- 1Rice Research Institute, Sichuan Agricultural University, Chengdu, China
- 2State Key Laboratory of Plant Genomics and National Center for Plant Gene Research, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China
- 3Key Laboratory of Biology and Genetic Resources of Rubber Tree, Rubber Research Institute, Chinese Academy of Tropical Agricultural Sciences, Danzhou, China
- 4Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing, China
Dof (DNA binding with one finger) proteins, a class of plant-specific transcription factors, are involved in plant growth and developmental processes and stress responses. However, their biological functions remain to be elucidated, especially in rice (Oryza sativa L.). Previously, we have reported that OsDof12 can promote rice flowering under long-day conditions. Here, we further investigated the other important agronomical traits of the transgenic plants overexpressing OsDof12 and found that overexpressing OsDof12 could lead to reduced plant height, erected leaf, shortened leaf blade, and smaller panicle resulted from decreased primary and secondary branches number. These results implied that OsDof12 is involved in rice plant architecture formation. Furthermore, we performed a series of Brassinosteroid (BR)-responsive tests and found that overexpression of OsDof12 could also result in BR hyposensitivity. Of note, in WT plants the expression of OsDof12 was found up-regulated by BR treatment while in OsDof12 overexpression plants two positive BR signaling regulators, OsBRI1 and OsBZR1, were significantly down-regulated, indicating that OsDof12 may act as a negative BR regulator in rice. Taken together, our results suggested that overexpression of OsDof12 could lead to altered plant architecture by suppressing BR signaling. Thus, OsDof12 might be used as a new potential genetic regulator for future rice molecular breeding.
Introduction
DOF (DNA binding with one finger) proteins are plant-specific transcription factors (Yanagisawa, 2002; Moreno-Risueno et al., 2007; Noguero et al., 2013). The Dof domain consists of 52 amino acid residues encompassing the CX2CX21CX2C motif (Yanagisawa, 2002; Umemura et al., 2004). Dof transcription factors, with the exception in pumpkin, usually regulate the expression of the target genes via binding a core DNA motif with an essential sequence of AAAG (Yanagisawa and Schmidt, 1999). Dof proteins are widespread and versatile regulators for various biological processes such as metabolism regulation, phytohormone response, seed germination and development, photoperiodic flowering and plant patterning in plants (Yanagisawa, 2002; Noguero et al., 2013). In carbohydrate metabolism, ZmDof1 and ZmDof2 acted antagonistically to control the expression of C4 phosphoenolpyruvate carboxylase (PEPC) in maize (Zea mays) (Yanagisawa, 2000). In Arabidopsis (Arabidopsis thaliana), AtDof1.1/OBP2 participated in regulation of indole glucosinolate biosynthesis (Skirycz et al., 2006). Overexpression of OsDof25 in Arabidopsis changed the nitrogen and carbon metabolism (Santos et al., 2012). In tobacco (Nicotiana tabacum), NtBBF1 (roiB domain B Factor) can bind to the rolB promotor in an auxin-regulated way to modulate its expression, which betters our understanding the mechanism underlying auxin induction (Baumann et al., 1999). Besides, several cases have documented that Dof genes are implicated in seed germination and development. The DOF gene Affecting Germination-1 (DAG1) and DOF gene Affecting Germination-2 (DAG2) controlled seed germination in Arabidopsis via a maternal switch (Papi et al., 2000; Gualberti et al., 2002). In rice, OsDof3 might enhance expression of type3 carboxypeptidase (CPD) under GA control in aleurones (Washio, 2001), and further investigation indicated that OsDof3 interacts with GAMYB to regulate synergically the expression of RAmy1A to mediate GA signaling during rice seed germination (Washio, 2003). RPBF (rice prolamin box binding factor) interplayed with the rice basic leucine zipper factor RISBZ1 to maintain proper expression of rice seed storage protein genes during seed development (Kawakatsu et al., 2009).
Moreover, Dof factors are involved in photoperiod flowering. In Arabidopsis, Cycling Dof Factor-1 (CDF1) binds to the COSTANS (CO) and FLOWERING LOCUS T (FT) promotor regions to block transactivation of this two flowering genes, whereas this inhibition could be released based on the GIGANTEA-FLAVIN-BINDING, KELCH REPEAT, F-BOX1(GI-FKF1) complex mediated degradation of CDF1 under long-day conditions (Imaizumi et al., 2005; Sawa et al., 2007; Song et al., 2012). Fornara et al. (2009) systematically studied a subset of Dof family related to CDF1 and found that CDF1, CDF2, CDF3, and CDF5 acted redundantly to repress flowering by decreasing the mRNA level of CO (Fornara et al., 2009). In rice, RDD1 (rice Dof daily fluctuations 1) was controlled by circadian clock, and repressing the expression of RDD1 led to delayed flowering time (Iwamoto et al., 2009). Furthermore, several studies have revealed the importance of Dof factors on plant patterning. In Arabidopsis, AtDof5.1 modulated leaf adaxial–abaxial polarity via binding to the promotor of Revoluta (REV) and enhancing expression of REV (Kim et al., 2010). AtDOF4.2 and AtDOF4.4 were engaged in regulating shoot branching and seed development (Skirycz et al., 2007; Zou et al., 2013). OBF-binding factor-1(OBP1) is a cell cycle regulator, and overexpressing OBP1 in Arabidopsis affects cell size and number, rendering dwarfish plant morphology (Skirycz et al., 2008).
To now, of the 36 predicted Dof homologous genes in whole Arabidopsis genome, 16 have been confirmed to participate in various biological processes (Noguero et al., 2013). However, in rice, to our knowledge, the studies deciphering Dof factors are rather limited; only 5 of the 30 predicted Dof members have been characterized in detail (Washio, 2003; Yamamoto et al., 2006; Li et al., 2008, 2009b; Iwamoto et al., 2009; Santos et al., 2012). In a previous study, we characterized the function of OsDof12 in rice (Li et al., 2008, 2009b). We found a pair of sense-antisense transcript at the locus of OsDof12 (LOC_Os03g07360), denoted as OsDof12 (sense transcript) and OsDof12os (antisense transcript), respectively. OsDof12 encodes a nuclear-localized protein of 440 amino acids (Li et al., 2008). Expression pattern analysis showed OsDof12 and OsDof12os were co-expressed but reciprocally regulated by each other (Li et al., 2008). Moreover, overexpression of OsDof12 promoted flowering in rice under long-day conditions by up-regulating rice florigen-encoding gene Hd3a and its downstream gene OsMADS14 (Li et al., 2009b). In wild type rice plants, the transcripts of OsDof12 were detected in various rice tissues at different development stages (Li et al., 2009b), which suggests OsDof12 might take part in various biological processes.
