microarray (i.e., similar to Northern blots), we achieved over 92% confirmation (Table 1). qRT-PCR data in conjunction with array data clearly show that TINY2 is responsive to only P. rapae, whereas SIMRAP2.4, ORA47, ERF11, and ERF104 were solely responsive to S. exigua feeding.
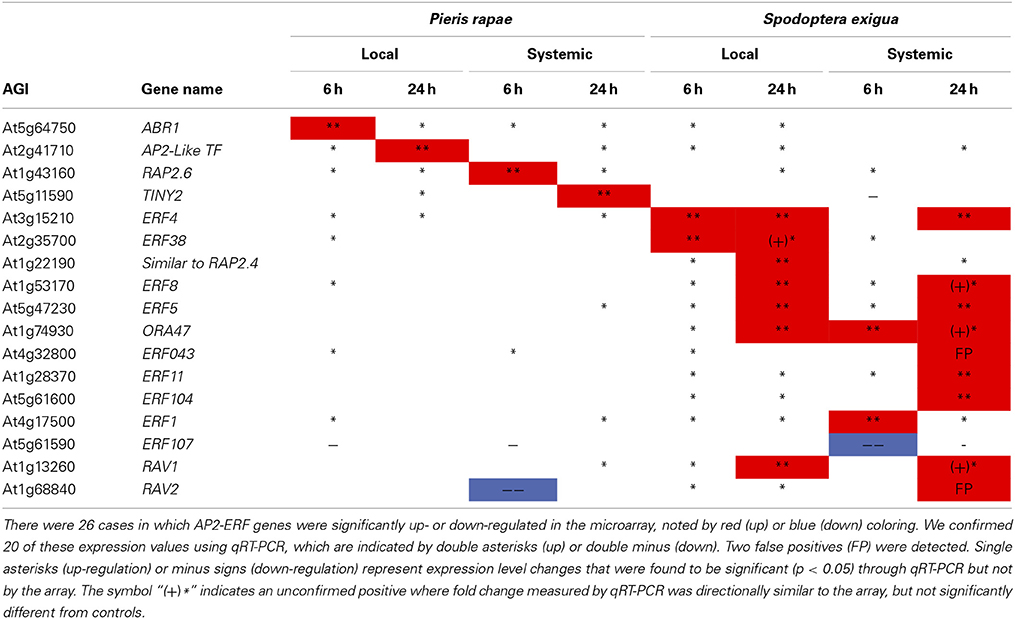
Table 1. qRT-PCR confirmation of AP2-ERF transcription factor genes identified by microarray analysis-ERF transcription factor genes affected by P. rapae and S. exigua in the array were amplified and quantified using qRT-PCR.
Insect Elicitation of Ethylene Release
The TF expression results led us ask whether insects induce ethylene production as a potential signaling mechanism in Arabidopsis. We used gas chromatography (GC) to measure ethylene levels emitted by locally attacked tissue at selected time points. Both P. rapae and S. exigua feeding induced the production of ethylene, but the times when levels became significantly different from controls differed. S. exigua induced significantly higher levels of ethylene than controls in Arabidopsis tissue by 30 min, while ethylene production by P. rapae -attacked plants did not differ from controls until after 2 h (Figure 1). Moreover, P. rapae-induced ethylene production remained higher than controls at later time points. Our results indicate that ethylene production induced by S. exigua occurs as a rapid burst shortly after the insect feeds on the plant, while ethylene production after P. rapae feeding is delayed. Ethylene levels in our analysis are lower than what has been typically seen with necrotroph and pathogen infestation (Penninckx et al., 1998; DeVos, 2006; Mur et al., 2009) or compared with other herbivore studies (DeVos et al., 2005). This may be due to our sampling technique in which damaged leaves were kept on the plants until their harvest periods during the time course. Therefore, ET was not allowed to accumulate in the headspace of the collection vials during the recovery period after herbivore removal.
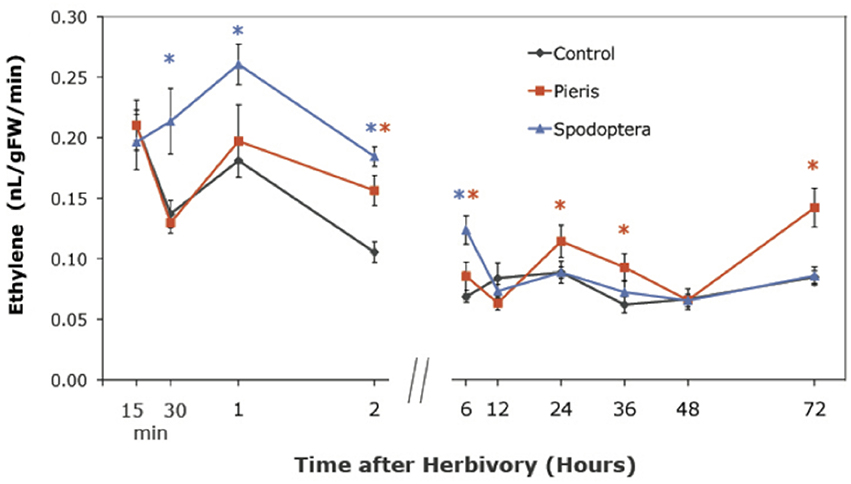
Figure 1. Ethylene production in WT Arabidopsis plants after short-term P. rapae and S. exigua feeding over a 72-h time course-Ethylene was measured by Gas Chromatography as nanoliters/gram fresh weight/minutes of incubation time. Asterisks represent data points that are significantly different than controls as determined by the GLM method in SAS (p < 0.05).
Insect Elicitation of Jasmonic Acid, Jasmonic Acid-Isoleucine, and Salicylic Acid
JA-IL and SA levels were measured using UPLC-MS/MS. We found no significant increases in SA in response to insect treatments (Supplementary Material, Figure 1). However, relative levels of JA (Figure 2) and JA-IL (Figure 3) in insect-treated plants as compared to controls varied between time points. Absolute levels of JA and JA-IL induction can be seen in the Supplementary Material (Figures S2, S3). S. exigua elicited a statistically significant increase in JA above controls at 0.5, 2, and 6 h after feeding. The higher mean at 1 h was not statistically significant, probably due to the larger standard deviation at this time point (p = 0.1104). In response to P. rapae, JA levels increased significantly immediately (after 15 min) and remained above controls until 24 h after treatment. Patterns of JA-IL production after S. exigua and P. rapae feeding matched those of JA.
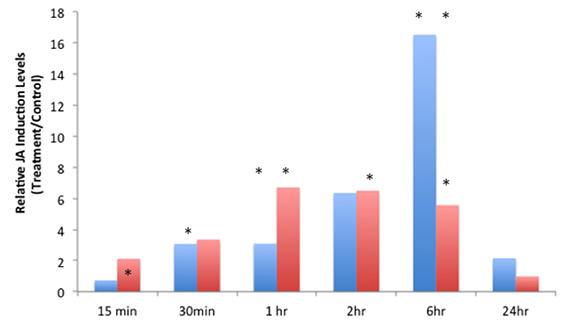
Figure 2. Relative Jasmonic Acid levels in WT Arabidopsis plants after S. exigua and P. rapae feeding over a 24-h time course-JA was measured by UPLC-MS/MS as pmol/g fresh weight. Blue bars represent JA levels in S. exigua treatments/controls. Red bars in represent JA levels in P. rapae treatments/controls. Asterisks represent time points where JA levels in the treatment plants were significantly greater than controls as determined by the GLM method in SAS (p < 0.05).
