- 1Department of Chemistry and Biotechnology, Graduate School of Engineering, Tottori University, Tottori, Japan
- 2RIKEN Center for Sustainable Resource Science, Yokohama, Japan
- 3Graduate School of Environmental and Life Science, Okayama University, Okayama, Japan
- 4Faculty of Agriculture, Tottori University, Tottori, Japan
Chitin, an N-acetyl-D-glucosamine polymer, is a component of fungal cell walls and a microbe/pathogen-associated molecular pattern that elicits plant defense responses. As polymeric chitin is difficult to handle due to its insolubility in water, many studies on chitin-induced immune responses have used water-soluble low-molecular weight chitin instead. Thus, it is unclear if polymeric chitin can induce resistance. Here, we examined the elicitor activity of chitin nanofiber (CNF) of submicron thickness prepared from polymeric chitin. CNF showed a high dispersing ability in water and induced both reactive oxygen species (ROS) production and chitin-induced defense-related gene expression in Arabidopsis thaliana seedlings. The Arabidopsis chitin elicitor receptor kinase 1 (Atcerk1) mutant, which is impaired in chitin perception, also failed to respond to CNF. CNF exposure triggered ROS generation in suspension-cultured cells from Oryza sativa. Furthermore, pre-treatment of Arabidopsis leaves with CNF effectively reduced pathogen infection by both the fungus Alternaria brassicicola and the bacterium Pseudomonas syringae pv. tomato DC3000. These results demonstrate that CNF has elicitor activity and will help define the role of polymeric chitin in plant immune responses.
Introduction
Chitin, an N-acetyl-D-glucosamine polymer, is a component of fungal cell walls and arthropod exoskeletons. As a biocompatible and biodegradable eco-friendly biopolymer, chitin has several promising applications in various fields. Due to its high nitrogen content and low C/N ratio, chitin can be used as a fertilizer or soil amendment to enhance crop growth. Moreover, chitin is expected to improve crop yields due to its ability to induce plant defense systems against pests and pathogens (Sharp, 2013).
Plant defense systems are activated in response to chitin in fungal (potential pathogen) cell walls, which is perceived as a microbe- or pathogen-associated molecular pattern (MAMP/PAMP). Bacterial flagellin, elongation factor Tu, lipopolysaccharides, and peptidoglycan are other examples of PAMPs, which are often highly conserved, constitutively expressed, and essential components of microbes (Antolin-Llovera et al., 2012). The recognition of PAMPs by pattern recognition receptors (PRRs) present at the plant cell surface induces PAMP-triggered immunity (PTI) (Antolin-Llovera et al., 2012).
The first plant PRR for chitin was identified in Oryza sativa (rice). Chitin elicitor binding protein (OsCEBiP) is a receptor-like protein (RLP) that contains an extracellular chitin-binding lysin motif (LysM) but lacks a known intracellular signaling domain (Kaku et al., 2006). OsCEBiP forms a complex with chitin elicitor receptor kinase1 (OsCERK1), a receptor-like kinase (RLK) that contains an active intracellular kinase domain, to initiate chitin signaling (Shimizu et al., 2010; Shinya et al., 2012). In Arabidopsis, AtCERK1 (also known as RLK1/LYK1) is essential for chitin signaling (Miya et al., 2007; Wan et al., 2008). AtCERK1 is an RLK that contains extracellular LysMs as well as an intracellular kinase domain. Chitin induces dimerization of AtCERK1 and activates immune responses, such as the generation of reactive oxygen species (ROS), activation of mitogen-activated protein kinases, and expression of defense-related genes (Miya et al., 2007; Wan et al., 2008). The biological activity of chitin elicitor depends on their size, with chitin heptamers to octamers showing high PAMP activity (Liu et al., 2012).
While several studies have examined chitin-induced PTI systems, chitins have not been widely used in practical applications. Because polymeric chitin is not soluble in most organic and inorganic solvents due to its high crystallinity (Pillai et al., 2009), many studies of chitin-induced PTI are based on water-soluble low-molecular weight chitin-oligosaccharides. Chitin only becomes soluble in water once it is costly degraded or chemically modified. Despite its huge availability, theses handling difficulties of polymeric chitin is a major obstacle for the utilization.
We recently developed chitin nanofiber (CNF) from polymeric chitin extracted from crab shell and mushrooms (Ifuku et al., 2009, 2011, 2012). Exoskeletons of crustaceans consist of CNFs. Chitin nanofibrils (∼3 nm in diameter) are embedded in a protein matrix and assemble into fibers (∼60 nm in diameter), and further these fibers assemble into micro-size bundles (Chen et al., 2008). Similarly, fungal cell walls consist of CNFs, which form a complex with glucans (Zivanovic et al., 2003). Extracted CNF has a highly uniform structure of 10–20 nm thickness and shows high dispersing ability in water due to its submicron size and high surface-to-volume ratio (Ifuku et al., 2011, 2012). In this study, we demonstrated that the polymeric CNF has elicitor activity in plants. We found that CNF induced ROS production and expression of defense genes and reduced pathogen infection in Arabidopsis and rice, similarly to chitin-oligosaccharide elicitors. We show that nanofibrillated chitin has useful applications for plant disease control.
Materials And Methods
Preparation of Chitin Nanofibers
Chitin powder from crab shell was purchased from Koyo Chemical (Tottori, Japan). CNFs were prepared without acetic acid as described previously (Ifuku et al., 2012). Briefly, dry chitin powder was dispersed in water at 1 wt.% and passed through a high pressure water-jet system (Star Burst Mini, HJP-25001S, Sugino Machine, Toyama, Japan) equipped with a ball-collision chamber for mechanical disintegration. Chitin-oligosaccharides (GlcNAc)2-6 and purified N-acetylchitohexaose (GlcNAc)6 were purchased from Yaizu Suisankagaku industry (Shizuoka, Japan).
