- 1Institut National de la Recherche Agronomique, Unité Mixtes de Recherche Biology and Genetics of Plant-Pathogen Interactions, Montpellier, France
- 2Centre de Coopération Internationale en Recherche Agronomique pour le Développement, Unité Mixtes de Recherche Biology and Genetics of Plant-Pathogen Interactions, Montpellier, France
- 3Agriculture Flagship, Commonwealth Scientific and Industrial Research Organisation, Canberra, ACT, Australia
- 4Commonwealth Scientific and Industrial Research Organisation, Digital Productivity and Service, Canberra, ACT, Australia
Plant immunity is often triggered by the specific recognition of pathogen effectors by intracellular nucleotide-binding, leucine-rich repeat receptors (NLR). Plant NLRs contain an N-terminal signaling domain that is mostly represented by either a Toll-interleukin1 receptor (TIR) domain or a coiled coil (CC) domain. In many cases, single NLR proteins are sufficient for both effector recognition and signaling activation. However, many paired NLRs have now been identified where both proteins are required to confer resistance to pathogens. Recent detailed studies on the Arabidopsis thaliana TIR-NLR pair RRS1 and RPS4 and on the rice CC-NLR pair RGA4 and RGA5 have revealed for the first time how such protein pairs function together. In both cases, the paired partners interact physically to form a hetero-complex receptor in which each partner plays distinct roles in effector recognition or signaling activation, highlighting a conserved mode of action of NLR pairs across both monocotyledonous and dicotyledonous plants. We also describe an “integrated decoy” model for the function of these receptor complexes. In this model, a plant protein targeted by an effector has been duplicated and fused to one member of the NLR pair, where it acts as a bait to trigger defense signaling by the second NLR upon effector binding. This mechanism may be common to many other plant NLR pairs.
Introduction
Plants, unlike animals do not possess circulating immune cells to intercept microbial signals. Thus, their innate immune system is cell autonomous, with each plant cell able to recognize microbial signals and respond accordingly. To prevent infection by viruses, bacteria, oomycetes, fungi or nematodes, plant possess two main types of immune receptors: pattern recognition receptors (PRRs) on the cell surface and intracellular nucleotide-binding and leucine-rich repeat receptors (NLR) (Monaghan and Zipfel, 2012; Jacob et al., 2013; Qi and Innes, 2013). PRRs are involved in the recognition of so called pathogen-associated molecular patterns (PAMPs) which are conserved molecules characteristic of entire groups of microbes, whereas NLR resistance (R) proteins perceive specific effectors called avirulence proteins (AVR) derived from pathogens (Jones and Dangl, 2006; Dodds and Rathjen, 2010). Such perception activates effector-triggered immunity that is often associated with a localized programmed cell death called the hypersensitive response (HR). NLR proteins are multidomain proteins and possess a conserved architecture including a C-terminal leucine-rich repeat (LRR) domain, a central nucleotide-binding (NB) domain and a variable N-terminal domain (Takken and Goverse, 2012). In most cases NLR proteins possess a Toll/interleukin-1 receptor (TIR) or coiled-coil (CC) domain at their N-terminus defining two major classes of R proteins: TIR-NLRs and CC-NLRs (Meyers et al., 1999; Pan et al., 2000).
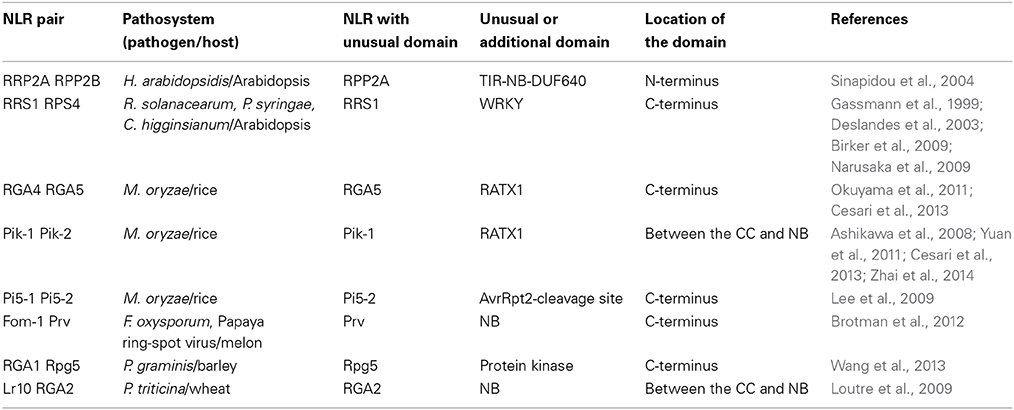
Flor's classical gene-for-gene concept of plant disease resistance states that “for each gene that conditions resistance in the host (R gene) there is a corresponding gene that conditions pathogenicity in the parasite (AVR gene)” (Flor, 1971). Indeed many cases of matching R and AVR genes have been described and, in most cases, individual NLR proteins mediate AVR effector recognition and confer resistance (Jones and Dangl, 2006; Bernoux et al., 2011a). However, an emerging picture is that disease resistance against a pathogen isolate, or recognition of an AVR protein requires, in certain cases, complementary pairs of NLR genes (Eitas and Dangl, 2010). In the first demonstration of this, two TIR-NLR proteins, RPP2A and RPP2B, were both shown to be necessary for Arabidopsis thaliana resistance to the oomycete pathogen Hyaloperonospora arabidopsidis (Sinapidou et al., 2004). More recently, a second pair of Arabidopsis TIR-NLR proteins, RPS4 and RRS1, was shown to be required for recognition of AvrRps4 from the bacterial pathogen Pseudomonas syringae, PopP2 from Ralstonia solanacearum and an as yet uncharacterized factor produced by the fungal pathogen Colletotrichum higginsianum (Gassmann et al., 1999; Deslandes et al., 2003; Birker et al., 2009; Narusaka et al., 2009). This example demonstrates that a single complementary pair of R proteins can mediate recognition of multiple pathogens. Complementary pairs of distinct CC-NLR proteins can also confer resistance as shown by the wheat Lr10 and RGA2 protein pair that mediate resistance to wheat leaf rust caused by Puccinia triticina (Loutre et al., 2009), barley Rpg5 and RGA1 that confer resistance to Puccinia graminis (Wang et al., 2013) and melon Prv and Fom-1 that function against Fusarium oxysporum and Papaya ring-spot virus (Brotman et al., 2012). Recent studies have also described several rice CC-NLRs acting as functional pairs to mediate resistance against M. oryzae, including RGA4/RGA5, Pik-1/Pik-2, and Pi5-1/Pi5-2 (Ashikawa et al., 2008; Lee et al., 2009; Okuyama et al., 2011; Yuan et al., 2011; Zhai et al., 2014). Thus, NLR pairs have been shown to function in the recognition of bacterial, viral, oomycete and fungal pathogens in both monocotyledonous and dicotyledonous plants, suggesting this is a common and widespread mechanism in plant immunity.
