- 1Graduate School of Life Sciences, Tohoku University, Sendai, Japan
- 2Biodiversity and Climate Research Center, Laboratory Center, Frankfurt am Main, Germany
- 3Frontier Research Institute for Interdisciplinary Sciences, Tohoku University, Sendai, Japan
- 4Faculty of Pharmaceutical Sciences, Josai University, Sakado, Japan
In Arabidopsis three basic region leucine zipper (bZIP) transcription factor genes, bZIP17, bZIP28, and bZIP60, play crucial roles in the unfolded protein response (UPR). Previously we found that bZIP60 is one of the spermine-induced genes. Consequently we further investigated the response of all the three bZIP genes to spermine. Expression of bZIP17, bZIP28, and bZIP60, and also their target genes was activated by spermine application as well as in plants with elevated endogenous spermine levels. Furthermore, spermine activated the splicing of the bZIP60 transcript mediated by the ribonuclease activity of inositol-requiring enzyme 1 and also recruited bZIP17 and bZIP60 proteins from endoplasmic reticulum to nucleus. We therefore propose that spermine is a novel UPR inducer. Moreover, induction of UPR by spermine required calcium-influx to the cytoplasm and the genes for mitogen-activated protein kinase kinase 9 (MKK9), mitogen-activated protein kinase 3 (MPK3) and MPK6. The result indicates that spermine-induced UPR is mediated by the MKK9-MPK3/MPK6 cascade in Arabidopsis.
Introduction
Polyamines (PAs) are low-molecular mass, aliphatic compounds and they are ubiquitously present in all living organisms (Tabor and Tabor, 1984; Cohen, 1998). In plants, major PAs are putrescine (Put), spermidine (Spd), spermine (Spm), and thermospermine (T-Spm; Knott et al., 2007; Takano et al., 2012). PAs have important roles in various physiological processes not only in embryogenesis, growth, development and aging/senescence but also in adaptation responses to abiotic and biotic stresses (Kusano et al., 2008; Alcázar et al., 2010; Takahashi and Kakehi, 2010; Tiburcio et al., 2014; Berberich et al., 2015).
Polyamine precursors are ornithine and/or arginine. Ornithine is converted to Put by ornithine decarboxylase (ODC), while arginine is converted to Put via agmatine by three steps of enzyme reactions; arginine decarboxylase (ADC), agmatine iminohydrolase, and N-carbamoylputrescine amidohydrolase. Put is converted to Spd by Spd synthase, in which another substrate, decarboxylated S-adenosylmethionine (dcSAM), is required. dcSAM is synthesized by S-adenosylmethionine decarboxylase (SAMDC) from S-adenosylmethionine (SAM). Spd is further converted either to Spm by Spm synthase (SPMS) or to T-Spm by T-Spm synthase, also called ACAULIS5 (ACL5). SPMS and ACL5 also require dcSAM as another substrate (Kusano et al., 2008; Alcázar et al., 2010; Takahashi and Kakehi, 2010). In Arabidopsis, the ODC gene is absent (Hanfrey et al., 2001). Thus Put is synthesized only by the ADC pathway in this organism. On the other hand, PA catabolism is governed by two enzymes: the one is copper-dependent amine oxidase (CuAO) and the other is polyamine oxidase (PAO; Kusano et al., 2015). In Arabidopsis, there are more than 10 CuAOs and 5 PAOs. All the latter enzymes are characterized (Kusano et al., 2015).
As mentioned, involvement of PAs and their metabolism in the defense responses against pathogens is described (Walters, 2003a,b). PAs and their oxidative products by CuAO and PAO play crucial roles in plant defense responses (Berberich et al., 2015, and the references therein). When Nicotiana tabacum carrying the resistance gene N was infected by Tobacco mosaic virus (TMV), host plant displayed a hypersensitive response (HR). Once HR occurs, it is known that the tissues surrounding the TMV infection sites were killed by a suicidal reaction to prevent the virus multiplication. Yamakawa et al. (1998) reported that Spm enriched in the apoplastic space during HR triggered by N. tabacum-TMV pathosystem. Subsequently we found that exogenously applied Spm stimulates the phosphorylation activities of two MAPKs, wound-induced protein kinase (WIPK, Seo et al., 1995) and salicylic acid-induced protein kinase (SIPK, Zhang and Klessig, 1997), which trigger the expression of downstream defense genes in N. tabacum (Takahashi et al., 2003, 2004), and proposed it to call ‘Spm-signaling pathway.’ Massive identification of the Spm-responsive genes using a super serial analysis of gene expression (SuperSAGE) approach was performed in Arabidopsis thaliana (Mitsuya et al., 2009). The identified Spm-responsive genes behaved similarly during cucumber mosaic virus (CMV)-induced HR in A. thaliana (Takahashi et al., 1994; Mitsuya et al., 2009). One of them was a gene (bZIP60) encoding a basic region leucine zipper 60 protein. bZIP60 was reported as a key transcription factor (TF) gene which is involved in the unfolded protein response (UPR; Iwata and Koizumi, 2005). UPR is induced, when unfolded or misfolded proteins are accumulated in the endoplasmic reticulum (ER), to refold or degrade the corresponding proteins (Walter and Ron, 2011). In plants, three bZIP-type proteins, bZIP17, bZIP28 as well as bZIP60, govern UPR in Arabidopsis (Iwata et al., 2008; Liu and Howell, 2010; Iwata and Koizumi, 2012; Howell, 2013). Those bZIP proteins reside in the ER under unstressed conditions, and once they sense ER stress, the former two proteins move to the Golgi apparatus and migrate to the nucleus after proteolytic processing by site 1 and site 2 proteases (S1P and S2P; Liu et al., 2007a,b). In contrast, bZIP60 transcript is unconventionally spliced by an RNase activity of inositol-requiring enzyme 1 (IRE1) and the product of the resulting spliced bZIP60 transcript is recruited to the nucleus. The Arabidopsis genome contains two IRE1 genes, AtIRE1A (At2g17520) and AtIRE1B (At5g24360; Liu and Howell, 2010; Deng et al., 2011; Nagashima et al., 2011; Iwata and Koizumi, 2012; Srivastava et al., 2013). Several chemicals such as tunicamycin (TM) and dithiothreitol (DTT) are known to be UPR inducers. TM inhibits the N-linked glycosylation and DTT inhibits the disulfide bond formation, both of which are important for protein maturation.
Here, we aim to reveal whether Spm only acts on bZIP60 or induces the whole UPR in Arabidopsis. We also address the upstream components in Spm-induced UPR pathways.
Materials and Methods
Plant Materials and Growth Condition
Arabidopsis thaliana ecotype Columbia-0 (Col-0) is used as wild type (WT). T-DNA insertion mutants, ire1a (SALK_018112, Deng et al., 2011, 2013), ire1b (SAIL_238_F07, Deng et al., 2011, 2013), bzip60 (SALK_050203, Deng et al., 2011, 2013), Atmpk6 (SALK_127507, Yoo et al., 2008; Zhou et al., 2009), Atmpk3 (SALK_151594, Yoo et al., 2008; Zhou et al., 2009) were obtained from Dr. Stephen H. Howell (Iowa State University) and Arabidopsis Biological Resource Center, while Atmkk9 (SAIL_60_H06, Xu et al., 2008) was kindly provided from Dr. Dongtao Ren (China Agricultural University). The double mutant, ire1aire1b, was obtained by crossing of ire1a and ire1b. Seeds were surface sterilized by 70% ethanol for 1min, then by 1% sodium hypochlorite solution containing 0.1% Tween-20 for 15 min, then followed by rinsing with sterilized water, three times. Sterilized seeds were sown on half strength MS medium (1% w/v agar, 1% w/v sucrose, 0.5 x MS salts, 0.05% vitamin B5, pH 5.6) and incubated in a growth chamber at 22°C under 16 h light/8 h dark conditions.
Various Treatments
Ten-days-old Arabidopsis seedlings were incubated in half strength MS solution supplemented with or without Spm (0.5 mM) for 12 h under continuous light condition. PAs treatment: 0.5 mM concentration of PAs was used unless otherwise mentioned and incubated for 12 h. DTT treatment: DTT was applied at 2 mM concentration for 5 h in half strength MS solution. Lanthanum chloride (LaCl3) treatment: 10-days-old seedlings were incubated with or without La3+ (0.5 mM) for 12 h.