Plant Materials
Arabidopsis thaliana, ecotype Columbia (Col-8) and cerk1-2 (GABI_096F09) were used. For inoculation tests, Arabidopsis plants were grown on sterilized soil [1:1 mixture of Supremix A (Sakata Seed Co., Yokohama, Japan), vermiculite] under controlled environmental conditions with 8 h light/16 h dark cycles at 22°C. For ROS assays and qRT-PCR, Arabidopsis seedlings were grown in liquid MGRL medium with 0.1% sucrose (Albert et al., 2006) at 22°C under continuous light for 10 days. Suspension-cultured rice cells derived from seed scutella of Oryza sativa japonica ‘Nipponbare’ were used. The rice cells were maintained using liquid L medium (Kuchitsu et al., 1993) on a rotary shaker at 25°C under dark conditions as described previously (Nakagami et al., 2010).
Oligomeric Chitin Analysis
Oligomeric chitin in CNF was detected by HPLC analysis as described by Sashiwa et al. (2003). The water-soluble fraction from a suspension of chitin powder in water (10 mg/mL) and the filtrate from a CNF dispersant (10 mg/mL) through a Millex-HA filter (Merk Millipore, Darmstadt, Germany) were analyzed. Chitin-oligosaccharides [(GlcNAc)2-6] (10 mg/mL) were dissolved in water and used as a positive control. HPLC analysis was performed using a Hitachi HPLC system (Hitachi, Tokyo, Japan) equipped with a L-7100 pump, L-7200 autosampler, and D-7400 UV detector and conducted on a Shodex Asahipak NH2P-50 column with CH3CN/H2O (7:3, v/v) with the following settings: injection, 0.1 mL sample/CH3CN (1:2, v/v); flow rate = 1.0 mL/min; and UV detection at 210 nm.
Chitinase Assay
Enzymatic degradation of chitin was analyzed by chitinase assay with Schales’ method as described by Ferrari et al. (2014). Un-nanofibrillated and nanofibrillated chitin (1 mg/mL) were incubated with chitinase (1.2 U, Wako Pure Chemicals Industries Ltd., Osaka, Japan) in 50 mM KPi buffer (pH6.0) at 30°C. Reactions were centrifuged at 4°C and 100 μL supernatant was mixed with 200 μL Schales’ regent (0.5 M sodium carbonate, 0.5 g/L potassium ferricyanide). The samples were incubated at 100°C for 15 min under dark conditions, and absorbance was then measured at 420 nm.
ROS Assay
Three 10-day-old Arabidopsis seedlings were incubated in liquid MGRL medium supplemented with 0.1% sucrose containing 100 μM L-012 (Wako, Japan) for 2 h at 22°C under darkness, and then transferred to liquid MGRL medium containing 0.1% sucrose and chitin-oligosaccharides or CNF. ROS production was determined by counting photons derived from L-012–mediated chemiluminescence using a TriStar LB942 microplate reader (Berthold technologies, Germany). Similarly, 40 mg rice cells was incubated with liquid L medium containing 1 mM L-012 for 2 h at 25°C under dark conditions, and then with liquid L medium containing chitin-oligosaccharides or CNF and horseradish peroxidase (final conc. 1 unit, Sigma–Aldrich, USA).
RNA Isolation and qRT-PCR Analysis
Arabidopsis seedlings (10-day-olds) were treated with 0.1 mg/mL CNF or water. Samples were harvested 1 h after treatment and frozen immediately. Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen, Netherlands) and cDNA was prepared using the ReverTra Ace Reverse Transcription Kit (Toyobo, Japan). Quantitative real-time PCR (qRT-PCR) was performed using the Mx3000P QPCR system (Agilent Technologies, Santa Clara, CA, USA) with Thunderbird SYBR qPCR Mix (Toyobo, Japan). Data were analyzed using an in-house script written in the R language as described by Tsuda et al. (2013). The gene-specific primers used were as follows: FRK1 (At2g19190) FW 5′-ACGGGCATAGTTCCACAAAG-3′, FRK1 RV 5′-CGTCAAAAGAACGACGATGA-3′; CYF81F (At5g57220) FW 5′-AATGGAGAGAGCAACACAATG-3′, CYF81F RV 5′-ATACTGAGCATGAGCCCTTTG-3′; WRKY22 (At4g01250) FW 5′-TCCTTCGGAGAGATTCGAGA-3′, WRKY22 RV 5′-CTGCTGCTACATGGCACACT-3′; ZAT10 (At1g27730) FW 5′-TGTCACGCAACTTCCTTCT-3′, ZAT10 RV 5′-TGGTGTCACTTTATGCTTATTC-3′; lectin-like protein gene (At3g16530) FW 5′-ACAATGCAGATTCACAAACTC-3′, lectin-like protein gene RV 5′-GCAAACGATACCTAGCCAA-3′; Actin-2 (At3g18780) FW 5′-GTTGGTGATGAAGCACAATCCAAG-3′, Actin-2 RV 5′-CTGGAACAAGACTTCTGGGCATCT-3′.
Pathogen Inoculation
Arabidopsis plants were sprayed with distilled water, 1 mg/mL chitin-oligosaccharides [(GlcNAc)2-6], or 0.1 or 1 mg/mL CNF (including 0.01% silwet L-77) 24 h before pathogen inoculation. Alternaria brassicicola isolate O-264 was maintained on potato dextrose agar medium. O-264 was incubated on V-8 juice agar for 2–3 days at 25°C in the dark and spores were obtained. Droplets (10 μL) of O-264 spore suspension (104 spores/mL) were placed on the leaf surface. Inoculated plants were kept under high humidity conditions in a moist chamber with a 10-h photoperiod at 22°C and lesion formation was observed 4 days post inoculation. Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) was grown on KB medium containing rifampicin (50 μg/mL). Prior to inoculation, bacteria were suspended in 10 mM MgCl2 to a density of OD600 = 0.0002. Arabidopsis leaves were syringe-infiltrated with bacterial suspension. Inoculated plants were incubated in a moist chamber under a 10-h photoperiod at 22°C. To determine the bacterial population, inoculated leaves were harvested and cut into 1-cm2 samples at 3 days post inoculation. Samples were homogenized in 10 mM MgCl2 and a series of diluted samples were plated on KB medium containing rifampicin and cycloheximide (50 μg/mL). The number of colonies per plate were counted.
