- Institute of Molecular Plant Sciences, School of Biological Sciences, University of Edinburgh, Edinburgh, UK
Plant immune responses against biotrophic pathogens are regulated by the signaling hormone salicylic acid (SA). SA establishes immunity by regulating a variety of cellular processes, including programmed cell death (PCD) to isolate and kill invading pathogens, and development of systemic acquired resistance (SAR) which provides long-lasting, broad-spectrum resistance throughout the plant. Central to these processes is post-translational modification of SA-regulated signaling proteins by ubiquitination, i.e., the covalent addition of small ubiquitin proteins. Emerging evidence indicates SA-induced protein ubiquitination is largely orchestrated by Cullin-RING ligases (CRLs), which recruit specific substrates for ubiquitination using interchangeable adaptors. Ligation of ubiquitin chains interlinked at lysine 48 leads to substrate degradation by the 26S proteasome. Here we discuss how CRL-mediated degradation of both nucleotide-binding/leucine-rich repeat domain containing immune receptors and SA-induced transcription regulators are critical for functional PCD and SAR responses, respectively. By placing these recent findings in context of knowledge gained in other eukaryotic model species, we highlight potential alternative roles for processive ubiquitination in regulating the activity of SA-mediated immune responses.
Introduction
Successful plant immune responses depend on the rapid recognition of the invading pathogen and subsequent local and systemic transmission of signals that induce resistance throughout all plant tissues. Pattern recognition receptors that recognize conserved pathogen-associated molecular patterns represent the first line of defense, leading to pattern-triggered immunity (Macho and Zipfel, 2014). To subvert immune responses, adapted pathogens have evolved an arsenal of effector proteins that suppress pattern-triggered immunity. The presence of these effector proteins can be sensed by intracellular nucleotide-binding/leucine-rich repeat domain containing (NLR) immune receptors, resulting in effector-triggered immunity (Jones and Dangl, 2006; van Ooijen et al., 2007). Effector-triggered immunity is characterized by rapid onset of programmed cell death (PCD) at the site of infection, which is thought to isolate and prevent proliferation of the invading pathogen. Following pathogen recognition, development of pattern- and effector-triggered immunity requires the immune signaling hormone salicylic acid (SA). Failure to accumulate SA upon pathogen attack results in severe disease susceptibility and inability to launch NLR receptor-mediated PCD (Delaney et al., 1994; Rairdan and Delaney, 2002). Additionally, SA accumulates in tissues adjacent and distant to the site of infection where it induces systemic acquired resistance (SAR), a long-lasting immune response effective against a broad -spectrum of pathogens (Spoel and Dong, 2012; Fu and Dong, 2013). A major function of SA is to initiate reprogramming of the transcriptome to prioritize immune responses over other cellular functions. Accordingly, SA fine-tunes the activity of a network of SA-responsive transcriptional regulators, the concerted action of which establishes disease resistance (Moore et al., 2011).
Recent work has highlighted an important role for the ubiquitin-mediated proteasome system in regulating many aspects of SA-dependent immunity. In eukaryotic cells, post-translational modification by a single or polymeric chain of ubiquitin modulates protein function and stability (Komander and Rape, 2012). Ubiquitin is a highly conserved, small protein (8.5 kDa) that is covalently attached to a target substrate in a multistep enzymatic pathway. First, a ubiquitin-activating E1 enzyme forms a high-energy thioester linkage to a ubiquitin moiety, which is then passed onto an active-site cysteine residue of a ubiquitin-conjugating E2 enzyme. The E2 enzyme works in physical partnership with an E3 ligase to attach ubiquitin onto a specific lysine (Lys) ε-amino group within the target substrate (Smalle and Vierstra, 2004; Komander and Rape, 2012). Compared to many other eukaryotes, plant genomes encode for disproportionally large numbers of E3 ligases; for example, the Arabidopsis genome contains over 1400 different predicted E3 ligase components (Vierstra, 2009), suggesting that protein ubiquitination plays critical roles in plant biology. E3 ligases selectively recruit substrates for ubiquitination and thus provide an important level of specificity to the ubiquitination machinery. E3 ligases can be categorized into different classes based on the presence of a RING, U-box, or HECT domain, leading to distinct ways of binding a partner E2 conjugating enzyme. In addition to single polypeptide E3 ligases, the modular multi-subunit family of Cullin-RING Ligases (CRLs) plays prominent roles in protein ubiquitination. The Cullin subunit of CRLs acts as a scaffold to bring together the RING domain-containing protein and a variable adaptor that recruits the target protein (Santner and Estelle, 2009; Vierstra, 2009; Sadanandom et al., 2012). Emerging evidence suggests that plant immune signaling is predominantly mediated by CRL1 (also known as SCF for SKP1/Cullin1/F-box) and CRL3 [also denoted as BC3B for BTB (Bric-à-brac, Tramtrack, and Broad complex)/Cullin3/BTB], which recruit substrate adaptors that contain F-box motifs or BTB domains, respectively.
Although substrate ubiquitination by E3 ligases can have various functions depending on chain topology and length (Komander and Rape, 2012; Walsh and Sadanandom, 2014), ubiquitin chain linkage via Lys48 signals for degradation of the substrate by the 26S proteasome, a large (2.5 MDa) ATP-dependent chambered protease containing over 30 distinct subunits (Pickart and Cohen, 2004).
Several excellent comprehensive reviews are available on the role of ubiquitination in plant immune signaling in general (Trujillo and Shirasu, 2010; Marino et al., 2012; Duplan and Rivas, 2014). Instead, here we specifically focus on recent advances in understanding the function of ubiquitination in SA-induced immune signaling. How processive ubiquitination and degradation of transcription activators may underpin SA-responsive gene expression in local and systemic immunity will be discussed, as well as how CRLs play an integral part of cellular decisions of life and death upon pathogen recognition.
Ubiquitin-Mediated Suppression of SA-Responsive Gene Transcription
Genetic screens for SA-insensitive Arabidopsis mutants have repeatedly identified npr1 (non-expresser of PR genes) mutant alleles (Cao et al., 1994; Delaney et al., 1995; Glazebrook et al., 1996; Shah et al., 1997). NPR1 encodes a transcription coactivator that in resting cells forms a high molecular weight oligomer in the cytoplasm through intermolecular disulfide bonds between conserved cysteine residues, preventing it from entering the nucleus. Pathogen-induced SA accumulation triggers transient cellular redox changes, resulting in reduction of these disulfide bonds, and release of NPR1 monomers (Mou et al., 2003; Tada et al., 2008). NPR1 monomer translocates to the nucleus where it controls the expression of over 2,200 genes in Arabidopsis (Kinkema et al., 2000; Wang et al., 2006), in part by physically interacting with and transactivating TGA transcription factors that associate with SA-responsive gene promoters (Zhang et al., 1999; Després et al., 2000; Zhou et al., 2000; Boyle et al., 2009). NPR1 protein contains an N-terminal BTB domain and a C-terminal ankyrin repeat domain (Cao et al., 1997; Ryals et al., 1997; Aravind and Koonin, 1999). Interestingly, the presence of these domains in a single protein is a typical feature of a substrate adaptor for CRL3, in which the BTB domain mediates interaction with Cullin 3, while the ankyrin repeat recruits substrates for ubiquitination (Petroski and Deshaies, 2005). However, yeast two-hybrid studies were unable to find direct physical interaction between Cullin 3 and NPR1 (Dieterle et al., 2005). Co-immunoprecipitation experiments nevertheless showed that NPR1 associates with a CRL3 in planta (Spoel et al., 2009). These results suggested that NPR1 may not be in the substrate adaptor position of this E3 ligase. Indeed, in Arabidopsis cells, monomeric NPR1 is itself subject to ubiquitination by a CRL3 and is subsequently degraded in the nucleus. Blocking NPR1 degradation pharmacologically with proteasome inhibitors or genetically by mutation of Cullin 3 resulted in accumulation of NPR1 monomer, moderate induction of NPR1 target genes, and elevated resistance to pathogen infection (Spoel et al., 2009). This indicated that constitutive degradation of NPR1 monomer by CRL3 prevents autoimmunity in absence of a pathogen threat. This suppressive effect of CRL3 and the proteasome probably impacts a large proportion of the immune transcriptome, as many genes are co-regulated by SA and proteasome inhibitor (Spoel et al., 2010).
Ubiquitin-mediated protein degradation plays a similar role in SA-dependent immune responses in rice. Analogous to the function of Arabidopsis NPR1, Oryza sativa WRKY45 is an SA-induced transcription activator of several hundred immune-related genes and confers resistance to bacterial and fungal pathogens (Shimono et al., 2007, 2012; Nakayama et al., 2013). Inhibition of the proteasome resulted in accumulation of polyubiquitinated OsWRKY45 in the nucleus and constitutive activation of its target genes in the absence of SA treatment (Matsushita et al., 2013). Although it remains unknown if OsWRKY45 is targeted for degradation by a CRL3, these findings indicate that constitutive turnover of this immune activator prevents autoimmune responses. SA also activates an NPR1-like protein, which functions in parallel with OsWRKY45 to regulate immune transcription in rice. By contrast to OsWRKY45, this OsNPR1 protein (also known as OsNH1) is thought to be predominantly involved in downregulation of gene expression, particularly those involved in photosynthetic activity (Sugano et al., 2010). Interestingly, OsNPR1 is not subject to constitutive proteasome-mediated degradation, intuitively suggesting that transcriptional repression does not require corepressor turnover. Hence, the presence of analogous proteasome-regulated modules consisting of unrelated transcription (co)activators in Arabidopsis and rice (i.e., NPR1 versus OsWRKY45) may reflect inherent constraints on how timely activation of SA-responsive immune genes can be achieved.
