- 1Department of Biological Sciences, University of North Texas, Denton, TX, USA
- 2Department of Biology, Colorado State University, Fort Collins, CO, USA
The principal components of plant productivity and nutritional value, from the standpoint of modern agriculture, are the acquisition and partitioning of organic carbon (C) and nitrogen (N) compounds among the various organs of the plant. The flow of essential organic nutrients among the plant organ systems is mediated by its complex vascular system, and is driven by a series of transport steps including export from sites of primary assimilation, transport into and out of the phloem and xylem, and transport into the various import-dependent organs. Manipulating C and N partitioning to enhance yield of harvested organs is evident in the earliest crop domestication events and continues to be a goal for modern plant biology. Research on the biochemistry, molecular and cellular biology, and physiology of C and N partitioning has now matured to an extent that strategic manipulation of these transport systems through biotechnology are being attempted to improve movement from source to sink tissues in general, but also to target partitioning to specific organs. These nascent efforts are demonstrating the potential of applied biomass targeting but are also identifying interactions between essential nutrients that require further basic research. In this review, we summarize the key transport steps involved in C and N partitioning, and discuss various transgenic approaches for directly manipulating key C and N transporters involved. In addition, we propose several experiments that could enhance biomass accumulation in targeted organs while simultaneously testing current partitioning models.
Introduction
The principal components of plant productivity and nutritional value, from the standpoint of modern agriculture, are the acquisition and partitioning of organic carbon (C) and nitrogen (N) compounds among the various organs of the plant. The initial step of C acquisition is the fixation of atmospheric C dioxide by photosynthesis followed by sugar partitioning to the non-photosynthetic tissues. N is primarily absorbed as inorganic salts from the soil solution and incorporated into amino acids in the root or shoot for subsequent transport to import-dependent tissues. These “sinks” are the primary consumers of newly assimilated sugars and amino acids, and include many essential organ systems such as expanding leaves, roots, seeds and fruits, storage tissue and organs, and secondary growth. As much as 80% of the C assimilated in mature leaves, and comparable amounts of amino acids, are exported in the phloem to satisfy the metabolic needs of the heterotrophic tissues (Kalttorres et al., 1987). Understanding the mechanisms and regulation of C and N partitioning to these heterotrophic tissues has always been a central theme to improving crop productivity. Moreover, human manipulation of C and N partitioning is evident from the earliest examples of crop domestication. A good example is the selective concentration of carbohydrate and storage proteins in the larger kernels and inflorescences of maize during domestication from ancestral teosinte as much as 10,000 years ago (Doebley, 2004). Likewise, the dramatic yield increases of the green revolution were, among other advances, based on the selection of smaller stature plants that invested more biomass in reproductive tissue versus vegetative growth. Indeed, yield trials of early twentieth century winter wheat versus late twentieth century cultivars grown in a common garden experiment showed that C assimilation per acre was the same for both varieties, yet the yields of the modern cultivars outperformed the early varieties by 40% because more C was directed into reproductive growth (seeds) versus vegetative tissues (Gifford et al., 1984). Yield enhancement of harvested organs continues to be a primary objective, whether the intent is edible organs for feed/food consumption, fiber for textiles and composites, wood for pulp, paper and building, or general biomass for biofuels. Nutritional and other quality enhancements of feed/food crops are also primary objectives that are in part dependent on C and N partitioning.
The last several decades have produced substantial biochemical and molecular advances in our understanding of cell-to-cell and long-distance partitioning of sugars and amino acids in plants. These advances have created novel opportunities to manipulate partitioning pathways to enhance yield and/or nutritional quality. The basic premise of these approaches is that altering the expression of the transporters for a desired sugar or amino acid will result in the preferential accumulation of that compound (or a derivative thereof) in a target organ or tissue. The success of these efforts are predicated on accurate knowledge of the physiological and biochemical processes involved to predict the outcome. The goal of this review is to briefly summarize current understanding of the functional contributions of sugar and amino acid transport systems to plant growth and then focus on the limited number of studies where ectopic expression has been attempted. In many of these, the desired outcome has been at least partially achieved, and it is apparent that identifying the limitation—that is, why the full desired effect was not obtained—is necessary to achieve greater gains. Thus, the application of existing knowledge for biotechnology gains has in many cases identified new avenues of biological discovery in the complex interaction of nutrient use and growth. In addition, this review ventures experimental approaches for improving and targeting biomass partitioning to specific organs.
Carbohydrates: Transport Mechanisms and Efforts to Manipulate Partitioning by Engineering Transport
Sucrose (Suc) is the primary form of reduced C transported long-distance in the vascular system of plants, and consequently has garnered the most attention. Suc is produced in the cytoplasm, either directly from the products of photosynthesis or from storage reserves. Once produced, it may remain in the cytoplasm to participate in other aspects of cellular metabolism or it may be compartmentalized, principally in the vacuole; it may pass through the symplasm—the collective cytoplasm of the plant—via plasmodesmata (PD); or it may undergo efflux to the apoplasm—the collective cell-wall space of the plant—and from there it may be taken up into the cytoplasm of an adjacent cell. As a relatively large polar solute, Suc requires protein transporters for efficient movement across membranes. Where the prevailing concentration gradients mandate energized transport, Suc/H+ symporters (Suc uptake transporters, SUT; or Suc uptake carriers, SUC; the SUT designation is used here, unless referring to a specific gene) that utilize the proton motive force for Suc/H+ symport are well characterized biochemically, genetically, and physiologically (Bush, 1993; Braun and Slewinski, 2009; Ayre, 2011). Those SUTs that are best characterized function at the plasma membrane to pump Suc into cells from the apoplasm, but members of one SUT sub-family localizes to the tonoplast to use the prevailing proton gradient to move Suc from inside the vacuole out to the cytoplasm (Endler et al., 2006; Reinders et al., 2008; Eom et al., 2011). Suc/H+ antiport to load Suc into vacuoles is supported by physiological studies and patch clamping with mutant Arabidopsis lines indicate a role for TMT1 and TMT2 in this process (Schulz et al., 2011). Under conditions where passive transport would suffice, a new family of sugar transporters, SWEETs, were recently identified that have mechanisms characteristic of facilitated diffusion (Chen et al., 2010, 2012; Chen, 2014).
The primary fate of photoassimilated C is long-distance transport from photoautotrophic source leaves to heterotrophic organs. C partitioning via long-distance transport of sugars in the phloem has been extensively and recently reviewed (Turgeon and Wolf, 2009; Ayre, 2011; Braun et al., 2014; Chen, 2014) and is thus only surveyed here. Among mesophyll cells, Suc appears to move cell to cell relatively freely through the PD of the symplasm, and then enters the minor-vein phloem for long-distance transport through the sieve tube elements by bulk flow (Figure 1). Bulk flow occurs when a hydrostatic pressure difference between source and sink tissues is large enough to drive flux through the sieve elements, with the pressure difference primarily energized by solute accumulation in source-leaf phloem. Establishing a sufficient solute concentration in source phloem is generically referred to as phloem loading, and three mechanisms are proposed: (1) In apoplasmic phloem loading, Suc exits the mesophyll symplasm in the vicinity of the phloem via SWEET proteins located principally on the plasma membrane of presumptive phloem parenchyma cells. Suc is then accumulated against a concentration gradient from the apoplasm into the companion cell/sieve element complex of the phloem by SUTs (Figure 2). Because uptake is energized by the proton motive force, Suc can accumulate to high levels such that the total solute concentration in source phloem can readily exceed 1 Osm. Sugar alcohols (polyols) are prominent transport sugars in some species, and appear to be loaded into the phloem from the apoplasm by proton symporters in a mechanism equivalent to Suc loading from the apoplasm (Noiraud et al., 2001; Gao et al., 2003; Reidel et al., 2009). (2) In the polymer trap mechanism, Suc diffuses from the mesophyll symplasm into the phloem through specialized, highly-branched PD, and a portion is then converted to raffinose family oligosaccharides (RFO), primarily the trisaccharide raffinose and tetrasaccharide stachyose, but also, in some species, longer chain oligosaccharides verbascose and ajugose. These highly branched PD, located between bundle sheath and intermediary cells (specialized companion cells), are proposed to have a precise size exclusion limit that allows diffusion of Suc but not larger RFOs (Figure 3). RFO synthesis in the intermediary cells thus maintains a diffusion-friendly Suc concentration while generating the high osmolarities necessary for the hydrostatic pressure that drives long-distance transport through the sieve tubes (Turgeon, 1996). (3) In passive loading, source leaf mesophyll cells accumulate high concentrations of Suc, which enters the phloem passively through regular PD (Figure 4). In this mechanism, there is not an energized step for concentrating solute into the companion cell/sieve element complex, and the high turgor required for bulk flow through the sieve elements is maintained throughout the leaf (Rennie and Turgeon, 2009; Slewinski and Braun, 2010). It should be noted that some species can employ multiple phloem loading pathways in the same vascular bundle (Voitsekhovskaja et al., 2009) and that different mechanisms can be favored depending on the plant’s specific needs (Gil et al., 2011).
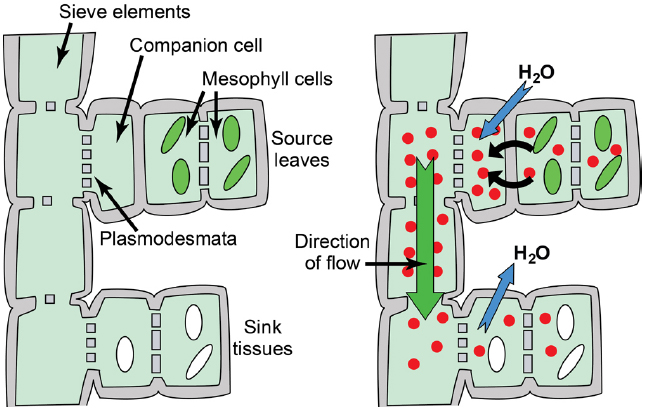
Figure 1. Phloem anatomy and osmotically generated-pressure flow. Left: Schematic of phloem connecting source leaves (represented as mesophyll cells with green chloroplasts) and heterotrophic sink tissues (represented as cells with white amyloplasts). Sieve elements are connected end to end to form sieve tubes, and companion cells are connected to sieve elements via plasmodesmata/pore units. The extent to which companion cells are connected to surrounding mesophyll/parenchyma cells varies substantially between loading strategies. Right: Hydrostatic pressure is established in source leaves by phloem loading—the accumulation of solute (red circles; primarily photoassimilate, but also amino acids, potassium, and others solutes) in the phloem—and the resulting osmosis of water. An exception is passive loading, in which solute and hydrostatic pressure is high throughout the source leaf (see Figure 4). Hydrostatic pressure drops in sink tissues as solute is used for growth and metabolism, and water dissipates. Bulk flow of water and nutrients through the sieve tubes follows the hydrostatic pressure gradient from source to sink.