Results
CNF Consists of Polymeric Chitin
We previously described the preparation of CNF using the Star Burst system, which employs high-pressure water jet technology (Ifuku et al., 2012). In this process, chitin powder dispersed in water is passed through the Star Burst system under high pressure and atomized via collision with a ceramic ball. After these mechanical treatments, CNF of 10–20 nm thickness is obtained in slurry form and is highly dispersed in water (Supplementary Figure S1). We performed high-performance liquid chromatography (HPLC) to assess whether chitin fibrils were reduced in length as well as thickness during this mechanical process. There were no obvious peaks of chitooligosaccharides from the aqueous fraction of un-nanofibrillated chitin powder (Figure 1A). Moreover, filtrate from CNF did not contain oligomeric chitin (Figure 1A). These results confirm that chitin was not disintegrated in length during preparation and that CNF consisted of polymeric chitin.
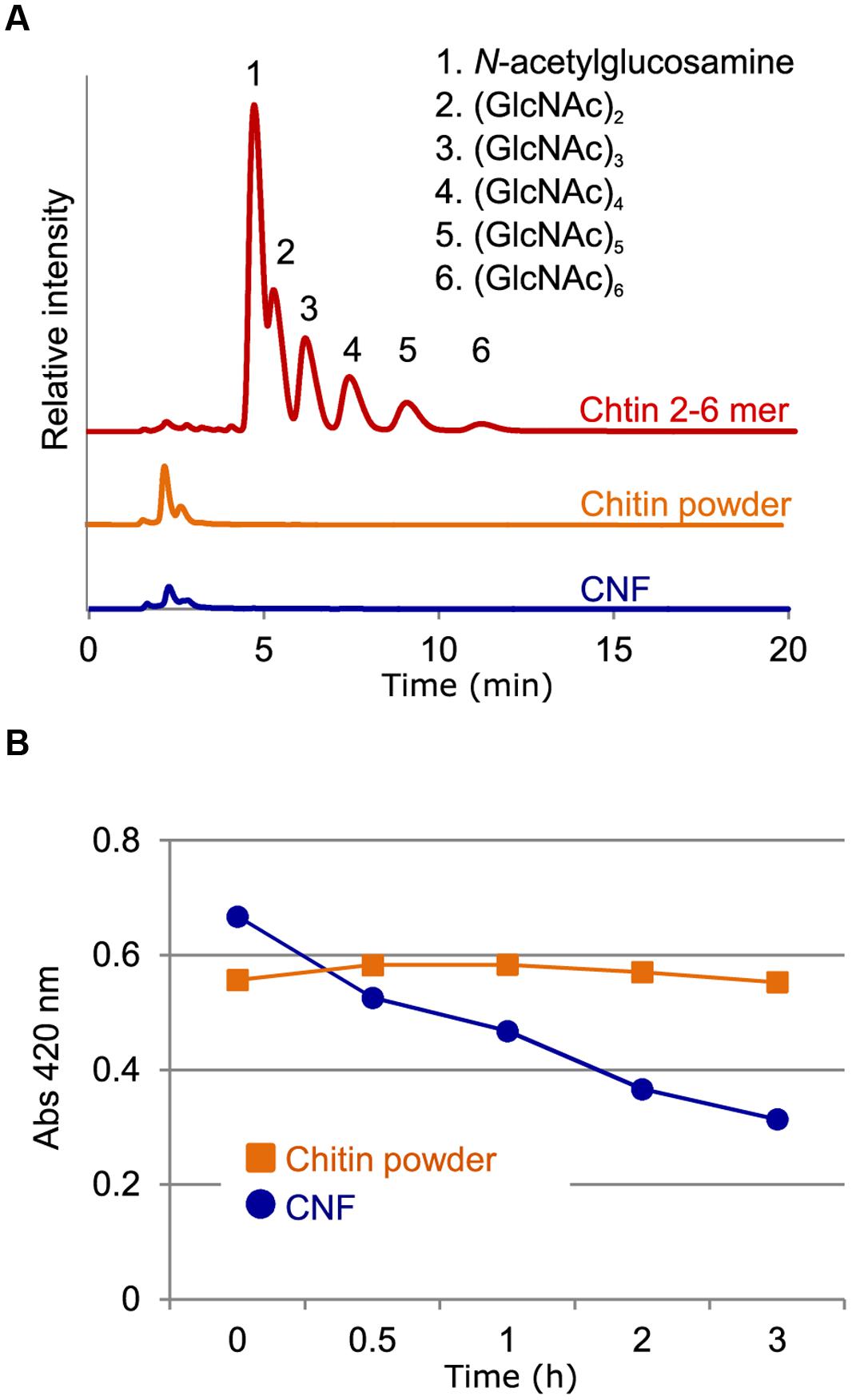
FIGURE 1. Characterization of chitin nanofiber. (A) Chitin nanofiber consists of long polymeric chitin. The water-soluble fraction from chitin powder suspension (chitin powder) and chitin nanofiber dispersant (CNF) were subjected to HPLC analysis. Chitin-oligosaccharides (GlcNAc)2-6 (Chitin 2-6 mer) were used as the positive control. (B) Comparison of chitinolytic enzyme sensitivity. Enzymatic degradation of chitin before (Chitin powder) and after nanofibrillation (CNF) was analyzed using a chitinase assay. Similar result obtained in four independent experiments and the data show one representative.
CNF is Rapidly Degraded by Chitinase
The unique properties of nanofibers come from their nanoscale size and high specific surface area, which provide greater access to the constituent molecules. We conducted chitinase assays to assess whether nanofibrillated chitin had increased sensitivity to chitinolytic enzymes. Whereas un-nanofibrillated chitin powder was not degraded over the course of 3 h, CNF was rapidly degraded (Figure 1B). These results suggest that in contrast to un-nanofibrillated chitin, which consists of chitin aggregates, CNF is composed of loosened chitin fibers that provide increased access to chitinase for degradation.
