- 1Jiangsu Key Laboratory for Bioresources of Saline Soils, Provincial Key Laboratory of Agrobiology, Institute of Agro-biotechnology, Jiangsu Academy of Agricultural Sciences, Nanjing, China
- 2Institute of Botany, Jiangsu Province and Chinese Academy of Sciences, Nanjing, China
- 3Department of Plant Physiology, Slovak Agricultural University, Nitra, Slovakia
- 4Key Laboratory of Coastal Biology and Bioresources Utilization, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai, China
Water is essential for plant growth and development. Water deficiency leads to loss of yield and decreased crop quality. To understand water transport mechanisms in plants, we cloned and characterized a novel tonoplast intrinsic protein (TIP) gene from soybean with the highest similarity to TIP2-type from other plants, and thus designated GmTIP2;3. The protein sequence contains two conserved NPA motifs and six transmembrane domains. The expression analysis indicated that this gene was constitutively expressed in all detected tissues, with higher levels in the root, stem and pod, and the accumulation of GmTIP2;3 transcript showed a significant response to osmotic stresses, including 20% PEG6000 (polyethylene glycol) and 100 μM ABA (abscisic acid) treatments. The promoter-GUS (glucuronidase) activity analysis suggested that GmTIP2;3 was also expressed in the root, stem, and leaf, and preferentially expressed in the stele of root and stem, and the core promoter region was 1000 bp in length, located upstream of the ATG start codon. The GUS tissue and induced expression observations were consistent with the findings in soybean. In addition, subcellular localization showed that GmTIP2;3 was a plasma membrane-localized protein. Yeast heterologous expression revealed that GmTIP2;3 could improve tolerance to osmotic stress in yeast cells. Integrating these results, GmTIP2;3 might play an important role in response to osmotic stress in plants.
Introduction
Lack of water resources is an important factor restricting the development of agriculture. Plant growth and development depend on water uptake and transport regulation across cellular membranes and tissues. In the past, it was thought that water moved across cell membranes by free diffusion through the lipid bilayer. However, its transport is now thought to be highly and selectively regulated by aquaporins. Aquaporins (AQPs) belong to the ancient major intrinsic protein (MIP) family found in animals, microbes, and plants. Since the discovery of AQP1 (Denker et al., 1988), many aquaporin genes have been found in plants including 35 AQPs in Arabidopsis (Quigley et al., 2002; Boursiac et al., 2005), 31 in Zea mays (Chaumont et al., 2001), and 33 in Oryza sativa (Sakurai et al., 2005). Guo et al. (2006) further analyzed the expression and function of the rice plasma membrane intrinsic protein (PIP) gene family. Other scholars found 23 AQPs in Physcomitrella patens (Danielson and Johanson, 2008), 37 in Solanum lycopersicum (Sade et al., 2009), 66 in soybean (Zhang et al., 2013), 47 in tomato (Reuscher et al., 2013), 71 in Gossypium hirsutum (Park et al., 2010), and 53 in Chinese cabbage (Tao et al., 2014). Plant AQPs can be categorized into major four subfamilies based on localization and expression patterns: plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), nodulin26-like intrinsic proteins (NIPs), small and basic intrinsic proteins (SIPs) (Chaumont et al., 2001; Kaldenhoff and Fischer, 2006), and uncategorized X intrinsic proteins (XIPs) (Danielson and Johanson, 2008).
AQPS play important roles in various physiological processes in plants, such as growth, development, and response to biotic and abiotic stresses. Srivastava et al. (2015) also reviewed the versatile functions of aquaporins as molecular conduits in the plant response to abiotic stresses. For example, Guenther and Roberts (2000) isolated two major intrinsic membrane proteins from Lotus japonicus, named LIMP1 and LIMP2. Functional analysis using the Xenopus oocytes system indicated that LIMP1 appeared to be a member of the TIP subfamily and LIMP2 was a nodulin 26 ortholog protein. Rodrigues et al. (2013) investigated a gene encoding a root-specific tonoplast intrinsic aquaporin (TIP) from Eucalyptus grandis named EgTIP2, whose expression was induced by PEG and mannitol treatments but was downregulated by abscisic acid, suggesting that EgTIP2 might be involved in the eucalyptus response to drought. Wang et al. (2014) cloned and characterized a tonoplast AQP gene (TsTIP1;2) from the halophyte Thellungiella salsuginea and reported that it mediated the transduction of both H2O and H2O2 across the membranes and might contribute to the survival of T. salsuginea under multiple stresses. Ligaba et al. (2011) studied the expression patterns of 7 MIP genes from barley under different abiotic stresses using quantitative real-time PCR (RT-PCR), indicating that abiotic stress modulates the expression of major intrinsic proteins in barley. Zelazny et al. (2007) by using FRET imaging analysis showed that plasma membrane aquaporins could interact to regulate their subcellular localization in living maize cells. Tomato SiTIP2;2 expressing in transgenic Arabidopsis could enhance the plant's tolerance to salt stress and interact with its homologous proteins SiTIP1;1 and SiTIP2;1 (Xin et al., 2014). Gao et al. (2010) overexpressed TaNIP, a putative aquaporin gene from wheat, and found that it could enhance salt tolerance in transgenic Arabidopsis. Wang et al. (2011) cloned the novel Glycine soja tonoplast intrinsic protein gene GsTIP2;1, and the overexpression of GsTIP2;1 in Arabidopsis repressed/reduced tolerance to salt and dehydration stress, suggesting that GsTIP2;1 might mediate stress sensitivity by enhancing water loss in plants.
