- 1Institute of Biochemistry and Biology, University of Potsdam, Potsdam, Germany
- 2Institute for Biology I, RWTH Aachen University, Aachen, Germany
The per-ARNT-sim (PAS) domain represents an ancient protein module that can be found across all kingdoms of life. The domain functions as a sensing unit for a diverse array of signals, including molecular oxygen, small metabolites, and light. In plants, several PAS domain-containing proteins form an integral part of the circadian clock and regulate responses to environmental change. Moreover, these proteins function in pathways that control development and plant stress adaptation responses. Here, we discuss the role of PAS domain-containing proteins in anticipation, and adaptation to environmental changes in plants.
Introduction
The evolution of the photosynthetic apparatus resulted in two major developments on earth. During photosynthesis, water is split into oxygen (O2), protons and electrons, which caused the oxygenation of the earth’s atmosphere for about 2.2 billion years ago (Knoll, 2003; Schippers et al., 2012). The first event, the release of O2, placed organisms under strong selection pressure to mitigate the reactive nature of highly toxic O2 derivatives, i.e., superoxide, hydroxyl radical and hydrogen peroxide commonly called reactive oxygen species (ROS; Raymond and Segrè, 2006; Schippers et al., 2012). The release of oxygen stimulated the evolution of aerobic metabolism using a superior electron acceptor, resulting in an increased energy availability, which might have accelerated the development of multicellular organisms (Thannickal, 2009; Schirrmeister et al., 2013). The development of the photosynthetic apparatus itself gave rise to autotrophic organisms whose energy metabolism is light dependent. Considering the rotational behavior of the Earth, energy production in plants is limited to a specific time-window during a single day. Both, light dependent metabolism as well as redox oscillations, have stimulated the emergence of a control system that directs cellular processes in a temporal manner, the circadian clock (Lai et al., 2012; Haydon et al., 2013). Interestingly, the acquisition of aerobic metabolism, photosynthesis and the evolution of circadian systems appear to have co-occurred (Loudon, 2012).
At the molecular level, the circadian clock consists of several transcriptional feedback loops (Chow and Kay, 2013). The central loop is formed by the morning-expressed CIRCADIAN CLOCK-ASSOCIATED1 and LATE ELONGATED HYPOCOTYL and the evening-expressed clock gene TIMING OF CAB EXPRESSION1 (TOC1), which act in a reciprocal manner (Gendron et al., 2012). In principle, the transcriptional loops allow for activating or deactivating specific genetic programs at different times of the day. However, to ensure anticipation of environmental conditions and accurate timing of cellular processes, circadian clocks are set to the correct time of day through the perception of environmental signals (Harmer, 2009; Sanchez et al., 2011; Lai et al., 2012; Haydon et al., 2013). Light is the best-studied zeitgeber (a signal that resets the clock), that entrains or synchronizes the clock (Alabadí et al., 2001). In addition, other external cues such as temperature (Michael et al., 2003), but also internal cues like energy status and redox homeostasis (Lai et al., 2012; Haydon et al., 2013; Zhou et al., 2015), all feed back into the clock to optimize plant growth and survival (Dodd et al., 2005). Especially for the adaptation against adverse environmental conditions a functional clock is a prerequisite (Lai et al., 2012; Kolmos et al., 2014; Gehan et al., 2015). Moreover, genes associated with abiotic stresses show rhythmic expression behavior, and their transcriptional response often depends on the time of the day at which the plant encounters the stress (Gehan et al., 2015; Matsuzaki et al., 2015).
Plants, as sessile organisms heavily rely on their ability to sense and adapt to changes in their environment. One particular protein domain, the per-ARNT-sim (PAS) domain, is a sensory module that can be found in all kingdoms of life (Nambu et al., 1991). The PAS domain can function as a chemoreceptor, a redox sensor, a photoreceptor, or a voltage sensor (Möglich et al., 2009), indicating its versatile role in signal transduction. In animals, the PAS-domain containing ARNT transcription factor regulates the response to environmental stimuli by heterodimerization with different protein interaction partners to initiate a specific transcriptional response (McIntosh et al., 2010). Moreover, both in animals and plants, several of the circadian timekeeping proteins contain a PAS domain, forming a direct link between sensing the environment and the circadian clock. Here, we focus on the role of PAS domain proteins in environmental adaptation, under the control of the circadian clock.
Structure of PAS Domains
The PAS domain is a sensory and protein–protein-interaction module, which can be found throughout all kingdoms of life. It was originally identified by sequence homology of three eukaryotic proteins: the circadian protein Period (per) and developmental regulator Sim (single-minded) of Drosophila and the vertebrate aryl hydrocarbon receptor nuclear transporter (ARNT), which comprise two PAS motifs each (Nambu et al., 1991). Conserved residues C-terminal to the PAS motif were at first assigned as PAC motif (Ponting and Aravind, 1997). However, the first three-dimensional structure of the PAS domain revealed that both, the PAS and PAC motif, form a globular fold made up of about 100 residues, thus redefining the PAS domain (Hefti et al., 2004). Although PAS domains share only a low sequence homology on the amino acid level (∼20%), the three-dimensional structure is highly conserved (Mei and Dvornyk, 2014). The PAS fold consists of an antiparallel five-stranded β-sheet in topological order 2-1-5-4-3 and several flanking α-helices (Figure 1), which are either packed on the core or extend from it (Möglich et al., 2009). In plants, PAS domains are combined in multidomain proteins with functionally diverse effector/regulatory domains such as Serine/Threonine kinases, F-Boxes or, HD-ZIP domains (Figure 2 and Table 1), thus mediating a plethora of cellular responses. Interestingly, the phytochrome and F-BOX containing PAS domain proteins were shown to interact (Jarillo et al., 2001a; Kim et al., 2007) and function either as an input to the clock and/or an integral component of the circadian oscillator.
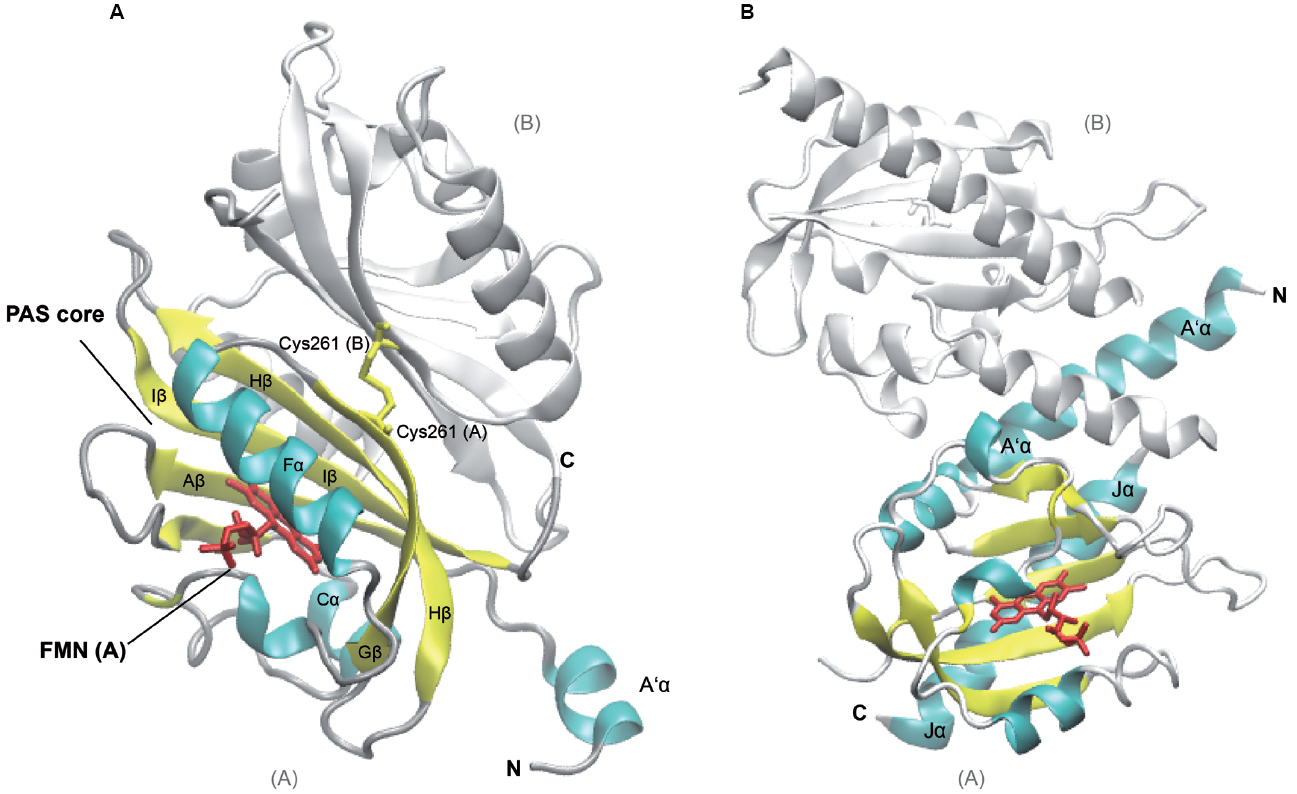
Figure 1. Structures of Arabidopsis thaliana LOV1 and LOV2 domains of phototropin 1 dark-adapted state. LOV1 domain (2Z6C; Nakasako et al., 2008), (A) and LOV2 of phototropin 1 in dark-adapted state (4HHD; Halavaty and Moffat, 2013), (B). Subunits B of LOV1 and LOV2 are shown in white, subunit A of the respective LOV domains is shown in colors according to secondary structure. Several α-helices (cyan) and β-strands (yellow) of the PAS core are labeled according to their nomenclature within the PAS domain as well as flanking helices, which are involved in dimerization and signaling. The Flavin mononucleotide chromophores (red) and the disulfide-bond-creating Cys at position 261 (Cys261, yellow) in the monomers of LOV2 domains, respectively, are depicted as sticks. C- and N-termini of subunits A and B are labeled (C, N). The graphic representation of the structure was generated using VMD (Humphrey et al., 1996).
