- 1State Key Laboratory of Biocontrol, Guangdong Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou, China
- 2Institute of Fruit Tree Research, Guangdong Academy of Agricultural Sciences, Guangzhou, China
Ceramidases hydrolyze ceramide into sphingosine and fatty acids and, although ceramidases function as key regulators of sphingolipid homeostasis in mammals, their roles in plants remain largely unknown. Here, we characterized the Arabidopsis thaliana ceramidase AtNCER1, a homolog of human neutral ceramidase. AtNCER1 localizes predominantly on the endoplasmic reticulum. The ncer1 T-DNA insertion mutants had no visible phenotype, but accumulated hydroxyceramides, and showed increased sensitivity to oxidative stress induced by methyl viologen. Plants over-expressing AtNCER1 showed increased tolerance to oxidative stress. These data indicate that the Arabidopsis neutral ceramidase affects sphingolipid homeostasis and oxidative stress responses.
Introduction
An important component of eukaryotic membranes, sphingolipids also function as key signaling molecules to regulate cell differentiation and death (Dickson and Lester, 2002; Hannun and Obeid, 2002; Spiegel and Milstien, 2003; Futerman and Hannun, 2004; Pyne et al., 2009). Genetic and biochemical analyses have identified several enzymes in the sphingolipid pathway. For example, accelerated cell death 5 (acd5) a ceramide kinase deficient mutant (Liang et al., 2003), accumulates ceramide at late developmental stages and shows an early defect in restricting Botrytis germination and growth (Bi et al., 2014). Mutation of inositolphosphorylceramide synthase (IPCS), an enzyme that converts ceramide to inositolphosphorylceramide, caused increased accumulation of salicylic acid and ceramide, and enhanced resistance to biotrophic pathogens (Wang et al., 2008). The ceramide synthases encoded by LAG ONE HOMOLOGUE (LOH) 1, LOH2, and LOH3 are responsible for the synthesis of ceramides and have different substrate preferences (Markham et al., 2011; Ternes et al., 2011).
Ceramidases degrade ceramides to sphingosine and fatty acids and regulate sphingolipid homeostasis in mammalian and yeast cells. Ceramidases can be divided into three categories: acidic, neutral, and alkaline ceramidases, based on their different catalytic pH values (Mao and Obeid, 2002). Acidic ceramidase mainly occurs in the lysosome, where it is responsible for the decomposition of ceramide (Monjusho et al., 2003). This enzyme has been identified and its full-length cDNA has been cloned from human (Koch et al., 1996) and mice (Li et al., 1998). A deficiency in human acidic ceramidase results in lysosomal storage disorders, commonly known as fat granulomatous disease (Koch et al., 1996). Neutral and alkaline ceramidases affect signal transduction and cell metabolism (Ohlsson et al., 2007). The alkaline ceramidase identified from Saccharomyces cerevisiae has a dual activity, catalyzing both hydrolysis and synthesis of yeast ceramide (Mao et al., 2000). Humans also have three alkaline ceramidases (Mao et al., 2001; Xu et al., 2006; Sun et al., 2007). Human alkaline ceramidase 2 can inhibit the synthesis of the subunit of bata1-integrin, thus affecting cell adhesion (Sun et al., 2009). In mammals, neutral ceramidase has been isolated from different tissues, including mouse liver (Tani et al., 2000), kidney (Mitsutake et al., 2001), brain (El Bawab et al., 1999), and intestine (Olsson et al., 2004), and from human intestinal tissue (Ohlsson et al., 2007). In other organisms, neutral ceramidase was also isolated from Dictyostelium discoideum (Monjusho et al., 2003), Drosophila melanogaster (Yoshimura et al., 2002), and zebra fish (Yoshimura et al., 2004). Although mice have no obvious phenotypic defects when the neutral ceramidase is mutated, they have defects in lipid metabolism in the intestine and increased susceptibility to intestinal cancer (Schmelz et al., 2001; Symolon et al., 2004), indicating that the neutral ceramidase affects ceramide and sphingosine levels in the gastrointestinal tract by regulating dietary sphingolipid metabolism (Kono et al., 2006).
In plants, no homologs of acid ceramidase have been identified, but the Arabidopsis thaliana genome encodes one alkaline ceramidase AtACER (At4G22330) and three neutral ceramidases, AtNCER1 (At1g07380), AtNCER2 (At2g38010), and AtNCER3 (At5g58980). A neutral ceramidase (OsCDase) was cloned from rice (Oryza sativa sp. japonica cv. Nipponbare). OsCDase localizes to the endoplasmic reticulum (ER) and Golgi and appears to use ceramide instead of phytoceramide as a substrate in vitro (Pata et al., 2008). Expression of Ta-CDase, a wheat neutral ceramidase, could be induced by fungal infection (Yu et al., 2011). However, there is no further investigation about the role of neutral ceramidase in regulation of sphingolipid homeostasis.
Here, we characterized Arabidopsis ceramidase 1 (AtNCER1), a homolog of human neutral ceramidase. Our results demonstrate that AtNCER1 affects sphingolipid homeostasis and plays a role in response to oxidative stress.
Materials and Methods
Mutant Analysis
Arabidopsis thaliana wild-type plants Columbia (Col-0) were used in this study and grown in a 16 h light/8 h dark cycle, as described previously (Bi et al., 2014). T-DNA insertion mutant seeds (SALK_054725, designated ncer1; SALK_020682, designated ncer3) were ordered from ABRC seed stock center (www.arabidopsis.org/). The primers used were: ncer1-LP, 5′-TCTCCACCAGTGTAAACGTCC-3′, ncer1-RP, 5′-TTTTCATTCTCAGCGTTCCTG-3′, ncer3-LP: 5′-TCTATCAGCTCCAGCAAATGG-3′, ncer3-RP: 5′-AGTAACGAGGATGCCATTTCC-3′, LBb1.3: 5′-ATTTTGCCGATTTCGGAAC-3′. The double mutant ncer1 ncer3 was also characterized.
Plasmid Construction and Subcellular Localization
To generate the GFP: AtNCER1 construct, we amplified the full-length AtNCER1 open reading frame from cDNA without the stop codon, using the following primer pair, which includes an AscI restriction site: F (5′-TTGGCGCGCCATGGAGCTATCTCTAGTCAGATTAT-3′) and R (5′-TTGGCGCGCCGTGTTACAACGAAAGCACTAGAA-3′). After AscI digestion, The PCR product was subcloned in frame with EGFP in a vector derived from pUC18. The P35S:eGFP:NCER1 plasmid was co-expressed with a plasmid expressing an ER marker (CD3-960), by transient expression in protoplasts. Protoplast transfection was performed as described previously (Bi et al., 2014). The transfected protoplasts were cultured under weak light (about 300 lux) for 16–24 h at room temperature and observed by confocal microscopy (LSM-780, Carl Zeiss). The excitation/emission wavelengths were: 488 nm/500–530 nm for green fluorescent protein (GFP), 561 nm/580–630 nm for mCherry, and 488 nm/650–750 nm for chlorophyll.
