- Saskatoon Research Centre, Agriculture and Agri-Food Canada, Saskatoon, SK, Canada
The fungus Leptosphaeria maculans (L. maculans) is the causal agent of blackleg disease of canola/oilseed rape (Brassica napus) worldwide. We previously reported cloning of the B. napus blackleg resistance gene, LepR3, which encodes a receptor-like protein. LepR3 triggers localized cell death upon recognition of its cognate Avr protein, AvrLm1. Here, we exploited the Nicotiana benthamiana model plant to investigate the recognition mechanism of AvrLm1 by LepR3. Co-expression of the LepR3/AvrLm1 gene pair in N. benthamiana resulted in development of a hypersensitive response (HR). However, a truncated AvrLm1 lacking its indigenous signal peptide was compromised in its ability to induce LepR3-mediated HR, indicating that AvrLm1 is perceived by LepR3 extracellularly. Structure-function analysis of the AvrLm1 protein revealed that the C-terminal region of AvrLm1 was required for LepR3-mediated HR in N. benthamiana and for resistance to L. maculans in B. napus. LepR3 was shown to be physically interacting with the B. napus receptor like kinase, SOBIR1 (BnSOBIR1). Silencing of NbSOBIR1 or NbSERK3 (BAK1) compromised LepR3-AvrLm1-dependent HR in N. benthamiana, suggesting that LepR3-mediated resistance to L. maculans in B. napus requires SOBIR1 and BAK1/SERK3. Using this model system, we determined that BnSOBIR1 and SERK3/BAK1 are essential partners in the LepR3 signaling complex and were able to define the AvrLm1 effector domain.
Introduction
Plants mainly rely on induced innate immune system to resist pathogen infection. Membrane-localized pattern-recognition receptors (PRRs) form the first layer of defense by detecting conserved microbe or pathogen-associated molecular patterns (MAMPs or PAMPs) and activating PAMP-triggered immunity (PTI; Jones and Dangl, 2006). One of the PRRs that has been studied extensively is FLAGELLIN SESING 2 (FLS2) that recognizes the bacterial PAMP flagellin 22 (flg22; Cook et al., 2015). FLS2 is a leucine-rich repeat receptor-like kinase (LRR-RLK) and requires another LRR-RLK, the brassinosteroid receptor BRASSINOSTEROID INSENSITIVE 1 -ASSOCIATED KINASE 1 (BAK1)/SERK3, for perception of flg22 (Ben Khaled et al., 2015). Specialized pathogens have evolved strategies to overcome PTI by secretion of effector proteins. Most effector proteins are translocated into the plant cell where they modulate basal immunity and promote pathogen infection (Feng and Zhou, 2012). To counteract their activity, plants have evolved resistance (R) proteins to detect pathogen effectors, which leads to the activation of effector-triggered immunity (ETI; Jones and Dangl, 2006; Thomma et al., 2011). The majority of R proteins are cytoplasmic, but there are several examples of receptor-like proteins (RLP) with trans-membrane and extracellular LRR domains. The most widely studied examples of RLPs are the tomato Cf proteins (e.g., Cf2, Cf4, Cf9) that recognize corresponding effector proteins (e.g., Avr2, Avr4, Avr9) secreted by the fungus Cladosporium fulvum, the cause of tomato leaf mold disease (Rivas and Thomas, 2005). Accumulating evidence suggests that overlap exists between the PRR perception of PAMPs and the RLP recognition of effectors secreted by the apoplastic fungi. A recent report by Postma et al. (2015) showed that upon induction by Avr4, Cf4, interacts with BAK1. The recently discovered transmembrane LRR-RLK, SUPPRESSOR OF BIR1-1 (SOBIR1), is also required for ETI initiated by RLPs (Liebrand et al., 2013). SOBIR1, which was initially identified as a suppressor of BIR1 (BAK1-interacting receptor-like kinase 1), is conserved throughout the plant kingdom (Gao et al., 2009). SOBIR1 is required for Cf2-, Cf4-, and Ve1-mediated HR in tobacco and resistance of tomato against the fungal pathogens C. fulvum and Verticillium dahlia, respectively (Gao et al., 2009; Liebrand et al., 2013).
Leptosphaeria maculans (L. maculans) is the causative agent for blackleg disease (phoma stem canker) on Brassica species and is a major pathogen of Brassica napus (oilseed rape, canola) crops worldwide (West et al., 2001). L. maculans ascospores are released from infected canola stubble and infect the leaves and cotyledons of canola seedlings (Rouxel and Balesdent, 2005). During leaf colonization, L. maculans remains extracellular, similar to C. fulvum infection on tomato. Resistance against L. maculans in canola seedlings stage is race-specific. To date, 16 race-specific resistance (R) genes against L. maculans have been identified from Brassica species (Raman et al., 2013), but only two R genes, LepR3, and Rlm2, have been cloned (Larkan et al., 2013, 2015). LepR3 recognizes the L. maculans effector AvrLm1 (Larkan et al., 2013) and the AvrLm2 gene which encodes the effector corresponding to Rlm2 has been cloned, (Ghanbarnia et al., 2015). LepR3 and Rlm2 are allelic and encode membrane-bound LRR-RLPs. We recently reported that B. napus Rlm2 interacts with the Arabidopsis thaliana SOBIR1 (Larkan et al., 2015), suggesting SOBIR1 is a component of LRR-RLP-mediated resistance against L. maculans.
Recognition of an AVR protein by its cognate R protein often culminates into a hypersensitive response (HR) around the infection site, a phenotype that can also be produced by co-expressing both genes in the model plant tobacco using agroinfiltration (Goodin et al., 2008; Ma et al., 2012). Such transient assays in tobacco have been used to analyze the function of R/AVR gene pairs. For example, co-expression of the gene pairs Cf2/Avr2, Cf4/Avr4, and Cf9/Avr9 from tomato-C. fulvum and Ve1/Ave1 from tomato-V. dahlia plant pathogen systems trigger HR in Nicotiana benthamiana leaves (van der Hoorn et al., 2000; Cai et al., 2001; Zhang et al., 2012). To overcome the limitation of B. napus for functional analysis of R/Avr proteins, we exploited the N. benthamiana model system to investigate newly discovered R/AVR gene pairs from the B. napus-L. maculans pathosystem. Here we use this system, to identify the AvrLm1effector domain. We provide evidence that B. napus SOBIR1 interacts with LepR3 and is required for LepR3-mediated cell death, indicating that BnSOBIR1 forms a signaling complex with LepR3 in B. napus to initiate the innate immunity response upon recognition of AvrLm1. Finally, we determined that LepR3-AvrLm1-mediated cell death in N. benthamiana is also dependent on SERK3/BAK1.
Materials and Methods
Plant Materials and L. maculans Isolate
The susceptible doubled-haploid (DH) line Topas DH16516 and the LepR3 transgenic line NLA8 that were used in this study have been described elsewhere (Larkan et al., 2013). L. maculans isolate 3R11 was used for pathology tests and transformation. The genotype of the single-spore L. maculans isolate 3R11 has been described by Ghanbarnia et al. (2015).
Generation of Transgenic L. maculans Strains
To generate a L. maculans GatewayTM compatible vector in which the designated gene was driven by AvrLm1 native promoter, the promoter region of AvrLm1 was amplified by PCR with the AvrLm1-Up-F and AvrLm1-Up-R primers (see Supplementary Table S1). The resulting amplicon was cloned into the Acc65I site of the GW-pPk2 vector (Larkan et al., 2013). The TubA terminator was amplified with the primer pair TubA-F/TubA-R and cloned into the PacI site of GW-pPk2. The resulting vector was named GW-pLM4. AvrLM1 truncated variants, AvrLm1 Δ40, Δ50, and CTΔ14, with GatewayTM attB linkers were synthesized by GenScript (GeneScript, USA). The synthesized genes were introduced into the entry vector pDONR/Zeo then into the binary vector GW-pLM4 following the GatewayTM protocol (Life Technologies, USA). The plasmids were transformed into Agrobacterium tumefaciens AGL1 and used for subsequent Agrobacterium-mediated transformation of pycnidiospores from the L. maculans isolate 3R11 (avrLm1) as described by Utermark and Karlovsky (2008). Supplementary Table S1 contains the sequences for all the primers used in this study.