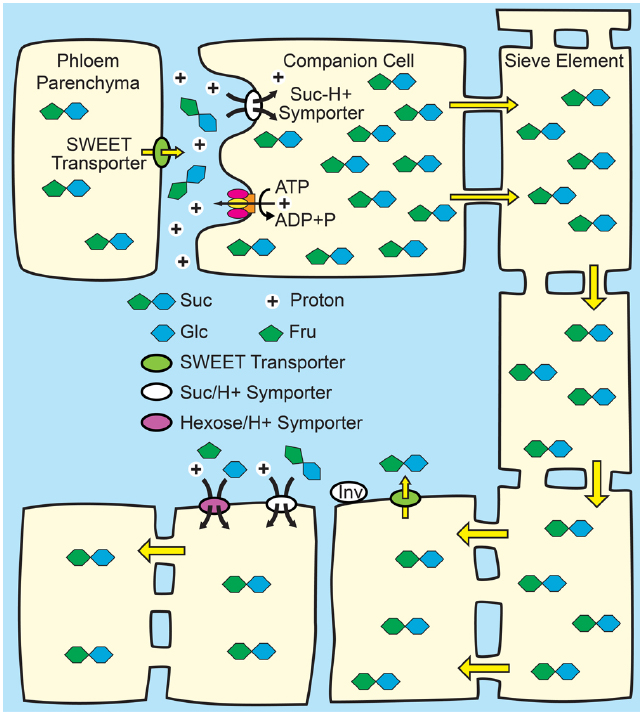
Figure 2. Phloem loading from the apoplasm. Top: Suc from mesophyll cells—principally phloem parenchyma cells—enters the apoplasm in the vicinity of the companion cell/sieve element complex via SWEET transporters. Suc, and in some species, polyols, is then loaded into the companion cell/sieve element complex up a thermodynamically unfavorable concentration gradient by Suc/H+ symporters (SUTs) energized by the proton motive force. The proton motive force is generated by the hydrolysis of ATP at the plasma membrane by proton pumping ATPases. Osmosis generates hydrostatic pressure that pushes the phloem sap toward sink tissues. Bottom: In sink tissues, solute is unloaded from the sieve elements. Post-phloem transport may be through the symplasm via plasmodesmata, or solute may efflux to the apoplasm for subsequent transmembrane transport in adjacent cells. Suc in the apoplasm of sink tissues may be taken up by SUTs, or maybe hydrolyzed to Glc and Fru by cell wall invertases (INV) and taken up by hexose/H+.
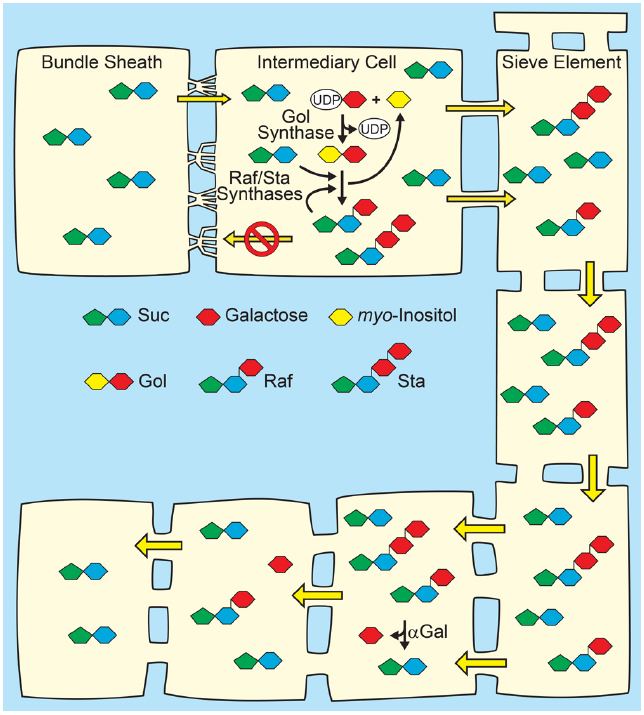
Figure 3. Polymer-trap model for loading through the symplasm. Top: Suc from mesophyll diffuses into intermediary cells (specialized companion cells) through highly branched plasmodesmata situated between the intermediary cells and bundle sheath cells, where a portion is converted to raffinose family oligosaccharides (RFO), raffinose (Raf) and stachyose (Sta), by the sequential action of Raf synthase and Sta synthase. The galactosyl donor for RFO synthesis is galactinol (Gol) created from UDP-galactose and myo-inositol by Gol synthase. The RFOs appear to be too large to diffuse back through the plasmodesmata into the mesophyll. Conversion of Suc to RFO favors the continued passive entry of Suc while RFO accumulation generates hydrostatic pressure by osmosis. Bottom: In sink organs, RFOs move through the post-phloem symplasm. Hydrolysis by alkaline α-galactosidases (αGal) and other enzymes convert RFOs to Suc and Glc, which can also partition via the apoplasm (not shown).
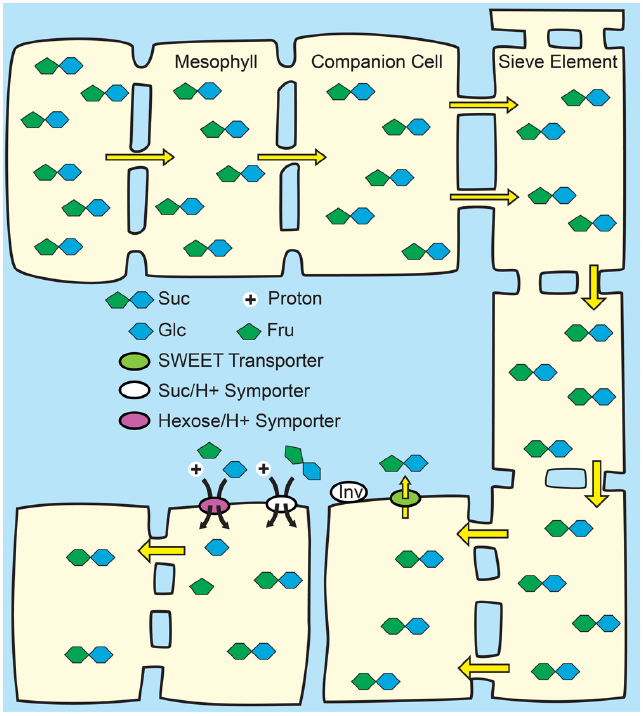
Figure 4. Passive loading through the symplasm. Top: Source leaves of species that use passive loading have regular plasmodesmata connections from the mesophyll cells through to the sieve elements and have high solute concentrations throughout the leaf. Consequently, solute is able to move relatively freely, and all cells have equally high hydrostatic pressures: there is not an energized concentrating step at the mesophyll/phloem interface. Bottom: Lower hydrostatic pressures in sink organs permit bulk flow through the sieve elements. Suc is unloaded from the sieve elements in sink organs through symplasmic or apoplasmic routes, as required by the specific tissues.
Phloem unloading occurs all along the transport path to nourish lateral tissues and storage reserves, and occurs extensively in the sink organs. Unloading may be by efflux across plasma membranes to the apoplasm, or through the symplasm via PD, or both routes may be used in combination. Within the recipient tissues, symplasmic continuity with the CC/SE complex can be extensive for efficient post-phloem partitioning via PD, or isolated domains may force apoplasmic transport (Patrick, 1997). Generalizing, growing vegetative tissues, such as young leaves and root tips, tend to have extensive symplasmic unloading (Oparka et al., 1999; Stadler et al., 2005b). This contrasts with developing seeds, where symplasmic isolation between maternal and filial tissues forces apoplasmic unloading: short distance symplasmic unloading occurs from the phloem strands entering the integument/testa tissue, from where nutrients enter the apoplasm for subsequent uptake into the embryo and endosperm (Wang and Fisher, 1994; Patrick and Offler, 2001; Stadler et al., 2005a). In addition, the principal route of unloading, and the extent of the symplasmic and apoplasmic domains in the post-phloem pathway, may change during organ development, as exemplified in maturing fruit and developing embryos (Ruan and Patrick, 1995; Ruan et al., 2001; Kim et al., 2005). Once released to the apoplasm, Suc may be retrieved back into the phloem or into adjacent cells by SUTs, or may be hydrolyzed by cell wall invertases, and recovered by hexose/H+ transporters.
The complexity of the loading and unloading pathways involved in C partitioning, and their importance to plant physiology, are in part reflected in the size of the gene families involved. Arabidopsis has nine SUTs arranged in three sub-families, and monocots have an additional one or two families not represented in dicots (Sauer, 2007; Braun and Slewinski, 2009; Reinders et al., 2012). The monosaccharide transporter-like (MST) gene family of Arabidopsis has 53 members, arranged in seven sub-families. The best characterized of these sub-families, and the one with members that localize to the plasma membrane and are thus most likely involved in intercellular transport, is the sugar transporter (STP) family, which contains 14 members (Büttner, 2007, 2010). The SWEET family of mono- and disaccharide transporters contains 17 genes in Arabidopsis and 21 in rice, arranged in four clades, with clade I mediating mainly Glc import and export, and clade III transporting mainly Suc (Chen et al., 2010, 2012). Plants have three types of invertases, localizing to the cell wall/apoplasm, cytoplasm, and vacuole, respectively, with six members in the cell wall invertase subfamily of Arabidopsis (Roitsch and Gonzalez, 2004). This list only includes the major candidate participants in membrane-mediated C partitioning, and not those involved in polymer synthesis or PD function.
