- 1Faculty of Biology, Institute of Genetics, University of Munich, Martinsried, Germany
- 2Department of Organismic Interactions, Max Planck Institute for Terrestrial Microbiology, Marburg, Germany
- 3Cluster of Excellence on Plant Sciences, Botanical Institute, University of Cologne, Cologne, Germany
Arbuscular mycorrhiza (AM) fungi (Glomeromycota) form symbiosis with and deliver nutrients via the roots of most angiosperms. AM fungal hyphae are taken up by living root epidermal cells, a program which relies on a set of plant common symbiosis genes (CSGs). Plant root epidermal cells are also infected by the plant growth-promoting fungus Piriformospora indica (Basidiomycota), raising the question whether this interaction relies on the AM-related CSGs. Here we show that intracellular colonization of root cells and intracellular sporulation by P. indica occurred in CSG mutants of the legume Lotus japonicus and in Arabidopsis thaliana, which belongs to the Brassicaceae, a family that has lost the ability to form AM as well as a core set of CSGs. A. thaliana mutants of homologs of CSGs (HCSGs) interacted with P. indica similar to the wild-type. Moreover, increased biomass of A. thaliana evoked by P. indica was unaltered in HCSG mutants. We conclude that colonization and growth promotion by P. indica are independent of the CSGs and that AM fungi and P. indica exploit different host pathways for infection.
Introduction
Plants form mutualistic interactions with fungi from different taxonomic groups. Arbuscular mycorrhiza (AM) is a widespread symbiosis between plants and fungi of the phylum Glomeromycota (Parniske, 2008; Smith and Read, 2008) and is considered a key factor that allowed plants to colonize land more than 400 million years ago (Schüßler and Walker, 2011). The ancestral nature of this interaction raises the question to what extent other and potentially younger interactions between plant roots and fungi evolved by co-opting the genetic framework for AM formation. Candidates for such interactions include endomycorrhizal interactions formed with fungi of the order Sebacinales of the phylum Basidiomycota (Selosse et al., 2009).
An experimental model for this group of fungi is Piriformospora indica, which infects various taxonomically unrelated hosts and can increase plant growth and biomass (Peškan-Berghöfer et al., 2004; Waller et al., 2005; Shahollari et al., 2007; Sherameti et al., 2008; Camehl et al., 2010, 2011; Hilbert et al., 2012; Nongbri et al., 2012; Lahrmann et al., 2013; Venus and Oelmüller, 2013).
Phythormones like ethylene, jasmonic acid and gibberellins seem to positively influence root colonization by P. indica, while salicylic acid has an inhibitory effect (Jacobs et al., 2011; Khatabi et al., 2012). Genes involved in the synthesis of indole-3-acetaldoxime-derived compounds restrict the growth of P. indica within Arabidopsis thaliana roots (Nongbri et al., 2012). Indole-3-acetaldoxime is also an intermediate in the biosynthesis of the phytohormone indole-3-acetic acid (IAA) (Mikkelsen et al., 2009; Burow et al., 2010), which has been implicated in the P. indica-induced host growth promotion, together with cytokinin (Vadassery et al., 2008).
In the initial stages of plant root colonization, P. indica invades living plant cells (Jacobs et al., 2011; Zuccaro et al., 2011; Lahrmann and Zuccaro, 2012; Lahrmann et al., 2013). This initial stage is followed by the death of colonized cells even though the plant host displays no macroscopic signs of disease (Deshmukh et al., 2006; Jacobs et al., 2011; Zuccaro et al., 2011; Lahrmann and Zuccaro, 2012; Qiang et al., 2012). In A. thaliana roots, P. indica continuously infects cells de novo and colonized living cells are found beside dying and dead colonized cells 3 days after infection (Qiang et al., 2012).
During the early stage of the interaction, a plant-derived membrane has been observed which surrounds and separates P. indica hyphae from the plant cytoplasm (Jacobs et al., 2011; Lahrmann and Zuccaro, 2012; Lahrmann et al., 2013). The structural arrangement of this interaction is similar to that of transcellular and arbuscular hyphae of AM fungi, which are equally surrounded by a peri-fungal membrane (Bonfante and Genre, 2010).
We explored whether these structural similarities are reflected by a shared genetic basis between both symbioses. The successful invasion of the outer cell layers by AM fungi requires an ancestral plant genetic program that is conserved among angiosperms and encompasses the common symbiosis genes (CSGs). In legumes, the CSGs are required for both AM and the root nodule symbiosis with nitrogen-fixing bacteria (Kistner et al., 2005; Gutjahr, 2014; Svistoonoff et al., 2014), and encode the symbiosis receptor-like kinase SYMRK (Antolín-Llovera et al., 2014; Ried et al., 2014), the nucleoporins of the NUP107–160/NUP84 subcomplex (Alber et al., 2007) NUP85, NUP133, and the SEC13 HOMOLOG1 (SEH1) NENA (Kanamori et al., 2006; Saito et al., 2007; Groth et al., 2010), CASTOR and POLLUX, cation channels localized at the nuclear envelope (Charpentier et al., 2008; Venkateshwaran et al., 2012), as well as the nucleoplasmatic complex formed by a calcium and calmodulin-dependent protein kinase (CCaMK) and CYCLOPS responsible for calcium signal decoding (Singh and Parniske, 2012; Singh et al., 2014). The signal transduction pathway involving the products of the CSGs leads from the perception of microbial signals at the plasma membrane to the transcriptional activation of genes in the nucleus (Gutjahr and Parniske, 2013). In the legume Lotus japonicus CSG mutants are all impaired in the intracellular accommodation of both rhizobia and AM fungi (Kistner et al., 2005).
A. thaliana is a member of the Brassicaceae, a family which lost the ability to establish AM symbiosis (Delaux et al., 2014). This loss is correlated with the absence of a specific set of CSGs, including CCaMK and CYCLOPS (Delaux et al., 2014). Despite this loss, A. thaliana retained homologs of common symbiosis genes (HCSGs). Based on phylogenetic and/or synteny analyses, orthologs or candidate orthologs of legume CSGs in A. thaliana have been identified for POLLUX (Ané et al., 2004; Delaux et al., 2013) and genes coding for members of the NUP107–160 subcomplex, including NUP133 and SEC13 (Wiermer et al., 2012; Binder and Parniske, 2013). Importantly, an AtPOLLUX version was able to restore nodulation in the non-nodulating mutant dmi1–4 of M. truncatula (Venkateshwaran et al., 2012), indicating relative conservation of its symbiotic activity in A. thaliana. SYMRK encodes a malectin-like domain leucine-rich repeat receptor-like kinase, a gene family that has expanded to 50 members in A. thaliana (Hok et al., 2011). However, based on synteny analysis (Kevei et al., 2005) and the lack of SYMRK-specific amino-acid sequence patterns in the kinase domain (Markmann et al., 2008), a SYMRK ortholog appears to be absent from the A. thaliana genome. ShRK1 (SYMRK-homologous Receptor-like Kinase 1) and ShRK2 are the closest SYMRK homologs in A. thaliana. The domain organization of ShRK1 and 2 is identical to that of L. japonicus SYMRK (Markmann et al., 2008).
To explore the role of L. japonicus CSGs and A. thaliana HCSGs in the interaction with P. indica, we investigated whether the fungus was able to penetrate root cells, complete its life cycle and promote host plant growth in the respective mutant backgrounds.