Interestingly, all of these resistance loci comprise two tightly linked NLR coding genes, most of which (except RPP2A/RPP2B) are transcribed in opposite directions with a relatively short intergenic region. This conserved genomic organization could be important for co-regulation of these NLR genes or to prevent recombination events leading to separation or inappropriate pairing of NLRs which could cause loss of resistance or spontaneous necrosis (Bomblies et al., 2007). Receptor-like kinase genes of the Lrk (Lr10 receptor-like kinase) and Tak (Triticum aestivum kinase) family also occur in pairs in barley (Hu and Wise, 2008) and wheat (Feuillet et al., 2001), possibly for similar reasons. A key question raised is how do two distinct NLR proteins act together to mediate AVR protein recognition? In the present review, we describe new insights into the functional mechanisms of paired NLR proteins focusing on the recently published models of RPS4/RRS1 and RGA4/RGA5. We will also introduce and discuss a novel model for the recognition of AVR proteins by pairs of co-acting NLRs: the “integrated-decoy model.”
Plant NLRs Play Distinct Roles within Co-Acting Pairs to Confer Resistance
In the absence of pathogens, NLR proteins are kept in an inactive state, whereas, after AVR-recognition, they are activated and induce disease resistance signaling. The molecular mechanisms occurring during the transition from “inactive” to “active” state and the downstream signaling partners are still largely unknown. However, since the first molecular characterization of an NLR-coding gene 20 years ago (Bent et al., 1994; Mindrinos et al., 1994), structure-function analyses have shown that individual domains and particular motifs of NLR proteins play critical roles in their auto-inhibition or activation mechanisms (Takken and Goverse, 2012; Qi and Innes, 2013). Current models of NLR function predict that, in the resting state, intramolecular interactions between different domains maintain R proteins in a closed auto-inhibited conformation. For a number of R proteins, this “off” state has been shown to be associated with ADP binding. It has been proposed that pathogen effector recognition favors a more open NLR protein structure allowing nucleotide exchange and binding to ATP (Williams et al., 2011). Since isolated TIR and CC domains have been shown to be sufficient to trigger an HR-like response (Maekawa et al., 2011; Bernoux et al., 2011b), it is proposed that in the activated state of NLRs, these N-terminal domains would become exposed to interact with signaling partners and initiate disease resistance responses. Structure-function analysis of the L6 TIR domain of flax and the MLA CC domain of barley showed that homotypic interactions of these signaling domains play an important role in the induction of HR (Bernoux et al., 2011b; Maekawa et al., 2011). However, these models based on NLRs functioning singly do not explain how two distinct NLR proteins co-operate in pathogen recognition and signaling.
Two recent studies provided the first analysis of how paired NLRs function together (Cesari et al., 2014; Williams et al., 2014). In one case, Williams et al. (2014) demonstrated by coupling crystal structure and functional analyses, that RPS4 and RRS1 TIR domains form homo- and hetero-dimers through a common conserved interface which includes a core serine-histidine (SH) motif. Mutation of the SH motif in the TIR domain of RRS1, RPS4 or both proteins abolished TIR hetero-dimer formation in solution and also prevented TIR homo-complex formation in yeast two-hybrid experiments. However, similar SH mutations introduced in the full length RPS4 and RRS1 did not prevent hetero-complex formation in co-immunoprecipitation experiments showing that other domains of the proteins, apart from the TIR domains, might also contribute to heterotypic interactions. Transient expression assays in tobacco revealed that the RPS4 TIR domain triggers an effector-independent cell death, which is dependent on the SH motif. Furthermore, a mutation within the RPS4 TIR domain (R30A) that strengthened its homodimerization also enhanced its autoactivity, indicating that the RPS4 TIR domain signals as a homo-dimer. Co-expression of the RRS1 TIR domain inhibited this RPS4 TIR autoactive cell death, and this was dependent on the RRS1 SH motif. This suggests that an inactive RRS1/RPS4 TIR hetero-dimer competes with formation of the active RPS4 TIR homo-dimer to inhibit signaling. Indeed, the structural data showed that both complexes involve a common interface and in vitro and in vivo experiments indicated that TIR domain hetero-dimers are more stable than homodimers. Thus, TIR domain hetero-interactions may play a crucial role in maintaining the full-length protein pair complex in an inactive state. Interestingly, TIR domain hetero-complex formation also seems to be required for AVR recognition since mutation of the SH motif in full length RRS1 impairs AvrRps4 and PopP2 recognition.
Both AvrRps4 and PopP2 are recognized by direct binding to the RRS1 protein, which contains a C-terminal WRKY domain, but do not interact with RPS4 (Deslandes et al., 2003; Tasset et al., 2010; Williams et al., 2014). Interestingly mutation of the P-loop motif required for nucleotide binding abolishes the function of RPS4, consistent with the standard NLR activation model, while P-loop mutations do not affect RRS1 function, suggesting a specialized receptor function that does not require the formation of a nucleotide-dependent activation state (Williams et al., 2014). Thus, Williams et al. proposed a model in which RPS4 and RRS1 form an inactive hetero-dimer in the absence of pathogen recognition. Binding of the AVRs to RRS1 causes the disruption of the RPS4/RRS1 TIR domain hetero-dimer, allowing formation of a signaling competent RPS4 TIR domain homo-dimer (Figure 1). Since recognition of the AVRs does not seem to lead to disruption of the RRS1/RPS4 full-length hetero-complex, the activated state may be a tetramer. Thus, RRS1 and RPS4 function as a hetero-complex receptor with each partner having a different role: RPS4 acting as an inducer of disease resistance signaling and RRS1 acting as an effector binding receptor and repressor of RPS4 TIR signaling activity.