Methyl Viologen Treatment
Surface-sterilized seeds were sown on ½ MS and grown vertically under 16 h light /8 h dark conditions in an incubator for 7 days and then transferred to half-strength Murashige and Skoog (½ MS) medium containing 1 μM methyl viologen (MV). Samples were collected at 7 days after treatment and extracted and analyzed for sphingolipid profiling.
Sphingolipid Analysis
Measurement of sphingolipids was performed and analyzed by Shimadzu UFLC-XR (Shimadzu, Japan) coupled with a hybrid quadrupole time-of-flight mass spectrometer (AB SCIEX Triple TOF 5600+, Foster City, CA, USA) using Phenomenex Luna C8 column (150 mm × 2.0 mm, 3 μm). Briefly, 30 mg of lyophilized samples was homogenized. The internal standards (C17 base D-erythro-sphingosine and C12-Ceramide) were added and extracted with the isopropanol/hexane/water (55:20:25 v/v/v) and incubated at 60°C for 15 min. After centrifugation, the supernatants were dried and de-esterified in methylamine in ethanol/water (70:30 v/v) as described previously (Wu et al., 2015). The sphingolipid species were analyzed using the software Multiquant (AB SCIEX).
Quantitative RT-PCR Analysis
Total RNA was extracted with E.Z.N.A. plant RNA kit (R6827-01, Omega Bio-tek). For each sample, 1 μg RNA was reverse transcribed into cDNA using PrimeScript® RT reagent kit (TAKARA, DRR047A). Real time PCR was performed with SYBR Premix Ex Taq || kit (Takara, RR820L) according to the manufacturer’s instructions, and quantitatively analyzed by StepOnePlusTM real-time PCR systems (AB SCIEX). ACT2 expression level was used as the internal control. All experiments were repeated at least three times. We used the 2−ΔΔCT method (Livak and Schmittgen, 2001) to determine the relative expression level of target genes according to the expression level of ACT2.
Results
Neutral Ceramidase Mutant and Overexpressing Plants Show Normal Growth and Development
To examine neutral ceramidase function, we first characterized the effect of loss or increase of AtNCER function. For loss of AtNCER function, we examined an AtNCER1 mutant (designated ncer1). The ncer1 mutant has a T-DNA insertion in the second exon of AtNCER1 (Figure 1A). To increase AtNCER function, we also generated transgenic plants with a 35S:AtNCER construct (NCER-OX). Both ncer1 and NCER-OX plants showed no apparent developmental differences from wild type (Figure 1B). However, the expression level of AtNCER1 reduced by 80% in ncer1 mutants and increased 12-fold in NCER-OX plants, when compared to wild-type plants (Figure 1C).
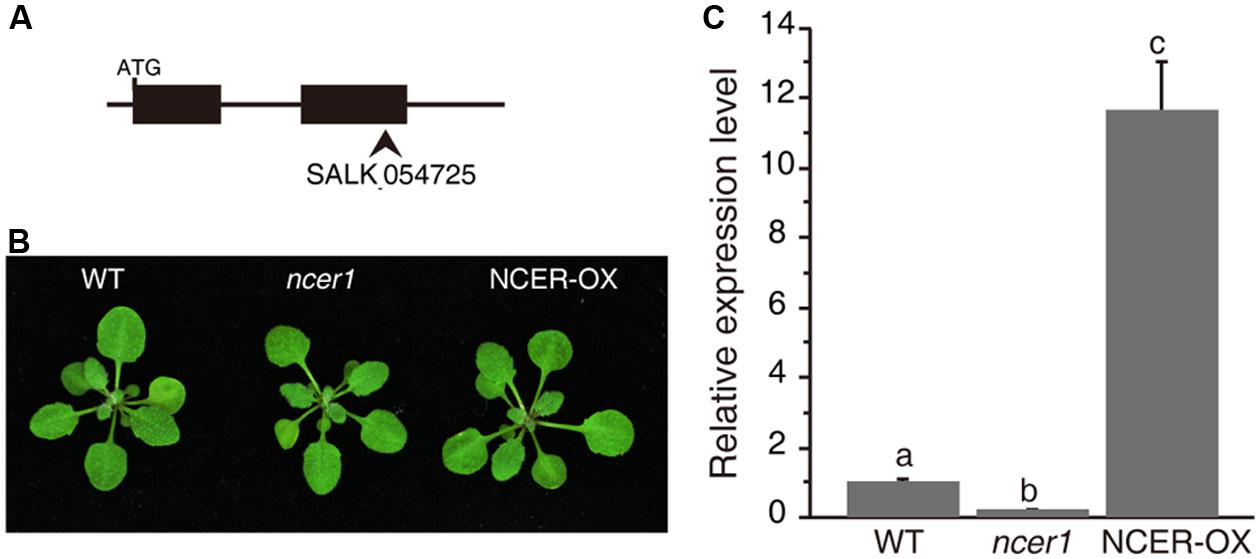
FIGURE 1. Phenotypes of the neutral ceramidase mutants. (A) Gene structure of AtNCER1. Rectangles show exons and the arrowhead indicates the T-DNA insertion site. (B) Phenotype of 3-week-old wild type, ncer1, and AtNCER1 over-expression line (NCER-OX). (C) Real time PCR qualification of AtNCER1 transcript levels in 3-week-old ncer1 and NCER-OX plants. ACT2 was used as the internal control. Gene expression values are presented relative to average wild-type levels, and error bars indicate SE from three technical replicates. This experiment was repeated three times with similar results. Data sets marked with different letters indicate significant differences assessed by a Student’s Newman–Keulst test (p < 0.05).
AtNCER1 Localizes on the ER
To examine the expression of AtNCER1, we used real-time PCR to quantify its transcript levels in flower, rosette leaf, cauline leaf, stem, silique, and root. AtNCER1 was expressed in all detected tissues, with relatively low expression in flowers (Figure 2A).