Engineering Partitioning by Manipulating Apoplasmic Loading
Strategic manipulation of C partitioning may lead to more productive plants and enable specific organs to be targeted for enhanced yield. Ectopic expression of SUTs from constitutive or tissue-specific promoters, in either sink or source organs, has been attempted in various plants. These studies demonstrate a clear potential, but also emphasize that the natural patterns of Suc flux need to be considered. In potato, constitutive over-expression of spinach SoSUT1 from the CaMV 35S promoter was used in an effort to increase transport to tubers (Leggewie et al., 2003). This gave higher levels of starch and lower levels of sugar in leaves, but had little impact on tubers yields. These effects were likely caused by futile cycling, in which Suc released to the apoplasm was recovered by mesophyll cells because the promoter drove expression in the mesophyll as well as in phloem. Ectopic expression of potato StSUT1 in excised pea cotyledon storage parenchyma enhanced Suc influx in a StSUT1-dependent fashion (roughly twofold). However, Suc uptake into intact cotyledons was only ∼23%, and when biomass accumulation was measured, a similar difference between excised and intact cotyledons was observed (Rosche et al., 2002). This study showed that ectopic SUT expression can be used for targeted yield enhancements, but the authors also argue that since most Suc is absorbed at the cotyledon surface by epidermal transfer cells, the full effect of overexpressing StSUT1 in storage parenchyma was not realized in intact seedlings since the “extra” transporters were not on the transfer cells exposed to apoplasmic Suc (Rosche et al., 2002). As another example of manipulating SUT activity for biotechnology, overexpression of a barley SUT (HvSUT1) using an endosperm-specific Hordein B1 promoter in wheat grains increased levels of storage protein, showing that enhanced Suc transport has positive impacts beyond carbohydrate alone (Weichert et al., 2010). Finally, over-expression of rice OsSUT5Z in potato using a tuber-specific, class-I patatin promoter was reported to enhance tuber yield by increasing tuber numbers rather than tuber size. This was not the anticipated outcome, and the authors suggest that altering Suc flux stimulated development of more stolons (Sun et al., 2011).
The above studies emphasize constitutive or sink-specific manipulation, and attempt to “pull” more Suc into the target tissue. Enhancing phloem transport by manipulating SUTs involved in phloem loading has been put forward as a “push” mechanism to improve plant productivity. For example, increasing source to sink transport was proposed as a means to enhance crop productivity since more carbohydrate would be sent to sink organs for growth and/or storage, and there would be less Suc-mediated product inhibition on photosynthesis (Ainsworth and Bush, 2011). In addition, Suc loading is modulated, both up and down, in response to the physiological and environmental needs (Vaughn et al., 2002), and it was proposed that heterologous promoters that are uncoupled from the natural regulation may be useful to keep loading rates constantly high (Srivastava et al., 2009).
Despite this, there are few published attempts to enhance phloem transport by over-expressing SUTs in the phloem. The cDNA of Arabidopsis AtSUC1 (Wippel and Sauer, 2012) and barley HvSUT1 (Reinders et al., 2012) were cloned downstream of the companion cell-specific AtSUC2 promoter, and both restored phloem loading in Arabidopsis Atsuc2 –/– mutants. These studies showed foreign transporters could restore WT growth to the mutant, but detailed analysis of C transport and impact on growth was not pursued. In a different approach, AtSUC2 cDNA was expressed from a companion cell-specific promoter derived from Commelina yellow mottle virus (CoYMVp) in a homozygous Atsuc2-4 background (Srivastava et al., 2009). CoYMVp is as strong as the natural AtSUC2 promoter, but is subject to different regulatory cascades: while SUT promoters naturally involved in phloem loading are repressed by Suc accumulation (Vaughn et al., 2002; Usadel et al., 2008), CoYMVp is activated by Suc (Dasgupta et al., 2014). This over-expression construct rescued Atsuc2-4 mutants, showing that foreign promoters can maintain high levels of SUT activity in conditions where expression might normally be repressed (Srivastava et al., 2009).
A more recent study fused CoYMVp to alternative SUTs, with the aim of keeping phloem loading levels high by uncoupling the natural transcriptional control via the foreign promoter and potential post-translational control with different proteins. Of the SUTs tested, AtSUC1, AtSUC2, and ZmSUT1 (a Zea mays gene from a different SUT subfamily) rescued phloem loading in the Atsuc2-4 mutant; several other SUT genes implicated in high affinity Suc uptake were unable to restore efficient phloem transport (Dasgupta et al., 2014). When SUTs that did rescue Atsuc2-4 were overexpressed in WT plants, enhanced phloem loading and transport to heterotrophic organs was evident, but improved growth and primary productivity was not observed. Rather, the plants were stunted. Separate research suggested a link between sugar transport and phosphate requirements (Lei et al., 2011), and this was tested in lines overexpressing SUTs from CoYMVp. The growth inhibition was accompanied by increased expression of phosphate-starvation induced genes, and was reversed by providing a higher supply of external phosphate (Dasgupta et al., 2014). These findings argue that “hyperloading” the phloem by overexpressing SUTs have the detrimental effect of disrupting C/phosphate homeostasis. The implications for agriculture are that efforts to enhance photosynthesis and growth by increasing phloem transport may be imperiled by the plant’s perception that it needs more P, unless the links between C and P homeostasis are better understood and uncoupled.
What future experiments could be done to enhance partitioning via apoplasmic loading? Molecular characterization of SWEET and SUT transporters provides, in principle, a complete framework to test hypotheses on the strategic release of Suc from one cell and loading into the adjacent cell. An obvious experiment is to overexpress SWEETs in phloem parenchyma cells, particularly Arabidopsis SWEET11 and SWEET12 (Chen et al., 2012), for enhanced Suc efflux to the apoplasm with simultaneous SUT overexpression in companion cells for increased uptake, and then test the impact on growth and whole plant C partitioning. Precise cell-specific deployment will be critical for accurate interpretation of the results, and a caveat to the success of these experiments is the resolution with which the outcome is measured (see below). Simply expecting larger plants in general, or larger target organs, is naïve, even though this may be the desired outcome from an applied standpoint. Instead, these experiments will more likely help identify rate-limiting steps in the overall process of whole-plant partitioning, and thus lead to new prospects (targets) for manipulation, and uncover unrecognized interactions of C homeostasis and other aspects of physiology. A corollary to these experiments aiming to enhance productivity are those that disrupt partitioning to test prevailing and alternative models of C allocation.
Manipulation of source-leaf phloem loading would energize the entire pathway, but by itself does not target resources to desired (i.e., harvested) organs. Targeted sink-specific manipulations may therefore be more fruitful for enhancing yield of specific organs, as evident from overexpression of StSUT1 in storage parenchyma of pea cotyledons and HvSUT1 in wheat endosperm, described above. But here too, the complete pathway of Suc unloading, from the sieve elements to the final recipient cells need to be considered (Patrick, 1997). For example, developing embryos and endosperm are symplasmically isolated from maternal tissues, and all nutrient transfer has two obligate membrane transport steps: efflux from maternal cells and influx into filial cells (Zhou et al., 2009). Tissue-specific SWEET over expression in integuments/testa of developing seeds may enhance sugar availability at the interface between maternal and filial tissues, but optimally positioned uptake carriers may be required to take advantage of the available resources.
Similarly, Suc unloading to the apoplasm in sink organs is associated with hydrolysis by cell wall invertase and uptake of the resulting hexose by hexose transporters. This is particularly prevalent in fruits undergoing rapid expansion and ripening (Ruan and Patrick, 1995; Patrick, 1997; Zhang et al., 2006, 2007; Jin et al., 2009). As the fruit ripens and accumulates more solutes (simple sugars, etc.), apoplasmic unloading prevents symplasmic “backflow.” In addition, hydrolysis of Suc to Glc and Fru doubles the osmolarity to benefit hydrostatic expansion; enhances the Suc gradient between symplasm and apoplasm to favor further passive unloading; and also prevents the retrieval of unloaded Suc back into the phloem by SUTs, which are expressed all along the phloem path.
Although the potential for manipulating the “pull” capacity of sinks is clear, little is known on how manipulation of hexose transporters could be advantageous to increase plant yield. Over-expression of sugar transporter AtSTP13 in Arabidopsis seedlings led to increases in Glc uptake, and also led to increases in the sugars and biomass throughout the plants (Schofield et al., 2009). In tomato, RNAi-mediated knockdown of the high affinity hexose transporter gene LeHT led to a 55% decrease in fruit hexose accumulation, implicating LeHT in driving accumulation of hexoses into storage parenchyma cells during tomato fruit ripening (McCurdy et al., 2010). Among those hexose transporters that have been characterized genetically, many of the knock out mutants have no visible phenotype. The expression patterns of Arabidopsis STP genes, as determined by classical hybridization and PCR approaches, as well as analysis of genome-wide transcriptome data, overwhelmingly place STP expression in various sink organs (Büttner, 2010) and this speaks strongly to their potential for targeting biomass to specific sinks by transgenic approaches. Genetic manipulation of additional sugar transporter genes using constitutive and tissue-specific promoters is needed to test this potential. Since manipulating one aspect of the transport system may have limited impact, coupling the targeted expression of genes encoding SWEETs, invertases, and hexose transporters may be more fruitful to capture the full potential of this strategy.
However, manipulating the transporters involved in apoplasmic phloem loading and unloading carries the inherent risk of increasing the apoplasmic concentrations of Suc and other nutrients used by pathogens. Pathogens have been shown to enhance SWEET expression to make reduced C more available (Chen et al., 2010, 2012; Chen, 2014). Several lines of genetic and metabolic evidence implicate sugar transporters in plant defense responses during plant-pathogen interactions. This includes reprogramming carbohydrate metabolism at the site of infection for reduced Suc export, increased Suc hydrolysis by cell wall invertase, and enhanced import of hexose by hexose transporters (Proels and Hückelhoven, 2014).