Genome DNA-Polymerase Chain Reaction (PCR) and Reverse Transcription (RT)-PCR Analyses
Plant genome DNA was prepared by the procedure described by Murray and Thompson (1980). Total RNA was extracted from the respective plants samples using Sepasol-RNA I Super (Nacalai Tesque, Kyoto, Japan). Total RNA was treated with DNaseI (Takara DNase, Japan). First strand cDNA was synthesized from the DNaseI-treated RNA with ReverTra Ace (Toyobo Co. Ltd., Osaka, Japan), oligo-dT primer and dNTPs. AtActin was amplified using a specific primer pair (Supplementary Table S1) and used as a loading control. The amplified DNA fragments were separated by agarose-gels and visualized by ethidium bromide-staining.
Quantitative Real-Time RT-PCR
Quantitative RT-PCR (qRT-PCR) analysis was performed by StepOne Real-Time PCR System (Applied Biosystems) using SYBR® Green RT-PCR Kit (FastStart Universal SYBR Green Master, ROX). A standard curve was constructed from different genes and the values were normalized to Actin levels. The primers used for qRT-PCR were described in Supplementary Table S1.
bZIP60 Splicing Assay
bZIP60 splicing assay was performed using the following primers; for detecting a unspliced form (SPU) and a spliced form (SPS), bZIP60-F primer and bZIP60-UB1 reverse primer and bZIP60-F primer and bZIP60-SB2 reverse primer, respectively, were used (Supplementary Table S1).
Construction of Green Fluorescent Protein (GFP) Fusion Plasmids, Biolistic Bombardments, and Microscopic Observation
The basal GFP vector was constructed as described previously (Ono et al., 2012). Briefly, a GFP coding fragment was amplified by PCR using pGFP2 (provided by Dr. N.-H. Chua). The fragment was double-digested with BamHI and SmaI, and subcloned into the pBI221 vector (Invitrogen), yielding pBI221GFP. Next, the internal two SacI sites of AtbZIP17 coding region were mutated without changing the amino acid sequence by two-step PCR. Then, a second PCR was performed on the respective mixtures of the first PCR products using the primers, AtbZIP17-SmaI-F and AtbZIP17-SacI-Rv (Supplementary Table S1). The coding region of AtbZIP60 was amplified by PCR. The resulting AtbZIP17 and AtbZIP60 fragments digested with SmaI and SacI were subcloned into the corresponding restriction enzyme sites of pBI221GFP, yielding pBI221GFP-AtbZIP17 and pBI221GFP-AtbZIP60, respectively. The resulting GFP-AtbZIPs’ constructs were bombarded into onion bulbs by particle bombardment. ER-targeting DsRED plasmid (Okamoto et al., 2008) or nuclear-targeting mCherry-VirD2_NLS plasmid (CD3-1106, purchased from ABRC) was co-bombarded as the ER- and nuclear-markers. After incubating the onion bulbs with or without Spm (or DTT) at 22°C for 16 h under darkness, onion epidermal cells were peeled and placed onto glass slides and observed with a fluorescence microscope (BX61; Olympus).
Arabidopsis Transgenic Plants Overexpressing SPMS
Three independent lines of the transgenic Arabidopsis overexpressing SPMS were used. The detailed procedure to generate the transgenics was described in Sagor et al. (2012a).
PA Analysis
Polyamine analysis was performed as described in Naka et al. (2010). In brief, plant samples (0.3–0.5 g per sample) were pulverized with a mortar and pestle under liquid nitrogen. Five volumes (2.5 mL per 0.5 g of plant sample) of 5% (v/v) cold perchloric acid were added to the resulting fine powders. The mixtures were transferred to plastic tubes and kept on ice for 1 h. After centrifugation at 15,000 × g for 30 min at 4°C, the supernatants were combined and filtered using a filter syringe (pore size, 0.2 μm). One milliliter of 2 N NaOH was added to 1.5 mL of plant extract and mixed thoroughly. Then 10 μL of benzoyl chloride was added and the mixture was incubated at room temperature for 20 min. After adding 2 mL of saturated NaCl and 2 mL of diethyl ester, samples were vigorously mixed and then centrifuged at 3,000 × g for 10 min at 4°C for phase-separation. An aliquot (1.5 mL) of the organic solvent phase was evaporated and the residue was resuspended in 50 μL of methanol. Benzoylated PAs were analyzed with a programmable Agilent 1200 liquid chromatograph using a reverse-phase column (4.6 mm × 250 mm, TSK-GEL ODS-80Ts, TOSOH, Tokyo, Japan) and detected at 254 nm. One cycle of the run took 60 min in total with a flow rate of 1 mL/min at 30°C, i.e., 42% acetonitrile for 25 min for PA separation, increased to 100% acetonitrile over 3 min, 100% acetonitrile for 20 min for washing, decreased to 42% acetonitrile over 3 min, and finally 42% acetonitrile for 9 min.
Statistical Analysis
Student’s t-tests were used for statistical analysis and were performed using Microsoft Excel statistical tools.
Results
Exogenously Applied Spm Induces bZIP17 and bZIP28 as Well as bZIP60 and their Target Genes
Ten-days-old Arabidopsis ecotype (Col-0) seedlings were treated with Spm or DTT (as positive treatment for UPR induction) and qRT-PCR was performed. Spm induced bZIP17, bZIP28 and bZIP60, and their respective target genes, HB-7, CNX1, and BiP3, respectively, as similar as DTT did (Figure 1, Supplementary Figure S1). The result clearly indicates that Spm is a novel UPR inducer.
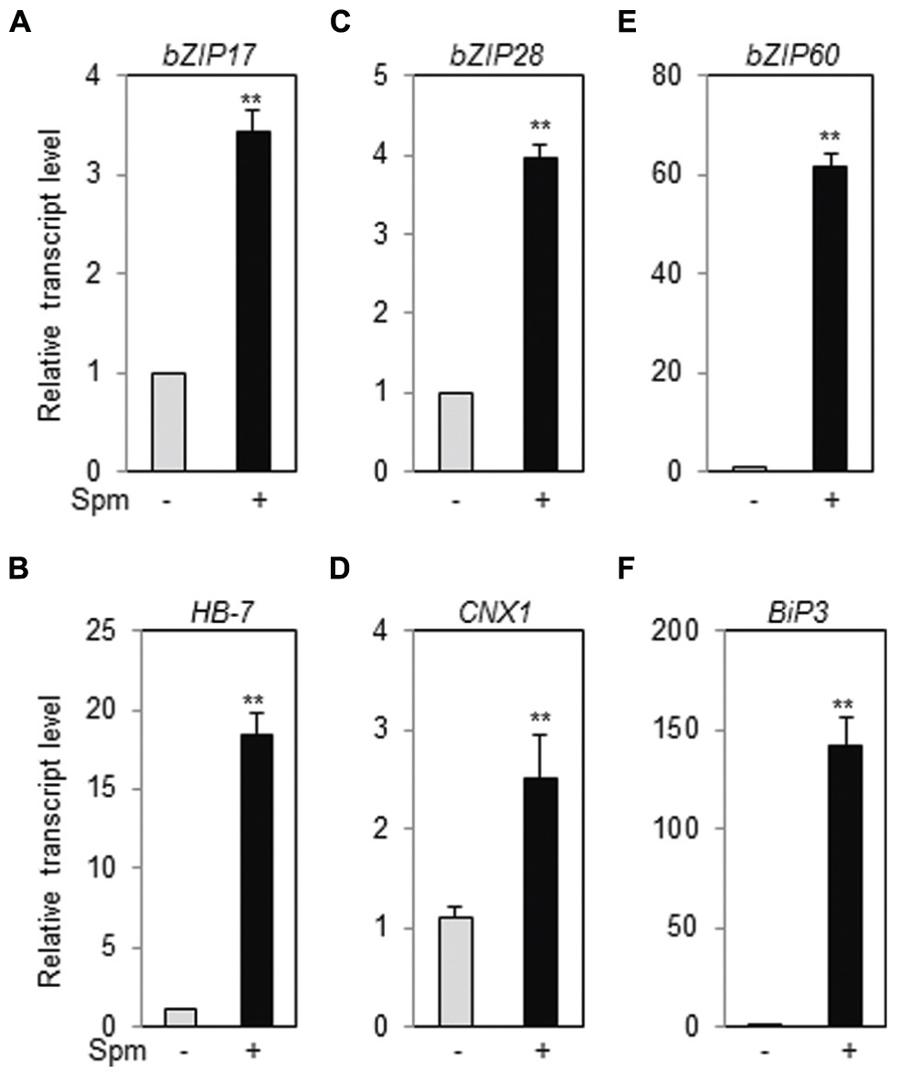
FIGURE 1. Exogenous Spm induces the expression of bZIP17, bZIP28, and bZIP60 and their target genes. qRT-PCR was performed using the cDNAs prepared from control- and Spm-treated Arabidopsis seedlings. (A) bZIP17; (B) HB-7; (C) bZIP28; (D) CNX1; (E) bZIP60; (F) Bip3. The transcript level in control Arabidopsis plant was set as 1 and relatively displayed. Values are mean (+SD) from three independent experiments. Asterisks indicate significant difference (∗∗P < 0.01).
