- 1 Department of Plant Pathology, University of Kentucky, Lexington, KY, USA
- 2 Department of Plant Biology and Ecology, College of Life Sciences, Nankai University, Tianjin, China
Salicylic acid (SA) is an important phytohormone that plays a vital role in a number of physiological responses, including plant defense. The last two decades have witnessed a number of breakthroughs related to biosynthesis, transport, perception and signaling mediated by SA. These findings demonstrate that SA plays a crictical role in both local and systemic defense responses. Systemic acquired resistance (SAR) is one such SA-dependent response. SAR is a long distance signaling mechanism that provides broad spectrum and long-lasting resistance to secondary infections throughout the plant. This unique feature makes SAR a highly desirable trait in crop production. This review summarizes the recent advances in the role of SA in SAR and discusses its relationship to other SAR inducers.
Introduction
Plants being sessile are constantly exposed to a number of pathogenic microbes, which based on their infectious lifestyles can be broadly divided into biotrophs and necrotrophs (Glazebrook, 2005; Mengiste, 2012; Lai and Mengiste, 2013). Biotrophic pathogens rely on nutrients from living host cells, whereas necrotrophic pathogens feed on dead cells. Plants employ distinct immune responses to counter these pathogens and this aspect has been covered in detail in several recent reviews (Spoel and Dong, 2012; Dangl et al., 2013). This first layer of host defense involves the recognition of pathogen (or microbe) associated-molecular patterns (PAMPs/MAMPs), such as bacterial flagellin, lipopolysaccharides, and peptidoglycans. PAMPs are recognized by specialized transmembrane proteins in the plant, termed pattern recognition receptors (PPRs). PRR-mediated recognition of PAMPs triggers downstream signaling leading to the activation of basal resistance termed PAMP-triggered immunity (PTI; Schwessinger and Ronald, 2012). PTI can be suppressed by pathogen encoded effector proteins commonly known as avirulence (avr) factors (Göhre and Robatzek, 2008; Cunnac et al., 2009; Bozkurt et al., 2011; Caillaud et al., 2012; Marrtin and Kamoun, 2012; Cheong et al., 2013; Cui et al., 2013; Dangl et al., 2013; Giraldo and Valent, 2013). The avr factors are in turn recognized by the host encoded resistance (R) proteins, which confer more durable and robust resistance termed R gene- or effector-triggered immunity (ETI; Bogdanove and Martin, 2000; Mackey et al., 2002, 2003; Gu et al., 2005; Jones and Dangl, 2006; Narusaka et al., 2009; Cesari et al., 2013). ETI is generally associated with programmed cell death (PCD) at the site of infection and this phenomenon is called hypersensitive response (HR; Holliday et al., 1981; Dangl et al., 1996; Morel and Dangl, 1997).
Induction of local responses is associated with the transport of defense signals throughout the plant resulting in broad-spectrum disease resistance against secondary infections. This phenomenon, known as systemic acquired resistance (SAR), is conserved among diverse plants and confers long-lasting resistance to unrelated pathogens (Chaturvedi et al., 2008; Dempsey and Klessig, 2012; Fu and Dong, 2013; Kachroo and Robin, 2013; Lucas et al., 2013; Shah and Zeier, 2013; Wendehenne et al., 2014). Several studies have shown that the establishment of SAR involves the generation and transport of signals via phloem to the uninfected distal tissues (Guedes et al., 1980; Tuzun and Kuc, 1985). Among the signals contributing to SAR are salicylic acid (SA) and several components of the SA pathway including the methylated derivative of SA (methyl SA,MeSA, Park et al., 2007). Additionally, the diterpenoid dehydroabietinal (DA, Chaturvedi et al., 2012), the nine carbon (C9) dicarboxylic acid azelaic acid (AzA, Jung et al., 2009), an amino acid derivative pipecolic acid (Pip; Návarová et al., 2012), auxin (Truman et al., 2010), the phosphorylated sugar glycerol-3-phosphate (G3P, Chanda et al., 2011; Mandal et al., 2012; Yu et al., 2013), the free radicals nitric oxide (NO) and reactive oxygen species (ROS; Wang et al., 2014a; El-Shetehy et al., 2015), galactolipids (Gao et al., 2014), factors contributing to cuticle formation (Xia et al., 2009, 2010, 2012) and the lipid transfer proteins (LTPs) DIR1 (Defective in Induced Resistance, Maldonado et al., 2002) and AZI1 (AzA insensitive, Jung et al., 2009), have all been proposed to serve as SAR signals. Here, we review the role of SA in SAR and discuss its relationship to these various SAR signals.
SA Biosysnthesis and SAR
Salicylic acid biosynthesis occurs via the shikimic acid pathway, which forms two distinct sub-branches both of which synthesize SA. These branched pathways, designated as isochorismate synthase (ICS)- and the phenylalanine ammonia-lyase (PAL)-derived pathways, utilize chorismate as the common precursor (Shah, 2003; Chen et al., 2009; Kachroo and Kachroo, 2009; Yu et al., 2010; An and Mou, 2011; Dempsey et al., 2011; Vlot et al., 2009; Singh et al., 2013; Figure 1). The first step of the PAL pathway involves conversion of phenylalanine (Phe) to trans-cinnamic acid and this reaction is catalyzed by PAL, a key enzyme of this pathway that is induced by pathogen infection. The Arabidopsis genome encodes four PAL isoforms and PAL quadruple mutants or wild-type plants treated with the PAL inhibitor, 2-aminoindan-2-phosphonic acid contain reduced SA, show increased susceptibility to pathogens and are unable to induce SAR (Yalpani et al., 1993; Mauch-Mani and Slusarenko, 1996; Pallas et al., 1996; Huang et al., 2010). Although relative contributions of PAL versus ICS branches toward SA biosynthesis vary between different plant species, at least in Arabidopsis majority of the pathogen-induced SA appears to be derived from the ICS branch. The ICS branch involves conversion of chorismate to isochorismate by ICS followed by coversion of isochorismate to SA by isochorismate pyruvate lyase (IPL). The Arabidopsis genome encodes two isoforms of ICS, of which ICS1 (SID2) accounts for ∼95% of basal- or pathogen-induced SA (Strawn et al., 2007; Garcion et al., 2008). A mutation in ICS1 also impairs SAR (Wildermuth et al., 2001; Jung et al., 2009; Chanda et al., 2011; Wang et al., 2014a), suggesting that SA contributed by both PAL- and ICS-pathways is critical for the induction and/or establishment of SAR. This together with the compromised SAR phenotype of transgenic plants expressing bacterial salicylate hydroxylase (NahG; Vernooij et al., 1994), an enzyme that catalyzes the conversion of SA to catechol, reemphasize the importance of SA in SAR. It is unclear what factors govern the specific recruitment of the PAL or ICS pathways for SA biosynthesis.
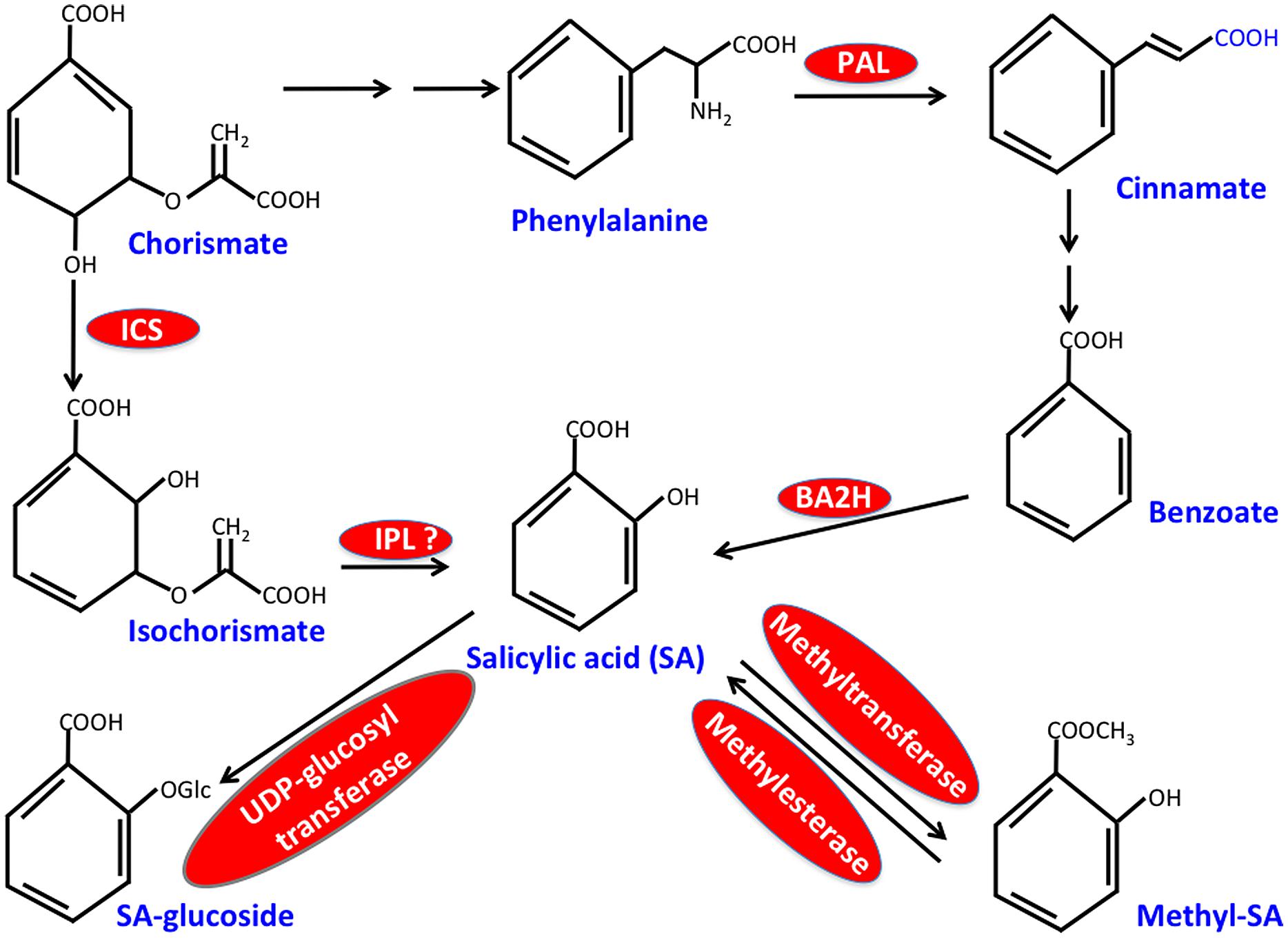
FIGURE 1. Simplified scheme for salicylic acid (SA) biosynthesis in plants. Critical enzymes are shown in red. ICS, isochorismate synthase; IPL, isochorismate pyruvate lyase; PAL, phenylalanine ammonia-lyase; BA2H, benzoic acid 2-hydroxylase.