Engineering Polymer Trapping Metabolism
Other efforts to enhance phloem loading and long-distant transport have relied on the fact that there are different phloem loading mechanisms, and have attempted to superimpose an alternative mechanism on top of the natural mechanism of the host plant. Specifically, polymer trapping biochemistry was introduced to two species, potato (Hannah et al., 2006) and Arabidopsis (Cao et al., 2013), both of which load from the apoplasm with SUTs. The aims of these experiments were twofold: (1) to assess the efficiency of RFO synthesis in the phloem and (2) to gage the efficiency with which RFO from the companion cells enters the translocation stream for long-distance transport. In addition, growth and development of the engineered plants were monitored. In both studies metabolic engineering was used to produce galactinol and raffinose in companion cells. In Arabidopsis, stachyose was also engineered. Both studies used companion cell-specific promoters to express genes encoding galactinol synthase and raffinose synthase, and in Arabidopsis, stachyose synthase. Thus, all the RFO biosynthesis occurred in the phloem after the SUT-mediated phloem loading step. Despite the high concentrations of Suc available in these cells, RFO synthesis and transport in both studies was low. When Arabidopsis was photosynthetically labeled with 14CO2 for 20 min followed by a 10 min chase period, only 2% of the label was incorporated into RFO, while Suc contained up to 80% and up to 30% was in Glc and Fru. This outcome is surprising and in sharp contrast to plants that transport RFO naturally, such as coleus (Turgeon and Gowan, 1992), cucurbits (Beebe and Turgeon, 1992), and catalpa (Otto, 1968; Turgeon and Medville, 2004), in which RFOs become quickly labeled to high specific activity.
Based on current models, this result is puzzling, and neither study had a satisfying explanation for the low rates of RFO synthesis other than to suggest that RFO biochemistry, especially in relation to phloem transport, was not straightforward. RFO synthesis should be efficient since apoplasmic loaders have ample reduced C in the companion-cell cytoplasm and the engineered proteins are thought to localize to the cytoplasm (Keller and Pharr, 1996); transport should be efficient because the PD-pore units between companion cells and sieve elements are open to diffusion of 10 kDa dextrans (Kempers and Van Bel, 1997; Knoblauch and Van Bel, 1998) and 67 kDa proteins (Stadler et al., 2005b). Further work on the biochemistry in companion cells of apoplasmic loaders and intermediary cells of polymer trap loaders is required to resolve why apoplasmic loaders do not effectively produce and transport RFOs. The precursors for galactinol, UDP-Gal and myo-inositol, or flux through the pathways leading to these, may be insufficient for higher level production.
Alternatively, the inability to produce high levels of RFO in companion cells may be a cell biology problem rather than a biochemical problem. In addition to many highly branched PD, intermediary cells of plants employing the polymer trap mechanism have many small vacuoles and extensive endomembrane systems, and are larger than companion cells found in similarly-sized leaf veins of plants that load from the apoplasm (Turgeon et al., 1993, 2001). A potential function for the extensive internal membranes and vacuoles in intermediary cells has not been put forward, but low rates of RFO synthesis in companion cells suggests a role for these membranes in RFO synthesis. Enzyme localization, stability, and/or interaction with cellular co-factors in companion cells relative to intermediary cells could therefore be a problem. It is worth noting that true intermediary cells and polymer trapping appear to have evolved independently multiple times, arguing that the internal structures are essential for function, and not species-specific characteristics (Turgeon et al., 2001).
If RFO synthesis in companion cells could be engineered to levels that contribute to hydrostatic pressure in the source leaf, then efficient RFO metabolism/utilization could be engineered in specific sinks for targeted partitioning (Figure 5). Expression of an alkaline α-galactosidase gene specifically in desired sink organs (Carmi et al., 2003), for example, would convert transported RFOs to Suc and galactose, which could then be converted to Glc (Barber et al., 2006; Dai et al., 2006) to enter “regular” pathways of primary metabolism. Since RFOs are not abundant transport sugars in apoplasmic loading species, competing, off-target sinks not engineered for efficient RFO catabolism may accumulate the RFO sugars (Figure 5). The principle here is to alter the hydrostatic pressure gradients that drive phloem transport throughout the plant: RFO utilization in target organs will result in low hydrostatic pressures and efficient nutrient import, whereas RFO accumulation in competing sinks that cannot metabolize the RFOs will have higher hydrostatic pressure that will decrease bulk flow to that tissue. With the strength of competing sinks reduced, more nutrients may be available for biomass accumulation in target sinks.
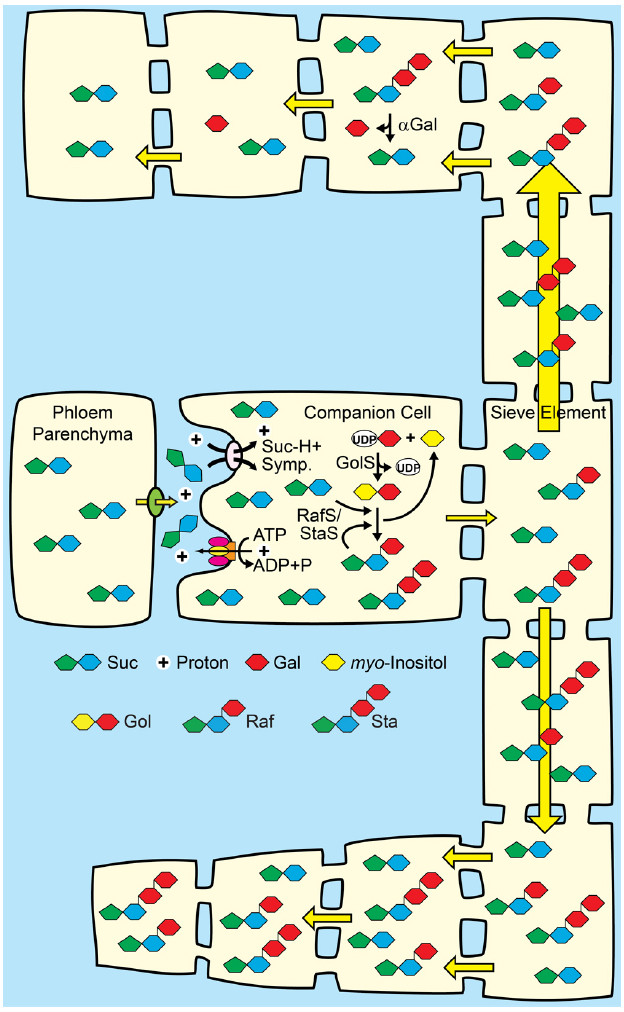
Figure 5. A proposal for combining apoplasmic loading with polymer trap biochemistry and sink-specific digestion of RFO sugars. Middle: Companion cells of source leaves loading from the apoplasm with Suc/H+ symporters are engineered to convert a portion of Suc to raffinose family oligosaccharides (RFO). See Figures 2 and 3 for details. Top: Target sinks engineered to efficiently catabolize RFOs using tissue-specific expression of genes encoding alkaline α-galactosidases (αGal) and galactose metabolizing enzymes. The hydrostatic pressure drops as the engineered sugars are catabolized. Bottom: Off-target sinks that are not engineered to catabolize RFOs accumulate the RFO and have a relatively high hydrostatic pressure. Direction of transport is based on hydrostatic pressure gradients, and engineered sinks are predicted to receive more sap (symbolized by relative arrow size). Engineered sinks are also predicted to compete better for resources and show better growth (symbolized by relative cell size).
As a corollary to superimposing polymer trap chemistry on apolasmic loaders, superimposing apoplasmic loading onto intermediary cells may enhance partitioning in species that load by the polymer trap mechanism. Species that phloem load by polymer trapping do not have an obligate need for SUTs or Suc in the apoplasm (McCaskill and Turgeon, 2007). Notwithstanding, Suc is present in the apoplasm and many species with intermediary cells also have “regular” companion cells that express genes encoding SUTs (McCaskill and Turgeon, 2007; Voitsekhovskaja et al., 2009). Cucurbits, and probably other polymer trap species, appear to be able to switch between apoplasmic and polymer trap loading under certain conditions, particularly virus infection (Gil et al., 2011). Engineering SWEET expression in phloem parenchyma and SUT expression in the intermediary cells may increase Suc availability for RFO synthesis and consequently improve phloem loading and transport. It would also test predictions of the polymer trap model. Polymer trapping proposes that the branched PD allow Suc to diffuse down its concentration gradient. If this is correct, it follows that Suc brought into intermediary cells by SUT mediated loading should be able to diffuse back out. If on the other hand, Suc accumulates in the intermediary cells, it would speak against the polymer trap model.
Engineering Passive Loading
Passive loading species are proposed to have the highest solute concentration in the mesophyll cells, and have PD with relatively large size-exclusion limits from the mesophyll all the way to the sieve tubes. Sugars and other nutrients are thus free to move along concentration gradients from the mesophyll into the sieve elements (Turgeon and Medville, 1998; Rennie and Turgeon, 2009). There is no energized concentrating step, and thus, speaking literally, there is no actual “loading” step. If this prevailing model is correct, specifically that PD with large size-exclusion limits connect the mesophyll and phloem cells to facilitate Suc movement without an apolasmic step nor a polymer trapping step, then it predicts that neither engineering a SWEET/SUT pairing to promote apoplasmic transport, nor polymer engineering to trap sugars in the phloem would improve loading and transport: The model predicts that any efforts to accumulate solute in the phloem would be thwarted by escape through the PD back to the mesophyll. Ironically, testing the passive-loading premise is exactly why these experiments should be conducted: If solutes do accumulate in the phloem after these manipulations, it would argue that the model is incorrect.