CNF Induces ROS Production
Chitin elicitor is known to elevate ROS levels in Arabidopsis (Miya et al., 2007). As shown in Figure 2A, treatment with chitin-oligosaccharides [(GlcNAc)2-6] induced ROS production in Arabidopsis seedlings. We found that CNF was also capable of inducing ROS generation and that the induction was faster and higher than with chitin oligomers in wild-type Col-8. CNF-induced ROS generation was abolished in the cerk1-2 mutant, which is impaired in chitin recognition (Figure 2A), indicating that CNF has elicitor activity mediated by the chitin receptor CERK1 in Arabidopsis. Maximal activation of innate immunity requires long-chain chitin oligomers (Liu et al., 2012). Whereas purified N-acetylchitohexaose (GlcNAc)6 induced the generation of more ROS than did CNF at the same concentration (Supplementary Figure S2), both CNF and (GlcNAc)6 induced ROS generation in a dose-dependent manner. Chitin-oligosaccharide elicitor induces biphasic generation of ROS in suspension-cultured rice cells (Yamaguchi et al., 2005). Here, we found that treatment of cultured rice cells with CNF induced biphasic ROS generation (Figure 2B). In contrast to our findings in Arabidopsis, ROS generation in cultured rice cells was higher for CNF than for (GlcNAc)6. These results indicate that polymeric CNF can be recognized by plants to trigger ROS production.
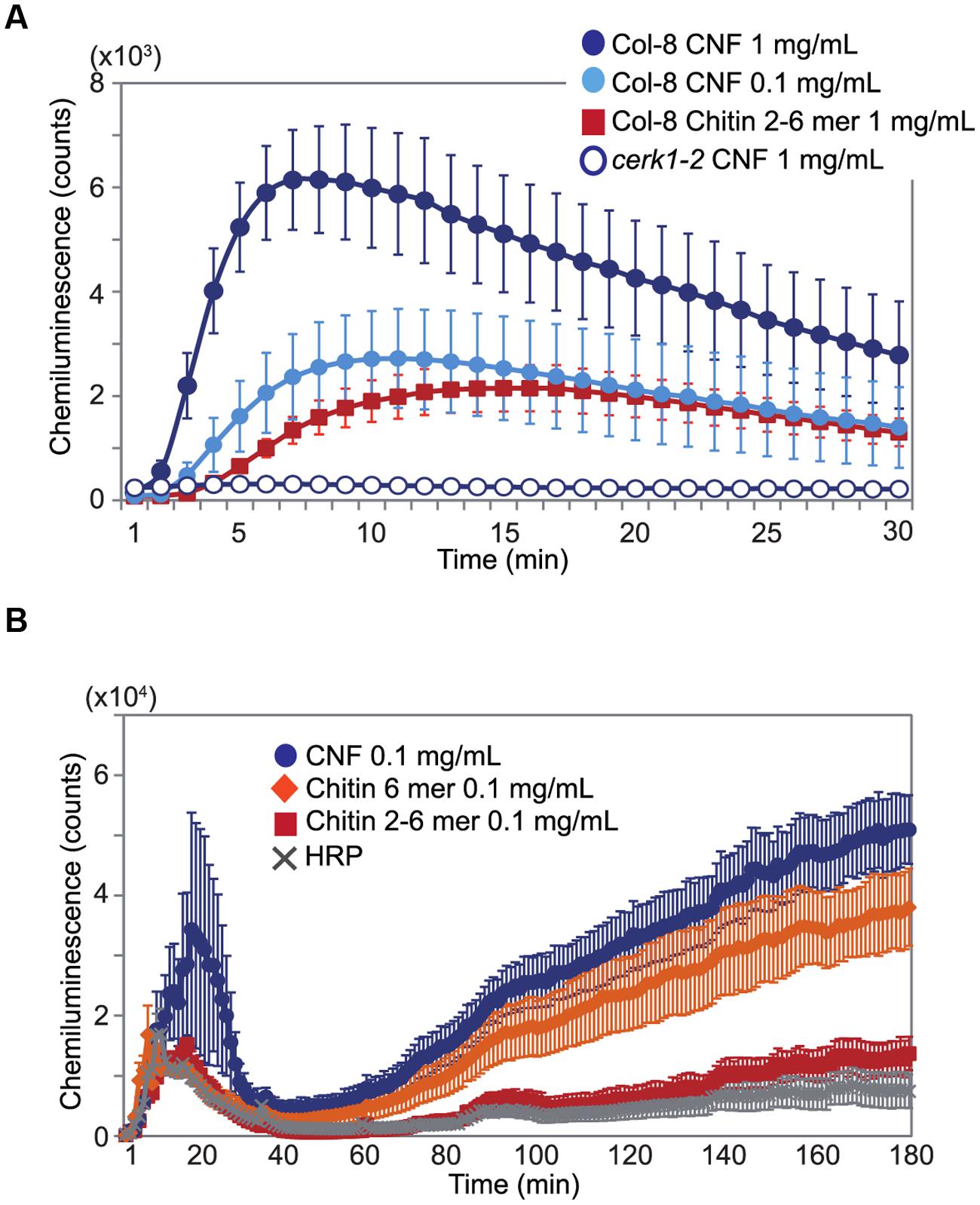
FIGURE 2. Generation of reactive oxygen species (ROS) by chitin nanofiber treatment. ROS production was measured by chemiluminescence mediated by L-012 after elicitor treatment. (A) ROS production in 10-day-old Arabidopsis seedlings. Wild-type Col-8 or cerk1-2 mutant plants was treated with 1 mg/mL or 0.1 mg/mL chitin nanofiber (CNF) or 1 mg/mL chitin-oligosaccharides (GlcNAc)2-6 (Chitin 2–6 mer). (B) ROS production in suspension-cultured rice cells treated with 0.1 mg/mL CNF, chitin-oligosaccharides (GlcNAc)2-6 (Chitin 2–6 mer) or N-acetylchitohexaose (Chitin 6 mer). HRP: horseradish peroxidase. The values represent the average and standard errors of six replicate experiments.
CNF Induces Chitin-inducible Gene Expression
Expression of defense-related genes such as lectin-like protein gene, Zat10, WRKY22, FRK, and CYP81F2 are up-regulated by chitin-oligosaccharide treatment (Ramonell et al., 2002; Zhang et al., 2002). To investigate whether long-chain polymeric chitin induces the expression of defense-related genes, we examined the expression of these genes in Arabidopsis seedlings 1 h after CNF treatment by quantitative reverse transcriptase-PCR (qRT-PCR). As shown in Figure 3, all selected chitin-responsive genes were significantly up-regulated by CNF treatment in Col-8. By contrast, the expression of these genes was not responsive to CNF treatment in the cerk1-2 mutant. These results indicate that CNF is an active elicitor capable of enhancing defense-related gene expression in a CERK1-dependent manner.