Salicylic acid synthesized in the chloroplasts is exported out to the cytosol via EDS5, a member of the MATE (Multidrug and Toxin Extrusion) transporter family, located in the chloroplast envelope (Nawrath et al., 2002; Serrano et al., 2013). Notably, a mutation in EDS5 results in complete shut down of SA biosynthesis rather than SA accumulation within the chloroplasts. Thus, mutations in ICS1 and EDS5 similarly affect SA levels and the corresponding mutants thereby exhibit overlapping defense defects. This is likely due to negative feed-back regulation of ICS1 by SA (Fragnière et al., 2011; Serrano et al., 2013). The triphosphate tunnel metalloenzyme 2 is a negative regulator of the SA feed-back loop and functions in defense signal amplification (Ung et al., 2014). Pathogen induced expression of ICS1 requires the binding of calmodulin binding protein CBP60g and its homolog, non-calmodulin binding SARD1 (SAR Deficient 1) to the ICS1 promoter. CBP60g and SARD1 specifically bind the GAAATTTTGG sequence in the ICS1 promoter (Truman and Glazebrook, 2012). The induction of ICS1 and thereby SA biosynthesis is inhibited in cbp60g sard1 double mutant, resulting in compromised SAR (Zhang et al., 2010).
Although a number of studies have demonstrated the critical requirement of SA in SAR, a specific requirement for SA accumulation beyond basal levels during SAR has not been established. For instance, plants lacking a functional R protein RPS2 accumulate normal levels of SA in their distal tissues in response to infection by Pseudomonas syringae pv. tomato expressing avrRpt2, yet these plants are compromised for SAR (Cameron et al., 1999). Additionally, exogenous application of either G3P or AzA, which induce SAR in wild-type plants, do not induce SA accumulation. However, neither G3P nor AzA can confer SAR in ics1 (sid2) mutant plants, which contain significantly reduced basal- and pathogen-induced SA. Thus, although SA is clearly critical for SAR, accumulation of SA alone is insufficient to establish SAR. Furthermore, although SA has been shown to accumulate to varying levels in the distal tissues of SAR induced plants (Table 1), there is no evidence suggesting that this accumulation is essential for SAR.
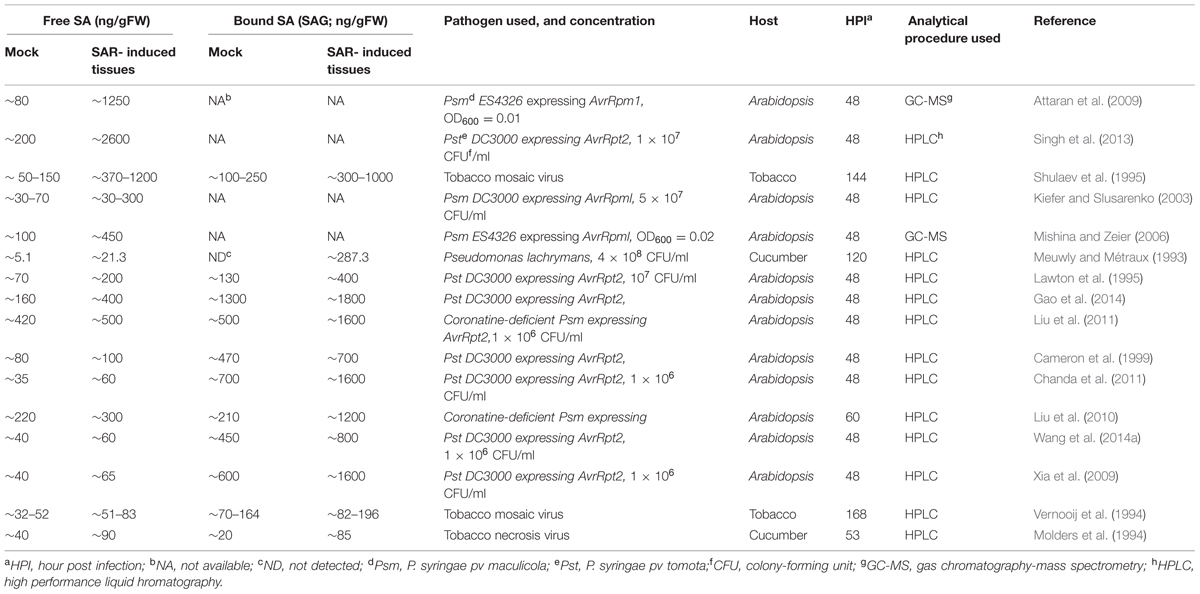
TABLE 1. Free and bound salicylic acid (SA) levels reported in distal tissues of mock-and pathogen-inoculated plants.
In comparison to local tissues, the distal tissues of SAR-induced plants have been shown to accumulate a broad range of SA ranging from as low as 10 ng/ g FW to ∼2.6 μg/g FW (Table 1; Rasmussen et al., 1991; Yalpani et al., 1991; Meuwly and Métraux, 1993; Molders et al., 1994; Vernooij et al., 1994; Lawton et al., 1995; Shulaev et al., 1995; Cameron et al., 1999; Kiefer and Slusarenko, 2003; Mishina and Zeier, 2006; Attaran et al., 2009; Liu et al., 2010, 2011; Xia et al., 2010; Chanda et al., 2011; Gao et al., 2014). The inability to accumulate SA in distal tissues has also been suggested to contribute to impaired SAR in ald1 (agd2-Like Defense response protein 1) and fmo1 (Flavin Monooxygenase 1) mutants, both of which accumulate normal SA in the local tissue (Song et al., 2004a,b; Mishina and Zeier, 2006). ALD1 encodes an aminotransferase that catalyzes the biosynthesis of the SAR inducer Pip, (Song et al., 2004b; Návarová et al., 2012) and FMO1 has been suggested to function downstream of Pip (Návarová et al., 2012). Thus, other factors besides SA might contribute to the SAR defect of ald1 and fmo1 mutants. One possibility is that SAR can be induced via SA-independent factors so long as a minimum basal level of SA can be maintained. Alternatively, SA accumulation in distal tissues might contribute to the priming process resulting in the activation of stronger defense responses upon secondary infections (Návarová et al., 2012; Gruner et al., 2013).
SA-Derivatives and SAR
A majority of the synthesized SA is converted and stored as biologically inactive derivatives via glucosylation, methylation and amino acid conjugation since accumulation of the acidic SA has adverse physiological consequences (Heil and Baldwin, 2002; Heidel et al., 2004). These include SA 2-O-β-D-glucose (SAG), SA glucose ester (SGE), methyl SA (MeSA), and SA-amino acid conjugates (Pierpoint, 1994; Vlot et al., 2009; Dempsey et al., 2011). Most recently, SA was shown to be derivatized to 2,3-dihydroxybenzoic acid (2,3-DHBA) and this reaction is catalyzed by SA 3-hydroxylase (S3H; Zhang et al., 2013). As predicted s3h knockout plants contain very high levels of SA, while plants expressing S3H gain-of-function mutations accumulate high amounts of 2,3-DHBA (Zhang et al., 2013). SA derivatives serve as storage forms that can be converted back to free SA (Hennig et al., 1993; Kawano et al., 2004; Kachroo and Kachroo, 2012). With the exception of MeSA however, the exact role of SA derivatives in SAR remains unclear.
Methyl SA is a volatile and phloem mobile SA derivative, which accumulates in infected and distal tissues in response to pathogen infection. Like MeSA, SA also accumulates in the phloem of tobacco leaves infected with tobacco mosaic virus or Colletotrichum lagenarium and in cucumber leaves infected with tobacco necrosis virus (Malamy et al., 1990; Métraux et al., 1990; Park et al., 2007). For SAR, MeSA must be converted to SA in the distal tissues between the 48–72 h period post primary infection. This time-frame correlates with that of pathogen-inducible MeSA accumulation in infected and systemic tissues. The biosynthesis of MeSA is catalyzed by SA methyltransferases (SAMT/BSMT), and the conversion of MeSA back to SA is mediated by methyl esterase (MES; Chen et al., 2003; Effmert et al., 2005; Koo et al., 2007). The tobacco MES was first identified based on its ability to bind SA, and therefore designated as SA-binding protein 2 (SABP2; Kumar and Klessig, 2003). Grafting studies in tobacco plants silenced for SABP2 have shown that SABP2 activity in scions, but not root-stocks is required for normal SAR (Park et al., 2007). Furthermore, the synthetic SA analog, 2,2,2,2′-tetra-fluoroacetophenone, which inhibits the esterase activity of SABP2, also inhibits SAR (Park et al., 2009). As in tobacco, homologs of SABP2 (AtMES9) and SAMT AtBSMT1 are required for SAR in Arabidopsis (Liu et al., 2011). Thus, the ability to derivatize SA to MeSA and reconvert MeSA back to SA are critical for SAR. Intriguingly, the requirement for AtBSMT1 in SAR can be bypassed by prolonged exposure to light after pathogen inoculation (Attaran et al., 2009; Liu et al., 2011). However, the role of light signaling in SAR and how it might compensate for MeSA is unclear. It is also not known whether MeSA merely functions to deliver SA to the distal tissues or has other function(s) in SAR. Notably, a certain percentage of SA is always transported from the inoculated to distal tissues (Meuwly et al., 1995; Kiefer and Slusarenko, 2003). The biological significance of this transport is unclear, particularly in view of the fact that SA is not considered to be the mobile SAR signal since wild-type tobacco scions grafted onto NahG root-stocks exhibit normal SAR (Vernooij et al., 1994; Meuwly et al., 1995; Kiefer and Slusarenko, 2003).