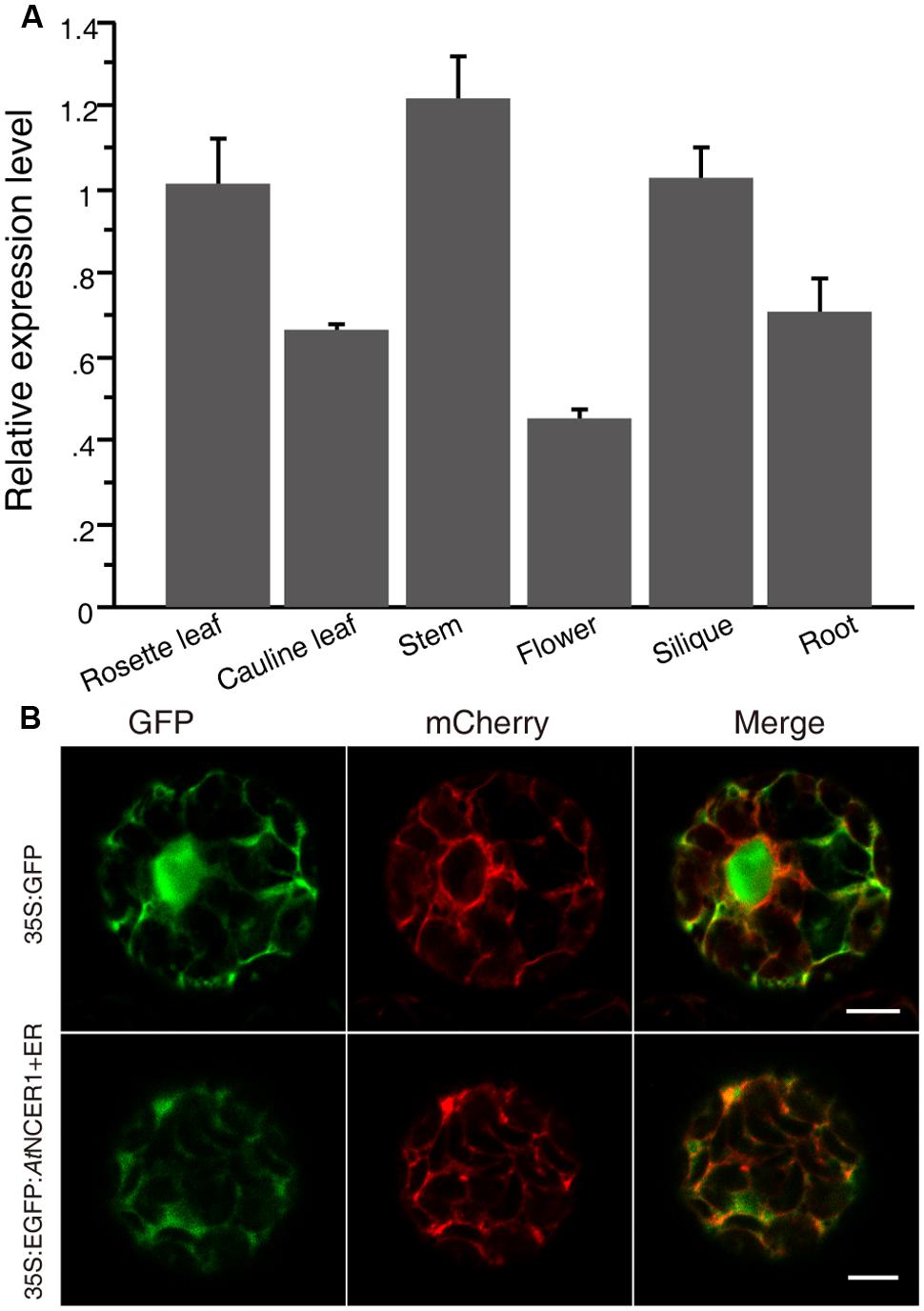
FIGURE 2. Expression of AtNCER and subcellular localization of AtNCER. (A) Expression of AtNCER1 in different tissues. Reverse transcription quantitative PCR was used to measure the relative transcript levels of AtNCER1 in the indicated tissues. The transcript level of ACT2 was used as the internal control. Gene expression values are presented relative to the rosette average leaf level, which was set as 1, and error bars indicate SE. (B) Subcellular localization of AtNCER1. The fusion construct 35S:eGFP:AtNCER1 was co-expressed with the endoplasmic reticulum (ER) mCherry marker (TAIR, CD3-960) by transient expression in protoplasts. The 35S:GFP construct and the ER mCherry marker was transformed as a control. The images were photographed by confocal microscopy after 16 h incubation. The experiments were repeated at least three times with similar results. Bar = 5 μm.
To investigate the subcellular localization of NCER1, we constructed a protein fusion to GFP, by constructing a 35S:eGFP:AtNCER1 expression cassette for transient expression in protoplasts with ER-mCherry as a marker. We observed that the GFP fluorescence co-localized with the ER marker, indicating that AtNCER1 is localized on the ER (Figure 2B).
Neutral Ceramidase Mutants have Increased Sensitivity to C2-Ceramide Induced Cell Death
C2-ceramide is a synthetic short-chain ceramide that can permeate cell membrane and induce cell death (Liang et al., 2003). Ceramidases catalyze hydrolysis of ceramides to generate sphingosine (Mao and Obeid, 2002; Wu et al., 2015). To investigate the sensitivity of ncer1 mutants to ceramide treatment, we isolated protoplasts from wild-type and ncer1 plants and treated them with 50 μM C2-ceramide. After 24 h C2 treatment, the viability of protoplasts in ncer1 remained at 80%, and showed no significant difference from wild type (Figure 3A). This trend was maintained to 48 h when the viability of both wild-type and ncer1 protoplasts decreased to about 40% (Figure 3B).
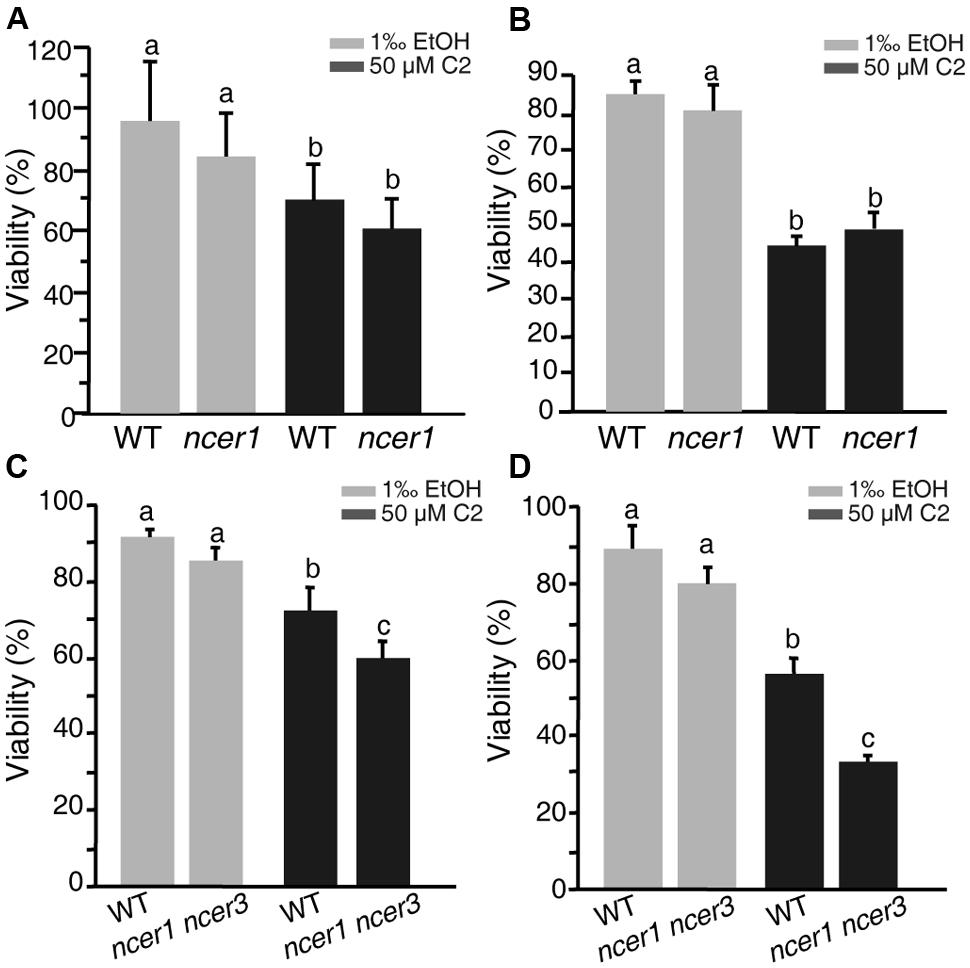
FIGURE 3. C2-ceramide treatments. Protoplasts isolated from 3-week-old wild-type, ncer1 mutant, and ncer1 ncer3 double mutant plants were treated with 50 μM C2-ceramide for 24 h (A,C) or 48 h (B,D). One thousandth of ethanol was used as the solvent control. Protoplast viability was measured by staining with fluorescein diacetate. Letters indicate statistically different values using a Student’s Newman–Keulst test (p < 0.05). For each treatment, at least 500 protoplasts were counted. Error bars indicate SD from three independent experiments.
