- Department of Life Science, Institute of Plant Biology, National Taiwan University, Taipei, Taiwan
Upon recognition of microbe-associated molecular patterns (MAMPs) such as the bacterial flagellin (or the derived peptide flg22) by pattern-recognition receptors (PRRs) such as the FLAGELLIN SENSING2 (FLS2), plants activate the pattern-triggered immunity (PTI) response. The L-type lectin receptor kinase-VI.2 (LecRK-VI.2) is a positive regulator of Arabidopsis thaliana PTI. Cysteine-rich receptor-like kinases (CRKs) possess two copies of the C-X8-C-X2-C (DUF26) motif in their extracellular domains and are thought to be involved in plant stress resistance, but data about CRK functions are scarce. Here, we show that Arabidopsis overexpressing the LecRK-VI.2-responsive CRK4, CRK6, and CRK36 demonstrated an enhanced PTI response and were resistant to virulent bacteria Pseudomonas syringae pv. tomato DC3000. Notably, the flg22-triggered oxidative burst was primed in CRK4, CRK6, and CRK36 transgenics and up-regulation of the PTI-responsive gene FLG22-INDUCED RECEPTOR-LIKE 1 (FRK1) was potentiated upon flg22 treatment in CRK4 and CRK6 overexpression lines or constitutively increased by CRK36 overexpression. PTI-mediated callose deposition was not affected by overexpression of CRK4 and CRK6, while CRK36 overexpression lines demonstrated constitutive accumulation of callose. In addition, Pst DC3000-mediated stomatal reopening was blocked in CRK4 and CRK36 overexpression lines, while overexpression of CRK6 induced constitutive stomatal closure suggesting a strengthening of stomatal immunity. Finally, bimolecular fluorescence complementation and co-immunoprecipitation analyses in Arabidopsis protoplasts suggested that the plasma membrane localized CRK4, CRK6, and CRK36 associate with the PRR FLS2. Association with FLS2 and the observation that overexpression of CRK4, CRK6, and CRK36 boosts specific PTI outputs and resistance to bacteria suggest a role for these CRKs in Arabidopsis innate immunity.
Introduction
Cell surface plant receptor-like kinases (RLKs) link perception of environmental stimuli and downstream signal transductions to trigger appropriate intracellular responses. The typical structure of RLKs consists of a N-terminal receptor domain in the extracellular region, a transmembrane domain, and a C-terminal intracellular protein kinase domain (Kacperska, 2004; De Smet et al., 2009). In Arabidopsis thaliana, RLKs belong to a large family with more than 600 members and are classified according to their extracellular domains (Shiu and Bleecker, 2001). The best-studied RLK sub-family is characterized by the presence of leucine-rich repeat (LRR) motifs in the extracellular region. Members of this sub-family are involved in a wide range of plant signaling initiation and activation processes (Walker, 1994; Kobe and Kajava, 2001). Major examples of LRR-RLKs with known functions are BRASSINOSTEROID INSENSITIVE 1 (BRI1) that is involved in the perception of the phytohormone brassinosteroid (Wang et al., 2001; Nam and Li, 2002), CLAVATA1 that recognizes the secreted peptide CLAVATA3 to control the size of the shoot apical meristem (Clark et al., 1997; Dievart et al., 2003; Ogawa, 2008), and PEP RECEPTOR 1 (PEPR1) and PEPR2 that sense the damage-associated molecular pattern peptide 1 (Pep1) (Krol et al., 2010; Yamaguchi et al., 2010). In addition, the pattern recognition receptors (PRRs) FLAGELLIN SENSING2 (FLS2) and EF-TU receptor (EFR) are LRR-RLKs that recognize bacterial flagellin (or the derived peptide flg22) and EF-Tu (or the derived peptides elf18/elf26), respectively (Gómez-Gómez and Boller, 2000; Zipfel et al., 2006). Cell surface-localized PRRs sense conserved microbial features called microbe-associated molecular patterns (MAMPs). Recognition of MAMPs induces the pattern-triggered immunity (PTI) response characterized by the production of reactive oxygen species (ROS), activation of the mitogen-activated protein kinases (MAPKs) cascade, expression of PTI-responsive genes, callose deposition and stomatal closure (Jones and Dangl, 2006; Melotto et al., 2006; Zhang et al., 2007; Pieterse et al., 2009; Zipfel, 2009, 2014; Desclos-Theveniau et al., 2012; Kadota et al., 2014; Monaghan et al., 2014). The best-characterized PRR involved in PTI is FLS2 that recognizes the conserved microbial peptide flg22 (Gómez-Gómez and Boller, 2000). Upon flg22 elicitation, FLS2 physically associates with another LRR-RLK termed BRI1-ASSOCIATED RECEPTOR-LIKE KINASE/SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE3 (BAK1/SERK3), and promotes PTI responses (Chinchilla et al., 2007; Heese et al., 2007; Roux et al., 2011). Furthermore, the malectin-like LRR-RLK IMPAIRED OOMYCETE SUSCEPTIBILITY1 (IOS1) enhances PTI through the regulation of FLS2-BAK1 association (Chen et al., 2014). Another LRR-RLK, BAK1-INTERACTING RECEPTOR-LIKE KINASE 2 (BIR2) acts by negatively regulating BAK1 function to maintain immune homeostasis through the control of BAK1-FLS2 complex formation (Halter et al., 2014). Current understanding suggests that PRRs are central components of multiprotein complexes and that a dynamic regulation exists among the members of PRR complexes to fine tune the different PTI outputs (Macho and Zipfel, 2014).
In addition to LRR-RLKs, L-type lectin-RLKs (LecRKs) that are characterized by an extracellular lectin domain are known to function in plant innate immunity (Bouwmeester and Govers, 2009; Singh and Zimmerli, 2013). For example, LecRK-I.9 is a mediator of cell wall/plasma membrane adhesions and is required for resistance to pathogens via enhancement of callose deposition (Bouwmeester et al., 2011). In addition, LecRK-I.9 also known as DORN1, is necessary for eATP recognition (Choi et al., 2014). LecRK-V.5 negatively regulates stomatal immunity (Desclos-Theveniau et al., 2012) and LecRK-VI.2 is a functional protein kinase critical for Arabidopsis resistance to hemi-biotrophic Pseudomonas syringae pv. tomato DC3000 and necrotrophic Pectobacterium carotovorum bacteria (Singh et al., 2012a,b). Recently, the Arabidopsis LecRK-VI.2 was found to associate with FLS2 and to prime the PTI response when ectopically expressed in Nicotiana benthamiana (Huang et al., 2014). A genome-wide microarray analysis of an Arabidopsis LecRK-VI.2 overexpression line revealed seven highly up-regulated cysteine-rich receptor-like kinases (CRKs) (Singh et al., 2012a). CRKs belong to a RLK sub-family with at least 44 members containing the motif with unknown function C-X8-C-X2-C (DUF26) in their extracellular domains (Chen, 2001; Wrzaczek et al., 2010). In the past decade, some CRKs were demonstrated to play critical roles in biotic and abiotic responses in Arabidopsis. For example, CRK45 also called ACRK1 was found to associate with CRK36 to negatively control ABA signaling (Tanaka et al., 2012; Zhang et al., 2013a). Besides its function in ABA-relative responses, ACRK1 also positively regulates disease resistance to Pst DC3000 (Zhang et al., 2013b). Similarly, CRK20 promotes conditions that are favorable for Pst DC3000 growth in Arabidopsis (Ederli et al., 2011). Furthermore, CRK6 and CRK7 mediate the Arabidopsis response to extracellular ROS production (Idanheimo et al., 2014), and overexpression of CRK5 increases disease resistant to Pst DC3000 and triggers hypersensitive response (HR)-like cell death (Chen et al., 2003). Similarly, overexpression of CRK4, CRK19, and CRK20 that are structurally-related to CRK5 induces HR-like cell death (Chen et al., 2004), and transgenic Arabidopsis plants overexpressing CRK13 also demonstrate enhanced resistance to Pst DC3000, HR-like cell death, and salicylic acid (SA) accumulation (Acharya et al., 2007). In addition to Arabidopsis CRKs, the wheat CRK TaCRK1 is more expressed in wheat varieties resistant to Rhizoctonia cerealis (Yang et al., 2013). Finally, a novel antifungal protein from the endosperm of Ginkgo seeds, ginkbilobin-2 (Gnk2) that contains a conserved C-X8-C-X2-C motif, inhibits the growth of the phytopathogenic fungus Fusarium oxysporum (Sawano et al., 2007).