Binary Vector Constructions
For transient expression, the LepR3 ORF was amplified using the primer pair LepR3-FB and LepR3-RB-S/+S. Full length AvrLm1 was amplified by PCR- from L. maculans-B. napus cDNA using primers AvrLm1-FB and AvrLm1-RB and RNA isolated from the young cotyledons of B. napus cv. Topas infected with L. maculans v23.1.3. Truncated AvrLm1 lacking the region encoding the signal peptide was amplified using primers ΔspAvrLm1-FB and AvrLm1-RB. The PR1a-AvrLm1 gene with GatewayTMattB linkers was synthesized by GenScript (GeneScript, USA). DNA fragments were introduced into the GatewayTM entry vector pDONR/Zeo (Life Technologies, USA) to produce pENTR/Zeo:LepR3, pENTR/Zeo:LepR3+S (with stop codon after the coding region), pENTR/Zeo:AvrLm1 and pENTR/Zeo:ΔspAvrLm1 constructs which were then transferred to the binary vector pEarleyGate100 (Earley et al., 2006). The resulting plasmids were transferred into the A. tumefaciens strain GV3101 (pMP90).
For co-immunoprecipitation (co-IP) studies, the pENTR/Zeo:LepR3 was recombined into the binary vector pGWB414 containing the C-terminal Human influenza hemagglutinin (HA) tag (Nakagawa et al., 2007) resulting in pGWB414:LepR3-HA. Full length BnSOBIR1-A3 and BnSOBIR1-C3 were amplified by PCR- from the L. maculans-B. napus cDNA using the primers BnSOBIR1A3C3-FB and BnSOBIR1-A3C3-RB-S. The amplified fragments were introduced into the GatewayTMentry vector pDONR/Zeo (Life Technologies, USA) resulting in the constructs pENTR/Zeo:BnSOBIR1-A3 and pENTR/Zeo:BnSOBIR1-C3. These were recombined into the binary vector pGWB417 which contains a C-terminal Myc tag. The binary plasmids were transferred into the A. tumefaciens strain GV3101 (pMP90).
Agrobacterium-mediated Transient Assay in N. benthamiana
Agrobacterium-mediated transient expression in N. benthamiana was performed according to the method described previously (Ma et al., 2013). Briefly, Agrobacterium was grown to an absorbance of 0.8 at OD600 in LB-mannitol medium supplemented with 20 μM acetosyringone and 10 mM MES (pH 5.6). Cells were pelleted by centrifugation at 3500 g for 20 min and then re-suspended in infiltration medium (1x MES, 10 mM MES pH 5.6, 2% w/v sucrose, 200 μM acetosyringone). The gene encoding the silencing suppressor, p19 from the Tomato bushy stunt virus (Voinnet et al., 2003), was co-expressed in all the transient expression assays. Leaves of 4–5 weeks old N. benthamiana plants were infiltrated with Agrobacterium culture at OD600 of 2.0 and the triple-infiltration was done with a 1:1:1 mix ratio of Agrobacterium. Total 30 leaves of N. benthamiana were infiltrated and occurrence of HR was calculated on the sites that PR1a-AvrLm1/LepR3 was co-infiltrated. Representative leaves showing HR were photographed 3 or 6 days after infiltration.
Co-immunoprecipitation and Western Blotting
Co-IP was performed as described by Liebrand et al. (2013). Briefly, total proteins were extracted from N. benthamiana leaves 48 h after infiltrating with a mixture of A. tumefaciens GV3101 containing either pGWB414:LepR3 or pGWB417:BnSOBIR1-A3 or pGWB417:BnSOBIR1-C3 or p19 in buffer [150 mM NaCl, 1.0% IGEPAL CA-630 (NP-40), 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris (pH 8.0) 1x complete protease inhibitor cocktail (Roche, USA)]. Extracts were centrifuged at 18,000 g, 4°C for 15 min, and 2 ml of supernatant was collected and applied to 50 μl of Anti-HA magnetic beads (Pierce, USA), which was then incubated for 2 h at 4°C in a rotator. After washing the beads four times with extraction buffer, immuno-precipitated proteins were separated on an 8% SDS-PAGE gel and transferred to overnight to a PVDF membrane using wet blotting (Bio-Rad, USA). Skimmed milk powder (5%) was used as a blocking agent. A 1:2000 dilution of anti-HA antibody (Pierce, USA) or 1:2000 diluted anti-Myc antibody (cMyc 9E10, sc-40-HRP, Santa Cruz) was used. The goat-anti-mouse secondary antibody (Pierce, USA) was used as a 1:15000 dilution. The luminescent signal was visualized using Immobilon Western Chemiluminescent HRP Substrate and BioMax MR film (Kodak).
TRV-mediated Gene Silencing in N. benthamiana Plants and Hypersensitive Response Assay
Virus-induced gene silencing (VIGS) was performed using the tobacco rattle virus (TRV)-mediated gene silencing vector pTRV1 (Ratcliff et al., 2001) and pTRV2 constructs: pTRV2:GFP (Burch-Smith et al., 2006), pTRV2:PDS (Liu et al., 2002), pTRV2:NbSBOBIR1 (Liebrand et al., 2013), and pTRV2:NbSERK3a/b (Chaparro-Garcia et al., 2011). The VIGS experiments in N. benthamiana were performed as described previously (Gabriels et al., 2006). In brief, cotyledons of 2-week-old N. benthamiana seedlings were infiltrated with the pTRV1 and pTRV2 constructs in a 1:1 ratio. Agroinfiltration was performed as described by Ma et al. (2012). For monitoring the development of HR, 3 weeks after TRV infiltration, mature leaves were co-infiltrated to express PR1a-AvrLm1/LepR3, PR1a-AvrLm1/GFP, LepR3/GFP, Bcl2-Associated proteinX(BAX; Lacomme and Santa Cruz, 1999)/GFP, respectively. All the co-infiltrations were performed in a 1:1 mix ratio of Agrobacterium containing the corresponding construct at OD600 = 1. Total 30 leaves of N. benthamiana were infiltrated and occurrence of HR was calculated on the sites that PR1a-AvrLm1/LepR3 was co-infiltrated. Six days after infiltration, leaves were examined for development of an HR and representative leaves were photographed. The experiment was repeated three times.
Quantitative RT-PCR Analysis
For qRT-PCR, total RNA was isolated from N. benthamiana at 2 weeks after agro-inoculation with the various VIGS constructs including TRV:GFP and TRV:NbSOBIR1. Wildtype (WT) N. benthamiana leaves without TRV inoculation was collected for the negative control as well. The N. benthamiana leaves were ground in liquid nitrogen. Total RNA from the samples was extracted with TRIzol LS reagent (Invitrogen, USA) and subsequently purified with RNAeasy Mini kit (Qiagen, USA). DNA was removed by on-column treatment with RNase-free DNase (Qiagen, USA). cDNA was synthesized using SuperScript III first-strand synthesis SuperMix kit according to the manufacture’s protocol (Invitrogen, USA). qRT-PCR was performed using a 7700 real-time PCR machine (Applied Biosystems) and SsoFast EvaGreen Supermix (BIO-RAD). RT-PCR was performed for the NbSBOIR1 gene with three biological samples. Expression of NbSOBIR1 was investigated using primer NbSOBIR1-to266 and NbSOBIR1-to267 (Liebrand et al., 2013). Expression of endogenous actin was used to calibrate the expression level of the query genes, as previously described (Liebrand et al., 2013). The primers used for qRT-PCR are described in Supplementary Table S1. Ct values were analyzed according to the 2-ΔΔCt method (Livak and Schmittgen, 2001). The statistical significance of differences was calculated using GraphPad Prism 6 (GraphPad Software, Inc., USA) with One-way ANOVA followed by the Turkey post-test to obtain the P-value. Data are shown as mean ± SEM of three biological replicates from one representative experiment. Significant differences between treatments and controls are represented by three asterisks (P < 0.001).
Results
AvrLm1 Triggers LepR3-mediated Cell Death in N. benthamiana
Transient expression of R genes and their cognate effectors (pathogen Avr genes) in N. benthamiana often leads to a HR and is a commonly used tool for the functional analysis of R-Avr genes (van der Hoorn et al., 2000; Ma et al., 2012). To determine if perception of AvrLm1 by LepR3 triggers HR in N. benthamiana, AvrLm1, and LepR3 were ectopically co-expressed in leaves using Agro-infiltration. L. maculans is confined to the apoplastic space, therefore to ensure efficient secretion of AvrLm1 into the apoplast the native signal peptide of AvrLm1 was replaced with the tobacco PR1a signal peptide (van Esse et al., 2006); this construct was designated PR1a-AvrLm1. The PR1a signal peptide has been widely used for secretion of plant pathogen effectors into the extracellular space (van Esse et al., 2007). The wild type AvrLm1 containing its indigenous signal peptide and the ΔspAvrLm1 construct encoding a truncated AvrLm1 protein without its signal peptide were included in this assay as well. A. tumefaciens harboring either PR1a-AvrLm1, wild type AvrLm1 or ΔspAvrLm1 was co-infiltrated with an A. tumefaciens strain carrying B. napus LepR3. Co-infiltration of either AvrLm1 or LepR3 with GFP served as the negative control. 80% of the sites infiltrated with PR1a-AvrLm1 showed a LepR3-dependent HR approximately 6 days after co-infiltration (Figure 1A). The wild type AvrLm1 and ΔspAvrLm1 did not trigger HR when co-expressed with LepR3 (Figures 1A,B). Expression of AvrLm1 or LepR3 alone did not cause HR. The lack of HR by the full length AvrLm1 (native signal peptide) is likely due to inefficient secretion of AvrLm1 as directed by its native signal peptide in N. benthamiana plants. These results indicate that AvrLm1 was able to activate LepR3 leading to the development of HR and that the recognition occurred outside the plant cell which is in accordance with LepR3 being a cell surface receptor.