Since Arabidopsis has three ceramidase-like genes, AtNCER1, AtNCER2, and AtNCER3, we therefore identified a T-DNA insertion mutant of AtNCER3 (designated ncer3) and crossed ncer1 and ncer3 plants to obtain double mutants. At 24 h, the viability of protoplasts from the double mutants significantly decreased when compared to wild type (Figure 3C). At 48 h, double mutant protoplasts showed clear sensitivity to ceramide treatment, compared with wild type (Figure 3D). These results indicate that AtNCER1 and AtNCER3 may function redundantly in Arabidopsis.
Alteration of Sphingolipids in ncer1 Mutants
To determine whether the sphingolipid profile changed in ncer1, we compared the sphingolipid contents of ncer1 and NCER-OX with wild type. We found that the ncer1 mutants showed significant increases in hydroxyceramides, but not ceramides, and no obvious changes in other sphingolipids, including the long chain bases (LCB), ceramides, and glucosylceramides (Figure 4A). Comparing hydroxyceramide with LCB moieties, we found a higher level of t18:0 and t18:1 hydroxyceramides in ncer1 than in wild type (Figure 4B). Comparing hydroxyceramides with different length fatty acid moieties, both hydroxyceramide-containing long chain (C16) and very long chain fatty acids (C24 and C26) accumulated (Figure 4C). No significant differences of sphingolipid contents were found between wild-type and NCER-OX plants (Figure 4). These results indicated that AtNCER1 may use hydroxyceramides other than ceramide for its substrate.
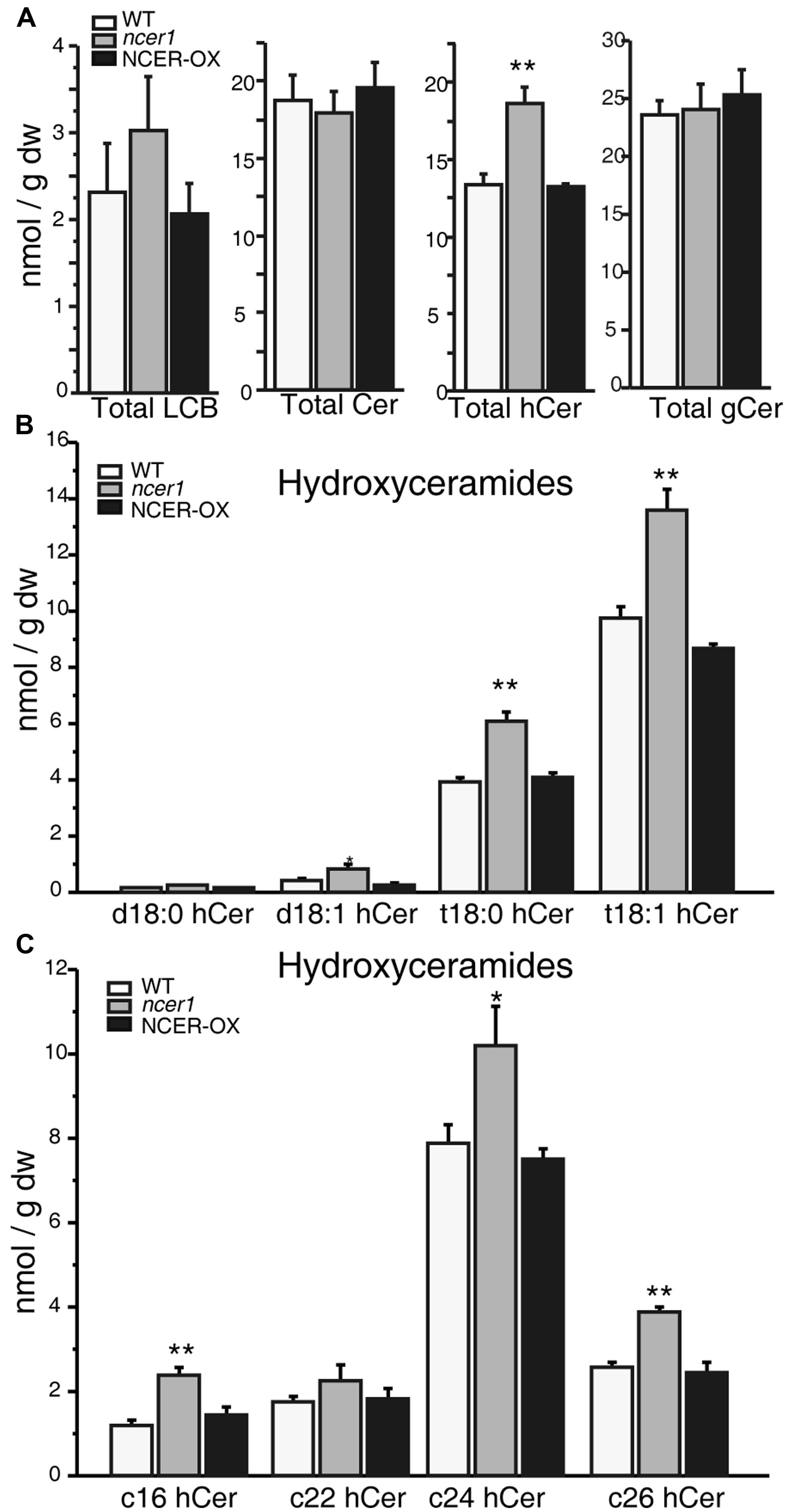
FIGURE 4. Sphingolipid analysis in ncer1 mutant plants. Sphingolipids were extracted from 21-day-old seedlings as described in Section “Materials and Methods.” The contents of free long chain bases (LCBs), ceramides (Cer), hydroxyceramides (hCer), and glucosylceramides (gCer) were quantified. The experiment was repeated at least three times using independent samples. Asterisks show a significant difference from the wild type using Student’s t-test (*p < 0.05, **p < 0.01). (A) Total LCBs, ceramides, hydroxyceramides, and glucosylceramides in the indicated plants. (B) Comparison of hydroxyceramides with the LCB moieties in the indicated plants. (C) Comparison of hydroxyceramides with the length of fatty acid moieties in the indicated plants.