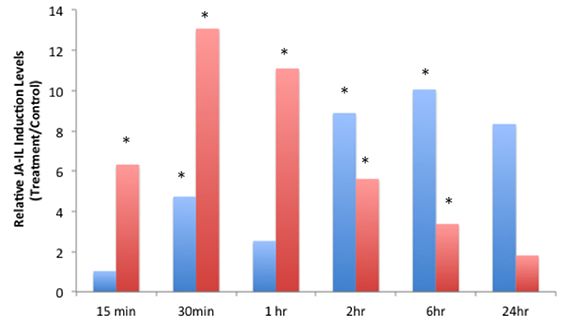
Figure 3. Relative Jasmonic-isoleucine (JA-IL) Acid levels in WT Arabidopsis plants after S. exigua and P. rapae feeding over a 24-h time course-JA-IL was measured by UPLC-MS/MS as pmol/g fresh weight. Blue bars represent JA-IL levels in S. exigua treatments/controls. Red bars in represent JA-IL levels in P. rapae treatments/controls. Asterisks represent time points where JA-IL levels in the treatment plants was significantly greater than controls as determined by the GLM method in SAS (p < 0.05).
ERF and Defense Gene Expression
To further understand the role of ET and JA signaling in response to feeding by caterpillars of the same two lepidopteran species, we measured differences in the expression of genes encoding ERF TFs and defense-related genes. We monitored gene expression patterns of ERFs and down-stream defense genes through time (Figure 4). Plants exposed to S. exigua feeding showed dramatic transcriptional responses. In general, the expression of both ERFs and defense-related genes was greater after feeding by S. exigua than by P. rapae. Only three TFs exhibited greater transcriptional changes in P. rapae-attacked tissue, namely ORA59, ERF5, and AtERF, all occurring at 15 min after treatment (p-value < 0.08). Every gene measured except MYC2, which is a JA-responsive gene, responded more strongly to S. exigua than to P. rapae at a given time point. This was especially true for ERF104, ERF8, PR4, PR3, and ERF11, whose expression increased in response to S. exigua, but declined in response to P. rapae. This suggests that these genes are important for the perception of, and response to, S. exigua, but not P. rapae. When gene expression levels were clustered using Cluster 3.0 (Eisen et al., 1998) by time point and treatment, most S. exigua (“Spod”) treatments clustered separately from P. rapae (“Pieris”) treatment, except in the case of MYC2 and PR4, indicating that defense gene responses to the two chewing insects are markedly different (Figure 5).
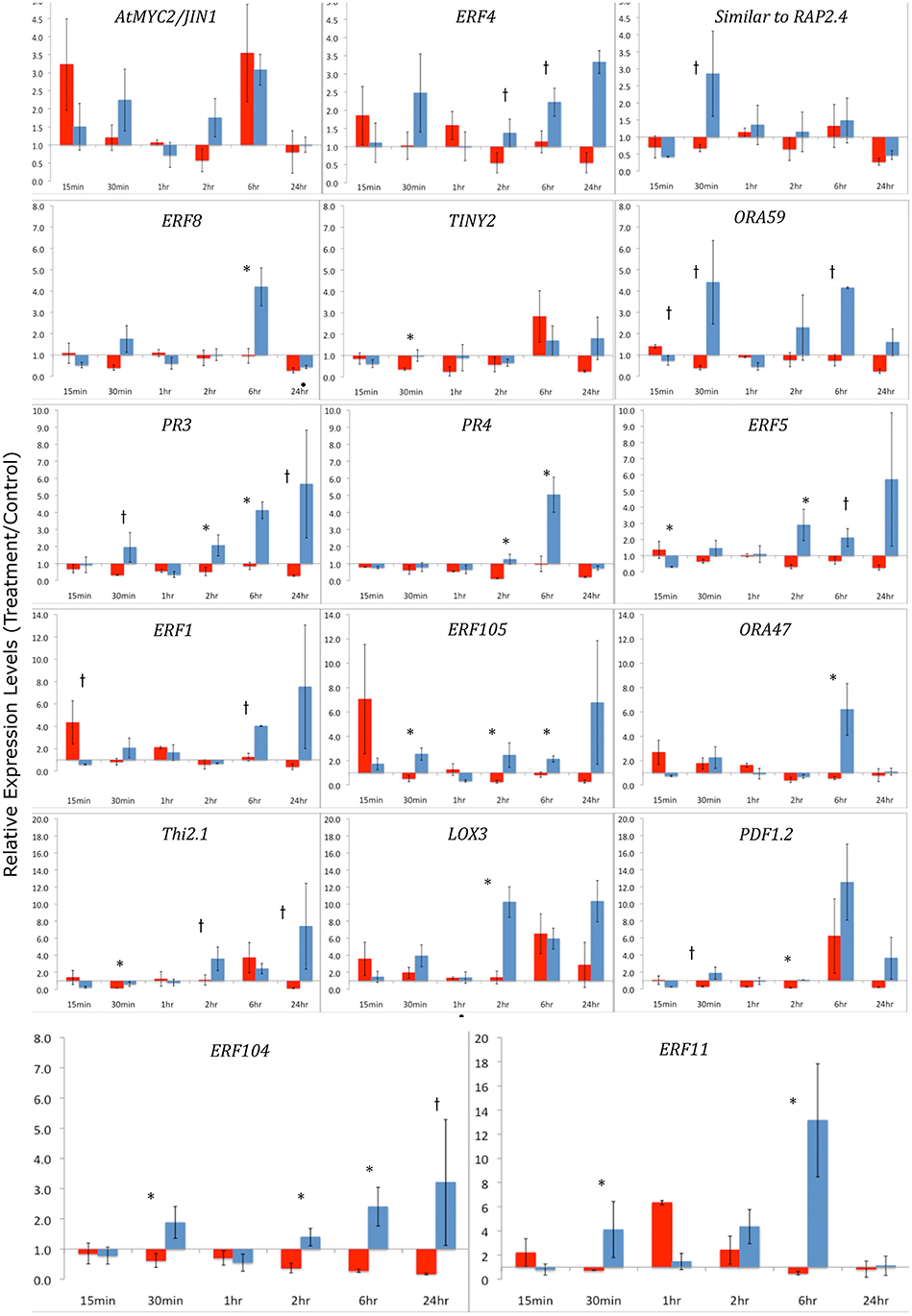
Figure 4. qRT-PCR of ERF Transcription Factors and defense-related genes after Herbivory-Y-axes represent fold changes of treatment/controls. Arabidopsis leaf tissue was collected 15 min, 30 min, 1 h, 2 h, 6 h, and 24 h after herbivory by S. exigua or P. rapae caterpillars. Red bars indicate Pieris treatments and blue bars represent Spodoptera treatments. qRT-PCR data were normalized to RNA quantity and the expression of an exogenous LUC spike. Fold changes differ in scale between each row. Control plants (cage only, no insects) were paired with treatment plants. Error bars represent the standard error of the means of the bioreplicates for each treatment and time point. Asterisks (p < 0.05) and lower case †'s (p < 0.08) indicate statistically significant differences between insect treatments as determined by Kruskal–Wallis ANOVA Analyses for ratios.

Figure 5. Cluster Analysis and Heat Map of Defense-Related Gene Expression by P. rapae vs. S. exigua. Gene expression values in Arabidopsis tissue after insect treatments were and clustered by time after feeding using the method described by Eisen et al. (1998). Red color indicates the up-regulation of a gene, while green shows down-regulation. A black box represents no change in expression.