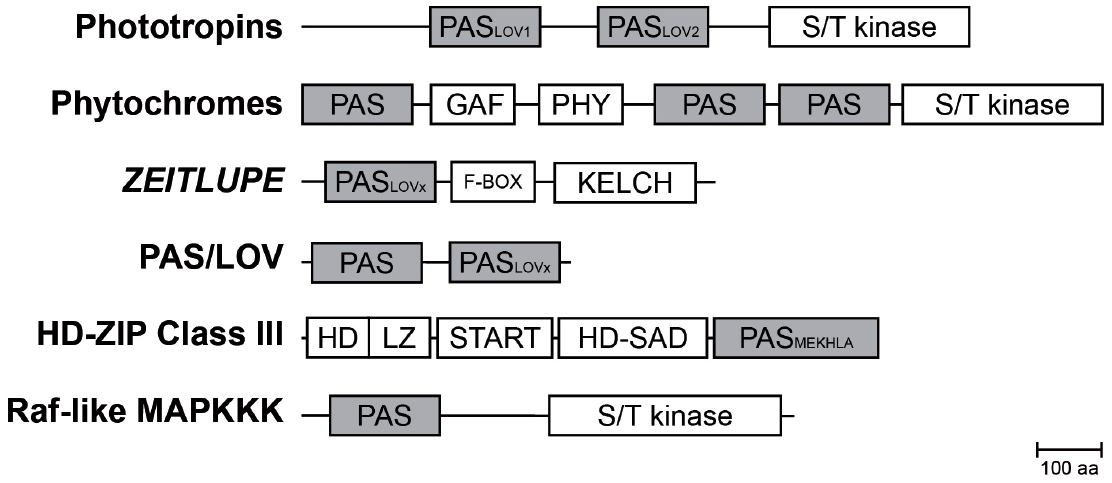
Figure 2. Composition of PAS containing multidomain proteins in plants. Schematic representation of PAS domain-containing proteins in Arabidopsis and their multidomain architecture (Mitchell et al., 2015). The PAS module is located in variable positions within the protein and associated with a variety of effector domains. Abbreviations: PAS, per-arnt-sim domain; PASLOV1/2, LOV (light oxygen, or voltage) subclass of PAS domain; PASMEKHLA, PAS-like MEKHLA domain; S/T kinase, Serine/Threonine kinase; F-BOX, domain interacting with SCF complex; KELCH, KELCH repeat; GAF, cGMP-specific phosphodiesterases, adenylyl cyclases and FhlA; PHY, GAF-like domain/Chromobillin binding; START, StAR-related lipid-transfer; HD-SAD, HD-START-associated domain; HD, homeodomain; LZ, Leucine zipper; aa, amino acids.
Structure of FMN-binding PAS-domain (LOV) in Plant Phototropins
The Arabidopsis thaliana genome contains two membrane-associated phototropin genes, Phot1 and Phot2 that encode serine/threonine kinases and serve as receptor for blue light (Kinoshita et al., 2001). They belong to a subclass of the PAS domain superfamily, termed LOV (light, oxygen, or voltage) domain. Defining feature of the LOV domain is the cofactor, a Flavin nucleotide (Huala et al., 1997). The photosensors harbor two N-terminal FMN-binding PAS domains, termed N- to C-terminal LOV1 and LOV2, which are coupled to the C-terminal kinase domain. The typical PAS fold consists of the five-stranded antiparallel β-sheet and four α-helices (β2α4β3: Aβ, Bβ, Cα, Dα, Eα, Fα, Gβ, Hβ, and Iβ; Figure 1). The core is flanked by amphiphatic α-helices (A’α, Jα) and mediates chromophore binding and signal transduction. Upon photoexcitation, the C4a of the oxidized FMN isoalloxazine ring forms a dark-reversible cysteyl-adduct with Cys39 within the LOV2 core domain (Salomon et al., 2000; Swartz et al., 2001) resulting in alteration of the hydrogen bonding in the binding pocket between the chromophore and the β-sheet, as well as the adjacent Jα helix (Crosson and Moffat, 2001; Halavaty and Moffat, 2013). Jα was shown to be required for kinase activity, and light-activated alterations of the interacting surface of the core domain leads to an intermediate partial unfolding of the α-helical structure. Together with the β-sheet it might constitute the interface involved in signal transmission by alleviating the repressive effect on the protein kinase domain of phototropins (Crosson and Moffat, 2002; Crosson et al., 2002; Corchnoy et al., 2003; Harper et al., 2003; Matsuoka and Tokutomi, 2005; Chen et al., 2007; Pfeifer et al., 2010). A’α and Jα helices both contribute to coiled coil dimerization of the phototropin (Figure 1; Halavaty and Moffat, 2013).
The two PAS cores share a high degree of similarity among each other and among the phototropins (Halavaty and Moffat, 2007; Nakasako et al., 2008) and exhibit identical photochemical cycles (Salomon et al., 2000; Kasahara et al., 2002). However, there are functional differences between the two domains: while LOV2 is required and sufficient for light sensing and the autophosphorylation resulting in phototropic hypocotyl growth (Christie et al., 2002), the LOV1 domain, mainly contributes to dimerization and regulating the sensitivity to blue-light activation (Matsuoka and Tokutomi, 2005; Nakasako et al., 2008; Tokutomi et al., 2008). The dimerization sites of LOV1 in the two phototropins are similar to the extent that they both form a face-to-face association of the β-scaffolds. They are stabilized by hydrophobic interactions of an N-terminal α-helix (relative to the PAS core) with the β-scaffolds of both subunits (Nakasako et al., 2008). While the LOV1 dimer of Phototropin 1 forms a stable disulfide bond [Cys261(A)—Cys261(B), Figure 1A], LOV1 of Phototropin 2 dimer is predominantly stabilized by hydrogen bonds of a threonine and methionine (Nakasako et al., 2008), underlining the structural rather than sequence conservation and giving a possible explanation for the different physiological roles of the two phototropins in plants.