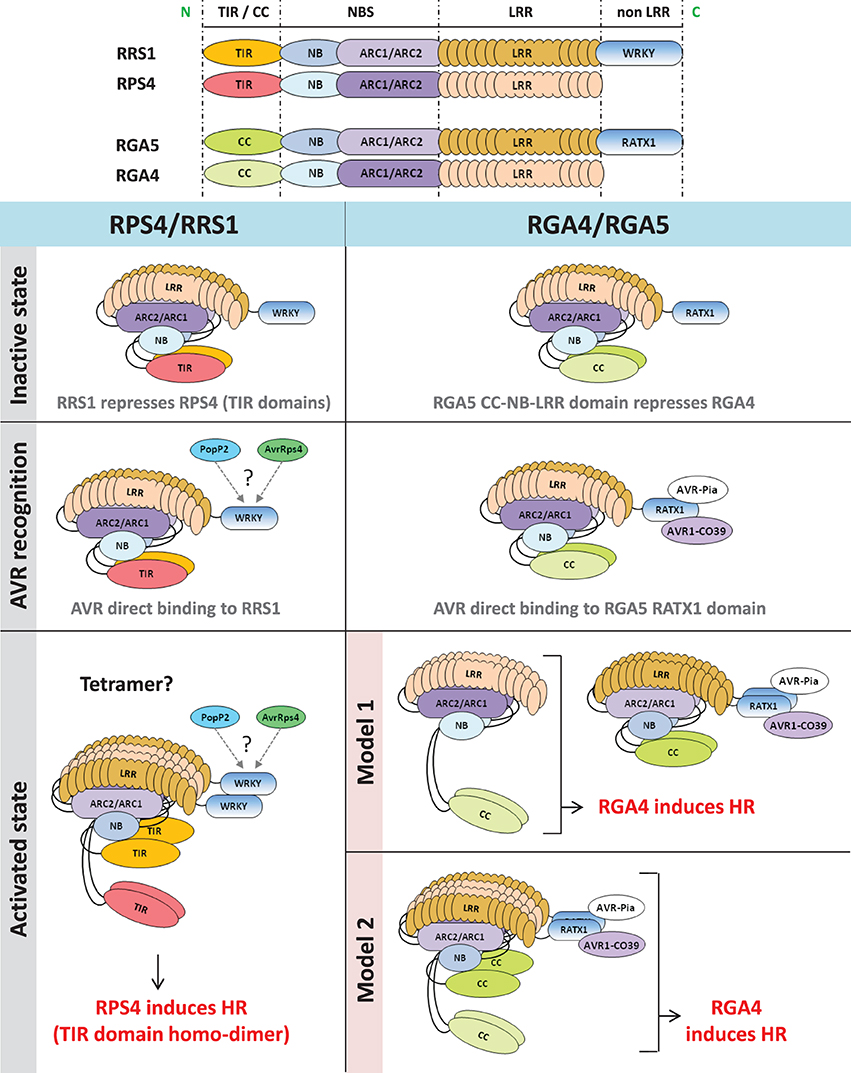
Figure 1. Functional models of RPS4/RRS1 and RGA4/RGA5 NLR pairs. In the inactive state, co-acting NLRs form a hetero-complex in which the signaling NLR (RPS4 or RGA4) is repressed by the AVR-receptor NLR (RRS1 or RGA5). After pathogen challenge, direct recognition of AVR proteins by the receptor NLR partner (RRS1 or RGA5) occurs. In the case of RGA5, AVR-binding involves the C-terminus containing the RATX1 domain while RRS1 interacts with PopP2 and AvrRps4 through its WRKY domain (Jones and Deslandes, personal communication). After AVR-recognition, NLR hetero-complexes are still present in both cases. Induction of HR by RPS4/RRS1 involves disruption of TIR domains hetero-interaction and subsequent homo-dimerization of RPS4 TIR domain. In the case of RGA4 and RGA5, we cannot rule out that induction of HR requires disruption of RGA4 and RGA5 hetero-complexes. Hence, two hypothetical models are proposed. According to the first model, HR is induced by RGA4 homo-complex freed from the RGA5/AVRs complex, whereas, in the second model, conformational changes within the RGA4/RGA5 hetero-complex would allow RGA4 to signal. N, N-terminal; C, C-terminal; TIR, Toll/interleukin1 receptor; CC, coiled coil; NBS, nucleotide binding site; NB, nucleotide binding; ARC, Apaf-1, R-protein and CED-4; LRR, leucine-rich repeats; HR, hypersensitive response.
In the second case, Cesari et al. (2014) investigated the mode of action of RGA4 and RGA5, two rice CC-NLRs. Together, RGA4 and RGA5 are necessary and sufficient to mediate Pia and Pi-CO39 resistances and recognize the M. oryzae effectors AVR-Pia and AVR1-CO39 (Cesari et al., 2013). Previous work showed that both effectors interact physically with RGA5, but do not bind to RGA4 suggesting that, similarly to RRS1, RGA5 acts as an AVR-receptor (Cesari et al., 2013). A domain at the C-terminus of RGA5 characterized by a heavy metal associated domain related to the cytoplasmic copper chaperone ATX1 from Saccharomyces cerevisiae (RATX1 domain) was identified as the AVR-binding domain in RGA5. Expression of the full length RGA4 protein, but not RGA5, in rice protoplasts and Nicotiana benthamiana leaves triggered cell death (Cesari et al., 2014). This autonomous activity of RGA4 was dependent on an intact P-loop motif and may be related to an unusual MHD motif within its NB domain in which the three usually highly conserved core amino acids are replaced by TYG. Mutational analysis showed that the glycine in this motif is crucial for the autoactivity of the protein as restoration of an aspartate residue at this position abolishes cell death. Yeast two-hybrid and co-immunoprecipitation assays indicate that RGA4 and RGA5 interact and form both homo- and hetero-complexes. These interactions are largely mediated by their CC domains although the RATX1 domain of RGA5 also self-interacts in yeast two-hybrid assay and could contribute to RGA5 homo-complex formation. Co-expression of RGA5 suppressed RGA4-mediated cell death indicating that formation of the RGA4/RGA5 hetero-complex might be crucial to regulate RGA4 activity in the absence of pathogen. Interestingly, the RATX1 domain of RGA5 is dispensable for RGA4 inhibition, showing that it probably serves primarily as an effector binding platform. Co-expression of AVR-Pia with RGA4 and RGA5 again triggered cell death. Hence, a functional model was proposed in which RGA4 acts as a signaling component regulated by its interaction with RGA5 which acts both as a repressor and a receptor that directly binds the AVR proteins (Figure 1).
From those discoveries, striking similarities are apparent between the RPS4/RRS1 and the RGA4/RGA5 functional models. Within a pair, distinct NLR proteins: (i) form homo- and hetero-complexes that are involved in important regulation processes both prior to and following pathogen recognition, (ii) cooperate to achieve one task, but are specialized and accomplish distinct functions within the complex (i.e., AVR-receptor or signal inducer), (iii) recognize multiple AVR proteins through direct binding to the “receptor” partner which possesses an unusual domain that is not conserved in other NLRs, (iv) rely on the N-terminal TIR or CC domain of the “signal inducer” NLR to translate recognition into activation of resistance responses. Recent analysis of the Pik-1 and Pik-2 CC-NLR pair showed that these proteins also form hetero-complexes through their CC domains (Zhai et al., 2014), with Pik-1 acting as a receptor mediating direct recognition of AVR-Pik whereas Pik-2 does not bind the effector but possesses HR signaling activity (Kanzaki et al., 2012; Cesari et al., 2013; Zhai et al., 2014). Thus, these functional models may be generally applicable to other NLR protein pairs.
An Integrated Decoy Model Describes AVR Recognition by NLR Pairs
NLR proteins mediate recognition of AVR effectors either by direct physical interaction (Dodds and Rathjen, 2010) or in an indirect manner (Dangl and Jones, 2001; van der Hoorn and Kamoun, 2008). In cases of indirect recognition, the NLR responds to modifications of another plant protein induced by the AVR protein. This plant protein may be either the operational target of the effector, in which case it is called a “guardee” since it is guarded by the NLR (Dangl and Jones, 2001), or a mimic of the operational target in which case it is called a “decoy” (van der Hoorn and Kamoun, 2008).