Sensitivity of ncer1 to Oxidative Stress Induced by MV Treatment
Reactive oxygen species (ROS) play an important role in the programmed cell death induced by ceramides (Bi et al., 2014). To examine oxidative stress tolerance in wild-type and mutant plants, we treated detached leaves with MV. After 3 days of treatment, we found significantly more leaves from ncer1 plants became bleached in both 1 and 3 μM MV treatments, compared with wild-type, indicating that the ncer1 mutants are more sensitive to oxidative stress (Figure 5A). We also performed a plate assay, in which surface-sterilized seeds were sown on 1/2x MS agar plates containing 1 μM MV and incubated for 10 days (Figure 5B). We found that about 80% of ncer1 mutant seedlings lost their green color and showed severe damage on plates containing 1 μM MV, compared with 55% of wild-type seedlings. Strikingly, NCER-OX seedlings showed increased tolerance to 1 μM MV (Figure 5C). These results showed that AtNCER1 affects Arabidopsis sensitivity to MV-induced oxidative stress.
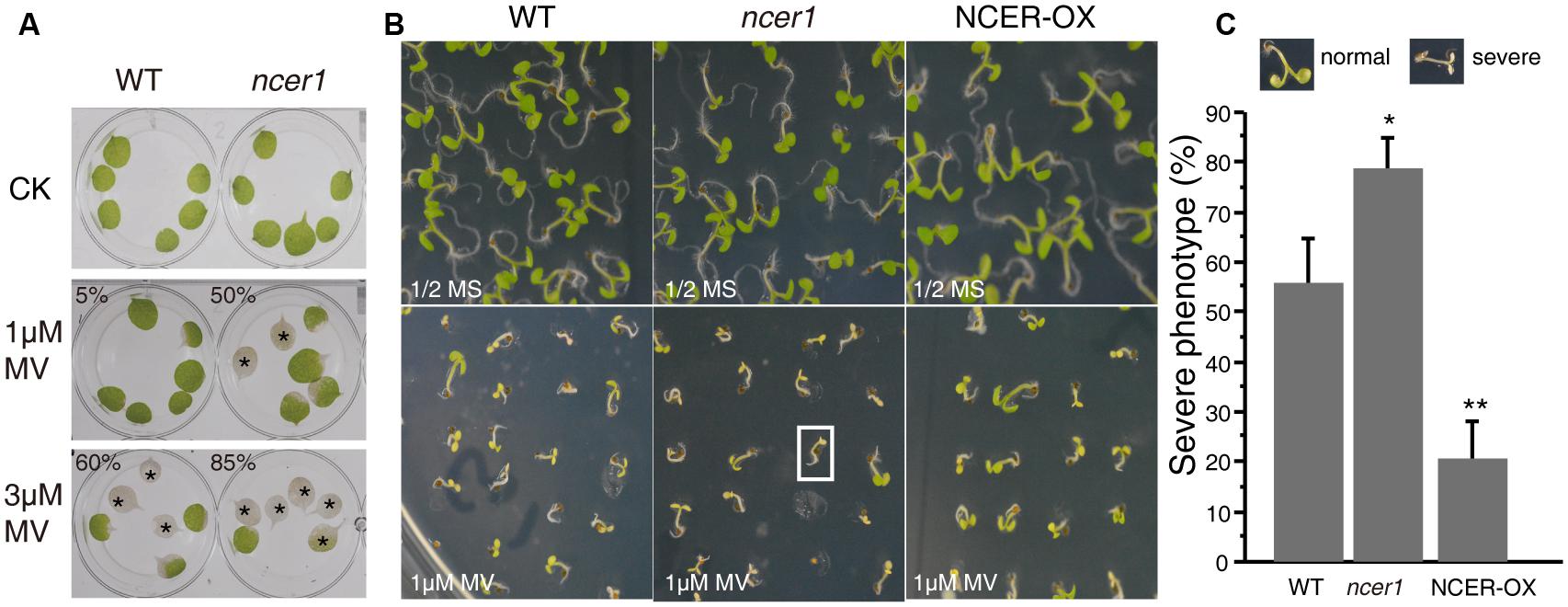
FIGURE 5. Sensitivity of ncer1 mutants to oxidative stress induced by MV. (A) Disks from the third and fourth leaves of 14-day-old wild-type and ncer1 mutant plants were incubated with or without 1 μM or 3 μM MV for 3 days and then photographed. Asterisks indicate completely bleached, damaged leaf disks. At least 12 leaves of wild-type and ncer1 were observed in each experiment. Two independent tests were done. The ratio of the bleached area and the whole leaf area showed significantly difference from the wild type using Student’s t-test (p < 0.05). (B) The phenotype of wild type, ncer1, and NCER-OX seedlings after MV treatment. Seedlings were grown on 1/2 MS containing or without 1 μM MV for 10 days and then photographed. The white square indicates one seedling with a severe phenotype. (C) Percentage of seedlings with a severe phenotype after 1 μM MV treatment. At least 250 seedlings were counted in each genotype. Error bars indicate SD from three independent experiments. Asterisks indicate significant differences by Student’s t-test (p* < 0.05; **p < 0.01).
To test whether MV oxidative stress affects the sphingolipid profile, we transferred 7-day-old seedlings to 1/2x MS agar plates containing 1 μM MV, then collected samples after 7 days. No significant difference in sphingolipid contents was shown in untreated control plants at early developmental stages (Figure 6). Compared with untreated controls, the amount of total LCB and hydroxyceramide increased significantly in response to MV treatments; also, ncer1 showed almost threefold more LCB and hydroxyceramide than non-treated control plants (Figure 6A). No significant changes were observed in the ceramides and glucosylceramides (Figure 6A). The amounts of hydroxyceramides containing both long chain and very long chain fatty acids showed significant accumulation in ncer1 compared with wild-type plants (Figure 6C). The wild-type and NCER-OX plants showed no significant differences in sphingolipid contents (Figure 6). A significantly high level of t18:0 and t18:1 hydroxyceramides accumulated in ncer1 mutants, compared with wild type, and NCER-OX plants, after MV treatments (Figure 6B), indicating that hydroxyceramides may be a potential substrate of AtNCER1.