Sensing-signaling Mechanisms
It was recently postulated that the general role for PAS domains is to modulate protein–protein interactions to form hetero- or homo-oligomers with other proteins (Möglich et al., 2009), or alter protein structures by rearranging interactions between domains in a single protein. As the PAS domain can act as a sensor, the occurrence of protein–protein interactions becomes signal dependent. This property provides specificity, allowing for complex spatial and temporal regulation of cellular signaling networks (Möglich et al., 2009). Here, the role of PAS domains in the modulation of protein activity and signaling is illustrated by several examples.
In contrast to other species, plant phytochromes contain three PAS domains (Hughes, 2013). It has been shown for PhyB, the most prominent photo-stable phytochrome, that the nuclear translocation depends on the tandem PAS domain (Chen et al., 2005). In its inactive form the tandem-repeat PAS domain interacts with the N-terminal sensory domain containing the other PAS motif (Figure 2). Upon red light, the protein is activated through a conformational change, exposing the nuclear-localization signal present in the tandem PAS motif, resulting in translocation to the nucleus. In addition, the tandem PAS domain is crucial for the red-light induced interaction between PhyB and PHY INTERACTING FACTORs (PIFs; Levskaya et al., 2009). Moreover, application of far-red light results in a reversal of the interaction between PhyB and PIFs (Ni et al., 1999). This photoreversibility of the protein interaction depends on the presence of the tandem PAS domain (Levskaya et al., 2009). Still, sensing of red light by phytochromes is established through the billin chromophore bound by the GAF domain within the N-terminal photosensory module (Galvão and Fankhauser, 2015). Although the function of the PAS domain in the photosensory module is unknown, it was shown to interact with and thereby stabilizing the chromophore binding GAF domain, which has also been implicated in PIF binding (Burgie et al., 2014). Thus, the PAS domains in phytochromes do not function as light sensors, and it is currently unclear, whether they have sensory roles themselves or mainly support conformational changes (Burgie and Vierstra, 2014).
The phototropins and ZEITLUPE family of LOV domain-containing proteins act as blue-light receptors. The ZTL family contains an N-terminal LOV domain followed by an F-box and six Kelch repeats (Figure 2), suggesting a role in light-regulated protein degradation (Demarsy and Fankhauser, 2009). F-box proteins are components of SCF-type (Skp1, Culin, and F-box) ubiquitin E3 ligases and ZTL was shown to interact with Arabidopsis Skp-like (ASK) proteins (Kevei et al., 2006). A mutation in the LOV domain of ZTL, as found in the ztl-21 mutant, was shown to interfere with the interaction with ASK1 (Kevei et al., 2006), indicating that light signals are required for ZTL to target proteins for degradation. In the light, photoexcitation of ZTL promotes interaction with GIGANTEA (GI), stabilizing ZTL during the day (Kim et al., 2007). Still, ZTL is not completely stabilized as photochemical characterization of ZTL reveals a half-life of about 4 h (Pudasaini and Zoltowski, 2013). It was postulated that this fast photocycle allows for estimating the timing of the day-night transition. Thus, the LOV domain of ZTL determines protein stability and directs protein–protein interactions.
So far, only a role for the PAS sensory domain in light receptors has been described, in other plant proteins the function of the PAS domain is mostly unknown and the signals they perceive are elusive. For example, the HD-ZIP III family of transcription factors contains a C-terminal PAS-like domain. A point mutation in this domain for CORONA/INCURVATA4 results in a bushy plant phenotype, suggesting a regulatory role for this domain (Duclercq et al., 2011). Furthermore, the PAS-like domain in HD-ZIP III member REVOLUTA (REV), was shown to modulate the DNA binding activity of the protein (Xie et al., 2014), however, the signal that modulates the regulatory function of the PAS-like domain in this case is unknown. Therefore, additional efforts are required to better understand the role of PAS domains in plants.
Plant PAS Domain Proteins
Plants as phototrophic sessile organisms are required to adapt especially to changes in light, but also other environmental cues. Hence, it is not surprising that many of the PAS sensory modules are coupled to light perception and signaling (Table 1). Among the five classes of photoreceptor families characterized in Arabidopsis, the phytochrome, cryptochrome, and ZEITLUPE family integrate the red/far-red and blue light information into the central oscillator (McClung, 2011).
Phototropin 1 and 2, Tandem LOV Domain Blue-light Receptors
The plant specific blue-light (and UVA) receptor protein kinase Phot1 was initially identified by a forward genetic screen for plants showing defects in the phototrophic response (Liscum and Briggs, 1995; Huala et al., 1997). Moreover, phototropins 1 and 2 are involved in blue-light induced stomata opening, and chloroplast migration (Jarillo et al., 2001b; Kinoshita et al., 2001; Sakai et al., 2001). This indicates that both receptors play an important role in sensing the light availability, for metabolic and physiological adaptation.
The structure of the two phototropins have been extensively studied (Christie et al., 2002; Nakasako et al., 2008; Halavaty and Moffat, 2013) and helped understanding the underlying mechanism of the structure-function relationship of PAS (LOV) domains. In the dark, each LOV domain non-covalently binds FMN, while blue light causes covalent binding of the chromophore, resulting in the release of its inhibitory effect on the C-terminal kinase domain — thereby facilitating autophosphorylation and subsequent activation of the phototropin signal transduction pathway (Tokutomi et al., 2008). Of note, this reaction is reversible allowing for a rapid switch of phototropin activation in a light-dependent manner.
Recently it was found that PIF4 and PIF5 act downstream of Phot1 to negatively modulate phototropism in Arabidopsis (Sun et al., 2013). Although no direct link between the circadian clock and Phot1/2 has been established, the dependence of PIF4 and PIF5 on Phot1 might present such an interaction. PIF4 and PIF5 expression are under the control of the evening complex, containing the proteins EARLY FLOWERING 3 (ELF3), ELF4, and LUX ARRHYTHMO (LUX) during hypocotyl elongation (Nusinow et al., 2011). Still, this interaction has so far not been described.
Phytochromes, Tandem PAS Domain Red-light Receptors
Phytochromes act as homodimeric red and far-red photoreceptors that are encoded by a small family of genes encompassing three clades, PHYA, PHYB, and PHYC (Casal, 2013). As stated above, they contain a single PAS motif within their phytochrome sensory domain, which is followed by a tandem PAS domain and histidine-kinase-like transmitter module (Hughes, 2013). In plants, phytochromes regulate a multitude of processes, including, seed germination, plant greening, cell expansion, and flowering time to regulate plant growth and development in response to environmental light cues (Galvão and Fankhauser, 2015).
Of note, PhyB mutants display a long circadian period, predominantly under continuous red light, while PhyA mutants display long period under constant red and blue light (Somers et al., 1998). PhyB has been shown to directly interact with ELF3 (Liu et al., 2001), forming a link to the clock. In addition, PIFs were found to negatively regulate PhyB levels by promoting the proteasome dependent degradation of the activated PhyB form (Jang et al., 2010; Ni et al., 2014). Interestingly, it was found that nuclear-localized PhyB is required for stabilizing the ELF3 protein (Nieto et al., 2015), which represses the PIF4 function by sequestering the protein. Moreover, PhyB is known to interact with another PAS domain containing clock protein, ZTL/ADO1 (Jarillo et al., 2001a), as described below. In contrast, no direct interaction between the other four phytochrome proteins with clock proteins have been reported so far.