Spm Activates the IRE1-Mediated bZIP60 Splicing
Canonical UPR inducers activate bZIP60 splicing, which is mediated by IRE1 (Deng et al., 2011, 2013). Thus we addressed this point. At 1 h after Spm treatment, the unspliced form (SPU) of bZIP60 transcripts was accumulated. At a little time gap after the SPU detection, the spliced form (SPS) of bZIP60 transcripts was detected, which followed by BiP3 induction (Figure 2). To further prove the IRE1 involvement in the process, we used ire1a, ire1b and ire1a ire1b (see Supplementary Figure S2) double mutant along with bzip60 mutant (Deng et al., 2011, 2013). In ire1a mutant, the levels of the SPS form of bZIP60 transcript and BiP3 transcript were slightly lowered compared to those of WT after Spm treatment. In ire1b and ire1a ire1b mutants, the transcriptional attenuation of the SPS-bZIP60 and BiP3 was strikingly observed (Figure 3). It shows that IRE1B is mainly participated in Spm-induced bZIP60 unconventional splicing.
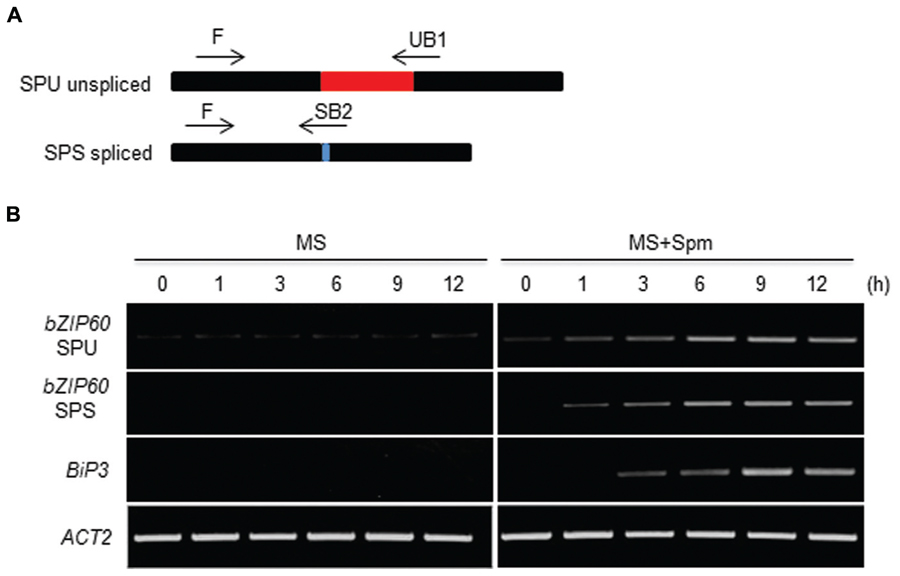
FIGURE 2. Time-course analysis of bZIP60 splicing upon Spm treatment. (A) A schematic representation of bZIP60 transcript with the position and orientation of primer pairs used for detecting the unspliced (SPU)- and spliced (SPS)-forms of bZIP60. (B) Time course RT-PCR analysis of SPU and SPS forms of bZIP60 transcript and its downstream target BiP3 transcript at indicated time intervals in control and Spm-treated seedlings. Actin was used as an internal control.
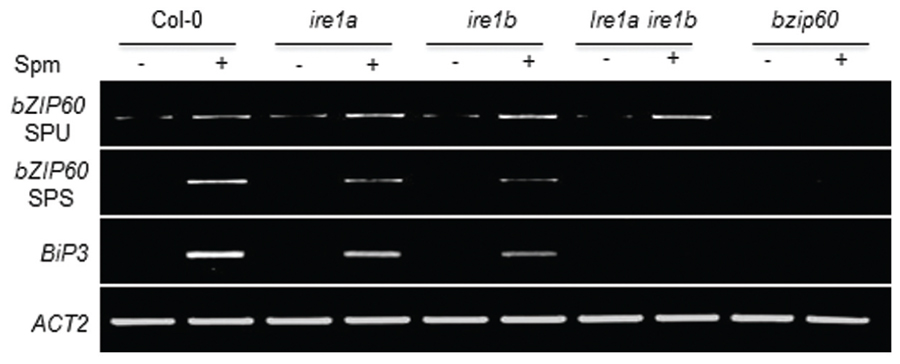
FIGURE 3. Spm-induced bZIP60 splicing occurs in an IRE1-dependent manner. Ten-days-old seedlings of Col-0 (control), ire1a (SALK_018112, Deng et al., 2011, 2013; Nagashima et al., 2011) mutant, ire1b (SAIL_238_F07) mutant, and ire1aire1b double mutant, and bzip60 (SALK_050203) mutant were treated with half strength MS solution as a control (-) and half strength MS solution containing 0.5 mM Spm (+). Results of RT-PCR for expression analysis of SPU and SPS forms of bZIP60 and its downstream gene BiP3 were displayed. Actin was used as an internal control.
Endogenous Spm Induces bZIP17, bZIP28, and bZIP60 and their Target Genes
We have generated the transgenic Arabidopsis overexpressing SPMS (Sagor et al., 2012a). The three independent lines, designated as SPMS OX_1, SPMS OX_15, and SPMS OX_21, were used in this study. In those transgenics, the SPMS transcripts were remarkably accumulated (Supplementary Figure S3A) and concomitantly Spm content increased significantly, in contrast, Spd content decreased (Supplementary Figure S3B). We examined the transcript levels of bZIP17, bZIP28, and bZIP60 in the Spm-enriched Arabidopsis plants. The levels of bZIP17, bZIP28, and bZIP60 transcripts became approximately 1.2∼1.3-fold, 2- to 3-fold, and 9- to 12-fold, respectively, in the SPMS OX transgenics compared those in WT (Figures 4A,C,E). The levels of BiP3 transcripts became clearly higher than those in WT (Figure 4F), whereas those of HB-7 and CNX1 were still higher than those in WT but not so much significant (Figures 4B,D). In the Spm-enriched plants, homeostatic regulation to suppress the constitutive UPR may be operated. Anyway, not only exogenously applied Spm but also high endogenous Spm are able to induce the UPR. PA specificity assay showed that T-Spm has a similar inducing activity as Spm does, and Spd has a weaker activity but Put has no such activity (Sagor et al., 2012b; Supplementary Figure S4, data not shown). Spm and T-Spm induced the expression of bZIP60 even at 100 μM (Supplementary Figure S4).