Materials and Methods
Fungal Strains and Growth Conditions
P. indica (Verma et al., 1998, DSM11827, Leibniz Institute DSMZ—German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany) and Piriformospora williamsii (Basiewicz et al., 2012) were grown at 28°C in the dark on plates with solid (1.5% agar) complete medium (CM) (Pham et al., 2004). For studies with plants grown in soil, fungal mycelium was propagated in liquid CM medium, in the dark, at RT and 120 rpm shaking. Mycelium was washed three times with sterile distilled water directly prior to mixing with soil substrate.
For experiments on plates, fungal chlamydospores were obtained from 4 to 6-week-old cultures, as follows: Tween water (0.002% Tween 20) was added to plates containing mycelium, which was rubbed off from the agar surface, and the resulting spore suspension was collected and centrifuged three times at 3000 g for washing. Chlamydospore concentration was estimated using a Fuchs–Rosenthal counting chamber and adjusted to the desired 5 × 105 ml−1 with Tween water.
Plant Genotypes and Growth Conditions
For co-cultivation with P. indica, L. japonicus seeds (Supplementary Table 1) were surface sterilized in a 2% sodium hypochlorite solution for 7 min, washed three times with sterile water for 5 min each, and imbibed in water overnight. Seeds were then put on plates with solid modified Hoagland's medium [HO, 5 mM KNO3; 5 mM Ca(NO3)2; 2 mM MgSO4; 4 mM KH2PO4; 0.03 g l−1 Sprint 138 iron chelate; 0.1% micronutrients solution containing 2.86 g l−1 H3BO3; 1.81 g l−1 MnCl2.4H2O; 0.08 g l−1 CuSO4.5H2O; 0.02 g l−1 85% MoO3.H2O; based on Hoagland and Arnon (1950)] and kept for 4 days at 24°C in the dark for germination.
A. thaliana seeds were obtained from “The Nottingham A. thaliana Stock Centre”—NASC (Scholl et al., 2000) (Supplementary Table 2). For co-cultivation with P. indica or P. williamsii and tests for plant growth promotion, A. thaliana seeds were sterilized by incubation for 5 min in 70% ethanol, 0.05% Tween 20 followed by 2 min in 100% ethanol, left to dry and stratified for 48 h at 4°C in the dark. Plants were then grown under long day conditions (16 h:8 h, light:dark, at 23°C, 85 μmol.m−2 s−1), on half-strength (½) Murashige and Skoog medium (MS, Murashige and Skoog, 1962) with or without sucrose (0.05%) or on modified HO solidified with gelrite 4 g l−1, and four times more phosphate than in the original recipe.
Seven to ten-day-old A. thaliana or four-day-old L. japonicus plants were mock-inoculated with 1 ml Tween water (control) or either P. indica or P. williamsii chlamydospore suspensions. Non-germinated seeds and retarded seedlings were removed from the plates before inoculation. Co-cultures or mock-inoculated plants were grown at 24°C under long day conditions (16 h:8 h, light:dark). Biomass of seedlings was determined using a digital microbalance 7 days post inoculation (dpi).
For analysis of growth promotion under different nutrient regimes, plants were grown on the following media: modified HO with no KNO3 or Ca(NO3)2, supplemented with 0.05 mM KNO3, 0.5 mM KNO3, or 5 mM KNO3 (standard concentration), modified HO depleted of phosphate or with 4 mM KH2PO4(standard concentration); and modified HO depleted of ammonium or with 10 or 20 mM NH4Cl. Where necessary, potassium and calcium concentrations were compensated with KCl and CaCl2, respectively.
For growth promotion experiments in soil, plants were grown on either low-nutrient soil (50–100 mg l−1 N, 50–100 mg l−1 P, 100–150 mg l−1 K) or high-nutrient soil (500 mg l−1 N, 500 mg l−1 P, 500 mg l−1 K) mixed with 1 g fresh or autoclaved mycelium per 100 g sterile substrate for inoculation and mock-inoculation, respectively. Plant height was determined 7, 11, 16, and 21 dpi.
Sequence Alignments
Complete protein sequences were obtained from The Arabidopsis Information Resource (TAIR—www.arabidopsis.org) for A. thaliana, and from the GenBank for L. japonicus. Alignments (Supplementary Figures 7–9) were performed with MAFFT 6.822 (Katoh et al., 2002) or ClustalW2 (Larkin et al., 2007; Goujon et al., 2010) with the default settings. Searches for conserved domains in the protein sequences were performed using ScanProsite (de Castro et al., 2006) and/or InterProScan (Jones et al., 2014), and based on data available from Kanamori et al. (2006), Markmann et al. (2008), and Groth et al. (2010).
Determination of Fungal Colonization by Microscopy
To observe P. indica intracellular sporulation in A. thaliana and L. japonicus roots, colonized roots at 14 dpi were incubated at 96°C for 1 (A. thaliana) or 10 min (L. japonicus) in 10% (w/v) KOH and double-stained in the dark at room temperature for 20 min with 10 μg ml−1 WGA-AF488 (Wheat Germ Agglutinin-Alexa Fluor 488) (Molecular Probes, Karlsruhe, Germany) to visualize fungal structures, and 10 μg ml−1 propidium iodide (PI) to visualize plant cell walls. Samples were analyzed with a Leica DMI6000B microscope using differential interference contrast or epifluorescence (GFP filter set for WGA-AF488: excitation 450–490 nm, emission 500–550 nm; TX2 filter settings for PI: 540–580 nm excitation, 608–683 nm emission).
Root colonization and plant cell viability were analyzed by confocal laser scanning microscopy (CLSM). Fungal cell walls within colonized roots at 4 (A. thaliana), and 3 dpi (L. japonicus) were affinity-labeled for 10 min with 10 μg ml−1 WGA-AF488 (Molecular Probes, Karlsruhe, Germany). Membranes were stained with 3 μM FM4-64 (Molecular Probes, Karlsruhe, Germany) for 4 min (at 260 mm Hg). Root samples were imaged with TCS-SP5 or TCS-SP8 confocal microscopes (Leica, Bensheim, Germany) with excitation at 488 nm for WGA-AF488 and detection at 500–540 nm. FM4-64 was excited at 633 nm and detected at 650–690 nm. Propidium iodide was excited at 540 nm and detected at 600–630 nm.
Quantification of Fungal Colonization by qPCR
Roots of A. thaliana and L. japonicus colonized with P. indica (14 dpi) were thoroughly washed to remove fungal hyphae from the root surface. Two hundred micrograms of root material were then used for DNA extraction according to the protocol of Doyle and Doyle (1987). Real-time qPCR analyses were performed from 10 ng DNA mixed with the appropriate primers: P. indica Transcription Elongation Factor (Butehorn et al., 2000); A. thaliana Ubiquitin (Khatabi et al., 2012); or L. japonicus Ubiquitin (Takeda et al., 2009) in 10 μl SYBRgreen Supermix (BIORAD) using the following amplification protocol: 2′–95°C; 40 × (30″–95°C; 30″–59°C; 30″–72°C); melting curve 95°C–60°C–95°C. Fungal colonisation was quantified by the 2−△Ct method (Livak and Schmittgen, 2001) by subtracting the raw threshold cycle (Ct) values of P. indica TEF from those of plant UBI to obtain ΔCt.