Ubiquitin-Mediated Activation of SA-Responsive Gene Transcription
Besides suppression of SA-responsive immune genes, the proteasome is also involved in gene activation. Pharmacological inhibition of the proteasome, genetic mutation of Cullin 3, and mutation of an NPR1 phosphorylation motif all stabilized the NPR1 protein but greatly reduced the SA-induced expression of its target genes in Arabidopsis (Spoel et al., 2009). Similarly, SA-induced transcriptional activity of OsWRKY45 in rice was impaired in the presence of proteasome inhibitor (Matsushita et al., 2013). Turnover of OsWRKY45 was dependent on a small 26 amino acid C-terminal region, which importantly was also required for its transactivation activity. Such overlap between transactivation domains and degradation motifs that signal ubiquitin-mediated proteasomal degradation has previously been discovered in transcription activators in both yeast and mammals (Salghetti et al., 2000). Fusion of well-defined degron motifs from yeast cyclin proteins to a DNA-binding domain even auto-activated gene transcription (Salghetti et al., 2000), suggesting that the intrinsic ability to activate transcription also makes activators a target for the ubiquitin-mediated proteasome. Additional work showed that like NPR1 and OsWRKY45, other activators also required turnover to unleash their full transcriptional potential (Spoel et al., 2010; Geng et al., 2012). This transcription process, sometimes dubbed ‘destruction–activation’, has been studied in more detail for GCN4 (General Control Non-inducible 4), a potent activator of genes involved in amino acid homeostasis. Upon amino acid starvation, the CDC4 F-box subunit of SCFCDC4 ligase targets GCN4 for ubiquitin-mediated degradation, a process required for recruitment of RNA Polymerase II (RNAPII) to GCN4 target genes (Lipford et al., 2005). Crucially, GCN4 was marked for degradation by the phosphorylative action of SRB10, a cyclin-dependent-kinase associated with the C-terminal domain of RNAPII (Liao et al., 1995; Chi et al., 2001). This indicates that when GCN4 initiates a round of transcription by recruiting RNAPII, it simultaneously triggers its own destruction. These results have led to the hypothesis that transcriptionally ‘spent’ activators may need to be cleared by the proteasome to reset target promoters and allow binding of ‘fresh’ activators (Figure 1; Lipford et al., 2005; Kodadek et al., 2006; Geng et al., 2012). A similar mode of regulation may control transcriptional activity of NPR1 and OsWRKY45 in plant immunity, as site-specific phosphorylation of a degron motif in NPR1was necessary for its ubiquitination and degradation, as well as for timely and sustained target gene expression (Spoel et al., 2009, 2010). Intriguingly, transcription initiation by MYC2, a transcription activator responsive to the developmental and immune hormone jasmonic acid, is also regulated by phosphorylation-induced proteasomal degradation (Zhai et al., 2013). These findings imply that proteasome-mediated regulation of transcription activators may be a general mechanism to control gene expression programs in plant immunity.
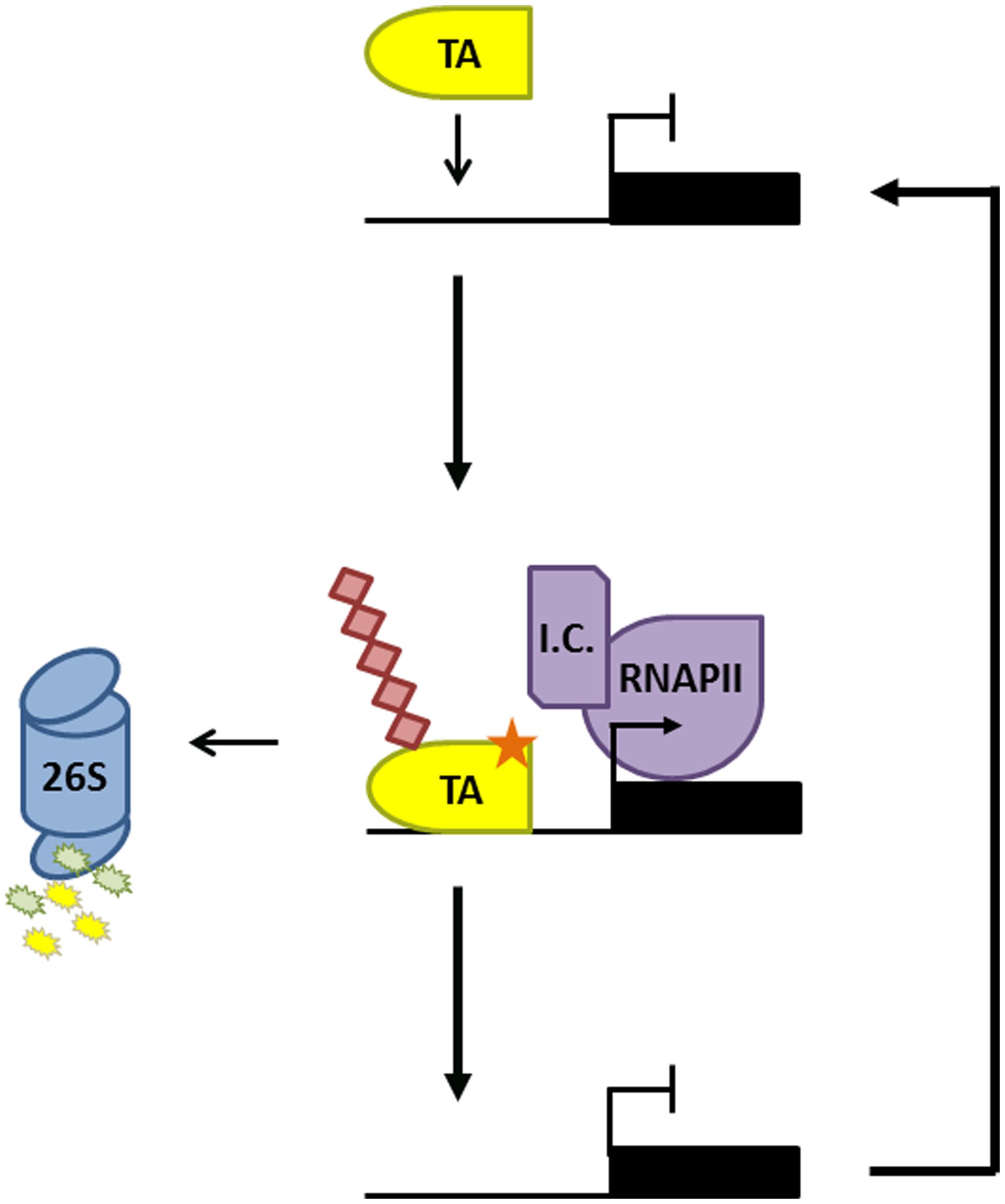
FIGURE 1. Proteasome-mediated activator turnover activates transcription. Promoter binding of a transcription activator (TA) results in recruitment of the transcription initiation complex (IC) and RNA Polymerase II (RNAPII). The TA is subsequently phosphorylated (orange star) by a kinase within the IC, marking it for ubiquitination (red diamonds) and degradation by the 26S proteasome. This allows a new TA to bind the promoter and reinitiate a new round of gene transcription.
Elegant studies on the estrogen receptor ERα in mammalian cells have shed more light on why activators are turned over in the process of activating gene transcription. Upon ligand binding, nuclear localized ERα forms a stable dimer, and associates with cofactors on estrogen-responsive DNA elements to trigger gene transcription. Not only did inhibition of ERα proteolysis suppress its transcriptional activity, vice versa inhibition of RNAPII prevented degradation of ERα, indicating that activator turnover and transcriptional activity were interdependent (Reid et al., 2003). By following ERα transactivation over fine time scales by chromatin immunoprecipitation, it was proposed that ERα-mediated transcription may have distinct cyclical phases in which the ubiquitin-mediated proteasome plays key roles (Metivier et al., 2003). In this model, the first cycle is transcriptionally non-productive but results in ERα-induced remodeling of the promoter to commit it to transcription. In subsequent cycles ERα orchestrates the ordered recruitment of cofactors, ultimately resulting in gene transcription via recruitment of RNAPII. Importantly, experimental data showed that the proteasome was recruited to an ERα target promoter toward the end of each cycle and preceded the clearance of ERα and general transcription cofactors. Thus, proteasome activity is thought to be vital to allow ERα-dependent promoters to move from the transcriptionally non-productive to productive phase and to permit productive cycles to continue until transcription is no longer required (Metivier et al., 2003; Zhou and Slingerland, 2014). If these findings indeed represent a general model for transcription regulation, then the proteasome could have additional roles in SA-responsive gene transcription in plants, including promoter remodeling and ordered cofactor degradation.