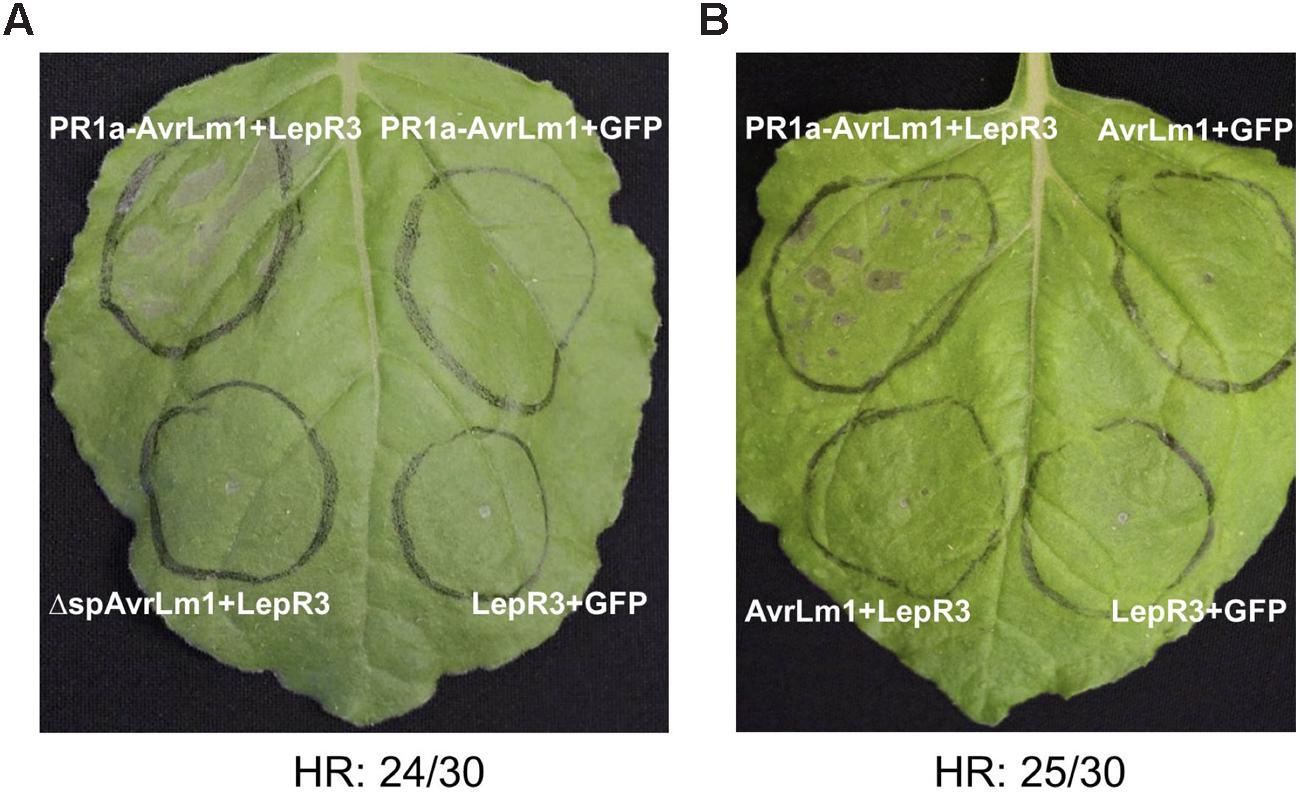
FIGURE 1. Transient expression of AvrLm1 triggers LepR3-mediated cell death in Nicotiana benthamiana leaves. (A) N. benthamiana leaves Agro-infiltrated with the LepR3, PR1a-AvrLm1, and GFP constructs in the order indicated on the image. To ensure secretion of AvrLm1 to the apoplastic space, the indigenous AvrLm1 signal peptide was replaced by the tobacco PR1a signal peptide and the construct was designated as PR1a-AvrLm1. Truncated AvrLm1 lacking its indigenous signal peptide (ΔspAvrLm1) was also included. Co-expression of LepR3 or PR1a-AvrLm1 with GFP served as negative controls. The experiment was performed three times and each time 10 plants were infiltrated. (B) N. benthamiana leaves Agro-infiltrated with PR1a-AvrLm1/LepR3, AvrLm1/LepR3, AvrLm1/GFP, and LepR3/GFP constructs in the order indicated in the image. The numbers under the panels indicate the total occurrence of HR within 30 sites co-infiltrated with PR1a-AvrLm1/LepR3. Images are from photographs taken 6 days after infiltration.
The N-terminal Region of AvrLm1 is Dispensable for LepR3-mediated Cell Death and Resistance
N-terminal and C-terminal truncated AvrLm1 constructs were generated to define the minimal region of AvrLm1 required to trigger LepR3-mediated cell death. Design of the AvrLm1 truncated constructs was aided by prediction of AvrLm1 secondary structure using PSIPRED (Buchan et al., 2010; Figure 2A). Based on the predicted secondary structure, three variants were constructed: AvrLm1-Δ40, in which the first predicted random coil after signal peptide was deleted; AvrLm1-Δ48, in which the deletion extended to the end of the predicted β-strand structure; AvrLm1-CTΔ14, which carried a deletion of the last β-strand at its C-terminus (Figure 2B). The tobacco PR1a signal peptide was fused to the N-terminus of the truncated constructs to allow efficient secretion of the AvrLm1 protein into the extracellular space. Each of the truncated AvrLm1 was co-expressed with the full length LepR3 in N. benthamiana leaves by Agro-infiltration. Infiltration of the PR1a-AvrLm1 together with LepR3 served as a positive control. AvrLm1-Δ40 induced a HR response similar to the full length PR1a-AvrLm1; however, the other truncated variants, AvrLm1-Δ48 and AvrLm1-CTΔ14, were impaired in inducing LepR3-dependent cell death (Figure 2C).