In current study, we demonstrated that the transgenic plants overexpressing OsDof12 indeed display pleiotropic phenotypes such as reduced plant height, shortened leaf length, more erected leaf, smaller panicle size and decreased grain yield. Further molecular biological analyses indicated that these changes in plant architecture could be ascribed to attenuated BR signaling in OsDof12 overexpression plants.
Materials and Methods
Plant Materials and Growth Conditions
To observe the influence of OsDof12 on plant architecture, the wild type cultivar Nipponbare (Oryza sativa L. ssp. japonica cv. Nipponbare) and the previous reported two OsDof12 overexpressing lines (Line OD2 and Line OD5, Nipponbare background) (Li et al., 2009b) were grown on the research field located in the Experimental Stations of the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, under nature field conditions. After mature stage (approximately 120 days after sowing), we measured the agronomic traits including plant height, internode length, leaf length, and panicle architecture.
For BR sensitivity tests, plants were grown on 1/2 Murashige and Skoog (MS) culture medium in a growth chamber under controlled conditions (16h/light, 30°C and 8h/dark, 26°C) for 7 or 8 days. For skotomorphogenesis analysis, the WT and OD plants were grown on 1/2 MS at 28°C in darkness for 2 weeks. For OsDof12 induction analysis, 2-week-old WT plants grown on 96-well PCR plates were treated with 100 nM Brassinolide (BL, the most bioactive BR compound, not being synthesized in rice, Yokota, 1997; Fujioka and Yokota, 2003; Kim et al., 2008; Vriet et al., 2013) solution, then the hole plants were sampled at different time points for expression assay. For expression levels analysis on BR related genes, 7-day-old plants of WT and OD were used.
Microscopic Observation
The middle part of the second internode from mature plants were harvested and fixed with 2.5% glutaraldehyde solution overnight at 4°C, and then dehydrated in order by 40, 50, 60, 70, 80, 90, 95, and 100% ethanol solutions. The samples got dried through critical point drying by liquid carbon dioxide. The dry specimens were mounted on a stub, gold-coated with an ion sputter coater (Hitachi, Tokyo, Japan) and then imaged with a scanning electron microscope (Hitachi, Tokyo, Japan).
BR Treatment
The seeds were dehusked and sterilized with 75% ethanol for 1 min, 3% NaClO for 25 min and then washed five times with sterile water. The seeds were grown on solid 1/2 MS medium containing a series of concentration of BL (WAKO, Japan) for 7 or 8 days. The lengths of second leaf sheath, second leaf, seedling height, root, and coleoptile were measured for further analyses.
BR Measurement
After sterilization, the seeds of WT and OD were sown and grown on 1/2 MS culture medium for 10 days, then about 1 gram of fresh seedlings were collected for measurement of Castasterone (CS, one of the biologically active BR compounds, a likely end product of brassinosteroid biosynthetic pathway in rice, Kim et al., 2008; Vriet et al., 2013) according to the method described previously (Ding et al., 2013).
Laminal Inclination Assay
The seeds were immersed for 2 days, and then the germinated seeds were sown on 1/2 MS supplied with different concentrations of BL. After incubation in a growth chamber for 8 days, the second lamina joints on WT and OD plants were imaged and measured by IMAGEJ according to the method described by Tong et al. (2009).
RNA Extraction and qRT-PCR Analysis
The samples were harvested and stored in liquid nitrogen until RNA extraction. The isolation of total RNA was performed applying Trizol reagent (Invitrogen, California, USA) with corresponding protocol. DNA digestion was accomplished by DnaseI (Takara, Japan), and first-strand cDNA was obtained by GoScript Reverse Transcription System (Promega, http://www.promega.com/). With a rice ubiquitin gene (UBQ, LOC_Os03g13170) set as the internal control, qRT-PCR was performed using EvaGreenq PCR MasterMix (Abm, Canada) on a real-time PCR System (Bio-Rad CFX96) with the specific primers listed in Table S2. The qRT-PCR program consists of 95°C for 3 min and 42 cycles of 95°C for 5 s, 60°C for 10 s. The relative expression level of each examined gene was quantified by a relative quantization method.
Results
Overexpression of OsDof12 Reduces Plant Height
In our previous study, we developed OsDof12 overexpressing lines in rice (hereafter named OD) (Li et al., 2009b). The OD lines, OD2 and OD5, and WT plants were grown in a paddy field in Beijing (40°10′N, 116°42′E) under nature field conditions. In order to investigate the effects of OsDof12 overexpression on rice agronomic traits, we traced the growth performance of OD and WT plants and found that the plant height of OD was almost the same as WT before heading stage. However, after the plant height was stable at mature stage, on average, the WT plant height reached approximate 83.7 cm while OD plant height was about 65.3 cm and 20% shorter than the WT (Figures 1A,C). Rice plant height consists of the internodes length and the panicle length. We measured and compared the internode and panicle lengths of OD plants with those of WT plants, respectively. Statistic data showed that the individual internode and panicle lengths in OD were remarkably shorter than those in WT (Figures 1B,D, 2B), which consequently led to decreased plant height of OD plant. Comparing the percentage length of each internode to the total culm in OD with that in WT, we found that the second and fourth internode lengths of OD were largely and slightly reduced (Figure 1E), respectively.
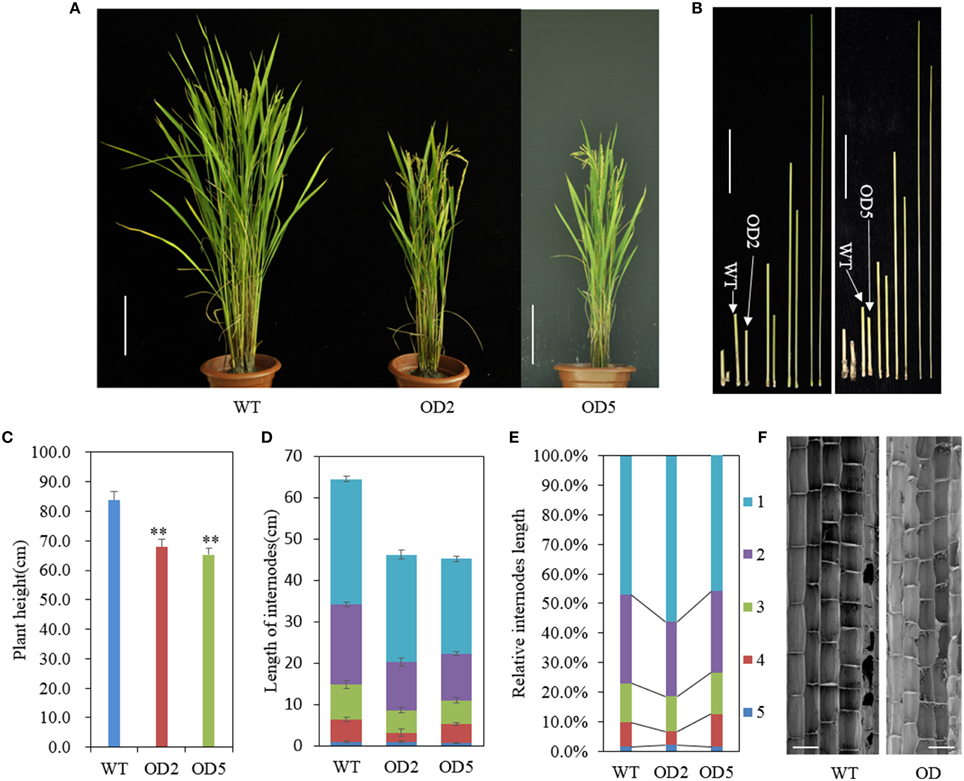
Figure 1. OsDof12 overexpression plants showed dwarf phenotype. (A) Gross morphology comparison between wild type (WT) and OsDof12 overexpressing transgenic plants (OD) at maturity stage (approximately 120 days after sowing). The OD plants showed significantly reduced plant height and more compacted plant architecture than WT plants. Bar = 15 cm. (B) Comparison of the internode length. Left: WT vs. OD2; Right: WT vs. OD5. Bar = 5 cm. (C) Statistical analysis of plant height. Values are mean ± sd (n = 30). Double asterisks (**) stands for P < 0.01 determined by student's t-test. (D) Schematic representation of each individual internode length. Values are mean ± sd, n = 15. (E) The percentage of each internode length accounting for the total culm length. (F) Microscopic observation by SEM on the longitudinal section of the middle part in the second internode. Bar = 100μm.