Regulation of SA Accumulation and SAR
Besides proteins that directly contribute to SA biosynthesis (ICS and PAL) or transport (EDS5), a number of other proteins have been identified that participate in pathogen induced SA accumulation and thereby SAR. These include EDS1 (Enhanced Disease Susceptibility 1), PAD4 (Phytoalexin Deficient 4), and NDR1 (Non-race-specific Disease Resistance 1; Century et al., 1995, 1997; Falk et al., 1999; Jirage et al., 1999; McDowell et al., 2000; Feys et al., 2001; Coppinger et al., 2004; Ishihara et al., 2008; Bhattacharjee et al., 2011; Cacas et al., 2011; Heidrich et al., 2011; Knepper et al., 2011; Lu et al., 2013). Unlike ICS1 and EDS5, mutations in EDS1, PAD4, or NDR1 cause partial reduction in SA levels. EDS1 and PAD4 are lipase-like proteins, which together with another lipase-like protein SAG101 (Senescence Associate Gene 101) form binary and ternary complexes (Feys et al., 2005; Zhu et al., 2011). EDS1 interacts with PAD4 in both cytosol and nucleus, and with SAG101 only in the nucleus. EDS1, PAD4, and SAG101 function cooperatively as well as independently in pathogen defense (Feys et al., 2005; Venugopal et al., 2009; Rietz et al., 2011; Zhu et al., 2011). For instance, all three proteins are required for R-mediated resistance against Turnip crinkle virus (TCV) but only PAD4 is required for the SA-mediated induction of the R gene which confers HR against TCV (HRT; Chandra-Shekara et al., 2004, 2006, 2007). Interestingly, EDS1, but not PAD4 or SAG101, interacts with HRT and potentiates HRT-mediated HR to TCV (Zhu et al., 2011). Similarly, only PAD4 is required for resistance to the green peach aphid, whereas EDS1 and SAG101 are not (Pegadaraju et al., 2005, 2007; Louis et al., 2010, 2012). Both nuclear and extranuclear localization of EDS1 is important for its defense function (García et al., 2010). However, the role of binary or ternary complex formation between EDS1, PAD4, and SAG101 proteins remains unknown. EDS1 was recently shown to participate in both SAR signal generation in the local tissues as well as perception in the distal leaves (Breitenbach et al., 2014).
The Arabidopsis genome encodes two isoforms of EDS1 that function redundantly and can compensate for each other (Zhu et al., 2011). However, some Arabidopsis ecotypes, such as Wassilewskija, Landsberg, and Dujon, contain only one functional EDS1 isoform, and this is sufficient for normal resistance in these ecotypes. Like Arabidopsis, soybean also contains two EDS1 isoforms. Interestingly, Arabidopsis eds1 mutant expressing the soybean EDS1 orthologs is only partially restored in SA levels, but completely restored in bacterial resistance (Wang et al., 2014b). This further questions the requirement for increased SA accumulation during defense activation and raises the possibility that a certain threshold of SA may be sufficient to induce appropriate defense responses. The soybean EDS1 orthologs are unable to potentiate TCV coat protein-derived activation of HRT even though they do interact with HRT (Wang et al., 2014b). This suggests that EDS1 orthologs in different plants may have evolved to perform overlapping as well as distinct functions.
SA Signaling Components
Salicylic acid-mediated signaling leading to SAR is dependent on the ankyrin repeat containing protein NPR1 [Non-expressor of Pathogenesis-Related (PR) genes] (Dong, 2004). Under basal or uninduced conditions, NPR1 exists as a cytosolic inactive oligomer formed by intermolecular disulfide bonding (Mou et al., 2003). Reducing conditions resulting from accumulation of SA cause dissociation of the NPR1 oligomer into active monomers and the monomeric form of NPR1 is translocated into the nucleus (Kinkema et al., 2000; Mou et al., 2003; Tada et al., 2008). Nuclear localization of NPR1 facilitates its interaction with members of the TGACG motif binding (TGA) transcription factors that belong to the basic leucine zipper (bZIP) protein family (Zhang et al., 1999; Després et al., 2000; Niggeweg et al., 2000; Zhou et al., 2000; Chern et al., 2001; Fan and Dong, 2002; Kim and Delaney, 2002). This in turn enhances binding of the TGA factors to promoter elements of NPR1-dependent target genes (Wang et al., 2006, 2011). Like NPR1, TGA factors are also required for SAR; the tga2 tga5 tga6 triple mutant is non-responsive to SA and is defective in SAR (Zhang et al., 2003). Recent studies have shown that NPR1 and TGA1 also undergo S-nitrosylation, which is necessary for the proper functioning of NPR1 in immunity and increases the DNA binding activity of TGA1 (Tada et al., 2008; Lindermayr et al., 2010). On the other hand, thiol S-nitrosylation has also been shown to promote NPR1 oligomerization and thereby its inactivation (Tada et al., 2008). The nuclear NPR1 is phosphorylated and degraded in a proteasome-dependent manner (Spoel et al., 2009), and the turnover of NPR1 is essential for SAR establishment. The Arabidopsis genome contains five paralogs of NPR1 (Liu et al., 2005). Like NPR1, NPR3, and NPR4 also interact with TGA proteins (Zhang et al., 2006). The npr3 npr4 mutant plants accumulate elevated levels of NPR1 and are consequently defective in SAR. NPR3 and NPR4 bind SA and function as adaptors of the Cullin 3 ubiquitin E3 ligase to mediate NPR1 degradation in an SA-dependent manner (Fu et al., 2012). However, the two differ in that NPR3 has higher affinity for SA than NPR4, and SA promotes the NPR1–NPR3 interaction but inhibits the NPR1–NPR4 interaction. These contrasting effects might offer a possible explanation for the nuances underlying NPR1-dependent immunity under different levels of SA. For instance, high concentration of SA in infected tissues might favor binding of NPR3 with SA, which would mediate degradation of the cell-death suppressor NPR1, and initiate PCD and local immunity. On the other hand, lower SA levels in the distal uninfected tissue would minimize NPR3-SA binding, thereby inhibiting PCD. Interestingly, in yet another study, NPR1 was also shown to bind SA via the transition metal copper (Wu et al., 2012; Manohar et al., 2015). The binding of SA to NPR was suggested to induce a conformational change in NPR1 (Wu et al., 2012), which in turn is important for NPR1-dependant PR1 expression.
NPR1 is also required for transgenerational SAR, which in turn involves epigenetic changes (Jaskiewicz et al., 2011; Luna et al., 2012). NPR1 othologs have been characterized from a number of plants including rice, tobacco, soybean, and cacao (Chern et al., 2001, 2005, 2014; Ekengren et al., 2003; Zwicker et al., 2007; Sandhu et al., 2009; Shi et al., 2010; Chen et al., 2013). Transgenic expression of Arabidopsis NPR1 confers enhanced resistance in heterologous plants (Lin et al., 2004; Shi et al., 2010; Chen et al., 2012). Conversely, transgenic expression of soybean orthologs can complement the Arabidopsis npr1 mutation (Sandhu et al., 2009). Overexpression of NPR1 also enhances pathogen resistance in monocots (Chern et al., 2005; Yuan et al., 2007). However, studies in rice and barley suggest that NPR1 function may not be fully conserved in monocots and dicots and that SA signaling and SAR in monocots might involve NPR1-independent pathways (Shimono et al., 2007; Dey et al., 2014). Transcription analysis in distal tissues revealed that bacteria-triggered SAR in barley was likely associated with jasmonic acid, ethylene and ABA, rather than SA. In contrast, SAR in maize is associated with SA accumulation in local and distal leaves (Balmer et al., 2013). Additionally, petiole exudates from pathogen infected Arabidopsis plants induced SAR in wheat (Chaturvedi et al., 2008). This suggests that SAR signaling in barley may not be similar to that in other monocots like maize and wheat.
The stability of NPR1 is dependent on Mediator (MED) 16 [allelic to Sensitive to Freezing (SFR) 6] (McKown et al., 1996; Warren et al., 1996), a subunit of the MED complex which functions as a bridge between transcription factors and the general RNA polymerase II transcriptional machinery (Zhang et al., 2012). A mutation in MED16 compromises SAR and SA-induced defense responses but does not affect SA levels or nuclear localization of NPR1. Thus, MED16 likely functions downstream of SA in the SAR pathway. Interestingly, MED16 is also required for jasmonic acid/ethylene-responsive gene expression and resistance to necrotrophic pathogens (Zhang et al., 2012). Thus, MED16 might function by relaying signals from transcription factors that are specific to the SA and JA/ethylene pathways. A mutation in another MED subunit, MED 15 (isolated in a screen for non-recognition-of-the SA analog, BTH, nrb4), also attenuates SAR and SA responsiveness (Canet et al., 2012). However, MED15 is not required for NPR1 stability or localization and likely functions downstream of NPR1.
SA Versus Other SAR Inducers
Systemic acquired resistance is a complex phenomenon that involves the interplay of a diverse group of chemicals and associated proteins, besides SA. Most of these molecules can now be placed in one of two main branches that comprise the SAR pathway. One branch involves SA and its signaling component NPR1, and the other branch involves the free radicals NO and ROS, which function directly upstream of AzA, which in turn is upstream of G3P (Wang et al., 2014a; Wendehenne et al., 2014; El-Shetehy et al., 2015). Unlike G3P and AzA, exogenous application of Pip or DA induces SA accumulation in the absence of pathogen infection (Chaturvedi et al., 2012; Návarová et al., 2012). Therefore, Pip and DA likely function in the SA branch of SAR. The presence of two SAR branches is supported by the fact that SA cannot restore SAR in mutants defective in NO, ROS, or G3P biosynthesis, while NO/ROS cannot confer SAR on mutants defective in SA synthesis or signaling. Furthermore, pharmacological inhibitors of NO synthesis or NO scavengers attenuate SA-induced SAR in tobacco (Song and Goodman, 2001). Interestingly, unlike SA, both NO and ROS function in a concentration dependent manner because they can confer SAR only when present at an optimal concentration (Wang et al., 2014a). Free radicals are well known to operate similarly in animal systems where too little or too much can produce opposing physiological effects (Delledonne et al., 1998; Besson-Bard et al., 2008; Wink et al., 2011). Free radicals are thought to participate in SAR by mediating the oxidation of carbon (C) 18 unsaturated fatty acids (FAs) containing a double bond on C 9. This results in the formation of 9-oxo nonanoic acid (ONA), which is converted to the di-carboxylic acid AzA by the addition of a carboxylic group. AzA is unable to confer SAR on mutants unable to synthesize G3P, indicating it functions upstream of G3P. Exogenous AzA increases the expression of the G3P synthesizing GLY1 and GLI1 genes, which encode G3P dehydrogenase and glycerol kinase, respectively. G3P operates in a feedback loop with the LTPs DIR1 and AZI1 such that lack of DIR1 or AZI1 impairs pathogen-induced G3P accumulation while lack of G3P results in reduced DIR1 and AZI1 transcripts (Yu et al., 2013). DIR1 and AZI1 form homo- and hetromers suggesting that a complex comprising these proteins might function in SAR. Perhaps such a complex or the individual LTPs serve in transporting SAR essential signal(s) to the distal tissues. G3P appears to be the logical choice for such a transported signal since it is a precursor for lipid biogenesis. However, no direct interaction could be detected between G3P and DIR1 raising the possibility that G3P may be derivatized and this derivative may then be transported from infected to distal tissues. Radiolabel feeding experiments showed that G3P is indeed converted to an as yet unidentified derivative which can translocate from infected to distal tissues in a DIR1-dependent manner (Chanda et al., 2011).