Nitrogen: Transport Mechanisms and Efforts to Manipulate Partitioning by Engineering Transport
Although nitrate and ammonia are the primary forms of N acquired from the soil solution by plants, “amino acids are the currency of N exchange” in the physiology of plant growth (Bush, 1999). Inorganic N taken up from the soil is either assimilated into amino acids in the root, or they are transported to photosynthetically active leaf tissue in the xylem via the transpiration stream. Amino acids synthesized from inorganic N in the root are used for basic metabolic needs of root cells, but a substantial amount of those amino acids are transported into the xylem where they make their way to the photosynthetically active leaves in the transpiration stream. In the leaf, amino acids are used for basic metabolism and protein synthesis. However, as generally mature organs, these leaves do not need a large percentage of those amino acids for metabolism. Thus, amino acids arriving from the roots are transported into the leaf phloem where they move to the many heterotrophic tissues of the plant, that include young leaves just building the photosynthetic machinery, roots, stem tissues, storage organs, and developing seeds. When inorganic N from the soil moves directly to the leaves in the xylem, it is assimilated into amino acids in the mesophyll and then, much like the amino acids arriving from the root, they are transported into the phloem for systemic distribution to import-dependent sinks. Taken together, it is clear that amino acids are the primary form of transported N supporting the growth and development of multicellular plants (Bush, 1999).
The systemic distribution of amino acids requires the combined activity of many amino acid transport proteins. Example transport steps include export from xylem parenchyma in the root into the xylem transpiration stream, transport into mesophyll cells, export from mesophyll cells, transport into and out of the phloem, and transport into the heterotrophic cells of all the import-dependent sinks. Remarkable advances in our understanding of the transport properties and identity of plant amino acid transporters in the past 25 years was initially enabled by detailed biochemical descriptions using purified plasma membrane vesicles (Bush, 1993) followed by the successful cloning of a wide array of amino acid transporter gene families (Fischer et al., 1998; Ortiz-Lopez et al., 2000; Tegeder, 2012). To date, at least six families of amino acid transporters have been identified in plants with more than 60 genes encoding putative amino acid transporters. These transporters are functionally differentiated by their transport properties (such as substrate specificity and transport mechanism) and expression patterns that are regulated by both developmental and environmental cues (Liu and Bush, 2006; Tegeder, 2012, 2014). The majority of the transporters described to date are involved with active uptake into the cell, and only two reports describe bidirectional transporters (BAT1 and SIAR1) that may be involved in amino acid export (Dundar and Bush, 2009; Ladwig et al., 2012). BAT1’s putative role in amino acid export from the cell is based on its activity as a facilitated carrier and on a vascular tissue-localized expression pattern of a GUS expressing gene-trap inserted in the BAT1 gene (Dundar and Bush, 2009). However, subsequent localization of the BAT1 protein using a BAT1::GFP reporter localized the carrier to the mitochondria, thereby suggesting a primary role in intracellular amino acid metabolism (Michaeli et al., 2011). In contrast, SIAR1 was localized to the plasma membrane of vascular tissue, thus supporting its role in amino acid export (Ladwig et al., 2012).
Given the complexity of amino acid circulation in the plant, a reasonable hypothesis suggests that altering the pattern and/or timing of the expression of one or more transporters could have a positive impact on plant growth and nutritional quality. For example, Rolletschek et al. (2005) documented a 20% increase in seed N content and seed size as a result of the ectopic expression of the Vicia faba VfAAP1 amino acid transporter gene in the developing seeds of pea and Vicia narbonensis using the seed specific legumin B4 promoter. These results suggest that amino acid transport into the storage parenchyma of the developing seed is rate limiting. Interestingly, there was also an overall increase in plant biomass, suggesting targeted changes in N allocation in one tissue can have a broader impact on plant growth. In a subsequent report, the transgenic peas were grown in field trials over two seasons (Weigelt et al., 2008). Seed N content was increased as before, but there were compensatory decreases in seed starch content and seed size. Transcript and metabolite profiling indicated the transgenic seeds were experiencing C limitations as more amino acids were being synthesized. Several pathways associated with coordinated C and N metabolism localized to the mitochondria were up-regulated, suggesting metabolic adjustments in response to a changing N:C ratio (Weigelt et al., 2008).
Tegeder and colleagues took a different approach in attempting to increase the sulfur content of legume seeds (Tan et al., 2010). In these experiments, the yeast S-methylmethionine permease 1 gene (MMP1) was expressed in transgenic pea under the control of the Arabidopsis AAP1 promoter which directs expression throughout the phloem and in the seeds. S-methylmethionine (SMM) transport was targeted because it is identified as a major form of phloem mobile S in plants (Bourgis et al., 1999). MMP1 expressing plants averaged a 33% increase in biomass, a 19% increase in seed number, a 31% increase in total seed N and a 19% increase in total seed S. There was also an average 67% increase in xylem and 60% increase in leaf SMM. Individual seed S content was unaltered. Analysis of the expression levels of genes involved in SMM biosynthesis suggested increased SMM synthesis in the root. That suggests SMM is transported to the leaf in the xylem, which was supported by the observed increased SMM xylem content. The increase in seed N content was driven by a 33% increase in phloem amino acid content. The surprising result in this experiment is the impact of altered SMM transport on N metabolism and yield. While S content per seed was unaltered, seed N content was increased, suggesting SMM abundance may have a regulatory impact on plant N metabolism. Indeed, SMM represents only 0.2% of xylem and phloem amino acids, suggesting it could be playing a role as a metabolic signal reporting on the S:N ratio in the plant. It is particularly interesting that two experiments that manipulate amino acid distribution in the plant (Weigelt et al., 2008; Tan et al., 2010) have had significant impacts on metabolic pathways that are associated with balancing the relative abundance of three important essential elements (N, C, and S). These observations are perhaps not surprising given the complex interactions between the functional roles of these central nutrients in plant metabolism.
Other approaches for manipulating N distribution in the plant might include ectopic expression of a desired amino acid transporter in the leaf phloem using a promoter that drives companion cell/sieve element specific expression. Under the expectation that anything loaded in the phloem moves with mass flow to actively growing sinks, this could increase the N content of harvested organs, such as mature seeds. Ectopic expression in the leaf phloem might be coupled to increased expression of a bidirectional transporter in the mesophyll that could increase the efflux of the desired amino acid into the apoplast. Likewise, one could simultaneously use a transgenic approach to increase the biosynthesis of a desired amino acid in the mesophyll, thereby increasing the pool size available for export. These approaches focus on the “push” side of the allocation pathway by attempting to increase the flux of an amino acid(s) to the sink tissue of the plant. Alternatively, one can increase the “pull” side of allocation by enhancing the uptake capacity of a desired sink, as Rolletschek et al. (2005) did with the ectopic expression of the VfAAP1 gene.
In a combined strategy, the Tegeder lab (Zhang et al., 2014) has applied both a push and pull approach by ectopically expressing the pea PsAAP1 gene in transgenic pea using the Arabidopsis AtAAP1 promoter. In pea, the Arabidopsis promoter drove expression in the leaf companion cell/sieve element complex as well as the epidermal cells of the seed cotyledons, thereby generating enhanced amino acid transport capacity in both the source and sink tissues (Figure 6). The transgenic peas had an average 24% increase in biomass, compared to controls, a 228% increase in leaf phloem amino acids, a 35% increase in seed yield and a 6% increase in seed N. As previously observed, the activity of the transgenes had a pleotropic effect on overall plant N metabolism with greater N acquisition and significant increases in both leaf and root N content.
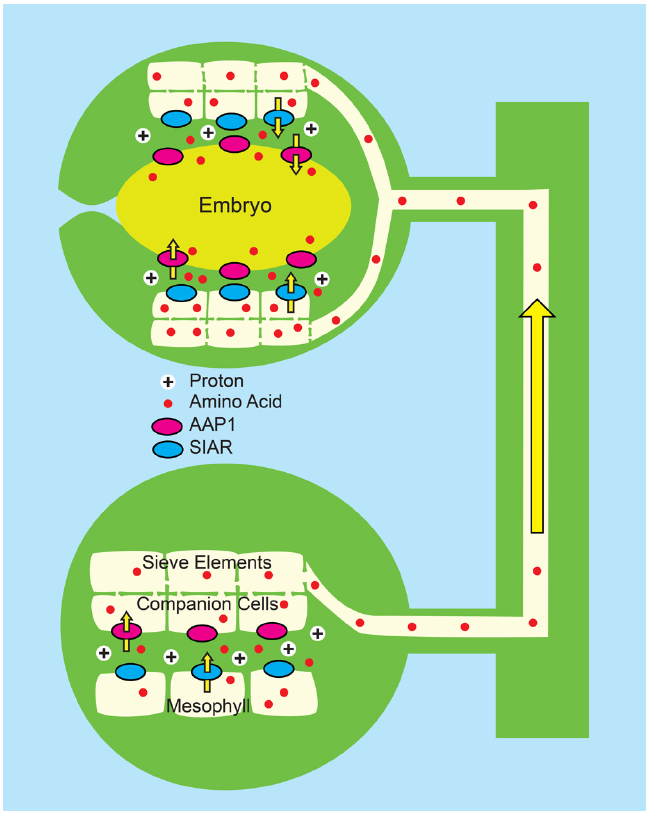
Figure 6. Combined “push” and “pull” strategy for enhancing N transport to developing pea seedling (Zhang et al., 2014). Pea plants (Pisum sativum) were engineered to overexpress PsAAP1 (encoding Amino Acid Permease 1, magenta ovals) using the Arabidopsis AtAAP1 promoter. In pea, the AtAAP1 promoter promotes expression in the companion cell/sieve element complex of leaves, and in epidermal transfer cells of seed cotyledons. Bottom: In source leaves, amino acids (red circles) enter the apoplasm, possibly via the bidirectional transporter encoded by SIAR1 (blue ovals). Uptake into the companion cell/sieve element complex is by endogenous PsAAP1, and is enhanced by PsAAP1 encoded from the transgene. Top: In the testa of developing seeds, amino acids unload from the sieve elements and the post-phloem symplasm most likely via SIAR1 encoded transporters. Uptake into epidermal transfer cells of the cotyledons is by endogenous PsAAP1, and is enhanced by PsAAP1 encoded from the transgene.
Transgenic manipulation of amino acid transport in plants has a real potential to increase the nutritional value of harvested tissue by increasing the overall N content and/or by targeted increases of specific amino acids, such as cysteine and methionine. However, it is clear from early transgenic experiments that altering amino acid distribution in the plant has a major impact on both N and C metabolism, which will require additional research to better understand the complex interactions between these macronutrients.