Circadian Clock Proteins ZTL, LKP2, and FKF1
The three paralogs in Arabidopsis, ZEITLUPE (ZTL), FLAVIN-KELCH-FBOX-1 (FKF1), and LOV-Kelch-Protein-2 (LKP2), share a high degree of amino acid identity of about 70–80% (Somers et al., 2000). They comprise a single N-terminal LOV-type PAS domain similar to that of phototropins binding oxidized FMN (Imaizumi et al., 2003), followed by an F-Box and a C-terminal Kelch-repeat domain, which mediate formation of SCF E3 ligase complexes and substrate binding for polyubiquitination, respectively (Somers et al., 2000; Andrade et al., 2001; Yasuhara et al., 2004; Figure 2). Blue light is perceived in a fluence-dependent manner via the PAS (LOV) domain and fed to the light-input of the circadian clock and regulation photoperiodic flowering control via protein–protein-interactions (Nelson et al., 2000; Imaizumi et al., 2003; Somers et al., 2004; Takase et al., 2011). The protein–protein-interaction either targets the partner to proteasomal degradation, such as the core clock components TOC1 and PRR5 by ZTL in the dark, or stabilizes the proteins, as shown for interactions of FKF1 with CONSTANS (CO) and CYCLING DOF FACTORS (CDFs), or all three proteins with GIGANTEA (GI) in blue light (Imaizumi et al., 2005; Kiba et al., 2007; Kim et al., 2007; Baudry et al., 2010; Song et al., 2014). Despite their structural similarity, ZTL, FKF1, and LKP2 exert distinct physiological roles. While ZTL is a major player in the evening loop of the circadian clock, the latter are mainly involved in the morning loop. This is achieved by a combination of different regulatory mechanisms, such as differential expression, protein stability and intracellular localization, as well as the photokinetic properties and fluence sensitivities of their light-sensing PAS domains (Somers et al., 2000, 2004; Kim et al., 2007; Baudry et al., 2010; Pudasaini and Zoltowski, 2013; Song et al., 2014).
ZTL was shown to interact with PhyB in vitro (Jarillo et al., 2001a), and several mutations in either of the domains of ZTL protein, including the PAS domain, did not abolish the interaction with the C-terminal part of PhyB, indicating that ztl phenotypes are not due to disrupted interaction between these two proteins (Kevei et al., 2006).
PAS/LOV Protein—Another Potential Plant Blue-light Receptor
Arabidopsis encodes yet another potential photoreceptor, termed PAS/LOV protein (PLP isoforms A to C). PLP harbors a PAS domain at either end of the protein, the C-terminal domain showing homology to FMN-binding LOV domains. Having no predicted effector domain, however, it is likely that they act in trans on other proteins (Crosson et al., 2002).
Although PLPs have not been studied in great detail and the exact function of both PAS domains are not resolved to date, they seem to be involved in the plant reaction to salt and dehydration stress, and protein interaction is modulated in a blue-light dependent manner in yeast (Ogura et al., 2008a,b). In addition, the PLP gene shows a clear diurnal expression pattern, with a maximum expression in the afternoon (Mockler et al., 2007), suggesting that it might act as a clock output.
PAS-like Domain in HD-ZIP Transcription Factors
REVOLUTA (REV), PHABULOSA (PHB), PHAVOLUTA (PHV), INCURATA4/CORONA (ICU4/CAN), and ATHB8 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 8) constitute the plant-specific Class III of homeodomain leucine zipper (HD-ZIP III) transcription factors. They are well studied major regulators of developmental processes and involved in establishing adaxial-abaxial polarity in lateral organs, meristem regulation and formation, vascular development, as well as embryonic patterning (Talbert et al., 1995; Zhong and Ye, 1999; McConnell et al., 2001; Otsuga et al., 2001; Prigge et al., 2005; Carlsbecker et al., 2010; Smith and Long, 2010). As a recurring pattern in plant PAS domain proteins of a family, they function not only redundantly, but display distinct, yet antagonistic function (Prigge et al., 2005).
The multidomain proteins consist of the N-terminally located eponymous HD-ZIP domain followed by two regulatory domains, the lipid ligand binding START and the START associated domain (HD-SAD, Mukherjee et al., 2009; Schrick et al., 2014). START/HD-SAD domains serve as regulatory domains recognized by microRNAs miR165 and miR166 (Emery et al., 2003). The C-terminus is constituted by a so-called MEKHLA domain, an approximately 150 amino acid long PAS-like domain (Mukherjee and Bürglin, 2006) that shares highest homology to bacterial sensor histidine kinases such as the oxygen sensor FixL (Hao et al., 2002; Mukherjee and Bürglin, 2006). It is not known, however, whether the transcription factors bind the same cofactor (i.e., heme) with its PAS-like domain because of the high sequence variability (Mukherjee and Bürglin, 2006). The PAS-like domain negatively regulates REV activity by inhibiting protein homodimerization (Magnani and Barton, 2011). Upon stimulation this inhibition is lifted and the REV protein is activated. Of note, HD-ZIP III proteins are known to be redox-sensitive (Comelli and Gonzalez, 2007; Xie et al., 2014), suggesting that the PAS-like domains in these proteins might also sense oxygen or ROS. A genome-wide binding analysis for REV revealed that PRR5 is a target gene, representing a link with the morning-loop of the circadian clock (Brandt et al., 2012).
MAPKKKs Harboring PAS Domains
Mitogen-activated protein kinase (MAPK) cascades are a common way of signal transduction and they are associated with integrating environmental cues into adequate responses (Romeis, 2001; Sinha et al., 2011). Arabidopsis encodes about 80 MAPKKK (MAP3K) genes. The subgroup B2 of Raf-like kinases consists of six members and comprises an N-terminal PAS domain (MAPK Group, 2002; Colcombet and Hirt, 2008). To date, knowledge of the physiological framework in which this subgroup of MAPKKKs exerts its role, the signaling mechanism and molecular function of the PAS domain is scarce.
MAP3Kδ4 was reported to be an auxin- and abscisic acid (ABA)-responsive kinase affecting plant growth, mediating tolerance to salt stress, and it is likely to be involved in other abiotic stress responses as well (Sasayama et al., 2011; Shitamichi et al., 2013). Recently, two functional redundant putative MAPKKK of the same group, Raf10 and Raf11, were identified likewise as positive regulators of ABA response and seed dormancy (Lee et al., 2015). As for MAP3Kδ4, the exact molecular function of the PAS domain within the MAPK signaling is not yet understood. Moreover, in animals it has been shown that a MAPK cascade can reset the circadian clock (Akashi and Nishida, 2000), whether such a scenario also exists in plant remains to be discovered.
Environmental Sensing and Plant Adaptation Through PAS Domain-containing Proteins
Light is the major clock input signal, which is used by plants to set the pace and phase of the oscillator, adjusts growth and development to changing environmental conditions. Most of the here described PAS domain-containing proteins sense light, and have been shown previously to be involved in entraining the circadian clock.
Although the PAS domain-containing phytochromes represent an important class of photoreceptors, they are not required for regulating circadian responses, per se (Yanovsky et al., 2000; Strasser et al., 2010). Still, under free-running conditions, in a light-intensity and quality dependent manner, phytochrome mutants were shown to exhibit a lengthened or reduced period and arrhythmic leaf movements (Somers et al., 1998; Strasser et al., 2010). Although light input is essential for clock periodicity, it has not been possible so far to relate this strongly to the presence of phytochromes, even though PhyB interacts with and stabilizes ELF3 in the light (Nieto et al., 2015). Potentially, the loss of phytochrome-dependent light signaling can be compensated for by other photoreceptors to sustain light entrainment of the clock. Indeed, it was previously shown that cryptochromes mediate phytochrome signaling to the clock in both red and blue light (Devlin and Kay, 2000). In addition, it has been established that the blue light cryptochromes differentially control circadian period and sustain rhythmicity across a physiological temperature range (Gould et al., 2013). Still, PhyB and ELF3 were shown to be negative regulators of dark-induced senescence (Sakuraba et al., 2014). Prolonged light-deprivation causes the inactivation of PhyB and the subsequent accumulation of PIF4 and PIF5 and the onset of leaf senescence. Under normal day-night rhythms, ELF3 represses PIF4 and PIF5 expression during the night. However, prolonged darkness overrides this control. Interestingly, short pulses of red light during 10 days of darkness prevent the onset of senescence in a PhyB dependent manner (Sakuraba et al., 2014).