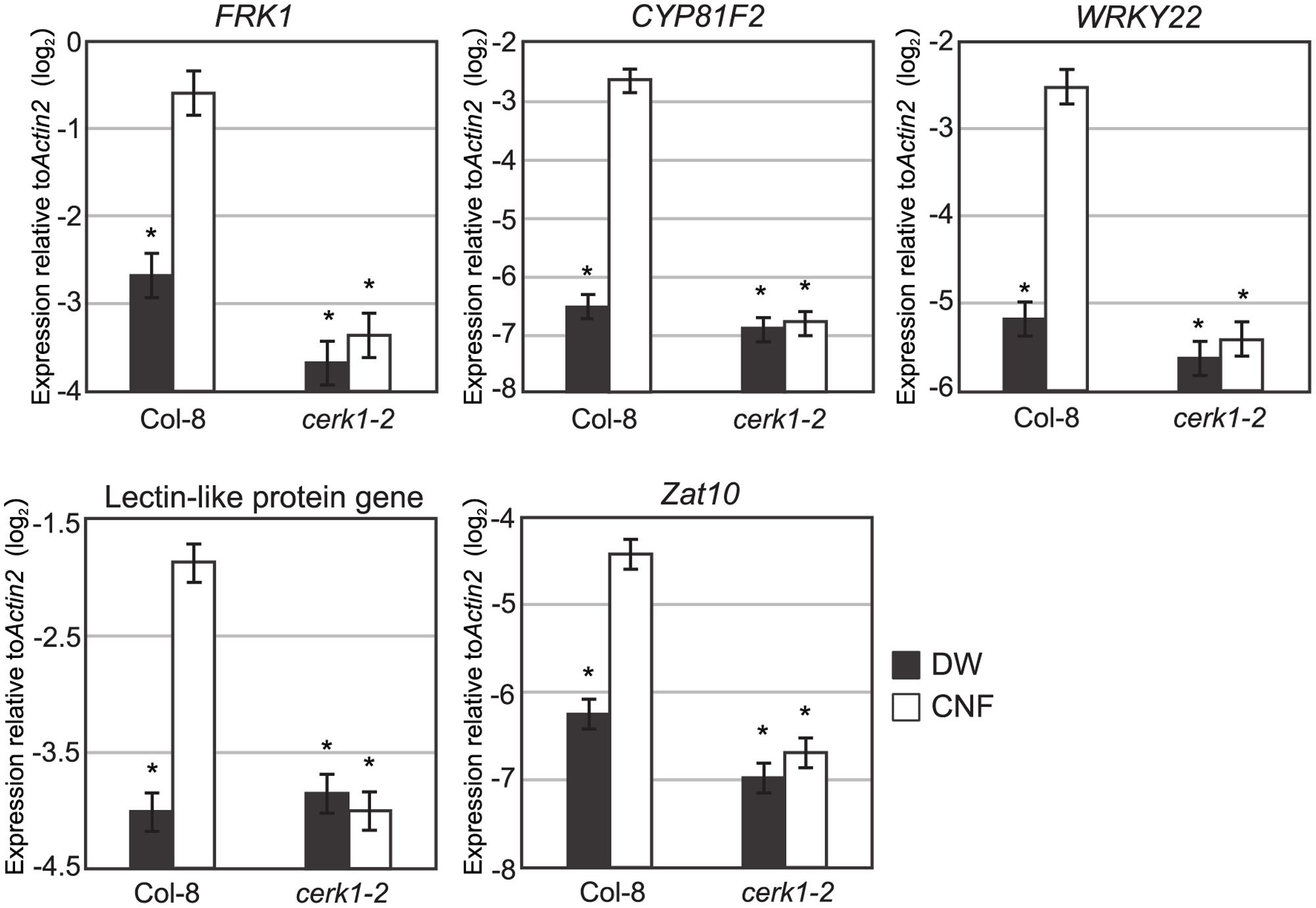
FIGURE 3. Chitin-responsive genes in Arabidopsis respond to chitin nanofiber. Ten-day-old seedlings of wild-type Col-8 or cerk1-2 mutant were treated with water (DW) or 0.1 mg/mL chitin nanofiber (CNF). Expression of chitin-responsive genes was measured by qRT-PCR. Data represent means and standard errors of two biological replicates calculated by the mixed linear model. Asterisks indicate significant differences from CNF-treated Col-8 (P < 0.01, two-tailed t-tests).
CNF Reduces Fungal and Bacterial Disease Symptoms
Chitin treatment induces resistance in host plants against both fungal and bacterial disease (Wan et al., 2008; Gimenez-Ibanez et al., 2009). To assess the effect of pre-treatment with long-chain polymeric CNF on pathogen infection, we inoculated Arabidopsis with the fungal pathogen A. brassicicola or bacterial pathogen Pst DC3000. Because of its high dispersing ability in water, we were able to apply CNF homogenously by spraying. Necrotic lesion formation upon A. brassicicola infection was reduced on leaves that were pre-treated with chitin-oligosaccharides [(GlcNAc)2-6] or CNF compared with control leaves (Figure 4A). Pre-treatment with 1 mg/mL or 0.1 mg/mL CNF significantly reduced lesion formation and the reduction was greater for the higher concentration of CNF.