In this study, we found that overexpression of the LecRK-VI.2-responsive CRK4, CRK6, and CRK36 increased Arabidopsis resistance to Pst DC3000. These CRKs were also shown to positively regulate flg22-triggered ROS production, PTI-responsive gene expression, and callose deposition, and stomatal immunity during Pst DC3000 infection. Exploratory experiments to identify protein-protein interaction by using co-immunoprecipitation (Co-IP) and bimolecular fluorescence complementation (BiFC) assays suggest that these CRKs are part of the PRR FLS2 protein complex. This study thus identified three CRKs positively modulating the Arabidopsis PTI response possibly through association with the PRR FLS2.
Materials and Methods
Biological Materials and Growth Conditions
A. thaliana ecotype Col-0 plants were grown in commercial potting soil/perlite (3:2) at 22–24°C day and 17–19°C night temperature under a 9-h-light/15-h-dark photoperiod. The lighting was supplied at an intensity of ~100 μE m−2s−1 by fluorescence tubes. T-DNA insertion mutants crk4-1 (SALK_063969), crk4-2 (SALK_089138), crk6-1 (SALK_205955), crk7-1 (WiscDsLox336D10), crk7-2 (WiscDsLox502A10), crk13-1 (SALK_085128), crk23-1 (SALK_076541), crk23-2 (SAIL_518_A03), crk36-1 (SALK_035659), crk36-2 (SALK_100834), crk37-1 (SALK_131604), and crk37-2 (SALK_012014) were obtained from the Arabidopsis Biological Resource Center (ABRC). Bacterial strains Pst DC3000 and Pst DC3000 hrcC− were provided by Kunkel (Washington University, St. Louis, Missouri, USA). Pst bacteria were cultivated at 28°C and 220 rpm in King's B medium containing 50 mg/mL rifampicin (DC3000) or rifampicin and kanamycin (DC3000 hrcC−).
Generation of Transgenic Plants
To generate transgenic Arabidopsis, the full length coding sequences (CDS) of CRKs were amplified by PCR with Phusion High-Fidelity DNA Polymerase (New England Biolabs) and PCR-amplified products were cloned in pCR8 TOPO TA cloning vector (Invitrogen) and TOPO-cDNA plasmids were recombined into the Gateway-compatible expression vectors pEarlyGate103 (ordered at ABRC). Pro35S: CRKs-GFP constructs were electroporated into Agrobacterium tumefaciens competent cells strain GV3101 and A. tumefaciens carrying Pro35S: CRKs-GFP were then used to transform Col-0 via floral drop-inoculation (Martinez-Trujillo et al., 2004). Transgenic plants were selected on MS medium containing 50 μM Glufosinate ammonium (Fluka). Multiple transgenic lines were obtained and raised to homozygotic T3 lines. All primer sequences are shown in Supplementary Table 1.
Pst DC3000 Bioassay
Five-week-old Arabidopsis plants were dipped in a bacterial suspension of 106 colony-forming units (CFU)/mL Pst DC3000 in 10 mM MgSO4 containing 0.01% Silwet L-77 (Lehle Seeds) for 15 min. After dipping, plants were kept at 100% relative humidity overnight. Disease symptoms were evaluated at 3 days post inoculation (dpi). For bacterial titers, leaf discs collected at 2 dpi were washed twice with sterile water and homogenized in 10 mM MgSO4. Quantification was done by plating appropriate dilutions on King's B agar containing rifampicin (50 mg/liter) as previously described (Zimmerli et al., 2000). Each biological repeat represents nine leaf discs (0.5 cm diameter) from three different plants.
Oxidative Burst Assay
Reactive Oxygen Species (ROS) evaluation was performed as previously described (Huang et al., 2012). Eight leaf discs of 0.25 cm2 from three 5-week-old Arabidopsis plants were incubated in 100 μL ddH2O overnight in 96-well plates. Water was then replaced by 100 μL of reaction solution containing 50 μM of luminol and 10 μg/mL of horseradish peroxidase (Sigma) supplemented with 50 nM of flg22 or water only for the mock controls. ROS measurements expressed as means of RLU (Relative Light Units) were conducted immediately after adding the solution with a Centro LIApcLB 692 plate luminometer (Berthold Technologies, Bad Wildbad, Germany). ROS evaluation was performed at a 2 min interval reading time for a period of 25 min.
Stomatal Movement Measurement
Epidermal peels from the abaxial side of fully expanded leaves were floated in the stomatal buffer solution (25 mM MES-KOH, pH 6.15, and 10 mM KCl) under light (100 μmol m−2s−1) for at least 3 h to open stomata before stomatal aperture evaluation was performed as described (Desclos-Theveniau et al., 2012). Briefly, after treatment with 10 mM MgSO4 (Mock control) or bacterial suspensions (108 CFU/mL Pst DC3000 in 10 mM MgSO4), pictures of stomata were taken of random regions at various time points using an Olympus BX51 microscope digital camera and application software DP2-BSW. Stomatal apertures were measured using the “measure” function of ImageJ (http://rsb.info.nih.gov/ij/). Three plants were used per biological replicates.
RNA Extraction and Gene Expression Analysis
Total RNA was extracted and purified using the MaestroZol reagent according to the manufacturer's instructions (Omics Biotechnology Co., Ltd) with the addition of PLUS reagent for polysaccharides and proteoglycans elimination. Genomic DNA contaminations were removed using Qiagen RNase-Free DNase Set. For cDNA synthesis, 2 μg of total RNA were prepared in a volume of 22 μL DEPC-treated H2O and denatured at 65°C for 5 min. Eighteen point 5 mL of master mix (16 μL M-MLV buffer, 1 mM dNTP, 5 mM OligoT, 100 U M-MLV reverse transcriptase, [Invitrogen]) was added into each tube and then incubated at 37°C for 1 h, 70°C for 10 min. The cDNA was then diluted 5-times before reverse transcription-PCR (RT-PCR) or quantitative RT-PCR (qRT-PCR) analyses. RT-PCR amplification was done with 1 μL of the first-strand cDNA as template, 10 μL of Taq PLUS PCR MasterMix (#KT205, Tiangen Biotech, http://www.tiangen.com/en/) and 1 μL of 10 mM forward and reverse primers in a total volume of 20 μL. The cycling conditions were 94°C for 3 min for one initial step followed by 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min, for 35 cycles. The PCR was terminated with one extra step at 72°C for 10 min. qRT-PCR were conducted on a CFX Real-Time PCR Detection System (Bio-Rad). SYBR Green fast qPCR master mix (Bio-Rad; 1 μL of cDNA, 5 μL SYBR Green supermix, 5 μL filtered sterile H2O, 0.5 μL of 10 mM forward and reverse primers, in a total volume of 12 μL per well) was employed for the analysis. The cycling conditions were composed of an initial 3 min denaturation step at 95°C, followed by 40 cycles at 95°C for 3 s and 60°C for 30 s. Melting curve was run from 65 to 95°C with 0.5-s time interval to ensure the specificity of product. Data were analyzed using Bio-Rad CFX manager software. UBQ10 was used as reference gene for normalization of gene expression levels in all samples. The wild-type (WT) without any treatment or mock treatment were considered as controls in each experiments (expression level = 1). Primer sequences are shown in Supplementary Table 1.