Concluding Remarks
From the few published reports on efforts to manipulate C and N partitioning, it is evident that there is potential for agricultural improvements but also unexpected pleiotropic effects which limit the desired gains, and in some cases, have unpredicted effects on plant vigor. Of particular interest from both an applied and a basic science perspective are those reporting an influence on another nutrient, such as HvSUT1 overexpression in endosperm increasing storage protein content (and consequently more N; Weichert et al., 2010); and conversely, VfAAP1 expression in seeds increasing seed biomass (and consequently more C; Weigelt et al., 2008); SUT overexpression in companion cells impacting P requirements (Dasgupta et al., 2014); and MMP1-mediated transport of SMM in phloem enhancing seed N content (Tan et al., 2010). These studies point to the complex cross talk between nutrients in specific organs and throughout the entire plant. Also in relation to nutrient cross talk, it would be very interesting to combine manipulated C, N and other nutrient transport systems to look for synergistic effects.
Studying the phloem, phloem transport, and partitioning of nutrients directly is notoriously difficult (Turgeon and Wolf, 2009), and this limitation hampers interpretation of overexpression studies. Analysis of gross morphology (organ size, etc.) is an indirect metric. However, labeling experiments to monitor nutrient transport pathways, either directly with radiolabeled or stable isotopes, or indirectly with dyes or fluorescent proteins, have been, and continue to be, informative and essential. Key technological breakthroughs would be those that allow high-resolution analysis of C and N metabolite concentrations in intact tissues, preferably in real time. Nuclear magnetic resonance imaging has been applied to whole plants, mostly in the context of water relations, but has limited resolution and utility for low-concentration metabolites (Van As et al., 2009). Fluorescence-activated cell sorting and laser capture microdissection allow cell-specific analysis of transcripts and proteins, but have limited utility for small metabolites because of their high solubility and rapid turnover. Imaging mass spectrometry using matrix-assisted laser desorption/ionization (MALDI-IMS) is an emerging technology for spatial metabolomics in tissue sections (Zaima et al., 2010; Horn et al., 2012; Horn and Chapman, 2014). MALDI-IMS resolution is currently at the multiple-cell level and could, in principle, resolve differences across loading/unloading zones—for example, between integuments and cotyledons of the embryo—but the tissue needs to be suitably fixed to overcome the problems of diffusion and metabolism. FRET (Förster resonance energy transfer) nanosensors are engineered proteins that change their fluorescence emission wavelength when a specific metabolite is bound, and have been used to monitor changes in Glc and Suc levels in living plant tissue in real time (Deuschle et al., 2006; Chaudhuri et al., 2008; Chen et al., 2012). Nanosensors also exist for various amino acids (Okumoto et al., 2005; Gruenwald et al., 2012; Zhang et al., 2013). Their continued improvement and deployment to plant apoplasmic and symplasmic compartments in plant tissues may help improve our ability to monitor metabolite levels after manipulating partitioning systems.
Although advances in our understanding of C and N partitioning systems allow us to test hypotheses by manipulating transporter streams from the “push,” “pull” and combined sides, it is also clear that our current understanding is limited when looking at the bigger picture of source/sink relationships. Moreover, complex metabolic interactions between assimilation and utilization of essential nutrients confounds attempts to engineer C and N partitioning. A systems biology approach that compares whole plant nutrient partitioning in high yielding domesticated plants and less productive ancestral varieties may shed light on the labyrinth of interconnected networks that were modified during crop domestication and improvement. Nested association mapping populations, genome wide association studies, and whole-genome re-sequencing are some of the tools that could identify genetic changes in key genes and quantitative trait loci (Meyer and Purugganan, 2013; Olsen and Wendel, 2013; Wallace et al., 2014). In addition, coordinated “omic” approaches could identify key changes in transcripts, proteins, and metabolites. In spite of the many advances in our understanding of the pathways of assimilate partitioning, we still lack fundamental insights into the control and regulation of this complex, system wide, physiological process in plants. Notwithstanding, the potential of developing targeted biomass strategies to optimize partitioning for practical yield gains in crops is an applied goal that supports and justifies continued basic science research in the fundamentals of C and N allocation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
BA and DB acknowledge the experimental and intellectual contributions of their respective laboratory personnel. Research on C partitioning in the laboratory of BA is currently supported by the National Science Foundation (grants 0922546, 1121819). Research in the laboratory of DB is supported by Department of Energy grant (DE-FG02-08ER64629) and USDA-AFRI grant (2011-67013-30056).
References
Ainsworth, E. A., and Bush, D. R. (2011). Carbohydrate export from the leaf: a highly regulated process and target to enhance photosynthesis and productivity. Plant Physiol. 155, 64–69. doi: 10.1104/pp.110.167684
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ayre, B. G. (2011). Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Mol. Plant 4, 377–394. doi: 10.1093/mp/ssr014
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Barber, C., Rosti, J., Rawat, A., Findlay, K., Roberts, K., and Seifert, G. J. (2006). Distinct properties of the five UDP-D-glucose/UDP-D-galactose 4-epimerase isoforms of Arabidopsis thaliana. J. Biol. Chem. 281, 17276–17285. doi: 10.1074/jbc.M512727200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Beebe, D. U., and Turgeon, R. (1992). Localization of galactinol, raffinose, and stachyose synthesis in Cucurbita pepo leaves. Planta 188, 354–361. doi: 10.1007/BF00192802
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bourgis, F., Roje, S., Nuccio, M. L., Fisher, D. B., Tarczynski, M. C., Li, C. J., et al. (1999). S-methylmethionine plays a major role in phloem sulfur transport and is synthesized by a novel type of methyltransferase. Plant Cell 11, 1485–1497. doi: 10.1105/tpc.11.8.1485
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Braun, D. M., and Slewinski, T. L. (2009). Genetic control of carbon partitioning in grasses: roles of sucrose transporters and TIE-DYED loci in phloem loading. Plant Physiol. 149, 71–81. doi: 10.1104/pp.108.129049
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Braun, D. M., Wang, L., and Ruan, Y.-L. (2014). Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signalling to enhance crop yield and food security. J. Exp. Bot. 65, 1713–1735. doi: 10.1093/jxb/ert416
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bush, D. R. (1993). Proton-coupled sugar and amino acid transporters in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 513–542. doi: 10.1146/annurev.pp.44.060193.002501
Bush, D. R. (1999). “Amino acid transport,” in Plant Amino Acids: Biochemistry and Biotechnology, ed. B. K. Singh (New York, NY: Marcel Dekker), 305–318.
Büttner, M. (2007). The monosaccharide transporter(-like) gene family in Arabidopsis. FEBS Lett. 581, 2318–2324. doi: 10.1016/j.febslet.2007.03.016
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Büttner, M. (2010). The Arabidopsis sugar transporter (AtSTP) family: an update. Plant Biol. (Stuttg.) 12, 35–41. doi: 10.1111/j.1438-8677.2010.00383.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cao, T., Lahiri, I., Singh, V., Louis, J., Shah, J., and Ayre, B. G. (2013). Metabolic engineering of raffinose-family oligosaccharides in the phloem reveals alterations in carbon partitioning and enhances resistance to green peach aphid. Front. Plant Sci. 4:263. doi: 10.3389/fpls.2013.00263
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Carmi, N., Zhang, G. F., Petreikov, M., Gao, Z. F., Eyal, Y., Granot, D., et al. (2003). Cloning and functional expression of alkaline α-galactosidase from melon fruit: similarity to plant SIP proteins uncovers a novel family of plant glycosyl hydrolases. Plant J. 33, 97–106. doi: 10.1046/j.1365-313X.2003.01609.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chaudhuri, B., Hormann, F., Lalonde, S., Brady, S. M., Orlando, D. A., Benfey, P., et al. (2008). Protonophore- and pH-insensitive glucose and sucrose accumulation detected by FRET nanosensors in Arabidopsis root tips. Plant J. 56, 948–962. doi: 10.1111/j.1365-313X.2008.03652.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, L. Q. (2014). SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 201, 1150–1155. doi: 10.1111/nph.12445
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, L. Q., Hou, B. H., Lalonde, S., Takanaga, H., Hartung, M. L., Qu, X. Q., et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532. doi: 10.1038/nature09606
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, L.-Q., Qu, X.-Q., Hou, B.-H., Sosso, D., Osorio, S., Fernie, A. R., et al. (2012). Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335, 207–211. doi: 10.1126/science.1213351