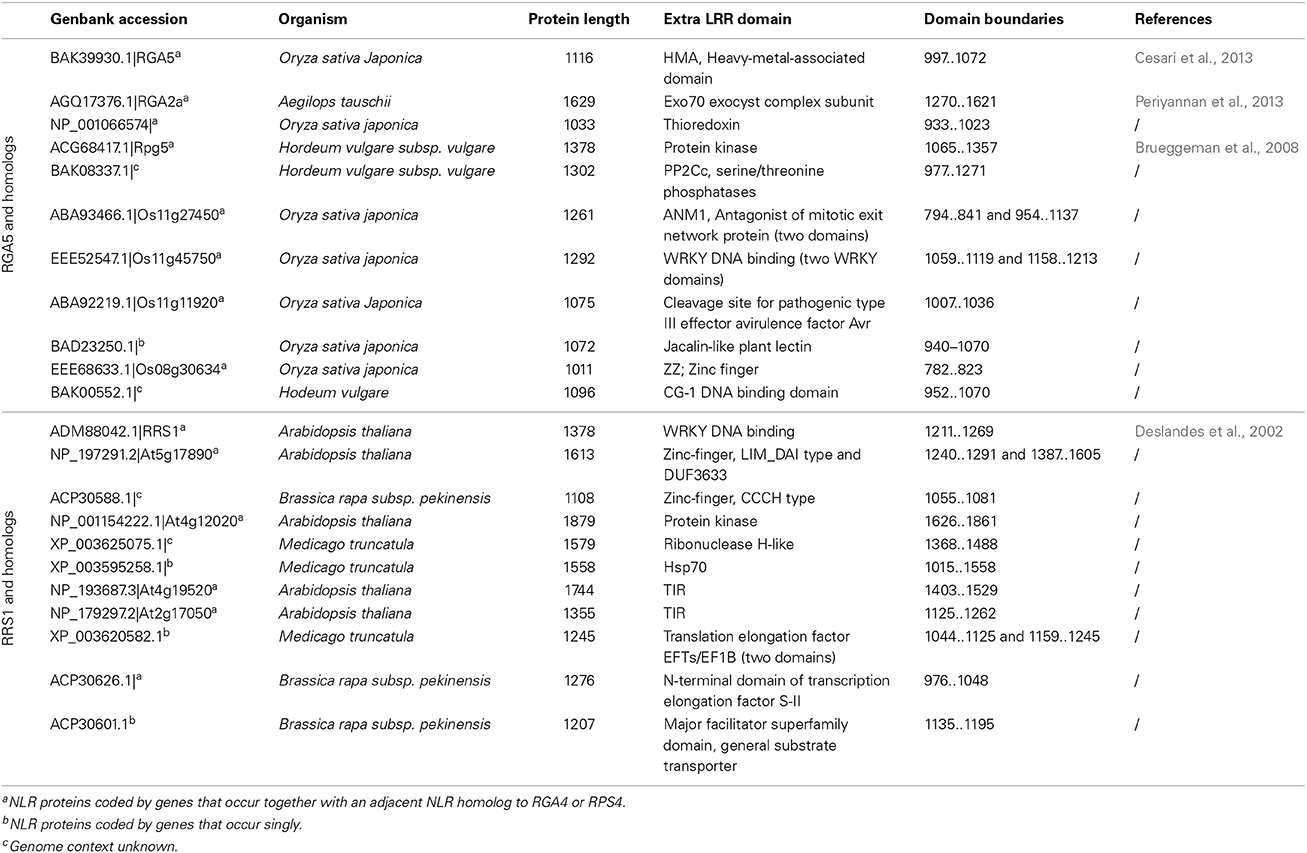
In the case of NLR pairs, all studied examples show direct binding between AVR effectors and the NLR acting as an AVR receptor. AVR1-CO39 and AVR-Pia bind to RGA5, AvrRps4, and PopP2 interact with RRS1 and AVR-Pik binds to Pik-1 (Kanzaki et al., 2012; Cesari et al., 2013; Williams et al., 2014; Zhai et al., 2014). However, one striking feature of these NLR receptor proteins is that, apart from their conserved multidomain NLR architecture, they all contain additional unusual non-conserved domains (Table 1). Indeed, RRS1 possesses a C-terminal WRKY domain (Deslandes et al., 2002, p. 1) and RGA5 and Pik-1 both display a similar RATX1 domain (Okuyama et al., 2011; Cesari et al., 2013). In other known NLR pairs, one partner also contains an additional domain: a C-terminal protein kinase domain in Rpg5 (Brueggeman et al., 2008), an N-terminal TIR-NB-DUF640 in RPP2A (Sinapidou et al., 2004), a C-terminal NB domain in Prv (Brotman et al., 2012), and a second NB domain following the first one in RGA2 (Loutre et al., 2009). In Pi5-2 we identified a C-terminal domain containing a cleavage site (VPPFGEW, amino acids 1022 to 1028) for the P. syringae type III effector AvrRpt2, similar to that found in RIN4. This observation raises a central question: What could be the function of those specific non-conserved domains? Their striking diversity and the fact that they are not conserved in the majority of NLRs suggests that they do not play a critical role in signaling or regulation within the pair. This is consistent with the observation that RGA5, Pik-1 and RRS1 do not function directly in resistance signaling and that the RATX1 domain of RGA5 is not required to regulate RGA4.
In the case of RGA4/RGA5, both AVR-Pia and AVR1-CO39 bind directly to the RATX1-containing domain at the C-terminus of RGA5 (Cesari et al., 2013). A similar RATX1 domain is also present in Pik-1 and this was shown to bind AVR-Pik (Kanzaki et al., 2012; Cesari et al., 2013; Zhai et al., 2014). However, in Pik-1, the RATX1 is located between the CC and the NB domains suggesting that this unusual AVR-binding domain has been acquired independently in the two CC-NLRs. In the light of these findings, we propose an “integrated decoy” model to explain effector recognition in these cases of paired NLRs (Figure 2). This is an extension of the decoy model (van der Hoorn and Kamoun, 2008) in which proteins that were initially targeted by pathogen effectors, have evolved into decoys guarded by NLRs. In this case however, the decoy is integrated in the R protein structure allowing direct recognition of AVR effectors. Support for the idea that the RATX1 domain present in RGA5 and Pik-1 could be a decoy comes from the observation that a similar domain is also present in the rice protein Pi21 which is required for full susceptibility to the rice blast fungus (Fukuoka et al., 2009). Thus, proteins containing this domain could be important virulence targets for M. oryzae effectors such as AVR1-CO39, AVR-Pia, and AVR-Pik. The C-terminal domains or motifs of RRS1 (WRKY), Pi5-2 (AvrRpt-cleavage site), and Rpg5 (protein kinase) all belong to families of proteins that have important roles in defense signaling (Pandey and Somssich, 2009; Deslandes and Rivas, 2012; Lin et al., 2013) supporting the hypothesis that they may represent integrated decoys. Consistent with this, recent unpublished work has suggested that AvrRps4 and PopP2 interact with the WRKY domain of RRS1 (Deslandes and Jones, personal communication). A key prediction of this model is that the virulence targets of these effectors are related to the recognition domains of the receptor NLR protein; RATX1 domain containing proteins in the case of AVR-Pia, AVR1-CO39 and AVR-Pik, and WRKY class transcription factors in the case of AvrRps4 and PopP2. Thus, the identification of the virulence targets of these effectors, which has not yet been reported, will be a critical test of the general validity of this model.