Callose Staining
Leaves of 5-week-old Arabidopsis were syringe infiltrated with 108 CFU/mL Pst DC3000 hrcC− suspended in 10 mM MgSO4 or 500 nM flg22 dissolved in H2O. Control plants were respectively infiltrated with 10 mM MgSO4or H2O only. After infiltration, nine leaf discs from three different plants were selected for analyses. Harvested leaf samples were cleared overnight by incubation in 95% ethanol at room temperature and then washed three times (0.5–1 h for each washing) with sterilized H2O. Cleared leaves were stained with 0.01% aniline blue in 0.15 M phosphate buffer, pH 8, for 1 h. Callose deposits were visualized under UV illumination using a Nikon Optiphot-2 microscope. Callose deposition was evaluated using the “analyze particles” function of ImageJ (http://rsb.info.nih.gov/ij/).
Subcellular Localization in Arabidopsis Protoplast
Subcellular localization assays were performed as previously described (Chen et al., 2014). Briefly, plasmids containing 35S: CRKs-GFP or 35S: GFP were co-transfected with the plasma membrane marker pm-rkCD3-1007 (Nelson et al., 2007) into Arabidopsis mesophyll protoplasts by polyethylene glycol (Sigma)(Yoo et al., 2007). The samples were visualized 30 h after transfection using a TCS SP5 confocal spectral microscope imaging system (Leica).
Bimolecular Fluorescence Complementation Assay in Arabidopsis Protoplast
For BiFC assays, plasmids of Pro35S: FLS2-YFPN (modified pEarleyGate201, Huang et al., 2014) and Pro35S: BAK1-YFPC (modified pEarleyGate202, Huang et al., 2014), Pro35S: FLS2-YFPN and Pro35S: CRKs-YFPC, Pro35S: RCI2B-YFPN and Pro35S: RCI2B-YFPc, or Pro35S: RCI2B-YFPN and Pro35S: CRKs-YFPC were transformed into Arabidopsis protoplasts by polyethylene glycol (Sigma) for transient expression (Yoo et al., 2007). The samples were visualized after treatment with 100 nM flg22 using a TCS SP5 confocal spectral microscope imaging system (Leica).
Co-immunoprecipitation Assay in Arabidopsis Protoplast
For Co-IP assays, the plasmids of Pro35S: FLS2-HA (modified pEarleyGate100 with a AvrII-3xHA-SpeI fragment introduced after the attR2 recombination site) and Pro35S: CRKs-GFP, Pro35S: FLS2-HA and Pro35S: GFP, or Pro35S: FLS2-HA and Pro35S: RBI2B-GFP were transformed into Arabidopsis protoplasts by polyethylene glycol (Sigma) for transient expression (Yoo et al., 2007). Total proteins were extracted with 0.5 mL protein extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 10 mM DTT, 10 mM EDTA, 1 mM NaF, 1 mM Na2MoO4·2H2O, 1% [w/v] polyvinylpyrrolidone, 1% [v/v] IGEPAL CA-630 [Sigma-Aldrich] and 1% [v/v] Roche protease inhibitor cocktail) and incubated with gentle shaking at 4°C for 1 h. Samples were then centrifuged at 14000 rpm for 15 min at 4°C. Proteins were separated by SDS–PAGE and then transferred to a polyvinylidine fluoride membrane (Immobilon-P; Millipore). Supernatants (1.5 mL) were adjusted to 2 mg/mL protein and incubated for 2 h at 4°C with 20 mL GFP Trap-A beads (Chromotek). Following incubation, beads were washed four times with TBS containing 0.5% (v/v) IGEPALCA-630. Proteins were separated by 8% SDS–PAGE and then transferred to a polyvinylidine fluoride membrane (Immobilon-P; Millipore). GFP and HA fusion proteins were detected by immunoblotting with anti-GFP and anti-HA primary antibodies, respectively.
Results
Lines Overexpressing CRK4, CRK6, and CRK36 are More Resistant to Virulent Bacteria
Overexpression of LecRK-VI.2, a positive regulator of PTI, induces a strong up-regulation (> 10 times) of CRK37 (At4g04500), CRK23 (At4g23310), CRK7 (At4g23150), CRK6 (At4g23140), CRK36 (At4g04490), CRK13 (At4g23210), and CRK4 (At3g45860) (Supplementary Table 2) (Singh et al., 2012a). To test the possible role of these CRKs in the Arabidopsis resistance response to bacterial pathogens, T-DNA insertion lines were ordered at ABRC and homozygotic mutants were tested for their resistance to the virulent bacterial pathogen Pst DC3000. As shown in Supplementary Figure 1, none of the tested T-DNA mutants demonstrated an altered resistance phenotype suggesting that either these CRKs are not involved in the Arabidopsis resistance to Pst DC3000 or functional redundancy hides CRK's role in defense against bacteria. Since most CRKs are localized in a cluster at chromosome 4, it was difficult to generate multiple CRK T-DNA insertion mutants. We thus tried to generate CRK37, CRK23, CRK7, CRK6, CRK36, CRK13, and CRK4 overexpression lines to evaluate their resistance responses to Pst DC3000. Stable Arabidopsis plants overexpressing CRK7 could not be obtained, possibly because of lethality as already observed for CRK13 (Acharya et al., 2007). Lines overexpressing CRK13, CRK23, and CRK37 demonstrated a WT disease phenotype at 3 dpi with Pst DC3000 (Supplementary Figure 2). By contrast, Arabidopsis overexpressing CRK4, CRK6, and CRK36 were more resistant to Pst DC3000 as illustrated by reduced disease symptoms and bacterial titers (Figure 1). These observations suggest that the LecRK-VI.2-responsive CRK4, CRK6, and CRK36 are involved in Arabidopsis resistance to Pst DC3000.
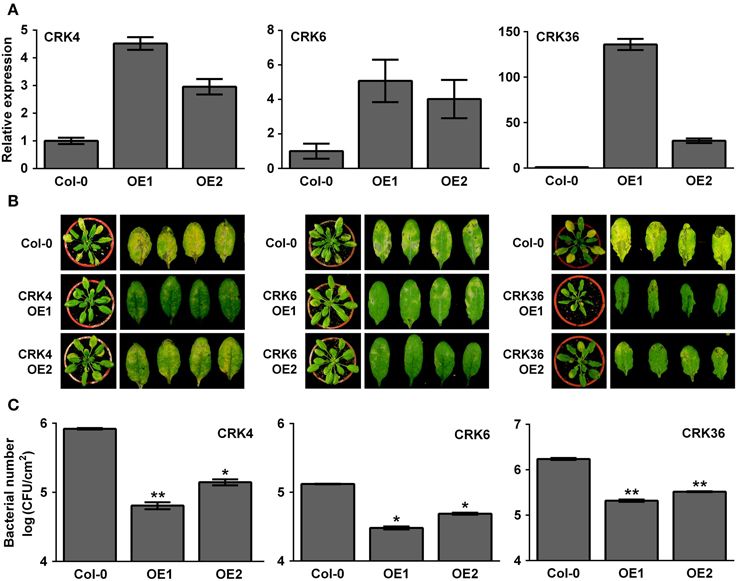
Figure 1. Arabidopsis overexpressing CRK4, CRK6, and CRK36 are more resistant to Pst DC3000.(A) Expression levels of CRK4, CRK6, and CRK36 in respective overexpression line 1 (OE1) and 2 (OE2). Total RNAs were extracted from 5-week-old Arabidopsis and CRK expression levels analyzed by qRT-PCR were expressed relatively to Col-0 WT (set at 1). UBQ10 expression levels were used for normalization. Results are average ± SE of three biological replicates each consisting of three technical repeats (n = 9). (B) Disease symptoms in Col-0 and respective CRK overexpression line 1 (OE1) and 2 (OE2). Five-week-old plants were dip-inoculated with 106 CFU/mL Pst DC3000 and pictures were taken 3 days later. (C) Bacterial growth. Pst DC3000 titers were evaluated at 2 dpi. Bacteria inoculation as in (B). Results are average ± SE of three biological replicates each consisting of nine leaf discs (n = 27). Asterisks represent a significant difference to Col-0 WT (t-test, *p < 0.05, **p < 0.01).