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dai, N., Petreikov, M., Portnoy, V., Katzir, N., Pharr, D. M., and Schaffer, A. A. (2006). Cloning and expression analysis of a UDP-galactose/glucose pyrophosphorylase from melon fruit provides evidence for the major metabolic pathway of galactose metabolism in raffinose oligosaccharide metabolizing plants. Plant Physiol. 142, 294–304. doi: 10.1104/pp.106.083634
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dasgupta, K., Khadilkar, A. S., Sulpice, R., Pant, B., Scheible, W.-R., Fisahn, J., et al. (2014). Expression of sucrose transporter cDNAs specifically in companion cells enhances phloem loading and long-distance transport of sucrose but leads to an inhibition of growth and the perception of a phosphate limitation. Plant Physiol. 165, 715–731. doi: 10.1104/pp.114.238410
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Deuschle, K., Chaudhuri, B., Okumoto, S., Lager, I., Lalonde, S., and Frommer, W. B. (2006). Rapid Metabolism of glucose detected with FRET glucose nanosensors in epidermal cells and intact roots of Arabidopsis RNA-silencing mutants. Plant Cell 18, 2314–2325. doi: 10.1105/tpc.106.044073
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Doebley, J. (2004). The genetics of maize evolution. Annu. Rev. Genet. 38, 37–59. doi: 10.1146/annurev.genet.38.072902.092425
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dundar, E., and Bush, D. R. (2009). BAT1, a bidirectional amino acid transporter in Arabidopsis. Planta 229, 1047–1056. doi: 10.1007/s00425-009-0892-8
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Endler, A., Meyer, S., Schelbert, S., Schneider, T., Weschke, W., Peters, S. W., et al. (2006). Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol. 141, 196–207. doi: 10.1104/pp.106.079533
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Eom, J. S., Cho, J. I., Reinders, A., Lee, S. W., Yoo, Y., Tuan, P. Q., et al. (2011). Impaired function of the tonoplast-localized sucrose transporter in rice, OsSUT2, limits the transport of vacuolar reserve sucrose and affects plant growth. Plant Physiol. 157, 109–119. doi: 10.1104/pp.111.176982
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fischer, W. N., Andre, B., Rentsch, D., Krolkiewicz, S., Tegeder, M., Breitkreuz, K., et al. (1998). Amino acid transport in plants. Trends Plant Sci. 3, 188–195. doi: 10.1016/s1360-1385(98)01231-x
Gao, Z., Maurousset, L., Lemoine, R., Yoo, S.-D., Van Nocker, S., and Loescher, W. (2003). Cloning, expression, and characterization of sorbitol transporters from developing sour cherry fruit and leaf sink tissues. Plant Physiol. 131, 1566–1575. doi: 10.1104/pp.102.016725
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gifford, R. M., Thorne, J. H., Hitz, W. D., and Giaquinta, R. T. (1984). Crop productivity and photoassimilate partitioning. Science 225, 801–808. doi: 10.1126/science.225.4664.801
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gil, L., Yaron, I., Shalitin, D., Sauer, N., Turgeon, R., and Wolf, S. (2011). Sucrose transporter plays a role in phloem loading in CMV-infected melon plants that are defined as symplastic loaders. Plant J. 66, 366–374. doi: 10.1111/j.1365-313X.2011.04498.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gruenwald, K., Holland, J. T., Stromberg, V., Ahmad, A., Watcharakichkorn, D., and Okumoto, S. (2012). Visualization of glutamine transporter activities in living cells using genetically encoded glutamine sensors. PLoS ONE 7:e38591. doi: 10.1371/journal.pone.0038591
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hannah, M. A., Zuther, E., Buchel, K., and Heyer, A. G. (2006). Transport and metabolism of raffinose family oligosaccharides in transgenic potato. J. Exp. Bot. 57, 3801–3811. doi: 10.1093/jxb/erl152
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Horn, P. J., and Chapman, K. D. (2014). Lipidomics in situ: insights into plant lipid metabolism from high resolution spatial maps of metabolites. Prog. Lipid. Res. 54, 32–52. doi: 10.1016/j.plipres.2014.01.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Horn, P. J., Korte, A. R., Neogi, P. B., Love, E., Fuchs, J., Strupat, K., et al. (2012). Spatial mapping of lipids at cellular resolution in embryos of cotton. Plant Cell 24, 622–636. doi: 10.1105/tpc.111.094581
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jin, Y., Ni, D.-A., and Ruan, Y.-L. (2009). Posttranslational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose level. Plant Cell 21, 2072–2089. doi: 10.1105/tpc.108.063719
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keller, F., and Pharr, D. M. (1996). “Metabolism of carbohydrates in sinks and sources: galactosyl-sucrose oligosaccharides,” in Photoassimilate Distribution in Plants and Crops: Source-Sink Relationships, eds E. Zamski and A. A. Schaffer (New York: Marcel Dekker), 157–183.
Kalttorres, W., Kerr, P. S., Usuda, H., and Huber, S. C. (1987). Diurnal changes in maize leaf photosynthesis. 1. Carbon exchange-rate, assimilate export rate, and enzyme-activities. Plant Physiol. 83, 283–288. doi: 10.1104/pp.83.2.283
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kempers, R., and Van Bel, A. J. E. (1997). Symplasmic connections between sieve element and companion cell in the stem phloem of Vicia faba L have a molecular exclusion limit of at least 10 kDa. Planta 201, 195–201.
Kim, I., Cho, E., Crawford, K., Hempel, F. D., and Zambryski, P. C. (2005). Cell-to-cell movement of GFP during embryogenesis and early seedling development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 102, 2227–2231. doi: 10.1073/pnas.0409193102
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Knoblauch, M., and Van Bel, A. J. E. (1998). Sieve tubes in action. Plant Cell 10, 35–50. doi: 10.1105/tpc.10.1.35.
Ladwig, F., Stahl, M., Ludewig, U., Hirner, A. A., Hammes, U. Z., Stadler, R., et al. (2012). Siliques are Red1 from Arabidopsis acts as a bidirectional amino acid transporter that is crucial for the amino acid homeostasis of siliques. Plant Physiol. 158, 1643–1655. doi: 10.1104/pp.111.192583
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Leggewie, G., Kolbe, A., Lemoine, R., Roessner, U., Lytovchenko, A., Zuther, E., et al. (2003). Overexpression of the sucrose transporter SoSUT1 in potato results in alterations in leaf carbon partitioning and in tuber metabolism but has little impact on tuber morphology. Planta 217, 158–167. doi: 10.1007/s00425-003-0975-x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lei, M. G., Liu, Y. D., Zhang, B. C., Zhao, Y. T., Wang, X. J., Zhou, Y. H., et al. (2011). Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiol. 156, 1116–1130. doi: 10.1104/pp.110.171736
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Liu, X., and Bush, D. R. (2006). Expression and transcriptional regulation of amino acid transporters in plants. Amino Acids 30, 113–120. doi: 10.1007/s00726-005-0248-z
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
McCaskill, A., and Turgeon, R. (2007). Phloem loading in Verbascum phoeniceum L. depends on the synthesis of raffinose-family oligosaccharides. Proc. Natl. Acad. Sci. U.S.A. 104, 19619–19624. doi: 10.1073/pnas.0707368104
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
McCurdy, D. W., Dibley, S., Cahyanegara, R., Martin, A., and Patrick, J. W. (2010). Functional characterization and RNAi-mediated suppression reveals roles for hexose transporters in sugar accumulation by tomato fruit. Mol. Plant 3, 1049–1063. doi: 10.1093/mp/ssq050
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Meyer, R. S., and Purugganan, M. D. (2013). Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 14, 840–852. doi: 10.1038/nrg3605
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Michaeli, S., Fait, A., Lagor, K., Nunes-Nesi, A., Grillich, N., Yellin, A., et al. (2011). A mitochondrial GABA permease connects the GABA shunt and the TCA cycle, and is essential for normal carbon metabolism. Plant J. 67, 485–498. doi: 10.1111/j.1365-313X.2011.04612.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Noiraud, N., Maurousset, L., and Lemoine, R. (2001). Identification of a mannitol transporter, AgMaT1, in celery phloem. Plant Cell 13, 695–705. doi: 10.1105/tpc.13.3.695
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Okumoto, S., Looger, L. L., Micheva, K. D., Reimer, R. J., Smith, S. J., and Frommer, W. B. (2005). Detection of glutamate release from neurons by genetically encoded surface-displayed FRET nanosensors. Proc. Natl. Acad. Sci. U.S.A. 102, 8740–8745. doi: 10.1073/pnas.0503274102
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Olsen, K. M., and Wendel, J. F. (2013). A bountiful harvest: genomic insights into crop domestication phenotypes. Annu. Rev. Plant. Biol. 64, 47–70. doi: 10.1146/annurev-arplant-050312-120048
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Oparka, K. J., Roberts, A. G., Boevink, P., Santa Cruz, S., Roberts, L., Pradel, K. S., et al. (1999). Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97, 743–754. doi: 10.1016/S0092-8674(00)80786-2
Ortiz-Lopez, A., Chang, H. C., and Bush, D. R. (2000). Amino acid transporters in plants. Biochim. Biophys. Acta 1465, 275–280. doi: 10.1016/s0005-2736(00)00144-9
Otto, K. (1968). “Carbon metabolism: nature and formation of end products,” in Harvesting the Sun—Photosynthesis in Plant Life, ed. A. San Pietro (New York, NY: Academic Press, Incorporated), 131–152.