But why would cyclical activation of transcription by unstable activators be advantageous over continuous activation by stable activators? Although the answer to this question remains at large, a recent mathematical and in silico analysis of proteasome involvement in transcription may have provided some clues (Lee et al., 2014). The gene targets of many mammalian transcription activators often include components of E3 ligases that promote proteolysis of that activator, generating a negative feedback loop to maintain appropriate levels of activator. Mathematical modeling of this feedback loop showed that cellular perturbations resulting in destabilization of the E3 ligase led to over-accumulation of activators and subsequent hyper-activation of gene expression. However, if the E3 ligase was modeled as a necessary transcription cofactor working in conjunction with the activator, a much more measured gene expression output was achieved upon cellular perturbation. These models suggest that the paradoxical involvement of E3 ligases in gene transcription activated by unstable activators may be necessary to provide a cellular safety mechanism. The authors of this work compared this to the principle of safety interlock devices in engineering, where a system will not function unless safety can be guaranteed (Lee et al., 2014). A similar system may be operational for NPR1- and OsWRKY45-dependent gene expression. Notably, interrogation of a list of NPR1-dependent genes provided by Wang et al. (2006) indicates that NPR1 activates the expression of genes encoding for its paralogues, NPR3, and NPR4. These BTB-containing proteins function as substrate adaptors that recruit NPR1 to CRL3 for ubiquitination and subsequent degradation (Fu et al., 2012). This suggests that similar to the mathematical system described above, a negative feedback loop may exist between NPR1 and CRL3NPR3/NPR4. As CRL3 has a supportive role in NPR1-dependent gene transcription (Spoel et al., 2009), it may be part of a cellular safety mechanism to keep NPR1 activity in check when cellular perturbations are encountered. In support of this hypothesis, although genetic perturbations of CRL3NPR3/NPR4 activity resulted in autoimmune phenotypes due to over-accumulation of NPR1 protein, this did not lead to over-activation of NPR1 target genes in the presence of SA (Spoel et al., 2009; Fu et al., 2012).
Processive Ubiquitination of Transcription Activators
In plants, research has mainly focused on polyubiquitination as a means of regulating protein degradation. However, recent advances in understanding processive ubiquitination in several eukaryotes have highlighted that ubiquitin may have additional important roles in the control of plant transcription factors. The notion that ubiquitin may be directly involved in transcription activation was first explored in yeast. Transcription induced by an artificial activator consisting of the yeast VP16 transactivation domain and the bacterial LexA DNA binding protein (LexA-VP16), was shown to require ubiquitination and degradation mediated by the F-box protein MET30. Strikingly, when ubiquitin was fused in-frame to LexA-VP16, the requirement for MET30 was completely bypassed (Salghetti et al., 2001), suggesting that ubiquitination has dual functions to both activate and destroy transcription activators. Subsequently, additional studies indicated roles for monoubiquitination in transcription activation (Bres et al., 2003; Greer et al., 2003; Burgdorf et al., 2004). Monoubiquitination does not usually signal for proteasome-mediated degradation, for which approximately four or more Lys48-linked ubiquitins are required (Thrower et al., 2000). Instead it was reported that promoter occupancy of the yeast prototypical transcription activator, GAL4, was stabilized by monoubiquitination (Ferdous et al., 2007; Archer et al., 2008b). Interestingly, unmodified GAL4 was destabilized by ATPase activity of the proteasome 19S regulatory particle, preventing transcription activation. Monoubiquitination limited the lifetime of physical interactions between the GAL4 activation domain and 19S subunits (Figure 2A; Archer et al., 2008a). This type of regulatory system likely extends to many other eukaryotes, as interactions between tumor suppressor protein p53, a transcription activator in mammalian cells, and its target promoters were also destabilized by 19S ATPases (Kim et al., 2009).
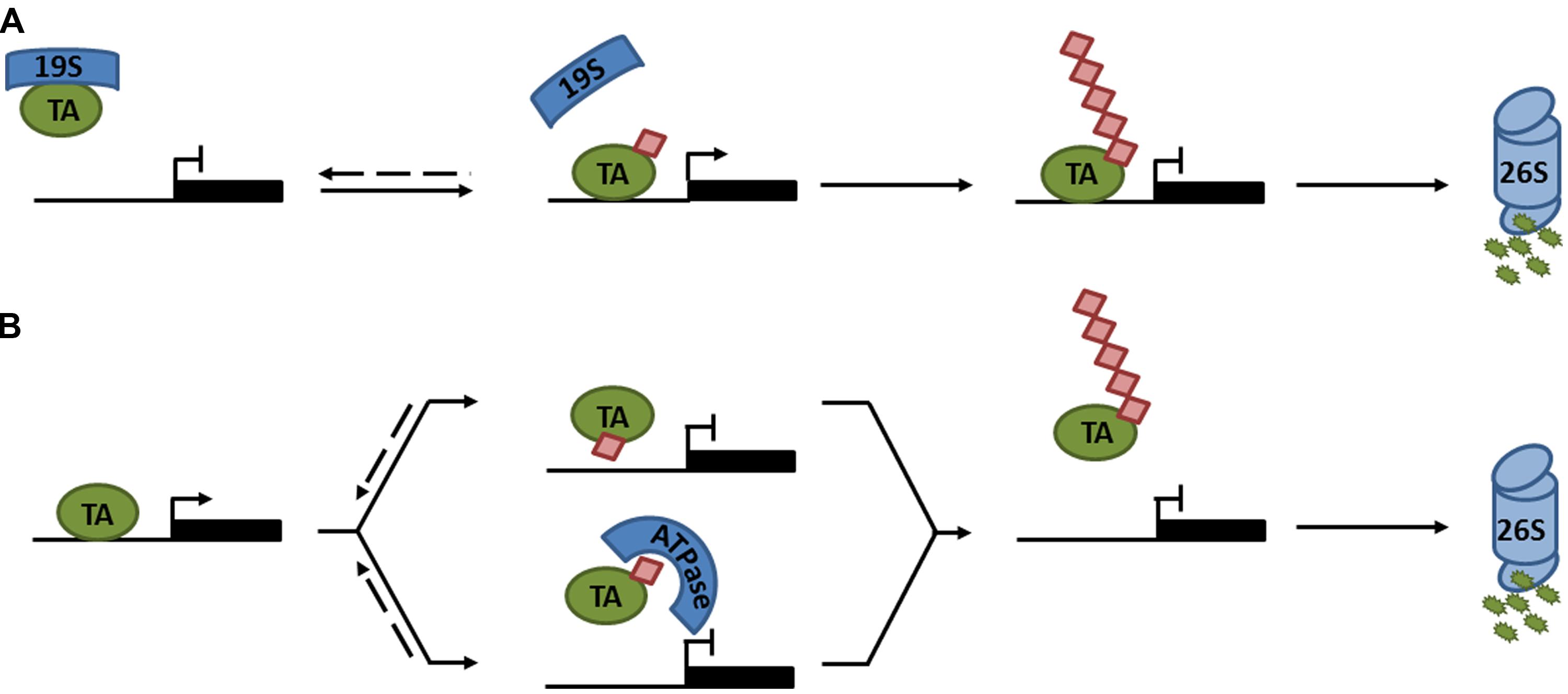
FIGURE 2. Regulation of transcription activator activity by monoubiquitination. (A) The 19S proteasome subcomplex binds an unmodified transcription activator (TA), preventing it from associating with its target promoter. Monoubiquitination of the TA disrupts association with the 19S subcomplex, allowing the TA to bind its target promoter and activate gene expression. Subsequent polyubiquitination marks the TA for degradation by the 26S proteasome. (B) Monoubiquitination of a TA prevents transcription either by sterically hindering the binding to its target promoter (top) or by recruiting an ATPase that prevents it from associating with its target promoter (bottom). Subsequent polyubiquitination marks the TA for degradation by the 26S proteasome. Dashed lines indicate reversible steps.
In contrast to these reports, examples of monoubiquitination leading to suppression of transcription activators have also emerged (Peloponese et al., 2004; Inui et al., 2011; Tang et al., 2011; Ndoja et al., 2014). Using the artificial LexA-VP16 activator described above, a recent report argued that in-frame fusion of ubiquitin to LexA-VP16 was susceptible to cleavage by deubiquitinases (DUBs). Preventing deubiquitination by introducing a non-cleavable ubiquitin-LexA-VP16 mutant resulted in suppression of transcriptional activity by the AAA+ ATPase, CDC48, which stripped this activator from its target promoter (Figure 2B; Ndoja et al., 2014). These findings were extended from artificial to native transcription activators. CDC48 was implicated in yeast sulfur metabolism by removing the monoubiquitinated transcriptional activator, MET4, from its target promoters upon ubiquitination by SCFMET30 (Ndoja et al., 2014). Moreover, monoubiquitination of mammalian receptor-activated SMADs (R-SMAD), involved in TGF-β-mediated embryonic development and tissue homeostasis, attenuated its transcriptional activity by two possible mechanisms: (i) monoubiquitination prevented either R-SMAD transcription complex formation or DNA binding by steric hindrance; or (ii) the CDC48 homolog, p97, actively removed monoubiquitinated R-SMADs from the promoter (Figure 2B; Inui et al., 2011; Tang et al., 2011; Ndoja et al., 2014). Taken together, all these reports clearly illustrate that monoubiquitination can directly regulate the activity of transcription activators through a variety of different mechanisms (Figure 2). Additionally, monoubiquitination may indirectly regulate the activities of some activators by modulating their nucleocytoplasmic localization (van der Horst et al., 2006).