Overexpression of CRK4, CRK6, and CRK36 Alters Early and Late PTI Responses
Since LecRK-VI.2 positively regulates PTI (Singh et al., 2012a), and overexpression of the LecRK-VI.2-responsive CRK4, CRK6, and CRK36 increased Arabidopsis resistance to Pst DC3000 (Figure 1), we hypothesized that these CRKs may positively regulate the Arabidopsis PTI response. The production of reactive oxygen species (ROS), an early PTI response (Kadota et al., 2014), was thus evaluated in lines overexpressing CRK4, CRK6, and CRK36. ROS production was increased in lines overexpressing these three CRKs after treatment with the MAMP flg22, but no constitutive ROS production was observed in mock-treated plants (Figure 2). This observation suggests priming of ROS accumulation upon PTI elicitation in CRK4, CRK6, and CRK36 overexpression lines.
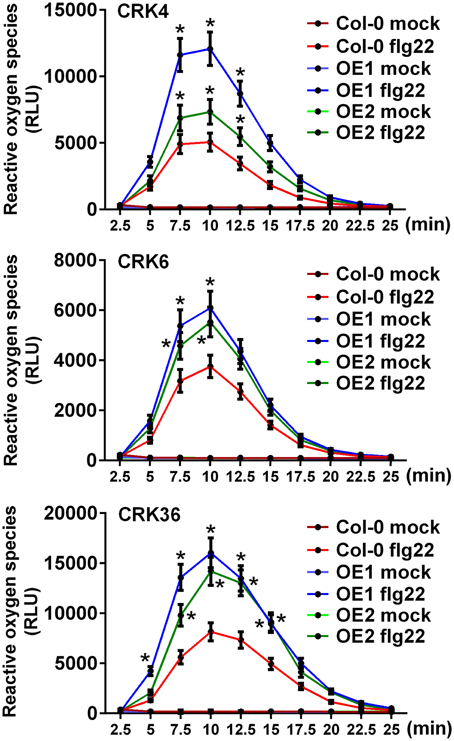
Figure 2. Priming of ROS accumulation upon flg22 treatment in lines overexpressing CRK4, CRK6, and CRK36. ROS production represented as Relative Light Units (RLU) was evaluated for 25 min from leaf discs of three 5-week-old Arabidopsis in response to 50 nM flg22. Results represent average values ± SE of three independent biological replicates each consisting of nine leaf discs (n = 27). Asterisks indicate a significant difference to respective Col-0 controls (mock or flg22-treated) (t-test, *p < 0.05).
Expression levels of the PTI-responsive FLG22-INDUCED RECEPTOR-LIKE 1 (FRK1), an intermediate PTI response (Asai et al., 2002), were also evaluated in CRK overexpression lines after flg22 treatment. Both CRK4 and CRK6 overexpression lines demonstrated a WT level of FRK1 expression before PTI elicitation and at 1 h after flg22 treatment (Supplementary Figure 3), while FRK1 up-regulation was significantly increased 3 h after flg22 treatment indicating potentiation of FRK1 expression upon PTI elicitation (Figure 3). By contrast, overexpression of CRK36 did increase FRK1 mRNA accumulation before and at 1 h after elicitation with flg22 (Figure 3), suggesting a direct effect of this CRK on FRK1 expression.
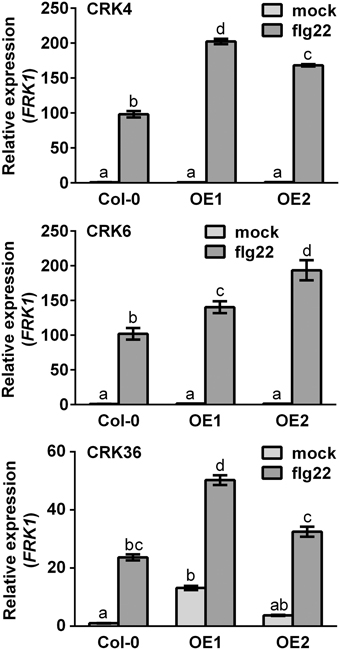
Figure 3. FRK1 gene expression in CRK4, CRK6, and CRK36 overexpression lines. Total RNA from 10-day-old Arabidopsis seedlings was extracted at 1 h (CRK36) or 3 h (CRK4 and CRK6) after treatment with 10 nM flg22. Relative gene expression levels were compared to mock-treated Col-0 WT (defined value of 1) by qRT-PCR analyses. UBQ10 was used for normalization. Results represent average values ± SE of three independent biological replicates each consisting of three plants (n = 9). Different letters represent a significant difference (ANOVA, p < 0.01).
Callose deposition, a typical late PTI response (Gómez-Gómez et al., 1999), was also monitored at 9 hpi with the strain Pst DC3000 hrcC−, a bacterial mutant that cannot repress the PTI response because being defective in delivering type-III effectors (Brooks et al., 2004). Overexpression of CRK4 or CRK6 did not affect PTI-mediated callose deposition (Figures 4A,B). However, bacteria-inoculated, water- and flg22-treated CRK36 overexpression lines accumulated more callose deposits then WT controls (Figure 4C). These observations suggest that CRK36 positively regulates callose deposition while CRK4 and CRK6 do not play a critical role in the modulation of this PTI output.
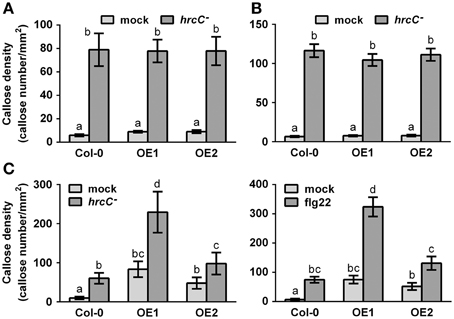
Figure 4. Callose accumulation in CRK4, CRK6, and CRK36 overexpression lines. Callose deposits per mm2 in CRK4 (A), CRK6 (B) and CRK36 (C) overexpression lines were determined in leaf discs of three 5-week-old plants 9 h after infiltration with 108 CFU/mL Pst DC3000 hrcC− (A–C) or 500 nM flg22 (C). Callose deposits were revealed by aniline blue staining. Quantification of callose accumulation was performed using the image J software. Results represent average values ± SE of three independent biological replicates each consisting of 12 leaf discs (n = 36). Different letters represent a significant difference (ANOVA, p < 0.01).
Enhanced Stomatal Immunity in Lines Overexpressing CRK4, CRK6, and CRK36
As part of the PTI response, Arabidopsis plants close stomata that are in contact with bacteria (Melotto et al., 2006; Zeng et al., 2010; Desclos-Theveniau et al., 2012; Singh et al., 2012a). To enter leaves, virulent bacteria such as Pst DC3000 reopen stomata in a coronatine (COR)-dependent manner (Melotto et al., 2006; Zeng et al., 2010). To clarify the possible role of CRK4, CRK6, and CRK36 in stomatal immunity, stomatal apertures of CRK transgenics were evaluated at 1 and 3 hpi with Pst DC3000. Lines overexpressing CRK4 or CRK36 demonstrated a defective Pst DC3000-mediated stomatal reopening at 3 hpi (Figure 5). Over-expression of CRK6 induced stomatal closure in the mock controls and at 3 hpi with Pst DC3000 (Figure 5). These data suggest that CRK4 and CRK36 counteract the COR-dependent reopening of stomata while CRK6 acts as a strong positive regulator of stomatal closure.
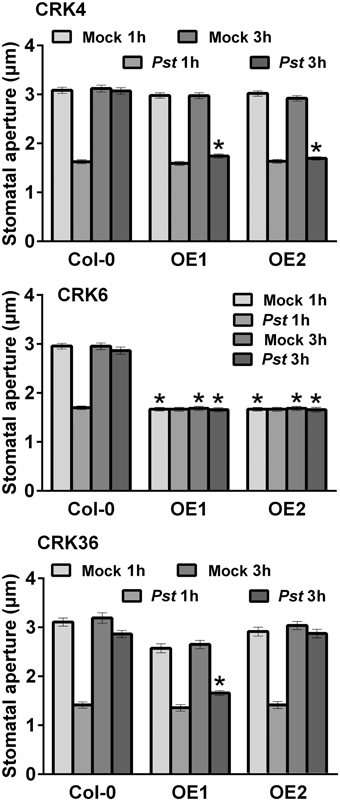
Figure 5. Alteration of stomatal immunity by overexpression of CRK4, CRK6, and CRK36. Stomatal apertures were evaluated in leaf epidermal peels of 5-week-old Arabidopsis exposed to MgSO4 buffer (Mock) or Pst DC3000 (108 CFU/mL) for 1 and 3 h. Results represent average values ± SE of three independent biological replicates each consisting of 100 technical repeats (n = 300). Asterisks represent a significant difference to respective Col-0 WT controls (t-test, *p < 0.001).