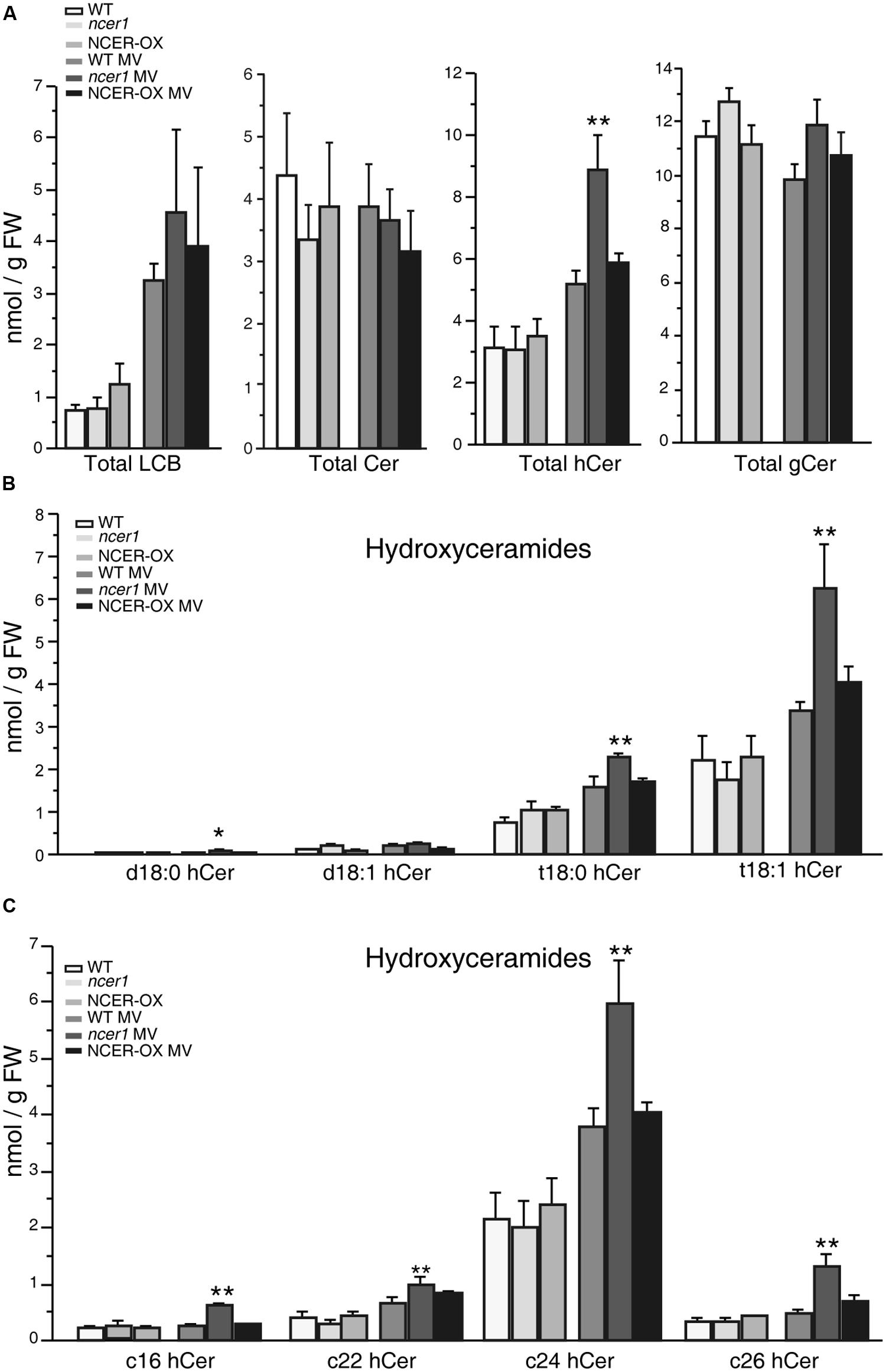
FIGURE 6. Sphingolipid analysis after MV treatment. Seven-day-old seedlings were transferred to 1/2x MS containing 1 μM MV. Sphingolipids were extracted after 7 days with or without MV treatment, as described in Section “Materials and Methods.” The contents of free LCBs, ceramides (Cer), hydroxyceramides (hCer), and glucosylceramides (gCer) were quantified. The experiment was repeated three times using independent samples. Data sets marked with asterisks indicate significant differences confirmed by Student’s t-test (*p < 0.05; **p < 0.01). (A) Total LCBs, ceramides, hydroxyceramides, and glucosylceramides after MV treatment. (B) Comparison of hydroxyceramides with the LCB moieties in the indicated plants after MV treatment. (C) Comparison of hydroxyceramides with the length of fatty acid moieties in the indicated plants after MV treatment.
Discussion
Sphingolipids are important signaling molecules involved in the regulation of cell physiological activities and in the mediation of plant responses to biotic and abiotic stress (Dickson and Lester, 2002; Hannun and Obeid, 2002; Spiegel and Milstien, 2003; Futerman and Hannun, 2004;Pyne et al., 2009). Plants have about 200 different kinds of sphingolipid molecules (Chen et al., 2009), with high diversity in the composition of the head groups, the degree of hydroxylation, and the position of the double bond (Chen et al., 2008). Sphingolipids have four hydroxylation sites: the C-1 and C-3 hydroxyl groups arise from Ser and palmitoyl-CoA precursors, respectively, and the third hydroxyl group at the C-4 position is synthesized by LCB C-4 hydroxylases encoded by SPHINGOID BASE HYDROXYLASE 1 (SBH1) and SBH2. The sbh1/2 mutants have a severely dwarfed phenotype and fail to progress from vegetative to reproductive growth, which may result from the accumulation of sphingolipids with the dihydroxy LCB/C16 fatty acid–containing ceramide backbone (Chen et al., 2008). The Arabidopsis thaliana genome harbors two FATTY ACID HYDROXYLASE genes (AtFAH1 and AtFAH2), which encode the enzymes that catalyze α-hydroxylation of the fatty acid moiety in plant sphingolipids. The fah1/fah2 mutants showed elevated levels of LCBs and ceramides and resistance to G. cichoracearum infections (Konig et al., 2012). These data suggest that sphingolipid hydroxylation has a very important role in regulation of the plant response to biotic and abiotic stress.
Few studies have examined the connection between elevated hydroxyceramides and induction of cell death in plants. However, in many mammalian cell types, evidence clearly shows that hydroxyceramide and hydroxy complex sphingolipids have unique functions in membrane homeostasis and cell signaling, and these functions cannot be substituted by their non-hydroxy counterparts (Kota and Hama, 2014). Trihydroxy LCBs (tLCBs) of 4-hydroxysphinganine, t16:0, t18:0, t19:0, and t20:0, and non-hydroxy fatty acids (NFAs) isolated from equine kidneys exhibited stronger apoptosis-inducing activities than dLCB-NFA toward tumor cell lines (Kyogashima et al., 2008). In plants, we previously found that acd5 mutants show a modest increase in the amount of hydroxyceramides, relative to wild type, even in 17-day-old plants. In addition, after Botrytis cinerea infections, hydroxyceramides in acd5 increased to a greater extent than in wild-type plants (Bi et al., 2014). These results suggest that hydroxyceramides are involved in programmed cell death and biotic stress. In this work, we found that total LCB levels increased after MV treatment, but found no significant difference between wild type and mutants. By contrast, without treatment, 3-week-old ncer1 plants accumulated more hydroxyceramide compared with wild type. Furthermore, when challenged by MV treatment, hydroxyceramide increased to higher levels in ncer1 plants than in wild type, indicating that hydroxyceramides may be involved in oxidative stress.