In our experiment with S. exigua, we observed several instances where gene expression was increased in control plants, especially during the initial time points, suggesting that thigmotropic stimuli while placing cages (without insects) on the control plant may have contributed to elevated gene expression. We also found that the starting control levels for JA in the S. exigua bioassay were higher than in the P. rapae assay (Figure 2). We conducted an experiment to determine whether cages put on plants elicited similar patterns on the expression of ERF8, ERF11, PDF1.2, and Thi2.1 as those found in our insect experiments. Our results suggest that touch may be a small contributing factor as gene expression in untouched plants was less than in touched plants, but the transcriptional response elicited by thigmotropic stimulation was not enough to explain the large expression changes in the insect experiments (Supplementary Material, Figure 2). It remains possible that the frequent movement typical of S. exigua larvae, which is not shown by P. rapae larvae (McCartney, 2007) produced the greater background.
Insect Feeding and Glucosinolate Assays
To assess the role of ERFs in resistance to insects, we conducted no-choice feeding assays with both P. rapae and S. exigua on WT, erf104, erf105, erf5, and erf5 Arabidopsis genotypes. As expected, P. rapae, which is adapted to feeding on glucosinolate-containing plants, maintained similar RCR, RGR, and efficiencies of conversion of ingested food on all the genotypes (Figure 6). In contrast, S. exigua had significantly lower RCR on erf104 and erf6. Although higher specific leaf masses can cause lower RCR because there is more nutrition per unit volume of leaf consumed, this was not the case here because the specific leaf masses did not differ statistically among the genotypes (data not shown). Despite eating significantly less of erf104 and erf6, S. exigua growth rates did not differ among genotypes. This compensatory feeding to maintain a constant growth rate is a common behavior of S. exigua. The quality of the erf mutants as food for growth also differed for S. exigua: ECI was significantly higher on erf104 than on the other mutants and WT.
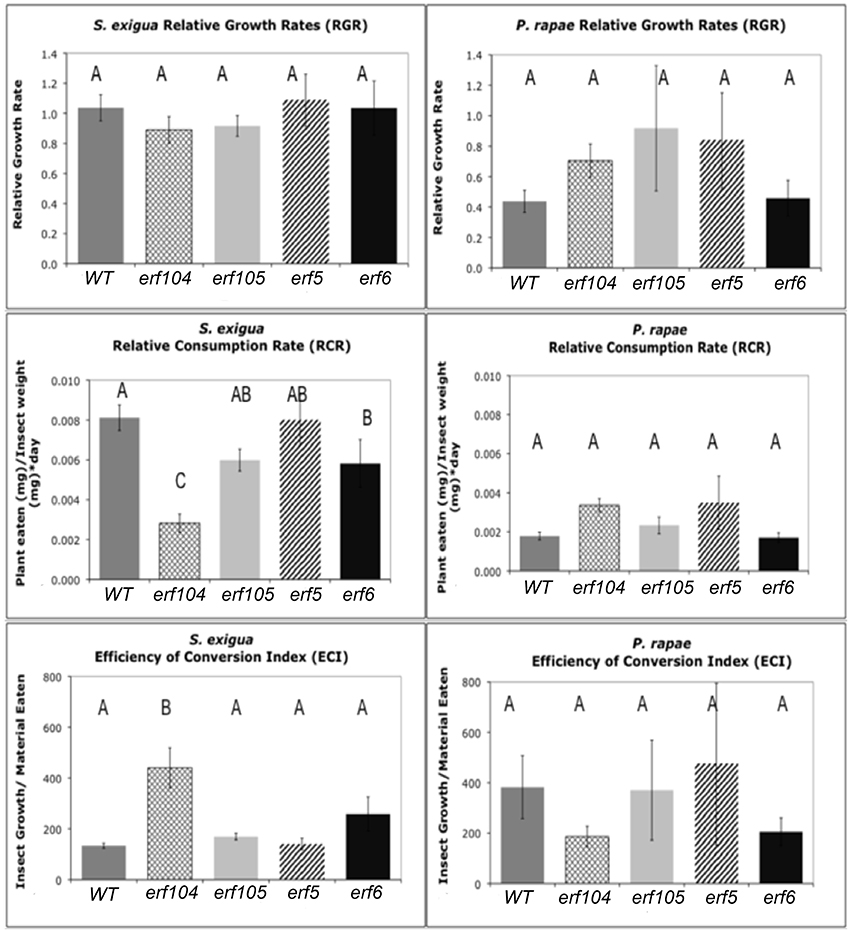
Figure 6. S. exigua and P. rapae relative growth rates (RGR), relative consumption rates (RCR), and efficiency of conversion of ingested food index (ECI) on WT and erf mutant plants. RGR represents the total weight gained by an insect relative to its initial weight and total feeding time. RCR is calculated as the total material (mg) eaten divided by initial insect weight (mg) multiplied by the feeding period (days). ECI is calculated as the difference in insect mass before and after the feeding assay divided by the total material (g) eaten. Sample sizes differed between treatments and ranged between 11 and 31 insects. Error bars represent standard errors of the mean. Letters above columns represent post-hoc Tukey values after using the GLM model in SAS. Different letters indicate significant differences between genotypes p < 0.05.
The ability of plants to respond to insect feeding by increasing indolyl glucosinolates did not depend on functional ERFs; both S. exigua and P. rapae feeding increased concentrations of indolyl glucosinolates in the youngest leaves in all genotypes (Supplementary Material, Table S5). In contrast, changes in aliphatic glucosinolates were heterogeneous. When data from all genotypes were combined, there was a significant correlative relationship between total indolyl GS levels and RCR that was negative for S. exigua and positive for P. rapae, consistent with their tolerance for GS; however in neither case was the relationship strong. Within genotypes, there were only 5 significant relationships between GS levels and RCR and all but one were all positive correlations.
Discussion
We examined the transcriptional reprogramming of Arabidopsis genes encoding TFs elicited by 2 different chewing herbivores or wounding. We identified 141 TF genes that were differentially expressed in at least one treatment. TFs are often points of cross-talk between signaling pathways and have been shown to be important for responses to biotic stresses, including herbivory (Li et al., 1999; Abe et al., 2003; Lorenzo et al., 2003; Fernandez-Calvo et al., 2011; Schweizer et al., 2013). The TF gene expression profiles differed considerably between responses to the two caterpillars, suggesting that responses to different stimuli are shaped by transcriptional activation of stimulus-specific TFs, as are responses to wounding.
Similarities in Arabidopsis Responses to Chewing Caterpillars and Wounding
Using a microarray analysis, we found that expression of JAZ, WRKY, and NAC TFs was elevated by both unrelated insects and wounding. This suggests a significant transcriptional role for them after diverse biotic stresses. Many genes in these families were up- or down-regulated across most caterpillar and many wounding treatments. WRKY, NAC, and Zinc-Finger TF induction after Spodoptera littoralis feeding was also observed by Schweizer et al. (2013), who suggested a JA-independent role of these TFs in plant defense. Both S. exigua and wounding, but not P. rapae elicited genes in the C2H2 gene family, including ZAT10 and ZAT5. ZAT10 modulates a plant's ability to adapt to heat, salinity, and osmotic stress (Mittler et al., 2006) indicating both treatments may be triggering similar abiotic stress-related signaling. However, wounding and insect-induced genes with the most consistent and widespread transcriptional response in our study belonged to the JAZ (ZIM) family, which are important transcriptional regulators in the JA-response. Other studies have reported significant JAZ transcription after wounding, P. rapae, S. exigua, or JA treatments (Reymond et al., 2004; DeVos et al., 2005; Chung et al., 2008). JAZ proteins are key regulators of the JA-signaling pathway (Chini et al., 2007; Thines et al., 2007; Fernandez-Calvo et al., 2011) and have been found to be activated in response to Malacosoma disstria feeding on poplar (Major and Constabel, 2006), and S. exigua feeding on Arabidopsis (Chung et al., 2008). These genes may not be involved in differential responses to attack by specific insects, but appear to play a critical role in generalized biotic stress or wound signaling.