Plasma Membrane Localization of CRK4, CRK6, and CRK36
Subcellular localization of CRK4, CRK6, and CRK36 that possess a transmembrane domain (Chen, 2001), was performed by transiently expressing CRKs-GFP fusion protein driven by the cauliflower mosaic virus 35S promoter in Arabidopsis mesophyll protoplasts. The fluorescence signal was mainly localized at the cell periphery with a pattern similar to the plasma membrane marker pm-rk CD3-1007 (Nelson et al., 2007) (Figure 6). By contrast, the control protoplasts expressing GFP alone showed a nuclear/cytoplasmic localization (Figure 6). These observations suggest that like ACRK1 (CRK45) (Tanaka et al., 2012), CRK4, CRK6, and CRK36 are localized at the plasma membrane.
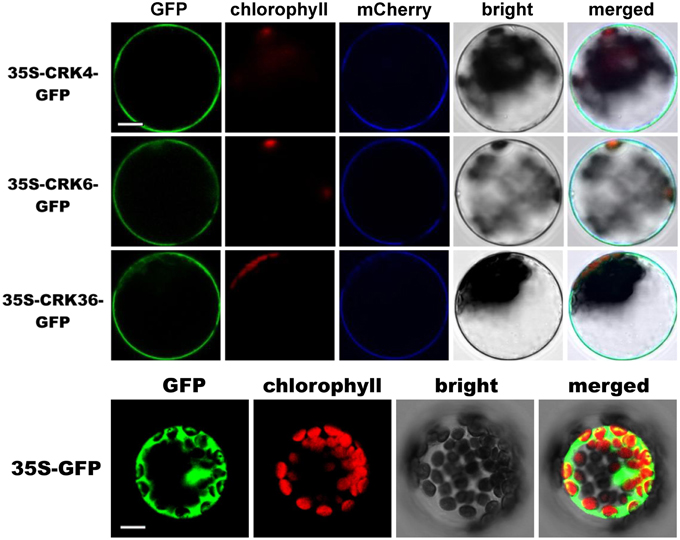
Figure 6. Membrane localization of CRK4, CRK6, and CRK36. Subcellular localization of CRK-GFP fusion proteins in Arabidopsis mesophyll protoplasts. CRK-GFP or GFP alone (negative control) constructs were transiently expressed under the cauliflower mosaic virus 35S promoter and the GFP signal was observed by confocal microscopy 16 h after transfection. The GFP fluorescence (green), the chlorophyll autofluorescence (red), the plasma membrane marker (pm-rk CD3-1007)-mCherry fluorescence localization (blue), the bright-field image, and the combined images are shown. This experiment was repeated three times with similar results. Scale bars represent 10 μm.
CRK4, CRK6, and CRK36 Associate with the PRR FLS2
Since overexpression of CRK4, CRK6, and CRK36 affects the early Arabidopsis PTI response, notably flg22-mediated ROS accumulation (Figure 2), we asked whether these CRKs belong to the PRR complex that recognizes the MAMP flg22. Toward this goal, we monitored CRKs association with FLS2, the PRR that recognizes flg22 (Gómez-Gómez and Boller, 2000; Chinchilla et al., 2007). Interactions were first evaluated by bimolecular fluorescence complementation (BiFC) assays (Walter et al., 2004) in Arabidopsis protoplasts. To demonstrate that our experimental conditions were appropriate, we analyzed the ligand-dependent interaction between BAK1 and FLS2 (Chinchilla et al., 2007; Heese et al., 2007; Roux et al., 2011). As expected, YFP signal was only observed after flg22 treatment (Figure 7A). When testing CRK4, CRK6, and CRK36 interactions with FLS2, the YFP fluorescence was observed in a ligand-independent manner (Figure 7A). The plasma membrane localized RARE-COLD-INDUCIBLE 2B (RCI2B)(Medina et al., 2007) also known as LTI6B (Cutler et al., 2000) was used to evaluate the specificity of CRK4, CRK6, and CRK36 association with FLS2. Importantly, both constructs were functional, as demonstrated by the clear YFP fluorescence observed in Arabidopsis protoplasts transfected with RCI2B-YFPN and RCI2B-YFPC (Figure 7B). However, no YFP fluorescence was observed when testing each CRK interactions with RCI2B suggesting that CRKs association with FLS2 observed by BiFC is specific (Figure 7B). Taken together, BiFC data suggest that CRK4, CRK6, and CRK36 associate with the PRR FLS2 independently of flg22 elicitation.
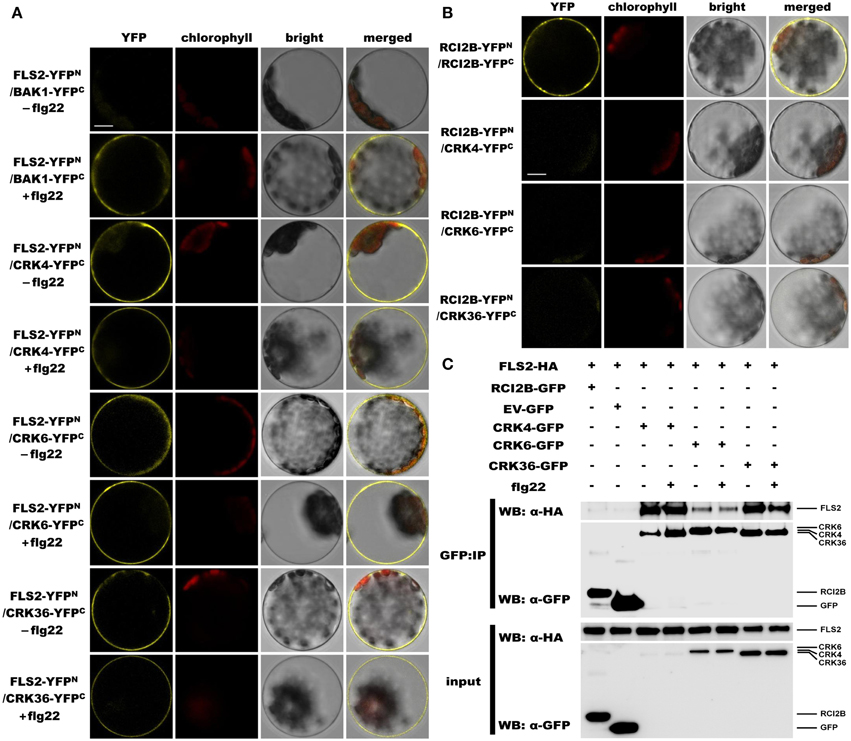
Figure 7. CRK4, CRK6, and CRK36 associate with the PRR FLS2 in a flg22-independent manner. (A) Analysis of associations between FLS2 and CRKs by BiFC assay. Arabidopsis protoplasts were co-transfected with FLS2-YFPN + BAK1-YFPC (positive control) or FLS2-YFPN + CRKs-YFPC plasmids and treated with (+) or without (−) 100 nM flg22 for 20 min. The YFP fluorescence (yellow), chlorophyll autofluorescence (red), bright field and the combined images were visualized under a confocal microscope 16 h after transfection. The scale bar represents 10 μm. (B) Association between RCI2B and CRKs by BiFC assay. Arabidopsis protoplasts were co-transfected with RCI2B-YFPN + RCI2B-YFPC (positive control), or RCI2B-YFPN + CRKs-YFPC plasmids and were observed by confocal microscopy 16 h after transfection. The scale bar represents 10 μm. (C) Analysis of association between FLS2 and CRKs by Co-IP assay. Arabidopsis protoplasts co-expressing RCI2B-GFP + FLS2-HA3, pG103 empty vector (EV-GFP) + FLS2-HA3, or CRKs-GFP + FLS2-HA3 constructs were treated with (+) or without (−) 100 nM flg22 for 10 min. Total protein extracts (input) and IP-proteins were detected using immunoblotting with an α-GFP or α-HA antibody. Experiments in (A–C) were repeated three times with similar results.