While monoubiquitination may play a regulatory role, processive ubiquitin chain elongation subsequently leads to activator turnover (Kodadek et al., 2006). This processive mono-to-polyubiquitination switch was explored in particular detail for the human Steroid Receptor Coactivator-3 (SRC-3). SRC-3 is an important developmental transcription coactivator, whose uncontrolled expression can lead to oncogenesis. SRC-3 was found to be subject to phosphorylation-dependent polyubiquitination by SCFFbw7α, resulting in its transcription-coupled degradation. However, SRC-3 was also multi (mono)-ubiquitinated by SCFFbw7α, which enhanced its transcriptional activity. Hence, it was proposed that biphasic, processive ubiquitination (i.e., transitioning from mono- to polyubiquitination) generates a timer for the functional lifetime of SRC-3 (Wu et al., 2007).
These intriguing findings relating to eukaryotic transcription indicate that ubiquitin-mediated control of transcription (co)activators in SA-dependent immunity is far more complex than generally appreciated. Current efforts in this field by several labs, including our own, may soon reveal additional roles for ubiquitin and ubiquitin ligases in the transcription activation of immune genes.
CRL3-Mediated Degradation of SA-Responsive Repressors?
In the past decade intimate relationships between plant hormone signaling and the ubiquitin-mediated proteasome have been uncovered. Recurring roles for CRL1 and CRL3 are found in jasmonic acid, ethylene, auxin, gibberellin, abscisic acid, strigolactone, and zeatin signaling (Kelley and Estelle, 2012). The role of CRL1 in jasmonic acid- and auxin-responsive gene expression is especially similar. In both cases the hormone facilitates physical interaction of the CRL1 F-box subunit with transcriptional repressors to form a hormone coreceptor complex. Hormone-dependent recruitment of repressors to CRL1 leads to their poly-ubiquitination and degradation, releasing the activity of transcriptional activators (Kelley and Estelle, 2012). A strikingly similar hormone perception mechanism regulates SA signaling, but instead utilizes CRL3. The CRL3 substrate adaptors NPR3 and NPR4 were shown to act as SA receptors. Whereas SA binding facilitated interaction between NPR3 and NPR1, it disrupted NPR4-NPR1 interaction. Moreover, genetic deletion of NPR3 and NPR4 severely impaired the ability to coimmunoprecipitate Cullin 3 and NPR1, indicating that NPR1 is the substrate of an SA-sensitive CRL3NPR3/NPR4 (Fu et al., 2012).
It is likely that CRL3 complexes exist with roles that extend beyond targeting NPR1. In analogy to jasmonic acid and auxin signaling, CRL3 could target a number of transcription (co)repressors described for SA-responsive genes. For example, TGA2 transcription factors act as repressors of PR genes (Zhang et al., 2003; Kesarwani et al., 2007). Moreover, NPR3 and NPR4 physically interact with TGA2 and other members of the TGA family (Liu et al., 2005; Zhang et al., 2006; Shi et al., 2013), implying that a CRL3NPR3/4 might target TGA factors for degradation to activate SA-responsive genes. Other conceivable targets of CRL3 include SNI1 (Suppressor of NPR1, Inducible), a corepressor of mostly NPR1-depenent genes (Mosher et al., 2006). SNI1 was recently shown to associate with CBNAC, a calmodulin-binding NAC transcription factor. Genetic analysis suggested that CBNAC is a transcription repressor of SA-dependent immune responses. Interestingly, SNI1 facilitated the binding of CBNAC to a DNA-binding motif in the SA-responsive PR-1 promoter (Kim et al., 2012). Finally, several NPR1-interacting NIMIN (NIM1/NPR1-Interacting) proteins act as corepressors, and their removal or inactivation is presumable necessary for activation of SA-responsive gene expression (Weigel et al., 2005). Thus, CRL3 targets could include SNI1, CBNAC, TGA factors, and NIMINs, but little is currently known about the stability of these (co)repressors. Analysis of transcription (co)factor interaction networks in rice between OsNPR paralogues, TGA factors, and NRR (Negative Regulator of Resistance) proteins that share limited homology to Arabidopsis NIMINs, paint a similar picture (Chern et al., 2014). All these factors formed a wide network of interactions in both yeast two-hydrid and split YFP assays, suggesting that involvement of CRL3 complexes in immunity may be functionally conserved in rice.
Alternative to direct targeting of (co)repressors by CRL3, a recent report suggests that these ubiquitin ligases can also promote the concurrent ubiquitination of multiple associated substrates. Upon light induction, the transcription factor PIF3 is recruited to CRL3LRB for ubiquitination. Strikingly, it was found that the PIF3 interaction partner, PhyB, was concomitantly recruited by CRL3LRB (Ni et al., 2014). CRL3 dimerisation through BTB domains might facilitate concurrent substrate degradation, essentially bringing together two active sites for substrate ubiquitination (Stogios et al., 2007). Hence, it plausible that CRL3NPR3/NPR4 simultaneously targets complexes consisting of NPR1 and the transcriptional repressors that physically interact with NPR1.
Peculiarly, unlike NPR3 and NPR4, NPR1 has not yet been observed in the substrate adaptor position of a CRL3 (Dieterle et al., 2005; Fu et al., 2012). However, computational predictions of NPR1 protein structure suggest that it forms a typical BTB domain fold that should allow interaction with Cullin 3 (Tada et al., 2008). Moreover, immediately C-terminal to the BTB domain, NPR1, NPR3, and NPR4 all contain key elements of a conserved helical 3-box structure that, analogous to the F-box motif, was shown to stimulate Cullin 3 interaction by packing tightly against its N-terminus (Zhuang et al., 2009; Canning et al., 2013). Reports that NPR1 itself may directly sense SA or may also be a SA receptor (Maier et al., 2011; Wu et al., 2012) further suggests that NPR1 could be part of a CRL analogous to other plant hormone pathways, although definitive proof for NPR1 as an SA receptor was not supported by another study (Fu et al., 2012).
If NPR1 does indeed reside in a substrate adaptor position of CRL3, this would have important implications for the role of its own turnover in SA-responsive gene expression. First, this would create an additional layer of complexity whereby a CRL3NPR3/NPR4 regulates the formation of a CRL3NPR1. Secondly, CRL substrate adaptors often paradoxically exhibit instability themselves. In absence of a substrate, both F-box and BTB adaptors have been shown to be subject to auto-ubiquitination within their respective CRLs (Bosu and Kipreos, 2008). The necessity of NPR1 turnover in activation of SA-responsive genes may therefore reflect a requirement to allow switching of diverse NPR substrate adaptors within core CRL3 complexes (Figure 3).