β-Glucuronidase (GUS) Staining Assays
GUS staining assays were performed with the L. japonicus symbiosis-reporter line T90 (Webb et al., 2000). Colonized roots at 3, 7, and 14 dpi with P. indica chlamydospores or Tween water were vacuum infiltrated with X-Gluc staining solution (100 mM sodium phosphate buffer pH 7.0; 10 mM EDTA; 0.1% Triton-X 100; 0.5 mg ml−1 X-Gluc; 1 mM K3[Fe(CN)6]; 1 mM K4[Fe(CN)6].3H2O) three times for 10 min, followed by incubation at 37°C for 18 h in the dark. Root systems were inspected for GUS staining with a Leica MZFLIII stereomicroscope.
The Mesorhizobium loti MAFF303099 strain constitutively expressing DsRED (Maekawa et al., 2009) was used to inoculate roots of the L. japonicus T90 line as a suspension in Fahraeus medium (Fahraeus, 1957) adjusted to an OD600 of 0.05.
Statistical Analyses
Statistical analyses were performed with R version 3.0.2 (2013-09-25) “Frisbee Sailing” (R Core Team, 2013) using the package “agricolae” (De Mendiburu, 2010). Pairwise or multiple comparisons of the different subsets of data were performed with the Kruskal–Wallis test followed by a Bonferroni–Holm correction using the mock-inoculated samples as control group. Two-sided, unpaired t-tests with equal variance were used to analyze relative amount of fungal DNA within plant roots.
Results and Discussion
Intracellular Infection and Sporulation by P. indica Occurs Independently of L. japonicus and A. thaliana Common Symbiosis Genes
We investigated whether the classical CSGs are involved in the interaction between roots of the legume L. japonicus and P. indica. In root epidermal cells of L. japonicus wild-type (ecotype “Gifu”), and in CSG mutants intracellular hyphae were detected at 7 dpi (Figure 1) and sporulation at 14 dpi (Supplementary Figure 1), evidencing that P. indica entered host cells and successfully completed its life cycle in all genotypes tested. These observations indicate that the CSGs are not required for the successful infection of host cells by P. indica.
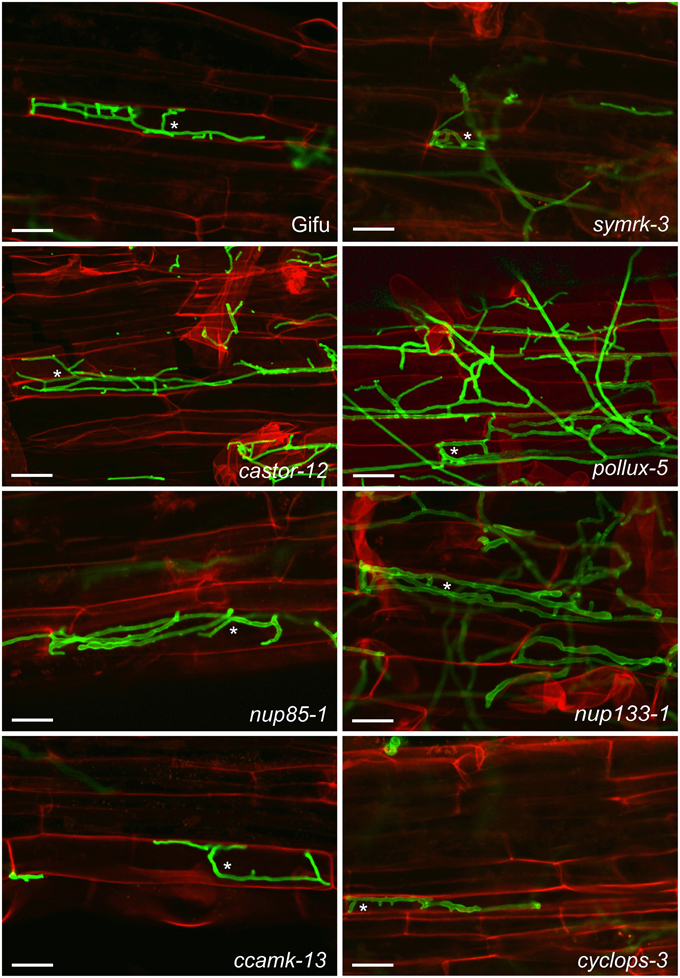
Figure 1. Colonization of L. japonicus root cells by P. indica. Wild-type (Gifu) and the indicated common symbiosis mutants were analyzed 7 dpi with P. indica chlamydospores. Intracellular hyphae were present in all genotypes (examples are indicated by asterisks). Roots were cleared and double stained with propidium iodide (red), for cell wall visualization, and WGA-AF488 (green), for fungal structures. Scale bar: 25 μm.
The L. japonicus symbiosis-reporter line T90 carries a promoter:GUS fusion and is activated in response to Mesorhizobium loti or nodulation factors, and AM fungi (Webb et al., 2000; Radutoiu et al., 2003; Kistner et al., 2005). T90 reporter activation requires L. japonicus CSGs (Kistner et al., 2005; Gossmann et al., 2012) and dominant active alleles of SYMRK and CCaMK are sufficient for T90 induction (Ried et al., 2014 and unpublished data). However, we did not detect GUS activity (blue staining) in P. indica or mock-inoculated roots, while blue staining was observed in M. loti DsRed inoculated roots (Supplementary Figure 2). These data indicate that the promoter:GUS fusion of the T90 line and the signal transduction pathway upstream are not activated during the interaction of L. japonicus with P. indica.
Moreover, we observed that A. thaliana wild-type (Col-0), which lacks some of the key CSGs (Delaux et al., 2014), supported intracellular root colonization and sporulation by P. indica (Figure 2 and Supplementary Figure 3). In addition, intracellular infection and sporulation also occurred in the roots of the A. thaliana HCSG mutants pollux, nup133, sec13, and the double mutants sec13 × nup133 and shrk1 × shrk2 (Figure 2 and Supplementary Figure 3), indicating that the fungus could successfully infect these mutants and complete its life cycle. These observations strongly support the conclusion that neither the CSGs which A. thaliana lost during its evolution nor the experimentally mutated HCSGs are required for the interaction with P. indica.
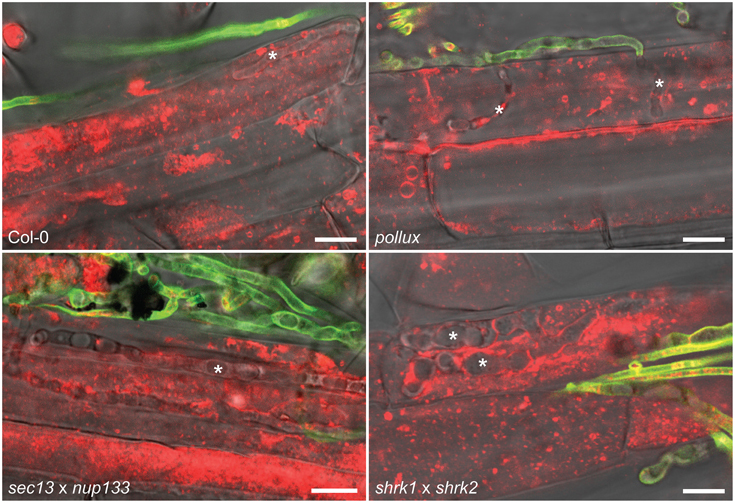
Figure 2. Colonization of A. thaliana root cells by P. indica. Hyphae (indicated by asterisks) were detected 4 dpi with P. indica chlamydospores within root cells of wild-type (Col-0) and the indicated HCSG mutants. Extracellular hyphae were stained with WGA-AF488 (green) but intracellular hyphae were not or weakly fluorescent, probably due to limited access of WGA-AF488 to the fungal cell wall within root cells. FM4-64-stained plant material (red) within invaded host cells is indicative of stage 1 or 2 of the infection process. Scale bar 10 μm.