Recent studies have shown that the C 18 FAs which serve as precusors for AzA are derived from the major plastidal lipids, monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG), which comprise ∼80% of the total lipids in plants (Zoeller et al., 2012; Gao et al., 2014). Thus, besides SA, NO, ROS and G3P, chloroplasts also serve as an important site for AzA biosynthesis. Notably, both galactose sugars in DGDG appear to be important for SAR since dgd1 plants producing α-glucose-β-galactose diacylglycerol via transgenic expression of a bacterial glucosyltransferase, are not restored in SAR even though they are partially restored in chloroplast function. Thus it appears that the position of the hydroxyl group on C 4 of galactose may be important for SAR since glucose and galactose are sterioisomeric sugars which differ only in the position of their axial hydroxyl group at C 4.
Cross Talk between SA and NO Pathways in SAR
Monogalactosyldiacylglycerol and DGDG galactolipids also serve additional functions in SAR. For instance, DGDG is required for SA and NO biosynthesis (Gao et al., 2014) and for AzA responsiveness. Interestingly, in spite of their impaired SA and NO synthesis, petiole exudates from pathogen-infected dgd1 plants were able to confer SAR in wild-type plants. This suggests that dgd1 plants can make signals that are capable of inducing SA- and NO-synthesis in plants with normal DGDG levels. These results show that SAR involves DGDG-dependent retrograde signaling between the chloroplast and nucleus and emphasizes the fact that the two branches of SAR are intricately linked (Gao et al., 2014).
In fact it is well known that there is cross talk between SA- and NO-mediated signaling. For example, NO mediated S-nitrosylation of NPR1 can result in the oligomerization and nuclear localization of NPR1 (Tada et al., 2008; Lindermayr et al., 2010). Moreover, SA has been suggested to regulate chloroplast structure since exogenous SA can cause swelling of grana thylakoids, coagulation of the stroma and increased chloroplast volume (Uzunova and Popova, 2000; Rivas-San Vicente and Plasencia, 2011). Regulation of SA and AzA levels by EDS1 is another case in point (Wittek et al., 2014). Together, these results suggest that the parallel operation of the interlinked SA- and NO-pathways might allow multiple points of regulation in fine tuning the optimal onset of SAR. This may be particularly relevant for signals like NO and ROS, which are functional within specific concentration ranges (Wang et al., 2014a).
Conclusion and Perspectives
Recent work on SAR has identified a number of chemical and protein signals and placed them in a common pathway that comprises at least two parallel branches (Figure 2). However, these studies also indicate the involvement of additional unknown signal(s) that function upstream of the branchpoint separating SA-NPR1- and NO-ROS-AzA-G3P-derived pathways. In addition, several chemical signals, including G3P and AzA, undergo derivatization into unknown compounds and at least one of the G3P-derivative is SAR bioactive (unpublished data). Identification of these signals should provide useful insights into signaling events leading to the induction and establishment of SAR. Another area of SAR research that has not received much attention is the transport and perception of signals in the distal tissues. Although cuticle was implicated in the perception of SAR signals (Xia et al., 2009), later studies on cuticle mutants have suggested that perception might relate to the severity of cuticular damage or perhaps other unknown factors (Xia et al., 2012). These aspects of SAR should provide exciting avenues for studying how SAR overlaps with basic physiological processes and the distinct events that decide the onset of SAR versus normal growth and development.
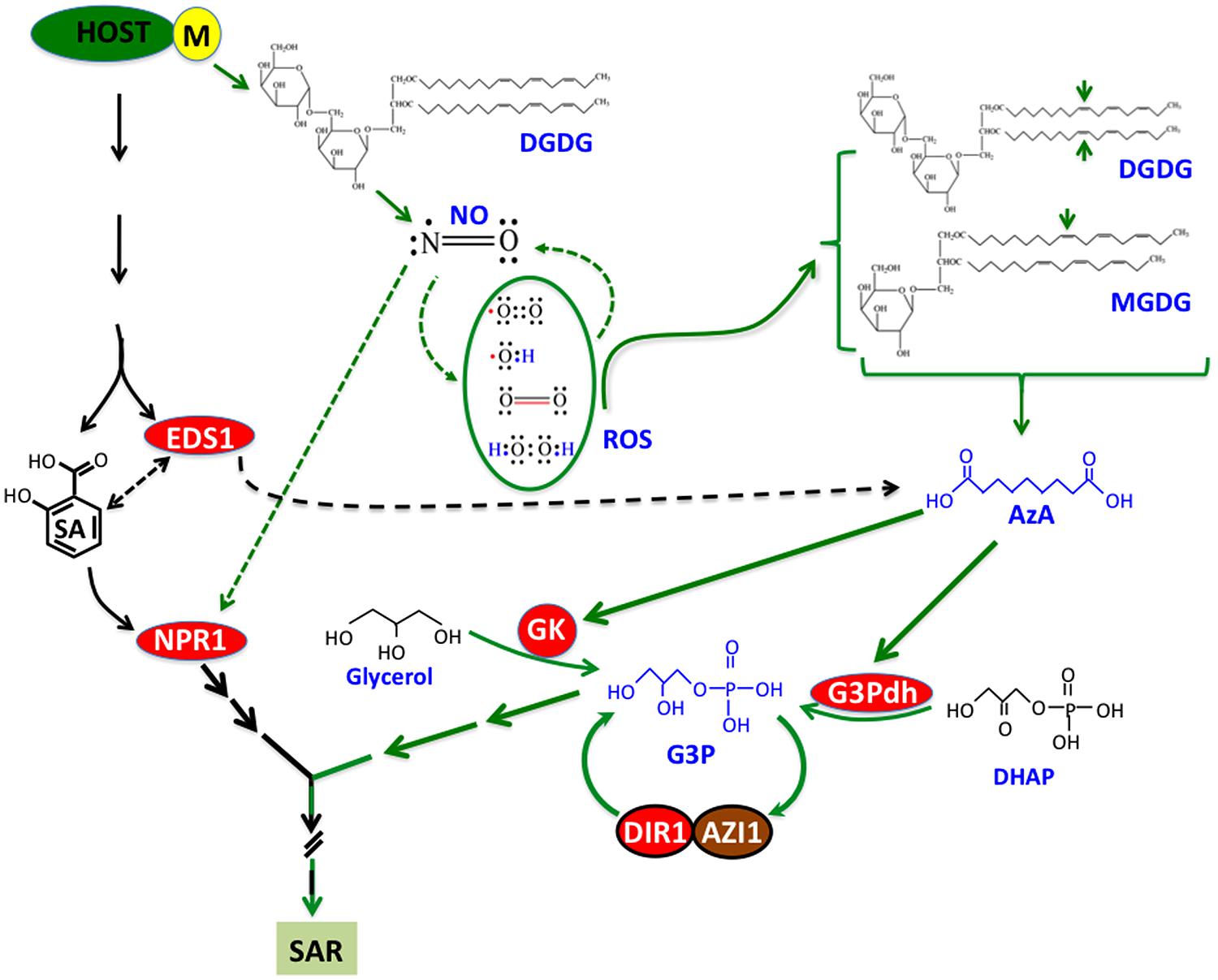
FIGURE 2. A simplified model showing pathways, chemicals, and proteins involved in SAR. Infection with avirulent (avr) pathogen induces accumulation of SA and nitric oxide (NO) in a digalactosyldiacylglycerol (DGDG)-dependent manner. NO operates in a feedback loop with reactive oxygen species (ROS), which catalyze oxidation of C18 unsaturated fatty acids (FA) present on monogalactosyldiacylglycerol (MGDG) and DGDG lipids. Oxidation of C18 FAs at C9 carbon (indicated by the arrows) generates azelaic acid (AzA), which triggers biosynthesis of glycerol-3-phosphate (G3P) via upregulation of genes encoding G3P biosynthetic enzymes (glycerol kinase, GK and G3P dehydrogenase, G3Pdh). G3P-mediated signaling is dependent on DIR1 and AZI1, which interact with each other and require G3P for the stability of their respective transcripts. Conversely, DIR1 and AZI1 are also required for G3P biosynthesis, suggesting that G3P and DIR1/AZI1 regulate SAR via a feedback loop. In the SA branch, EDS1 regulates both SA and AzA levels. NPR1 is a key downstream component in the SA branch which is nitrosylated by NO.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all members (past and present) of the Kachroo (AK and PK) laboratories who contributed to SA and SAR related work. Work in our laboratories is funded from the National Science Foundation (IOS# 0749731, #051909), Kentucky Soybean Board and the Kentucky Science and Engineering Foundation (KSF-2923-RDE-016). The information reported in this article (No. 15-12-039) is part of a project of the Kentucky Agricultural Experiment Station and is published with the approval of the Director.