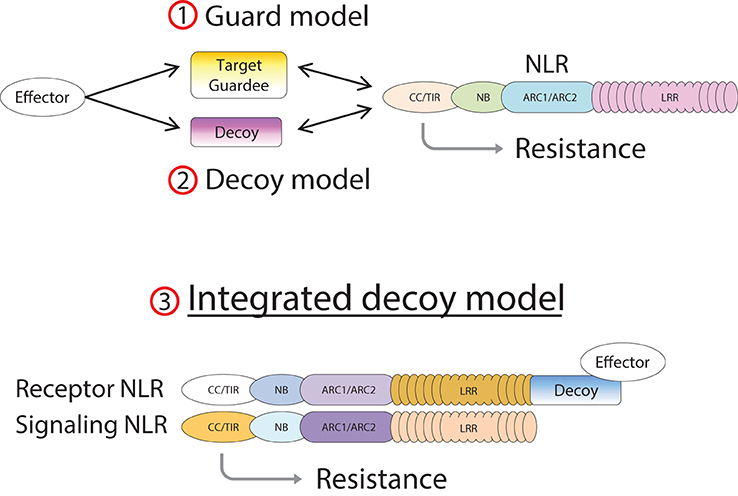
Figure 2. Model of integrated decoys in NLR protein pairs. Pathogen effectors target host proteins for manipulation in order to promote infection. Indirect recognition of these effectors occurs when these target proteins are guarded by host NLR proteins (1), or if duplicated target genes evolve into decoy proteins monitored by host NLRs (2). Alternatively the decoy may be integrated into the structure of the receptor component of an NLR pair (3), allowing AVR recognition by direct binding.
A genome search revealed that rice cultivar Nipponbare contains nine homologous gene pairs to RGA4/RGA5, which are all arranged as divergently transcribed genes with one exception arranged in a head-to-tail orientation. Interestingly, despite high sequence similarity in the CC-NLR domains, the RATX1 domain is not conserved among RGA5 rice homologs, nor in a wider set of RGA5 homologs identified in cereals, and in fact this C-terminal region is substituted in many cases by different protein domains that may also act as integrated decoys (Table 2, Supplemental Tables 1, 2). Consistent with this idea, many of these domains belong to protein families that are involved in disease resistance, such as protein kinases (i.e., MAPKs), WRKYs, or AvrRpt-cleavage site containing proteins. Likewise, Arabidopsis contains eight homologous pairs to RPS4/RRS1 (Narusaka et al., 2009), and a BLAST analysis performed with the RRS1 protein also identified many homologs in other plants in which the WRKY domain is replaced by various C-terminal domains, such as protein kinases, TIR, Zinc-finger, transcription elongation factor or different WRKY domains (Table 2, Supplemental Tables 3, 4). Blast2GO analysis of the C-terminal domains of the RGA5 and RRS1 homologs showed very similar Gene Ontology (GO) enrichment results. In terms of biological processes, both sets show a significant enrichment for “defense response” (Supplemental Figure 1) while in terms of molecular function, they both show a significant enrichment for catalytic activities such as “kinase activity” or “phosphatase activity” and for binding activity such as “DNA binding” (Supplemental Figure 2). This observation suggests that the protein domains that have been integrated in the structure of RGA5 and RRS1 homologs share similar functions and are involved in similar biological processes, particularly defense responses.
These observations suggest that diverse protein domains have been integrated in the NLR protein acting as a receptor in order to sense and physically bind AVR-effectors. In evolutionary terms, duplication of paired NLR genes may be followed by acquisition of new domains through random genome re-arrangements, with selection favoring chimeric NLR-decoy proteins that confer novel resistance phenotypes. Why these events seem to occur predominantly in paired NLRs, rather than in single NLRs is an intriguing question. Perhaps this architecture offers a unique functional model to exploit this integration because the mode of action only requires that interaction of the AVR with the receptor NLR disrupts suppression of the signaling NLR.
Under the integrated decoy model the “direct” vs. “indirect” recognition dichotomy is no longer appropriate to describe AVR recognition by NLR pairs, which have apparently evolved a direct recognition function via incorporation of a decoy molecule into their structure. Nevertheless, other examples of direct recognition, such as those mediated by the R proteins RPP1 in Arabidopsis, L6 and M in flax or N in tobacco, involve single NLR proteins with no additional domains (Dodds et al., 2006; Ueda et al., 2006; Catanzariti et al., 2010; Krasileva et al., 2010). As observed for these directly interacting single NLRs, the decoy component of the NLR pair may be in an evolutionary arms race with the corresponding effectors. Consistent with this, analysis of polymorphisms within Pik-1 indicates that diversifying selection occurs at the RATX1 domain and that these polymorphisms are responsible for recognition specificity of AVR-Pik alleles by the different Pik-1 alleles (Kanzaki et al., 2012).
Striking Similarities and Differences Between Plant and Animal Co-Acting NLRs
In an interesting parallel, several cytosolic mammalian NLR immune receptors have also been shown to function in heteromeric complexes (von Moltke et al., 2013). One of them, NAIP5 (NLR family, apoptosis inhibitory proteins 5), was shown to be essential to restrict the pathogenic bacteria Legionella pneumophila in mice macrophages (Beckers et al., 1995; Dietrich et al., 1995) and to respond to flagellin, inducing a caspase-1 dependent pyroptotic cell death (Lightfield et al., 2008; Bergsbaken et al., 2009). However, NAIP5 lacks a signaling CARD (caspase activation and recruitment domain) and caspase-1 activation relies on a second member of the NLR family, called NLRC4, which contains an N-terminal CARD (Amer et al., 2006; Franchi et al., 2006; Miao et al., 2006).
Two elegant studies shed light on the role and function of NAIP5 and NLRC4 upon flagellin perception (Kofoed and Vance, 2011; Zhao et al., 2011). Co-expression of flagellin with NLRC4 and NAIP5 in human cells lacking NAIP5 and NLRC4 homologs results in the formation of an inflammasome, a high molecular complex that contains the three proteins and activates the caspase-1 signaling pathway. Within the complex, flagellin interacts directly and specifically with NAIP5, but not with NLRC4. In this system, NAIP5 acts as the direct receptor for flagellin while NLRC4 orchestrates downstream signaling responses.
Interestingly, the NLCR4 inflammasome also responds to another conserved bacterial motif present in the type III secretion system of many bacterial pathogens: the inner rod protein (i.e., PrgJ in Salmonella and BsaK in Burkholderia) (Miao et al., 2010). However, NAIP5 is not required for PrgJ-mediated NLRC4 activation (Lightfield et al., 2011) and screening other members of the NAIP family revealed that NAIP2 binds directly and specifically to BsaK and PrgJ (Kofoed and Vance, 2011; Zhao et al., 2011). Co-expression of NAIP2, PrgJ and NLRC4, results in the formation of an inflammasome and caspase-1 mediated signaling activation.