Growth of P. indica within L. japonicus and A. thaliana Root Cells
Fungal structures were detectable within the boundaries of root epidermal cells in both L. japonicus (Figure 1 and Supplementary Figure 1) and A. thaliana (Supplementary Figure 3). We investigated whether these hyphae would penetrate into living plant cells. To determine the vitality status of the root cells, we used the lipophilic stain FM4-64 in combination with time-lapse imaging. After FM4-64 staining, P. indica hyphae were observed within cells containing mobile structures including possible vesicles in A. thaliana (Figure 2). In this host, we could detect three states of colonized root cells that differed in the mobility or presence of intracellular content. In the first state, vesicle-like structures moved at a speed similar to that observed within neighboring non-infected cells (Supplementary Movie 1) or cells of non-colonized roots (Supplementary Movie 3). In exceptional cases, some of these cells contained small, intact vacuoles. In the second state, the FM4-64-stained material showed very little or no movement (Supplementary Movie 2), and occasionally collapsed vacuoles were observed. We also observed invaded cells with no cytoplasmic content, probably representing a third state of cellular infection, during which the fungus grows within likely dead cells (Supplementary Figure 4, Supplementary Movie 2). This observation supports a three-stage model of cellular infection, in which P. indica first colonizes individual roots cells that show vesicular movement. In stage 2, vesicular movement has undergone at least partial arrest. In stage 3 the cellular content has disappeared, possibly through autophagocytosis or consumption by the fungus (Supplementary Figure 4). Immobilized, irregular fluorescent structures are also present within probably dying cells of non-colonized roots (Supplementary Movie 3, white arrowhead). Such dying cells have been attributed to a developmental program implicated in developmental events such as the removal of root cap cells (Fendrych et al., 2014).
Movement of FM4-64-labeled material in P. indica-infected cells could also be documented in the A. thaliana HSCG mutant pollux (Supplementary Movie 4), and the double mutants sec13 × nup133 and shrk1 × shrk2, indicating that similar stages of cell activity occur independently of HCSGs. Because of the technically demanding process of obtaining such movies, a quantitative comparison of colonization stages between the wild-type and the HCSG mutants was not performed.
Increased Relative Fungal Biomass within Roots of L. japonicus Common Symbiosis Mutants
In order to quantify the relative fungal biomass within host roots, we determined the ratio between fungal DNA and plant DNA. Interestingly, in L. japonicus, we detected a tendency for an increased relative amount of fungal DNA in most of the tested common symbiosis mutants, with a significant difference to the wild-type in nup85-1, ccamk-13, and cyclops-3 (Figure 3). This higher ratio of fungal to plant DNA may be the result of increased fungal proliferation, reduced root growth, and/or plant cell death in the mutants. Curiously, this effect was only observed on L. japonicus but not on A. thaliana mutants (Figure 3). Our results reveal that CSG-mediated pathways affect the relative P. indica colonization level in L. japonicus, and that this regulation is not operational in A. thaliana. Interestingly, CSGs potentially have a cell protecting effect (Esseling et al., 2004; Genre et al., 2009; Evangelisti et al., 2014). Apart from its signaling role in symbiosis (Antolín-Llovera et al., 2014; Ried et al., 2014), SYMRK has been implicated in desensitization of root hair cells against mechanic stress (Esseling et al., 2004), and CCaMK increased tolerance against the cell killing effect of Colletotrichum (Genre et al., 2009). In a ccamk mutant of M. truncatula infected by the hemi-biotrophic fungal pathogen Colletotrichum trifolii, the switch from biotrophy to necrotrophy occurred earlier (Genre et al., 2009).
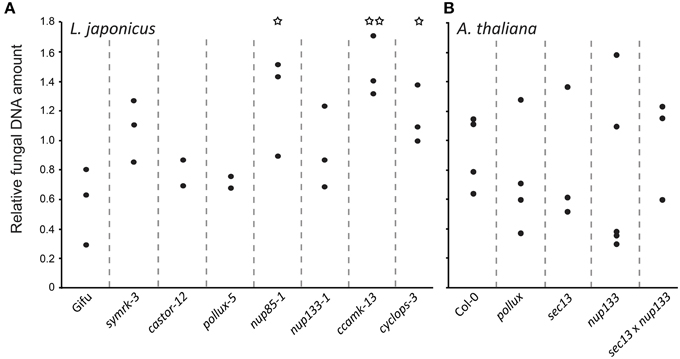
Figure 3. Quantification of P. indica in L. japonicus (A) and A. thaliana (B) wild-type and mutant roots. Real-time qPCR was used to quantify DNA from surface-washed P. indica-colonized roots at 14 dpi grown on modified HO medium using primers for the fungal gene Transcription Elongation Factor (TEF) and for A. thaliana and L. japonicus Ubiquitin (UBI) genes. Differences between the wild-type and mutants were investigated with a two-sided, unpaired t-test. *P < 0.05, **P < 0.01.
In barley, a dense colonization by P. indica is associated with root cell death (Deshmukh et al., 2006; Camehl et al., 2010; Nongbri et al., 2012). There is evidence that excessive P. indica proliferation is associated with cell death and/or with detrimental effects on the growth of the plant host (Deshmukh et al., 2006; Camehl et al., 2010; Nongbri et al., 2012). It is therefore possible that L. japonicus ccamk mutants suffer from an earlier onset of the necrotrophic phase of P. indica colonization, and that CSGs contribute to the maintenance of plant cellular integrity after fungal invasion. On the other hand, the CSGs are critical for lipochitooligo-saccharide (LCO)-induced lateral root emergence (Oláh et al., 2005; Maillet et al., 2011). While it is unclear whether P. indica stimulates lateral root emergence and, if so, whether it does it via the common symbiosis pathway, the altered fungal to plant biomass ratio of the CSG mutants could be due to a reduction in host root proliferation.
Plant Growth Promotion by P. indica is influenced by Nutrient Availability
In order to obtain an experimental system for the genetic dissection of the P. indica-mediated growth promotion (Peškan-Berghöfer et al., 2004; Shahollari et al., 2007; Sherameti et al., 2008; Camehl et al., 2010, 2011; Nongbri et al., 2012; Lahrmann et al., 2013; Venus and Oelmüller, 2013), we explored the influence of the substrate and nutrient availability. We evaluated the effect of P. indica on L. japonicus and A. thaliana grown in soil with two different nutrient concentrations. In both plant species, co-cultivation with P. indica in soil with high nutrient concentrations had little or no effect on the mean stem height. However, in soil with lower nutrient contents, P. indica inoculation roughly doubled the mean stem height of A. thaliana plants at 21 dpi, whereas there was only a small but significant effect on L. japonicus (Figure 4).