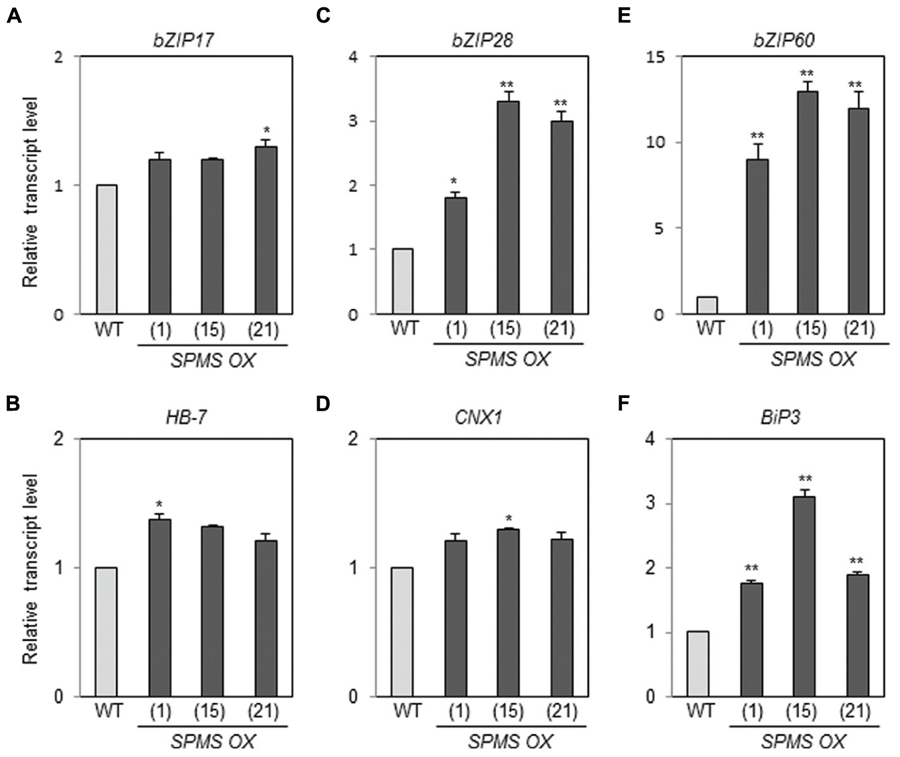
FIGURE 4. Endogenous Spm induces the expression of bZIP17, bZIP28, and bZIP60 and their target genes. The levels of bZIP17 (A), bZIP28 (C), and bZIP60 (E) transcripts, and their respective target gene transcripts [HB-7 (B), CNX1 (D), and BiP3 (F)] in the SPMS_OX transgenic lines were quantitatively determined by qRT-PCR analysis using the primer pairs listed in Supplementary Table S1. Values are mean (+SD) from three independent experiments. Asterisks indicate significant difference (∗P < 0.05 and ∗∗P < 0.01).
Spm Recruits bZIP17 and bZIP60 from ER to Nucleus
bZIP17, bZIP28, and bZIP60 encode the bZIP-type TF proteins. To accomplish their roles as TFs, their products have to reach to nuclei. Here we generated the GFP:bZIP17 and GFP:bZIP60 fusion plasmids and delivered them to onion epidermal cells. Before Spm treatment, both the fusion proteins seemed to locate in ER because the green fluorescent signals overlapped with the red fluorescent signals emitted by the ER-positive marker (Figures 5Aa–c,Ba–c). Upon Spm treatment, the green fluorescent signals from both the fusion proteins were merged to the red fluorescent signals emitted from the nucleus-targeting marker plasmid, indicating that bZIP17 and bZIP60 proteins were recruited to nucleus after Spm treatment (Figures 5Ag–i, Bg–i) as well as after DTT treatment (Figures 5Ad–f,Bd–f).
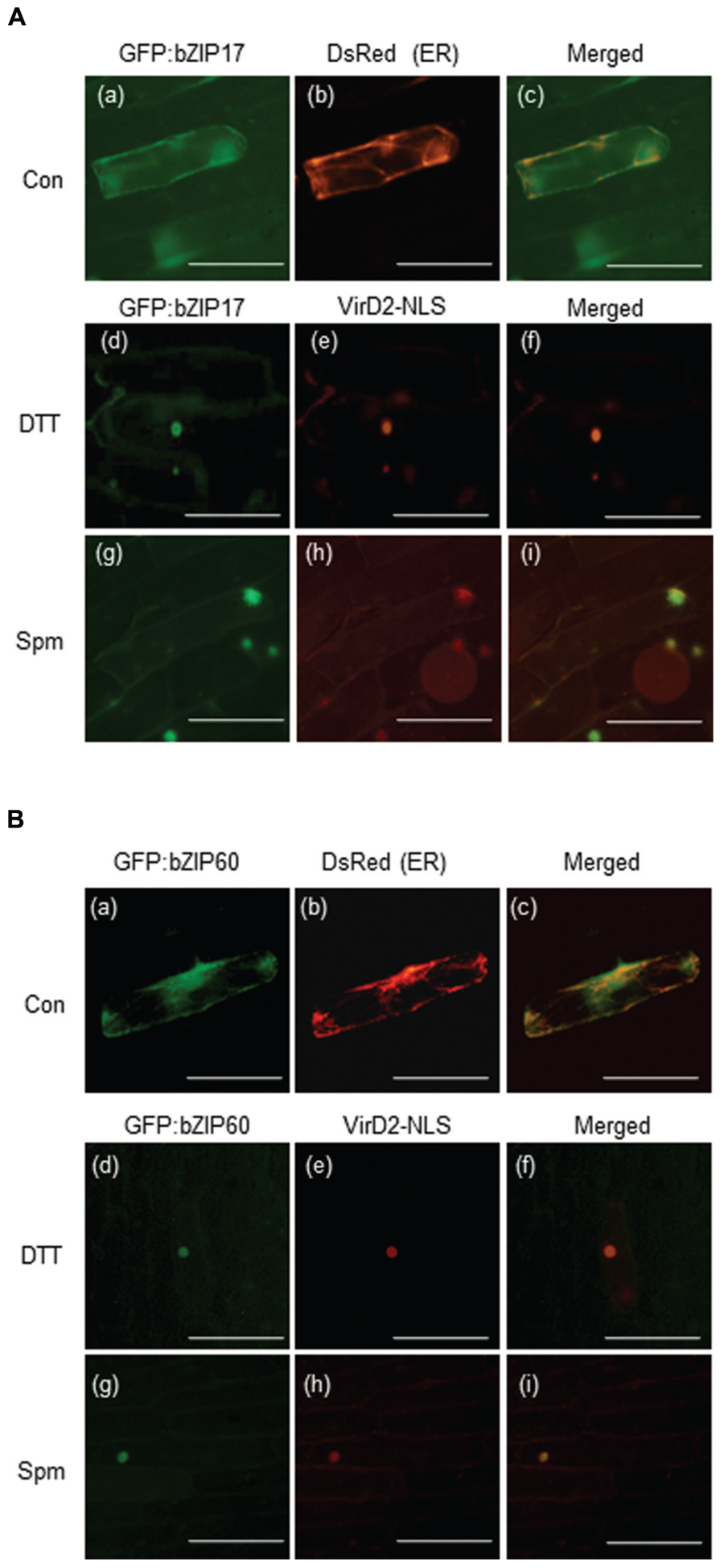
FIGURE 5. Recruitment of bZIP17 and bZIP60 proteins from ER to nucleus upon Spm treatment. (A) GFP:bZIP17 fusion protein under normal condition (a–c); DTT treatment (d–f); and Spm treatment (g–i). (B) GFP:bZIP60 fusion protein under normal condition (a–c); DTT treatment (d–f); and Spm treatment (g–i). The plasmids encoding GFP:bZIP17 and GFP:bZIP60 proteins were co-bombarded to onion bulbs with the plasmid encoding an ER marker peptide tagged with Ds-Red or with the plasmid encoding a nuclear marker protein VirD2_NLS using a particle gun. The bombarded samples were treated with or without Spm for 16 h under darkness at 22°C. The samples were also treated with DTT as a positive reference. Then onion epidermal cell layers were peeled off and observed by a fluorescence microscope (BX61; Olympus). Bar indicates 10 μm.
Ca2+-Influx is One of the Upstream Components of Spm-Induced UPR
In tobacco plant, we have demonstrated that Ca2+-influx to cytoplasm and the activation of the mitogen-activated protein kinase (MAPK) cascade consisting of NtMEK2-SIPK/WIPK are prerequisite for triggering a Spm-signaling pathway (Takahashi et al., 2003, 2004). First, therefore, we addressed whether Ca2+-influx to cytoplasm is required for UPR induction by Spm treatment. We applied a Ca2+-channel blocker, La3+, to the plant samples when Spm was applied. Upregulation of bZIP17, bZIP28, and bZIP60 was totally alleviated in the plants co-treated with Spm and La3+ (Figures 6A,C,E). Induction of the target genes, HB-7, CNX1 and BiP3, of bZIP17, bZIP28, and bZIP60 was also blocked by La3+ treatment (Figures 6B,D,F). The results indicate that Spm-induced UPR pathway requires the enhanced Ca2+ influx to cytoplasm.