The Co-IP approach in Arabidopsis protoplasts was further used to evaluate the in vivo association of CRK4, CRK6, and CRK36 with FLS2. Toward this goal, equal amounts of CRK4, CRK6, and CRK36 were pulled down with GFP-Trap beads from Arabidopsis protoplasts transiently co-expressing CRKs-GFP and HA3 epitope-tagged FLS2 and detection of FLS2-HA3 through anti-HA immunoblotting revealed FLS2 before and after flg22 treatment (Figure 7C). It is relevant to note that the FLS2 signal after CRK6 immunoprecipitation was weaker than after CRK4 and CRK36 immunoprecipitation (Figure 7C). As a negative control, we immunoprecipitated the plasma membrane localized RCI2B protein (Medina et al., 2007) with GFP-trap beads and analyzed the presence of FLS2-HA3 by anti-HA immunoblotting. FLS2 could not be detected (Figure 7C). Furthermore, no signals were detected when FLS2-HA3 was expressed alone (Figure 7C), confirming that FSL2 does not bind aspecifically to anti-GFP magnetic beads. Together, these data suggest that CRK4, CRK6, and CRK36 associate with FLS2 in a ligand-independent manner in Arabidopsis protoplast.
Discussion
Plant PRRs are cell-surface RLKs that trigger the PTI response through recognition of MAMPs to restrict invasion of potentially dangerous microbes (Jones and Dangl, 2006; Pieterse et al., 2009; Zipfel, 2009, 2014). Here, we characterized the function of Arabidopsis CRKs in plant innate immunity by gain-of-function analyses. The CRKs form a RLK subfamily of 44 members that are transcriptionally regulated by pathogens, ROS and SA (Chen et al., 2003, 2004; Acharya et al., 2007; Wrzaczek et al., 2010; Ederli et al., 2011). A recent genome-wide microarray analysis revealed seven CRK genes (CRK4, CRK6, CRK7, CRK13, CRK23, CRK36, and CRK37) that are highly up-regulated in an Arabidopsis line overexpressing the positive regulator of PTI, LecRK-VI.2 (Singh et al., 2012a). To evaluate the role of these CRKs in plant defense, we screened T-DNA insertion mutants for possible altered resistance to the virulent bacterial pathogen Pst DC3000. None of the T-DNA mutants analyzed demonstrated a disease phenotype after Pst DC3000 inoculation. Since CRKs are known to compose a relatively large family, it is likely that CRKs have overlapping functions among individual members (Chen et al., 2003, 2004; Acharya et al., 2007). Therefore, no disease phenotypes in CRK T-DNA insertion mutants may be explained by functional redundancy. Transgenic plants overexpressing CRK4, CRK6, CRK13, CRK23, CRK36, and CRK37 were generated to further evaluate whether these CRKs are involved in Arabidopsis disease resistance to bacteria. Arabidopsis overexpressing CRK4, CRK6, and CRK36 demonstrated an enhanced disease resistance to Pst DC3000. By contrast, CRK13, CRK23, and CRK37 overexpression did not alter Arabidopsis resistance to Pst DC3000. CRK13, CRK23, and CRK37 may not be involved in Arabidopsis resistance to bacteria. Surprisingly, we obtained lines stably overexpressing CRK13 (albeit only lines with relatively low overexpression levels) while a previous work shows that lines overexpressing CRK13 under the 35S promoter are stunted and do not survive to maturity because of lethality (Acharya et al., 2007). High overexpression of Arabidopsis CRK13 through a steroid-inducible Gal4 promoter induces programmed cell death (Acharya et al., 2007). Stable overexpression of CRK13 may thus induce cell death and affects plant development and survival when highly overexpressed or under certain environment conditions. Different overexpression levels or growth conditions such as different light intensities may therefore explain the discrepancy between our results and Acharya et al. (2007) observations. Overexpression of CRK4, CRK5, CRK19, and CRK20 under the control of a steroid-inducible promoter also induces spontaneous cell death (Chen et al., 2003, 2004). By contrast, none of the CRK overexpression lines analyzed in this study demonstrated a spontaneous cell death phenotype (Figure 1; Supplementary Figure 2). Possibly, only lines with low overexpression levels of CRKs involved in the regulation of program cell death were obtained in this study, since constitutive high overexpression may be lethal. For example, we could only obtain CRK4 and CRK6 overexpression lines with very low overexpression levels (Figure 1), suggesting a role for these CRKs in the regulation of cell death. Confirming this assumption, CRK4 is known to regulate program cell death (Chen et al., 2003, 2004). Taken together, these results support the idea that CRK4, CRK6, and CRK36 are potential players in Arabidopsis disease resistance to Pst DC3000.
To further characterize the role of Arabidopsis CRK4, CRK6, and CRK36 in innate immunity, we evaluated different PTI outputs in CRK4, CRK6, and CRK36 overexpression lines. ROS accumulation is considered as an early PTI response by acting as an anti-microbial agent and/or as a secondary messenger that triggers downstream defense responses, including stomatal closure and up-regulation of PTI marker genes (Melotto et al., 2006; Kadota et al., 2014). Overexpression of CRK4, CRK6, and CRK36 primed ROS production in response to flg22 treatment suggesting a role for these CRKs early during the activation of the PTI response. In addition, both up-regulation of FRK1 and stomatal innate immunity were enhanced in lines overexpressing CRK4, CRK6, and CRK36. PTI-responsive gene up-regulation and stomatal immunity are considered as moderately early PTI outputs (Melotto et al., 2006; Singh et al., 2012a), further suggesting that these CRKs play a role at or downstream of MAMP perception. Overexpression of CRK36 induced constitutive up-regulation of the PTI marker gene FRK1 and callose deposition, while FRK1 expression was primed after flg22 treatment and WT callose deposition levels were observed in CRK4 and CRK6 overexpression lines. CRK36 may thus function differently from CRK4 and CRK6 in the PTI signaling cascade or alternatively, higher CRK36 relative overexpression levels may explain this divergent behavior (Figure 1). These observations suggest that like LecRK-VI.2 (Singh et al., 2012a), CRK4, CRK6, and CRK36 positively regulate some outputs of the Arabidopsis PTI response.
Early findings indicate that during pathogen infection, the Arabidopsis PRR FLS2 recognizes bacterial flagellin (or the derived peptide flg22) and induces flg22-triggered immunity (Gómez-Gómez and Boller, 2000). Currently, PRRs are believed to be central components of membrane-located multiprotein complexes that are tightly regulated for timely modulation of diverse PTI outputs (Macho and Zipfel, 2014). In this study, we identified CRK4, CRK6, and CRK36 as potential membrane-localized protein that regulate flg22-mediated PTI. We thus asked whether these CRKs are part of the PRR FLS2 complex. CRK4, CRK6, and CRK36 were found to associate in vivo with FLS2 in a flg22-independent manner when analyzed by BiFC and Co-IP. This observation combined with the fact that overexpression of CRK4, CRK6, and CRK36 enhanced the production of flg22-triggered ROS, an early PTI response, suggest that these three CRKs act early during flg22-mediated PTI signaling, possibly at the PRR FLS2 complex. Protein activation by phosphorylation to regulate diverse signaling pathways is common in plants (Hardie, 1999; Nam and Li, 2002; Lu et al., 2010). Typically, BAK1 phosphorylates BIK1 after association with FLS2 and then BIK1 trans-phosphorylates FLS2 and BAK1 to activate PTI signaling (Lu et al., 2010). Since, CRK6 and CRK36 are active kinases (Tanaka et al., 2012; Idanheimo et al., 2014), they may participate in the phosphorylation cascade involved in the regulation of the PRR FSL2 complex. Since CRK proteins possess two copies of the C-X8-C-X2-C motif (Chen, 2001), CRKs could form inter-/intra-molecular disulfide bonds to strengthen the structural stability of the PRR FLS2 complex, as observed for the anti-fungal protein ginkbilobin-2 that maintains the structural stability of Gnk2 through its C-X8-C-X2-C motif by forming three intra-molecular disulfide bridges (Miyakawa et al., 2009). Collectively, this work suggests that CRK4, CRK6, and CRK36 overexpression enhances Arabidopsis resistance to Pst DC3000 bacteria through a positive role in the PTI response by association with the PRR FLS2. Further studies are necessary to investigate mechanistically CRK4, CRK6, and CRK36 functions at PRR complexes.