A neutral rice ceramidase appears to use ceramide instead of phytoceramide as a substrate in the CDase mutant stain of yeast (Pata et al., 2008). Our results indicated AtNCER1 may favor hydroxyceramides as its substrates. Our future work will look for potential AtNCER1 substrates, especially t18:0 and t18:1 hydroxyceramides.
Ceramide, as a bioactive lipid key player, involves in various cellular processes (Hannun and Obeid, 2002; Chen et al., 2009). Recent studies have demonstrated that sphingolipids can trigger the generation of ROS in both mammalian and plant cells (Hannun and Obeid, 2002; Shi et al., 2007). Certain ceramides can directly inhibit the activity of mitochondrial complex IV, leading to ROS production and oxidative stress (Kogot-Levin and Saada, 2014). In this work, we show that AtNCER1 regulates the plant response to oxidative stress. We speculate that the turbulence of sphingolipid in ncer1 may change intracellular redox state or effect of antioxidant systems. Further study may provide insights into the understanding of interplay between ROS and sphingolipids.
Conclusion
In this report, we characterized an Arabidopsis neutral ceramidase mutant (ncer1). Our results indicate that ncer1 accumulated high levels of hydroxyceramides in normal conditions, but showed no visible phenotype. We also found that ncer1 mutants were more sensitive to oxidative stress induced by MV and had higher levels of hydroxyceramides, indicating that AtNCER1 regulates the plant response to oxidative stress. Further study may provide insights into the understanding of interplay between ROS and sphingolipids.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the ABRC for providing Arabidopsis T-DNA insertion line for AtNCER and organelle markers (CD3-960). This work was supported by National Natural Science Foundation of China (31170247), the National Key Basic Science 973 program (2012CB114006) and the Fundamental Research Funds for the Central Universities (13lgjc27).
References
Bi, F.-C., Liu, Z., Wu, J.-X., Liang, H., Xi, X.-L., Fang, C., et al. (2014). Loss of ceramide kinase in Arabidopsis impairs defenses and promotes ceramide accumulation and mitochondrial H2O2 bursts. Plant Cell 26, 3449–3467. doi: 10.1105/tpc.114.127050
Chen, M., Cahoon, E. B., Saucedo-García, M., Plasencia, J., and Gavilanes-Ruíz, M. (2009). “Plant sphingolipids: structure, synthesis and function,” in: Lipids in Photosynthesis: Essential and Regulatory Functions, eds H. Wada and N. Murata (Dordrecht: Springer), 77–115. doi: 10.1007/978-90-481-2863-1_5
Chen, M., Markham, J. E., Dietrich, C. R., Jaworski, J. G., and Cahoon, E. B. (2008). Sphingolipid long-chain base hydroxylation is important for growth and regulation of sphingolipid content and composition in Arabidopsis. Plant Cell 20, 1862–1878. doi: 10.1105/tpc.107.057851
Dickson, R. C., and Lester, R. L. (2002). Sphingolipid functions in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1583, 13–25. doi: 10.1016/S1388-1981(02)00210-X
El Bawab, S., Bielawska, A., and Hannun, Y. A. (1999). Purification and characterization of a membrane-bound nonlysosomal ceramidase from rat brain. J. Biol. Chem. 274, 27948–27955. doi: 10.1074/jbc.274.39.27948
Futerman, A. H., and Hannun, Y. A. (2004). The complex life of simple sphingolipids. EMBO Rep. 5, 777–782. doi: 10.1038/sj.embor.7400208
Hannun, Y. A., and Obeid, L. M. (2002). The Ceramide-centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind. J. Biol. Chem. 277, 25847–25850. doi: 10.1074/jbc.R200008200
Koch, J., Gaertner, S., Li, C. M., Quintern, L. E., Bernardo, K., Levran, O., et al. (1996). Molecular cloning and characterization of a full-length complementary DNA encoding human acid ceramidase. Identification of the first molecular lesion causing Farber disease. J. Biol. Chem. 271, 33110–33115. doi: 10.1074/jbc.271.51.33110
Kogot-Levin, A., and Saada, A. (2014). Ceramide and the mitochondrial respiratory chain. Biochimie 100, 88–94. doi: 10.1016/j.biochi.2013.07.027
Konig, S., Feussner, K., Schwarz, M., Kaever, A., Iven, T., Landesfeind, M., et al. (2012). Arabidopsis mutants of sphingolipid fatty acid alpha-hydroxylases accumulate ceramides and salicylates. New Phytol. 196, 1086–1097. doi: 10.1111/j.1469-8137.2012.04351.x
Kono, M., Dreier, J. L., Ellis, J. M., Allende, M., Kalkofen, D. N., Sanders, K. M., et al. (2006). Neutral ceramidase encoded by the Asah2 gene is essential for the intestinal degradation of sphingolipids. J. Biol. Chem. 281, 7324–7331. doi: 10.1074/jbc.M508382200
Kota, V., and Hama, H. (2014). 2’-Hydroxy ceramide in membrane homeostasis and cell signaling. Adv. Biol. Regul. 54, 223–230. doi: 10.1016/j.jbior.2013.09.012
Kyogashima, M., Tadano-Aritomi, K., Aoyama, T., Yusa, A., Goto, Y., Tamiya-Koizumi, K., et al. (2008). Chemical and apoptotic properties of hydroxy-ceramides containing long-chain bases with unusual alkyl chain lengths. J. Biochem. 144, 95–106. doi: 10.1093/jb/mvn050
Li, C. M., Hong, S. B., Kopal, G., He, X., Linke, T., Hou, W. S., et al. (1998). Cloning and characterization of the full-length cDNA and genomic sequences encoding murine acid ceramidase. Genomics 50, 267–274. doi: 10.1006/geno.1998.5334
Liang, H., Yao, N., Song, J. T., Luo, S., Lu, H., and Greenberg, J. T. (2003). Ceramides modulate programmed cell death in plants. Genes Dev. 17, 2636–2641. doi: 10.1101/gad.1140503
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mao, C., and Obeid, L. M. (2002). “Ceramidases: regulators of turnover of ceramide and ceramide-mediated responses,” in: Ceramide Signaling, ed. A. H. Futerman (Austin, TX: Landes Bioscience), 29–40. doi: 10.1007/978-1-4419-9272-7_4
Mao, C., Xu, R., Bielawska, A., and Obeid, L. M. (2000). Cloning of an alkaline ceramidase from Saccharomyces cerevisiae. J. Biol. Chem. 275, 6876–6884. doi: 10.1074/jbc.275.10.6876