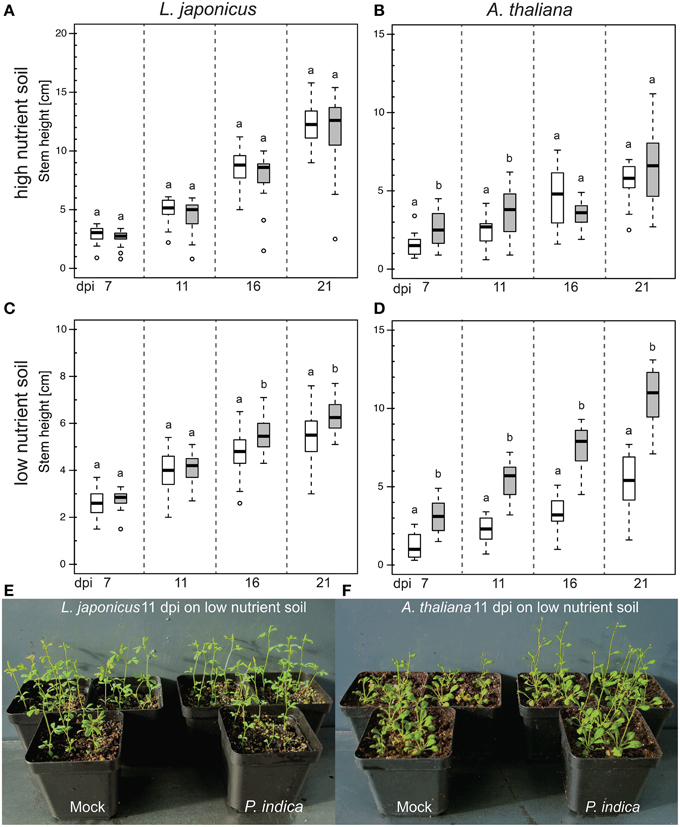
Figure 4. Effect of P. indica on the stem height of L. japonicus and A. thaliana grown in high or low nutrient soil. Box-plots represent the stem height of ca. 20 plants per treatment at the indicated dpi. (A,B) In high-nutrient soil (500 mg l−1 N, 500 mg l−1 P, 500 mg l−1 K), inoculation with P. indica did not change plant stem height at 21 dpi (p > 0.05). (C,D) On low-nutrient soil (50–100 mg l−1 N, 50–100 mg l−1 P, 100–150 mg l−1 K), P. indica inoculation led to an increase in the mean stem height at 21 dpi (p < 0.05). Statistical analyses were performed with a Kruskal–Wallis test followed by a Bonferroni–Holm correction using the mock-inoculated plants as control group. For each mock/P. indica-inoculated pair, box-plots sharing the same letter do not significantly differ (at the 5% significance level). White boxes: mock; gray boxes: P. indica-inoculated; open circles: outliers. (E,F) Exemplary pictures of P. indica- and mock-inoculated plants grown in low nutrient soil. Experiments were performed three times with similar results.
When A. thaliana plants were grown on agar plates with ½MS medium and no sugar, and inoculated with chlamydospores of P. indica, a growth promoting effect was observed. In contrast, when 0.05% sucrose was added to the medium, the plants were generally bigger and no increase was observed in the mean fresh weight of P. indica-inoculated plants compared to mock-treated plants (Supplementary Figure 5).
Since A. thaliana plants grown in the presence of P. indica exhibit increased uptake of nitrogen and phosphate (Shahollari et al., 2007; Kumar et al., 2011; Das et al., 2014), we investigated whether the concentration of nitrate [supplied as Ca(NO3)2 and/or KNO3], phosphate (KH2PO4), or ammonium (NH4Cl) on modified HO medium influenced the growth promotion of A. thaliana plants by P. indica. Co-cultivation with the fungus had a positive effect on the mean fresh weight of the plants under all nutrient conditions tested, except on a medium that limited plant growth due to the lack of a nitrogen source (Supplementary Figure 6). For the subsequent experiments, including those already reported in Lahrmann et al. (2013), we used modified HO medium, which consistently supported the growth-promoting effect by P. indica.
A. thaliana homologs of Common Symbiosis Genes are not required for P. indica-induced Growth Promotion
We investigated the influence of A. thaliana HCSGs on the host growth-promoting effect of P. indica. As a control, we included the closely related sebacinoid fungus P. williamsii (Basiewicz et al., 2012; Lahrmann et al., 2013), which did not induce or induced very little growth promotion of A. thaliana Col-0 (Lahrmann et al., 2013). Wild-type and mutant plants inoculated with P. indica had a significant higher mean fresh weight than control or P. williamsii-inoculated plants (Figure 5). Importantly, wild-type and mutant roots did not differ in their biomass upon P. indica inoculation. We conclude that the HCSGs POLLUX, NUP133, and SEC13 are not required for the growth promotion of A. thaliana by P. indica, confirming previous observations with atpollux mutants (Shahollari et al., 2007).
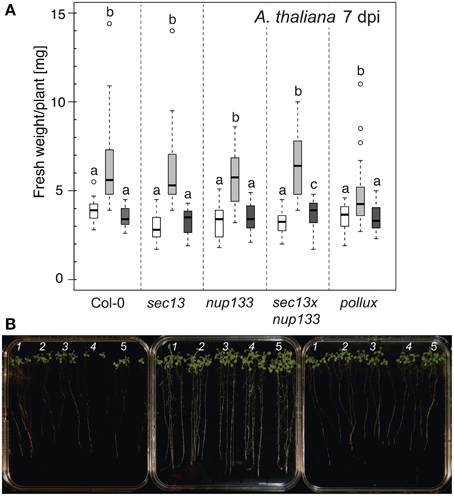
Figure 5. Effect of P. indica or P. williamsii on the biomass of A. thaliana. (A) Box-plots show the fresh weight of the indicated A. thaliana genotypes (ca. 15 plants/genotype) grown in the presence or absence of P. indica or P. williamsii. All plant genotypes accumulated more biomass upon co-cultivation with P. indica but not with P. williamsii. White box: mock; light gray box: P. indica; dark gray box: P. williamsii. Open circles: outliers. Statistical analyses were performed using a Kruskal–Wallis test followed by a Bonferroni–Holm correction using the mock-inoculated samples as control group. Groups that do not share the same letter are significantly different (at the 5% significance level). Comparisons between the three treatments were made for each genotype separately. Plant biomass was determined 7 dpi. (B) Representative plates showing sets of 14-day-old plants grown on modified HO medium are shown 7 days after mock-treatment with Tween water (left) or inoculation with either P. indica (center) or P. williamsii (right) chlamydospores. 1: pollux; 2: sec13 × nup133; 3: nup133; 4: sec13; 5: Col-0.