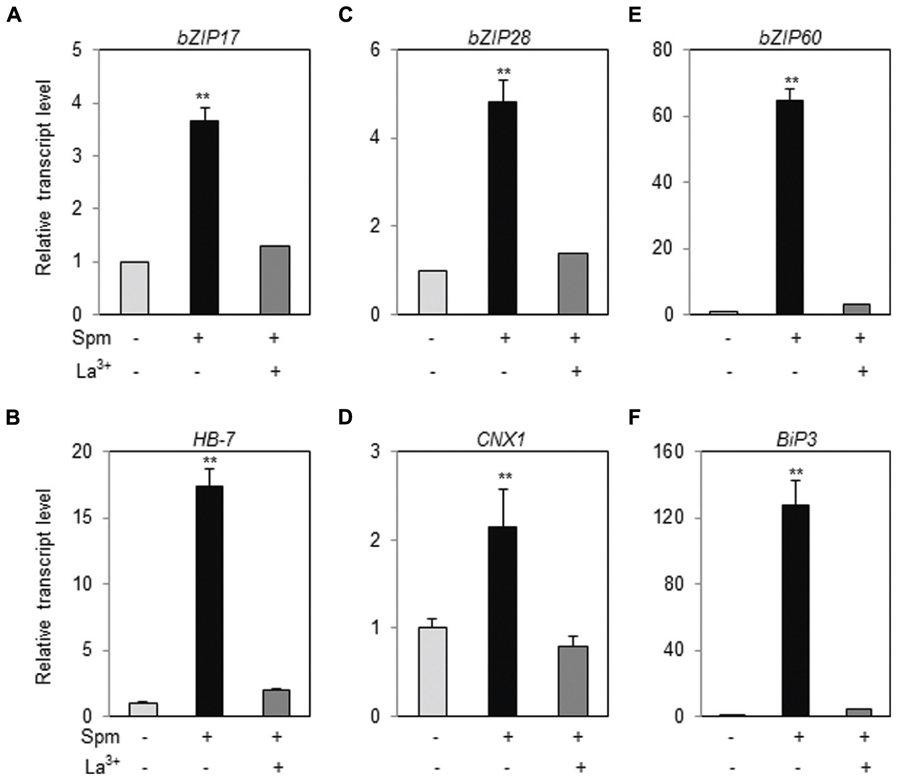
FIGURE 6. Ca2+-channel blocker alleviates the UPR induction by Spm. qRT-PCR was performed using the cDNAs prepared from control-, Spm-treated, and Spm and La3+-co-treated Arabidopsis seedlings. (A) bZIP17; (B) HB-7; (C) bZIP28; (D) CNX1; (E) bZIP60; (F) Bip3. The transcript level in control Arabidopsis plant was set as 1 and relatively displayed. Values are mean (+SD) from three independent experiments. Asterisks indicate significant difference (∗∗P < 0.01).
A MAPK Cascade is Another Upstream Component of Spm-Induced UPR
In A. thaliana, there are more than 60 MAPK kinase kinases (MAP3K), 10 MAPKKs (MKK), and 20 MAPKs (MAPK Group, 2002). Therefore, we examined whether any MKKs are up-regulated by exogenously applied Spm. Of 10 MKKs, MKK9 was strikingly induced by Spm (Figure 7A). The levels of MKK4 and MKK5 transcripts were also accumulated at the lesser extent compared to those of MKK9 (Figure 7A). The previous research showed that the downstream MAPKs of MKK9 are MPK6 (SIPK ortholog) and/or MPK3 (WIPK ortholog; see review Colcombet and Hirt, 2008; Yoo et al., 2008; Zhou et al., 2009; Rodriguez et al., 2010). In addition to MKK9, MPK3 was identified as Spm-responsive gene in a SuperSAGE analysis (Mitsuya et al., 2009). So we tested the expressional response of MPK3 and MPK6 upon Spm treatment. The qRT-PCR result showed that expression of both MPK3 and MPK6 was up-regulated by Spm treatment (Figure 7B). Furthermore, we quantified the transcript levels of MKK9, MPK3, and MPK6 in the Spm-enriched transgenic plants, and found that MKK9 and MPK3 transcripts were distinctly accumulated and MPK6 transcripts increased to approximately 1.2–1.4-fold levels in the SPMS OX plants compared to those in WT (Figure 7C).
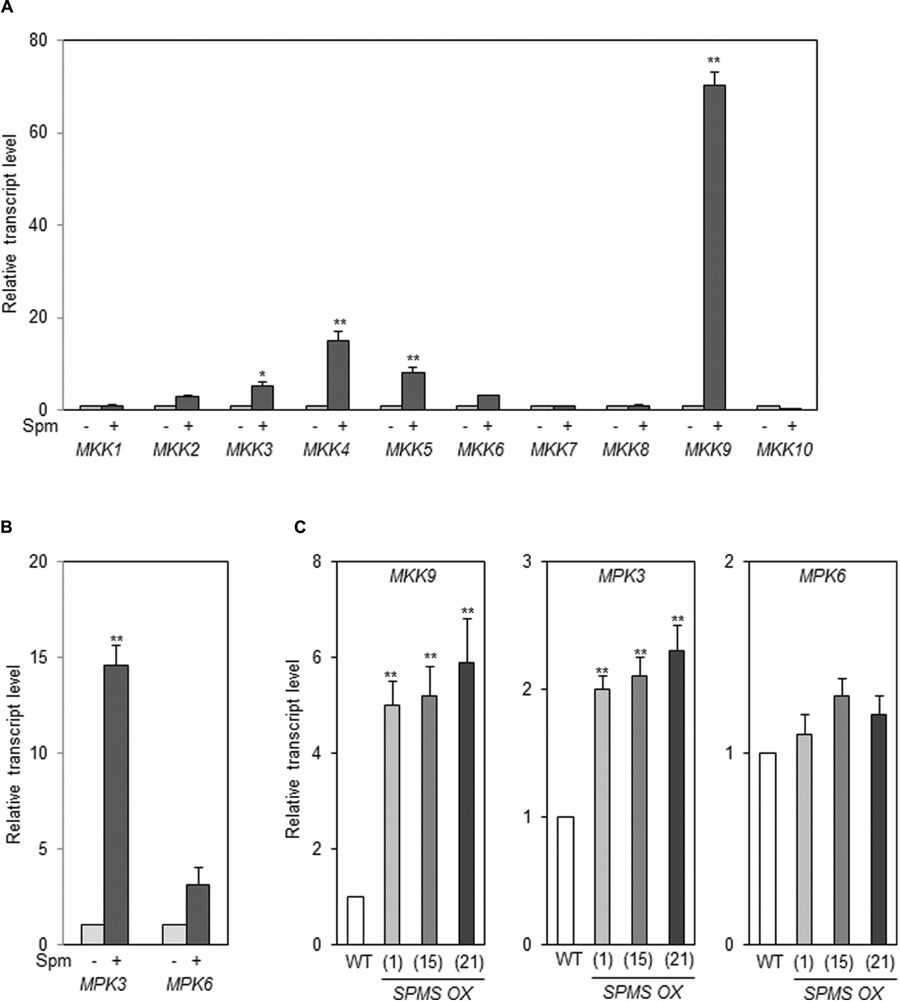
FIGURE 7. Spm upregulates MKK9, MPK3, and MPK6. (A) Exogenous Spm upregulated MKK9 among 10 MKKs. (B) Exogenous Spm upregulated MPK3 and MPK6. (C) The transcript levels of MKK9, MPK3, and MPK6 in the SPMS_OX transgenic lines. The levels of MKK9, MPK3, and MPK6 transcripts were quantitatively determined by qRT-PCR using the primer pairs listed in Supplementary Table S1. Values are mean (+SD) from three independent experiments. Asterisks indicate significant difference (∗P < 0.05 and ∗∗P < 0.01).