The LOV domain-containing ZTL was shown to interact with PhyB as well as with CRY1, linking both phytochrome and cryptochrome with the circadian clock (Jarillo et al., 2001a). The ztl mutant is described as a clock mutant with altered period in a light-dependent manner (Somers et al., 2000). Therefore, ZTL might regulate the integration of light signals into the clock. In Arabidopsis, the blue-light receptor CRY2 interacts with and activates the transcription factor CRYPTOCHROME-INTERACTING basic helix–loop–helix 1 (CIB1). CIB1 regulates the floral integrator gene FLOWERING LOCUS T (FT), which in turn controls floral initiation (Liu et al., 2008). In blue light, ZTL/LKP2 protect CIB1 from proteasomal degradation (Liu et al., 2013) emphasizing the convergence of more than one, evolutionary distinct PAS domain-containing light receptors involved in photoperiodic regulation. Next to a role of CIB1 in regulating flowering time, it was found to have a negative impact on plant immunity (Malinovsky et al., 2014). In line with this, ZTL was found to affect the plant defense response (Wang et al., 2011).
In addition, the ZTL family target PRR5 was shown to repress the expression of genes involved in abiotic stress (Nakamichi et al., 2009, 2012). Furthermore, a triple loss of function prr5 prr7 prr9 mutant displays enhanced cold, drought and salinity tolerance, placing the clock PRR proteins in a central position mediating adaptation to environmental cues. Interestingly, overexpression of LKP2 improves plant drought tolerance by positively regulating dehydration-inducible genes such as DREB1A-C, RD29A, as well as DEHYDRIN (Miyazaki et al., 2015). Moreover, these genes overlap with those that show elevated expression in the prr5 prr7 prr9 mutant (Nakamichi et al., 2012), in agreement with a role of LKP2 in promoting the degradation of PRR5 (Baudry et al., 2010), and thereby promoting plant adaptation to a changing environment.
The HD-ZIPIII transcription factor REV was shown to control oxidative stress tolerance in Arabidopsis (Xie et al., 2014), aside from its role in plant development. This indicates that the HD-ZIP III family might be an important player integrating environmental signals for growth regulation. However, still little is known regarding the emerging role of this family in adaptive growth. Initial findings suggest a rather complex network embedding the transcription factors requiring thorough future investigations.
Conclusion
The PAS domain fold is a common protein module found throughout the kingdoms of life. Although the domain has been implicated in sensing a multitude of signals, in plants only evidence for a role in light signaling has been obtained so far. Many of the PAS domain-containing proteins are intimately connected to the circadian clock, suggesting that they might contribute to linking environmental conditions with clock entrainment.
Here, we presented the emerging links between PAS domain proteins, time keeping and the response of the plant to the environment. So far, a clear implication for light signaling and plant adaptation through PAS domain proteins has been established by predominantly altering protein–protein interaction properties of their proteins. Still, plenty of effort is needed to dissect the biochemical and mechanistic functions of most PAS domain proteins in plants. The role of most of these proteins in response to the environment is unexplored. In addition, the stimulus upon which they exert their function remains elusive. An appealing aim would be to characterize the family of MAPKKK proteins further, which seem to be linked with abiotic stress tolerances. It would be intriguing to reveal whether the PAS domain controls their signaling activity.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Akashi, M., and Nishida, E. (2000). Involvement of the MAP kinase cascade in resetting of the mammalian circadian clock. Genes Dev. 14, 645–649. doi: 10.1101/gad.14.6.645
Alabadí, D., Oyama, T., Yanovsky, M. J., Harmon, F. G., Mas, P., and Kay, S. A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883. doi: 10.1126/science.1061320
Andrade, M. A., González-Guzmán, M., Serrano, R., and Rodríguez, P. L. (2001). A combination of the F-box motif and kelch repeats defines a large Arabidopsis family of F-box proteins. Plant Mol. Biol. 46, 603–614. doi: 10.1023/A:1010650809272
Baima, S., Possenti, M., Matteucci, A., Wisman, E., Altamura, M. M., Ruberti, I., et al. (2001). The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 126, 643–655. doi: 10.1104/pp.126.2.643
Baudry, A., Ito, S., Song, Y. H., Strait, A. A., Kiba, T., Lu, S., et al. (2010). F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22, 606–622. doi: 10.1105/tpc.109.072843
Brandt, R., Salla-Martret, M., Bou-Torrent, J., Musielak, T., Stahl, M., Lanz, C., et al. (2012). Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J. 72, 31–42. doi: 10.1111/j.1365-313X.2012.05049.x
Burgie, E. S., and Vierstra, R. D. (2014). Phytochromes: an atomic perspective on photoactivation and signaling. Plant Cell 26, 4568–4583. doi: 10.1105/tpc.114.131623
Burgie, E. S., Bussell, A. N., Walker, J. M., Dubiel, K., and Vierstra, R. D. (2014). Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc. Natl. Acad. Sci. U.S.A. 111, 10179–10184. doi: 10.1073/pnas.1403096111
Casal, J. J. (2013). Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 64, 403–427. doi: 10.1146/annurev-arplant-050312-120221
Carlsbecker, A., Lee, J.-Y., Roberts, C. J., Dettmer, J., Lehesranta, S., Zhou, J., et al. (2010). Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316–321. doi: 10.1038/nature08977
Chen, E., Swartz, T. E., Bogomolni, R. A., and Kliger, D. S. (2007). A LOV story: the signaling state of the Phot1 LOV2 photocycle involves chromophore-triggered protein structure relaxation, as probed by Far-UV time-resolved optical rotatory dispersion spectroscopy. Biochemistry 46, 4619–4624. doi: 10.1021/bi602544n
Chen, M., Tao, Y., Lim, J., Shaw, A., and Chory, J. (2005). Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr. Biol. 15, 637–642. doi: 10.1016/j.cub.2005.02.028
Chow, B. Y., and Kay, S. A. (2013). Global approaches for telling time: omics and the Arabidopsis circadian clock. Semin. Cell Dev. Biol. 24, 383–392. doi: 10.1016/j.semcdb.2013.02.005
Christie, J. M., Swartz, T. E., Bogomolni, R. A., and Briggs, W. R. (2002). Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 32, 205–219. doi: 10.1046/j.1365-313X.2002.01415.x
Colcombet, J., and Hirt, H. (2008). Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem. J. 413, 217–226. doi: 10.1042/BJ20080625
Corchnoy, S. B., Swartz, T. E., Lewis, J. W., Szundi, I., Briggs, W. R., and Bogomolni, R. A. (2003). Intramolecular proton transfers and structural changes during the photocycle of the LOV2 domain of phototropin 1. J. Biol. Chem. 278, 724–731. doi: 10.1074/jbc.M209119200
Comelli, R. N., and Gonzalez, D. H. (2007). Conserved homeodomain cysteines confer redox sensitivity and influence the DNA binding properties of plant class III HD-Zip proteins. Arch. Biochem. Biophys. 467, 41–47. doi: 10.1016/j.abb.2007.08.003
Crosson, S., and Moffat, K. (2001). Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. U.S.A. 98, 2995–3000. doi: 10.1073/pnas.051520298
Crosson, S., and Moffat, K. (2002). Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell 14, 1067–1075. doi: 10.1105/tpc.010475