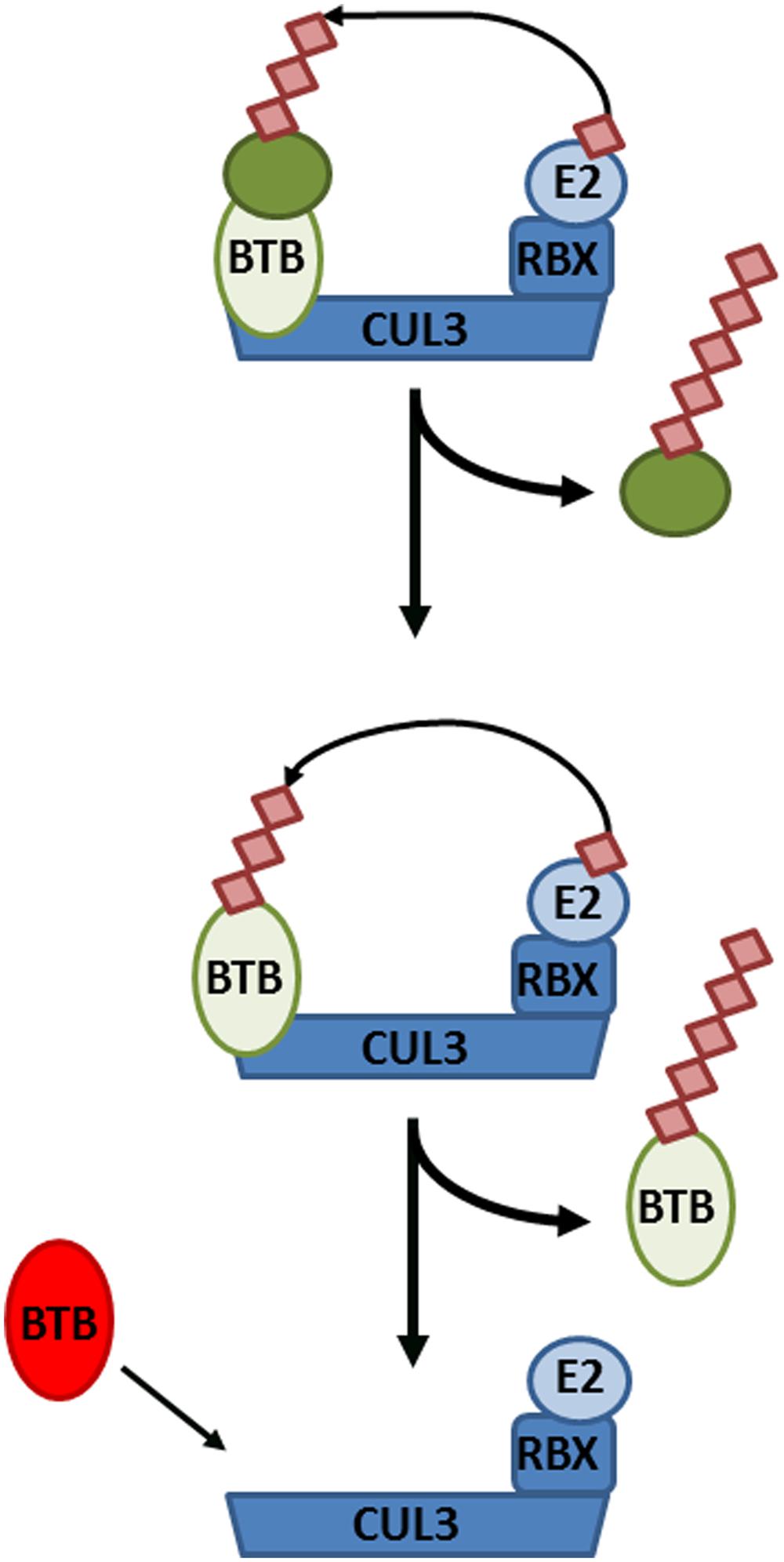
FIGURE 3. Cullin-RING ligase 3 (CRL3) autoubiquitination and adaptor switching. (Top) A target substrate (dark green circle) is polyubiquitinated and targeted for degradation by CRL3, consisting of the CUL3 backbone, Bric-à-brac, Tramtrack, and Broad complex (BTB) domain-containing adaptor, RING-Box protein (RBX), and an E2 conjugating enzyme. (Middle) After all available substrates have been polyubiquitinated and degraded, the BTB adaptor itself becomes subject to autoubiquitination and degradation. (Bottom) Consequently, the CRL3 can now switch to a new BTB adaptor in order to polyubiquitinate different substrates.
CRLs in SA-Mediated Programmed Cell Death and Survival
Salicylic acid is an agonist of PCD responses induced by NLR immune receptors upon intracellular detection of pathogen effectors. In some cases cellular decisions to live or die upon pathogen infection are shaped by the activities of CRLs. Mutation of CRL3NPR3/NPR4 components suggested that the stability of its substrate, NPR1, is an important determinant in PCD induced by the NLR receptors RPS2 and RPM1 (Fu et al., 2012). Indeed, analysis of pathogen-induced PCD in npr1 mutants previously revealed that NPR1 suppressed PCD induced by these NLR receptors (Rate and Greenberg, 2001). Moreover, mutation of NPR1 partially restored RPS2- and RPM1-induced PCD in npr3 and npr4 mutants (Fu et al., 2012). These results indicate that elevated levels of NPR1 promote cell survival and that its removal by CRL3NPR3/NPR4 is required for successful PCD induced by at least some NLR receptor classes (Figure 4). In agreement with its role in promoting cell survival, genetic experiments have indicated that the presence of NPR1 is not essential for successful NLR receptor-induced PCD and immunity (Rairdan and Delaney, 2002).
In contrast to NPR1’s pro-survival role, NLR receptors instigate PCD responses upon perception of pathogen effectors. In the absence of pathogen threats, NLR receptors must be kept tightly controlled to avoid autoimmune responses. Overexpression of the tomato NLR receptor, Prf, resulted in strong autoimmune phenotypes, including elevated SA levels and expression of SA-responsive genes in absence of pathogen attack (Oldroyd and Staskawicz, 1998). Moreover, overexpression of NLR receptors due to genomic duplication in the Arabidopsis bal variant also led to constitutive SA responses in absence of a pathogen as well as morphological defects such as severely stunted growth, highlighting the trade-off between growth and defense (Stokes et al., 2002; Yi and Richards, 2009). Similarly, mutation of the potential transcription corepressor, SRFR1, resulted in autoimmunity due to transcriptional upregulation of the co-regulated NLR receptors SNC1, RPS2, and RPS4 (Kwon et al., 2009; Kim et al., 2010; Li et al., 2010). Collectively these examples illustrate that the cellular protein levels of some NLR receptors are linked to their immune activities.
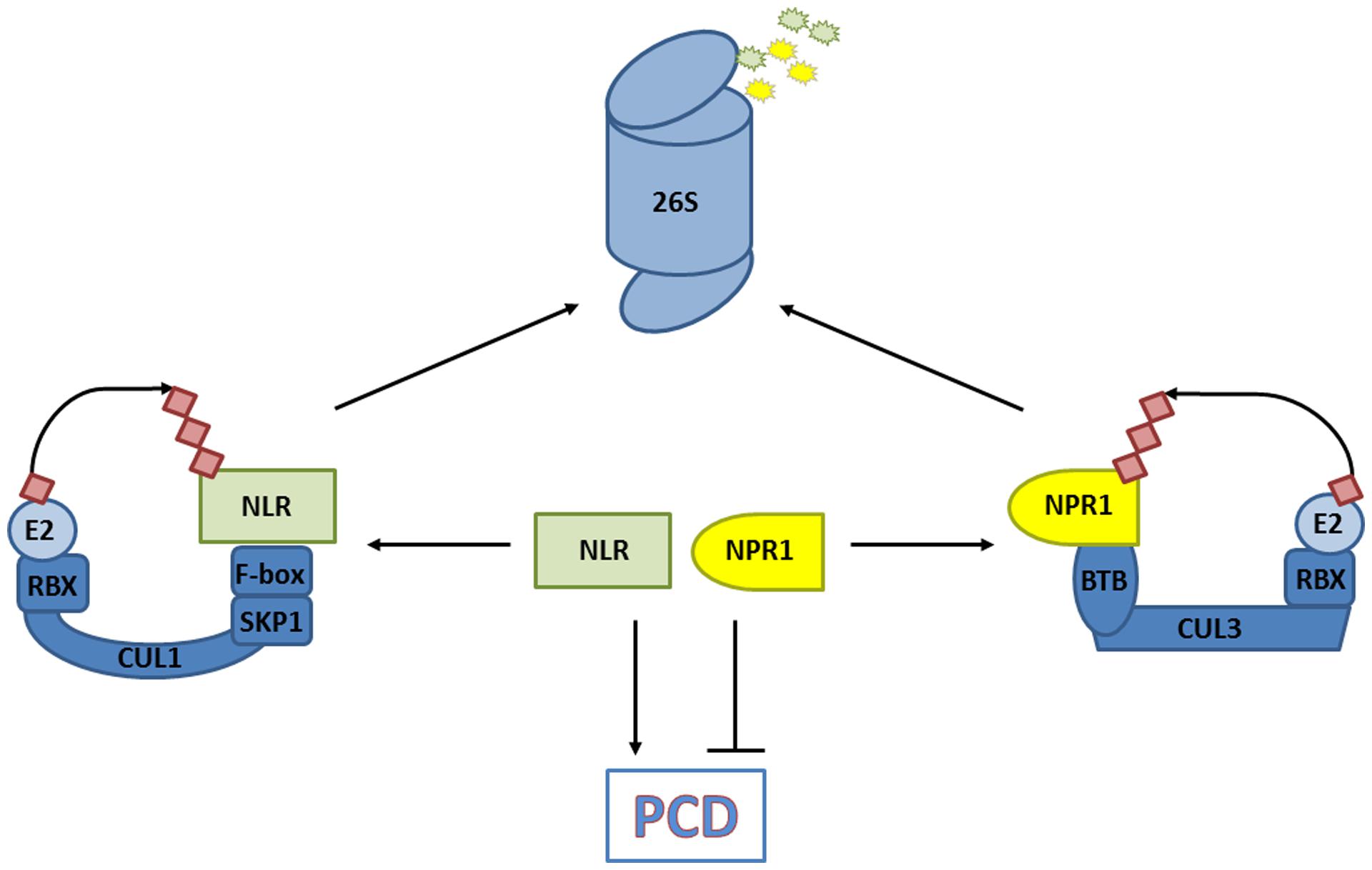
FIGURE 4. CRLs decide on cell fate. The degradation of substrates that promote programmed cell death (PCD) in response to a pathogen [e.g., nucleotide-binding/leucine-rich repeat domain containing (NLR) immune receptors], and those that prevent PCD (e.g., non-expresser of PR genes, NPR1) are controlled by CRLs. The balance of substrate abundance between promoters and suppressors of PCD dictates cell fate, and is regulated by their ubiquitination and 26S proteasome-mediated degradation.