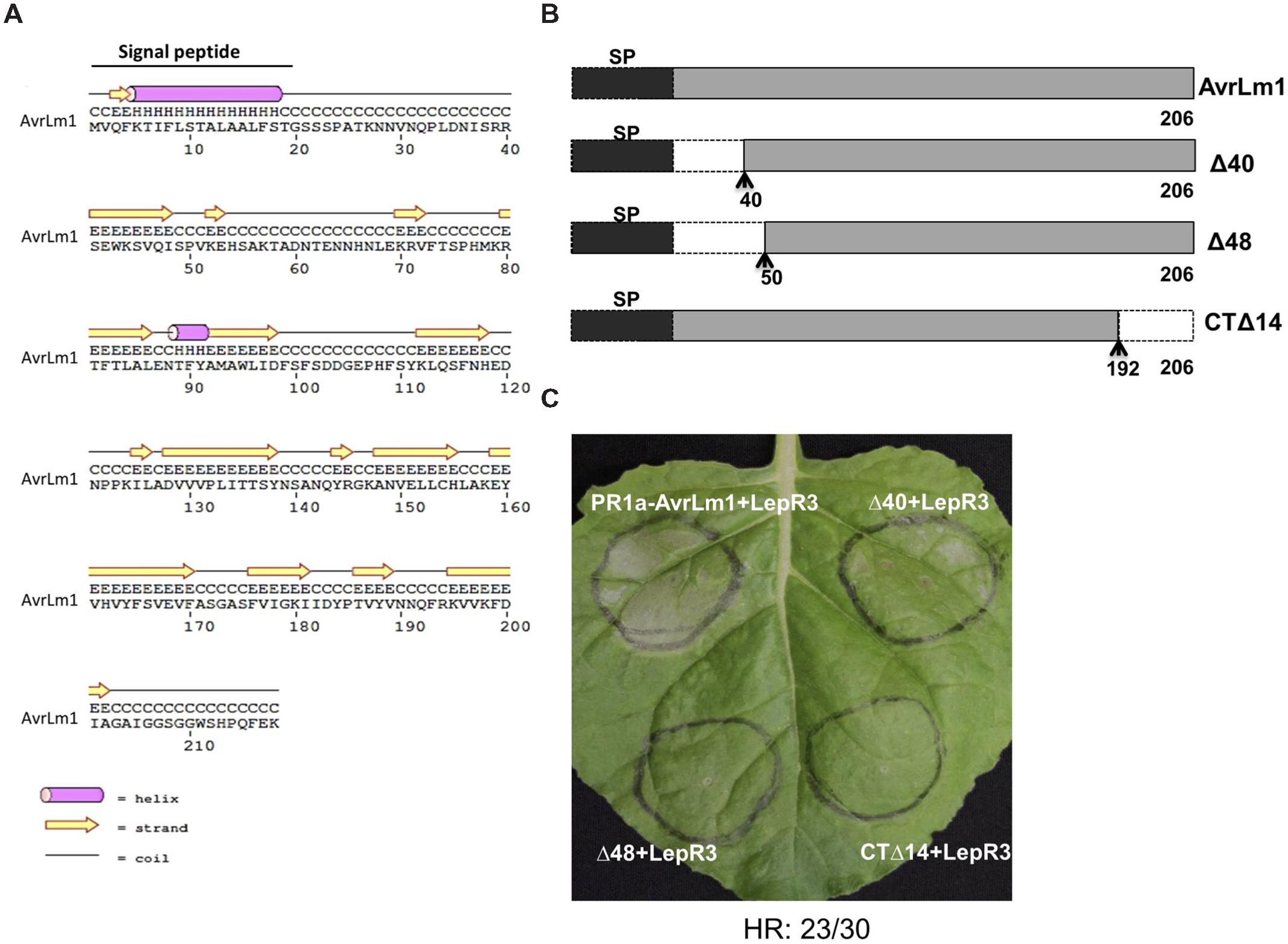
FIGURE 2. A small N-terminal region of AvrLm1 is dispensable for LepR3-mediated cell death. (A) Secondary structure prediction of the full length AvrLm1 protein. (B) Schematic diagram showing AvrLm1 truncations and their sizes. The signal peptide is shown as black boxes. The N- and C-terminal truncation sites are indicated by dashed lines with arrows showing the position of amino acid at the boundary between the deleted fragment and its adjacent remaining peptide segment. (C) N. benthamiana leaves were co-infiltrated with Agrobacterium cultures containing truncated AvrLm1 and LepR3 constructs. Images were taken at 6 days after infiltration. H means protein secondary structure helix, C means coli, and E means strand. The experiment was performed three times and each time with 10 plants. The numbers below the panels indicate the occurrence the occurrence of HR observed within 30 sites co-infiltrated with PR1a-AvrLm1/LepR3 or Δ40/LepR3.
The truncated and full length AvrLm1 with native signal peptide were transferred into the L. maculans isolate 3R11, which lacks the entire AvrLm1 ORF to investigate the role of these truncated variants in LepR3-mediated resistance in the natural host plant B. napus. It should be noted that all the truncated AvrLm1 constructs used for transformation of 3R11 contained the AvrLm1 native signal peptide and were driven by the native AvrLm1 promoter. Six transformants were selected for analysis from each transformation event. Transgenic 3R11 lines were inoculated on the cotyledons of 1-week-old seedlings of the B. napus cultivars Topas DH16516 (susceptible to L. maculans) and LepR3-transgenic NLA8 (a Topas DH16516–LepR3 transgenic line described by Larkan et al. (2013). Disease development was scored 14-days after inoculation. All of the L. maculans transgenic 3R11 were virulent and caused fully expanded lesions (similar to the wild type 3R11) on the susceptible Topas DH16516 plants (Figure 3) confirming that transformation of 3R11 did not affect its overall virulence. When tested on the LepR3-transgenic NLA8 plants, transgenic 3R11 carrying either wild type AvrLm1 or AvrLm1-Δ40 were avirulent. However, the transgenic 3R11 carrying AvrLm1-Δ48 or AvrLm1-CTΔ14 were virulent and caused extensive lesions similar to the response of susceptible control lines, confirming that both of these truncated AvrLm1 proteins failed to activate LepR3-mediated resistance. These findings confirmed the functional assay of AvrLm1 constructs in N. benthamiana. Taken together, these results showed that N-terminal region of AvrLm1 (or at least the first 18 amino acids of the mature AvrLm1 protein) is not required for LepR3-mediated resistance.
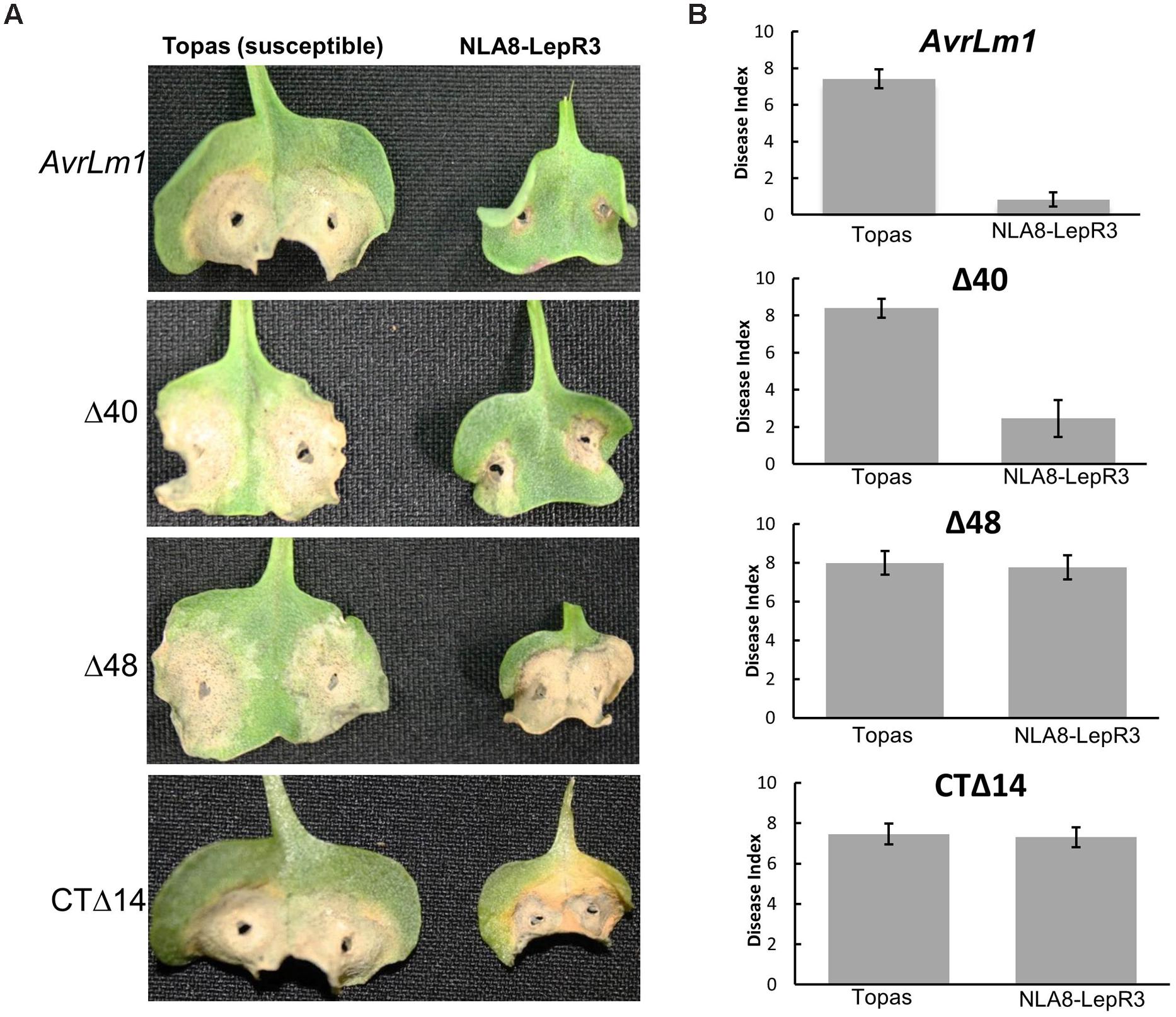
FIGURE 3. A small N-terminal region of AvrLm1 is not required for LepR3-mediated resistance in Brassica napus. Cotyledons of 7-day-old seedlings of susceptible B. napus Topas and LepR3-trasgenic NLA8 line were inoculated with a transgenic 3R11 Leptosphaeria maculans isolate carrying AvrLm1 or its truncated variants (Δ40, Δ48 and CTΔ14). Lesion development (disease index from 0 to 9 with 0 showing no lesion) was scored at 14 days after inoculation. Error bars indicate standard deviation. (A) Images taken from infected seedlings 14 days after inoculation. (B) Average disease index representing data collected from 12 cotyledons of the susceptible B. napus cv Topas and the Topas LepR3-transgenic plants (NLA8).