Conclusions
Despite the similarities between colonization of plant roots by AM fungi and P. indica, our data indicate that CSGs which are essential for AM development (Gutjahr and Parniske, 2013) are not required for root colonization by P. indica. In the AM symbiosis, signal transduction for the initiation of the intracellular accommodation program is mediated by the products of CSGs (Takeda et al., 2012). Since P. indica intracellular colonization was observed in the absence of individual CSGs or existing homologs in A. thaliana, we conclude that alternative pathways must exist that support intracellular accommodation of P. indica. Conceptually this could be achieved through the manipulation of general programs such as polarized secretion, endocytosis, plant immunity, and/or phytohormone signaling (Schäfer et al., 2009; Dörmann et al., 2014; Evangelisti et al., 2014). Identification of such compatibility programs is of prime interest because they might offer entry ports not only for beneficial fungi like P. indica but also to hyphal pathogens with similar infection strategies. This is in agreement with the recent observation that CSG mutants of M. truncatula show unaltered infection and haustorial development by the phytopathogenic oomycete Phytophthora palmivora (Rey et al., 2014). However, little is known about plant factors that are directly involved in the intracellular accommodation of P. indica. Tubby-like proteins, implicated in vesicle trafficking in mammals (Mukhopadhyay and Jackson, 2011), are required for normal colonization of A. thaliana roots by P. indica, and have been pinpointed as possible compatibility factors during the early stages of plant–fungus interaction (Reitz et al., 2012, 2013).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Andreas Binder for taking part in double blind experiments related to the plant growth responses. Part of this work was supported by a grant from the German Research Foundation (DFG) to MP (PA 493/8-3) in the frame of the Research unit FOR 964 “Calcium signaling via protein phosphorylation in plant model cell types during environmental stress adaption.” AZ and YD acknowledge support from the Max-Planck-Gesellschaft and AZ acknowledges support from the Cluster of Excellence on Plant Science (CEPLAS, EXC 1028).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00667
References
Alber, F., Dokudovskaya, S., Veenhoff, L. M., Zhang, W., Kipper, J., Devos, D., et al. (2007). The molecular architecture of the nuclear pore complex. Nature 450, 695–701. doi: 10.1038/nature06405
Ané, J. M., Kiss, G. B., Riely, B. K., Penmetsa, R. V., Oldroyd, G. E., Ayax, C., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303, 1364–1367. doi: 10.1126/science.1092986
Antolín-Llovera, M., Ried, M. K., and Parniske, M. (2014). Cleavage of the SYMBIOSIS RECEPTOR-LIKE KINASE ectodomain promotes complex formation with Nod factor receptor 5. Curr. Biol. 24, 422–427. doi: 10.1016/j.cub.2013.12.053
Basiewicz, M., Weiss, M., Kogel, K. H., Langen, G., Zorn, H., and Zuccaro, A. (2012). Molecular and phenotypic characterization of Sebacina vermifera strains associated with orchids, and the description of Piriformospora williamsii sp. nov. Fungal Biol. 116, 204–213. doi: 10.1016/j.funbio.2011.11.003
Binder, A., and Parniske, M. (2013). Analysis of the Lotus japonicus nuclear pore NUP107-160 subcomplex reveals pronounced structural plasticity and functional redundancy. Front. Plant Sci. 4:552. doi: 10.3389/fpls.2013.00552
Bonfante, P., and Genre, A. (2010). Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nat. Commun. 1, 48. doi: 10.1038/ncomms1046
Burow, M., Halkier, B. A., and Kliebenstein, D. J. (2010). Regulatory networks of glucosinolates shape Arabidopsis thaliana fitness. Curr. Opin. Plant Biol. 13, 348–353. doi: 10.1016/j.pbi.2010.02.002
Butehorn, B., Rhody, D., and Franken, P. (2000). Isolation and characterisation of Pitef1 encoding the translation elongation factor EF-1 alpha of the root endophyte Piriformospora indica. Plant Biol. 2, 687–692. doi: 10.1055/s-2000-16647
Camehl, I., Drzewiecki, C., Vadassery, J., Shahollari, B., Sherameti, I., Forzani, C., et al. (2011). The OXI1 kinase pathway mediates Piriformospora indica-induced growth promotion in Arabidopsis. PLoS Pathog. 7:e1002051. doi: 10.1371/journal.ppat.1002051
Camehl, I., Sherameti, I., Venus, Y., Bethke, G., Varma, A., Lee, J., et al. (2010). Ethylene signalling and ethylene-targeted transcription factors are required to balance beneficial and nonbeneficial traits in the symbiosis between the endophytic fungus Piriformospora indica and Arabidopsis thaliana. New Phytol. 185, 1062–1073. doi: 10.1111/j.1469-8137.2009.03149.x
Charpentier, M., Bredemeier, R., Wanner, G., Takeda, N., Schleiff, E., and Parniske, M. (2008). Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20, 3467–3479. doi: 10.1105/tpc.108.063255
Das, J., Ramesh, K. V., Maithri, U., Mutangana, D., and Suresh, C. K. (2014). Response of aerobic rice to Piriformospora indica. Indian J. Exp. Biol. 52, 237–251.
de Castro, E., Sigrist, C. J., Gattiker, A., Bulliard, V., Langendijk-Genevaux, P. S., Gasteiger, E., et al. (2006). ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–W365. doi: 10.1093/nar/gkl124
Delaux, P. M., Séjalon-Delmas, N., Bécard, G., and Ané, J. M. (2013). Evolution of the plant-microbe symbiotic ‘toolkit’. Trends Plant Sci. 18, 298–304. doi: 10.1016/j.tplants.2013.01.008
Delaux, P. M., Varala, K., Edger, P. P., Coruzzi, G. M., Pires, J. C., and Ané, J. M. (2014). Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genet. 10:e1004487. doi: 10.1371/journal.pgen.1004487
De Mendiburu, F. (2010). Agricolae: Statistical Procedures for Agricultural Research. Thesis, National Engineering University (UNI), Lima-Peru.
Deshmukh, S., Hückelhoven, R., Schäfer, P., Imani, J., Sharma, M., Weiss, M., et al. (2006). The root endophytic fungus Piriformospora indica requires host cell death for proliferation during mutualistic symbiosis with barley. Proc. Natl. Acad. Sci. U.S.A. 103, 18450–18457. doi: 10.1073/pnas.0605697103
Dörmann, P., Kim, H., Ott, T., Schulze-Lefert, P., Trujillo, M., Wewer, V., et al. (2014). Cell-autonomous defense, re-organization and trafficking of membranes in plant-microbe interactions. New Phytol. 204, 815–822. doi: 10.1111/nph.12978
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19, 11–15.
Esseling, J. J., Lhuissier, F. G., and Emons, A. M. (2004). A nonsymbiotic root hair tip growth phenotype in NORK-mutated legumes: implications for nodulation factor-induced signaling and formation of a multifaceted root hair pocket for bacteria. Plant Cell 16, 933–944. doi: 10.1105/tpc.019653
Evangelisti, E., Rey, T., and Schornack, S. (2014). Cross-interference of plant development and plant-microbe interactions. Curr. Opin. Plant Biol. 20C, 118–126. doi: 10.1016/j.pbi.2014.05.014
Fahraeus, G. (1957). The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J. Gen. Microbiol. 16, 374–381. doi: 10.1099/00221287-16-2-374
Fendrych, M., van Hautegem, T., van Durme, M., Olvera-Carrillo, Y., Huysmans, M., Karimi, M., et al. (2014). Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Curr. Biol. 24, 931–940. doi: 10.1016/j.cub.2014.03.025
Genre, A., Ortu, G., Bertoldo, C., Martino, E., and Bonfante, P. (2009). Biotic and abiotic stimulation of root epidermal cells reveals common and specific responses to arbuscular mycorrhizal fungi. Plant Physiol. 149, 1424–1434. doi: 10.1104/pp.108.132225
Gossmann, J. A., Markmann, K., Brachmann, A., Rose, L. E., and Parniske, M. (2012). Polymorphic infection and organogenesis patterns induced by a Rhizobium leguminosarum isolate from Lotus root nodules are determined by the host genotype. New Phytol. 196, 561–573. doi: 10.1111/j.1469-8137.2012.04281.x
Goujon, M., McWilliam, H., Li, W., Valentin, F., Squizzato, S., Paern, J., et al. (2010). A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699. doi: 10.1093/nar/gkq313
Groth, M., Takeda, N., Perry, J., Uchida, H., Dräxl, S., Brachmann, A., et al. (2010). NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 22, 2509–2526. doi: 10.1105/tpc.109.069807
Gutjahr, C. (2014). Phytohormone signaling in arbuscular mycorhiza development. Curr. Opin. Plant Biol. 20C, 26–34. doi: 10.1016/j.pbi.2014.04.003
Gutjahr, C., and Parniske, M. (2013). Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu. Rev. Cell Dev. Biol. 29, 593–617. doi: 10.1146/annurev-cellbio-101512-122413
Hilbert, M., Voll, L. M., Ding, Y., Hofmann, J., Sharma, M., and Zuccaro, A. (2012). Indole derivative production by the root endophyte Piriformospora indica is not required for growth promotion but for biotrophic colonization of barley roots. New Phytol. 196, 520–534. doi: 10.1111/j.1469-8137.2012.04275.x
Hoagland, D. R., and Arnon, D. I. (1950). The Water-culture Method for Growing Plants without Soil. Berkeley, CA: University of California.