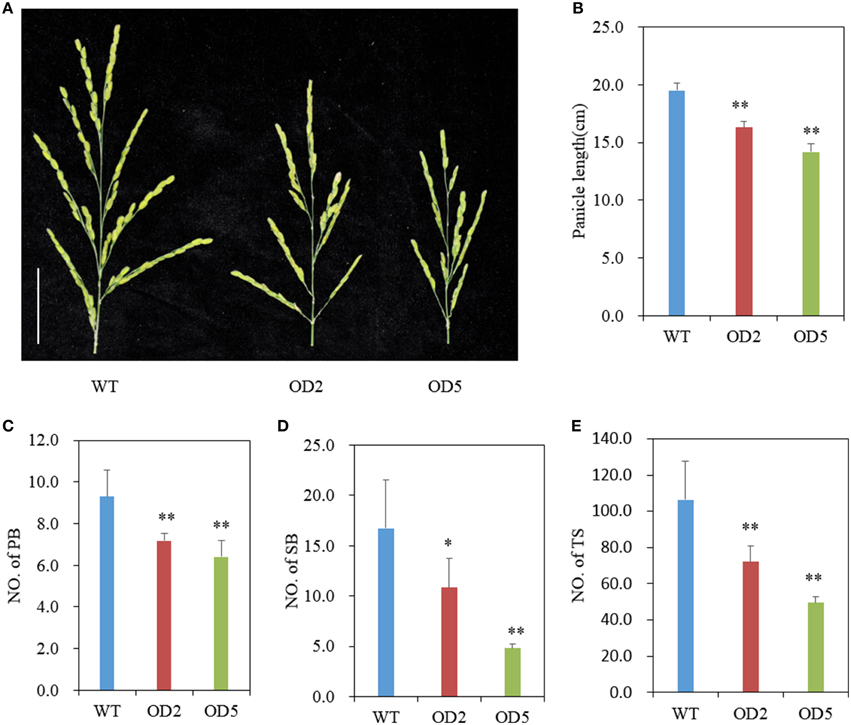
Figure 2. Comparison of panicle structure. (A) Panicle morphology comparison between WT and OD plants. Bar = 5 cm. (B-E) Statistical results of panicle length (B), number of primary branches (C), secondary branches (D), and total spikelets per panicle (E) in WT and OsDof12 overexpression plants. PB, primary branches; SB, secondary branches; TS, total spikelets. Values are mean ± sd (n = 30). Single asterisk (*) and double asterisks (**) stand for P < 0.05 and P < 0.01, respectively, determined by student's t-test.
Internode elongation is determined by cell division activity in the intercalary meristem, followed by cell elongation in the elongation zone (Yamamuro et al., 2000). To investigate whether those two factors caused the dwarfish morphology of OD, we observed the longitudinal cell morphology of the OD and WT internodes. After heading stage, we collected and fixed the middle sections of the second internodes, then observed the cell length under scanning electron microscope (SEM). As shown in Figure 1F, there was no obvious difference in the longitudinal cell length between WT and OD, thus the reduction in longitudinal cell number on the elongation zone may account for the shortage of the internodes in OD.
OsDof12 Overexpression Plants Produce Smaller Panicles
Besides plant height, we also analyzed the panicle structure and seed size in OD plants. As shown in Figures 2A,B, the panicles of OD are shorter and smaller than that of WT. We further investigated the panicle index including numbers of primary branches, secondary branches, and total spikelets, and found that the number of primary branches in OD was remarkably reduced (Figure 2C), meanwhile OD plants produced significantly less secondary branches (Figure 2D). And, expectedly, there was very significant reduction in total spikelet number per panicle in OD as compared with that in WT (Figure 2E). Nevertheless, no obvious differences of seed size and weight appeared between OD and wild type (Figure S1). These results imply that overexpression of OsDof12 could lead to smaller panicle structure with reduced primary branches, secondary branches and total spikelets.
Abnormal Leaf Morphology in OsDof12 Overexpression Plants
Aside from the alternations on plant height and panicle structure, OD also exhibited some other characteristic phenotypes. In comparison with WT plants, OD plants generated more compacted plant architecture with more erected leaves (Figure 1A). As shown in Figure 3, obviously the leaf joint angle on OD plants was smaller than that on WT plants. Moreover, we measured the length of flag leaves, penultimate leaves and antepenultimate leaves of OD and WT plants. Statistical analysis indicated that the leaves in OD plants were remarkably shorter than those in WT plants (Figure S2).
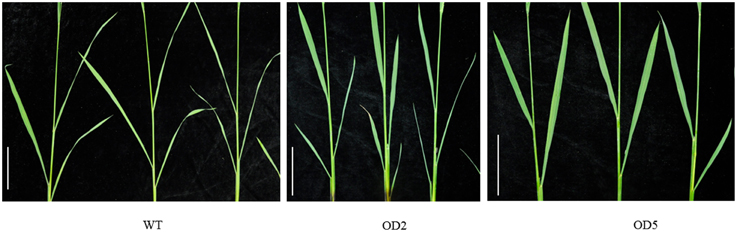
Figure 3. Comparison of the lamina joint zones. The leaf angle in OD2 and OD5 plants is less enlarged than WT plants. Bar = 10 cm.