LepR3 Interacts with B. napus AtSOBIR1 Homologs in planta
Recently, the A. thaliana LRR-receptor-like kinase (LRR-RLKs). Suppressor of Bir1-1 (AtSobir1) and its homologs from tomato were shown to interact specifically with a number of LRR-receptor like proteins (LRR-RLPs), such as tomato Cf-4 and Ve1, and play a role in LRR-RLP-mediated resistance against the corresponding fungal pathogen (Liebrand et al., 2013, 2014). We searched the genome sequence of the B. napus cultivar ‘Darmor-bzh’ (Chalhoub et al., 2014) using the AtSOBIR1 as a query and six predicted B. napus SOBIR1 homologs (BnSOBIR1s) were identified. Based on phylogenetic analysis of the six BnSOBIR1s, as well as AtSBOIR1 and NbSOBIR1 (Supplementary Figure S1), two of the B. napus orthologs of SOBIR1, BnSOBIR1-A3 (BnaA03g1476) and BnSOBIR1-C3 (BnaC03g17800D), had the closest homology to AtSOBIR1. BnSOBIR1-A3 and BnSOBIR1-C3 were subsequently cloned to examine their interaction with LepR3. BnSOBIR1 proteins fused at the C-terminus to the Myc epitope tag and LepR3 fused at the C-terminus to the HA epitope tag were generated and transiently co-expressed in N. benthamiana leaves to perform co-immunopurification experiments. Myc-tagged BnSobir1-A3 and BnSobir1-C3 were co-immunopurified with LepR3 when HA-tagged LepR3 was used to capture the SOBIR1 complex (Figure 4).
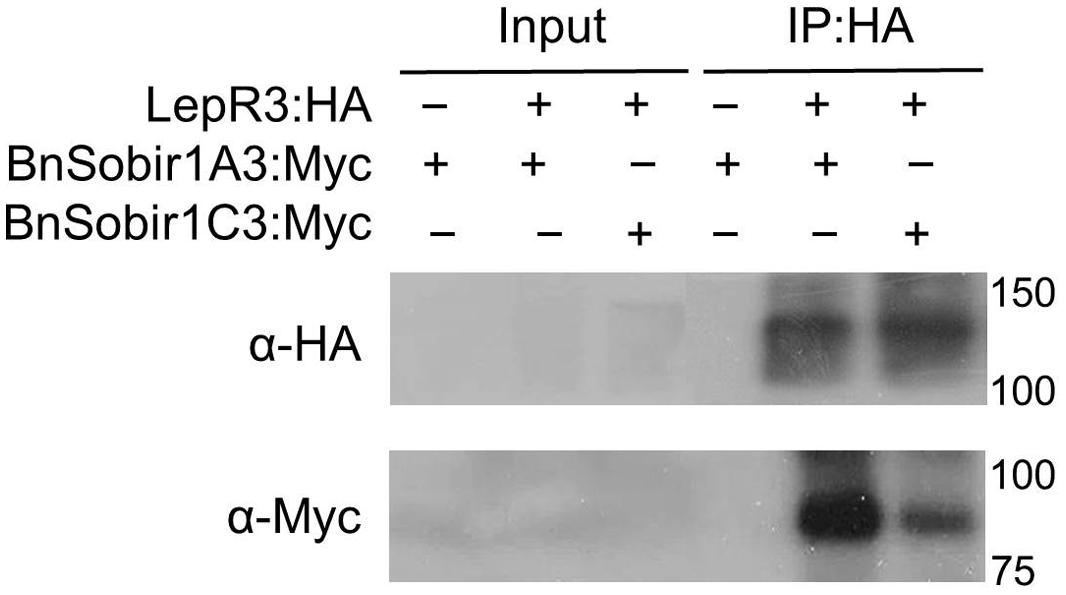
FIGURE 4. LepR3 interacts with BnSOBIR1s in planta. HA-tagged LepR3 and Myc-tagged BnSOBIR1s (BnSOBIR1-A3 and BnSOBIR1-C3) were co-expressed in N. benthamiana. Proteins were extracted after 48 h and subjected to immuno-precipitation by HA-Trap_M beads. Total proteins (input) and immuno-purified proteins (IP) were separated on an SDS-PAGE gel followed by blotting. Anti-HA antibody was used to detect the immuno-precipitated HA fusion proteins and anti-Myc antibody was used to detect co-immuno-precipitated Myc-BnSOBIR1 proteins, respectively.
NbSOBIR1 and NbSERK3 (BAK1) are Required for AvrLm1/LepR3-Induced Cell Death Response in N. benthamiana
The observation that the two SOBIR1 homologs from B. napus interact with LepR3 (Figure 4) suggests that both proteins play a role in LepR3-mediated defense signaling in plants. It has been reported that the AtSOBIR1 homolog from N. benthamiana (NbSOBIR1) was required for Avr4/Cf4 and Ave1/Ve1-triggered HR in tobacco plants (Liebrand et al., 2013; Fradin et al., 2014). We hypothesized that the NbSOBIR1, which exhibits closest homology with BnSOBIR1-A3 and -C3 (Supplementary Figure S1), is also required for AvrLm1/LepR3-triggered HR in N. benthamiana. To test this hypothesis, a recombinant TRV-based construct containing NbSOBIR1 was used to knockdown the expression of NbSOBIR1 homologs by VIGS. The same construct for NbSOBIR1 as reported by Liebrand et al. (2013) was used and the function of this silencing construct to knockdown the expression of NbSOBIR1 gene homologs has been shown to block Cf4-Avr4-mediated HR in N. benthamiana (Liebrand et al., 2013). A silencing construct targeting the tobacco phytoene desaturase (PDS) was included to monitor VIGS silencing efficiency and progression (VIGS of PDS causes photo-bleaching). A TRV:GFP construct was used as a negative control. Three weeks after viral inoculations, photobleaching symptoms were observed in all of the PDS silenced N. benthamiana plants confirming the onset of VIGS (Supplementary Figure S2). The N. benthamiana NbSOBIR1-silenced (TRV:NbSOBIR1) plants were infiltrated to transiently co-express PR1a-AvrLm1/LepR3; PR1a-AvrLm1/GFP; and LepR3/GFP. In TRV:NbSOBIR1 plants, PR1a-AvrLm1/LepR3-triggered HR was severely compromised (Figure 5A). However, in the control plant (TRV:GFP), HR induced by the co-expression PR1a-AvrLm1/LepR3 was not affected (Figure 5A). In addition, qRT-PCR confirmed that NbSOBIR1 expression levels were ∼90% reduced upon inoculation with TRV:NbSOBIR1 compared with the one inoculated with TRV:GFP or the wildtype (WT) N. benthamiana without TRV inoculation (Figure 5B). Furthermore, to verify TRV:NbSOBIR1 plants were not compromised in their ability to mount programmed cell death, the proapoptotic factor Bcl2-Associated protein X (BAX) was transiently expressed in TRV:NbSOBIR1 plants (Lacomme and Santa Cruz, 1999). Expression of BAX triggered a strong cell death in SOBIR1-silenced plants (Figure 5A), confirming that the ability of these plants to mount programmed cell death was not compromised.