Thus, similarly to plant co-acting NLRs, NAIPs, and NLRC4 act as a dual immune receptor in which one partner (NAIP) specifically recognizes and binds to pathogen molecules whereas the other partner (NLRC4) translates immune signaling. However, Naip and Nlrc4 genes are not co-located in the mouse genome, as seen for the plant NLR gene pairs. In addition, NAIP proteins are involved in the recognition of PAMPs whereas plant NLR proteins recognize specific effectors. Another striking difference is that NAIPs are monomeric in the absence of ligand and associate with NLRC4 only upon ligand recognition (Halff et al., 2012; Tenthorey et al., 2014) whereas the plant paired NLRs associate before and after effector recognition (Cesari et al., 2014; Williams et al., 2014). Thus, NLRC4 seems to act as a downstream signaling partner for various NAIP receptors. In contrast, no other NLRs have been shown to bind to RGA4 or RPS4, so they do not appear to be common signaling partners, but rather act as part of a specific receptor complex. On the other hand two plant NLRs, Arabidopsis ADR1 and tomato NRC1 have been described to act as a signaling hub or NB-LRR helper for the function and defense signaling activation of certain immune receptors (Gabriëls et al., 2007; Roberts et al., 2013), and thus could function in a similar manner to NLRC4. However, there is not yet any evidence that ADR1 or NRC1 interact physically with other NLRs. It is possible that the genetically paired NLRs may have evolved from such helper interactions through physical coupling of co-evolved receptor and helper NLR genes.
Concluding Remarks and Future Directions
The molecular characterization of the first NLR genes was achieved 20 years ago with the cloning of RPS2 and N (Bent et al., 1994; Mindrinos et al., 1994; Whitham et al., 1994). Since then, remarkable progress in our understanding of NLR proteins' mode of action have been made, unraveling a high level of complexity in the way these receptors operate to perceive pathogen effectors in plant cells. Both “direct” and “indirect” AVR-recognition have been observed and the “integrated decoy” model described here could be regarded as an intermediate between these recognition modes. This model implies that some instances of direct recognition are derived from guard/decoy type indirect recognition. Numerous potential integrated decoys occur in RRS1 and RGA5 homologs and if they represent domains targeted by effector proteins, their identification and analysis should provide valuable information about cellular processes targeted by pathogens during infection. In addition, integrated decoys could also be used as baits to screen for interacting AVR proteins. Ideally, the widespread nature of integrated decoys in NLR protein pairs should pave the way toward R protein engineering allowing effector targets to be fused to a receptor NLR operating within a pair to create an “effector trap.”
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the CSIRO Bioinformatics core and Annette McGrath for their help in the Interproscan and Blast2GO analyses. We thank Laurent Deslandes and Jonathan Jones for sharing unpublished data. Stella Cesari was supported by the program “contrat jeune scientifique” of INRA and Maud Bernoux was a recipient of a Discovery Early Career Award (DE130101292) from the Australian Research Council.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00606/abstract
Supplemental Figure 1. Domains integrated in RGA5 and RRS1 homologs are involved in similar biological processes, particularly defense responses. (A) Five hundred homologs of RGA5 were identified by Blastp against the NCBI nr database. Their sequences were aligned and a cut-off between the conserved N-terminus and the variable C-terminus was defined in the multiple alignment. C-terminal sequences (after the cut-off) were analyzed by Blast2GO and total number of GO annotations corresponding to each biological process is displayed. (B) Same procedure as in (A) was applied for RRS1 homologs (cut-off 1). GO, gene ontology; BP, biological process; #seqs, number of sequences; #GO, GO terms.
Supplemental Figure 2. Domains integrated in RGA5 and RRS1 homologs show a significant enrichment for similar molecular functions. (A) Blast2GO analysis for RGA5 homologs' C-terminal domains (molecular function). (B) Same as in (A) for RRS1 homologs' C-terminal domains (cut-off 1). GO, gene ontology.
Supplemental Table 1. Manually identified C-terminal domains in RGA5 homologs.
Supplemental Table 2. Interproscan identified C-terminal domains in RGA5 homologs.
Supplemental Table 3. Interproscan identified C-terminal domains in RRS1 homologs (cut-off L850).
Supplemental Table 4. Interproscan identified C-terminal domains in RRS1 homologs (cut-off Y894).
References
Amer, A., Franchi, L., Kanneganti, T.-D., Body-Malapel, M., Ozoren, N., Brady, G., et al. (2006). Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J. Biol. Chem. 281, 35217–35223. doi: 10.1074/jbc.M604933200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ashikawa, I., Hayashi, N., Yamane, H., Kanamori, H., Wu, J., Matsumoto, T., et al. (2008). Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics 180, 2267–2276. doi: 10.1534/genetics.108.095034
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Beckers, M.-C., Yoshida, S.-I., Morgan, K., Skamene, E., and Gros, P. (1995). Natural resistance to infection with Legionella pneumophila: chromosomal localization of the Lgn1 susceptibility gene. Mamm. Genome 6, 540–545. doi: 10.1007/BF00356173
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bent, A. F., Kunkel, B. N., Dahlbeck, D., Brown, K. L., Schmidt, R., Giraudat, J., et al. (1994). RPS2 of Arabodopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265, 1856–1860. doi: 10.1126/science.8091210
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bergsbaken, T., Fink, S. L., and Cookson, B. T. (2009). Pyroptosis: host cell death and inflammation. Nat. Rev. Microbiol. 7, 99–109. doi: 10.1038/nrmicro2070
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bernoux, M., Ellis, J. G., and Dodds, P. N. (2011a). New insights in plant immunity signaling activation. Curr. Opin. Plant Biol. 14, 512–518. doi: 10.1016/j.pbi.2011.05.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bernoux, M., Ve, T., Williams, S., Warren, C., Hatters, D., Valkov, E., et al. (2011b). Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9, 200–211. doi: 10.1016/j.chom.2011.02.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Birker, D., Heidrich, K., Takahara, H., Narusaka, M., Deslandes, L., Narusaka, Y., et al. (2009). A locus conferring resistance to Colletotrichum higginsianum is shared by four geographically distinct Arabidopsis accessions. Plant J. 60, 602–613. doi: 10.1111/j.1365-313X.2009.03984.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bomblies, K., Lempe, J., Epple, P., Warthmann, N., Lanz, C., Dangl, J. L., et al. (2007). Autoimmune response as a mechanism for a dobzhansky-muller-type incompatibility syndrome in plants. PLoS Biol. 5:e236. doi: 10.1371/journal.pbio.0050236
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brotman, Y., Normantovich, M., Goldenberg, Z., Zvirin, Z., Kovalski, I., Stovbun, N., et al. (2012). Dual resistance of melon to Fusarium oxysporum Races 0 and 2 and to Papaya ring-spot virus is controlled by a pair of head-to-head-oriented NB-LRR genes of unusual architecture. Mol. Plant 6, 235–238. doi: 10.1093/mp/sss121