Overexpression of OsDof12 Altered BR Sensitivity
Brassinosteroids (BRs) play pivotal roles in regulating plant growth and development (Müssig and Altmann, 2001; Fujioka and Yokota, 2003; Salas Fernandez et al., 2009; Tong and Chu, 2012; Wang et al., 2012; Zhu et al., 2013). By now, many BR metabolic and signaling-related genes have been well characterized (Yamamuro et al., 2000; Hong et al., 2002, 2003, 2005; Bai et al., 2007; Tanaka et al., 2009; Tong et al., 2009, 2012; Li et al., 2009a; Sakamoto et al., 2011; Thornton et al., 2011; Zhang et al., 2012). Notably, almost all of the corresponding mutants or misexpressors display abnormal plant height and leaf inclination phenotypes. Considering that the plants overexpressing OsDof12 exhibit reduced plant height and erect leaf morphology, we assumed that OsDof12 might be involved in BR metabolism or signal transduction. To test this hypothesis, we designed and conducted a series of experiments to evaluate the BR response of OD2 plants.
Firstly, we measured the endogenous BR levels by quantifying the CS content in WT and OD2 plants, respectively. As a matter of fact, no obvious difference in BR levels between WT and OD plants was observed (Table S1), suggesting that OsDof12 overexpression may not affect the metabolism of BRs, which was further confirmed by expression level analysis on BR metabolism related genes (Figure S3).
Then, we performed lamina joint bending assay by applying BL on the plants. Notably, the lamina joints of OD plants were remarkably less enlarged than that of WT when both subjected to a mock treatment without BL (Figure 4A). When treated with the increasing doses of BL, though the angles of lamina joint on both OD and WT plants became relatively larger, the lamina inclination curve of OD ascended much slower than that of WT (Figure 4B), indicating that overexpression of OsDof12 may cause defects on the BR signal transduction pathway leading to impaired bending of leaves.
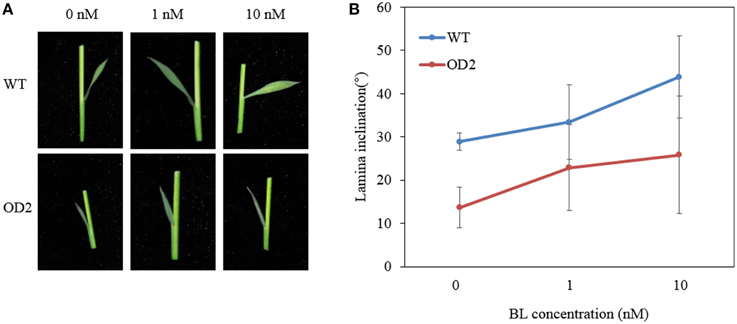
Figure 4. Lamina joint bending analysis. (A) The second lamina joint morphology in WT and OD2 plants grown on BL contained half-strength MS for 7 days. (B) Quantification of the second lamina angel bending as shown in (A), values are mean ± sd, n = 20.
Besides OD plants exhibiting lamina joint abnormality, we also found that overexpression of OsDof12 affected other aspects of BR-related morphology. For instance, the second leaf sheath length of OD plants was apparently shorter than that of WT plants. In WT plants, when treated with 1 nM BL, the second leaf sheath length slightly but not significantly increased than those in the mock treatment, however, 10 nM or higher concentrations of BL treatments gave rise to evidently shorter second leaf sheath length in WT plants (Figure 5A), suggesting that relative higher level of exogenous BR inhibits the elongation of the leaf sheath. In contrast, various concentrations of BL treatment on OD could hardly altered the elongation of the second leaf sheath (Figure 5A), indicating that the elongation of the second leaf sheath in OD was hyposensitive to exogenous BR treatment. The similar situation also happened to the elongation of the second leaf and seedling height in WT and OD plants, respectively. The second leaf length and seedling height in OD plants was apparently shorter than those in WT. Relative higher concentrations of BL decreased evidently the elongation of the second leaf and seedling height in WT plants (Figures 5B,C), while various levels of BL treatment could hardly affect the elongation of the second leaf length and seedling height in OD plants (Figures 5B,C), indicating OD plants may have lower sensitivity in response to exogenous BR.
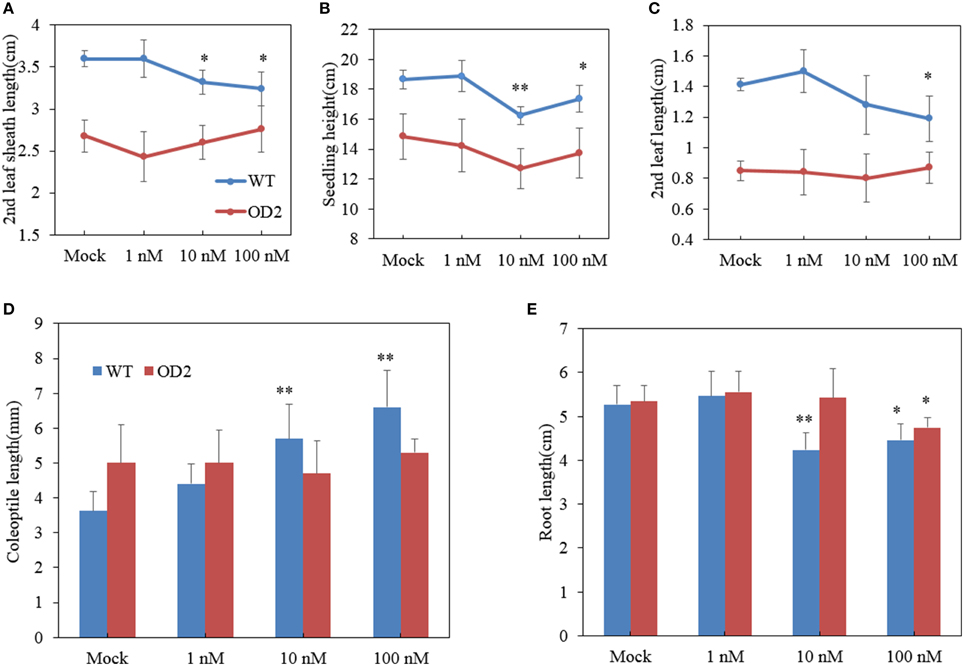
Figure 5. Altered BR sensitivity in overexpressing transgenic plants. (A–E) Statistical analysis of the 2nd leaf sheath length (A), seedling height (B), 2nd leaf length (C), coleoptile length (D), and root length (E) in plants treated by various levels of BL. Values are mean ± sd, n = 15. Single asterisk and double asterisks respectively stand for P < 0.05 and P < 0.01 determined by student's t-test.