Spm-Induction of UPR Pathway is Mediated by the MKK9-MPK3/MPK6 Cascade
Then the question arose whether all the paths of bZIP17, bZIP28, and bZIP60 in the UPR are positioned downstream of the MKK9-MPK3/MPK6 cascade. To answer the issue, we used the T-DNA insertion mutants of MKK9, MPK3, and MPK6. The T-DNA insertion sites in MKK9, MPK3, and MPK6 were displayed in Supplementary Figure S5A. The homogeneity of the T-DNA insertion in the mutants, mkk9, mpk3, and mpk6, was confirmed by the genome DNA-PCR (Supplementary Figures 5B–D; Xu et al., 2008; Yoo et al., 2008; Zhou et al., 2009). The levels of MKK9-, MPK3-, and MPK6-transcripts in the mkk9, mpk3, and mpk6 mutants were almost null in relative to those of WT, respectively (Supplementary Figures 5E–G). Using the loss-of-function mutants, we examined the levels of bZIP17, bZIP28, and bZIP60 transcripts upon Spm treatment. The expression of bZIP17, bZIP28, and bZIP60 was not enhanced by Spm treatment in the mkk9 mutant (Figure 8A). The expression of their target genes was also not induced by Spm in the mkk9 mutant (Figure 8B). The expression of the bZIP genes and their target genes was more or less similarly induced by Spm in the mpk3 and mpk6 mutants. The latter result may be explained that MPK3 and MPK6 have redundant function to transduce the Spm-signal. It should be noted that Spm-signaling pathway was blocked in Nicotiana benthamiana in which both, SIPK and WIPK, were simultaneously silenced but neither in SIPK-silencing nor in WIPK-silencing N. benthamiana (Takahashi et al., 2004). Collectively it indicates that the induction of bZIP17, bZIP28, and bZIP60 depends on the MKK9-MPK3/MPK6 cascade.
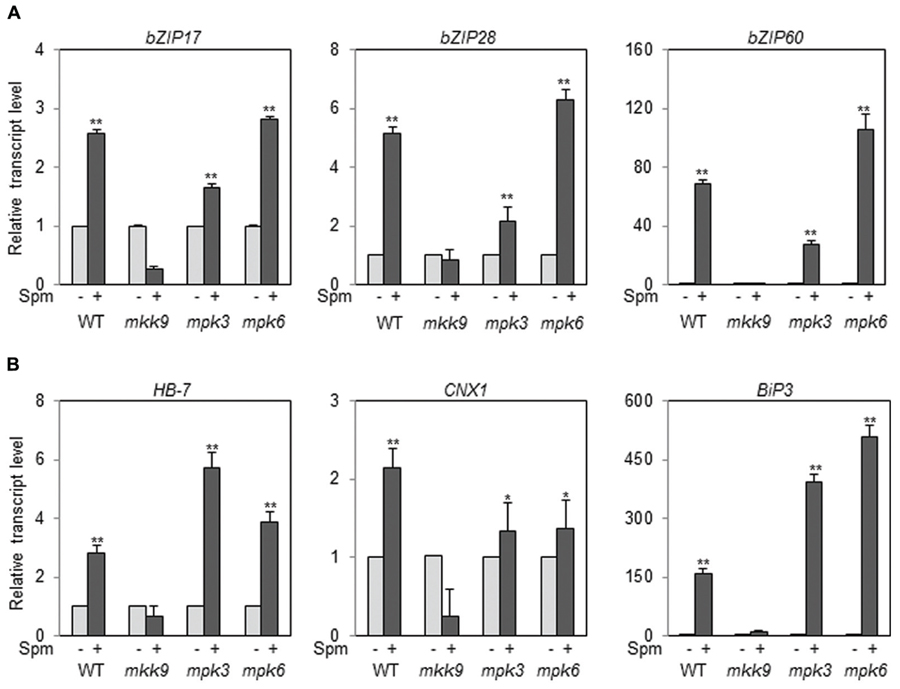
FIGURE 8. The expression of bZIP17, bZIP28, and bZIP60 (A) and their target genes (B) in the loss-of-function mutants, mkk9, mpk3, and mpk6, upon Spm treatment. The expression levels of the three bZIP genes and their target genes in mkk9, mpk3, and mpk6 were determined by qRT-PCR. The experiments were performed using three independent biological samples. The mean + SD were displayed. Asterisks indicate significant difference (∗P < 0.05 and ∗∗P < 0.01).
Discussion
Spm Activates Three Pivotal bZIP Genes and their Target Genes in UPR
Previous research showed that exogenous Spm induces the expression of Arabidopsis bZIP60 and its ortholog, NtbZIP60, of N. tabacum (Tateda et al., 2008; Mitsuya et al., 2009). We addressed whether Spm induces bZIP60 specifically or rather induces the whole UPR process. Here, we showed that Spm induces not only bZIP60 but also bZIP17 and bZIP28 expression (Figures 1 and 4). In the case of bZIP60, an IRE1-dependent bZIP60 splicing process was also activated by Spm (Figures 2 and 3). We speculate that the proteolytic bZIP17 and bZIP28 processing by S1P and S2P is activated by Spm because their downstream target genes such as HB-7, RD20, CNX1, and CRT2 were concomitantly upregulated (Figure 1, data not shown). In addition, in Spm-enriched Arabidopsis plants, the three bZIP genes were induced (Figure 4). We further showed that Spm can recruit bZIP17 and bZIP60 proteins to nuclei in plant cells (Figure 5). Taken together, we conclude that Spm is a novel inducer of UPR.
Spm-Induced UPR Pathway is Mediated by the MKK9-MPK3/MPK6 Cascade
Previous work showed that Spm activates two kinds of MAPKs, SIPK and WIPK, and this activation requires the production of reactive oxygen species and Ca2+ influx in tobacco leaves (Takahashi et al., 2003, 2004). Our result showed that MKK9-MPK3/MPK6 is the MAPK cascade for Spm-induced UPR in Arabidopsis. The Arabidopsis MAPK signaling pathways are involved in various biological processes (Colcombet and Hirt, 2008; Rodriguez et al., 2010). The MKK9-MPK3/MPK6 cascade was used in ethylene signaling (Yoo et al., 2008) and was also involved in the phytoalexin production, and in salt stress response in Arabidopsis (Xu et al., 2008). Furthermore, our result showed that, in mkk9 mutant, Spm-induced bZIP17, bZIP28, and bZIP60 expression was significantly alleviated (Figure 8). Moreover, the Spm-induced expression of the respective target genes, HB-7, CNX1, and BiP3, was also abrogated in the mkk9 mutant (Figure 8). The result suggests that Spm-evoked Ca2+-elevation activates the MKK9-MPK3/MPK6 cascade, which triggers the bZIP17, bZIP28, and bZIP60 induction. It should be noted that, in mammalian system, IRE1 activates JNK protein kinases (Urano et al., 2000). To know whether Arabidopsis IRE1 can activate the MKK9-MPK3/MPK6 pathway is of interest.
How does Spm Induce UPR?
We noticed that the Spm-induced expression profile of three UPR key bZIP genes and their target genes differs from the one induced by the typical UPR inducers, DTT and TM. Martinez and Chrispeels (2003) described that the respective UPR agents have different effects; i.e., the genes induced by TM and DTT were different in Arabidopsis. It is quite reasonable because TM affects N-glycosylation whereas DTT changes the cellular redox state (Liu and Howell, 2010). Then the question arose how Spm induces the UPR. Spm-induced expression of bZIP17, bZIP28, and bZIP60 genes was blocked by a Ca2+ channel blocker (Figure 6). Spm- and T-Spm-deficient Arabidopsis plants cannot grow well in Ca2+-depleted MS medium (Yamaguchi et al., 2006), suggesting that the tetraamines are involved in Ca2+ dynamics and homeostasis. PAs are known to modulate Ca2+ dynamics via control of cation channel activity (Dobrovinskaya et al., 2000; Pottosin and Shabala, 2014). Furthermore, the action potential to regulate cation channels is the order of Spm > Spd ≥ Put, which may explain the PA specificity to evoke UPR induction. As exogenously applied T-Spm also induces Spm-responsive genes at the higher degree compared to Spm (Sagor et al., 2012b), T-Spm may possess a similar or the higher action potential in terms of modulating cation channels in relative to Spm. It is known that thapsigargin, another UPR inducer, affects Ca2+ homeostatic balance. Thapsigargin leads to ER Ca2+ depletion due to inhibition of the Sarco/ER Ca2+ ATPase (SERCA; Xu et al., 2008). Taken together, we hypothesize that Spm induces UPR pathway through affecting Ca2+ dynamics. Of course further study is needed to substantiate the hypothesis.