Transcriptional factor gene expression was attenuated after wounding in comparison to insect feeding. However, we observed TFs known to play a role as DAMPs including WIPK (Reviewed by Heil and Land, 2014, this edition) and several ZATs (Mittler et al., 2006). The up-regulation of JA-regulated genes, such as JAZ (Chung et al., 2008) and NAC TFs (reviewed by Bu et al., 2008; reviewed by Wasternack and Hause, 2013) suggest a mechanism for the plants' ability to perceive wound damage and trigger responses similar to herbivory. Interestingly, only wounding induced the expression of LOB TFs. LOBs encode a diverse, plant-specific class of proteins that control new growth during root, leaf, shoot, and xylem development (Shuai et al., 2002; Soyano et al., 2008). Fan et al. (2012) showed LOBS to critical players downstream of auxin signaling during callus formation. Taken together, this suggests that LOBs could play a major role in plant recovery or organ regeneration after wounding, but not after herbivore-related damage.
The Major Differences in Plant Responses to the Two Caterpillars Involved the Regulation of ERF Transcription Factors
We observed dramatic differences between the expression profiles of AP2/ERF TFs that were either elicited by either P. rapae or S. exigua when we conducted both microarray analysis and qRT-PCR. AP2/ERF TFs comprise about 120 members (Nakano et al., 2006), are exclusive to plants, and consist of ERF or B3 DNA binding domains and several sub-families including AP2 and RAV (McGrath et al., 2005). Many ERFs are responsive to the hormones ET and JA (Lorenzo and Solano, 2005; Pre et al., 2008), although individual gene responses to either ET or JA can differ. For example, expression of ERF1 is compromised in both coi1 and ein1 plants (Berrocal-Lobo et al., 2004), which are deficient in JA- and ET-signaling respectively, and the expression of ERFs is rapidly induced by exogenous application of both hormones in WT plants (Brown et al., 2003). Fujimoto et al. (2000) showed that ERFs directly activate the transcription of defense-related genes such as PDF1.2, B-chitinase (PR3), and Hevein-like protein (PR4) by binding to GCC-boxes in their promoters. Alternatively, Lorenzo et al. (2004) found the JA-inducible gene AtMYC acts to repress the expression of ERFs, while activating other wound-responsive genes such as VSP2, Thi2.1, and LOX3. Although our subsequent qPCR experiments found some of these ERF TFs to be affected by both caterpillars, we found TINY2, to be specifically up-regulated by P. rapae whereas ORA47, SIMRAP2.4, ERF11, and ERF104 were only up-regulated by S. exigua (Table 1). Each of these genes appears to be important in plant stress responses. For example, TINY2 transcription increases in response to ABA, drought, salt, cold, wounding, and SA treatment (slightly), but not ethylene (Wei et al., 2005). ORA47 expression was increased by insect regurgitant from T. ni larvae (Walley et al., 2007) and its over-expression increased the expression of the wound-responsive VSP2 but not LOX3 (Wang et al., 2008). The RAP2.4 homolog SIMRAP2.4 is responsive to JA, wounding, heat, and water stress and functions in osmoregulation (Walley et al., 2007; Wang et al., 2008; Rae et al., 2011). ERF11 is an ethylene-inducible transcriptional repressor with an EAR motif (Yang et al., 2005) and is highly induced by chitin treatment (Libault et al., 2007), MeJA application and Alternaria brassicicola infection (McGrath et al., 2005). S. exigua also elicited the expression of ERF104, which is a MAPK6 target required for FLG22-induced ET signaling (Bethke et al., 2009). Plants over-expressing ERF104 had increased transcripts of pathogenesis-related genes that are not induced by ERF1 activation or JA and ET treatment, so ERF104 signaling may represent a novel TF response specific to insect attack. We found that S. exigua larva that fed erf104 knock-out mutant Arabidopsis plants in no-choice assays did not have reduced growth rates, but did consume less plant mass than other genotypes, including Col-0 WT, erf5, erf6, and erf105 (Figure 6). However, mutant erf104 plants did not have reduced glucosinolate production (Table S5). This suggests that ERF104 may be a negative regulator of plant defenses and knocking out the gene confers a resistance in Arabidopsis plants in a GS-independent manner. Several ERFs (ERF11, ERF3, and ERF4) contain an EAR motif that functions in negative regulation of ethylene-responsive genes via the GCC box (Fujimoto et al., 2000; Brown et al., 2003; McGrath et al., 2005). However, ERF104 does not appear to contain this domain. Interestingly, all four of the ERF TFs uniquely up-regulated by S. exigua were found to be highly chitin-responsive by Libault et al. (2007) and may suggest a role in Arabidopsis's differential hormonal and transcriptional response to S. exigua vs. P. rapae feeding.
Differences in Hormone Signaling After Caterpillar Feeding
Signaling after herbivory involves the crosstalk among JA, ET, SA and other hormones as well as the regulation of TFs and defense related genes (Reviewed by Reymond et al., 2004; Zhu-Salzman et al., 2004; Mewis et al., 2006; Vogel et al., 2007; Wasternack and Hause, 2013). In this study we show that herbivory by two different insects elicits increases in both ET and JA, but not SA. However, the timing of ET and JA responses and the total concentrations induced by the insects were different. In most cases, S. exigua elicited stronger, and often earlier, responses, which may shape downstream responses. This is highlighted by the differential expression of ERF TFs and PR genes in response to insect feeding.
Increased ethylene emissions after insect herbivory are well-documented (for review see Von Dahl and Baldwin, 2007). In our study, ethylene production in Arabidopsis plants after S. exigua attack occurred as a rapid burst and peaked after 1 h (Figure 1). ET levels continued to remain above control levels until 6 h, after which they attenuated. Conversely, P. rapae feeding did not induce ET levels that were significantly different from controls until after 2 h, and they remained elevated throughout most of the time course. Our results suggest that ET could serve as an important signal in defense responses to S. exigua as well as P. rapae, and that the timing of peak ethylene production may be crucial to organizing different down-stream responses to each attacker. These results are consistent with these species' feeding behaviors. S. exigua changes feeding sites and/or plants at least every hour, while P. rapae larvae may feed on the same leaf for up to a day (McCartney, 2007).
It is well-known that JA production is an important component in plant defense responses, especially after wounding or herbivory. Levels of JA as well as the pre-cursor oxylipins OPDA and dnOPDA gradually increased over a 24-h time course after P. rapae feeding (Reymond et al., 2004). DeVos et al. (2006) reported an increase in JA production after P. rapae feeding that peaked at 48 h after feeding. Herbivory by S. exigua also increased JA levels in Zea mays (Schmelz et al., 2003). In this study we found that both insect species increased the production of JA at levels that were significantly different from controls at early time points (S. exigua, 30 min; P. rapae, 15 min) and which then tapered off after 24 h. Our results suggest that in response to S. exigua and P. rapae, the timing and ratios of ET and JA may comprise a regulatory mechanism for differential response.
