A novel conserved mechanism for plant NLR protein pairs: the ‘integrated decoy’ hypothesis
- 1The Sainsbury Laboratory, Norwich Research Park, Norwich, UK
- 2The Genome Analysis Centre, Norwich Research Park, Norwich, UK
- 3Department of Biological Chemistry, John Innes Centre, Norwich Research Park, Norwich, UK
- 4Iwate Biotechnology Research Center, Kitakami, Japan
A commentary on
A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy” hypothesis
by Cesari, S., Bernoux, M., Moncuquet, P., Kroj, T., and Dodds, P. N. (2014). Front. Plant Sci. 5: 606. doi: 10.3389/fpls.2014.00606
Our conceptual and mechanistic understanding of how plant nucleotide-binding leucine-rich repeat (NLR or NB-LRR) proteins perceive pathogens continues to advance. NLRs are intracellular multidomain proteins that recognize pathogen-derived effectors either directly or indirectly (Jones and Dangl, 2006; Van Der Hoorn and Kamoun, 2008; Dodds and Rathjen, 2010; Cesari et al., 2014). In the direct model, the NLR protein binds a pathogen effector or serves as a substrate for the effector's enzymatic activity. In the indirect model, the NLR recognizes modifications of additional host protein(s) targeted by the effector. Such intermediate host protein(s) are often called effector targets (ETs). However, given that effectors can act on multiple host targets, the specific protein that mediates recognition by the NLR may not be the effector's operative target and may have evolved to function as a decoy dedicated to pathogen detection. This “decoy” model contrasts with the “guard” model in which the NLR perceives the effector via its action on its operative target (Van Der Hoorn and Kamoun, 2008).
In a recent article, Cesari et al. (2014) elegantly synthesized the literature to propose a novel model of how NLRs recognize effectors termed the “integrated decoy” hypothesis. Based on new data from several pathosystems, it appears that some NLRs recognize pathogen effectors through extraneous domains that have evolved by duplication of an ET followed by fusion into the NLR. This NLR-integrated domain mimics the effector binding/substrate property of the original ET to enable pathogen detection. In addition, these “receptor” or “sensor” NLRs typically partner with NLR proteins with a classic architecture that function as signaling partners required for the resistance response (Eitas and Dangl, 2010; Cesari et al., 2013, 2014; Williams et al., 2014).
Here, we expand on the Cesari et al. (2014) model and introduce the possibility that NLR-integrated domains do not have to be decoys (as in defective mimics) of the effector's operative target. Indeed, in addition to binding effectors or serving as their substrates, operative targets carry a biochemical activity that is modulated by the effector. The perturbation of this activity by the effector leads to effector-triggered susceptibility, an activity often related to immunity (Boller and He, 2009; Dodds and Rathjen, 2010; Win et al., 2012). Clearly NLR-integrated domains must retain the “sensor” activity of the ancestral ET, but they could also retain their biochemical activity, continuing to function in the effector-targeted pathway even as an extraneous domain within a classic NLR architecture. At present, this possibility cannot be discounted given that the biochemical activities of the ancestral ETs and their NLR-integrated counterparts are generally unknown. Additionally, when NLR-fusions occurred recently, there may not have been enough time for the integrated ET to lose its original function and evolve into a decoy. We therefore propose to refer to the extraneous domains of classic NLR proteins described by Cesari et al. (2014) as sensor domains (SD), a term that is agnostic to any potential biochemical activities of the integrated module.
How to test whether or not SDs are decoys? We propose a straightforward genetic test that can reject the decoy hypothesis. Isogenic plants either carrying or lacking the NLR-SD can be challenged with a pathogen strain that lacks the matching avirulence effector (Figure 1). There are several possible outcomes. If the NLR-SD isogenic lines do not differ in their response to the pathogen without the matching effector, the result is inconclusive and the null decoy hypothesis cannot be rejected. If the presence of NLR-SD without the known matching effector shows higher levels of resistance, and there are no signs of typical effector-triggered immunity, then the SD is likely to have retained the ET biochemical activity and contributes to basal immunity in a manner analogous to the ancestral ET. An even more interesting result would be if in the absence of the matching effector, the NLR-SD line is more susceptible as has been shown for several ETs (Van Schie and Takken, 2014). In this scenario, another (unrecognized) effector might still be targeting the original biochemical activity of the SD domain. It would be conceptually fascinating if an NLR that functions as a resistance (R) gene against certain strains of a pathogen becomes a susceptibility (S) gene when exposed to other strains. Once again, this concept emphasizes how the outcome of plant-pathogen interactions is so critically dependent on the genotypes of the interacting organisms—a gene that has a certain impact in a particular genetic combination can have the exact opposite effect in another (Jones and Dangl, 2006; Van Der Hoorn and Kamoun, 2008; Dodds and Rathjen, 2010; Win et al., 2012).