Author Contributions
LZ and YY designed the experiments. YY, YC, PH, and JH performed the experiments. LZ and YY analyzed the results and wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the ABRC for providing seeds and Barbara Kunkel for the pathogens. We acknowledge the staff of Technology Commons, College of Life Science, National Taiwan University for their help in microscopy and for qRT-PCR equipment. This work was supported by the National Science Council of Taiwan grant 102-2628-B-002-011-MY3 (to LZ) and the Frontier and Innovative Research grant of the National Taiwan University code numbers 101R7846 and 102R7846 (to LZ).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00322/abstract
References
Acharya, B. R., Raina, S., Maqbool, S. B., Jagadeeswaran, G., Mosher, S. L., Appel, H. M., et al. (2007). Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J. 50, 488–499. doi: 10.1111/j.1365-313X.2007.03064.x
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W. L., Gómez-Gómez, L., et al. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. doi: 10.1038/415977a
Bouwmeester, K., De Sain, M., Weide, R., Gouget, A., Klamer, S., Canut, H., et al. (2011). The Lectin Receptor Kinase LecRK-I.9 is a novel phytophthora resistance component and a potential host target for a RXLR effector. PLoS Pathog. 7:e1001327. doi: 10.1371/journal.ppat.1001327
Bouwmeester, K., and Govers, F. (2009). Arabidopsis L-type lectin receptor kinases: phylogeny, classification, and expression profiles. J. Exp. Bot. 60, 4383–4396. doi: 10.1093/jxb/erp277
Brooks, D. M., Hernández-Guzmán, G., Kloek, A. P., Alarcón-Chaidez, F., Sreedharan, A., Rangaswamy, V., et al. (2004). Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 17, 162–174. doi: 10.1094/MPMI.2004.17.2.162
Chen, C. W., Panzeri, D., Yeh, Y. H., Kadota, Y., Huang, P. Y., Tao, C. N., et al. (2014). The Arabidopsis malectin-like leucine-rich repeat receptor-like kinase IOS1 associates with the pattern recognition receptors FLS2 and EFR and is critical for priming of pattern-triggered immunity. Plant Cell 26, 3201–3219. doi: 10.1105/tpc.114.125682
Chen, K., Du, L., and Chen, Z. (2003). Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol. Biol. 53, 61–74. doi: 10.1023/B:PLAN.0000009265.72567.58
Chen, K., Fan, B., Du, L., and Chen, Z. (2004). Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Mol. Biol. 56, 271–283. doi: 10.1007/s11103-004-3381-2
Chen, Z. X. (2001). A superfamily of proteins with novel cysteine-rich repeats. Plant Physiol. 126, 473–476. doi: 10.1104/pp.126.2.473
Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nurnberger, T., Jones, J. D. G., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500. doi: 10.1038/nature05999
Choi, J., Tanaka, K., Cao, Y., Qi, Y., Qiu, J., Liang, Y., et al. (2014). Identification of a plant receptor for extracellular ATP. Science 343, 290–294. doi: 10.1126/science.343.6168.290
Clark, S. E., Williams, R. W., and Meyerowitz, E. M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. doi: 10.1016/S0092-8674(00)80239-1
Cutler, S. R., Ehrhardt, D. W., Griffitts, J. S., and Somerville, C. R. (2000). Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. U.S.A. 97, 3718–3723. doi: 10.1073/pnas.97.7.3718
Desclos-Theveniau, M., Arnaud, D., Huang, T. Y., Lin, G. J., Chen, W. Y., Lin, Y. C., et al. (2012). The Arabidopsis lectin receptor kinase LecRK-V.5 represses stomatal immunity induced by Pseudomonas syringae pv. tomato DC3000. PLoS Pathog. 8:e1002513. doi: 10.1371/journal.ppat.1002513
De Smet, I., Voss, U., Jürgens, G., and Beeckman, T. (2009). Receptor-like kinases shape the plant. Nat. Cell Biol. 11, 1166–1173. doi: 10.1038/ncb1009-1166
Dievart, A., Dalal, M., Tax, F. E., Lacey, A. D., Huttly, A., Li, J. M., et al. (2003). CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell 15, 1198–1211. doi: 10.1105/tpc.010504
Ederli, L., Madeo, L., Calderini, O., Gehring, C., Moretti, C., Buonaurio, R., et al. (2011). The Arabidopsis thaliana cysteine-rich receptor-like kinase CRK20 modulates host responses to Pseudomonas syringae pv. tomato DC3000 infection. J. Plant Physiol. 168, 1784–1794. doi: 10.1016/j.jplph.2011.05.018
Gómez-Gómez, L., and Boller, T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. doi: 10.1016/S1097-2765(00)80265-8
Gómez-Gómez, L., Felix, G., and Boller, T. (1999). A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 18, 277–284. doi: 10.1046/j.1365-313X.1999.00451.x
Halter, T., Imkampe, J., Mazzotta, S., Wierzba, M., Postel, S., Bucherl, C., et al. (2014). The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol. 24, 134–143. doi: 10.1016/j.cub.2013.11.047
Hardie, D. G. (1999). Plant protein serine threonine kinases: classification and functions. Annu. Rev. Plant Physiol. 50, 97–131. doi: 10.1146/annurev.arplant.50.1.97
Heese, A., Hann, D. R., Gimenez-Ibanez, S., Jones, A. M. E., He, K., Li, J., et al. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. U.S.A. 104, 12217–12222. doi: 10.1073/pnas.0705306104
Huang, P. Y., Yeh, Y. H., Liu, A. C., Cheng, C. P., and Zimmerli, L. (2014). The Arabidopsis LecRK-VI.2 associates with the pattern-recognition receptor FLS2 and primes Nicotiana benthamiana pattern-triggered immunity. Plant J. 79, 243–255. doi: 10.1111/tpj.12557
Huang, T. Y., Desclos-Theveniau, M., Chien, C. T., and Zimmerli, L. (2012). Arabidopsis thaliana transgenics overexpressing IBR3 show enhanced susceptibility to the bacterium Pseudomonas syringae. Plant Biol. 15, 832–840. doi: 10.1111/j.1438-8677.2012.00685.x
Idanheimo, N., Gauthier, A., Salojarvi, J., Siligato, R., Brosche, M., Kollist, H., et al. (2014). The Arabidopsis thaliana cysteine-rich receptor-like kinases CRK6 and CRK7 protect against apoplastic oxidative stress. Biochem. Biophys. Res. Commun. 445, 457–462. doi: 10.1016/j.bbrc.2014.02.013
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kacperska, A. (2004). Sensor types in signal transduction pathways in plant cells responding to abiotic stressors: do they depend on stress intensity? Physiol. Plant. 122, 159–168. doi: 10.1111/j.0031-9317.2004.00388.x
Kadota, Y., Sklenar, J., Derbyshire, P., Stransfeld, L., Asai, S., Ntoukakis, V., et al. (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54, 43–55. doi: 10.1016/j.molcel.2014.02.021
Kobe, B., and Kajava, A. V. (2001). The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11, 725–732. doi: 10.1016/S0959-440X(01)00266-4
Krol, E., Mentzel, T., Chinchilla, D., Boller, T., Felix, G., Kemmerling, B., et al. (2010). Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J. Biol. Chem. 285, 13471–13479. doi: 10.1074/jbc.M109.097394