Defense Gene Expression After Insect Feeding
Gene expression of ERF TFs and defense-related genes was very different in response to the two insect treatments most likely as a result of varied hormone signaling. S. exigua feeding activated the transcription of ERF TFs. Fold change increases in ERF4, ERF104, ERF8, SIMRAP2.4, ORA59, ERF5, ERF105, ORA47, ERF1, and ERF11 were significantly higher in S. exigua treatments. In fact, in only 2 cases, ORA59, and ERF1 at 15 min, is gene expression significantly higher in P. rapae-treated plants. This difference is particularly notable in the expression of ERF104 and ERF11, which are significantly increased by S. exigua, and often repressed by P. rapae at these time points. Furthermore, ERF1, which was also chitin-responsive, is increased by S. exigua, but not by P. rapae after 6 h.
We observed a similar pattern with the defense-related genes we analyzed. S. exigua, but not P. rapae elicited the increased expression of PR3, PR4 (HEL), PDF1.2, LOX3, and Thi2.1, which are JA-responsive genes (Lorenzo et al., 2003, 2004; Koornneef and Pieterse, 2008). In fact, except for LOX3, during the various time points, P. rapae down-regulated the expression of these genes compared to S. exigua. DeVos et al. (2005) also found that P. rapae did not significantly increase PDF1.2 or PR4 (HEL) transcription, although an increase in PDF1.2::GUS activity at the periphery of P.-damaged tissue was seen. Although both insects elicited the production of JA and ET, P. rapae did not increase the transcription of these JA-inducible genes, suggesting that P. rapae may be suppressing defense-related signaling. In our study, hormonal and transcriptional responses to S. exigua were consistently increased while responses to P. rapae were attenuated or in some cases, absent. This, in conjunction with significant JA induction and rapid ET elicitation induced by S. exigua vs. P. rapae suggests that P. rapae may be suppressing host responses or evading detection compared with S. exigua. When compared to other biotic stresses, an attenuated transcriptional response in Arabidopsis after P. rapae attack has previously been reported. For example, Reymond et al. (2000) found that wounding induced far more genes than P. rapae, including water-stress related genes and suggested that this may be due to a feeding strategy that reduces overall leaf damage. In a similar study, Bodenhausen and Reymond (2007) found comparable trends with P. rapae feeding on Arabidopsis coi1-1 plants. By using a stress gene-specific microarray, the authors observed more SA-related and disease resistance genes transcribed in the coi1-1 mutants than in WT plants after P. rapae feeding, suggesting a putative mechanism for JA-dependent gene suppression. Interestingly, the same response was not seen after Spodoptera littoralis feeding. In our study, we found that this potential suppression of responses is not likely due to an accumulation of SA, but could be a by-product of weakened ET and JA elicitation compared to S. exigua.
The cross-talk among the ET, JA, ABA, and SA signaling-pathways in response to pathogens and insects is complex and involves antagonisms between the ABA/JA branch via MYC2 and the ET/JA branch via ERFs (reviewed by Wasternack and Hause, 2013). Both of these branches require activation of JAZ proteins, which were up-regulated in both insect treatments. One hypothesis posed by Verhage et al. (2011) states that Arabidopsis plants quickly perceive P. rapae feeding and suppress responses by activating a JA-dependent MYC2 pathway vs. a JA/ET-dependent ERF pathway. A previous study by DeVos et al. (2006) found that wounding + Pieris OS suppressed PDF1.2 likely through the ABA-activation of AtMYC2. Recently, Vos et al. (2013) found that induction of AtMYC2 by P. rapae is largely controlled by ABA and JA, which act together to prime the plant's defenses against future herbivores. Our results further support the hypothesis that P. rapae elicits the ABA/JA-MYC2 pathway, thus by-passing ET/JA-ERF signaling. Conversely, S. exigua clearly elicits a rapid JA and ET “burst” (Von Dahl et al., 2007), which most likely has downstream effects on ERF transcription and JA-mediated responses. In many ways, we observed that responses to S. exigua were more aligned with pathways activated by necrotrophic pathogens because of its activation of the ERF-branch of the biotic stress response (reviewed by Pieterse et al., 2012).
Interestingly, when Verhage et al. (2011) used P. rapae wounding + OS, the ERF pathway was activated. Yet, the application of S. littoralis and Pieris brassicae OS suppressed several wound-responsive genes in Arabidopsis (Consales et al., 2012), including an ERF-family TF. It is tempting to speculate that something about Pieris feeding behavior attenuates wound signaling and that the differences in defense responses, including hormone release and TF transcription, originate at the feeding site. This suggests that an insect's ability to elicit responses divergent from wound responses may be a key element of host plant specialization. In our comparison and results of others, responses to S. exigua, an herbivore with a very broad diet, more closely resemble responses to necrotophic pathogens than do responses to the more specialized P. rapae. We observed that at the whole plant level, P. rapae feeds in one location on the edge of a leaf and continues to remove tissue from that site. Conversely, S. exigua eats small amounts throughout the plant, creating several small holes throughout the leaves and distributing their damage across the leaf and/or plant (McCartney, 2007). In a previous study (Rehrig et al., 2011), we found that herbivory by S. exigua was particularly distressing to Arabidopsis plants and was most likely affecting both primary and secondary metabolism. The similar results reported here suggest that the trauma inflicted by S. exigua may be a due to a combination of HAMPs and DAMPs elicited after feeding. Furthermore, due to the elicitation of ET, JA, and chitin-responsive genes, S. exigua feeding activates responses that are more similar to microbe-associated molecular responses (MAMPs) than herbiviory.