Hok, S., Danchin, E. G., Allasia, V., Panabières, F., Attard, A., and Keller, H. (2011). An Arabidopsis (malectin-like) leucine-rich repeat receptor-like kinase contributes to downy mildew disease. Plant Cell Environ. 34, 1944–1957. doi: 10.1111/j.1365-3040.2011.02390.x
Jacobs, S., Zechmann, B., Molitor, A., Trujillo, M., Petutschnig, E., Lipka, V., et al. (2011). Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica. Plant Physiol. 156, 726–740. doi: 10.1104/pp.111.176446
Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Kanamori, N., Madsen, L. H., Radutoiu, S., Frantescu, M., Quistgaard, E. M., Miwa, H., et al. (2006). A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. U.S.A. 103, 359–364. doi: 10.1073/pnas.0508883103
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kevei, Z., Seres, A., Kereszt, A., Kalò, P., Kiss, P., Tóth, G., et al. (2005). Significant microsynteny with new evolutionary highlights is detected between Arabidopsis and legume model plants despite the lack of macrosynteny. Mol. Genet. Genomics 274, 644–657. doi: 10.1007/s00438-005-0057-9
Khatabi, B., Molitor, A., Lindermayr, C., Pfiffi, S., Durner, J., von Wettstein, D., et al. (2012). Ethylene supports colonization of plant roots by the mutualistic fungus Piriformospora indica. PLoS ONE 7:e35502. doi: 10.1371/journal.pone.0035502
Kistner, C., Winzer, T., Pitzschke, A., Mulder, L., Sato, S., Kaneko, T., et al. (2005). Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17, 2217–2229. doi: 10.1105/tpc.105.032714
Kumar, M., Yadav, V., Kumar, H., Sharma, R., Singh, A., Tuteja, N., et al. (2011). Piriformospora indica enhances plant growth by transferring phosphate. Plant Signal. Behav. 6, 723–725. doi: 10.4161/psb.6.5.15106
Lahrmann, U., Ding, Y., Banhara, A., Rath, M., Hajirezaei, M. R., Döhlemann, S., et al. (2013). Host-related metabolic cues affect colonization strategies of a root endophyte. Proc. Natl. Acad. Sci. U.S.A. 110, 13965–13970. doi: 10.1073/pnas.1301653110
Lahrmann, U., and Zuccaro, A. (2012). Opprimo ergo sum–evasion and suppression in the root endophytic fungus Piriformospora indica. Mol. Plant Microbe Interact. 25, 727–737. doi: 10.1094/MPMI-11-11-0291
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Maekawa, T., Maekawa-Yoshikawa, M., Takeda, N., Imaizumi-Anraku, H., Murooka, Y., and Hayashi, M. (2009). Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J. 58, 183–194. doi: 10.1111/j.1365-313X.2008.03774.x
Maillet, F., Poinsot, V., André, O., Puech-Pagès, V., Haouy, A., Gueunier, M., et al. (2011). Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469, 58–63. doi: 10.1038/nature09622
Markmann, K., Giczey, G., and Parniske, M. (2008). Functional adaptation of a plant receptor-kinase paved the way for the evolution of intracellular root symbioses with bacteria. PLoS Biol. 6:e68. doi: 10.1371/journal.pbio.0060068
Mikkelsen, M. D., Fuller, V. L., Hansen, B. G., Nafisi, M., Olsen, C. E., Nielsen, H. B., et al. (2009). Controlled indole-3-acetaldoxime production through ethanol-induced expression of CYP79B2. Planta 229, 1209–1217. doi: 10.1007/s00425-009-0907-5
Mukhopadhyay, S., and Jackson, P. K. (2011). The tubby family proteins. Genome Biol. 12:225. doi: 10.1186/gb-2011-12-6-225
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nongbri, P. L., Johnson, J. M., Sherameti, I., Glawischnig, E., Halkier, B. A., and Oelmüller, R. (2012). Indole-3-acetaldoxime-derived compounds restrict root colonization in the beneficial interaction between Arabidopsis roots and the endophyte Piriformospora indica. Mol. Plant Microbe Interact. 25, 1186–1197. doi: 10.1094/MPMI-03-12-0071-R
Oláh, B., Brière, C., Bécard, G., Dénarié, J., and Gough, C. (2005). Nod factors and a diffusible factor from arbuscular mycorrhizal fungi stimulate lateral root formation in Medicago truncatula via the DMI1/DMI2 signalling pathway. Plant J. 44, 195–207. doi: 10.1111/j.1365-313X.2005.02522.x
Parniske, M. (2008). Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 6, 763–775. doi: 10.1038/nrmicro1987
Peškan-Berghöfer, T., Shahollari, B., Giong, P. H., Hehl, S., Markert, C., Blanke, V., et al. (2004). Association of Piriformospora indica with Arabidopsis thaliana roots represents a novel system to study beneficial plant–microbe interactions and involves early plant protein modifications in the endoplasmic reticulum and at the plasma membrane. Physiol. Plant. 122, 465–477. doi: 10.1111/j.1399-3054.2004.00424.x
Pham, G., Kumari, R., Singh, A., Malla, R., Prasad, R., Sachdev, M., et al. (2004). “Axenic culture of symbiotic fungus Piriformospora indica,” in Plant Surface Microbiology, eds A. Varma, L. Abbott, D. Werner and R. Hampp (Berlin Heidelberg: Springer), 593–613.