Elongation of the Coleoptiles and Root in Response to BL
The coleoptile length and root elongation are respects to evaluate BR sensitivity of plants (Yamamuro et al., 2000). Thus, we measured the coleoptile length of 8-day-old seedlings. As shown in Figure 5D, the coleoptile elongation of WT was promoted by BL treatment in dose-dependent manner. However, the coleoptile elongation of OD plants showed no difference between BL and mock treatment.
Similar root growth pattern was shared by OD and WT plants when grown in 0 nM or 1 nM BL medium, 1 nM BL slightly increased the root elongation (Figure 5E). When treated with 10 nM or 100 nM BL, the WT plants generated apparently shorter root. However, for OD plants, 10 nM BL could not obviously reduce root elongation, only up to 100 nM BL treatment could inhibit the root elongation (Figure 5E). The results also suggested that overexpression of OsDof12 may result in lower BR sensitivity.
Skotomorphogenic Phenotypes of OsDof12 Overexpressing Plants
The phenotypes of mesocotyl and internode elongation in darkness have been employed as a good criteria to determine whether the dwarf phenotype is related to BR (Hong et al., 2002, 2003). To test whether overexpression of OsDof12 affects the elongation of mesocotyl and internode, we grew the seeds on half-strength MS under totally dark conditions. Two weeks later, we observed the phenotypes of WT and OD plants and found that the elongation of mesocotyl and first internode in OD plants was similar to those in WT plants (Figures 6A–C), however, the second internode length in OD plants was much less than that in WT plants (Figures 6A,D), indicating overexpression of OsDof12 specifically inhibits the elongation of the internode in dark conditions.
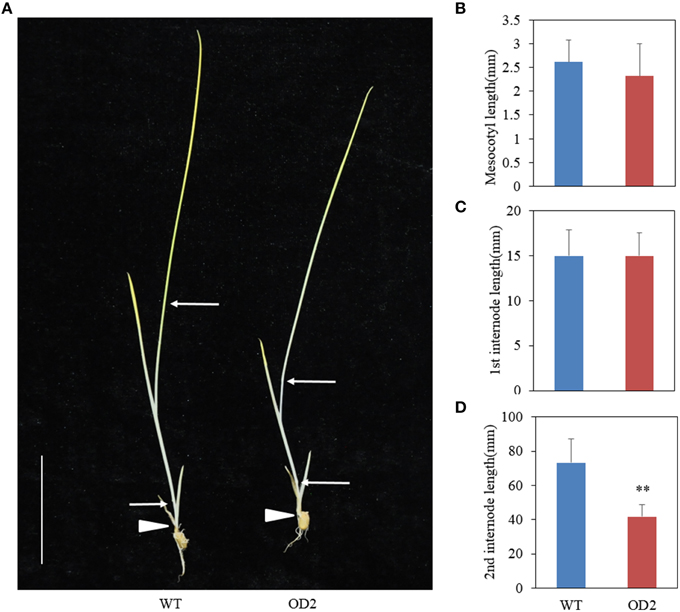
Figure 6. Skotomorphogy analysis. (A) Two-week-old seedlings grown on half-strength MS in dark conditions. The mesocotyl (B) and first internode (C) length of OD2 is comparable to WT, whereas the second internode (D) was obviously shortened in OD2.Values are mean ± sd, n = 12. Double asterisks stand for P < 0.01 determined by student's t-test. Arrows and arrowheads indicate the nodes and mesocotyls, respectively. Bar = 25 mm.
Expression Patterns of BR Signaling Related Genes Were Changed in Transgenic Plants Overexpressing OsDof12
As demonstrated above, OsDof12 may be a regulator for maintaining normal BR signaling. We then test whether OsDof12 responds to BR treatment. As shown in Figure 7A, the OsDof12 transcripts rapidly accumulated after BL treatment for 1 h; although OsDof12 gradually descended from 2 to 4 h, it was still higher than that before BR treatment; after BR treatment the transcriptional level peak of OsDof12 appeared at 8 h. This result suggests OsDof12 could be induced by exogenous BR, which indicates OsDof12 might be involved in BR regulation.
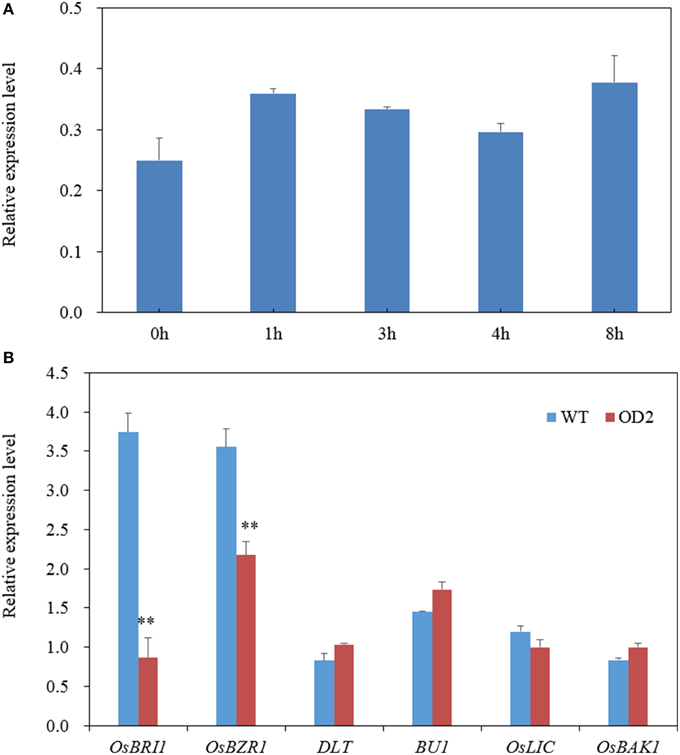
Figure 7. Gene expression analysis. (A) OsDof12 induction analysis. Two-week-old WT seedlings were subjected to exogenous 100 nM BL treatment, then whole plants were collected at different time point for gene expression analysis. (B) qRT-PCR analysis of BR signaling genes. Seven-day-old seedlings grown on half-strength MS were used for expression analysis. Values are mean ± sd, n = 3. Double asterisks stand for P < 0.01 determined by student's t-test.
Using qRT-PCR, we further analyzed the expression level of six major BR signaling components. As shown in Figure 7B, OsBRI1 and OsBZR1 were down-regulated, while OsBU1, OsLIC, and DLT were not affected, suggesting that OsDof12 is possibly involved in BR signaling pathway.
Discussion
Dof genes have been known involved in various processes at different development stages in plants (Yanagisawa, 2002; Noguero et al., 2013). In previous study, we focused on overexpression of OsDof12 promoting flowering in rice under long-day conditions. In the present study, we demonstrate that overexpression of OsDof12 can alter the rice plant architecture. Further evidences indicate OsDof12 may participate in BR signaling to modulate rice architecture. All these clearly show that OsDof12 is a pleiotropic regulator for plant growth and development.