Author Contributions
Erin M. Rehrig—Prepared manuscript, including tables and figures, analyzed and interpreted data, conducted qRT-PCR and all insect rearing and bioassays. Heidi M. Appel—Edited manuscript, interpreted data, managed microarray project, oversaw insect and plant growth, co-authored funding for project. A. Daniel Jones—Conducted UPLC-MS/MS analysis on JA, SA, and JA-IL on plant samples, edited manuscript and provided critical feedback. Jack C. Schultz—Edited manuscript, assisted with the analysis and interpretation of data, conducted statistics, procured funding for project
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Lucy Rubino, Clayton Coffman, and Dr. Abbie Ferrieri for their help with conducting the insect feeding experiments. We also offer our appreciation to Dr. Shuqun Zhang for his technical assistance with the GC ethylene analysis and donation of erf mutant Arabidopsis seeds. We also thank Dr. Irmgard Seidl-Adams for her invaluable support and guidance with the qRT-PCR, and Ms. Lijun Chen of the Michigan State University Mass Spectrometry and Metabolomics Core for technical assistance with UPLC-MS/MS analyses. This research was supported by grants from the National Science Foundation, DEB 0313492 (Jack Schultz, Primary Investigator) and IOS 0614890 (Heidi Appel and Jack Schultz, PIs). The Quattro Premier XE UPLC-MS/MS system was purchased using funds from National Science Foundation Major Research Instrumentation grant DBI 0619489 (A. Daniel Jones, PI). A. Daniel Jones acknowledges support from Michigan AgBioResearch project MICL02143.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00407/abstract
References
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. doi: 10.1105/tpc.006130
Alborn, H. T., Hansen, T. V., Jones, T. H., Bennett, D. C., Tumlinson, J. H., Schmelz, E. A., et al. (2007). Disulfooxy fatty acids from the American bird grasshopper Schistocerca americana, elicitors of plant volatiles. Proc. Natl. Acad. Sci. U.S.A. 104, 12976–12981. doi: 10.1073/pnas.0705947104
Babst, B. A., Ferrieri, R. A., Gray, D. W., Lerdau, M., Schlyer, D. J., Schueller, M., et al. (2005). Jasmonic acid induces rapid changes in carbon transport and partitioning in Populus. New Phytol. 167, 63–72. doi: 10.1111/j.1469-8137.2005.01388.x
Beckers, G. J., and Spoel, S. H. (2006). Fine-tuning plant defence signalling: salicylate versus jasmonate. Plant Biol. (Stuttg). 8, 1–10. doi: 10.1055/s-2005-872705
Berrocal-Lobo, M., Molina, A., Solano, R., and Plant, J. (2004). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Mol. Plant-Microbe Interact. 29, 23–32. doi: 10.1046/j.1365-313x.2002.01191.x
Bethke, G., Unthan, T., Uhrig, J. F., Poschl, Y., Gust, A. A., Scheel, D., et al. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 8067–8072. doi: 10.1073/pnas.0810206106
Bodenhausen, N., and Reymond, P. (2007). Signaling pathways controlling induced resistance to insect herbivores in Arabidopsis. Mol. Plant Microbe Interact. 20, 1406–1420. doi: 10.1094/MPMI-20-11-1406
Boter, M., Ruiz-Rivero, O., Abdeen, A., and Prat, S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes. Dev. 18, 1577–1591. doi: 10.1101/gad.297704
Brown, R. L., Kazan, K., Mcgrath, K. C., Maclean, D. J., and Manners, J. M. (2003). A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 132, 1020–1032. doi: 10.1104/pp.102.017814
Bu, Q., Jiang, H., Li, C. B., Zhai, Q., Zhang, J., Wu, X., et al. (2008). Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 18, 756–767. doi: 10.1038/cr.2008.53
Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Choi, J., Choi, D., Lee, S., Ryu, C. M., and Hwang, I. (2011). Cytokinins and plant immunity: old foes or new friends? Trends Plant Sci. 16, 388–394. doi: 10.1016/j.tplants.2011.03.003
Chung, H. S., Koo, A. J. K., Gao, X., Jayanty, S., Thines, B., Jones, A. D., et al. (2008). Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 146, 952–964. doi: 10.1104/pp.107.115691
Consales, F., Schweizer, F., Erb, M., Gouhier-Darimont, C., Bodenhausen, N., Bruessow, F., et al. (2012). Insect oral secretions suppress wound-induced responses in Arabidopsis. J. Exp. Bot. 63, 727–737. doi: 10.1093/jxb/err308
Davuluri, R. V., Sun, H., Palaniswamy, S. K., Matthews, N., Molina, C., Kurtz, M., et al. (2003). AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4:25. doi: 10.1186/1471-2105-4-25
Delessert, C., Wilson, I. W., Van Der Straeten, D., Dennis, E. S., and Dolferus, R. (2004). Spatial and temporal analysis of the local response to wounding in Arabidopsis leaves. Plant Mol. Biol. 55, 165–181. doi: 10.1007/s11103-004-0112-7
DeVos, M. (2006). Signal Signature, Transcriptomics, and Effectiveness of Induced Pathogen and Insect Resistance in Arabidopsis. Ph.D., University of Utrecht, Utrecht.
DeVos, M., Van Oosten, V. R., Van Poecke, R. M. P., Van Pelt, J. A., Pozo, M. J., Mueller, M. J., et al. (2005). Signal dignature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant-Microbe Interact. 18, 923–937. doi: 10.1094/MPMI-18-0923
DeVos, M., Van Zaanen, W., Koornneef, A., Korzelius, J. P., Dicke, M., Van Loon, L. C., et al. (2006). Herbivore-induced resistance against microbial pathogens in Arabidopsis. Plant Physiol. 142, 352–363. doi: 10.1104/pp.106.083907
Dombrecht, B., Xue, G. P., Sprague, S. J., Kirkegaard, J. A., Ross, J. J., Reid, J. B., et al. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19, 2225–2245. doi: 10.1105/tpc.106.048017
Ehlting, J., Chowrira, S., Mattheus, N., Aeschliman, D., Arimura, G.-I., and Bohlmann, J. (2008). Comparative transcriptome analysis of Arabidopsis thaliana infested by diamond back moth (Plutella xylostella) larvae reveals signatures of stress response, secondary metabolism, and signalling. BMC Genomics 9:154. doi: 10.1186/1471-2164-9-154
Ehlting, J., Mattheus, N., Aeschliman, D. S., Li, E., Hamberger, B., Cullis, I. F., et al. (2005). Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J. 42, 618–640. doi: 10.1111/j.1365-313X.2005.02403.x
Eisen, M. B., Spellman, P. T., Brown, P. O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868. doi: 10.1073/pnas.95.25.14863
Fan, M., Xu, C., Xu, K., and Hu, Y. (2012). LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 22, 1169–1180. doi: 10.1038/cr.2012.63
Fernandez-Calvo, P., Chini, A., Fernandez-Barbero, G., Chico, J. M., Gimenez-Ibanez, S., Geerinck, J., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715. doi: 10.1105/tpc.110.080788
Fujimoto, S. Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12, 393–404. doi: 10.1105/tpc.12.3.393
Galis, I., Gaquerel, E., Pandey, S. P., and Baldwin, I. T. (2009). Molecular mechanisms underlying plant memory in JA-mediated defence responses. Plant Cell Environ. 32, 617–627. doi: 10.1111/j.1365-3040.2008.01862.x
Green, J. M., Appel, H., Rehrig, E. M., Harnsomburana, J., Chang, J. F., Balint-Kurti, P., et al. (2012). PhenoPhyte: a flexible affordable method to quantify 2D phenotypes from imagery. Plant Methods 8:45. doi: 10.1186/1746-4811-8-45
Grunewald, W., Vanholme, B., Pauwels, L., Plovie, E., Inze, D., Gheysen, G., et al. (2009). Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep. 10, 923–928. doi: 10.1038/embor.2009.103