Patrick, J. W. (1997). Phloem unloading: sieve element unloading and post-sieve element transport. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 191–222. doi: 10.1146/annurev.arplant.48.1.191
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Patrick, J. W., and Offler, C. E. (2001). Compartmentation of transport and transfer events in developing seeds. J. Exp. Bot. 52, 551–564. doi: 10.1093/jexbot/52.356.551
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Proels, R. K., and Hückelhoven, R. (2014). Cell-wall invertases, key enzymes in the modulation of plant metabolism during defence responses. Mol. Plant Pathol. 15, 858–864. doi: 10.1111/mpp.12139
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Reidel, E. J., Rennie, E. A., Amiard, V., Cheng, L., and Turgeon, R. (2009). Phloem loading strategies in three plant species that transport sugar alcohols. Plant Physiol. 149, 1601–1608. doi: 10.1104/pp.108.134791
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Reinders, A., Sivitz, A. B., Starker, C. G., Gantt, J. S., and Ward, J. M. (2008). Functional analysis of LjSUT4, a vacuolar sucrose transporter from Lotus japonicus. Plant Mol. Biol. 68, 289–299. doi: 10.1007/s11103-008-9370-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Reinders, A., Sivitz, A. B., and Ward, J. M. (2012). Evolution of plant sucrose uptake transporters (SUTs). Front. Plant Sci. 3:22. doi: 10.3389/fpls.2012.00022
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rennie, E. A., and Turgeon, R. (2009). A comprehensive picture of phloem loading strategies. Proc. Natl. Acad. Sci. U.S.A. 106, 14162–14167. doi: 10.1073/pnas.0902279106
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Roitsch, T., and Gonzalez, M. C. (2004). Function and regulation of plant invertases: sweet sensations. Trends Plant Sci. 9, 606–613. doi: 10.1016/j.tplants.2004.10.009
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rolletschek, H., Hosein, F., Miranda, M., Heim, U., Gotz, K. P., Schlereth, A., et al. (2005). Ectopic expression of an amino acid transporter (VfAAP1) in seeds of Vicia narbonensis and pea increases storage proteins. Plant Physiol. 137, 1236–1249. doi: 10.1104/pp.104.056523
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rosche, E., Blackmore, D., Tegeder, M., Richardson, T., Schroeder, H., Higgins, T. J. V., et al. (2002). Seed-specific overexpression of a potato sucrose transporter increases sucrose uptake and growth rates of developing pea cotyledons. Plant J. 30, 165–175. doi: 10.1046/j.1365-313X.2002.01282.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ruan, Y. L., Llewellyn, D. J., and Furbank, R. T. (2001). The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell 13, 47–60. doi: 10.1105/tpc.13.1.47
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ruan, Y. L., and Patrick, J. W. (1995). The cellular pathway of post-phloem sugar-transport in developing tomato fruit. Planta 196, 434–444. doi: 10.1007/BF00203641
Sauer, N. (2007). Molecular physiology of higher plant sucrose transporters. FEBS Lett. 581, 2309–2317. doi: 10.1016/j.febslet.2007.03.048
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schofield, R. A., Bi, Y. M., Kant, S., and Rothstein, S. J. (2009). Over-expression of STP13, a hexose transporter, improves plant growth and nitrogen use in Arabidopsis thaliana seedlings. Plant Cell Environ. 32, 271–285. doi: 10.1111/j.1365-3040.2008.01919.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schulz, A., Beyhl, D., Marten, I., Wormit, A., Neuhaus, E., Poschet, G., et al. (2011). Proton-driven sucrose symport and antiport are provided by the vacuolar transporters SUC4 and TMT1/2. Plant J. 68, 129–136. doi: 10.1111/j.1365-313X.2011.04672.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Slewinski, T. L., and Braun, D. M. (2010). Current perspectives on the regulation of whole-plant carbohydrate partitioning. Plant Sci. 178, 341–349. doi: 10.1016/j.plantsci.2010.01.010
Srivastava, A. C., Ganesan, S., Ismail, I. O., and Ayre, B. G. (2009). Effective carbon partitioning driven by exotic phloem-specific regulatory elements fused to the Arabidopsis thaliana AtSUC2 sucrose-proton symporter gene. BMC Plant Biol. 9:7. doi: 10.1186/1471-2229-9-7
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Stadler, R., Lauterbach, C., and Sauer, N. (2005a). Cell-to-cell movement of green fluorescent protein reveals post-phloem transport in the outer integument and identifies symplastic domains in Arabidopsis seeds and embryos. Plant Physiol. 139, 701–712. doi: 10.1104/pp.105.065607
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Stadler, R., Wright, K. M., Lauterbach, C., Amon, G., Gahrtz, M., Feuerstein, A., et al. (2005b). Expression of GFP-fusions in Arabidopsis companion cells reveals non-specific protein trafficking into sieve elements and identifies a novel post-phloem domain in roots. Plant J. 41, 319–331. doi: 10.1111/j.1365-313X.2004.02298.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sun, A., Dai, Y., Zhang, X., Li, C., Meng, K., Xu, H., et al. (2011). A transgenic study on affecting potato tuber yield by expressing the rice sucrose transporter genes OsSUT5Z and OsSUT2M. J. Integr. Plant Biol. 53, 586–595. doi: 10.1111/j.1744-7909.2011.01063.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tan, Q., Zhang, L., Grant, J., Cooper, P., and Tegeder, M. (2010). Increased phloem transport of S-methylmethionine positively affects sulfur and nitrogen metabolism and seed development in pea plants. Plant Physiol. 154, 1886–1896. doi: 10.1104/pp.110.166389
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tegeder, M. (2012). Transporters for amino acids in plant cells: some functions and many unknowns. Curr. Opin. Plant Biol 15, 315–321. doi: 10.1016/j.pbi.2012.02.001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tegeder, M. (2014). Transporters involved in source to sink partitioning of amino acids and ureides: opportunities for crop improvement. J. Exp. Bot. 65, 1865–1878. doi: 10.1093/jxb/eru012
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Turgeon, R. (1996). Phloem loading and plasmodesmata. Trends Plant Sci. 1, 418–423. doi: 10.1016/S1360-1385(96)10045-5
Turgeon, R., Beebe, D. U., and Gowan, E. (1993). The intermediary cell—minor vein anatomy and raffinose oligosaccharide synthesis in the Scrophulariaceae. Planta 191, 446–456. doi: 10.1007/BF00195746
Turgeon, R., and Gowan, E. (1992). Sugar synthesis and phloem loading in Coleus blumei leaves. Planta 187, 388–394. doi: 10.1007/BF00195663
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Turgeon, R., and Medville, R. (1998). The absence of phloem loading in willow leaves. Proc. Natl. Acad. Sci. U.S.A. 95, 12055–12060. doi: 10.1073/pnas.95.20.12055
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Turgeon, R., and Medville, R. (2004). Phloem loading. A reevaluation of the relationship between plasmodesmatal frequencies and loading strategies. Plant Physiol. 136, 3795–3803. doi: 10.1104/pp.104.042036
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Turgeon, R., Medville, R., and Nixon, K. C. (2001). The evolution of minor-vein phloem and phloem loading. Am. J. Bot. 88, 1331–1339. doi: 10.2307/3558441
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Turgeon, R., and Wolf, S. (2009). Phloem transport: cellular pathways and molecular trafficking. Annu. Rev. Plant Biol. 60, 207–221. doi: 10.1146/annurev.arplant.043008.092045
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Usadel, B., Bläsing, O. E., Gibon, Y., Retzlaff, K., Hoehne, M., Günther, M., et al. (2008). Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol. 146, 1834–1861. doi: 10.1104/pp.107.115592
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Van As, H., Scheenen, T., and Vergeldt, F. J. (2009). MRI of intact plants. Photosyn. Res. 102, 213–222. doi: 10.1007/s11120-009-9486-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vaughn, M. W., Harrington, G. N., and Bush, D. R. (2002). Sucrose-mediated transcriptional regulation of sucrose symporter activity in the phloem. Proc. Natl. Acad. Sci. U.S.A. 99, 10876–10880. doi: 10.1073/pnas.172198599
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Voitsekhovskaja, O. V., Rudashevskaya, E. L., Demchenko, K. N., Pakhomova, M. V., Batashev, D. R., Gamalei, Y. V., et al. (2009). Evidence for functional heterogeneity of sieve element-companion cell complexes in minor vein phloem of Alonsoa meridionalis. J. Exp. Bot. 60, 1873–1883. doi: 10.1093/jxb/erp074
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wallace, J. G., Larsson, S. J., and Buckler, E. S. (2014). Entering the second century of maize quantitative genetics. Heredity 112, 30–38. doi: 10.1038/hdy.2013.6
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, N., and Fisher, D. B. (1994). The use of fluorescent tracers to characterize the post-phloem transport pathway in maternal tissues of developing wheat grains. Plant Physiol. 104, 17–27. doi: 10.1104/pp.104.1.17
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Weichert, N., Saalbach, I., Weichert, H., Kohl, S., Erban, A., Kopka, J., et al. (2010). Increasing sucrose uptake capacity of wheat grains stimulates storage protein synthesis. Plant Physiol. 152, 698–710. doi: 10.1104/pp.109.150854
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Weigelt, K., Kuster, H., Radchuk, R., Muller, M., Weichert, H., Fait, A., et al. (2008). Increasing amino acid supply in pea embryos reveals specific interactions of N and C metabolism, and highlights the importance of mitochondrial metabolism. Plant J. 55, 909–926. doi: 10.1111/j.1365-313X.2008.03560.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wippel, K., and Sauer, N. (2012). Arabidopsis SUC1 loads the phloem in suc2 mutants when expressed from the SUC2 promoter. J. Exp. Bot. 63, 669–679. doi: 10.1093/jxb/err255
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zaima, N., Goto-Inoue, N., Hayasaka, T., and Setou, M. (2010). Application of imaging mass spectrometry for the analysis of Oryza sativa rice. Rapid Commun. Mass Spectrom. 24, 2723–2729. doi: 10.1002/rcm.4693
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, C., Wei, Z. H., and Ye, B. C. (2013). Imaging and tracing of intracellular metabolites utilizing genetically encoded fluorescent biosensors. Biotechnol. J. 8, 1280–1291. doi: 10.1002/biot.201300001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, L., Garneau, M. G., Majumdar, R., Grant, J., and Tegeder, M. (2014). Improvement of pea biomass and seed productivity by simultaneous increase of phloem and embryo loading with amino acids. Plant J. 81, 134–146. doi: 10.1111/tpj.12716
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, W.-H., Zhou, Y., Dibley, K. E., Tyerman, S. D., Furbank, R. T., and Patrick, J. W. (2007). Nutrient loading of developing seeds. Funct. Plant Biol. 34, 314–331. doi: 10.1071/FP06271
Zhang, X.-Y., Wang, X.-L., Wang, X.-F., Xia, G.-H., Pan, Q.-H., Fan, R.-C., et al. (2006). A shift of phloem unloading from symplasmic to apoplasmic pathway is involved in developmental onset of ripening in grape berry. Plant Physiol. 142, 220–232. doi: 10.1104/pp.106.081430
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhou, Y. C., Chan, K., Wang, T. L., Hedley, C. L., Offler, C. E., and Patrick, J. W. (2009). Intracellular sucrose communicates metabolic demand to sucrose transporters in developing pea cotyledons. J. Exp. Bot. 60, 71–85. doi: 10.1093/jxb/ern254
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: assimilate partitioning, sugar transport in plants, amino acid transport, crop yield, nutritional value
Citation: Yadav UP, Ayre BG and Bush DR (2015) Transgenic approaches to altering carbon and nitrogen partitioning in whole plants: assessing the potential to improve crop yields and nutritional quality. Front. Plant Sci. 6:275. doi: 10.3389/fpls.2015.00275
Received: 19 December 2014; Accepted: 06 April 2015;
Published: 22 April 2015.
Edited by:
Susan Gibson, University of Minnesota, USAReviewed by:
John M. Ward, University of Minnesota, USAPaulo Arruda, Universidade Estadual de Campinas, Brazil
Copyright © 2015 Yadav, Ayre and Bush. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brian G. Ayre, Department of Biological Sciences, University of North Texas, 1155 Union Circle, Denton, TX 76203, USA brian.ayre@unt.edu; Daniel R. Bush, Department of Biology, Colorado State University, Lake Street, Fort Collins, CO 80523, USA dbush@colostate.edu
 Umesh P. Yadav
Umesh P. Yadav