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brueggeman, R., Druka, A., Nirmala, J., Cavileer, T., Drader, T., Rostoks, N., et al. (2008). The stem rust resistance gene Rpg5 encodes a protein with nucleotide-binding-site, leucine-rich, and protein kinase domains. Proc. Natl. Acad. Sci. U.S.A. 105, 14970–14975. doi: 10.1073/pnas.0807270105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Catanzariti, A. M., Dodds, P. N., Ve, T., Kobe, B., Ellis, J. G., and Staskawicz, B. J. (2010). The AvrM effector from flax rust has a structured C-terminal domain and interacts directly with the M resistance protein. Mol. Plant Microbe Interact. 23, 49–57. doi: 10.1094/MPMI-23-1-0049
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cesari, S., Kanzaki, H., Fujiwara, T., Bernoux, M., Chalvon, V., Kawano, Y., et al. (2014). The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 33, 1941–1959. doi: 10.15252/embj.201487923
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cesari, S., Thilliez, G., Ribot, C., Chalvon, V., Michel, C., Jauneau, A., et al. (2013). The Rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25, 1463–1481. doi: 10.1105/tpc.112.107201
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dangl, J. L., and Jones, J. D. G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Deslandes, L., Olivier, J., Peeters, N., Feng, D. X., Khounlotham, M., Boucher, C., et al. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. U.S.A. 100, 8024–8029. doi: 10.1073/pnas.1230660100
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Deslandes, L., Olivier, J., Theulières, F., Hirsch, J., Feng, D. X., Bittner-Eddy, P., et al. (2002). Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. U.S.A. 99, 2404–2409. doi: 10.1073/pnas.032485099
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Deslandes, L., and Rivas, S. (2012). Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci. 17, 644–655. doi: 10.1016/j.tplants.2012.06.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dietrich, W. F., Damron, D. M., Isberg, R. R., Lander, E. S., and Swanson, M. S. (1995). Lgn1, a gene that determines susceptibility to Legionella pneumophila, maps to mouse chromosome 13. Genomics 26, 443–450. doi: 10.1016/0888-7543(95)80161-E
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dodds, P. N., Lawrence, G. J., Catanzariti, A. M., Teh, T., Wang, C. I., Ayliffe, M. A., et al. (2006). Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. U.S.A. 103, 8888–8893. doi: 10.1073/pnas.0602577103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eitas, T. K., and Dangl, J. L. (2010). NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr. Opin. Plant Biol. 13, 472–477. doi: 10.1016/j.pbi.2010.04.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Feuillet, C., Penger, A., Gellner, K., Mast, A., and Keller, B. (2001). Molecular evolution of receptor-like kinase genes in hexaploid wheat. Independent evolution of orthologs after polyploidization and mechanisms of local rearrangements at paralogous loci. Plant Phys. 125, 1304–1313. doi: 10.1104/pp.125.3.1304
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Flor, H. H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. doi: 10.1146/annurev.py.09.090171.001423
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Franchi, L., Amer, A., Body-Malapel, M., Kanneganti, T.-D., Özören, N., Jagirdar, R., et al. (2006). Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1β in salmonella-infected macrophages. Nat. Immunol. 7, 576–582. doi: 10.1038/ni1346
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fukuoka, S., Saka, N., Koga, H., Ono, K., Shimizu, T., Ebana, K., et al. (2009). Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325, 998–1001. doi: 10.1126/science.1173275
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gabriëls, S. H. E. J., Vossen, J. H., Ekengren, S. K., van Ooijen, G., Abd-El-Haliem, A. M., Berg, G. C. M., et al. (2007). An NB-LRR protein required for HR signalling mediated by both extra- and intracellular resistance proteins: NRC1 is required for HR signaling. Plant J. 50, 14–28. doi: 10.1111/j.1365-313X.2007.03027.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gassmann, W., Hinsch, M. E., and Staskawicz, B. J. (1999). The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 20, 265–277. doi: 10.1046/j.1365-313X.1999.t01-1-00600.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Halff, E. F., Diebolder, C. A., Versteeg, M., Schouten, A., Brondijk, T. H. C., and Huizinga, E. G. (2012). Formation and structure of a NAIP5-NLRC4 inflammasome induced by direct interactions with conserved N- and C-terminal regions of flagellin. J. Biol. Chem. 287, 38460–38472. doi: 10.1074/jbc.M112.393512
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hu, P., and Wise, R. P. (2008). Diversification of the Lrk/Tak kinase gene clusters is associated with subfunctionalisation and cultivar specific transcript accumulation in barley. Funct. Itegr. Genomics 8, 199–209. doi: 10.1007/s10142-008-0077-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jacob, F., Vernaldi, S., and Maekawa, T. (2013). Evolution and conservation of plant NLR functions. Front. Immunol. 4:297. doi: 10.3389/fimmu.2013.00297
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kanzaki, H., Yoshida, K., Saitoh, H., Fujisaki, K., Hirabuchi, A., Alaux, L., et al. (2012). Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions: Co-evolution of fungal and rice genes. Plant J. 72, 894–907. doi: 10.1111/j.1365-313X.2012.05110.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kofoed, E. M., and Vance, R. E. (2011). Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477, 592–595. doi: 10.1038/nature10394
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Krasileva, K. V., Dahlbeck, D., and Staskawicz, B. J. (2010). Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 22, 2444–2458. doi: 10.1105/tpc.110.075358
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, S.-K., Song, M.-Y., Seo, Y.-S., Kim, H.-K., Ko, S., Cao, P.-J., et al. (2009). Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two Coiled-Coil-Nucleotide-Binding-Leucine-Rich Repeat genes. Genetics 181, 1627–1638. doi: 10.1534/genetics.108.099226
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lightfield, K. L., Persson, J., Brubaker, S. W., Witte, C. E., von Moltke, J., Dunipace, E. A., et al. (2008). Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat. Immunol. 9, 1171–1178. doi: 10.1038/ni.1646
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lightfield, K. L., Persson, J., Trinidad, N. J., Brubaker, S. W., Kofoed, E. M., Sauer, J.-D., et al. (2011). Differential requirements for NAIP5 in activation of the NLRC4 inflammasome. Infect. Immun. 79, 1606–1614. doi: 10.1128/IAI.01187-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lin, W., Ma, X., Shan, L., and He, P. (2013). Big roles of small kinases: the complex functions of receptor-like cytoplasmic kinases in plant immunity and development: multiple functions of RLCKs. J. Integr. Plant Biol. 55, 1188–1197. doi: 10.1111/jipb.12071
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Loutre, C., Wicker, T., Travella, S., Galli, P., Scofield, S., Fahima, T., et al. (2009). Two different CC-NBS-LRR genes are required for Lr10-mediated leaf rust resistance in tetraploid and hexaploid wheat. Plant J. 60, 1043–1054. doi: 10.1111/j.1365-313X.2009.04024.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Maekawa, T., Cheng, W., Spiridon, L. N., Töller, A., Lukasik, E., Saijo, Y., et al. (2011). Coiled-Coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9, 187–199. doi: 10.1016/j.chom.2011.02.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Meyers, B. C., Dickerman, A. W., Michelmore, R. W., Sivaramakrishnan, S., Sobral, B. W., and Young, N. D. (1999). Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 20, 317–332. doi: 10.1046/j.1365-313X.1999.t01-1-00606.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miao, E. A., Alpuche-Aranda, C. M., Dors, M., Clark, A. E., Bader, M. W., Miller, S. I., et al. (2006). Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat. Immunol. 7, 569–575. doi: 10.1038/ni1344