Higher plants have developed a complex of metabolic mechanisms, which include biosynthetic and catabolic pathway, to maintain BR homeostasis for normal growth and development (Tanaka et al., 2005; Vriet et al., 2013). The abnormal phenotypes in OD plants are similar with the phenotypes of BR-deficient mutants. Therefore, we investigated this possible clues that OsDof12 might be involved in BR metabolism. Measurement of the endogenous BR levels indicated that the BR content in plants overexpressing OsDof12 was not affected. Meanwhile, we quantified expression levels of BR metabolic genes, such as D2, D11, OsDWARF, OsDWARF1, OsDWARF4, CYP734As and no obvious difference were found between OD and WT plants. These results excluded the possibility that OsDof12 might modulate plant architecture by affecting BR metabolism pathway.
Notably, a range of BR response tests on OsDof12 overexpression plants, including lamina joint assay, sheath, root, coleoptile elongation pattern analysis, and skotomorphogenesis analysis all suggest that overexpression of OsDof12 reduces the BR sensitivity. Besides, the OsDof12 expression was induced by exogenous BL treatment and the expression patterns of two BR signaling components were suppressed in OsDof12 overexpression plants, which further suggest that OsDof12 is involved in coordinating rice BR signaling.
Rice plant architecture is composed of tiller number, internode elongation, leaf angle and panicle structure, and favorable architecture is able to facilitate improving the yield (Wang and Li, 2006, 2008; Yang and Hwa, 2008). Plant height is one of the most important agronomic traits (Sakamoto and Matsuoka, 2004; Wang and Li, 2008). Rice dwarf phenotypes have been well categorized into several patterns (Yamamuro et al., 2000), and BR-associated mutants usually display dm-type dwarfism where the second internode is specifically shortened. The OsDof12 overexpression lines, OD2 and OD5, both generated shortened internodes, among which the relative length of the second internode were shortened significantly, therefore, we speculate that they belong to dm-type dwarfism. As suggested by our microscopic observation in the elongation zone, being different from the two BR mutants, d61-2 (Yamamuro et al., 2000) and dlt (Tong et al., 2009) where the elongation of longitudinal cell is severely hampered, the cell length in OD plants is not affected, which might indicate that the cell number decrease should be the major cause for the dwarfism in OD. These results suggest that the regulatory mechanism underlying cell elongation and plant height appear to be quite complicated, thus the detail mechanism in OsDOf12 regulation of plant height should be further studied.
Erect leaf pattern is another desirable trait for ideal plant architecture. Generally, the erect leaves in dense plantings allow more light penetrating through the upper leaves layer to make the lower leaves layer capture more light for photosynthesis and assimilation (Sinclair and Sheehy, 1999). Indeed, several cases have witnessed the effect of erect leaf on improving yield (Morinaka et al., 2006; Sakamoto et al., 2006). For example, the weakest allele of OsBRI1 (d61-7), the counterpart of Arabidopsis BR receptor BRI1, produced erect leaf and generate greater biomass under high density planting conditions (Morinaka et al., 2006). The erect leaf trait of OsDof12 overexpression lines is favorable for ideal plant architecture. However, similar to d61-7 (Morinaka et al., 2006), at least under normal planting density, OsDof12 overexpression lines would produce decreased spikelets. Therefore, whether the harvest index of OsDof12 overexpression lines increase or not under different higher planting densities need to be further investigated.
Overexpression of OsDof12 leads to shorter leaves and less grain yield under normal planting dense, which is unfavorable for utilization in plant breeding. On the contrary, whether the plants down-expressing OsDof12 induce longer leaves and higher grain yield should be further validated. Indeed, we have tried in this direction but failed to obtain OsDof12 suppressed lines with obviously distinguishable phenotypes from WT (Li et al., 2009b). We speculated this might result from functional redundancy in rice Dof genes, and the functional redundancy in Dofs have been demonstrated in Arabidopsis (Ahmad et al., 2013). Therefore, in our future study, we will try to better understand the full-version function of OsDof12 by utilizing CRISPR/Cas9 (Belhaj et al., 2015) and Chimeric Repressor gene Silencing Technology (CRES-T) (Mitsuda et al., 2011).
In summary, we show the involvement of OsDof12 in BR signaling coordination in rice and the effects of overexpression of OsDof12 on rice plant architecture, and the findings imply OsDof12 might be a potential genetic module for future rice breeding strategy.
Author Contributions
SL and LZ conceived and designed the experiments. QW and DaL performed most of the experiments. QW, DaL, SL, and LZ wrote the manuscript. DeL and XZ performed phenotypes observation and statistic analysis. XuL and XiL performed microscopic observation. All authors have read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the grants from the National Natural Science Foundation of China (31471475), Ministry of Agriculture of China (2014ZX08009-001), and the State Key Laboratory of Plant Genomics, China (2015B0129-03). We thank Mr. Yanbao Tian (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, China) for technical assistance with SEM observation, and we also thank Dr. Yi Xu (Rutgers University, USA) for critical reading on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00833
Figure S1. Comparison of the grain size between wild type and OD lines. (A) There was no obvious difference between WT and OsDof12 overexpression lines in grain size. Bar = 1.5 mm. (B) Statistical data of 1000-grain weight. Values are mean ± sd, n = 1000.
Figure S2. Morphology and statistical analysis of the leaf length. (A) Gross morphology of flag leaves, penultimate leaves and antepenultimate leaves. Bar = 8 cm. (B)The length of flag leaves, penultimate leaves and antepenultimate leaves in OsDof12 overexpression plants was shorter than WT plants. Values are mean ± sd (n = 30). Single asterisk and double asterisks stand for P < 0.05 and P < 0.01 determined by student's t-test, respectively.
Figure S3. Expression analysis of BR metabolism related genes in WT plants and transgenic plants overexpressing OsDof12. Expression level of OsDof12 in OD plants was significantly higher than in WT plants. D11 and CYP734A6 was slightly but not obviously down-regulated in OD plants, while expression levels of the other BR metabolism related genes were comparable in both plants. n.d means not detected. Double asterisks stand for P < 0.01 determined by student's t-test.
Table S1. Quantification of CS in WT and OD2. Means ± SD of two replicates are shown (ng·g-1 F.W.).
Table S2. The primers used for qRT-PCR in this study.