Mao, C., Xu, R., Szulc, Z. M., Bielawska, A., Galadari, S. H., and Obeid, L. M. (2001). Cloning and characterization of a novel human alkaline ceramidase: a mammalian enzyme that hydrolyzes phytoceramide. J. Biol. Chem. 276, 26577–26588. doi: 10.1074/jbc.M102818200
Markham, J. E., Molino, D., Gissot, L., Bellec, Y., Hematy, K., Marion, J., et al. (2011). Sphingolipids containing very-long-chain fatty acids define a secretory pathway for specific polar plasma membrane protein targeting in Arabidopsis. Plant Cell 23, 2362–2378. doi: 10.1105/tpc.110.080473
Mitsutake, S., Tani, M., Okino, N., Mori, K., Ichinose, S., Omori, A., et al. (2001). Purification, characterization, molecular cloning, and subcellular distribution of neutral ceramidase of rat kidney. J. Biol. Chem. 276, 26249–26259. doi: 10.1074/jbc.M102233200
Monjusho, H., Okino, N., Tani, M., Maeda, M., Yoshida, M., and Ito, M. (2003). A neutral ceramidase homologue from Dictyostelium discoideum exhibits an acidic pH optimum. Biochem. J. 376, 473–479. doi: 10.1042/BJ20030652
Ohlsson, L., Palmberg, C., Duan, R. D., Olsson, M., Bergman, T., and Nilsson, A. (2007). Purification and characterization of human intestinal neutral ceramidase. Biochimie 89, 950–960. doi: 10.1016/j.biochi.2007.03.009
Olsson, M., Duan, R.-D., Ohlsson, L., and Nilsson, A. (2004). Rat intestinal ceramidase: purification, properties, and physiological relevance. Am. J. Physiol. Gastrointest. Liver Physiol. 287, 929–937. doi: 10.1152/ajpgi.00155.2004
Pata, M. O., Wu, B. X., Bielawski, J., Xiong, T. C., Hannun, Y. A., and Ng, C. K. (2008). Molecular cloning and characterization of OsCDase, a ceramidase enzyme from rice. Plant J. 55, 1000–1009. doi: 10.1111/j.1365-313X.2008.03569.x
Pyne, N. J., Long, J. S., Lee, S. C., Loveridge, C., Gillies, L., and Pyne, S. (2009). New aspects of sphingosine 1-phosphate signaling in mammalian cells. Adv. Enzyme Regul. 49, 214–221. doi: 10.1016/j.advenzreg.2009.01.011
Schmelz, E. M., Roberts, P. C., Kustin, E. M., Lemonnier, L. A., Sullards,M. C., and Dillehay, D. L. et al. (2001). Modulation of intracellular β-Catenin localization and intestinal tumor genesis in vivo and in vitro by sphingolipids. Cancer Res. 61, 6723–6729.
Shi, L., Bielawski, J., Mu, J.,Dong, H., Teng, C., Zhang, J., et al. (2007). Involvement of sphingoid bases in mediating reactive oxygen intermediate production and programmed cell death in Arabidopsis. Cell Res. 17, 1030–11040. doi: 10.1038/cr.2007.100
Spiegel, S., and Milstien, S. (2003). Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat. Rev. Mol. Cell Biol. 4, 397–407. doi: 10.1038/nrm1103
Sun, W., Hu, W., Xu, R., Jin, J., Szulc, Z. M., Zhang, G., et al. (2009). Alkaline ceramidase 2 regulates beta1 integrin maturation and cell adhesion. FASEB J. 23, 656–666. doi: 10.1096/fj.08-115634
Sun, W., Xu, R., Hu, W., Jin, J., Crellin, H. A., Bielawski, J., et al. (2007). Up-regulation of the human alkaline ceramidase 1 and acidceramidase mediates calcium-induced differentiation of epidermal keratinocytes. J. Invest. Dermatol. 128, 389–397. doi: 10.1038/sj.jid.5701025
Symolon, H., Schmelz, E. M., Dillehay, D. L., and Merrill, A. H. Jr. (2004). Dietary soy sphingolipids suppress tumorigenesis and gene expression in 1,2-dimethylhydrazine-treated CF1 mice and ApcMin/+Mice. J. Nutr. 134, 1157–1161.
Tani, M., Okino, N., Mori, K., Tanigara, T., Izu, H., and Ito, M. (2000). Molecular cloning of the full-length cDNA encoding mouse neutral ceramidase. J. Biol. Chem. 275, 11229–11234. doi: 10.1074/jbc.275.15.11229
Ternes, P., Feussner, K., Werner, S., Lerche, J., Iven, T., Heilmann, I., et al. (2011). Disruption of the ceramide synthase LOH1 causes spontaneous cell death in Arabidopsis thaliana. New Phytol. 192, 841–854. doi: 10.1111/j.1469-8137.2011.03852.x
Wang, W., Yang, X., Tangchaiburana, S., Ndeh, R., Markham, J. E., Tsegaye, Y., et al. (2008). An inositolphosphorylceramide synthase is involved in regulation of plant programmed cell death associated with defense in Arabidopsis. Plant Cell 20, 3163–3179. doi: 10.1105/tpc.108.060053
Wu, J.-X, Li, J., Liu, Z., Yin, J., Chang, Z.-Y., Rong, C., et al. (2015). The Arabidopsis ceramidase AtACER functions in disease resistance and salt tolerance. Plant J. 81, 767–780. doi: 10.1111/tpj.12769
Xu, R., Jin, J., Hu, W., Sun, W., Bielawski, J., Szulc, Z., et al. (2006). Golgi alkaline ceramidase regulates cell proliferation and survival by controlling levels of sphingosine and S1P. FASEB J. 20, 1813–1825. doi: 10.1096/fj.05-5689com
Yoshimura, Y., Okino, N., Tani, M., and Ito, M. (2002). Molecular clonng and characterization of a secretory neutral ceramidase of Drosophila melanogaster. J. Biochem. 132, 229–236. doi: 10.1093/oxfordjournals.jbchem.a003215
Yoshimura, Y., Tani, M., Okino, N., Iida, H., and Ito, M. (2004). Molecular cloning and functional analysis of zebrafish neutral ceramidase. J. Biol. Chem. 279, 44012–44022. doi: 10.1074/jbc.M405598200
Keywords: Arabidopsis neutral ceramidases, oxidative stress, sphingolipids, hydroxyceramides, ceramides
Citation: Li J, Bi F-C, Yin J, Wu J-X, Rong C, Wu J-L and Yao N (2015) An Arabidopsis neutral ceramidase mutant ncer1 accumulates hydroxyceramides and is sensitive to oxidative stress. Front. Plant Sci. 6:460. doi: 10.3389/fpls.2015.00460
Received: 03 January 2015; Accepted: 08 June 2015;
Published: 19 June 2015
Edited by:
Richard Sayre, New Mexico Consortium at Los Alamos National Labs, USAReviewed by:
Ian S. Wallace, The University of Nevada, Reno, USAXia Wu, University of Washington, USA
Copyright © 2015 Li, Bi, Yin, Wu, Rong, Wu and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nan Yao, State Key Laboratory of Biocontrol, Guangdong Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou 510275, China, yaonan@mail.sysu.edu.cn
 Jian Li1
Jian Li1 Fang-Cheng Bi
Fang-Cheng Bi Jian Yin
Jian Yin Jian-Xin Wu
Jian-Xin Wu Nan Yao
Nan Yao