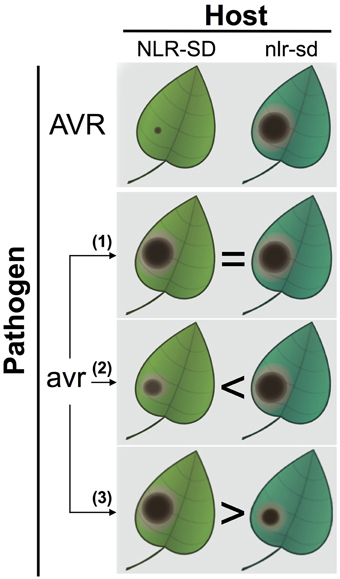
Figure 1. A genetic test to inform whether NLR-SD proteins have retained a biochemical activity independent of perception of an avirulence effector. In the top panel, isogenic plants either carrying or lacking the NLR-SD display differential resistance to a pathogen strain carrying the AVR (avirulence) effector (top panel, NLR-SD plants displaying full resistance to the avirulent pathogen strain). To challenge the decoy hypothesis, the differential NLR-SD lines are challenged with a pathogen strain that lacks the AVR effector (avr) and is isogenic to the AVR strain. In these experiments, three outcomes can be expected. (1) No differences between the NLR-SD lines are observed resulting in inconclusive results—the null decoy hypothesis cannot be rejected. The reason the result is inconclusive is because it is now accepted that effectors have other activities than suppression of immunity (nutrition, development, epigenetics etc.), and therefore the targeted host proteins do not necessarily modulate susceptibility/resistance phenotypes. (2) The plants carrying the NLR-SD are more resistant to the avr pathogen strain that lacks the AVR effector. (3) The plants carrying the NLR-SD are more susceptible to the avr pathogen strain that lacks the AVR effector. In these two cases, the SD is likely to have retained the biochemical activity of its ancestral host protein and the decoy hypothesis can be rejected. In scenario (2), the higher levels of resistance to the avr pathogen conferred by the NLR-SD are consistent with a role of the SD in basal immunity analogous to the ancestral target. In scenario (3), however, the NLR-SD is more susceptible to its isogenic line possibly because the SD is targeted by another (unrecognized) effector. In such a case, the NLR-SD resistance (R) gene becomes a susceptibility (S) gene depending on the genotype of the pathogen it is challenged with.
Our goal is not to engage in an exercise in semantics. However, we wish to avoid conceptually restrictive terminology and urge the plant-microbe interactions community to test a rich spectrum of models and hypotheses. The proposed sensor domain terminology would accommodate this breadth of ideas. Ultimately, it may very well turn out that the majority, if not all, of the NLR integrated domains have lost their biochemical activities and have evolved into decoys. Also, it is possible that the sensor domain has already evolved into a decoy prior to recombination into a NLR. Nonetheless, further genetic and biochemical experiments are required to determine whether sensor domains of NLR-SDs are decoys or biochemically functional duplicates of their ancestral ETs.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members of the Kamoun Lab, Peter Dodds, and Yan Ma for their comments on the concepts discussed here. CHW and SK are funded by the Biotechnology and Biological Science Research Council (BBSRC), the European Research Council (ERC), and the Gatsby Charitable Foundation. KVK is funded by the BBSRC and the Gatsby Charitable Foundation. MJB is funded by the BBSRC (grant BB/J00453), the ERC, and the John Innes Foundation. RT is funded by MEXT (Scientific Research on Innovative Areas 23113009), Japan.
References
Boller, T., and He, S. Y. (2009). Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324, 742–744. doi: 10.1126/science.1171647
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cesari, S., Bernoux, M., Moncuquet, P., Kroj, T., and Dodds, P. N. (2014). A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy” hypothesis. Front. Plant Sci. 5:606. doi: 10.3389/fpls.2014.00606
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cesari, S., Thilliez, G., Ribot, C., Chalvon, V., Michel, C., Jauneau, A., et al. (2013). The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25, 1463–1481. doi: 10.1105/tpc.112.107201
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eitas, T. K., and Dangl, J. L. (2010). NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr. Opin. Plant Biol. 13, 472–477. doi: 10.1016/j.pbi.2010.04.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Van Der Hoorn, R. A., and Kamoun, S. (2008). From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20, 2009–2017. doi: 10.1105/tpc.108.060194
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Van Schie, C. C., and Takken, F. L. (2014). Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52, 551–581. doi: 10.1146/annurev-phyto-102313-045854
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Williams, S. J., Sohn, K. H., Wan, L., Bernoux, M., Sarris, P. F., Segonzac, C., et al. (2014). Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344, 299–303. doi: 10.1126/science.1247357
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Win, J., Chaparro-Garcia, A., Belhaj, K., Saunders, D. G., Yoshida, K., Dong, S., et al. (2012). Effector biology of plant-associated organisms: concepts and perspectives. Cold Spring Harb. Symp. Quant. Biol. 77, 235–247. doi: 10.1101/sqb.2012.77.015933
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: NLR protein pairs, integrated decoy, pathogen recognition, plant immunity, Arabidopsis thaliana, rice, sensor domain, decoy
Citation: Wu C-H, Krasileva KV, Banfield MJ, Terauchi R and Kamoun S (2015) The “sensor domains” of plant NLR proteins: more than decoys? Front. Plant Sci. 6:134. doi: 10.3389/fpls.2015.00134
Received: 20 January 2015; Accepted: 19 February 2015;
Published: 05 March 2015.
Edited by:
Adi Avni, Tel Aviv University, IsraelReviewed by:
Matthieu Joosten, Wageningen University, NetherlandsCopyright © 2015 Wu, Krasileva, Banfield, Terauchi and Kamoun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sophien Kamoun, sophien.kamoun@tsl.ac.uk
 Chih-Hang Wu
Chih-Hang Wu Ksenia V. Krasileva
Ksenia V. Krasileva Mark J. Banfield
Mark J. Banfield Ryohei Terauchi
Ryohei Terauchi Sophien Kamoun
Sophien Kamoun