References
Ahmad, M., Rim, Y., Chen, H., and Kim, J. Y. (2013). Functional characterization of Arabidopsis Dof transcription factor AtDof4.1. Russ. J. Plant Physiol. 60, 116–123. doi: 10.1134/S1021443712060027
Bai, M. Y., Zhang, L. Y., Gampala, S. S., Zhu, S. W., Song, W. Y., Chong, K., et al. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. U.S.A. 104, 13839–13844. doi: 10.1073/pnas.0706386104
Baumann, K., De Paolis, A., Costantino, P., and Gualberti, G. (1999). The DNA binding site of the Dof protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the rolB oncogene in plants. Plant Cell 11, 323–333. doi: 10.1105/tpc.11.3.323
Belhaj, K., Chaparro-Garcia, A., Kamoun, S., Patron, N. J., and Nekrasov, V. (2015). Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 32, 76–84. doi: 10.1016/j.copbio.2014.11.007
Ding, J., Mao, L. J., Wang, S. T., Yuan, B. F., and Feng, Y. Q. (2013). Determination of endogenous brassinosteroids in plant tissues using solid-phase extraction with double layered cartridge followed by high-performance liquid chromatography-tandem mass spectrometry. Phytochem. Anal. 24, 386–394. doi: 10.1002/pca.2421
Fornara, F., Panigrahi, K. C. S., Gissot, L., Sauerbrunn, N., Rühl, M., Jarillo, J. A., et al. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17, 75–86. doi: 10.1016/j.devcel.2009.06.015
Fujioka, S., and Yokota, T. (2003). Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 54, 137–164. doi: 10.1146/annurev.arplant.54.031902.134921
Gualberti, G., Papi, M., Bellucci, L., Ricci, L., Bouchez, D., Camilleri, C., et al. (2002). Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell 14, 1253–1263. doi: 10.1105/tpc.010491
Hong, Z., Ueguchi-Tanaka, M., Fujioka, S., Takatsuto, S., Yoshida, S., Hasegawa, Y., et al. (2005). The Rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell 17, 2243–2254. doi: 10.1105/tpc.105.030973
Hong, Z., Ueguchi-Tanaka, M., Shimizu-Sato, S., Inukai, Y., Fujioka, S., Shimada, Y., et al. (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32, 495–508. doi: 10.1046/j.1365-313X.2002.01438.x
Hong, Z., Ueguchi-Tanaka, M., Umemura, K., Uozu, S., Fujioka, S., Takatsuto, S., et al. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15, 2900–2910. doi: 10.1105/tpc.014712
Imaizumi, T., Schultz, T. F., Harmon, F. G., Ho, L. A., and Kay, S. A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297. doi: 10.1126/science.1110586
Iwamoto, M., Higo, K., and Takano, M. (2009). Circadian clock- and phytochrome-regulated Dof-like gene, Rdd1, is associated with grain size in rice. Plant Cell and Environ. 32, 592–603. doi: 10.1111/j.1365-3040.2009.01954.x
Kawakatsu, T., Yamamoto, M. P., Touno, S. M., Yasuda, H., and Takaiwa, F. (2009). Compensation and interaction between RISBZ1 and RPBF during grain filling in rice. Plant J. 59, 908–920. doi: 10.1111/j.1365-313X.2009.03925.x
Kim, B. K., Fujioka, S., Takatsuto, S., Tsujimoto, M., and Choe, S. (2008). Castasterone is a likely end product of brassinosteroid biosynthetic pathway in rice. Biochem. Biophys. Res. Commun. 374, 614–619. doi: 10.1016/j.bbrc.2008.07.073
Kim, H.-S., Kim, S. J., Abbasi, N., Bressan, R. A., Yun, D.-J., Yoo, S.-D., et al. (2010). The DOF transcription factor Dof5.1 influences leaf axial patterning by promoting Revoluta transcription in Arabidopsis. Plant J. 64, 524–535. doi: 10.1111/j.1365-313X.2010.04346.x
Li, D., Wang, L., Wang, M., Xu, Y. Y., Luo, W., Liu, Y. J., et al. (2009a). Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol. J. 7, 791–806. doi: 10.1111/j.1467-7652.2009.00444.x
Li, D., Yang, C., Li, X., Gan, Q., Zhao, X., and Zhu, L. (2009b). Functional characterization of rice OsDof12. Planta 229, 1159–1169. doi: 10.1007/s00425-009-0893-7
Li, D., Yang, C., Li, X., Ji, B., and Zhu, L. (2008). Sense and antisense OsDof12 transcripts in rice. BMC Mol. Biol. 9:80. doi: 10.1186/1471-2199-9-80
Mitsuda, N., Matsui, K., Ikeda, M., Nakata, M., Oshima, Y., Nagatoshi, Y., et al. (2011). CRES-T, an effective gene silencing system utilizing chimeric repressors. Methods Mol. Biol. 754, 87–105. doi: 10.1007/978-1-61779-154-3_5
Moreno-Risueno, M., Martínez, M., Vicente-Carbajosa, J., and Carbonero, P. (2007). The family of DOF transcription factors: from green unicellular algae to vascular plants. Mol. Genet. Genomics 277, 379–390. doi: 10.1007/s00438-006-0186-9
Morinaka, Y., Sakamoto, T., Inukai, Y., Agetsuma, M., Kitano, H., Ashikari, M., et al. (2006). Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol. 141, 924–931. doi: 10.1104/pp.106.077081
Müssig, C., and Altmann, T. (2001). Brassinosteroid signaling in plants. Trends Endocrinol. Metab. 12, 398–402. doi: 10.1016/S1043-2760(01)00477-5
Noguero, M., Atif, R. M., Ochatt, S., and Thompson, R. D. (2013). The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci. 209, 32–45. doi: 10.1016/j.plantsci.2013.03.016
Papi, M., Sabatini, S., Bouchez, D., Camilleri, C., Costantino, P., and Vittorioso, P. (2000). Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Genes Dev. 14, 28–33. doi: 10.1101/gad.14.1.28
Sakamoto, T., Kawabe, A., Tokida-Segawa, A., Shimizu, B., Takatsuto, S., Shimada, Y., et al. (2011). Rice CYP734As function as multisubstrate and multifunctional enzymes in brassinosteroid catabolism. Plant J. 67, 1–12. doi: 10.1111/j.1365-313X.2011.04567.x
Sakamoto, T., and Matsuoka, M. (2004). Generating high-yielding varieties by genetic manipulation of plant architecture. Curr. Opin. Biotechnol. 15, 144–147. doi: 10.1016/j.copbio.2004.02.003
Sakamoto, T., Morinaka, Y., Ohnishi, T., Sunohara, H., Fujioka, S., Ueguchi-Tanaka, M., et al. (2006). Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 24, 105–109. doi: 10.1038/nbt1173
Salas Fernandez, M. G., Becraft, P. W., Yin, Y., and Lubberstedt, T. (2009). From dwarves to giants? Plant height manipulation for biomass yield. Trends Plant Sci. 14, 454–461. doi: 10.1016/j.tplants.2009.06.005