Guo, A., He, K., Liu, D., Bai, S., Gu, X., Wei, L., et al. (2005). DATF: a database of Arabidopsis transcription factors. Bioinformatics 21, 2568–2569. doi: 10.1093/bioinformatics/bti334
Heil, M. (2009). Damaged-self recognition in plant herbivore defence. Trends Plant Sci. 14, 356–363. doi: 10.1016/j.tplants.2009.04.002
Hou, X., Lee, L. Y., Xia, K., Yan, Y., and Yu, H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19, 884–894. doi: 10.1016/j.devcel.2010.10.024
Husbands, A., Bell, E. M., Shuai, B., Smith, H. M., and Springer, P. S. (2007). LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 35, 6663–6671. doi: 10.1093/nar/gkm775
Kaku, H., Nishizawa, Y., Ishii-Minami, N., Akimoto-Tomiyama, C., Dohmae, N., Takio, K., et al. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 11086–11091. doi: 10.1073/pnas.0508882103
Koornneef, A., and Pieterse, C. M. J. (2008). Cross talk in defense signaling. Plant Physiol. 146, 839–844. doi: 10.1104/pp.107.112029
Larionov, A., Krause, A., and Miller, W. (2005). A standard curve based method for relative real time PCR data processing. BMC Bioinformatics 6:62. doi: 10.1186/1471-2105-6-62
Li, J., Brader, G., and Palva, E. T. (1999). The WRKY70 transcription factor: a mode of convergence for jasmonate-mediated and salicylate mediated signals in plant defense. Plant Cell 16, 319–331. doi: 10.1105/tpc.016980
Libault, M., Wan, J., Czechowski, T., Udvardi, M., and Stacey, G. (2007). Identification of 118 Arabidopsis transcription factor and 30 Ubiquitin-Ligase genes responding to chitin, a plant-defense elicitor. Mol. Plant-Microbe Interact. 20, 900–911. doi: 10.1094/MPMI-20-8-0900
Lorenzo, O., Chico, J. M., Sanchez-Serrano, J. J., and Solano, R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16, 1938–1950. doi: 10.1105/tpc.022319
Lorenzo, O., Piqueras, R., Sanchez-Serrano, J. J., and Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178. doi: 10.1105/tpc.007468
Lorenzo, O., and Solano, R. (2005). Molecular players regulating the jasmonate signalling network. Curr. Opin. Plant Biol. 8, 532–540. doi: 10.1016/j.pbi.2005.07.003
Lu, J., Ju, H., Zhou, G., Zhu, C., Erb, M., Wang, X., et al. (2011). An EAR-motif-containing ERF transcription factor affects herbivore-induced signaling, defense and resistance in rice. Plant J. 68, 583–596. doi: 10.1111/j.1365-313X.2011.04709.x
Major, I. T., and Constabel, C. P. (2006). Molecular analysis of poplar defense against herbivory: comparison of wound- and insect elicitor-induced gene expression. New Phytol. 172, 617–635. doi: 10.1111/j.1469-8137.2006.01877.x
Mattiacci, L., Dicke, M., and Posthumus, M. A. (1995). beta-Glucosidase: an elicitor of herbivore-induced plant odor that attracts host-searching parasitic wasps. Proc. Natl. Acad. Sci. U.S.A. 92, 2036–2040. doi: 10.1073/pnas.92.6.2036
McCartney, N. (2007). Analysis of Pieris rapae and Spodoptera exigua Feeding Site Preferences on Arabidopsis thaliana “Columbia” Ecotype in Relation to Within-Plant Chemical and Structural Heterogeneity. University Park, PA: M.S., The Pennsylvania State University.
McGrath, K. C., Dombrecht, B., Manners, J. M., Schenk, P. M., Edgar, C. I., Maclean, D. J., et al. (2005). Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 139, 949–959. doi: 10.1104/pp.105.068544
Mewis, I., Appel, H. M., Hom, A., Raina, R., and Schultz, J. C. (2005). Major signaling pathways modulate Arabidopsis glucosinolate accumulation and response to both phloem-feeding and chewing insects. Plant Physiol. 138, 1149–1162. doi: 10.1104/pp.104.053389
Mewis, I., Tokuhisa, J. G., Schultz, J. C., Appel, H. M., Ulrichs, C., and Gershenzon, J. (2006). Gene expression and glucosinolate accumulation in Arabidopsis thaliana in response to generalist and specialist herbivores of different feeding guilds and the role of defense signaling pathways. Phytochemistry 67, 2450–2462. doi: 10.1016/j.phytochem.2006.09.004
Mittler, R., Kim, Y., Song, L., Coutu, J., Coutu, A., Ciftci-Yilmaz, S., et al. (2006). Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 580, 6537–6542. doi: 10.1016/j.febslet.2006.11.002
Mur, L. A., Lloyd, A. J., Cristescu, S. M., Harren, F. J., Hall, M. A., and Smith, A. R. (2009). Biphasic ethylene production during the hypersensitive response in Arabidopsis: a window into defense priming mechanisms? Plant Signal. Behav. 4, 610–613. doi: 10.4161/psb.4.7.8904
Nakano, T., Suzuki, K., Ohtsuki, N., Tsujimoto, Y., Fujimura, T., and Shinshi, H. (2006). Identification of genes of the plant-specific transcription-factor families cooperatively regulated by ethylene and jasmonate in Arabidopsis thaliana. J. Plant Res. 119, 407–413. doi: 10.1007/s10265-006-0287-x
Pare, P. W., and Tumlinson, J. H. (1999). Plant volatiles as a defense against insect herbivores. Plant Physiol. 121, 325–332. doi: 10.1104/pp.121.2.325
Pauwels, L., Barbero, G. F., Geerinck, J., Tilleman, S., Grunewald, W., Perez, A. C., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791. doi: 10.1038/nature08854
Penninckx, I. A., Thomma, B. P., Buchala, A., Metraux, J. P., and Broekaert, W. F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113. doi: 10.1105/tpc.10.12.2103
Pieterse, C. M., Van Der Does, D., Zamioudis, C., Leon-Reyes, A., and Van Wees, S. C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Pillai, K. C. S., and Tienzo, P. (1959). On the distribution of the extreme studentized deviate from the sample mean. Biometrika 46, 467–472. doi: 10.1093/biomet/46.3-4.467
Pre, M., Atallah, M., Champion, A., De Vos, M., Pieterse, C. M. J., and Memelink, J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147, 1347–1357. doi: 10.1104/pp.108.117523
Pylatuik, J., and Fobert, P. (2005). Comparison of transcript profiling on Arabidopsis microarray platform technologies. Plant Mol. Biol. 58, 609–624. doi: 10.1007/s11103-005-6506-3
Rae, L., Lao, N. T., and Kavanagh, T. A. (2011). Regulation of multiple aquaporin genes in Arabidopsis by a pair of recently duplicated DREB transcription factors. Planta 234, 429–444. doi: 10.1007/s00425-011-1414-z
Ramakers, C., Ruijter, J. M., Deprez, R. H. L., and Moorman, A. F. M. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66. doi: 10.1016/S0304-3940(02)01423-4
Rehrig, E. M., Appel, H. M., and Schultz, J. C. (2011). Measuring ‘normalcy’ in plant gene expression after herbivore attack. Mol. Ecol. Resour. 11, 294–304. doi: 10.1111/j.1755-0998.2010.02929.x
Reymond, P., Bodenhausen, N., Van Poecke, R. M. P., Krishnamurthy, V., Dicke, M., and Farmer, E. E. (2004). A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell 16, 3132–3147. doi: 10.1105/tpc.104.026120
Reymond, P., and Farmer, E. E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1, 404–411. doi: 10.1016/S1369-5266(98)80264-1
Reymond, P., Weber, H., Damond, M., and Farmer, E. E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–720. doi: 10.1105/tpc.12.5.707
Riechmann, J. L., Heard, J., Martin, G., Reuber, L.-Z., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Robert-Seilaniantz, A., Grant, M., and Jones, J. D. (2011). Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. doi: 10.1146/annurev-phyto-073009-114447
Rozen, S., and Skaletsky, H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386.