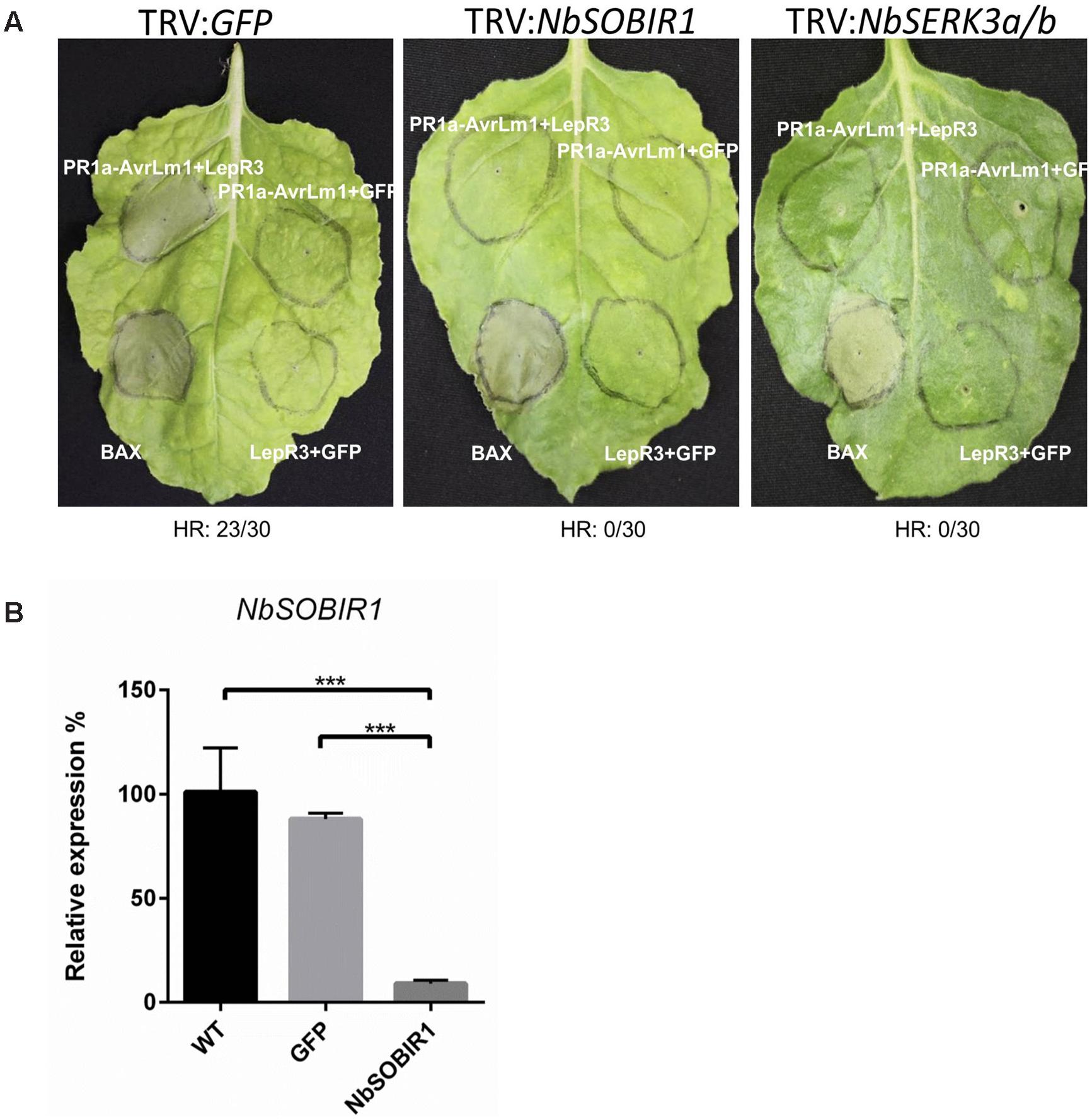
FIGURE 5. NbSOBIR1 and NbSERK3 (BAK1) are required for LepR3-mediated development of cell death. (A) 2-week-old N. benthamiana seedlings were subjected to virus-induced gene silencing (VIGS) by inoculation with the TRV:NbSOBIR1, TRV: NbSERK3(BAK1), and TRV:GFP, respectively. TRV:GFP served as a control. Three weeks after TRV inoculation, PR1a-AvrLm1/LepR3, PR1a-AvrLm1/GFP, and LepR3/GFP were transiently co-expressed in the order indicated on the image. Cell death caused by Transiently expressed BAX served as a control. Leaves were photographed 6 days after infiltration. The experiment was repeated three times with 10 plants for each TRV construct. Numbers below the panels indicate the occurrence or the absence of HR within 30 sites co-infiltrated with PR1a-AvrLm1/LepR3. (B) qRT-PCR analysis showing NbSOBIR1 expression in N. benthamiana inoculated with the indicated TRV VIGS constructs (TRV:GFP and TRV:NbSOBIR1) and in wildtype (WT) N. benthamiana without TRV inoculation. NbSOBIR1 gene expression levels are normalized to that of actin. Values are means ± SE of triplicate reactions of three independent biological samples. Significant differences are represented by three asterisks (P < 0.001). Statistical differences (P <= 0.001) between groups were calculated by a one-way ANOVA and different groupings are indicated. Error bars represent the standard deviation.
Recently, it has been reported that BRI1-ASSOCIATED KINASE 1 (BAK1)/SOMATIC EMBRYOGENESIS RECEPTOR KINASE (SERK)3 associates with Cf-4 and Cf-9 upon elicitation with the matching effector ligands Avr4 and Avr9 (Postma et al., 2015). In addition, BAK1-silenced N. benthamiana plants were compromised in Avr4-triggered endocytosis and effector-triggered cell death. BAK1 is also required for resistance of Cf-4 tomato plants against C. fulvum carrying Avr4 (Postma et al., 2015). To determine if the components of Cf4 receptor complex are also common to the LepR3 receptor complex, the requirement for BAK1/SERK3 in AvrLm1/LepR3-triggered cell death in N. benthamiana plants was investigated. The AvrLm1/LepR3-triggered cell death was examined by co-expression of these constructs in N. benthamiana NbSERK3-silenced plants. The effectiveness of the silencing construct TRV2: SERK3a/b (BAK1) to knockdown the expression of NbSERK3a/b homologs in N. benthamiana has been shown (Chaparro-Garcia et al., 2011; Postma et al., 2015). The TRV:GFP construct was used as a negative control. Three weeks after viral inoculation, the tobacco NbSERK3a/b-silenced (TRV:NbSERK3) plants were infiltrated to transiently co-express PR1a-AvrLm1/LepR3, PR1a-AvrLm1/GFP, and LepR3/GFP. The PR1a-AvrLm1/LepR3-triggered cell death was compromised in NbSERK3a/b-silenced plants, as compared to TRV:GFP control plants (Figure 5A).
Discussion
Race-specific resistance against L. maculans remains the only practical approach to control blackleg disease of canola (Raman et al., 2013). The genetics of this race-specific resistance have been studied in detail; however, the molecular mechanism of L. maculans perception by B. napus remained unknown until the cloning of LepR3 and Rlm2, two B. napus R genes against blackleg (Larkan et al., 2013; Larkan et al., 2015). Here we presented further studies to unravel the components of LepR3-AvrLm1 recognition complex. Since most of the tools developed for the functional analysis of R-Avr genes cannot be applied to B. napus, transient expression in N. benthamiana was used to study AvrLm1-LepR3 interaction. Using this model plant, two LRR-RLK proteins, SOBIR1 and BAK1 were identified as components of the LepR3 recognition complex. The requirement of SOBIR1 for plant immunity initiated by RLP proteins has been shown for the tomato Cf4, Cf9, and Ve1 which are effective against C. fulvum and V. dahlia, respectively (Liebrand et al., 2014). The kinase domain of SOBIR1 is suggested to be involved in downstream signaling. Indeed, both the kinase and LRR domains of SOBIR1 are required for Cf4/Avr4-induced HR, although they are dispensable for interaction with Cf4 (Bi et al., 2015). In addition several RLPs from A. thaliana form a complex with the AtSOBIR1. SOBIR1 bind to the AtRLP23, an Arabidopsis LRR-RLP that perceives the Necrosis and ethylene-inducing peptide 1-like proteins (NLPs), a conserved protein in many prokaryotic and eukaryotic microorganisms (Albert et al., 2015). AtRLP42 that was identified as RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1 (RBPG1) was shown to bind AtSOBIR1 (Zhang et al., 2014). Another Arabidopsis immune receptor is RLP30 that perceives the SCLEROTINIA CULTURE FILTRATE ELICITOR1 (SCFE1) from Sclerotinia sclerotiorum and is dependent on SOBIR1 for its function (Zhang et al., 2013). Six paralogs of SOBIR1 (BnSOBIR1s) are present in B. napus and LepR3 interacted with two BnSOBIR1 which had closest similarity to AtSOBIR1. This findings indicates that a degree of functional redundancy exists and also that other BnSOBIR1 are likely part of receptor complexes for the presently unknown RLP against L. maculans. The interaction of LepR3 with SOBIR1 in the absence of AvrLm1 suggests that this interaction is not ligand-dependent, as has also been reported for the interaction of Cf proteins with SOBIR1 (Postma et al., 2015). It has been documented that NbSOBIR1 is required for Cf4- or Ve1-mediated cell death in N. benthamiana plants (Liebrand et al., 2013). These findings show the presence of conserved mechanisms for the perception of apoplastic effectors in two distinct host-pathogen systems.