References
An, C., and Mou, Z. (2011). Salicylic acid and its function in plant immunity. J. Integr. Plant Biol. 53, 412–428. doi: 10.1111/j.1744-7909.2011.01043.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Attaran, E., Zeier, T. E., Griebel, T., and Zeier, J. (2009). Methyl salicylate production and jasmonate signaling are not essential for systemic acquired resistance in Arabidopsis. Plant Cell 21, 954–971. doi: 10.1105/tpc.108.063164
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Balmer, D., de Papajewski, D. V., Planchamp, C., Glauser, G., and Mauch-Mani, B. (2013). Induced resistance in maize is based on organ-specific defence responses. Plant J. 74, 213–225. doi: 10.1111/tpj.12114
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Besson-Bard, A., Pugin, A., and Wendehenne, D. (2008). New insights into nitric oxide signaling in plants. Annu. Rev. Plant Biol. 59, 21–39. doi: 10.1146/annurev.arplant.59.032607.092830
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bhattacharjee, S., Halane, M. K., Kim, S. H., and Gassmann, W. (2011). Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 334, 1405–1408. doi: 10.1126/science.1211592
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bogdanove, A. J., and Martin, G. B. (2000). AvrPto-dependent Pto-interacting proteins and AvrPto-interacting proteins in tomato. Proc. Natl. Acad. Sci. U.S.A. 97, 8836–8840. doi: 10.1073/pnas.97.16.8836
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bozkurt, T. O., Schornack, S., Win, J., Shindo, T., Ilyas, M., Oliva, R., et al. (2011). Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl. Acad. Sci. U.S.A. 108, 20832–20837. doi: 10.1073/pnas.1112708109
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Breitenbach, H. H., Wenig, M., Wittek, F., Jordá, L., Maldonado-Alconada, A. M., Sarioglu, H., et al. (2014). Contrasting roles of the apoplastic aspartyl protease apoplastic, enhanced disease susceptibility1-dependent1 and legume lectin-like protein1 in arabidopsis systemic acquired resistance. Plant Physiol. 165, 791–809. doi: 10.1104/pp.114.239665
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cacas, J. L., Petitot, A. S., Bernier, L., Estevan, J., Conejero, G., Mongrand, S., et al. (2011). Identification and characterization of the non-race specific disease resistance 1 (NDR1) orthologous protein in coffee. BMC Plant Biol. 11:144. doi: 10.1186/1471-2229-11-144
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Caillaud, M. C., Piquerez, S. J., Fabro, G., Steinbrenner, J., Ishaque, N., Beynon, J., et al. (2012). Subcellular localization of the Hpa RxLR effector repertoire identifies a tonoplast-associated protein HaRxL17 that confers enhanced plant susceptibility. Plant J. 69, 252–265. doi: 10.1111/j.1365-313X.2011.04787.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cameron, R., Paiva, N. L., Lamb, C. J., and Dixon, R. A. (1999). Accumulation of salicylic acid and PR-1 gene transcripts in relation to the systemic acquired resistance (SAR) response induced by Pseudomonas syringae pv. tomato in Arabidopsis. Physiol. Mol. Plant Pathol. 55, 121–130. doi: 10.1006/pmpp.1999.0214
Canet, J. V., Dobón, A., and Tornero, P. (2012). Non-recognition-of-BTH4, an Arabidopsis mediator subunit homolog, is necessary for development and response to salicylic acid. Plant Cell 24, 4220–4235. doi: 10.1105/tpc.112.103028
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Century, K. S., Holub, E. B., and Staskawicz, B. J. (1995). NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. U.S.A. 92, 6597–6601. doi: 10.1073/pnas.92.14.6597
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Century, K. S., Shapiro, A. D., Repetti, P. P., Dahlbeck, D., Holub E., and Staskawicz, B. J. (1997). NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278, 1963–1965. doi: 10.1126/science.278.5345.1963
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cesari, S., Thillieza, G., Ribota, C., Chalvona, V., Michela, C., Jauneauc, A., et al. (2013). The rice resistance protein pair RGA4/RGA5 recognizes the magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25, 1463–1481. doi: 10.1105/tpc.112.107201
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chanda, B., Xia, Y., Mandal, M. K., Sekine, K. T., Gao, Q. M., Selote, D., et al. (2011). Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nat. Genet. 43, 421–427. doi: 10.1038/ng.798
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chandra-Shekara, A. C., Gupte, M, Navarre, D., Raina, S., Raina, R., Klessig, D. F., et al. (2006). Light-dependent hypersensitive response and resistance signaling to turnip crinkle virus in Arabidopsis. Plant J. 45, 320–334. doi: 10.1111/j.1365-313X.2005.02618.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chandra-Shekara, A. C., Navarre, D., Kachroo, A., Kang, H. G., Klessig, D. F., and Kachroo, P. (2004). Signaling requirements and role of salicylic acid in HRT- and rrt-mediated resistance to turnip crinkle virus in Arabidopsis. Plant J. 40, 647–659. doi: 10.1111/j.1365-313X.2004.02241.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chandra-Shekara, A. C., Venugopal, S. C., Barman, S. R., Kachroo, A., and Kachroo, P. (2007). Plastidial fatty acid levels regulate resistance gene-dependent defense signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 7277–7282. doi: 10.1073/pnas.0609259104
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chaturvedi, R., Krothapalli, K., Makandar, R., Nandi, A., Sparks, A., Roth, M., et al. (2008). Plastid omega3-fatty acid desaturase-dependent accumulation of a systemic acquired resistance inducing activity in petiole exudates of Arabidopsis thaliana is independent of jasmonic acid. Plant J. 54, 106–117. doi: 10.1111/j.1365-313X.2007.03400.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chaturvedi, R., Venables, B., Petros, R. A., Nalam, V., Li, M., Wang, X., et al. (2012). An abietane diterpenoid is a potent activator of systemic acquired resistance. Plant J. 71, 161–172. doi: 10.1111/j.1365-313X.2012.04981.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, F., D’Auria, J. C., Tholl, D., Ross, J. R., Gershenzon, J., Noel, J. P., et al. (2003). An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J. 36, 577–588. doi: 10.1046/j.1365-313X.2003.01902.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, J. C., Lu, H. C., Chen, C. E., Hsu, H. F., Chen, H. H., and Yeh, H. H. (2013). The NPR1 ortholog PhaNPR1 is required for the induction of PhaPR1 in Phalaenopsis aphrodite. Bot. Stud. 54, 31. doi: 10.1186/1999-3110-54-31
Chen, X. K., Zhang, J. Y., Zhang, Z., Du, X. L., Du, B. B., and Qu, S. C. (2012). Overexpressing MhNPR1 in transgenic Fuji apples enhances resistance to apple powdery mildew. Mol. Biol. Rep. 39, 8083–8089. doi: 10.1007/s11033-012-1655-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, Z., Zheng, Z., Huang, J., Lai, Z., and Fan, B. (2009). Biosynthesis of salicylic acid in plants. Plant Signal. Behav. 4, 493–496. doi: 10.4161/psb.4.6.8392
Cheong, H., Kim, C. Y., Jeon, J. S., Lee, B. M., Moon S. J., and Hwang, I. (2013). Xanthomonas oryzae pv. oryzae type III effector XopN targets OsVOZ2 and a putative thiamine synthase as a virulence factor in rice. PLoS ONE 8:e73346. doi: 10.1371/journal.pone.0073346
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chern, M., Bai, W., Ruan, D., Oh, T., Chen, X., and Ronald, P. C. (2014). Interaction specificity and coexpression of rice NPR1 homologs 1 and 3 (NH1 and NH3), TGA transcription factors and Negative Regulator of Resistance (NRR) proteins. BMC Genomics 15:461. doi: 10.1186/1471-2164-15-461
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chern, M., Fitzgerald, H. A., Canlas, P. E., Navarre, D. A., and Ronald, P. C. (2005). Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol. Plant Microbe Interact. 6, 511–520. doi: 10.1094/MPMI-18-0511
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chern, M. S., Fitzgerald, H. A., Yadav, R. C., Canlas, P. E., Dong, X., and Ronald, P. C. (2001). Evidence for a disease-resistance pathway in rice similar to the NPR1-mediated signaling pathway in Arabidopsis. Plant J. 27, 101–113. doi: 10.1046/j.1365-313x.2001.01070.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Coppinger, P., Repetti, P. P., Day, B., Dahlbeck, D., Mehlert, A., and Staskawicz, B. J. (2004). Overexpression of the plasma membrane-localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana. Plant J. 40, 225–237. doi: 10.1111/j.1365-313X.2004.02203.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cui, F., Wu, S., Sun, W., Coaket, G., Kunkel, B., He, P., et al. (2013). The Pseudomonas syringae type III effector AvrRpt2 promotes pathogen virulence via stimulating Arabidopsis auxin/indole acetic acid protein turnover. Plant Physiol. 162, 1018–1029. doi: 10.1104/pp.113.219659
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cunnac, S., Lindeberg, M., and Collmer, A. (2009). Pseudomonas syringae type III secretion system effectors: repertoires in search of functions. Curr. Opin. Microbiol. 12, 53–60. doi: 10.1016/j.mib.2008.12.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dangl, J. L., Dietrich, R. A., and Richberg, M. H. (1996). Death don’t have nomercy: cell death programs in plant-microbe interactions. Plant Cell 8, 1793–1807. doi: 10.1105/tpc.8.10.1793
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dangl, J. L., Horvath, D. M., and Staskawicz, B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science 341, 746–751. doi: 10.1126/science.1236011
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Delledonne, M., Xia, Y., Dixon, R. A., and Lamb, C. (1998). Nitric oxide functions as a signal in plant disease resistance. Nature 394, 585–588. doi: 10.1038/29087
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dempsey, D. A., and Klessig, D. F. (2012). SOS-too many signals for systemic acquired resistance? Trends Plant Sci. 17, 538–545. doi: 10.1016/j.tplants.2012.05.011
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dempsey, D. A., Vlot, A. C., Wildermuth, M. C., and Klessig, D. F. (2011). Salicylic acid biosynthesis and metabolism. Arabidopsis Book 9, e0156. doi: 10.1199/tab.0156
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Després, C., DeLong, C., Glaze, S., Liu, E., and Fobert, P. R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12, 279–290. doi: 10.1105/tpc.12.2.279
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dey, S., Wenig, M., Langen, G., Sharma, S., Kugler, K. G., Knappe, C., et al. (2014). Bacteria-triggered systemic immunity in barley is associated with WRKY and ETHYLENE RESPONSIVE FACTORs but not with salicylic acid. Plant Physiol. 166, 2133–2151. doi: 10.1104/pp.114.249276
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dong, X. (2004). NPR1, all things considered. Curr. Opin. Plant Biol. 7, 547–552. doi: 10.1016/j.pbi.2004.07.005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Effmert, U., Saschenbrecker, S., Ross, J., Negre, F., Fraser, C. M., Noel, J. P., et al. (2005). Floral benzenoid carboxyl methyltransferases: from in vitro to in planta function. Phytochemistry 66, 1211–1230. doi: 10.1016/j.phytochem.2005.03.031
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ekengren, S. K., Liu, Y., Schiff, M., Dinesh-Kumar, S. P., and Martin, G. B. (2003). Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J. 36, 905–917. doi: 10.1046/j.1365-313X.2003.01944.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
El-Shetehy, M., Wang, C., Shine, M. B., Yu, K., Navarre, D., Wendehenne, D., et al. (2015). Nitric oxide and reactive oxygen species are required for systemic acquired resistance in plants. Plant Signal. Behav. In Press.