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miao, E. A., Mao, D. P., Yudkovsky, N., Bonneau, R., Lorang, C. G., Warren, S. E., et al. (2010). Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. U.S.A. 107, 3076–3080. doi: 10.1073/pnas.0913087107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mindrinos, M., Katagiri, F., Yu, G.-L., and Ausubel, F. M. (1994). The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78, 1089–1099. doi: 10.1016/0092-8674(94)90282-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Monaghan, J., and Zipfel, C. (2012). Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15, 349–357. doi: 10.1016/j.pbi.2012.05.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Narusaka, M., Shirasu, K., Noutoshi, Y., Kubo, Y., Shiraishi, T., Iwabuchi, M., et al. (2009). RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 60, 218–226. doi: 10.1111/j.1365-313X.2009.03949.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Okuyama, Y., Kanzaki, H., Abe, A., Yoshida, K., Tamiru, M., Saitoh, H., et al. (2011). A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 66, 467–479. doi: 10.1111/j.1365-313X.2011.04502.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pan, Q., Wendel, J., and Fluhr, R. (2000). Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J. Mol. Evol. 50, 203–213. doi: 10.1007/s002399910023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pandey, S. P., and Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. doi: 10.1104/pp.109.138990
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Periyannan, S., Moore, J., Ayliffe, M., Bansal, U., Wang, X., Huang, L., et al. (2013). The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science 341, 786–788. doi: 10.1126/science.1239028
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Qi, D., and Innes, R. W. (2013). Recent advances in plant NLR structure, function, localization, and signaling. Front. Immunol. 4:348. doi: 10.3389/fimmu.2013.00348
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roberts, M., Tang, S., Stallmann, A., Dangl, J. L., and Bonardi, V. (2013). Genetic requirements for signaling from an autoactive plant NB-LRR intracellular innate immune receptor. PLoS Genet. 9:e1003465. doi: 10.1371/journal.pgen.1003465
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sinapidou, E., Williams, K., Nott, L., Bahkt, S., Tör, M., Crute, I., et al. (2004). Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J. 38, 898–909. doi: 10.1111/j.1365-313X.2004.02099.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Takken, F. L., and Goverse, A. (2012). How to build a pathogen detector: structural basis of NB-LRR function. Curr. Opin. Plant Biol. 15, 375–384. doi: 10.1016/j.pbi.2012.05.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tasset, C., Bernoux, M., Jauneau, A., Pouzet, C., Brière, C., Kieffer-Jacquinod, S., et al. (2010). Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PLoS Pathog. 6:e1001202. doi: 10.1371/journal.ppat.1001202
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tenthorey, J. L., Kofoed, E. M., Daugherty, M. D., Malik, H. S., and Vance, R. E. (2014). Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes. Mol. Cell 54, 17–29. doi: 10.1016/j.molcel.2014.02.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ueda, H., Yamaguchi, Y., and Sano, H. (2006). Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Mol. Biol. 61, 31–45. doi: 10.1007/s11103-005-5817-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
van der Hoorn, R. A. L., and Kamoun, S. (2008). From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20, 2009–2017. doi: 10.1105/tpc.108.060194
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
von Moltke, J., Ayres, J. S., Kofoed, E. M., Chavarría-Smith, J., and Vance, R. E. (2013). Recognition of bacteria by inflammasomes. Annu. Rev. Immunol. 31, 73–106. doi: 10.1146/annurev-immunol-032712-095944
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, X., Richards, J., Gross, T., Druka, A., Kleinhofs, A., Steffenson, B., et al. (2013). The rpg4-mediated resistance to wheat stem rust (Puccinia graminis) in barley (Hordeum vulgare) requires Rpg5, a second NBS-LRR gene, and an actin depolymerization factor. Mol. Plant Microbe Interact. 26, 407–418. doi: 10.1094/MPMI-06-12-0146-R
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Whitham, S., Dinesh-Kumar, S. P., Choi, D., Hehl, R., Corr, C., and Baker, B. (1994). The product of the tobacco mosaic virus resistance gene N: similarity to Toll and the interleukin-1 receptor. Cell 78, 1101–1115. doi: 10.1016/0092-8674(94)90283-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Williams, S. J., Sohn, K. H., Wan, L., Bernoux, M., Sarris, P. F., Segonzac, C., et al. (2014). Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344, 299–303. doi: 10.1126/science.1247357
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Williams, S. J., Sornaraj, P., deCourcy-Ireland, E., Menz, R. I., Kobe, B., Ellis, J. G., et al. (2011). An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, whereas wild-type M protein binds ADP. Mol. Plant Microbe Interact. 24, 897–906. doi: 10.1094/MPMI-03-11-0052
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yuan, B., Zhai, C., Wang, W., Zeng, X., Xu, X., Hu, H., et al. (2011). The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC-NBS-LRR genes. Theor. Appl. Genet. 122, 1017–1028. doi: 10.1007/s00122-010-1506-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhai, C., Zhang, Y., Yao, N., Lin, F., Liu, Z., Dong, Z., et al. (2014). Function and interaction of the coupled genes responsible for Pik-h encoded rice blast resistance. PLoS ONE 9:e98067. doi: 10.1371/journal.pone.0098067
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhao, Y., Yang, J., Shi, J., Gong, Y.-N., Lu, Q., Xu, H., et al. (2011). The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477, 596–600. doi: 10.1038/nature10510
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: NLR protein pairs, integrated decoy, pathogen recognition, plant immunity, Arabidopsis thaliana, rice
Citation: Cesari S, Bernoux M, Moncuquet P, Kroj T and Dodds PN (2014) A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy” hypothesis. Front. Plant Sci. 5:606. doi: 10.3389/fpls.2014.00606
Received: 15 August 2014; Paper pending published: 11 September 2014;
Accepted: 17 October 2014; Published online: 25 November 2014.
Edited by:
Gitta Coaker, University of California, Davis, USAReviewed by:
Murray Grant, University of Exeter, UKRoger Wise, United States Department of Agriculture-Agricultural Research Service and Iowa State University, USA
Copyright © 2014 Cesari, Bernoux, Moncuquet, Kroj and Dodds. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Kroj, Institut National de la Recherche Agronomique, Unité Mixtes de Recherche Biology and Genetics of Plant-Pathogen Interactions, TA A-54/K, Campus International de Baillarguet, 34398 Montpellier, France e-mail:kroj@supagro.inra.fr;
Peter N. Dodds, CSIRO Agriculture, Clunies Ross Street, GPO Box 1600, Canberra, ACT 2601, Australia e-mail:peter.dodds@csiro.au
 Stella Cesari
Stella Cesari