Santos, L. A., De Souza, S. R., and Fernandes, M. S. (2012). OsDof25 expression alters carbon and nitrogen metabolism in Arabidopsis under high N-supply. Plant Biotechnol. Rep. 6, 327–337. doi: 10.1007/s11816-012-0227-2
Sawa, M., Nusinow, D. A., Kay, S. A., and Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265. doi: 10.1126/science.1146994
Sinclair, T. R., and Sheehy, J. E. (1999). Erect leaves and photosynthesis in rice. Science 283, 1456–1457. doi: 10.1126/science.283.5407.1455c
Skirycz, A., Jozefczuk, S., Stobiecki, M., Muth, D., Zanor, M. I., Witt, I., et al. (2007). Transcription factor AtDOF4;2 affects phenylpropanoid metabolism in Arabidopsis thaliana. New Phytol. 175, 425–438. doi: 10.1111/j.1469-8137.2007.02129.x
Skirycz, A., Radziejwoski, A., Busch, W., Hannah, M. A., Czeszejko, J., Kwasniewski, M., et al. (2008). The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant J. 56, 779–792. doi: 10.1111/j.1365-313X.2008.03641.x
Skirycz, A., Reichelt, M., Burow, M., Birkemeyer, C., Rolcik, J., Kopka, J., et al. (2006). DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 47, 10–24. doi: 10.1111/j.1365-313X.2006.02767.x
Song, Y. H., Smith, R. W., To, B. J., Millar, A. J., and Imaizumi, T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336, 1045–1049. doi: 10.1126/science.1219644
Tanaka, A., Nakagawa, H., Tomita, C., Shimatani, Z., Ohtake, M., Nomura, T., et al. (2009). BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol. 151, 669–680. doi: 10.1104/pp.109.140806
Tanaka, K., Asami, T., Yoshida, S., Nakamura, Y., Matsuo, T., and Okamoto, S. (2005). Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol. 138, 1117–1125. doi: 10.1104/pp.104.058040
Thornton, L. E., Peng, H., and Neff, M. M. (2011). Rice CYP734A cytochrome P450s inactivate brassinosteroids in Arabidopsis. Planta 234, 1151–1162. doi: 10.1007/s00425-011-1464-2
Tong, H., Jin, Y., Liu, W., Li, F., Fang, J., Yin, Y., et al. (2009). DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 58, 803–816. doi: 10.1111/j.1365-313X.2009.03825.x
Tong, H., Liu, L., Jin, Y., Du, L., Yin, Y., Qian, Q., et al. (2012). DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 24, 2562–2577. doi: 10.1105/tpc.112.097394
Tong, H., and Chu, C. (2012). Brassinosteroid signaling and application in rice. J. Genet. Genomics 39, 3–9. doi: 10.1016/j.jgg.2011.12.001
Umemura, Y., Ishiduka, T., Yamamoto, R., and Esaka, M. (2004). The Dof domain, a zinc finger DNA-binding domain conserved only in higher plants, truly functions as a Cys2/Cys2 Zn finger domain. Plant J. 37, 741–749. doi: 10.1111/j.1365-313X.2003.01997.x
Vriet, C., Russinova, E., and Reuzeau, C. (2013). From squalene to brassinolide: the steroid metabolic and signaling pathways across the plant kingdom. Mol. Plant 6, 1738–1757. doi: 10.1093/mp/sst096
Wang, Y., and Li, J. (2008). Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59, 253–279. doi: 10.1146/annurev.arplant.59.032607.092902
Wang, Y., and Li, J. (2006). Genes controlling plant architecture. Curr. Opin. Biotechnol. 17, 123–129. doi: 10.1016/j.copbio.2006.02.004
Wang, Z. Y., Bai, M. Y., Oh, E., and Zhu, J. Y. (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46, 701–724. doi: 10.1146/annurev-genet-102209-163450
Washio, K. (2001). Identification of Dof proteins with implication in the gibberellin-regulated expression of a peptidase gene following the germination of rice grains. Biochim. Biophys. Acta 1520, 54–62. doi: 10.1016/S0167-4781(01)00251-2
Washio, K. (2003). Functional dissections between GAMYB and dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol. 133, 850–863. doi: 10.1104/pp.103.027334
Yamamoto, M. P., Onodera, Y., Touno, S. M., and Takaiwa, F. (2006). Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes. Plant Physiol. 141, 1694–1707. doi: 10.1104/pp.106.082826
Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., et al. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12, 1591–1605. doi: 10.1105/tpc.12.9.1591
Yanagisawa, S. (2000). Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J. 21, 281–288. doi: 10.1046/j.1365-313x.2000.00685.x
Yanagisawa, S. (2002). The Dof family of plant transcription factors. Trends Plant Sci. 7, 555–560. doi: 10.1016/S1360-1385(02)02362-2
Yanagisawa, S., and Schmidt, R. J. (1999). Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 17, 209–214. doi: 10.1046/j.1365-313X.1999.00363.x
Yang, X. C., and Hwa, C. M. (2008). Genetic modification of plant architecture and variety improvement in rice. Heredity 101, 396–404. doi: 10.1038/hdy.2008.90
Yokota, T. (1997). The structure, biosynthesis and function of brassinosteroids. Trends Plant Sci. 2, 137–143. doi: 10.1016/S1360-1385(97)01017-0
Zhang, C., Xu, Y., Guo, S., Zhu, J., Huan, Q., Liu, H., et al. (2012). Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genet. 8:e1002686. doi: 10.1371/journal.pgen.1002686
Zhu, J. Y., Sae-Seaw, J., and Wang, Z. Y. (2013). Brassinosteroid signalling. Development 140, 1615–1620. doi: 10.1242/dev.060590
Keywords: OsDof12, Dof transcription factor, plant architecture, rice (Oryza sativa L.)
Citation: Wu Q, Li D, Li D, Liu X, Zhao X, Li X, Li S and Zhu L (2015) Overexpression of OsDof12 affects plant architecture in rice (Oryza sativa L.). Front. Plant Sci. 6:833. doi: 10.3389/fpls.2015.00833
Received: 17 August 2015; Accepted: 23 September 2015;
Published: 08 October 2015.
Edited by:
Rodomiro Ortiz, Swedish University of Agricultural Sciences, SwedenReviewed by:
Chenglin Chai, Louisiana State University-Baton Rouge, USAHao Peng, Washington State University, USA
Copyright © 2015 Wu, Li, Li, Liu, Zhao, Li, Li and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shigui Li, Rice Research Institute, Sichuan Agricultural University, No. 211 Huimin Road, Chengdu 611130, China, lishigui@sicau.edu.cn;
Lihuang Zhu, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, No.1 West Beichen Road, Chaoyang District, Beijing 100101, China, lhzhu@genetics.ac.cn
†These authors have contributed equally to this work.
 Qi Wu
Qi Wu Dayong Li
Dayong Li Dejun Li3
Dejun Li3 Xiaobing Li
Xiaobing Li Lihuang Zhu
Lihuang Zhu