Falk, A., Feys, B. J., Frost, L. N., Jonathan, D. G., Jones, D. G., Michael, J., et al. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. U.S.A. 96, 3292–3297. doi: 10.1073/pnas.96.6.3292
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fan, W., and Dong, X. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14, 1377–1389. doi: 10.1105/tpc.001628
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Feys, B. J., Moisan, L. J., Newman, M. A., and Parker, J. E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20, 5400–5411. doi: 10.1093/emboj/20.19.5400
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Feys, B. J., Wiermer, M., Bhat, R. A., Moisan, L. J., Medina-Escobar, N., Neu, C., et al. (2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17, 2601–2613. doi: 10.1105/tpc.105.033910
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fragnière, C., Serrano, M., Abou-Mansour, E., Métraux, J. P., and L’Haridon, F. (2011). Salicylic acid and its location in response to biotic and abiotic stress. FEBS Lett. 585, 1847–1852. doi: 10.1016/j.febslet.2011.04.039
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fu, Z. Q., and Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fu, Z. Q., Yan, S., Saleh, A., Wang, W., Ruble, J., Oka, N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232. doi: 10.1038/nature11162
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gao, Q. M., Xia, Y., Yu, K., Navarre, D., Kachroo, A., and Kachroo, P. (2014). Galactolipids are required for nitric oxide biosynthesis and systemic acquired resistance. Cell Rep. 9, 1681–1691. doi: 10.1016/j.celrep.2014.10.069
García, A. V., Blanvillain-Baufumé, S., Huibers, R. P., Wiermer, M., Li, G., Gobbato, E., et al. (2010). Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog. 6:e1000970. doi: 10.1371/journal.ppat.1000970
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Garcion, C., Lohmann, A., Lamodière, E., Catinot, J., Buchala, A., Doermann, P., et al. (2008). Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 147, 1279–1287. doi: 10.1104/pp.108.119420
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Giraldo, M., and Valent, B. (2013). Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 11, 800–814. doi: 10.1038/nrmicro3119
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Göhre, V., and Robatzek, S. (2008). Breaking the barriers: microbial effector molecules subvert plant immunity. Annu. Rev. Phytopathol. 46, 189–215. doi: 10.1146/annurev.phyto.46.120407.110050
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gruner, K., Griebel, T., Návarová, H., Attaran, E., and Zeier, J. (2013). Reprogramming of plants during systemic acquired resistance. Front. Plant Sci. 4:252. doi: 10.3389/fpls.2013.00252
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gu, K., Yang, B., Tian, D., Wu, L., Wang, D., Sreekala, C., et al. (2005). R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435, 1122–1125. doi: 10.1038/nature03630
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Guedes, M. E. M., Richmond, S., and Kuc, J. (1980). Induced systemic resistance to anthracnose in cucumber as influenced by the location of the inducer inoculation with Colletotrichum lagenarium and the onset of flowering and fruiting. Physiol. Plant Pathol. 17, 229–233. doi: 10.1016/0048-4059(80)90056-9
Heidel, A. J., Clarke, J. D., Antonovics, J., and Dong, X. (2004). Fitness costs of mutations affecting the systemic acquired resistance pathway in Arabidopsis thaliana. Genetics 168, 2197–2206. doi: 10.1534/genetics.104.032193
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Heidrich, K., Wirthmueller, L., Tasset, C., Pouzet, C., Deslandes, L., and Parker, J. E. (2011). Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science 334, 1401–1404. doi: 10.1126/science.1211641
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Heil, M., and Baldwin, I. T. (2002). Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci. 7, 61–66. doi: 10.1016/S1360-1385(01)02186-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hennig, J., Malamy, J., Grynkiewicz, G., Indulski, J., and Klessig, D. (1993). Interconversion of the salicylic acid signal and its glucoside in tobacco. Plant J. 4, 593–600. doi: 10.1046/j.1365-313X.1993.04040593.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Holliday, M. J., Keen, N. T., and Long, M. (1981). Cell death patterns and accumulation of fluorescent material in the hypersensitive response of soybean leaves to Pseudomonas syringae pv. glycinea. Physiol. Plant Pathol. 18, 279–287. doi: 10.1016/S0048-4059(81)80079-3
Huang, J., Gu, M., Lai, Z., Fan, B., Shi, K., Zhou, Y. H., et al. (2010). Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 153, 1526–1538. doi: 10.1104/pp.110.157370
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ishihara, T., Sekine, K. T., Hase, S., Kanayama, Y., Seo, S., Ohashi, Y., et al. (2008). Overexpression of the Arabidopsis thaliana EDS5 gene enhances resistance to viruses. Plant Biol. 10, 451–461. doi: 10.1111/j.1438-8677.2008.00050.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jaskiewicz, M., Conrath, U., and Peterhänsel, C. (2011). Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 12, 50–55. doi: 10.1038/embor.2010.186
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jirage, D., Tootle, T. L., Reuber, T. L., Frost, L. N., Feys, B. J., Parker, J. E., et al. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 96, 13583–13588. doi: 10.1073/pnas.96.23.13583
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jung, H. W., Tschaplinkski, T. J., Wang, L., Glazebrook, J., and Greenberg, J. T. (2009). Priming in systemic plant immunity. Science 324, 89–91. doi: 10.1126/science.1170025
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kachroo, A., and Kachroo, P. (2009). Fatty acid-derived signals in plant defense. Annu. Rev. Phytopathol. 47, 153–176. doi: 10.1146/annurev-phyto-080508-081820
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kachroo, A., and Robin, G. P. (2013). Systemic signaling during plant defense. Curr. Opin. Plant Biol. 16, 527–533. doi: 10.1016/j.pbi.2013.06.019
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kachroo, P., and Kachroo, A. (2012). “The roles of salicylic acid and jasmonic acid in plant immunity,” in Molecular Plant Immunity, ed. G. Sessa (Oxford: Wiley-Blackwell). doi: 10.1002/9781118481431.ch4
Kawano, T., Furuichi, T., and Muto, S. (2004). Controlled free salicylic acid levels and corresponding signaling mechanisms in plants. Plant Biotechnol. 21, 319–335. doi: 10.5511/plantbiotechnology.21.319
Kiefer, I. W., and Slusarenko, A. J. (2003). The pattern of systemic acquired resistance induction within the Arabidopsis rosette in relation to the pattern of translocation. Plant Physiol. 132, 840–847. doi: 10.1104/pp.103.021709
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kim, H. S., and Delaney, T. P. (2002). Over-expression of TGA5, which encodes a bZIP transcription factor that interacts with NIM1/NPR1, confers SAR-independent resistance in Arabidopsis thaliana to Peronospora parasitica. Plant J. 32, 151–163. doi: 10.1046/j.1365-313X.2001.01411.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kinkema, M., Fan, W., and Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350. doi: 10.1105/tpc.12.12.2339
Knepper, C., Savory, E. A., and Day, B. (2011). Arabidopsis NDR1 is an integrin-like protein with a role in fluid loss and plasma membrane-cell wall adhesion. Plant Physiol. 156, 286–300. doi: 10.1104/pp.110.169656
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Koo, Y. J., Kim, M. A., Kim, E. H., Song, J. T., Jung, C., Moon, J. K., et al. (2007). Overexpression of salicylic acid carboxyl methyltransferase reduces salicylic acid-mediated pathogen resistance in Arabidopsis thaliana. Plant Mol. Biol. 64, 1–15. doi: 10.1007/s11103-006-9123-x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kumar, D., and Klessig, D. F. (2003). High-affinity salicylic acid-binding protein 2 is required for plant innate immunity and has salicylic acid stimulated lipase activity. Proc. Natl. Acad. Sci. U.S.A. 100, 16101–16106. doi: 10.1073/pnas.0307162100
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lai, Z., and Mengiste, T. (2013). Genetic and cellular mechanisms regulating plant responses to necrotrophic pathogens. Curr. Opin. Plant Biol. 16, 505–512. doi: 10.1016/j.pbi.2013.06.014
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lawton, K., Weymann, K., Friedrich, L., Vernooij, B., Uknes, S., and Ryals, J. (1995). Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol. Plant Microbe Interact. 8, 863–870. doi: 10.1094/MPMI-8-0863
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lin, W. C., Lu, C. F., Wu, J. W., Cheng, M. L., Lin, Y. M., Yang, N. S., et al. (2004). Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Res. 13, 567–581. doi: 10.1007/s11248-004-2375-9
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lindermayr, C., Sell, S., Müller, B., Leister, D., and Durner, J. (2010). Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22, 2894–2907. doi: 10.1105/tpc.109.066464
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Liu, G., Holub, E. B., Alonso, J. M., Ecker, J. R., and Fobert, P. R. (2005). An Arabidopsis NPR1-like gene, NPR4, is required for disease resistance. Plant J. 41, 304–318. doi: 10.1111/j.1365-313X.2004.02296.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Liu, P. P., von Dahl, C. C., and Klessig, D. F. (2011). The extent to which methyl salicylate is required for signaling systemic acquired resistance is dependent on exposure to light after infection. Plant Physiol. 157, 2216–2226. doi: 10.1104/pp.111.187773
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Liu, P. P., Yang, Y., Pichersky, E., and Klessig, D. F. (2010). Altering expression of benzoic acid/salicylic acid carboxyl methyltransferase 1 compromises systemic acquired resistance and PAMP-triggered immunity in Arabidopsis. Mol. Plant Microbe Interact. 23, 82–90. doi: 10.1094/MPMI-23-1-0082
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Louis, J., Gobbato, E., Mondal, H. A., Feys, B. J., Parker, J. E., and Shah, J. (2012). Discrimination of Arabidopsis PAD4 activities in defense against green peach aphid and pathogens. Plant Physiol. 158, 1860–1872. doi: 10.1104/pp.112.193417
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Louis, J., Leung, Q., Pegadaraju, V., Reese, J., and Shah, J. (2010). PAD4-dependent antibiosis contributes to the ssi2-conferred hyper-resistance to the green peach aphid. Mol. Plant Microbe Interact. 23, 618–627. doi: 10.1094/MPMI-23-5-0618
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lu, H., Zhang, C., Albrecht, U., Shimizu, R., Wang, G., and Bowman, K. D. (2013). Overexpression of a citrus NDR1 ortholog increases disease resistance in Arabidopsis. Front. Plant Sci. 4:157. doi: 10.3389/fpls.2013.00157
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lucas, W. J., Groover, A., Lichtenberger, R., Furuta, K., Yadav, S. R., Helariutta, Y., et al. (2013). The plant vascular system: evolution, development and functions. J. Integr. Plant Biol. 55, 294–388. doi: 10.1111/jipb.12041
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Luna, E., Bruce, T. J., Roberts, M. R., Flors, V., and Ton, J. (2012). Next-generation systemic acquired resistance. Plant Physiol. 158, 844–853. doi: 10.1104/pp.111.187468
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mackey, D., Belkhadir, Y., Alonso, J. M., Ecker, J. R., and Dangl, J. L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379–389. doi: 10.1016/S0092-8674(03)00040-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mackey, D., Holt, B. F., Wiig, A., and Dangl, J. L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754. doi: 10.1016/S0092-8674(02)00661-X
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Malamy, J., Carr, J. P., Klessig, D. F., and Raskin, I. (1990). Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250, 1002–1004. doi: 10.1126/science.250.4983.1002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Maldonado, A. M., Doerner, P., Dixon, R. A., Lamb, C. J., and Cameron, R. K. (2002). A putative lipid transfer protein involved in systemic resistance signaling in Arabidopsis. Nature 419, 399–403. doi: 10.1038/nature00962
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mandal, M. K., Chandra-Shekara, A. C., Jeong, R. D., Yu, K., Zhu, S., Chanda, B., et al. (2012). Oleic acid-dependent modulation of NITRIC OXIDE ASSOCIATED1 protein levels regulates nitric oxide-mediated defense signaling in Arabidopsis. Plant Cell 24, 1654–1674. doi: 10.1105/tpc.112.096768
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Manohar, M., Tian, M., Moreau, M., Park, S. W., Choi, H. W., Fei, Z., et al. (2015). Identification of multiple salicylic acid-binding proteins using two high throughput screens. Front. Plant Sci. 5:777. doi: 10.3389/fpls.2014.00777
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Marrtin, F., and Kamoun, S. (2012). Effectors in Plant-microbe Interactions. Chichester; Ames, IA: Wiley-Blackwell.