Physiological Relevance of Spm-Induced UPR
It was demonstrated that the polyamine Spm, exogenously applied or endogenously enriched, has a protective role in the defense response of plants against both abiotic and biotic stresses like salt, drought and heat stresses, and attack by viral pathogens (Yamaguchi et al., 2006, 2007; Mitsuya et al., 2009; Sagor et al., 2012a; Tiburcio et al., 2014; Berberich et al., 2015). A protective role was also shown for components of the UPR. When the bZIP60 ortholog of N. benthamiana was silenced by a virus-induced gene silencing approach, the host plant became hypersensitive to non-host bacterial pathogen (Tateda et al., 2008). bZIP60 was upregulated when the avirulent CMV-Y strain, but not the virulent CMV-B2 strain, was used to infect Arabidopsis ecotype C24 which carries the resistance gene, RCY1, to CMV-Y (Takahashi et al., 1994; Mitsuya et al., 2009). In parallel Spm synthase gene was upregulated and Spm content increased in the C24-CMV-Y pathosystem (Mitsuya et al., 2009). We assume a link inasmuch as the accumulated Spm activates UPR. The protection against viral infection could be accomplished indirectly by degradation of the excess Spm by polyamine oxidases which produce H2O2. H2O2 would then be transmitted to the surrounding tissue of the infection site, thereby inducing the UPR in such cells. Since the virus is unable to replicate properly while UPR takes place the tissue is protected. A distinct role of UPR in plant immunity mediated by IRE1/bZIP60 has been shown by Moreno et al. (2012). Those authors demonstrated that SA, an important phytohormone in immune response, can induce IRE1/bZIP60-mediated UPR. Nagashima et al. (2014) further showed that SA activates two signaling arms, namely bZIP28 and bZIP60, of the UPR in Arabidopsis. Those results suggest that bZIP60 (or its ortholog) has a defensive role against avirulent pathogens. In contrast, bZIP17 and bZIP28 were found to be rather associated with abiotic stresses such as drought, ABA and high salinity, and heat stress, respectively (Soderman et al., 1996; Liu et al., 2007b; Liu and Howell, 2010; Howell, 2013), for which a defensive role of Spm has also been demonstrated as mentioned above. In conclusion, the broad effect of Spm in the stress response of plants could originate, at least partly, in the capacity of Spm as an UPR inducer.
Funding
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) to TK (26⋅04081, 15K14705) and to DK (25⋅5682). SGHM is a recipient of JSPS foreign researcher fellowship.
Conflict of Interest Statement
The Guest Associate Editor Taku Takahashi declares that, despite having previously collaborated with the authors Masaru Niitsu and Tomonobu Kusano, the review process was handled objectively. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Drs N.-H. Chua and D. Ren and Arabidopsis Biological Resource Center for providing a GFP plasmid, Atmkk9, Atmpk3, and Atmpk6 mutant seeds, respectively. Dr. Stephen H. Howell is also acknowledged for providing ire1a, ire1b, and bzip60 mutant seeds.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00687
References
Alcázar, R., Altabella, T., Marco, F., Bortolotti, C., Reymond, M., Koncz, C., et al. (2010). Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta 231, 1237–1249. doi: 10.1007/s00425-010-1130-0
Berberich, T., Sagor, G. H. M., and Kusano, T. (2015). “Polyamines in plant stress response,” in Polyamine: A Universal Molecular Nexus for Growth, Survival and Specialized Metabolism, eds T. Kusano and H. Suzuki (Tokyo: Springer), 155–168.
Colcombet, J., and Hirt, H. (2008). Arabidopsis MAPKs: a complex signaling network involved in multiple biological processes. Biochem. J. 413, 217–226. doi: 10.1042/BJ20080625
Deng, Y., Humbert, S., Liu, J. X., Srivastava, R., Rothstein, S. J., and Howell, S. H. (2011). Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 7247–7252. doi: 10.1073/pnas.1102117108
Deng, Y., Srivastava, R., and Howell, S. H. (2013). Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 19633–19638. doi: 10.1073/pnas.1314749110
Dobrovinskaya, O. R., Muniz, J., and Pottosin, I. I. (2000). Asymmetric block of the plant vacuolar Ca2+-permeable channel by organic cations. Eur. Biophys. J. 28, 552–563. doi: 10.1007/s002490050237
Hanfrey, C., Sommer, S., Mayer, M. J., Burtin, D., and Michael, A. J. (2001). Arabidopsis polyamine biosynthesis: absence of ornithine decarboxylase and the mechanism of arginine decarboxylase activity. Plant J. 27, 551–560. doi: 10.1046/j.1365-313X.2001.01100.x
Howell, S. H. (2013). Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 64, 477–499. doi: 10.1146/annurev-arplant-050312-120053
Iwata, Y., Fedoroff, N. V., and Koizumi, N. (2008). Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20, 3107–3121. doi: 10.1105/tpc.108.061002
Iwata, Y., and Koizumi, N. (2005). An Arabidopsis transcription factor, AtbZIP60, regulated the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl. Acad. Sci. U.S.A. 102, 5280–5285. doi: 10.1073/pnas.0408941102
Iwata, Y., and Koizumi, N. (2012). Plant transducers of the endoplasmic reticulum unfolded protein response. Trends Plant Sci. 12, 720–727. doi: 10.1016/j.tplants.2012.06.014
Knott, J. M., Romer, P., and Sumper, M. (2007). Putative spermine synthases from Thalassiosira pseudonana and Arabidopsis thaliana synthesize thermospermine rather than spermine. FEBS Lett. 581, 3081–3086. doi: 10.1016/j.febslet.2007.05.074
Kusano, T., Berberich, T., Tateda, C., and Takahashi, Y. (2008). Polyamines: essential factors for growth and survival. Planta 228, 367–381. doi: 10.1007/s00425-008-0772-7
Kusano, T., Kim, D. W., Liu, T., and Berberich, T. (2015). “Polyamine catabolism in plants,” in Polyamine: A Universal Molecular Nexus for Growth, Survival and Specialized Metabolism, eds T. Kusano and H. Suzuki (Tokyo: Springer), 77–88.