Qiang, X., Zechmann, B., Reitz, M. U., Kogel, K. H., and Schäfer, P. (2012). The mutualistic fungus Piriformospora indica colonizes Arabidopsis roots by inducing an endoplasmic reticulum stress-triggered caspase-dependent cell death. Plant Cell 24, 794–809. doi: 10.1105/tpc.111.093260
Radutoiu, S., Madsen, L. H., Madsen, E. B., Felle, H. H., Umehara, Y., Grønlund, M., et al. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425, 585–592. doi: 10.1038/nature02039
R Core Team. (2013). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Reitz, M. U., Bissue, J. K., Zocher, K., Attard, A., Hückelhoven, R., Becker, K., et al. (2012). The subcellular localization of Tubby-like proteins and participation in stress signaling and root colonization by the mutualist Piriformospora indica. Plant Physiol. 160, 349–364. doi: 10.1104/pp.112.201319
Reitz, M. U., Pai, S., Imani, J., and Schäfer, P. (2013). New insights into the subcellular localization of Tubby-like proteins and their participation in the Arabidopsis-Piriformospora indica interaction. Plant Signal Behav. 8:e25198. doi: 10.4161/psb.25198
Rey, T., Chatterjee, A., Buttay, M., Toulotte, J., and Schornack, S. (2014). Medicago truncatula symbiosis mutants affected in the interaction with a biotrophic root pathogen. New Phytol. 206, 497–500. doi: 10.1111/nph.13233
Ried, M. K., Antolin-Llovera, M., and Parniske, M. (2014). Spontaneous symbiotic reprogramming of plant roots triggered by receptor-like kinases. Elife 3:e03891. doi: 10.7554/elife.03891
Saito, K., Yoshikawa, M., Yano, K., Miwa, H., Uchida, H., Asamizu, E., et al. (2007). NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19, 610–624. doi: 10.1105/tpc.106.046938
Schäfer, P., Pfiffi, S., Voll, L. M., Zajic, D., Chandler, P. M., Waller, F., et al. (2009). Phytohormones in plant root-Piriformospora indica mutualism. Plant Signal. Behav. 4, 669–671. doi: 10.4161/psb.4.7.9038
Scholl, R. L., May, S. T., and Ware, D. H. (2000). Seed and molecular resources for Arabidopsis. Plant Physiol. 124, 1477–1480. doi: 10.1104/pp.124.4.1477
Schüler, A., and Walker, C. (2011). “Evolution of the ‘plant-symbiotic’ fungal phylum, Glomeromycota,” in Evolution of fungi and fungal-like organisms, eds. S. Pöggeler and J. Wöstemeyer (Berlin Heidelberg: Springer), 163–185.
Selosse, M. A., Dubois, M. P., and Alvarez, N. (2009). Do Sebacinales commonly associate with plant roots as endophytes? Mycol. Res. 113, 1062–1069. doi: 10.1016/j.mycres.2009.07.004
Shahollari, B., Vadassery, J., Varma, A., and Oelmüller, R. (2007). A leucine-rich repeat protein is required for growth promotion and enhanced seed production mediated by the endophytic fungus Piriformospora indica in Arabidopsis thaliana. Plant J. 50, 1–13. doi: 10.1111/j.1365-313X.2007.03028.x
Sherameti, I., Tripathi, S., Varma, A., and Oelmüller, R. (2008). The root-colonizing endophyte Pirifomospora indica confers drought tolerance in Arabidopsis by stimulating the expression of drought stress-related genes in leaves. Mol. Plant Microbe Interact. 21, 799–807. doi: 10.1094/MPMI-21-6-0799
Singh, S., Katzer, K., Lambert, J., Cerri, M., and Parniske, M. (2014). CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15, 139–152. doi: 10.1016/j.chom.2014.01.011
Singh, S., and Parniske, M. (2012). Activation of calcium- and calmodulin-dependent protein kinase (CCaMK), the central regulator of plant root endosymbiosis. Curr. Opin. Plant Biol. 15, 444–453. doi: 10.1016/j.pbi.2012.04.002
Svistoonoff, S., Hocher, V., and Gherbi, H. (2014). Actinorhizal root nodule symbioses: what is signalling telling on the origins of nodulation? Curr. Opin. Plant Biol. 20, 11–18. doi: 10.1016/j.pbi.2014.03.001
Takeda, N., Maekawa, T., and Hayashi, M. (2012). Nuclear-localized and deregulated calcium- and calmodulin-dependent protein kinase activates rhizobial and mycorrhizal responses in Lotus japonicus. Plant Cell 24, 810–822. doi: 10.1105/tpc.111.091827
Takeda, N., Sato, S., Asamizu, E., Tabata, S., and Parniske, M. (2009). Apoplastic plant subtilases support arbuscular mycorrhiza development in Lotus japonicus. Plant J. 58, 766–777. doi: 10.1111/j.1365-313X.2009.03824.x
Vadassery, J., Ritter, C., Venus, Y., Camehl, I., Varma, A., Shahollari, B., et al. (2008). The role of auxins and cytokinins in the mutualistic interaction between Arabidopsis and Piriformospora indica. Mol. Plant Microbe Interact. 21, 1371–1383. doi: 10.1094/MPMI-21-10-1371
Venkateshwaran, M., Cosme, A., Han, L., Banba, M., Satyshur, K. A., Schleiff, E., et al. (2012). The recent evolution of a symbiotic ion channel in the legume family altered ion conductance and improved functionality in calcium signaling. Plant Cell 24, 2528–2545. doi: 10.1105/tpc.112.098475
Venus, Y., and Oelmüller, R. (2013). Arabidopsis ROP1 and ROP6 influence germination time, root morphology, the formation of F-actin bundles, and symbiotic fungal interactions. Mol. Plant 6, 872–886. doi: 10.1093/mp/sss101
Verma, S., Varma, A., Rexer, K.-H., Hassel, A., Kost, G., Sarbhoy, A., et al. (1998). Piriformospora indica, gen. et sp. nov., a new root-colonizing fungus. Mycologia 90, 896–903. doi: 10.2307/3761331
Waller, F., Achatz, B., Baltruschat, H., Fodor, J., Becker, K., Fischer, M., et al. (2005). The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc. Natl. Acad. Sci. U.S.A. 102, 13386–13391. doi: 10.1073/pnas.0504423102
Webb, K. J., Skøt, L., Nicholson, M. N., Jørgensen, B., and Mizen, S. (2000). Mesorhizobium loti increases root-specific expression of a calcium-binding protein homologue identified by promoter tagging in Lotus japonicus. Mol. Plant Microbe Interact. 13, 606–616. doi: 10.1094/MPMI.2000.13.6.606
Wiermer, M., Cheng, Y. T., Imkampe, J., Li, M., Wang, D., Lipka, V., et al. (2012). Putative members of the Arabidopsis Nup107-160 nuclear pore sub-complex contribute to pathogen defense. Plant J. 70, 796–808. doi: 10.1111/j.1365-313X.2012.04928.x
Keywords: Piriformospora indica, Lotus japonicus, Arabidopsis thaliana, biotrophy, common symbiosis genes, intracellular colonization, growth promotion
Citation: Banhara A, Ding Y, Kühner R, Zuccaro A and Parniske M (2015) Colonization of root cells and plant growth promotion by Piriformospora indica occurs independently of plant common symbiosis genes. Front. Plant Sci. 6:667. doi: 10.3389/fpls.2015.00667
Received: 13 January 2015; Accepted: 13 August 2015;
Published: 17 September 2015.
Edited by:
Ralph Panstruga, RWTH Aachen University, GermanyReviewed by:
Andrea Genre, University of Turin, ItalyPatrick Schäfer, University of Warwick, UK
Sébastien Duplessis, Institut National de la Recherche Agronomique, France
Copyright © 2015 Banhara, Ding, Kühner, Zuccaro and Parniske. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin Parniske, Genetics, Faculty of Biology, University of Munich (LMU), Großhaderner Strasse 4, 82152 Martinsried, Germany, parniske@lmu.de
†Present Address: Yi Ding, Boyce Thompson Institute for Plant Research, Ithaca, NY, USA
 Aline Banhara
Aline Banhara Yi Ding
Yi Ding Regina Kühner
Regina Kühner Alga Zuccaro
Alga Zuccaro Martin Parniske
Martin Parniske