Recent work revealed that protein levels of several NLR receptors are tightly controlled by CRL activities. An early screen for mutations leading to SA-mediated autoimmune phenotypes identified the cpr1 (constitutive expressor of PR genes) mutant (Bowling et al., 1994). Importantly, protein levels of the NLR receptors SNC1 and RPS2 were inversely correlated with CPR1 activity, and loss-of-function mutations in SNC1 largely suppressed the autoimmune phenotype of mutant cpr1 plants. Cloning of CPR1 revealed it encodes an F-box protein, suggesting it controls the abundance of specific NLR receptors by targeting them for proteasome-mediated degradation. Indeed, CPR1 directly interacted with SNC1 and RPS2, and in case of SNC1 this appeared to lead to its polyubiquitination and degradation by the proteasome (Cheng et al., 2011; Gou et al., 2012). NLR receptor signaling probably involves other CRL1 ubiquitin ligases as well but with distinct functions. Rather than eliciting autoimmunity, silencing of the F-box protein ACIF1 in tobacco and tomato compromised NLR receptor-mediated PCD and immunity (van den Burg et al., 2008). ACIF1 interacted with other CRL1 subunits, suggesting it can form a functional ubiquitin ligase but its direct targets remain unknown. Notably, several non-CRL ubiquitin ligases that regulate NLR accumulation or signaling have also been identified and are discussed in other excellent reviews (Marino et al., 2012; Duplan and Rivas, 2014). Hence, ubiquitin ligases – and CRLs in particular – play an integral role in cellular decisions of life and death by controlling the level of NLR receptors and PCD suppressors (Figure 4).
In addition to canonical ubiquitination pathway enzymes (E1, E2, E3), an E4 class of ubiquitin ligases has been described (Koegl et al., 1999). E4 ligases also polyubiquitinate substrates, but contrary to E3 ligases, they largely lack substrate specificity and rather function to elongate existing ubiquitin chains, thereby potentially promoting recognition of substrates by the proteasome. A recent forward genetic screen for mutants that enhanced autoimmunity of snc1, a mutation that renders this NLR receptor constitutively active, identified the E4 ligase MUSE3 (Mutant snc1-Enhancing; Huang et al., 2014). Mutant muse3 plants exhibited elevated levels of SNC1 and RPS2, while overexpression of MUSE3 in itself did not lead to reduction of NLR receptor accumulation. However, coexpression of MUSE3 together with the F-box protein CPR1 resulted in a greater decrease in NLR receptor accumulation than observed with CPR1 expression alone, indicating that MUSE3 and SCFCPR1 function cooperatively to destabilize NLR receptors. In case of SNC1 but not RPS2, a direct physical association with MUSE3 was indeed found, suggesting that MUSE3 may recognize NLR receptors via distinct mechanisms. Thus, an intricate set of cooperative ubiquitin ligases underpin SA-dependent NLR receptor signaling to prevent autoimmunity and promote timely activation of immune responses.
The Road Ahead…
In this review we have discussed the emerging roles of ubiquitin ligases in aspects of SA-mediated immune signaling, including transcriptional reprogramming and cellular decisions of life and death. Similar to other hormone signaling pathways, members of the CRL class of ubiquitin ligases appear to fulfill particularly important tasks, although the targets of these CRLs still remain largely unknown. In SA-induced gene transcription the precise role of CRL3-mediated turnover of NPR1 requires further investigation into processive ubiquitination events and it remains to be discovered if immune-induced CRL3 targets substrates other than NPR1 for proteasome-mediated degradation. Although the role of CRLs in controlling the accumulation of specific NLR receptors is becoming increasingly clear, it remains poorly understood why protein abundance is a key factor in determining NLR receptor activity. Conformational control of NLR receptors by highly conserved eukaryotic chaperone complexes is thought to keep receptors in a recognition-competent state and facilitate their activation upon pathogen perception (Shirasu, 2009; van Ooijen et al., 2010). It is plausible that uncontrolled accumulation of some NLR receptors could result in a shortage of available chaperones and consequent conformation-induced auto-activation of NLR receptors. Finally, many E3 ligases construct ubiquitin chain topologies distinct from proteasome-recognized Lys48-linkages. These alternative chain topologies serve a wide variety of different cellular signaling functions in eukaryotes, yet little is known about their existence and roles in plant biology (Walsh and Sadanandom, 2014). Hence, much remains to be discovered in the exciting field of plant ubiquitin signaling in general and in SA-mediated immune responses in particular.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or? financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Gerben van Ooijen and Dr. Michael Skelly for critically reading the manuscript and apologize to colleagues whose work we were not able to include. JJF was supported by a studentship from the Biotechnology and Biological Sciences Research Council (BBSRC) and SHS by a BBSRC grant (BB/L006219/1) and Royal Society University Research Fellowship (UF090321).
References
Aravind, L., and Koonin, E. V. (1999). Fold prediction and evolutionary analysis of the POZ domain: structural and evolutionary relationship with the potassium channel tetramerization domain. J. Mol. Biol. 285, 1353–1361. doi: 10.1006/jmbi.1998.2394
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Archer, C. T., Burdine, L., Liu, B., Ferdous, A., Johnston, S. A., and Kodadek, T. (2008a). Physical and functional interactions of monoubiquitylated transactivators with the proteasome. J. Biol. Chem. 283, 21789–21798. doi: 10.1074/jbc.M803075200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Archer, C. T., Delahodde, A., Gonzalez, F., Johnston, S. A., and Kodadek, T. (2008b). Activation domain-dependent monoubiquitylation of Gal4 protein is essential for promoter binding in vivo. J. Biol. Chem. 283, 12614–12623. doi: 10.1074/jbc.M801050200
Bosu, D. R., and Kipreos, E. T. (2008). Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 3, 7. doi: 10.1186/1747-1028-3-7
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bowling, S. A., Guo, A., Cao, H., Gordon, A. S., Klessig, D. F., and Dong, X. (1994). A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6, 1845–1857. doi: 10.1105/tpc.6.12.1845
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Boyle, P., Le Su, E., Rochon, A., Shearer, H. L., Murmu, J., Chu, J. Y.,et al. (2009). The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. Plant Cell 21, 3700–3713. doi: 10.1105/tpc.109.069971
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bres, V., Kiernan, R. E., Linares, L. K., Chable-Bessia, C., Plechakova, O., Treand, C.,et al. (2003). A non-proteolytic role for ubiquitin in Tat-mediated transactivation of the HIV-1 promoter. Nat. Cell Biol. 5, 754–761. doi: 10.1038/ncb1023
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Burgdorf, S., Leister, P., and Scheidtmann, K. H. (2004). TSG101 interacts with apoptosis-antagonizing transcription factor and enhances androgen receptor-mediated transcription by promoting its monoubiquitination. J. Biol. Chem. 279, 17524–17534. doi: 10.1074/jbc.M313703200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Canning, P., Cooper, C. D. O., Krojer, T., Murray, J. W., Pike, A. C., Chaikuad, A.,et al. (2013). Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases. J. Biol. Chem. 288, 7803–7814. doi: 10.1074/jbc.M112.437996
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cao, H., Bowling, S. A., Gordon, A. S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. doi: 10.1105/tpc.6.11.1583
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cao, H., Glazebrook, J., Clarke, J. D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. doi: 10.1016/S0092-8674(00)81858-9
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cheng, Y. T., Li, Y., Huang, S., Huang, Y., Dong, X., Zhang, Y.,et al. (2011). Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc. Natl. Acad. Sci. U.S.A. 108, 14694–14699. doi: 10.1073/pnas.1105685108
Chern, M., Bai, W., Ruan, D., Oh, T., Chen, X., and Ronald, P. C. (2014). Interaction specificity and coexpression of rice NPR1 homologs 1 and 3 (NH1 and NH3), TGA transcription factors and Negative Regulator of Resistance (NRR) proteins. BMC Genomics 15:461. doi: 10.1186/1471-2164-15-461
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chi, Y., Huddleston, M. J., Zhang, X., Young, R. A., Annan, R. S., Carr, S. A.,et al. (2001). Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 15, 1078–1092. doi: 10.1101/gad.867501
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Delaney, T. P., Friedrich, L., and Ryals, J. A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. U.S.A. 92, 6602–6606. doi: 10.1073/pnas.92.14.6602
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Delaney, T. P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D.,et al. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. doi: 10.1126/science.266.5188.1247
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Després, C., DeLong, C., Glaze, S., Liu, E., and Fobert, P. R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12, 279–290. doi: 10.1105/tpc.12.2.279
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dieterle, M., Thomann, A., Renou, J. P., Parmentier, Y., Cognat, V., Lemonnier, G.,et al. (2005). Molecular and functional characterization of Arabidopsis Cullin 3A. Plant J. 41, 386–399. doi: 10.1111/j.1365-313X.2004.02302.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Duplan, V., and Rivas, S. (2014). E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front. Plant Sci. 5:42. doi: 10.3389/fpls.2014.00042
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ferdous, A., Sikder, D., Gillette, T., Nalley, K., Kodadek, T., and Johnston, S. A. (2007). The role of the proteasomal ATPases and activator monoubiquitylation in regulating Gal4 binding to promoters. Genes Dev. 21, 112–123. doi: 10.1101/gad.1493207
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fu, Z. Q., and Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Fu, Z. Q., Yan, S., Saleh, A., Wang, W., Ruble, J., Oka, N.,et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232. doi: 10.1038/nature11162
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Geng, F., Wenzel, S., and Tansey, W. P. (2012). Ubiquitin and proteasomes in transcription. Annu. Rev. Biochem. 81, 177–201. doi: 10.1146/annurev-biochem-052110-120012
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Glazebrook, J., Rogers, E. E., and Ausubel, F. M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982.