Crosson, S., Sudarshan Rajagopal, A., and Moffat, K. (2002). The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry 42, 2–10. doi: 10.1021/bi026978l
Demarsy, E., and Fankhauser, C. (2009). Higher plants use LOV to perceive blue light. Curr. Opin. Plant Biol. 12, 69–74. doi: 10.1016/j.pbi.2008.09.002
Delvin, P. F., Patel, S. R., and Whitelam, G. C. (1998). Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10, 1479–1487
Devlin, P. F., and Kay, S. A. (2000). Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12, 2499–2509. doi: 10.1105/tpc.12.12.2499
Dodd, A. N., Salathia, N., Hall, A., Kevei, E., Toth, R., Nagy, F., et al. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309, 630–633. doi: 10.1126/science.1115581
Duclercq, J., Assoumou Ndong, Y. P., Guerineau, F., Sangwan, R. S., and Catterou, M. (2011). Arabidopsis shoot organogenesis is enhanced by an amino acid change in the ATHB15 transcription factor. Plant Biol. (Stuttg). 13, 317–324. doi: 10.1111/j.1438-8677.2010.00363.x
Emery, J. F., Floyd, S. K., Alvarez, J., Eshed, Y., Hawker, N. P., Izhaki, A., et al. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774. doi: 10.1016/j.cub.2003.09.035
Franklin, K. A., Praekelt, U., Stoddart, W. M., Billingham, O. E., Halliday, K. J., and Whitelam, G. C. (2003). Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol. 131, 1340–1346. doi: 10.1104/pp.102.015487
Galvão, V. C., and Fankhauser, C. (2015). Sensing the light environment in plants: photoreceptors and early signaling steps. Curr. Opin. Neurobiol. 34C, 46–53. doi: 10.1016/j.conb.2015.01.013
Gehan, M. A., Greenham, K., Mockler, T. C., and McClung, C. R. (2015). Transcriptional networks – crops, clocks, and abiotic stress. Curr. Opin. Plant Biol. 24, 39–46. doi: 10.1016/j.pbi.2015.01.004
Gendron, J. M., Pruneda-Paz, J. L., Doherty, C. J., Gross, A. M., Kang, S. E., and Kay, S. A. (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 109, 3167–3172. doi: 10.1073/pnas.1200355109
Gould, P. D., Ugarte, N., Domijan, M., Costa, M., Foreman, J., MacGregor, D., et al. (2013). Network balance via CRY signalling controls the Arabidopsis circadian clock over ambient temperatures. Mol. Syst. Biol. 9, 650. doi: 10.1038/msb.2013.7
Halavaty, A. S., and Moffat, K. (2007). N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry 46, 14001–14009. doi: 10.1021/bi701543e
Halavaty, A. S., and Moffat, K. (2013). Coiled-coil dimerization of the LOV2 domain of the blue-light photoreceptor phototropin 1 from Arabidopsis thaliana. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 69, 1316–1321. doi: 10.1107/S1744309113029199
Hao, B., Isaza, C., Arndt, J., Soltis, M., and Chan, M. K. (2002). Structure-based mechanism of O2 sensing and ligand discrimination by the FixL heme domain of Bradyrhizobium japonicum. Biochemistry 41, 12952–12958. doi: 10.1021/bi020144l
Harmer, S. L. (2009). The circadian system in higher plants. Annu. Rev. Plant. Biol. 60, 357–377. doi: 10.1146/annurev.arplant.043008.092054
Harper, S. M., Neil, L. C., and Gardner, K. H. (2003). Structural basis of a phototropin light switch. Science 301, 1541–1544. doi: 10.1126/science.1086810
Haydon, M. J., Mielczarek, O., Robertson, F. C., Hubbard, K. E., and Webb, A. A. (2013). Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 502, 689–692. doi: 10.1038/nature12603
Hefti, M. H., Françoijs, K.-J., de Vries, S. C., Dixon, R., and Vervoort, J. (2004). The PAS fold. Eur. J. Biochem. 271, 1198–1208. doi: 10.1111/j.1432-1033.2004.04023.x
Huala, E., Oeller, P. W., Liscum, E., Han, I. S., Larsen, E., and Briggs, W. R. (1997). Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science 278, 2120–2123.
Hughes, J. (2013). Phytochrome cytoplasmic signaling. Annu. Rev. Plant Biol. 64, 377–402. doi: 10.1146/annurev-arplant-050312-120045
Humphrey, W., Dalke, A., and Schulten, K. (1996). VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38. doi: 10.1016/0263-7855(96)00018-5
Imaizumi, T., Schultz, T. F., Harmon, F. G., Ho, L. A., and Kay, S. A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297. doi: 10.1126/science.1110586
Imaizumi, T., Tran, H. G., Swartz, T. E., Briggs, W. R., and Kay, S. A. (2003). FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426, 302–306. doi: 10.1038/nature02090
Jang, I. C., Henriques, R., Seo, H. S., Nagatani, A., and Chua, N. H. (2010). Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22, 2370–2383. doi: 10.1105/tpc.109.072520
Jarillo, J. A., Capel, J., Tang, R. H., Yang, H. Q., Alonso, J. M., Ecker, J. R., et al. (2001a). An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature 410, 487–490. doi: 10.1038/35068589
Jarillo, J. A., Gabrys, H., Capel, J., Alonso, J. M., Ecker, J. R., and Cashmore, A. R. (2001b). Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature 410, 952–954. doi: 10.1038/35073622
Kasahara, M., Swartz, T. E., Olney, M. A., Onodera, A., Mochizuki, N., Fukuzawa, H., et al. (2002). Photochemical properties of the flavin mononucleotide-binding domains of the phototropins from Arabidopsis, rice, and Chlamydomonas reinhardtii. Plant Physiol. 129, 762–773. doi: 10.1104/pp.002410
Kevei, E., Gyula, P., Hall, A., Kozma-Bognár, L., Kim, W. Y., Eriksson, M. E., et al. (2006). Forward genetic analysis of the circadian clock separates the multiple functions of ZEITLUPE. Plant Physiol. 140, 933–945. doi: 10.1104/pp.105.074864
Kiba, T., Henriques, R., Sakakibara, H., and Chua, N.-H. (2007). Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19, 2516–2530. doi: 10.1105/tpc.107.053033
Kim, W. Y., Fujiwara, S., Suh, S. S., Kim, J., Kim, Y., Han, L., et al. (2007). ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449, 356–360. doi: 10.1038/nature06132
Kinoshita, T., Doi, M., Suetsugu, N., Kagawa, T., Wada, M., and Shimazaki, K. (2001). Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414, 656–660. doi: 10.1038/414656a
Knoll, A. H. (2003). The geological consequences of evolution. Geobiology 1, 3–14. doi: 10.1046/j.1472-4669.2003.00002.x
Kolmos, E., Chow, B. Y., Pruneda-Paz, J. L., and Kay, S. A. (2014). HsfB2b-mediated repression of PRR7 directs abiotic stress responses of the circadian clock. Proc. Natl. Acad. Sci. U.S.A. 111, 16172–16177. doi: 10.1073/pnas.1418483111
Lai, A. G., Doherty, C. J., Mueller-Roeber, B., Kay, S. A., Schippers, J. H. M., and Dijkwel P. P. (2012). CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci. U.S.A. 109, 17129–17134. doi: 10.1073/pnas.1209148109
Lee, S.-J., Lee, M. H., Kim, J. -I., and Kim, S. Y. (2015). Arabidopsis Putative MAP Kinase Kinase Kinases Raf10 and Raf11 are Positive Regulators of Seed Dormancy and ABA Response. Plant Cell Physiol. 56, 84–97. doi: 10.1093/pcp/pcu148
Levskaya, A., Weiner, O. D., Lim, W. A., and Voigt, C. A. (2009). Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001. doi: 10.1038/nature08446
Liscum, E., and Briggs, W. R. (1995). Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell 7, 473–485. doi: 10.1105/tpc.7.4.473
Liu, H., Yu, X., Li, K., Klejnot, J., Yang, H., Lisiero, D., et al. (2008). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535–1539. doi: 10.1126/science.1163927
Liu, H., Wang, Q., Liu, Y., Zhao, X., Imaizumi, T., Somers, D. E., et al. (2013). Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 110, 17582–17587. doi: 10.1073/pnas.1308987110