Mauch-Mani, B., and Slusarenko, A. J. (1996). Production of salicylic acid precursors is a major function of phenylalanine ammonia-lyase in the resistance of Arabidopsis to Peronospora parasitica. Plant Cell 8, 203–212. doi: 10.1105/tpc.8.2.203
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
McDowell, J. M., Cuzick, A., Can, C., Beynon, J., Dangl, J. L., and Holub, E. B. (2000). Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1 and salicylic acid accumulation. Plant J. 22, 523–529. doi: 10.1046/j.1365-313x.2000.00771.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
McKown, R., Kuroki, G., and Warren, G. (1996). Cold responses of Arabidopsis mutants impaired in freezing tolerance. J. Exp. Bot. 47, 1919–1925. doi: 10.1093/jxb/47.12.1919
Mengiste, T. (2012). Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50, 267–294. doi: 10.1146/annurev-phyto-081211-172955
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Métraux, J. P., Signer, H., Ryals, J., Ward, E., Wyss-Benz, M., Gaudin, J., et al. (1990). Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250, 1004–1006. doi: 10.1126/science.250.4983.1004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Meuwly, P., and Métraux, J. P. (1993). Ortho-anisic acid as internal standard for the simultaneous quantitation of salicylic acid and its putative biosynthetic precursors in cucumber leaves. Anal. Biochem. 214, 500–505. doi: 10.1006/abio.1993.1529
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Meuwly, P., Molders, W., Buchala, A., and Metraux, J. P. (1995). Local and systemic biosynthesis of salicylic acid in infected cucumber plants. Plant Physiol. 109, 1107–1114.
Mishina, T. E., and Zeier, J. (2006). The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 141, 1666–1675. doi: 10.1104/pp.106.081257
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Molders, W., Meuwly, P., Summermatter, K., and Métraux, J. P. (1994). Salicylic acid content in cucumber plants infected with Pseudomonas lachrymans and tobacco necrosis virus. Acta Hortic. 381, 375–378.
Morel, J. B., and Dangl, J. L. (1997). The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 4, 671–683. doi: 10.1038/sj.cdd.4400309
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mou, Z., Fan, W., and Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. doi: 10.1016/S0092-8674(03)00429-X
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Narusaka, M., Shirasu, K., Noutoshi, Y., Kubo, Y., Shiraishi, T., Iwabuchi, M., et al. (2009). RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 60, 218–226. doi: 10.1111/j.1365-313X.2009.03949.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Návarová, H., Bernsdorff, F., Döring, A. C., and Zeier, J. (2012). Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24, 5123–5141. doi: 10.1105/tpc.112.103564
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nawrath, C., Heck, S., Parinthawong, N., and Metraux, J. P. (2002). EDS5, an essential component of salicylic acid dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14, 275–286. doi: 10.1105/tpc.010376
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Niggeweg, R., Thurow, C., Kegler, C., and Gatz, C. (2000). Tobacco transcription factor TGA2.2 is the main component of as-1-binding factor ASF-1 and is involved in salicylic acid- and auxin-inducible expression of as-1-containing target promoters. J. Biol. Chem. 275, 19897–19905. doi: 10.1074/jbc.M909267199
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pallas, J. A., Paiva, N. L., Lamb, C., and Dixon, R. A. (1996). Tobacco plants epigenetically suppressed in phenylalanine ammonia-lyase expression do not develop systemic acquired resistance in response to infection by tobacco mosaic virus. Plant J. 10, 281–293. doi: 10.1046/j.1365-313X.1996.10020281.x
Park, S. W., Kaiyomo, E., Kumar, D., Mosher, S. L., and Klessig, D. F. (2007). Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318, 113–116. doi: 10.1126/science.1147113
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Park, S. W., Liu, P. P., Forouhar, F., Vlot, A. C., Tong, L., Tietjen, K., et al. (2009). Use of a synthetic salicylic acid analog to investigate the roles of methyl salicylate and its esterases in plant disease resistance. J. Biol. Chem. 284, 7307–7317. doi: 10.1074/jbc.M807968200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pegadaraju, V., Knepper, C., Reese, J., and Shah, J. (2005). Premature leaf senescence modulated by the Arabidopsis PHYTOALEXIN DEFICIENT 4 gene is associated with defense against the phloem-feeding green peach aphid1. Plant Physiol. 139, 1927–1934. doi: 10.1104/pp.105.070433
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pegadaraju, V., Louis, J., Singh, V., Reese, J. C., Bautor, J., Feys, B. J., et al. (2007). Premature leaf senescence modulated by the Arabidopsis PHYTOALEXIN DEFICIENT4 gene is associated with defense against the phloem-feeding green peach aphid1. Plant J. 52, 332–341. doi: 10.1111/j.1365-313X.2007.03241.x
Pierpoint, W. S. (1994). Salicylic acid and its derivatives in plants: medicines, metabolites and messenger molecules. Bot. Res. 20, 163–235. doi: 10.1016/S0065-2296(08)60217-7
Rasmussen, J. B., Hammerschmidt, R., and Zook, M. N. (1991). Systemic induction of salicylic acid accumulation in cucumber after inoculation with Pseudomonas syringae pv syringae. Plant Physiol. 97, 1342–1347. doi: 10.1104/pp.97.4.1342
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rietz, S., Stamm, A., Malonek, S., Wagner, S., Becker, D., Medina-Escobar, N., et al. (2011). Different roles of enhanced disease susceptibility1 (EDS1) bound to and dissociated from phytoalexin deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 191, 107–119. doi: 10.1111/j.1469-8137.2011.03675.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rivas-San Vicente, M., and Plasencia, J. (2011). Salicylic acid beyond defence: its role in plant growth and development. J. Exp. Bot. 62, 3321–3338. doi: 10.1093/jxb/err031
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sandhu, D., Tasma, I. M., Frasch, R., and Bhattacharyya, M. K. (2009). Systemic acquired resistance in soybean is regulated by two proteins, Orthologous to Arabidopsis NPR1. BMC Plant Biol. 9:105. doi: 10.1186/1471-2229-9-105
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schwessinger, B., and Ronald, P. C. (2012). Plant innate immunity: perception of conserved microbial signatures. Annu. Rev. Plant Biol. 63, 451–482. doi: 10.1146/annurev-arplant-042811-105518
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Serrano, M., Wang, B., Aryal, B., Garcion, C., Abou-Mansour, E., Heck, S., et al. (2013). Export of salicylic acid from the chloroplast requires the MATE-like transporter EDS5. Plant Physiol. 162, 1815–1821. doi: 10.1104/pp.113.218156
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shah, J. (2003). The salicylic acid loop in plant defense. Curr. Opin. Plant Biol. 6, 365–371. doi: 10.1016/S1369-5266(03)00058-X
Shah, J., and Zeier, J. (2013). Long-distance communication and signal amplification in systemic acquired resistance. Front. Plant Sci. 4:30. doi: 10.3389/fpls.2013.00030
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shi, Z., Maximova, S. N., Liu, Y., Verica, J., and Guiltinan, M. J. (2010). Functional analysis of the Theobroma cacao NPR1 gene in Arabidopsis. BMC Plant Biol. 10:248. doi: 10.1186/1471-2229-10-248
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shimono, M., Sugano, S., Nakayama, A., Jiang, C. J., Ono, K., Toki, S., et al. (2007). Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19, 2064–2076. doi: 10.1105/tpc.106.046250
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shulaev, V., Leon, J., and Raskin, I. (1995). Is salicylic acid a translocated signal of systemic acquired resistance in tobacco? Plant Cell 7, 1691–1701. doi: 10.1105/tpc.7.10.1691
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Singh, V., Roy, S., Giri, M. K., Chaturvedi, R., Chowdhury, Z., Shah, J., et al. (2013). Arabidopsis thaliana FLOWERING LOCUS D is required for systemic acquired resistance. Mol. Plant Microbe Interact. 26, 1079–1088. doi: 10.1094/MPMI-04-13-0096-R
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Song, F., and Goodman, R. M. (2001). Activity of nitric oxide is dependent on, but is partially required for function of, salicylic acid in the signaling pathway in tobacco systemic acquired resistance. Mol. Plant Microbe Interact. 14, 1458–1462. doi: 10.1094/MPMI.2001.14.12.1458
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Song, J. T., Lu, H., McDowell, J. M., and Greenberg, J. T. (2004a). A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J. 40, 200–212. doi: 10.1111/j.1365-313X.2004.02200.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Song, J., Lu, H., and Greenberg, J. (2004b). Divergent roles in Arabidopsis thaliana development and defense of two homologous genes, ABERRANT GROWTH AND DEATH2 and AGD2-LIKE DEFENSE RESPONSE PROTEIN1, encoding novel aminotransferases. Plant Cell 16, 353–366. doi: 10.1105/tpc.019372
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Spoel, S., and Dong, X. (2012). How do plants achieve immunity? Defence withoug specialized immune cells. Nat. Rev. Immunol. 12, 89–100. doi: 10.1038/nri3141
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Spoel, S. H., Mou, Z., Tada, Y., Spivey, N. W., Genschik, P., and Dong, X. (2009). Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual role in regulating plant immunity. Cell 137, 860–872. doi: 10.1016/j.cell.2009.03.038
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Strawn, M. A., Marr, S. K., Inoue, K., Inada, N., Zubieta, C., and Wildermuth, M. C. (2007). Arabidopsis isochorismate synthase functional in pathogen-induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J. Biol. Chem. 282, 5919–5933. doi: 10.1074/jbc.M605193200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tada, Y., Spoel, S. H., Pajerowska-Mukhtar, K., Mou, Z., Song, J., Wang, C., et al. (2008). Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956. doi: 10.1126/science.1156970