BAK1 was demonstrated to be another component of the LepR3-SOBIR1 recognition complex. The requirement of BAK1 for HR initiated by LepR3/AvrLm1 supports similar findings reported for the recognition of C. fulvum apoplastic effectors Avr4/9 by the tomato RLPs Cf4/9 (Postma et al., 2015). We recently reported that SOBIR1 interacts with the second B. napus R protein Rlm2 that recognizes L. maculans AvrLm2 effector proteins (Ghanbarnia et al., 2015; Larkan et al., 2015). It is likely that BAK1 is also required for recognition of AvrLm2 by Rlm2. BAK1 is best known as being required for the perception of the bacterial PAMP, flg22, through interaction with the FLS2 receptor complex (Chinchilla et al., 2007). However, data that we have presented here and that by Postma et al. (2015) clearly shows the importance of BAK1 in ETI against the apoplastic fungi and highlights the common mechanisms involved in the recognition of PAMPs and the recognition of effectors from extracellular fungi. Accumulating evidence suggests that common features shared between PTI and ETI blur the distinction between these two immunity systems (Thomma et al., 2011). Several PAMPs and effectors are discussed in a recent review by Cook et al. as examples that deviate from the currently accepted definition of PTI/ETI (Cook et al., 2015). A more relevant example to our study is the case of Cf4/Avr4. It was suggested that Cf4 is a PRR that recognizes conserved chitin binding domains present in C. fulvum Avr4 and its homolog in Mycosphaerella fijiensis, a pathogen that causes the black leaf streak disease in banana (Stergiopoulos et al., 2010). However, AvrLm1, and AvrLm2 both lack any known functional or conserved domains, a feature common to most effector proteins. In addition, AvrLm1 and AvrLm2 have other features typical of fungal and oomycete effectors, such as being located in the AT-rich block of the genome, and showing allelic variation caused by point mutation (AvrLm2) or complete deletion (AvrLm1; Gout et al., 2006; Ghanbarnia et al., 2015).
We also exploited transient expression in N. benthamina to identify the effector domain of AvrLm1 and the results validated by expressing the various truncated AvrLm1 constructs in L. maculans and conducting pathology assays on the host plant B. napus. Based on these analyses, only the first 18 amino acids of the AvrLm1 mature protein is dispensable for its function to induce LepR3-dependent HR. Deletion in the C-terminal part of AvrLm1 disrupts the LepR3 mediated HR. The C-terminal region of effectors from plant pathogenic oomycetes and fungi has been shown to act as effector domain. The AvrLm1 gene is located in a large AT-rich, heterochromatin-like region that is mostly devoid of other L. maculans coding sequences (Rouxel et al., 2011). Furthermore, the genome environment surrounding AvrLm1 is enriched with transposons elements (TEs), which is thought to contribute to allele diversification. The AvrLm1 allele is entirely deleted in L. maculans isolates virulent on B. napus cultivars containing LepR3 or Rlm1 (Gout et al., 2006; Rouxel et al., 2011). AvrLm1 contains only one cysteine residue, which is unusual for effectors secreted to the apoplast of host plants. However, we demonstrated by transient expression in N. benthamiana and transformation of L. maculans that AvrLm1 must be secreted to the host apoplast to be perceived by LepR3. Still, it is possible that AvrLm1 is translocated at some stage into the plant host cells to modulate host immunity. Another B. napus R gene (Rlm1) encoding Rlm1 that also recognizes AvrLm1 product but is physically distinct from LepR3. We have not been able to detect the direct interaction between AvrLm1 and LepR3 (unpublished data) pointing to the possibility that AvrLm1 interacts with a presently unknown host protein that is commonly guarded by Rlm1 and LepR3. Determining the structure of AvrLm1 could help to determine surface residues required for the interaction with host plant targets.
The information presented here underlines the importance of understanding the molecular function of B. napus receptor complexes against L. maculans in developing durable resistance against this rapidly evolving pathogen. We demonstrated for the first time that N. benthamiana model system can be used to facilitate functional characterisation of R/Avr gene pairs from the B. napus-L. maculans pathogen system which is less amenable to genetic manipulation. These findings provide invaluable tools to analyze the function of additional R and Avr genes from the B. napus-L. maculans pathogen system that will be identified in future.
Author Contributions
LM conducted the experiments. LM and MB designed the experiments, analyzed the data and wrote the manuscript. Both authors provided intellectual input, approved the manuscript and are accountable for accuracy and integrity of this study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Thomas Liebrand and Dr. Matthieu Joosten (Wageningen University, the Netherlands) for providing the pGWB20-At-SOBIR1–Myc, pTRV::GFP, and pTRV2::NbSOBIR1 constructs, Dr. Sophien Kamoun (The Sainsbury Laboratory, UK) for providing the pTRV2::SERK3 (BAK1) construct and Dr. Haiyan Wang (Agricultural University of Hebei, China) for assisting with the transient assay. We also thank Dr. Dwayne D. Hegedus (AAFC, Saskatoon Research Centre) for critical review of the manuscript. Funding for this project was provided by ADF (Saskatchewan Ministry of Agriculture), SaskCanola and Western Grain Research Foundation.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00933
FIGURE S1 | Virus-induced gene silencing (VIGS) of phytoene desaturase (PDS) gene in Nicotiana benthamiana. VIGS of the phytoene desaturase (PDS) gene leads to photobleaching in leaves of N. benthamiana plant. Pictures were taken at 3 weeks after inoculation.
FIGURE S2 | Phylogenetic tree of SOBIR1 from Arabidopsis, Brassica napus and N. benthamiana. The protein sequences of AtSBOIR1, BnSOBIR1-A3, -C3,-A4a,-C4a,-A4b,-C4b, and NbSOBIR1 were aligned and analyzed with CLC for the phylogenetic tree.
References
Albert, A., Böhm, H., Albert, M., Feiler, C. E., Imkampe, J., Wallmeroth, N., et al. (2015). An RLP23–SOBIR1–BAK1 complex mediates NLP-triggered immunity. Nat. Plants 1:15140. doi: 10.1038/nplants.2015.140
Ben Khaled, S., Postma, J., and Robatzek, S. (2015). A moving view: subcellular trafficking processes in pattern recognition receptor–triggered plant immunity. Annu. Rev. Phytopathol. 53, 379–402. doi: 10.1146/annurev-phyto-080614-120347
Bi, G., Liebrand, T. W. H., Bye, R. R., Postma, J., Van Der Burgh, A. M., Robatzek, S., et al. (2015). SOBIR1 requires the GxxxG dimerization motif in its transmembrane domain to form constitutive complexes with receptor-like proteins. Mol. Plant Pathol. doi: 10.1111/mpp.12266 [Epub ahead of print].