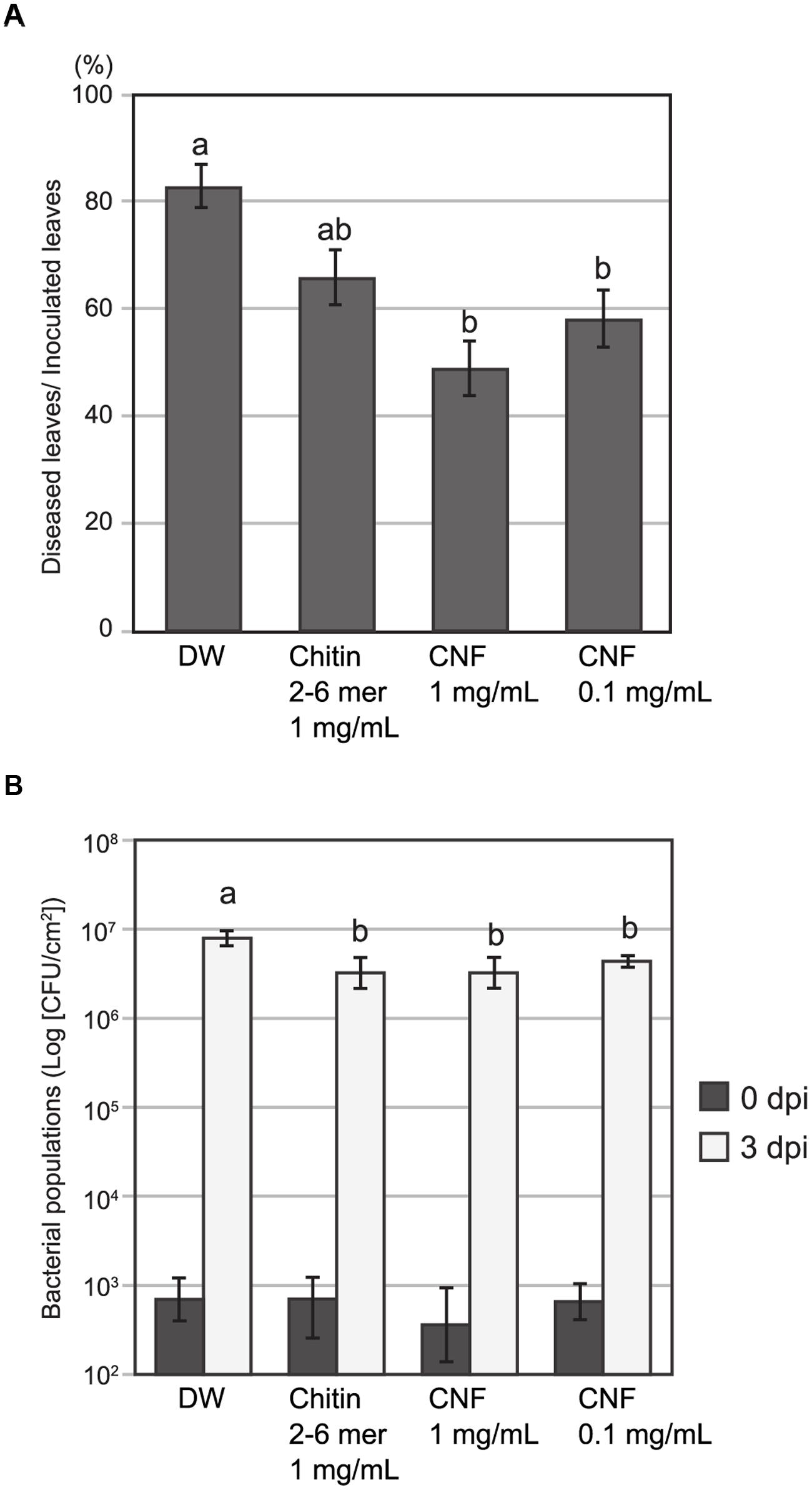
FIGURE 4. Pre-treatment with chitin nanofiber reduces fungal and bacterial disease in Arabidopsis. Plants were sprayed 24 h before pathogen inoculation with chitin nanofiber (CNF) (1 or 0.1 mg/mL), chitin-oligosaccharides (GlcNAc)2-6 (Chitin 2–6 mer) (1 mg/mL), or distilled water (DW). (A) Pre-treated Arabidopsis leaves were inoculated with fungal pathogen A. brassicicola. Lesion formation was observed 4 days after inoculation. Data indicate the rate of diseased leaves per all inoculated leaves and represent the mean and standard errors of six independent experiments. Means with the same letter are not significantly different according to Tukey’s test (P < 0.05). (B) Pre-treated Arabidopsis leaves were infiltrated with bacterial pathogen Pseudomonas syringae pv. tomato DC3000. Leaf disks were collected 0 and 3 days after inoculation to determine bacterial growth. Data represent the means and standard errors of three independent experiments. Means with the same letter at 3 dpi are not significantly different according to Tukey’s test (P < 0.1).
To examine resistance to bacterial pathogens, we pre-treated Arabidopsis leaves with oligomeric chitin or polymeric CNF before infiltrating them with bacterial suspension. The population of Pst DC3000 at day 0 was not different and increased in all leaves 3 days post inoculation. However, the bacterial population in leaves pre-treated with chitin elicitors was slightly, but significantly, lower than that in the control leaves (Figure 4B). These results suggest that CNF is an effective elicitor for reducing infection of both fungal and bacterial pathogens.
Discussion
Chitin elicitor triggers ROS generation, defense gene expression, ion flux, phytoalexin production, and disease resistance in both dicot and monocot plants (Shibuya and Minami, 2001). In this study, we showed that polymeric CNF is capable of inducing ROS generation and chitin-responsive gene expression (Figures 2 and 3) as well as resistance against pathogen infection in Arabidopsis (Figure 4). These results indicate that plants can recognize and respond to long chain polymeric chitin. Previous studies have reported that AtCERK1 binds to polymeric chitin and plays an essential role in chitin signaling in Arabidopsis (Petutsching et al., 2010; Wan et al., 2012). In our study, the ROS generation and gene expression responses to CNF were impaired in the cerk1-2 mutant (Figures 2 and 3). These findings demonstrate that CNF induces PTI through CERK1, similarly to chitin-oligosaccharide. Although AtCERK1 was proposed to function both in chitin perception and signaling, despite its low chitin binding affinity (Liu et al., 2012; Cao et al., 2014), AtLYK5, which has a higher chitin binding affinity, was recently shown to be the primary chitin receptor (Cao et al., 2014). AtLYK5 is required for AtCERK1 dimerization and phosphorylation in a chitin-dependent manner (Cao et al., 2014). Polymeric chitin was bound to AtLYK5 in vitro (Petutsching et al., 2010), and it will be interesting to explore the use of CNF as a ligand in future research.