Gou, M., Shi, Z., Zhu, Y., Bao, Z., Wang, G., and Hua, J. (2012). The F-box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant J. 69, 411–420. doi: 10.1111/j.1365-313X.2011.04799.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Greer, S. F., Zika, E., Conti, B., Zhu, X. S., and Ting, J. P. (2003). Enhancement of CIITA transcriptional function by ubiquitin. Nat. Immunol. 4, 1074–1082. doi: 10.1038/ni985
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Huang, Y., Minaker, S., Roth, C., Huang, S., Hieter, P., Lipka, V.,et al. (2014). An E4 ligase facilitates polyubiquitination of plant immune receptor resistance proteins in Arabidopsis. Plant Cell 26, 485–496. doi: 10.1105/tpc.113.119057
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Inui, M., Manfrin, A., Mamidi, A., Martello, G., Morsut, L., Soligo, S.,et al. (2011). USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat. Cell Biol. 13, 1368–1375. doi: 10.1038/ncb2346
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kelley, D. R., and Estelle, M. (2012). Ubiquitin-mediated control of plant hormone signaling. Plant Physiol. 160, 47–55. doi: 10.1104/pp.112.200527
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kesarwani, M., Yoo, J., and Dong, X. (2007). Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 144, 336–346. doi: 10.1104/pp.106.095299
Kim, H. S., Park, H. C., Kim, K. E., Jung, M. S., Han, H. J., Kim, S. H.,et al. (2012). A NAC transcription factor and SNI1 cooperatively suppress basal pathogen resistance in Arabidopsis thaliana. Nucleic Acids Res. 40, 9182–9192. doi: 10.1093/nar/gks683
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kim, S. H., Gao, F., Bhattacharjee, S., Adiasor, J. A., Nam, J. C., and Gassmann, W. (2010). The Arabidopsis resistance-like gene SNC1 is activated by mutations in SRFR1 and contributes to resistance to the bacterial effector AvrRps4. PLoS Pathog. 6:e1001172. doi: 10.1371/journal.ppat.1001172
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kim, Y. C., Wu, S. Y., Lim, H. S., Chiang, C. M., and Kodadek, T. (2009). Non-proteolytic regulation of p53-mediated transcription through destabilization of the activator∙promoter complex by the proteasomal ATPases. J. Biol. Chem. 284, 34522–34530. doi: 10.1074/jbc.M109.017277
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kinkema, M., Fan, W., and Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350. doi: 10.1105/tpc.12.12.2339
Kodadek, T., Sikder, D., and Nalley, K. (2006). Keeping transcriptional activators under control. Cell 127, 261–264. doi: 10.1016/j.cell.2006.10.002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Koegl, M., Hoppe, T., Schlenker, S., Ulrich, H. D., Mayer, T. U., and Jentsch, S. (1999). A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96, 635–644. doi: 10.1016/S0092-8674(00)80574-7
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Komander, D., and Rape, M. (2012). The ubiquitin code. Annu. Rev. Biochem. 81, 203–229. doi: 10.1146/annurev-biochem-060310-170328
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kwon, S. I., Kim, S. H., Bhattacharjee, S., Noh, J. J., and Gassmann, W. (2009). SRFR1, a suppressor of effector-triggered immunity, encodes a conserved tetratricopeptide repeat protein with similarity to transcriptional repressors. Plant J. 57, 109–119. doi: 10.1111/j.1365-313X.2008.03669.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lee, D., Kim, M., and Cho, K. H. (2014). A design principle underlying the paradoxical roles of E3 ubiquitin ligases. Sci. Rep. 4, 5573. doi: 10.1038/srep05573
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Li, Y., Li, S., Bi, D., Cheng, Y. T., Li, X., and Zhang, Y. (2010). SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog. 6:e1001111. doi: 1001110.1001371/journal.ppat.1001111
Liao, S. M., Zhang, J., Jeffery, D. A., Koleske, A. J., Thompson, C. M., Chao, D. M.,et al. (1995). A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature 374, 193–196. doi: 10.1038/374193a0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lipford, J. R., Smith, G. T., Chi, Y., and Deshaies, R. J. (2005). A putative stimulatory role for activator turnover in gene expression. Nature 438, 113–116. doi: 10.1038/nature04098
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Liu, G., Holub, E. B., Alonso, J. M., Ecker, J. R., and Fobert, P. R. (2005). An Arabidopsis NPR1-like gene, NPR4, is required for disease resistance. Plant J. 41, 304–318. doi: 10.1111/j.1365-313X.2004.02296.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Macho, A. P., and Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272. doi: 10.1016/j.molcel.2014.03.028
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Maier, F., Zwicker, S., Hückelhoven, A., Meissner, M., Funk, J., Pfitzner, A. J. P.,et al. (2011). NONEXPRESSOR OF PATHOGENESIS-RELATED PROTEINS1 (NPR1) and some NPR1-related proteins are sensitive to salicylic acid. Mol. Plant Pathol. 12, 73–91. doi: 10.1111/j.1364-3703.2010.00653.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Marino, D., Peeters, N., and Rivas, S. (2012). Ubiquitination during plant immune signaling. Plant Physiol. 160, 15–27. doi: 10.1104/pp.112.199281
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Matsushita, A., Inoue, H., Goto, S., Nakayama, A., Sugano, S., Hayashi, N.,et al. (2013). Nuclear ubiquitin proteasome degradation affects WRKY45 function in the rice defense program. Plant J. 73, 302–313. doi: 10.1111/tpj.12035
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Metivier, R., Penot, G., Hubner, M. R., Reid, G., Brand, H., Kos, M.,et al. (2003). Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115, 751–763. doi: 10.1016/S0092-8674(03)00934-6
Moore, J. W., Loake, G. J., and Spoel, S. H. (2011). Transcription dynamics in plant immunity. Plant Cell 23, 2809–2820. doi: 10.1105/tpc.111.087346
Mosher, R. A., Durrant, W. E., Wang, D., Song, J., and Dong, X. (2006). A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell 18, 1750–1765. doi: 10.1105/tpc.105.039677
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mou, Z., Fan, W., and Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. doi: 10.1016/S0092-8674(03)00429-X
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nakayama, A., Fukushima, S., Goto, S., Matsushita, A., Shimono, M., Sugano, S.,et al. (2013). Genome-wide identification of WRKY45-regulated genes that mediate benzothiadiazole-induced defense responses in rice. BMC Plant Biol. 13:150. doi: 10.1186/1471-2229-13-150
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ndoja, A., Cohen, R. E., and Yao, T. (2014). Ubiquitin signals proteolysis-independent stripping of transcription factors. Mol. Cell 53, 893–903. doi: 10.1016/j.molcel.2014.02.002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ni, W., Xu, S. L., Tepperman, J. M., Stanley, D. J., Maltby, D. A., Gross, J. D.,et al. (2014). A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344, 1160–1164. doi: 10.1126/science.1250778
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Oldroyd, G. E., and Staskawicz, B. J. (1998). Genetically engineered broad-spectrum disease resistance in tomato. Proc. Natl. Acad. Sci. U.S.A. 95, 10300–10305. doi: 10.1073/pnas.95.17.10300
Peloponese, J. M. Jr., Iha, H., Yedavalli, V. R. K., Miyazato, A., Li, Y., Haller, K.,et al. (2004). Ubiquitination of human T-cell leukemia virus type 1 tax modulates its activity. J. Virol. 78, 11686–11695. doi: 10.1128/JVI.78.21.11686-11695.2004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Petroski, M. D., and Deshaies, R. J. (2005). Function and regulation of Cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20. doi: 10.1038/nrm1547
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pickart, C. M., and Cohen, R. E. (2004). Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 5, 177–187. doi: 10.1038/nrm1336
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rairdan, G. J., and Delaney, T. P. (2002). Role of salicylic acid and NIM1/NPR1 in race-specific resistance in Arabidopsis. Genetics 161, 803–811.
Rate, D. N., and Greenberg, J. T. (2001). The Arabidopsis aberrant growth and death2 mutant shows resistance to Pseudomonas syringae and reveals a role for NPR1 in suppressing hypersensitive cell death. Plant J. 27, 203–211. doi: 10.1046/j.0960-7412.2001.1075
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Reid, G., Hubner, M. R., Metivier, R., Brand, H., Denger, S., Manu, D.,et al. (2003). Cyclic, proteasome-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol. Cell 11, 695–707. doi: 10.1016/S1097-2765(03)00090-X
Ryals, J., Weymann, K., Lawton, K., Friedrich, L., Ellis, D., Steiner, H. Y.,et al. (1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor IkB. Plant Cell 9, 425–439.