Buchan, D. W. A., Ward, S. M., Lobley, A. E., Nugent, T. C. O., Bryson, K., and Jones, D. T. (2010). Protein annotation and modelling servers at University College London. Nucleic Acids Res. 38, W563–W568. doi: 10.1093/nar/gkq427
Burch-Smith, T. M., Schiff, M., Liu, Y., and Dinesh-Kumar, S. P. (2006). Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol. 142, 21–27. doi: 10.1104/pp.106.084624
Cai, X., Takken, F. L. W., Joosten, M. H. A. J., and De Wit, P. J. G. M. (2001). Specific recognition of AVR4 and AVR9 results in distinct patterns of hypersensitive cell death in tomato, but similar patterns of defence-related gene expression. Mol. Plant Pathol. 2, 77–86. doi: 10.1046/j.1364-3703.2001.00053.x
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A. P., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chaparro-Garcia, A., Wilkinson, R. C., Gimenez-Ibanez, S., Findlay, K., Coffey, M. D., Zipfel, C., et al. (2011). The receptor-like kinase SERK3/BAK1 is required for basal resistance against the late blight pathogen Phytophthora infestans in Nicotiana benthamiana. PLoS ONE 6:e16608. doi: 10.1371/journal.pone.0016608
Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nurnberger, T., Jones, J. D. G., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500. doi: 10.1038/nature05999
Cook, D. E., Mesarich, C. H., and Thomma, B. P. H. J. (2015). Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 53, 541–563. doi: 10.1146/annurev-phyto-080614-120114
Earley, K. W., Haag, J. R., Pontes, O., Opper, K., Juehne, T., Song, K., et al. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. doi: 10.1111/j.1365-313X.2005.02617.x
Feng, F., and Zhou, J. M. (2012). Plant-bacterial pathogen interactions mediated by type III effectors. Curr. Opin. Plant Biol. 15, 469–476. doi: 10.1016/j.pbi.2012.03.004
Fradin, E. F., Zhang, Z., Rovenich, H., Song, Y., Liebrand, T. W. H., Masini, L., et al. (2014). Functional analysis of the tomato immune receptor Ve1 through domain swaps with its non-functional homolog Ve2. PLoS ONE 9:e88208. doi: 10.1371/journal.pone.0088208
Gabriels, S. H., Takken, F. L., Vossen, J. H., De Jong, C. F., Liu, Q., Turk, S. C., et al. (2006). CDNA-AFLP combined with functional analysis reveals novel genes involved in the hypersensitive response. Mol. Plant Microbe Interact. 19, 567–576. doi: 10.1094/MPMI-19-0567
Gao, M., Wang, X., Wang, D., Xu, F., Ding, X., Zhang, Z., et al. (2009). Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6, 34–44. doi: 10.1016/j.chom.2009.05.019
Ghanbarnia, K., Fudal, I., Larkan, N. J., Links, M. G., Balesdent, M.-H., Profotova, B., et al. (2015). Rapid identification of the Leptosphaeria maculans avirulence gene AvrLm2 using an intraspecific comparative genomics approach. Mol. Plant Pathol. 16, 699–709. doi: 10.1111/mpp.12228
Goodin, M. M., Zaitlin, D., Naidu, R. A., and Lommel, S. A. (2008). Nicotiana benthamiana: its history and future as a model for plant–pathogen interactions. Mol. Plant Microbe Interact. 21, 1015–1026. doi: 10.1094/MPMI-21-8-1015
Gout, L., Fudal, I., Kuhn, M.-L., Blaise, F., Eckert, M., Cattolico, L., et al. (2006). Lost in the middle of nowhere: the AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans. Mol. Microbiol. 60, 67–80. doi: 10.1111/j.1365-2958.2006.05076.x
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Lacomme, C., and Santa Cruz, S. (1999). Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc. Natl. Acad. Sci. U.S.A. 96, 7956–7961. doi: 10.1073/pnas.96.14.7956
Larkan, N. J., Lydiate, D. J., Parkin, I. A. P., Nelson, M. N., Epp, D. J., Cowling, W. A., et al. (2013). The Brassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol. 197, 595–605. doi: 10.1111/nph.12043
Larkan, N. J., Ma, L., and Borhan, M. H. (2015). The Brassica napus receptor-like protein RLM2 is encoded by a second allele of the LepR3/Rlm2 blackleg resistance locus. Plant Biotechnol. J. 13, 983–992. doi: 10.1111/pbi.12341
Liebrand, T. W., Van Den Berg, G. C., Zhang, Z., Smit, P., Cordewener, J. H., America, A. H., et al. (2013). Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci. U.S.A. 110, 10010–10015. doi: 10.1073/pnas.1220015110
Liebrand, T. W. H., Van Den Burg, H. A., and Joosten, M. H. A. J. (2014). Two for all: receptor-associated kinases SOBIR1 and BAK1. Trends Plant Sci. 19, 123–132. doi: 10.1016/j.tplants.2013.10.003
Liu, Y., Schiff, M., Marathe, R., and Dinesh-Kumar, S. P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. doi: 10.1046/j.1365-313X.2002.01297.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, L., Cornelissen, B. J. C., and Takken, F. L. W. (2013). A nuclear localization for Avr2 from Fusarium oxysporum is required to activate the tomato resistance protein I-2. Front. Plant Sci. 4:94. doi: 10.3389/fpls.2013.00094
Ma, L., Lukasik, E., Gawehns, F., and Takken, F. L. (2012). The use of agroinfiltration for transient expression of plant resistance and fungal effector proteins in Nicotiana benthamiana leaves. Methods Mol. Biol. 835, 61–74. doi: 10.1007/978-1-61779-501-5_4
Nakagawa, T., Suzuki, T., Murata, S., Nakamura, S., Hino, T., Maeo, K., et al. (2007). Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71, 2095–2100. doi: 10.1271/bbb.70216
Postma, J., Liebrand, T. W. H., Bi, G., Evrard, A., Bye, R. R., Mbengue, M., et al. (2015). The Cf-4 receptor-like protein associates with the BAK1 receptor-like kinase to initiate receptor endocytosis and plant immunity. BioRxiv doi: 10.1101/019471
Raman, H., Raman, R., and Larkan, N. (2013). “Genetic dissection of blackleg resistance loci in rapeseed (Brassica napus L.),” in Plant Breeding from Laboratories of Fields, ed. S. B. Andersen (Rijeka: InTech.), 85–120.
Ratcliff, F., Martin-Hernandez, A. M., and Baulcombe, D. C. (2001). Technical advance: tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25, 237–245. doi: 10.1046/j.0960-7412.2000.00942.x
Rivas, S., and Thomas, C. M. (2005). Molecular interactions between tomato and the leaf mold pathogen Cladosporium fulvum. Annu. Rev. Phytopathol. 43, 395–436. doi: 10.1146/annurev.phyto.43.040204.140224
Rouxel, T., and Balesdent, M. H. (2005). The stem canker (blackleg) fungus, Leptosphaeria maculans, enters the genomic era. Mol. Plant Pathol. 6, 225–241. doi: 10.1111/j.1364-3703.2005.00282.x
Rouxel, T., Grandaubert, J., Hane, J. K., Hoede, C., Van De Wouw, A. P., Couloux, A., et al. (2011). Effector diversification within compartments of the Leptosphaeria maculans genome affected by Repeat-Induced Point mutations. Nat. Commun. 2:202. doi: 10.1038/ncomms1189
Stergiopoulos, I., Van Den Burg, H. A., Ökmen, B., Beenen, H. G., Van Liere, S., Kema, G. H. J., et al. (2010). Tomato Cf resistance proteins mediate recognition of cognate homologous effectors from fungi pathogenic on dicots and monocots. Proc. Natl. Acad. Sci. U.S.A. 107, 7610–7615. doi: 10.1073/pnas.1002910107
Thomma, B. P., Nurnberger, T., and Joosten, M. H. (2011). Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23, 4–15. doi: 10.1105/tpc.110.082602
Utermark, J., and Karlovsky, P. (2008). Genetic transformation of filamentous fungi by Agrobacterium tumefaciens. Protoc. Exch. doi: 10.1038/nprot.2008.83
van der Hoorn, R. A., Laurent, F., Roth, R., and De Wit, P. J. (2000). Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol. Plant Microbe Interact. 13, 439–446. doi: 10.1094/MPMI.2000.13.4.439
van Esse, H. P., Bolton, M. D., Stergiopoulos, I., De Wit, P. J., and Thomma, B. P. (2007). The chitin-binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant Microbe Interact. 20, 1092–1101. doi: 10.1094/MPMI-20-9-1092
van Esse, H. P., Thomma, B. P., Van ’T Klooster, J. W., and De Wit, P. J. (2006). Affinity-tags are removed from Cladosporium fulvum effector proteins expressed in the tomato leaf apoplast. J. Exp. Bot. 57, 599–608. doi: 10.1093/jxb/erj044
Voinnet, O., Rivas, S., Mestre, P., and Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33, 949–956. doi: 10.1046/j.1365-313X.2003.01676.x
West, J. S., Kharbanda, P. D., Barbetti, M. J., and Fitt, B. D. L. (2001). Epidemiology and management of Leptosphaeria maculans (phoma stem canker) on oilseed rape in Australia. Can. Eur. Plant Pathol. 50, 10–27. doi: 10.1046/j.1365-3059.2001.00546.x
Zhang, L., Kars, I., Essenstam, B., Liebrand, T. W. H., Wagemakers, L., Elberse, J., et al. (2014). Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the Arabidopsis receptor-like protein responsiveness to botrytis polygalacturonases1. Plant Physiol. 164, 352–364. doi: 10.1104/pp.113.230698
Zhang, W., Fraiture, M., Kolb, D., Löffelhardt, B., Desaki, Y., Boutrot, F. F. G., et al. (2013). Arabidopsis RECEPTOR-LIKE PROTEIN30 and Receptor-Like Kinase SUPPRESSOR OF BIR1-1/EVERSHED Mediate Innate Immunity to Necrotrophic Fungi. Plant Cell 25, 4227–4241. doi: 10.1105/tpc.113.117010
Keywords: receptor-like protein, LepR3, SOBIR1, BAK1/SERK3, Brassica napus, Leptosphaeria maculans
Citation: Ma L and Borhan MH (2015) The receptor-like kinase SOBIR1 interacts with Brassica napus LepR3 and is required for Leptosphaeria maculans AvrLm1-triggered immunity. Front. Plant Sci. 6:933. doi: 10.3389/fpls.2015.00933
Received: 03 September 2015; Accepted: 15 October 2015;
Published: 29 October 2015.
Edited by:
Simone Ferrari, Sapienza Università di Roma, ItalyReviewed by:
Mahmut Tör, University of Worcester, UKAndrea A. Gust, University of Tübingen, Germany
Copyright © 2015 Ma and Borhan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. Hossein Borhan, hossein.borhan@agr.gc.ca
 Lisong Ma
Lisong Ma M. Hossein Borhan
M. Hossein Borhan