Lu, D. P., Wu, S. J., Gao, X. Q., Zhang, Y. L., Shan, L. B., and He, P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. P Natl Acad Sci U.S.A. 107, 496–501. doi: 10.1073/pnas.0909705107
Macho, A. P., and Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272. doi: 10.1016/j.molcel.2014.03.028
Martinez-Trujillo, M., Limones-Briones, V., Cabrera-Ponce, J. L., and Herrera-Estrella, L. (2004). Improving transformation efficiency of Arabidopsis thaliana by modifying the floral dip method. Plant Mol. Biol. Rep. 22, 63–70. doi: 10.1007/BF02773350
Medina, J., Ballesteros, M. L., and Salinas, J. (2007). Phylogenetic and functional analysis of Arabidopsis RCI2 genes. J. Exp. Bot. 58, 4333–4346. doi: 10.1093/jxb/erm285
Melotto, M., Underwood, W., Koczan, J., Nomura, K., and He, S. Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126, 969–980. doi: 10.1016/j.cell.2006.06.054
Miyakawa, T., Miyazono, K., Sawano, Y., Hatano, K., and Tanokura, M. (2009). Crystal structure of ginkbilobin-2 with homology to the extracellular domain of plant cysteine-rich receptor-like kinases. Proteins 77, 247–251. doi: 10.1002/prot.22494
Monaghan, J., Matschi, S., Shorinola, O., Rovenich, H., Matei, A., Segonzac, C., et al. (2014). The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16, 605–615. doi: 10.1016/j.chom.2014.10.007
Nam, K. H., and Li, J. M. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. doi: 10.1016/S0092-8674(02)00814-0
Nelson, B. K., Cai, X., and Nebenfuhr, A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x
Ogawa, M. (2008). Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319, 294. doi: 10.1126/science.1150083
Pieterse, C. M. J., Leon-Reyes, A., Van Der Ent, S., and Van Wees, S. C. M. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. doi: 10.1038/nchembio.164
Roux, M., Schwessinger, B., Albrecht, C., Chinchilla, D., Jones, A., Holton, N., et al. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23, 2440–2455. doi: 10.1105/tpc.111.084301
Sawano, Y., Miyakawa, T., Yamazaki, H., Tanokura, M., and Hatano, K. (2007). Purification, characterization, and molecular gene cloning of an antifungal protein from Ginkgo biloba seeds. Biol. Chem. 388, 273–280. doi: 10.1515/BC.2007.030
Shiu, S. H., and Bleecker, A. B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. U.S.A. 98, 10763–10768. doi: 10.1073/pnas.181141598
Singh, P., Chien, C. C., Mishra, S., Tsai, C. H., and Zimmerli, L. (2012b). The Arabidopsis LECTIN RECEPTOR KINASE-VI.2 is a functional protein kinase and is dispensable for basal resistance to Botrytis cinerea. Plant Signal. Behav. 8:e22611. doi: 10.4161/psb.22611
Singh, P., Kuo, Y. C., Mishra, S., Tsai, C. H., Chien, C. C., Chen, C. W., et al. (2012a). The Lectin Receptor Kinase-VI.2 is required for priming and positively regulates Arabidopsis pattern-triggered immunity. Plant Cell 24, 1256–1270. doi: 10.1105/tpc.112.095778
Singh, P., and Zimmerli, L. (2013). Lectin receptor kinases in plant innate immunity. Front. Plant Sci. 7:124. doi: 10.3389/fpls.2013.00124
Tanaka, H., Osakabe, Y., Katsura, S., Mizuno, S., Maruyama, K., Kusakabe, K., et al. (2012). Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. Plant J. 70, 599–613. doi: 10.1111/j.1365-313X.2012.04901.x
Walker, J. C. (1994). Structure and function of the receptor-like protein-kinases of higher-plants. Plant Mol. Biol. 26, 1599–1609. doi: 10.1007/BF00016492
Walter, M., Chaban, C., Schütze, K., Batistic, O., Weckermann, K., Näke, C., et al. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438. doi: 10.1111/j.1365-313X.2004.02219.x
Wang, Z. Y., Seto, H., Fujioka, S., Yoshida, S., and Chory, J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383. doi: 10.1038/35066597
Wrzaczek, M., Brosche, M., Salojarvi, J., Kangasjarvi, S., Idanheimo, N., Mersmann, S., et al. (2010). Transcriptional regulation of the CRK/DUF26 group of Receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 10:95. doi: 10.1186/1471-2229-10-95
Yamaguchi, Y., Huffaker, A., Bryan, A. C., Tax, F. E., and Ryan, C. A. (2010). PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22, 508–522. doi: 10.1105/tpc.109.068874
Yang, K., Rong, W., Qi, L., Li, J., Wei, X., and Zhang, Z. (2013). Isolation and characterization of a novel wheat cysteine-rich receptor-like kinase gene induced by Rhizoctonia cerealis. Sci. Rep. 3, 3021. doi: 10.1038/srep03021
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zeng, W., Melotto, M., and He, S. Y. (2010). Plant stomata: a checkpoint of host immunity and pathogen virulence. Curr. Opin. Biotechnol. 21, 599–603. doi: 10.1016/j.copbio.2010.05.006
Zhang, J., Shao, F., Cui, H., Chen, L. J., Li, H. T., Zou, Y., et al. (2007). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-Induced immunity in plants. Cell Host Microbe 1, 175–185. doi: 10.1016/j.chom.2007.03.006
Zhang, X. J., Han, X. M., Shi, R., Yang, G. Y., Qi, L. W., Wang, R. G., et al. (2013b). Arabidopsis cysteine-rich receptor-like kinase 45 positively regulates disease resistance to Pseudomonas syringae. Plant Physiol. Biochem. 73, 383–391. doi: 10.1016/j.plaphy.2013.10.024
Zhang, X. J., Yang, G. Y., Shi, R., Han, X. M., Qi, L. W., Wang, R. G., et al. (2013a). Arabidopsis cysteine-rich receptor-like kinase 45 functions in the responses to abscisic acid and abiotic stresses. Plant Physiol. Biochem. 67, 189–198. doi: 10.1016/j.plaphy.2013.03.013
Zimmerli, L., Jakab, C., Metraux, J. P., and Mauch-Mani, B. (2000). Potentiation of pathogen-specific defense mechanisms in Arabidopsis by beta-aminobutyric acid. Proc. Natl. Acad. Sci. U.S.A. 97, 12920–12925. doi: 10.1073/pnas.230416897
Zipfel, C. (2009). Early molecular events in PAMP-triggered immunity. Curr. Opin. Plant Biol. 12, 414–420. doi: 10.1016/j.pbi.2009.06.003
Zipfel, C. (2014). Plant pattern-recognition receptors. Trends Immunol. 35, 345–351. doi: 10.1016/j.it.2014.05.004
Keywords: plant, Arabidopsis thaliana, cysteine-rich receptor-like kinase, innate immunity, pattern-triggered immunity, bacteria, flagellin, stomatal immunity
Citation: Yeh Y-H, Chang Y-H, Huang P-Y, Huang J-B and Zimmerli L (2015) Enhanced Arabidopsis pattern-triggered immunity by overexpression of cysteine-rich receptor-like kinases. Front. Plant Sci. 6:322. doi: 10.3389/fpls.2015.00322
Received: 19 March 2015; Accepted: 23 April 2015;
Published: 12 May 2015.
Edited by:
Simone Ferrari, Sapienza Università di Roma, ItalyCopyright © 2015 Yeh, Chang, Huang, Huang and Zimmerli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laurent Zimmerli, Institute of Plant Biology, National Taiwan University, No. 1, Sec. 4, Roosevelt Road, Taipei 106, Taiwan, lauzim2@ntu.edu.tw
 Yu-Hung Yeh
Yu-Hung Yeh Yu-Hsien Chang
Yu-Hsien Chang Pin-Yao Huang
Pin-Yao Huang Jing-Bo Huang
Jing-Bo Huang Laurent Zimmerli
Laurent Zimmerli