Liu, J. X., and Howell, S. H. (2010). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22, 782–796. doi: 10.1105/tpc.109.072173
Liu, J. X., Srivastava, R., Che, P., and Howell, S. H. (2007a). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19, 4111–4117. doi: 10.1105/tpc.106.050021
Liu, J. X., Srivastava, R., Che, P., and Howell, S. H. (2007b). Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signalling. Plant J. 51, 897–909. doi: 10.1111/j.1365-313X.2007.03195.x
MAPK Group. (2002). Mitogen-activated protein kinase cascades in plants. A new nomenclature. Trends Plant Sci. 7, 301–308. doi: 10.1016/S1360-1385(02)02302-6
Martinez, I. M., and Chrispeels, M. J. (2003). Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15, 561–576. doi: 10.1105/tpc.007609
Mitsuya, Y., Takahashi, Y., Berberich, T., Miyazaki, A., Matsumura, H., Takahashi, H., et al. (2009). Spermine plays a significant role in the defense response of Arabidopsis thaliana to cucumber mosaic virus. J. Plant Physiol. 166, 626–643. doi: 10.1016/j.jplph.2008.08.006
Moreno, A. A., Mukhtar, M. S., Blanco, F., Boatwright, J. L., Moreni, I., Jordan, M. R., et al. (2012). IRE1/bZIP60-mediated unfolded protein response plays a distinct roles in plant immunity and abiotic stress responses. PLoS ONE 7:e31944. doi: 10.1371/journal.pone.0031944
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res. 19, 4321–4326. doi: 10.1093/nar/8.19.4321
Nagashima, Y., Iwata, Y., Ashida, M., Mishiba, K., and Koizumi, N. (2014). Exogenous salicylic acid activates two signalling arms of the unfolded protein response in Arabidopsis. Plant Cell Physiol. 55, 1772–1778. doi: 10.1093/pcp/pcu108
Nagashima, Y., Mishiba, K., Suzuki, E., Shimada, Y., Iwata, Y., and Koizumi, N. (2011). Arabidopsis IRE1 catalysis unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci. Rep. 1, 29. doi: 10.1038/srep00029
Naka, Y., Watanabe, K., Sagor, G. H. M., Niitsu, M., Pillai, A., Kusano, T., et al. (2010). Quantitative analysis of plant polyamines including thermospermine during growth and salinity stress. Plant Physiol. Biochem. 48, 527–533. doi: 10.1016/j.plaphy.2010.01.013
Okamoto, K., Nagano, H., Iwakawa, H. O., Mizumoto, H., Takeda, A., Kaido, M., et al. (2008). cis-preferential requirement of a-1 frameshift product p88 for the replication of Red clover necrotic mosaic virus RNA1. Virology 375, 205–212. doi: 10.1016/j.virol.2008.02.004
Ono, Y., Kim, D. W., Watanabe, K., Sasaki, A., Niitsu, M., Berberich, T., et al. (2012). Constitutively and highly expressed Oryza sativa polyamine oxidases localize in peroxisomes and catalyze polyamine back conversion. Amino Acids 42, 867–876. doi: 10.1007/s00726-011-1002-3
Pottosin, I., and Shabala, S. (2014). Polyamine control of cation transport across plant membranes: implications for ion homeostasis and abiotic stress signaling. Front. Plant Sci. 5:154. doi: 10.3389/fpls.2014.00154
Rodriguez, M. C., Petersen, M., and Mundy, J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621–649. doi: 10.1146/annurev-arplant-042809-112252
Sagor, G. H. M., Berberich, T., Takahashi, Y., Niitsu, M., and Kusano, T. (2012a). The polyamine spermine protects Arabidopsis from heat stress-induced damage by increasing expression of heat shock-related genes. Transgenic Res. 22, 595–605. doi: 10.1007/s11248-012-9666-9663
Sagor, G. H. M., Takahashi, H., Niitsu, M., Takahashi, Y., Berberich, T., and Kusano, T. (2012b). Exogenous thermospermine has an activity to induce a subset of the defence genes and restrict cucumber mosaic virus multiplication in Arabidopsis thaliana. Plant Cell Rep. 31, 1227–1232. doi: 10.1007/s00299-012-1243-y
Seo, S., Okamoto, M., Seto, H., Ishizuka, K., Sano, H., and Ohashi, Y. (1995). Tobacco MAP kinase: a possible mediator in wound signal transduction pathways. Science 270, 1988–1992. doi: 10.1126/science.270.5244.1988
Soderman, E., Mattsson, J., and Engström, P. (1996). The Arabidopsis homeobox gene AtHB-7 is induced by water deficit and by abscisic acid. Plant J. 10, 375–381. doi: 10.1046/j.1365-313X.1996.10020375.x
Srivastava, R., Deng, Y., Shah, S., Rao, A. G., and Howell, S. H. (2013). binding protein is a master regulator of the endoplasmic reticulum stress sensor/transducer bZIP28 in Arabidopsis. Plant Cell 25, 1416–1429. doi: 10.1105/tpc.113.110684
Tabor, C. W., and Tabor, H. (1984). Polyamines. Annu. Rev. Biochem. 53, 749–790. doi: 10.1146/annurev.bi.53.070184.003533
Takahashi, H., Goto, N., and Ehara, Y. (1994). Hypersensitive response in cucumber mosaic virus-inoculated Arabidopsis thaliana. Plant J. 6, 369–377. doi: 10.1046/j.1365-313X.1994.06030369.x
Takahashi, T., and Kakehi, J.-I. (2010). Polyamines: ubiquitous polycations with unique roles in growth and stress responses. Ann. Bot. 105, 1–6. doi: 10.1093/aob/mcp259
Takahashi, Y., Berberich, T., Miyazaki, A., Seo, S., Ohashi, Y., and Kusano, T. (2003). Spermine signalling in tobacco: activation of mitogen-activated protein kinases by spermine is mediated through mitochondrial dysfunction. Plant J. 36, 820–829. doi: 10.1046/j.1365-313X.2003.01923.x
Takahashi, Y., Uehara,Y., Berberich, T., Ito, A., Saitoh, H., Miyazaki, A., et al. (2004). A subset of hypersensitive response marker genes, including HSR203J, is the downstream target of a spermine signal transduction pathway in tobacco. Plant J. 40, 586–595. doi: 10.1111/j.1365-313X.2004.02234.x
Takano, A., Kakehi, J. I., and Takahashi, T. (2012). Thermospermine is not a minor polyamine in the plant kingdom. Plant Cell Physiol. 53, 606–616. doi: 10.1093/pcp/pcs019
Tateda, C., Ozaki, R., Onodera, Y., Takahashi, Y., Yamaguchi, K., Berberich, T., et al. (2008). NtbZIP60, an endoplasmic reticulum-localized transcription factor, plays a role in defense response against bacterial pathogen in tobacco. J. Plant Res. 121, 603–611. doi: 10.1007/s10265-008-0185-185
Tiburcio, A. F., Altabella, T., Bitrián, M., and Alcázar, R. (2014). The roles of polyamines during the lifespan of plants: from development to stress. Planta 240, 1–18. doi: 10.1007/s00425-014-2055-9
Urano, F., Wang, X., Bertolotti, A., Zhang, Y., Chung, P., Harding, H. P., et al. (2000). Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287, 664–666. doi: 10.1126/science.287.5453.664
Walters, D. (2003a). Resistance to plant pathogens: possible roles for free polyamines and polyamine catabolism. New Phytol. 159, 109–115. doi: 10.1046/j.1469-8137.2003.00802.x
Walters, D. R. (2003b). Polyamines and plant disease. Phytochemistry 64, 97–107. doi: 10.1016/S0031-9422(03)00329-7
Walter, P., and Ron, D. (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086. doi: 10.1126/science.1209038
Xu, J., Li, Y., Wang, Y., Liu, H., Lei, L., Yang, H., et al. (2008). Activation of MAPK kinase 9 induces ethylene and calnexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 283, 26996–27006. doi: 10.1074/jbc.M801392200
Yamaguchi, K., Takahashi, Y., Berberich, T., Imai, A., Miyazaki, A., Takahashi, T., et al. (2006). The polyamine spermine protects against high salt stress in Arabidopsis thaliana. FEBS Lett. 580, 6783–6788. doi: 10.1016/j.febslet.2006.10.078
Yamaguchi, K., Takahashi, Y., Berberich, T., Imai, A., Takahashi, T., Michael, A., et al. (2007). A protective role for the polyamine spermine against drought stress in Arabidopsis. Biochem. Biophys. Res. Commun. 352, 486–490. doi: 10.1016/j.bbrc.2006.11.041
Yamakawa, H., Kamada, H., Satoh, M., and Ohashi, Y. (1998). Spermine is a salicylate-independent endogenous inducer for both tobacco acidic pathogenesis-related proteins and resistance against Tobacco mosaic virus infection. Plant Physiol. 118, 1213–1222. doi: 10.1104/pp.118.4.1213
Yoo, S. D., Cho, Y. H., Tena, G., Xiong, Y., and Sheen, J. (2008). Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451, 789–795. doi: 10.1038/nature06543
Zhang, S., and Klessig, D. F. (1997). Salicylic acid activates a 48-kDa MAP kinase in tobacco. Plant Cell 9, 809–824. doi: 10.1105/tpc.9.5.809
Keywords: calcium, mitogen activated protein kinase cascade, polyamine, spermine, unfolded protein response, UPR inducer
Citation: Sagor GHM, Chawla P, Kim DW, Berberich T, Kojima S, Niitsu M and Kusano T (2015) The polyamine spermine induces the unfolded protein response via the MAPK cascade in Arabidopsis. Front. Plant Sci. 6:687. doi: 10.3389/fpls.2015.00687
Received: 20 July 2015; Accepted: 20 August 2015;
Published: 10 September 2015.
Edited by:
Taku Takahashi, Okayama University, JapanCopyright © 2015 Sagor, Chawla, Kim, Berberich, Kojima, Niitsu and Kusano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tomonobu Kusano, Graduate School of Life Sciences, Tohoku University, 2-1-1 Katahira, Aoba, Sendai 980-8577, Japan, kusano@ige.tohoku.ac.jp
†Present address: Dong W. Kim, RIKEN Center for Sustainable Resource Science, 1-7-22 Suehiro-cho, Tsurumi-Ku, Yokohama, Kanagawa 230-0045, Japan
‡ These authors have contributed equally to this work.
 G. H. M. Sagor
G. H. M. Sagor Pratima Chawla
Pratima Chawla Dong W. Kim
Dong W. Kim Thomas Berberich
Thomas Berberich Seiji Kojima
Seiji Kojima Masaru Niitsu
Masaru Niitsu Tomonobu Kusano
Tomonobu Kusano