Chitin-oligosaccharide elicitor induces resistance to the fungi A. brassicicola and Erysiphe cichorasearum in Arabidopsis (Wan et al., 2008) and to Magnaporthe grisea in rice (Kouzai et al., 2014). Furthermore, chitin-oligosaccharide elicitor is also effective against bacterial pathogens (Wan et al., 2008); even though bacterial pathogens do not contain chitin, chitin elicitor signaling through AtCERK1 has an apparent effect on PTI against Pst DC3000 infection (Gimenez-Ibanez et al., 2009). We showed here that CNF efficiently reduced both A. brassiciola and Pst DC3000 infection (Figure 4). Despite the diversity of PAMPs and its corresponding PRRs, PTI events are largely overlapping (Antolin-Llovera et al., 2012). PTI is generally effective against non-specific and wide range of pathogens. Because public concern for environmental and biological systems is growing, ideal disease management should be safe for human and animals and eco-friendly. MAMPs are candidate substances for sustainable crop protection (Burketova et al., 2015). We show that nanofibrillated chitin could be practical material for plant disease control.
While the ability of chitin to induce resistance in plants is evident, chitin has not been widely used in agricultural applications. There have been a few attempts to use chitinous waste from edible mushrooms and crustaceans in agriculture for nutrition or soil amendment to enhance crop growth (Sharp, 2013). However, the application of chitinous compost in open fields had no discernable effect on disease control. Chitin must first be released from complex with protein or glucans in chitinous waste and nanofibrous structure or oligomeric fragment of chitin can be recognized by plants. Some reports have suggested that degradation of polymeric chitin to oligomeric chitin is required for recognition by PRRs (Stacey and Shibuya, 1997; Shibuya and Minami, 2001). As it would take time for chitin fragments to be released from compost, any effects of chitinous compost on disease resistance would likely be slow-acting. We demonstrated here that CNF was degraded by chitinase more rapidly than was un-nanofibrillated chitin (Figure 1B). Chitin fragments could be released and recognized by PRRs soon after CNF treatment; therefore, CNF may be a useful fast-acting elicitor. In addition, it was suggested that smaller fragments of chitin are not absolutely required for chitin recognition on account of strong binding of polymeric chitin to AtCERK1 (Petutsching et al., 2010; Cao et al., 2014). The timing of ROS generation induced by CNF was comparable with that induced by chitin-oligosaccharides (Figure 2), which suggests that polymeric CNF could be directly recognized by plant PRR. These findings indicate that the CNF nanostructure allows PRRs rapid access to polymeric CNF for initiation of PTI.
In summary, we have demonstrated that nanofibrillated polymeric chitin shows elicitor activity to induce ROS production and defense-related gene expression. Further, CNF effectively reduced the symptoms of both fungal and bacterial infection. Thus, using nanofibrillation to produce CNF of submicron size and high surface-to-volume ratio, and therefore much greater dispersibility in water, makes it possible to elucidate the elicitor activity of polymeric chitin. Our results also show that nanofibrillated chitin could be a useful and practical material for plant disease control in agriculture. Further study is needed to improve the material properties of CNF to enhance its elicitor activity for a broad range of host plants.
Author Contributions
ME and HM wrote the main manuscript text. ME, HM, SI, HN, and HK conceived and designed the experiments. ME, HM, TU, and SO performed research. All authors reviewed and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Hiroshi Otani (Tottori University) and Jeffery L. Dangl (University of North Carolina at Chapel Hill) for providing the fungal and bacterial strains, Naoto Shibuya (Meiji University) for providing the cerk1-2 mutant, and Kenichi Tsuda (Max Planck Institute for Plant Breeding Research) for technical assistance with the qRT-PCR analysis. This work was partially supported by the Adaptable and Seamless Technology Transfer Program through Target-driven R&D (A-step) of JST, The Creating STart-ups from Advanced Research and Technology (START) project of JST and by JSPS KAKENHI Grant Numbers 25850034 to HM, 24688007 and 15H01247 to HN.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.01098
References
Albert, P., Miya, A., Hiratsuka, K., Kawakami, N., and Shibuya, N. (2006). A high-throughput evaluation system for Arabidopsis mutants for defense signaling. Plant Biotechnol. 23, 459–466. doi: 10.5511/plantbiotechnology.23.459
Antolin-Llovera, M., Ried, M. K., Binder, A., and Parniske, M. (2012). Receptor kinase signaling pathways in plat-microbe interactions. Annu. Rev. Phytopathol. 50, 451–473. doi: 10.1146/annurev-phyto-081211-173002
Burketova, L., Trda, L., Ott, P. G., and Valentova, O. (2015). Bio-based resistance inducers for sustainable plant protection against pathogens. Biotechnol. Adv. 33, 994–1004. doi: 10.1016/j.biotechadv.2015.01.004
Cao, Y., Liang, Y., Tanaka, K., Nguyen, C. T., Jedrzejczak, R. P., Joachimiak, A., et al. (2014). The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. Elife 3, e03766. doi: 10.7554/eLife.03766
Chen, P.-U., Lin, A. Y.-M., McKittrick, J., and Meyers, A. (2008). Structure and mechanical properties of crab exoskeletons. Acta Biomater. 4, 587–596. doi: 10.1016/j.actbio.2007.12.010
Ferrari, A. R., Gaber, Y., and Fraaije, M. W. (2014). A fast, sensitive and easy colorimetric assay for chitinase and cellulose activity detection. Biotechnol. Biofuels 7, 37. doi: 10.1186/1754-6834-7-37
Gimenez-Ibanez, S., Hann, D. R., Ntoukakis, V., Petutschnig, E., Lipka, V., and Rathjen, J. P. (2009). AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19, 423–429. doi: 10.1016/j.cub.2009.01.054
Ifuku, S., Nogi, M., Abe, K., Yoshioka, M., Morimoto, M., and Saimoto, H. (2009). Preparation of chitin nanofibers with a uniform width as alpha-chitin from crab shells. Biomacromolecules 10, 1584–1588.