Liu, X. L., Covington, M. F., Fankhauser, C., Chory, J., and Wagner, D. R. (2001). ELF3 encodes a circadian clock–regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13, 1293–1304. doi: 10.1105/tpc.13.6.1293
Loudon, A. S. (2012). Circadian biology: a 2.5 billion year old clock. Curr. Biol. 22, R570–R571. doi: 10.1016/j.cub.2012.06.023
Magnani, E., and Barton, M. K. (2011). A per-ARNT-sim-like sensor domain uniquely regulates the activity of the homeodomain leucine zipper transcription factor REVOLUTA in Arabidopsis. Plant Cell 23, 567–582. doi: 10.1105/tpc.110.080754
Malinovsky, F. G., Batoux, M., Schwessinger, B., Youn, J. H., Stransfeld, L., Win, J., et al. (2014). Antagonistic regulation of growth and immunity by the Arabidopsis basic helix-loop-helix transcription factor homolog of brassinosteroid enhanced expression2 interacting with increased leaf inclination1 binding bHLH1. Plant Physiol. 164, 1443–1455. doi: 10.1104/pp.113.234625
MAPK Group. (2002). Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 7, 301–308. doi: 10.1016/S1360-1385(02)02302-6
Matsuoka, D., and Tokutomi, S. (2005). Blue light-regulated molecular switch of Ser/Thr kinase in phototropin. Proc. Natl. Acad. Sci. U.S.A. 102, 13337–13342. doi: 10.1073/pnas.0506402102
Matsuzaki, J., Kawahara, Y., and Izawa, T. (2015). Punctual Transcriptional Regulation by the Rice Circadian Clock under Fluctuating Field Conditions. Plant Cell 27, 633–648. doi: 10.1105/tpc.114.135582
McClung, C. R. (2011). The genetics of plant clocks. Adv. Genet. 74, 105–139. doi: 10.1016/B978-0-12-387690-4.00004-0
McConnell, J. R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M. K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. doi: 10.1038/35079635
McIntosh, B. E., Hogenesch, J. B., and Bradfield, C. A. (2010). Mammalian Per-Arnt-Sim Proteins in Environmental Adaptation. Annu. Rev. Physiol. 72, 625–645. doi: 10.1146/annurev-physiol-021909-135922
Mei, Q., and Dvornyk, V. (2014). Evolution of PAS domains and PAS-containing genes in eukaryotes. Chromosoma. 123, 385–405. doi: 10.1007/s00412-014-0457-x
Michael, T. P., Salome, P. A., and McClung, C. R. (2003). Two Arabidopsis circadian oscillators can be distinguishedby differential temperature sensitivity. Proc. Natl. Acad. Sci. U.S.A. 100, 6878–6883. doi: 10.1073/pnas.1131995100
Mitchell, A., Chang, H.-Y., Daugherty, L., Fraser, M., Hunter, S., Lopez, R., et al. (2015). The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43, D213–D221. doi: 10.1093/nar/gku1243
Miyazaki, Y., Abe, H., Takase, T., Kobayashi, M., and Kiyosue, T. (2015). Overexpression of LOV KELCH PROTEIN 2 confers dehydration tolerance and is associated with enhanced expression of dehydration-inducible genes in Arabidopsis thaliana. Plant Cell Rep. 4, 843–852. doi: 10.1007/s00299-015-1746-4
Mockler, T. C., Michael, T. P., Priest, H. D., Shen, R., Sullivan, C. M., Givan, S. A., et al. (2007). THE DIURNAL PROJECT: diurnal and circadian expression profiling, model-based pattern matching and promoter analysis. Cold Spring Harb. Symp. Quant. Biol. 72, 353–363. doi: 10.1101/sqb.2007.72.006
Möglich, A., Ayers, R. A., and Moffat, K. (2009). Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17, 1282–1294. doi: 10.1016/j.str.2009.08.011
Mukherjee, K., and Bürglin, T. R. (2006). MEKHLA, a novel domain with similarity to PAS domains, is fused to plant homeodomain-leucine zipper III proteins. Plant Physiol. 140, 1142–1150. doi: 10.1104/pp.105.073833
Mukherjee, K., Brocchieri, L., and Bürglin, T. R. (2009). A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 26, 2775–2794. doi: 10.1093/molbev/msp201
Nakamichi, N., Kiba, T., Kamioka, M., Suzuki, T., Yamashino, T., Higashiyama, T., et al. (2012). Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. U.S.A. 109, 17123–17128. doi: 10.1073/pnas.1205156109
Nakamichi, N., Kusano, M., Fukushima, A., Kita, M., Ito, S., Yamashino, T., et al. (2009). Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 50, 447–462. doi: 10.1093/pcp/pcp004
Nakasako, M., Zikihara, K., Matsuoka, D., Katsura, H., and Tokutomi, S. (2008). Structural Basis of the LOV1 Dimerization of Arabidopsis Phototropins 1 and 2. J. Mol. Biol. 381, 718–733. doi: 10.1016/j.jmb.2008.06.033
Nambu, J. R., Lewis, J. O., Wharton, K. A., and Crews, S. T. (1991). The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell 67, 1157–1167.
Nelson, D. C., Lasswell, J., Rogg, L. E., Cohen, M. A., and Bartel, B. (2000). FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101, 331–340. doi: 10.1016/S0092-8674(00)80842-9
Ni, M., Tepperman, J. M., and Quail, P. H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400, 781–784.
Ni, W., Xu, S. L., Tepperman, J. M., Stanley, D. J., Maltby, D. A., Gross, J. D., et al. (2014). A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344, 1160–1164. doi: 10.1126/science.1250778
Nieto, C., López-Salmerón, V., Davière, J. M., and Prat, S. (2015). ELF3-PIF4 interaction regulates plant growth independently of the evening complex. Curr. Biol. 25, 187–193. doi: 10.1016/j.cub.2014.10.070
Nusinow, D. A., Helfer, A., Hamilton, E. E., King, J. J., Imaizumi, T., Schultz, T. F., et al. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475, 398–402. doi: 10.1038/nature10182
Ochando, I., González-Reig, S., Riopli, J. J., Vera, A., and Martínez-Laborda, A. (2008). Alteration of the shoot radial pattern in Arabidopsis thaliana by a gain-of-function allele of the class III HD-Zip gene INCURVATA4. Int. J. Dev. Biol. 52, 953–961. doi: 10.1387/ijdb.072306io
Ogura, Y., Komatsu, A., Zikihara, K., Nanjo, T., Tokutomi, S., Wada, M., et al. (2008a). Blue light diminishes interaction of PAS/LOV proteins, putative blue light receptors in Arabidopsis thaliana with their interacting partners. J. Plant Res. 121, 97–105. doi: 10.1007/s10265-007-0118-8
Ogura, Y., Tokutomi, S., Wada, M., and Kiyosue, T. (2008b). PAS/LOV proteins. Plant Signal. Behav. 3, 966–968. doi: 10.4161/psb.6150
Otsuga, D., DeGuzman, B., Prigge, M. J., Drews, G. N., and Clark, S. E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25, 223–236. doi: 10.1046/j.1365-313x.2001.00959.x
Pfeifer, A., Mathes, T., Lu, Y., Hegemann, P., and Kottke, T. (2010). Blue light induces global and localized conformational changes in the kinase domain of full-length phototropin. Biochemistry 49, 1024–1032. doi: 10.1021/bi9016044
Ponting, C. P., and Aravind, L. (1997). PAS: a multifunctional domain family comes to light. Curr. Biol. 7, R674–R677.
Prigge, M. J., Otsuga, D., Alonso, J. M., Ecker, J. R., Drews, G. N., and Clark, S. E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17, 61–76. doi: 10.1105/tpc.104.026161
Pudasaini, A., and Zoltowski, B. D. (2013). Zeitlupe senses blue-light fluence to mediate circadian timing in Arabidopsis thaliana. Biochemistry 52, 7150–7158. doi: 10.1021/bi401027n
Qin, M., Kuhn, R., Moran, S., and Quail, P. H. (1997). Overexpressed phytochrome C has similar photosensory specificity to phytochrome B but a distinctive capacity to enhance primary leaf expansion. Plant J. 12, 1163–1172.
Raymond, J., and Segrè, D. (2006). The effect of oxygen on biochemical networks and the evolution of complex life. Science 311, 1764–1767. doi: 10.1126/science.1118439
Reed, J. W., Nagatani, A., Elich, T. D., Fagan, M., and Chory, J. (1994). Phytochrome A and Phytochrome B Have Overlapping but Distinct Functions in Arabidopsis Development. Plant Physiol. 104, 1139–1149.