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Truman, W. M., Bennett, M. H., Turnbull, C. G., and Grant, M. R. (2010). Arabidopsis auxin mutants are compromised in systemic acquired resistance and exhibit aberrant accumulation of various indolic compounds. Plant Physiol. 152, 1562–1573. doi: 10.1104/pp.109.152173
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Truman, W., and Glazebrook, J. (2012). Co-expression analysis identifies putative targets for CBP60g and SARD1 regulation. BMC Plant Biol. 12:216. doi: 10.1186/1471-2229-12-216
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tuzun, S., and Kuc, J. (1985). Movement of a factor in tobacco infected with Peranospora tabacina Adam which systemically protects against blue mold. Physiol. Plant Pathol. 26, 321–330. doi: 10.1016/0048-4059(85)90007-4
Ung, H., Moeder, W., and Yoshioka, K. (2014). Arabidopsis triphosphate tunnel metalloenzyme2 is a negative regulator of the salicylic acid-mediated feedback amplification loop for defense responses. Plant Physiol. 166, 1009–1021. doi: 10.1104/pp.114.248757
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Uzunova, A. N., and Popova, L. P. (2000). Effect of salicylic acid on leaf anatomy and chloroplast ultrastructure of barley plants. Photosynthetica 38, 243–250. doi: 10.1023/A:1007226116925
Venugopal, S. C., Jeong, R. D., Mandal, M., Zhu, S., Chandra-Shekara, A. C., Xia, Y., et al. (2009). ENHANCED DISEASE SUSCEPTIBILITY and salicylic acid act redundantly to regulate resistance gene-mediated signaling. PLoS Genet. 5:e1000545. doi: 10.1371/journal.pgen.1000545
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vernooij, B., Friedrich, L., Morse, A., Reist, R., Kolditz-Jawhar, R., Ward, E., et al. (1994). Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell 6, 959–965. doi: 10.1105/tpc.6.7.959
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic Acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, C., El-Shetehy, M., Shine, M. B., Yu, K., Navarre, D., Wendehenne, D., et al. (2014a). Free radicals mediate systemic acquired resistance. Cell Rep. 7, 348–355 doi: 10.1016/j.celrep.2014.03.032
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, J., Shine, M. B., Gao, Q. M., Navarre, D., Jiang, W., Liu, C., et al. (2014b). Enhanced Disease Susceptibility1 mediates pathogen resistance and virulence function of a bacterial effector in soybean. Plant Physiol. 165, 1269–1284. doi: 10.1104/pp.114.242495
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, D., Amornsiripanitch, N., and Dong, X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2:e123. doi: 10.1371/journal.ppat.0020123
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, W., Barnaby, J. Y., Tada, Y., Li, H., Tör, M., Caldelari, D., et al. (2011). Timing of plant immune responses by a central circadian regulator. Nature 470, 110–114. doi: 10.1038/nature09766
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Warren, G., McKown, R., Marin, A., and Teutonico, R. (1996). Isolation of mutations affecting the development of freezing tolerance in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 111, 1011–1019. doi: 10.1104/pp.111.4.1011
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wendehenne, D., Gao, Q. M., Kachroo, A., and Kachroo, P. (2014). Free radical-mediated systemic immunity in plants. Curr. Opin. Plant Biol. 20, 127–134. doi: 10.1016/j.pbi.2014.05.012
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wildermuth, M. C., Dewdney, J., Wu, G., and Ausubel, F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. doi: 10.1038/35107108
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wink, D. A., Hines, H. B., Cheng, R. Y. S., Switzer, C. H., Flores-Santana, W., Vitek, M. P., et al. (2011). Nitric oxide and redox mechanisms in the immune response. J. Leukoc. Biol. 89, 873–891. doi: 10.1189/jlb.1010550
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wittek, F., Hoffmann, T., Kanawati, B., Bichlmeier, M., Knappe, C., Wenig, M., et al. (2014). Arabidopsis ENHANCED DISEASE SUSCEPTIBILITY1 promotes systemic acquired resistance via azelaic acid and its precursor 9-oxo nonanoic acid. J. Exp. Bot. 65, 5919–5931. doi: 10.1093/jxb/eru331
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wu, Y., Zhang, D., Chu, J. Y., Boyle, P., Wang, Y., Brindle, I. D., et al. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 6, 639–647. doi: 10.1016/j.celrep.2012.05.008
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Xia, Y., Gao, Q. M., Yu, K., Navarre, D., Hildebrand, D., Kachroo, A., et al. (2009). An intact cuticle in distal tissues is essential for the induction of systemic acquired resistance in plants. Cell Host Microbe. 5, 151–165. doi: 10.1016/j.chom.2009.01.001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Xia, Y., Yu, K., Gao, Q. M., Wilson, E. V., Navarre, D., Kachroo, P., et al. (2012). Acyl CoA binding proteins are required for cuticle formation and plant responses to microbes. Front. Plant Sci. 3:224. doi: 10.3389/fpls.2012.00224
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Xia, Y., Yu, K., Navarre, D., Seebold, K., Kachroo, A., and Kachroo, P. (2010). The glabra1 mutation affects cuticle formation and plant responses to microbes. Plant Physiol. 154, 833–846. doi: 10.1104/pp.110.161646
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yalpani, N., Leon, J., Lawton, M. A., and Raskin, I. (1993). Pathway of salicylic acid biosynthesis in healthy and virus-inoculated tobacco. Plant Physiol. 103, 315–321.
Yalpani, N., Silverman, P., Wilson, T. M., Kleier, D. A., and Raskin, I. (1991). Salicylic acid is a systemic signal and an inducer of pathogenesis-related proteins in virus-infected tobacco. Plant Cell 3, 809–818. doi: 10.1105/tpc.3.8.809
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yu, J., Gao, J., Wang, X. Y., Wei, Q., Yang, L. F., Qiu, K., et al. (2010). The pathway and regulation of salicylic acid biosynthesis in probenazole-treated Arabidopsis. J. Plant Biol. 53, 417–424. doi: 10.1007/s12374-010-9130-y
Yu, K., Soares, J. M., Mandal, M. K., Wang, C., Chanda, B., Gifford, A. N., et al. (2013). A feedback regulatory loop between G3P and lipid transfer proteins DIR1 and AZI1 mediates azelaic-acid-induced systemic immunity. Cell Rep. 3, 1266–1278. doi: 10.1016/j.celrep.2013.03.030
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yuan, Y., Zhong, S., Li, Q., Zhu, Z., Lou, Y., Wang, L., et al. (2007). Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol. J. 5, 313–324. doi: 10.1111/j.1467-7652.2007.00243.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, K., Halitschke, R., Yin, C., Liu, C. J., and Gan, S. S. (2013). Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc. Natl. Acad. Sci. U.S.A. 110, 14807–14812. doi: 10.1073/pnas.1302702110
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, X., Wang, C., Zhang, Y., Sun, Y., and Mou, Z. (2012). The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24, 4294–4309. doi: 10.1105/tpc.112.103317
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, Y., Cheng, Y. T., Qu, N., Zhao, Q., Bi, D., and Li, X. (2006). Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J. 48, 647–656. doi: 10.1111/j.1365-313X.2006.02903.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. U.S.A. 96, 6523–6528. doi: 10.1073/pnas.96.11.6523
Zhang, Y., Tessaro, M. J., Lassner, M., and Li, X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15, 2647–2653. doi: 10.1105/tpc.014894
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, Y., Xu, S., Ding, P., Wang, D., Cheng, Y. T., He, J., et al. (2010). Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. U.S.A. 107, 18220–18225. doi: 10.1073/pnas.1005225107
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhou, J. M., Trifa, Y., Silva, H., Pontier, D., Lam, E., Shah, J., et al. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant Microbe Interact. 13, 191–202. doi: 10.1094/MPMI.2000.13.2.191
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhu, S., Jeong, R. D., Venugopal, S. C., Lapchyk, L., Navarre, D., Kachroo, A., et al. (2011). SAG101 forms a ternary complex with EDS1 and PAD4 and is required for resistance signaling against Turnip crinkle virus. PLoS Pathog. 7:e1002318. doi: 10.1371/journal.ppat.1002318
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zoeller, M., Stingl, N., Krischke, M., Fekete, A., Waller, F., Berger, S., et al. (2012). Lipid profiling of the Arabidopsis hypersensitive response reveals specific lipid peroxidation and fragmentation processes: biogenesis of pimelic and azelaic acid. Plant Physiol. 160, 365–378. doi: 10.1104/pp.112.202846
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zwicker, S., Mast, S., Stos, V., Pfitzner, A. J., and Pfitzner, U. M. (2007). Tobacco NIMIN2 proteins control PR gene induction through transient repression early in systemic acquired resistance. Mol. Plant Pathol. 4, 385–400. doi: 10.1111/j.1364-3703.2007.00399.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: systemic resistance, plant defense, glycerol-3-phosphate, lipids, reactive oxygen species
Received: 23 January 2015; Accepted: 23 March 2015;
Published online: 13 April 2015.
Edited by:
Hua Lu, University of Maryland, Baltimore County, USAReviewed by:
Kemal Kazan, Commonwealth Scientific and Industrial Research Organization, AustraliaSteven H. Spoel, University of Edinburgh, UK
Citation:Gao Q-M, Zhu S, Kachroo P and Kachroo A (2015) Signal regulators of systemic acquired resistance. Front. Plant Sci. 6:228. doi: 10.3389/fpls.2015.00228
Copyright © 2015 Gao, Zhu, Kachroo and Kachroo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aardra Kachroo, Department of Plant Pathology, University of Kentucky, 201F Plant Science Building, 1405 Veterans drive, Lexington, KY 40546, USA apkach2@uky.edu
†These authors have contributed equally to this work.
 Qing-Ming Gao
Qing-Ming Gao Shifeng Zhu
Shifeng Zhu Pradeep Kachroo
Pradeep Kachroo