Saldanha, A. J. (2004). Java Treeview–extensible visualization of microarray data. Bioinformatics 20, 3246–3248. doi: 10.1093/bioinformatics/bth349
Schmelz, E. A., Alborn, H. T., and Tumlinson, J. H. (2003). Synergistic interactions between volicitin, jasmonic acid and ethylene mediate insect-induced volatile emission in Zea mays. Physiol. Plant. 117, 403–412. doi: 10.1034/j.1399-3054.2003.00054.x
Schmelz, E. A., Carroll, M. J., Leclere, S., Phipps, S. M., Meredith, J., Chourey, P. S., et al. (2006). Fragments of ATP synthase mediate plant perception of insect attack. Proc. Natl. Acad. Sci. U.S.A. 103, 8894–8899. doi: 10.1073/pnas.0602328103
Schweizer, F., Bodenhausen, N., Lassueur, S., Masclaux, F. G., and Reymond, P. (2013). Differential contribution of transcription factors to Arabidopsis thaliana defense against Spodoptera littoralis. Front. Plant Sci. 4:13. doi: 10.3389/fpls.2013.00013
Shuai, B., Reynaga-Pena, C. G., and Springer, P. S. (2002). The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 129, 747–761. doi: 10.1104/pp.010926
Slansky, F., and Scriber, J. M. (1985). “Food consumption and utilization,” in Comprehensive Insect Physiology, Biochemistry and Pharmacology, eds G. A. Kerkut and L. I. Gilbert (New York, NY: Pergamon Press), 87–163.
Soyano, T., Thitamadee, S., Machida, Y., and Chua, N. H. (2008). ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell 20, 3359–3373. doi: 10.1105/tpc.108.061796
Storey, J. D., and Tibshirani, R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 100, 9440–9445. doi: 10.1073/pnas.1530509100
Stork, W., Diezel, C., Halitschke, R., Galis, I., and Baldwin, I. T. (2009). An ecological analysis of the herbivory-elicited JA burst and its metabolism: plant memory processes and predictions of the moving target model. PLoS ONE 4:e4697. doi: 10.1371/journal.pone.0004697
Stotz, H. U., Pittendrigh, B. R., Kroymann, J., Weniger, K., Fritsche, J., Bauke, A., et al. (2000). Induced plant defense responses against chewing insects. Ethylene signaling reduces resistance of Arabidopsis against Egyptian cotton worm but not diamondback moth. Plant Physiol. 124, 1007–1018. doi: 10.1104/pp.124.3.1007
Thaler, J. S., Farag, M. A., Pare, P. W., and Dicke, M. (2002). Jasmonate-deficient plants have reduced direct and indirect defences against herbivores. Ecol. Lett. 5, 764–774. doi: 10.1046/j.1461-0248.2002.00388.x
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665.
Thompson, G. A., and Goggin, F. L. (2006). Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. J. Exp. Bot. 57, 755–766. doi: 10.1093/jxb/erj135
Verhage, A., Vlaardingerbroek, I., Raaymakers, C., Van Dam, N. M., Dicke, M., Van Wees, S. C., et al. (2011). Rewiring of the jasmonate signaling pathway in Arabidopsis during insect herbivory. Front. Plant Sci. 2:47. doi: 10.3389/fpls.2011.00047
Vogel, H., Kroymann, J., and Mitchell-Olds, T. (2007). Different transcript patterns in response to specialist and generalist herbivores in the wild Arabidopsis relative Boechera divaricarpa. PLoS ONE 2:e1081. doi: 10.1371/journal.pone.0001081
Von Dahl, C., and Baldwin, I. (2007). Deciphering the role of ethylene in plant–herbivore interactions. J. Plant Growth Regul. 26, 201–209. doi: 10.1007/s00344-007-0014-4
Von Dahl, C. C., Winz, R. A., Halitschke, R., Kuhnemann, F., Gase, K., and Baldwin, I. T. (2007). Tuning the herbivore-induced ethylene burst: the role of transcript accumulation and ethylene perception in Nicotiana attenuata. Plant J. 51, 293–307. doi: 10.1111/j.1365-313X.2007.03142.x
Vos, I. A., Verhage, A., Schuurink, R. C., Watt, L. G., Pieterse, C. M. J., and Van Wees, S. C. M. (2013). Onset of herbivore-induced resistance in systemic tissue primed for jasmonate-dependent defenses is activated by abscisic acid. Front. Plant Sci. 4:539. doi: 10.3389/fpls.2013.00539
Walley, J. W., Coughlan, S., Hudson, M. E., Covington, M. F., Kaspi, R., Banu, G., et al. (2007). Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet. 3:e172. doi: 10.1371/journal.pgen.0030172
Wang, Z., Cao, G., Wang, X., Miao, J., Liu, X., Chen, Z., et al. (2008). Identification and characterization of COI1-dependent transcription factor genes involved in JA-mediated response to wounding in Arabidopsis plants. Plant Cell Rep. 27, 125–135. doi: 10.1007/s00299-007-0410-z
Wasternack, C., and Hause, B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. doi: 10.1093/aob/mct067
Wei, G., Pan, Y., Lei, J., and Zhu, Y.-X. (2005). Molecular cloning, phylogenetic analysis, expressional profiling and in vitro studies of TINY2 from Arabidopsis thaliana. J. Biochem. Mol. Biol. 38, 440–446. doi: 10.5483/BMBRep.2005.38.4.440
Winz, R. A., and Baldwin, I. T. (2001). Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. IV. Insect-Induced ethylene reduces jasmonate-induced nicotine accumulation by regulating putrescine N-methyltransferase transcripts. Plant Physiol. 125, 2189–2202. doi: 10.1104/pp.125.4.2189
Wittstock, U., Agerbirk, N., Stauber, E. J., Olsen, C. E., Hippler, M., Mitchell-Olds, T., et al. (2004). Successful herbivore attack due to metabolic diversion of a plant chemical defense. Proc. Natl. Acad. Sci. U.S.A. 101, 4859–4864. doi: 10.1073/pnas.0308007101
Wu, J., and Baldwin, I. T. (2009). Herbivory-induced signalling in plants: perception and action. Plant Cell Environ. 32, 1161–1174. doi: 10.1111/j.1365-3040.2009.01943.x
Yadav, V., Mallappa, C., Gangappa, S. N., Bhatia, S., and Chattopadhyay, S. (2005). A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17, 1953–1966. doi: 10.1105/tpc.105.032060
Yang, Z., Tian, L., Latoszek-Green, M., Brown, D., and Wu, K. (2005). Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 58, 585–596. doi: 10.1007/s11103-005-7294-5
Zeng, W., Brutus, A., Kremer, J. M., Withers, J. C., Gao, X., Jones, A. D., et al. (2011). A genetic screen reveals Arabidopsis stomatal and/or apoplastic defenses against Pseudomonas syringae pv. tomato DC3000. PLoS Pathog. 7:e1002291. doi: 10.1371/journal.ppat.1002291
Keywords: Arabidopsis, herbivory, transcription factors, ERFs, ethylene, jasmonate, wounding
Citation: Rehrig EM, Appel HM, Jones AD and Schultz JC (2014) Roles for jasmonate- and ethylene-induced transcription factors in the ability of Arabidopsis to respond differentially to damage caused by two insect herbivores. Front. Plant Sci. 5:407. doi: 10.3389/fpls.2014.00407
Received: 27 May 2014; Accepted: 31 July 2014;
Published online: 19 August 2014.
Edited by:
Martin Heil, Centro de Investigación y de Estudios Avanzados del I.P.N. - Unidad Irapuato, MexicoReviewed by:
Richard Bostock, University of California, Davis, USACorné M. J. Pieterse, Utrecht University, Netherlands
Copyright © 2014 Rehrig, Appel, Jones and Schultz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erin M. Rehrig, Department of Biology and Chemistry, Fitchburg State University, 160 Pearl St. Fitchburg, MA 01564, USA e-mail: erehrig@fitchburgstate.edu
 Erin M. Rehrig
Erin M. Rehrig Heidi M. Appel
Heidi M. Appel A. Daniel Jones
A. Daniel Jones Jack C. Schultz
Jack C. Schultz