Romeis, T. (2001). Protein kinases in the plant defence response. Curr. Opin. Plant Biol. 4, 407–414. doi: 10.1016/S1369-5266(00)00193-X
Sakai, T., Kagawa, T., Kasahara, M., Swartz, T. E., Christie, J. M., Briggs, W. R., et al. (2001). Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. U.S.A. 98, 6969–6974. doi: 10.1073/pnas.101137598
Sakuraba, Y., Jeong, J., Kang, M. Y., Kim, J., Paek, N. C., and Choi, G. (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5, 4636. doi: 10.1038/ncomms5636
Salomon, M., Christie, J. M., Knieb, E., Lempert, U., and Briggs, W. R. (2000). Photochemical and Mutational Analysis of the FMN-Binding Domains of the Plant Blue Light Receptor, Phototropin. Biochemistry 39, 9401–9410. doi: 10.1021/bi000585
Sanchez, A., Shin, J., and Davis, S. J. (2011). Abiotic stress and the plant circadian clock. Plant. Signal. Behav. 6, 223–231. doi: 10.4161/psb.6.2.14893
Sasayama, D., Matsuoka, D., Oka, M., Shitamichi, N., Furuya, T., Azuma, T., et al. (2011). MAP3Kδ4, an Arabidopsis Raf-like MAP3K, regulates plant growth and shoot branching. Plant Biotech. 28, 463–470. doi: 10.5511/plantbiotechnology.11.1027a
Schippers, J. H. M., Nguyen, H. M., Lu, D., Schmidt, R., and Mueller-Roeber, B. (2012). ROS homeostasis during development: an evolutionary conserved strategy. Cell. Mol. Life Sci. 69, 3245–3257. doi: 10.1007/s00018-012-1092-4
Schirrmeister, B. E., de Vos, J. M., Antonelli, A., and Bagheri, H. C. (2013). Evolution of multicellularity coincided with increased diversification of cyanobacteria and the Great Oxidation Event. Proc. Natl. Acad. Sci. U.S.A. 110, 1791–1796. doi: 10.1073/pnas.1209927110
Schrick, K., Bruno, M., Khosla, A., Cox, P. N., Marlatt, S. A., Roque, R. A., et al. (2014). Shared functions of plant and mammalian StAR-related lipid transfer (START) domains in modulating transcription factor activity. BMC Biol. 12:70. doi: 10.1186/s12915-014-0070-8
Schultz, T. F., Kiyosue, T., Yanovsky, M., Wada, M., and Kay, S. A. (2001). A Role for LKP2 in the Circadian Clock of Arabidopsis. Plant Cell 13, 2659–2670. doi: 10.1105/tpc.010332
Shitamichi, N., Matsuoka, D., Sasayama, D., Furuya, T., and Nanmori, T. (2013). Over-expression of MAP3Kδ4, an ABA-inducible Raf-like MAP3K that confers salt tolerance in Arabidopsis. Plant Biotech. 30, 111–118. doi: 10.5511/plantbiotechnology.13.0108a
Sinha, A. K., Jaggi, M., Raghuram, B., and Tuteja, N. (2011). Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal. Behav. 6, 196–203. doi: 10.4161/psb.6.2.14701
Smith, Z. R., and Long, J. A. (2010). Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 464, 423–426. doi: 10.1038/nature08843
Somers, D. E., Devlin, P. F., and Kay, S. A. (1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282, 1488–1490.
Somers, D. E., Kim, W. Y., and Geng, R. (2004). The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16, 769–782. doi: 10.1105/tpc.016808
Somers, D. E., Schultz, T. F., Milnamow, M., and Kay, S. A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101, 319–329. doi: 10.1016/S0092-8674(00)80841-7
Song, Y. H., Estrada, D. A., Johnson, R. S., Kim, S. K., Lee, S. Y., MacCoss, M. J., et al. (2014). Distinct roles of FKF1, Gigantea, and Zeitlupe proteins in the regulation of Constans stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. U.S.A. 111, 17672–17677. doi: 10.1073/pnas.1415375111
Strasser, B., Sánchez-Lamas, M., Yanovsky, M. J., Casal, J. J., and Cerdán, P. D. (2010). Arabidopsis thaliana life without phytochromes. Proc. Natl. Acad. Sci. U.S.A. 107, 4776–4781. doi: 10.1073/pnas.0910446107
Sun, J., Qi, L., Li, Y., Zhai, Q., and Li, C. (2013). PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis. Plant Cell 25, 2102–2114. doi: 10.1105/tpc.113.112417
Swartz, T. E., Corchnoy, S. B., Christie, J. M., Lewis, J. W., Szundi, I., Briggs, W. R., et al. (2001). The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J. Biol. Chem. 276, 36493–36500. doi: 10.1074/jbc.M103114200
Takase, T., Nishiyama, Y., Tanihigashi, H., Ogura, Y., Miyazaki, Y., Yamada, Y., et al. (2011). LOV KELCH PROTEIN2 and ZEITLUPE repress Arabidopsis photoperiodic flowering under non-inductive conditions, dependent on FLAVIN-BINDING KELCH REPEAT F-BOX1. Plant J. 67, 608–621. doi: 10.1111/j.1365-313X.2011.04618.x
Talbert, P. B., Adler, H. T., Parks, D. W., and Comai, L. (1995). The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121, 2723–2735.
Thannickal, V. J. (2009). Oxygen in the evolution of complex life and the price we pay. Am. J. Respir. Cell Mol. Biol. 40, 507–510. doi: 10.1165/rcmb.2008-0360PS
Tokutomi, S., Matsuoka, D., and Zikihara, K. (2008). Molecular structure and regulation of phototropin kinase by blue light. Biochim. Biophys. Acta 1784, 133–142. doi: 10.1016/j.bbapap.2007.09.010
Wang, W., Barnaby, J. Y., Tada, Y., Li, H., Tör, M., Caldelari, D., et al. (2011). Timing of plant immune responses by a central circadian regulator. Nature 470, 110–114. doi: 10.1038/nature09766
Xie, Y., Huhn, K., Brandt, R., Potschin, M., Bieker, S., Straub, D., et al. (2014). REVOLUTA and WRKY53 connect early and late leaf development in Arabidopsis. Development 141, 4772–4783. doi: 10.1242/dev.117689
Yanovsky, M. J., Mazzella, M. A., and Casal, J. J. (2000). A quadruple photoreceptor mutant still keeps track of time. Curr. Biol. 10, 1013–1015. doi: 10.1016/S0960-9822(00)00651-5
Yasuhara, M., Mitsui, S., Hirano, H., Takanabe, R., Tokioka, Y., Ihara, N., et al. (2004). Identification of ASK and clock-associated proteins as molecular partners of LKP2 (LOV kelch protein 2) in Arabidopsis. J. Exp. Bot. 55, 2015–2027. doi: 10.1093/jxb/erh226
Zhong, R., and Ye, Z. H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11, 2139–2152.
Keywords: PAS domain, circadian clock, signal transduction, environmental stress response, growth adaptation
Citation: Vogt JHM and Schippers JHM (2015) Setting the PAS, the role of circadian PAS domain proteins during environmental adaptation in plants. Front. Plant Sci. 6:513. doi: 10.3389/fpls.2015.00513
Received: 15 April 2015; Accepted: 25 June 2015;
Published: 09 July 2015.
Edited by:
Seth J. Davis, Max Planck Institute for Plant Breeding Research, GermanyReviewed by:
Christian Heintzen, University of Manchester, UKJianping Yang, Chinese Academy of Agricultural Sciences, China
Copyright © 2015 Vogt and Schippers. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jos H. M. Schippers, Institute for Biology I, RWTH Aachen University, Worringerweg 1, 52074 Aachen, Germany, schippers@bio1.rwth-aachen.de
 Julia H. M. Vogt
Julia H. M. Vogt