Sadanandom, A., Bailey, M., Ewan, R., Lee, J., and Nelis, S. (2012). The ubiquitin-proteasome system: central modifier of plant signalling. New Phytol. 196, 13–28. doi: 10.1111/j.1469-8137.2012.04266.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Salghetti, S. E., Caudy, A. A., Chenoweth, J. G., and Tansey, W. P. (2001). Regulation of transcriptional activation domain function by ubiquitin. Science 293, 1651–1653. doi: 10.1126/science.1062079
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Salghetti, S. E., Muratani, M., Wijnen, H., Futcher, B., and Tansey, W. P. (2000). Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc. Natl. Acad. Sci. U.S.A. 97, 3118–3123. doi: 10.1073/pnas.97.7.3118
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Santner, A., and Estelle, M. (2009). Recent advances and emerging trends in plant hormone signalling. Nature 459, 1071–1078. doi: 10.1038/nature08122
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shah, J., Tsui, F., and Klessig, D. F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant Microbe Interact. 10, 69–78. doi: 10.1094/MPMI.1997.10.1.69
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shi, Z., Maximova, S., Liu, Y., Verica, J., and Guiltinan, M. J. (2013). The salicylic acid receptor NPR3 is a negative regulator of the transcriptional defense response during early flower development in Arabidopsis. Mol. Plant 6, 802–816. doi: 10.1093/mp/sss091
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shimono, M., Koga, H., Akagi, A., Hayashi, N., Goto, S., Sawada, M.,et al. (2012). Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Mol. Plant Pathol. 13, 83–94. doi: 10.1111/j.1364-3703.2011.00732.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shimono, M., Sugano, S., Nakayama, A., Jiang, C. J., Ono, K., Toki, S.,et al. (2007). Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19, 2064–2076. doi: 10.1105/tpc.106.046250
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shirasu, K. (2009). The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 60, 139–164. doi: 10.1146/annurev.arplant.59.032607.092906
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Smalle, J., and Vierstra, R. D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55, 555–590. doi: 10.1146/annurev.arplant.55.031903.141801
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Spoel, S. H., and Dong, X. N. (2012). How do plants achieve immunity? defence without specialized immune cells. Nat. Rev. Immunol. 12, 89–100. doi: 10.1038/nri3141
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Spoel, S. H., Mou, Z., Tada, Y., Spivey, N. W., Genschik, P., and Dong, X. (2009). Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137, 860–872. doi: 10.1016/j.cell.2009.03.038
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Spoel, S. H., Tada, Y., and Loake, G. J. (2010). Post-translational protein modification as a tool for transcription reprogramming. New Phytol. 186, 333–339. doi: 10.1111/j.1469-8137.2009.03125.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Stogios, P. J., Chen, L., and Prive, G. G. (2007). Crystal structure of the BTB domain from the LRF/ZBTB7 transcriptional regulator. Protein Sci. 16, 336–342. doi: 10.1110/ps.062660907
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Stokes, T. L., Kunkel, B. N., and Richards, E. J. (2002). Epigenetic variation in Arabidopsis disease resistance. Genes Dev. 16, 171–182. doi: 10.1101/gad.952102
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sugano, S., Jiang, C. J., Miyazawa, S. I., Masumoto, C., Yazawa, K., Hayashi, N.,et al. (2010). Role of OsNPR1 in rice defense program as revealed by genome-wide expression analysis. Plant Mol. Biol. 74, 549–562. doi: 10.1007/s11103-010-9695-3
Tada, Y., Spoel, S. H., Pajerowska-Mukhtar, K., Mou, Z., Song, J., Wang, C.,et al. (2008). Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956. doi: 10.1126/science.1156970
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tang, L.-Y., Yamashita, M., Coussens, N. P., Tang, Y., Wang, X., Li, C.,et al. (2011). Ablation of Smurf2 reveals an inhibition in TGF-β signalling through multiple mono-ubiquitination of Smad3. EMBO J. 30, 4777–4789. doi: 10.1038/emboj.2011.393
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Thrower, J. S., Hoffman, L., Rechsteiner, M., and Pickart, C. M. (2000). Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102. doi: 10.1093/emboj/19.1.94
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Trujillo, M., and Shirasu, K. (2010). Ubiquitination in plant immunity. Curr. Opin. Plant Biol. 13, 402–408. doi: 10.1016/j.pbi.2010.04.002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
van den Burg, H. A., Tsitsigiannis, D. I., Rowland, O., Lo, J., Rallapalli, G., Maclean, D.,et al. (2008). The F-box protein ACRE189/ACIF1 regulates cell death and defense responses activated during pathogen recognition in tobacco and tomato. Plant Cell 20, 697–719. doi: 10.1105/tpc.107.056978
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
van der Horst, A., de Vries-Smits, A. M. M., Brenkman, A. B., van Triest, M. H., van den Broek, N., Colland, F.,et al. (2006). FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat. Cell Biol. 8, 1064–1073. doi: 10.1038/ncb1469
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
van Ooijen, G., Lukasik, E., Van Den Burg, H. A., Vossen, J. H., Cornelissen, B. J. C., and Takken, F. L. W. (2010). The small heat shock protein 20 RSI2 interacts with and is required for stability and function of tomato resistance protein I-2. Plant J. 63, 563–572. doi: 10.1111/j.1365-313X.2010.04260.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
van Ooijen, G., van den Burg, H. A., Cornelissen, B. J. C., and Takken, F. L. W. (2007). Structure and function of resistance proteins in solanaceous plants. Annu. Rev. Phytopathol. 45, 43–72. doi: 10.1146/annurev.phyto.45.062806.094430
Vierstra, R. D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10, 385–397. doi: 10.1038/nrm2688
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Walsh, C. K., and Sadanandom, A. (2014). Ubiquitin chain topology in plant cell signaling: a new facet to an evergreen story. Front. Plant Sci. 5:122. doi: 10.3389/fpls.2014.00122
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, D., Amornsiripanitch, N., and Dong, X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2:e123. doi: 10.1371/journal.ppat.0020123
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Weigel, R. R., Pfitzner, U. M., and Gatz, C. (2005). Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell 17, 1279–1291. doi: 10.1105/tpc.104.027441
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wu, R. C., Feng, Q., Lonard, D. M., and O’Malley, B. W. (2007). SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell 129, 1125–1140. doi: 10.1016/j.cell.2007.04.039
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wu, Y., Zhang, D., Chu, J. Y., Boyle, P., Wang, Y., Brindle, I. D.,et al. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1, 639–647. doi: 10.1016/j.celrep.2012.05.008
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yi, H., and Richards, E. J. (2009). Gene duplication and hypermutation of the pathogen resistance gene SNC1 in the Arabidopsis bal variant. Genetics 183, 1227–1234. doi: 10.1534/genetics.109.105569
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhai, Q., Yan, L., Tan, D., Chen, R., Sun, J., Gao, L.,et al. (2013). Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genet. 9:e1003422. doi: 1003410.1001371/journal.pgen.1003422
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, Y., Cheng, Y. T., Qu, N., Zhao, Q., Bi, D., and Li, X. (2006). Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J. 48, 647–656. doi: 10.1111/j.1365-313X.2006.02903.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. U.S.A. 96, 6523–6528. doi: 10.1073/pnas.96.11.6523
Zhang, Y., Tessaro, M. J., Lassner, M., and Li, X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15, 2647–2653. doi: 10.1105/tpc.014894
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhou, J. M., Trifa, Y., Silva, H., Pontier, D., Lam, E., Shah, J.,et al. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant Microbe Interact. 13, 191–202. doi: 10.1094/MPMI.2000.13.2.191
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhou, W., and Slingerland, J. M. (2014). Links between oestrogen receptor activation and proteolysis: relevance to hormone-regulated cancer therapy. Nat. Rev. Cancer 14, 26–38. doi: 10.1038/nrc3622
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhuang, M., Calabrese, M. F., Liu, J., Waddell, M. B., Nourse, A., Hammel, M.,et al. (2009). Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol. Cell 36, 39–50. doi: 10.1016/j.molcel.2009.09.022
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: Cullin-RING ligase (CRL), ubiquitin ligase, salicylic acid (SA), NPR1, plant immunity, proteasome, transcription activator, gene expression
Citation: Furniss JJ and Spoel SH (2015) Cullin-RING ubiquitin ligases in salicylic acid-mediated plant immune signaling. Front. Plant Sci. 6:154. doi: 10.3389/fpls.2015.00154
Received: 09 December 2014; Paper pending published: 27 January 2015;
Accepted: 26 February 2015; Published online: 13 March 2015.
Edited by:
Loreto Holuigue, Pontificia Universidad Católica de Chile, ChileRoger W. Innes, Indiana University Bloomington, USA
Copyright © 2015 Furniss and Spoel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steven H. Spoel, Institute of Molecular Plant Sciences, School of Biological Sciences, University of Edinburgh, Edinburgh EH9 3JR, UK steven.spoel@ed.ac.uk
 James J. Furniss
James J. Furniss Steven H. Spoel
Steven H. Spoel