Ifuku, S., Nomura, R., Morimoto, M., and Saimoto, H. (2011). Preparation of chitin nanofibers from mushrooms. Materials 4, 1417–1425. doi: 10.3390/molecules191118367
Ifuku, S., Yamada, K., Morimoto, M., and Saimoto, H. (2012). Nanofibrillation of dry chitin powder by star burst system. J. Nanomater. 2012, 1–7. doi: 10.1155/2012/645624
Kaku, H., Nishizawa, Y., Ishii-Minami, N., Akimoto-Tomiyama, C., Dohmae, N., and Takio, K. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 11086–11091. doi: 10.1073/pnas.0508882103
Kouzai, Y., Nakajima, K., Hayafune, M., Ozawa, K., Kaku, H., Shibuya, N., et al. (2014). CEBiP is the major chitin oligomer-binding protein in rice and plays a main role in the perception of chitin oligomers. Plant Mol. Biol. 84, 519–528. doi: 10.1007/s11103-013-0149-6
Kuchitsu, K., Kikuyama, M., and Shibuya, N. (1993). N-Acetylchitooligosaccharides, biotic elicitor for phytoalexin production, induce transient membrane depolarization in suspension-cultured rice cells. Protoplasma 174, 79–81. doi: 10.1007/BF01404046
Liu, T., Liu, Z., Song, C., Hu, Y., Han, Z., She, J., et al. (2012). Chitin-induced dimerization activates a plant immune receptor. Science 336, 1160–1164. doi: 10.1126/science.1218867
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., and Shirasu, K. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 19613–19618. doi: 10.1073/pnas.0705147104
Nakagami, H., Sugiyama, N., Mochida, K., Daudi, A., Yoshida, Y., Toyoda, T., et al. (2010). Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 153, 1161–1174. doi: 10.1104/pp.110.157347
Petutsching, E. K., Jones, A. M. E., Serazetdinova, L., Lipka, U., and Lipka, V. (2010). The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 285, 28902–28911. doi: 10.1074/jbc.M110.116657
Pillai, C. K. S., Paul, W., and Sharma, C. P. (2009). Chitin and chitosan polymers: chemistry, solubility and fiber formation. Prog. Polym. Sci. 34, 641–678. doi: 10.1016/j.progpolymsci.2009.04.001
Ramonell, K. M., Zhang, B., Ewing, R. M., Chen, Y., Xu, D., Stacey, G., et al. (2002). Microarray analysis of chitin elicitation in Arabidopsis thaliana. Mol. Plant Pathol. 3, 301–311. doi: 10.1046/j.1364-3703.2002.00123.x
Sashiwa, H., Fujishima, S., Yamano, N., Kawasaki, N., Nakayama, A., Muraki, E., et al. (2003). Enzymatic production of N-acetyl-D-glucosamine from chitin. Degradation study of N-acetylchitooligosaccharide and the effect of mixing of crude enzymes. Carbohydr. Polym. 51, 391–395. doi: 10.1016/S0144-8617(02)00192-3
Sharp, R. G. (2013). A review of the application of chitin and its derivatives in agriculture to modify plant-microbial interactions and improve crop yields. Agronomy 3, 757–793. doi: 10.3390/agronomy3040757
Shibuya, N., and Minami, E. (2001). Oligosaccharide signalling for defence responses in plant. Physiol. Mol. Plant Pathol. 59, 223–233. doi: 10.1006/pmpp.2001.0364
Shimizu, T., Nakano, T., Takamizawa, D., Desaki, Y., Ishii-Minami, N., and Nishizawa, Y. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214. doi: 10.1111/j.1365-313X.2010.04324.x
Shinya, T., Motoyama, N., Ikeda, A., Wada, M., Kamiya, K., and Hayafune, M. (2012). Functional characterization of CEBiP and CERK1 homologs in Arabidopsis and rice reveals the presence of different chitin receptor systems in plants. Plant Cell Physiol. 53, 1696–1706. doi: 10.1093/pcp/pcs113
Stacey, G., and Shibuya, N. (1997). Chitin recognition in rice and legumes. Plant Soil 194, 161–169. doi: 10.1023/A:1004244104972
Tsuda, K., Mine, A., Bethke, G., Igarashi, D., Botanga, C. J., Tsuda, Y., et al. (2013). Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana. PLoS Genet. 9:e1004015. doi: 10.1371/journal.pgen.1004015
Wan, J., Tanaka, K., Zhang, X.-C., Son, G. H., Brechenmacher, L., Nguyen, T. H. N., et al. (2012). LYK4, a lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol. 160, 396–406. doi: 10.1104/pp.112.201699
Wan, J., Zhang, X.-C., Neece, D., Ramonell, K. M., Clough, S., and Kim, S.-Y. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20, 471–481. doi: 10.1105/tpc.107.056754
Yamaguchi, T., Minami, E., Ueki, J., and Shibuya, N. (2005). Elicitor-induced activation of phospholipases plays an important role for the induction of defense responses in suspension-cultured rice cells. Plant Cell Physiol. 46, 579–587. doi: 10.1093/pcp/pci065
Zhang, B., Ramonell, K., Somerville, S., and Stacey, G. (2002). Characterization of early, chitin-induced gene expression in Arabidopsis. Mol. Plant Microbe Interact. 15, 963–970. doi: 10.1094/MPMI.2002.15.9.963
Keywords: chitin, nanofiber, elicitor, Arabidopsis thaliana, rice, Alternaria brassicicola, Pseudomonas syringae pv. tomato DC3000
Citation: Egusa M, Matsui H, Urakami T, Okuda S, Ifuku S, Nakagami H and Kaminaka H (2015) Chitin Nanofiber Elucidates the Elicitor Activity of Polymeric Chitin in Plants. Front. Plant Sci. 6:1098. doi: 10.3389/fpls.2015.01098
Received: 24 September 2015; Accepted: 22 November 2015;
Published: 09 December 2015.
Edited by:
Andrea Genre, University of Turin, ItalyReviewed by:
Naoya Takeda, National Institute for Basic Biology, JapanEiichi Minami, National Institute of Agrobiological Sciences, Japan
Copyright © 2015 Egusa, Matsui, Urakami, Okuda, Ifuku, Nakagami and Kaminaka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hironori Kaminaka, kaminaka@muses.tottori-u.ac.jp
†These authors have contributed equally to this work.
 Mayumi Egusa
Mayumi Egusa Hidenori Matsui
Hidenori Matsui Takeshi Urakami1
Takeshi Urakami1 Hirofumi Nakagami
Hirofumi Nakagami Hironori Kaminaka
Hironori Kaminaka