In this study, a novel tonoplast intrinsic aquaporin from soybean, GmTIP2;3, was cloned and characterized. Protein structure analysis showed that GmTIP2;3 possesses typical aquaporin characteristics, such as six transmembrane domains and NPA motifs. The expression analysis indicated that it was constitutively expressed in all tissues tested, especially in the root, stem, and pod, and exhibited responses to ABA and PEG treatments at certain time points. Subcellular localization showed it to be localized in the cell plasma membrane. The promoter activity assay demonstrated that the core sequence for this gene was 1000 bp upstream from the ATG start codon. Yeast heterologous expression revealed that GmTIP2;3 could improve osmotic tolerance in yeast cells. Integrating these results, GmTIP2;3 plays an important role in response to osmotic stress in plants.
Materials and Methods
Plant Materials
Glycine max var. Willimas 82 was selected for the experiments, which included growth of seedlings, flowering, podding, extracting total RNA for GmTIP2;3 cloning, and tissue expression and induced expression analysis. Lotus japonicus was used to transfer the promoter sequence for activity testing and Arabidopsis ecotype Col-0 was used for transformation. Protoplasts were grown in a 7:2:1 (v/v/v) mixture of vermiculite:soirite:perlite under a 16-h light/8-h dark regime, and the day and night temperatures were 23°C / 20°C, respectively. The plants were watered every week.
Gene Cloning and Sequence Analysis
The gene primers were designed based on the full-length coding sequences, and RT-PCR (reverse transcriptase-polymerase chain reaction) was performed to isolate the genes from soybean tissues. The neighbor-joining (NJ) tree was constructed from soybean GmTIP2;3 and from other plant TIPs based on alignment using the Clustalx and MEGA 5.0 software, and used to explore the evolutionary relationships of soybean and other plant TIPs.
qRT-PCR Analysis
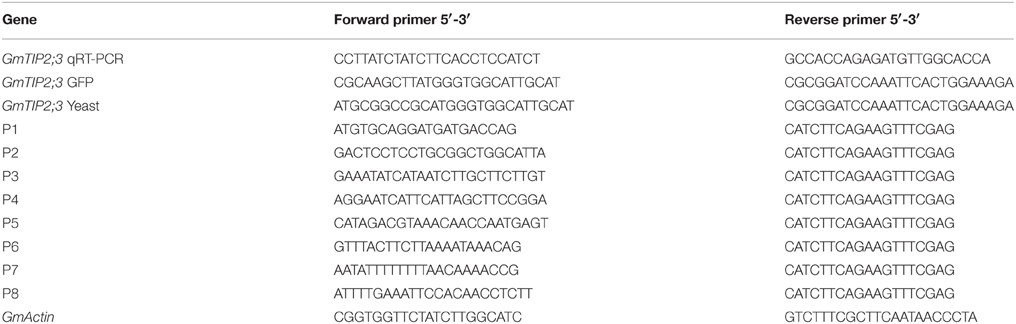
Soybean samples from the seedling, flowering, and podding stages (root, stem, leaf at young seedling stage; root, stem, leaf, and flower at flowering stage; and root, stem, leaf, and pod at podding stage) were harvested and frozen in liquid nitrogen for RNA extraction. Soybean roots were collected from plants treated with PEG6000 (polyethylene glycol) for 0, 2, 4, 12, and 48 h and with 100 μ M ABA (abscisic acid) for 10, 20, 30, 45, 60, 90, and 120 min. The total RNA for all samples used in this study was isolated using TRIzol® reagent (Invitrogen) following the manufacturer's instructions and used for qRT-PCR analysis. The qRT-PCR analysis was conducted according to the method described by Zhang et al. (2013) and was repeated three times. The primers used are given in Table 1.
Subcellular Localization
PCR-generated Hind III-BamH1 fragments containing the open reading frame of GmTIP2;3 were subcloned upstream of the GFP gene in plasmid pJIT166GFP. All constructs were validated by sequencing. The primers used are listed in Table 1.
Arabidopsis protoplasts were isolated according to Yoo et al. (2007). The CDS of GmTIP2;3 without stop codons was used to create an in-frame fusion with GFP gene inserted into the pJIT166-GFP vector. The resulting fusion construct and an empty vector as a control (p35S::GFP) were introduced into Arabidopsis protoplasts by the PEG4000-mediated method (Abel and Theologis, 1994). After incubation of transformed Arabidopsis protoplasts for 18–24 h at room temperature, GFP signal was detected by confocal fluorescence microscopy (Zeiss, LSM510 Meta, Carl Zeiss AG).
Promoter Analysis
A 2081 bp-long region (named P1) located upstream of the ATG start codon was cloned from soybean genome DNA using primers described in Table 1, and sequence analysis was performed using the PlantCARE online software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). To find the core promoter region of GmTIP2;3, seven truncated fragments (P2-P8) were cloned from P1 and transformed into A. rhizogenes strain K599 for GUS activity detection. Soybean hairy root transformation was performed according to the method given by Subramanian et al. (2005). The primer pairs are listed in Table 1.
Histochemical and Fluorometric GUS Assay
For histochemical staining of GUS, fresh tissue samples including whole transgenic lotus plants, soybean hairy roots, and dissected leaves from positive plants that had undergone osmotic stress (20% PEG6000 and 100 μMABA), salinity (200 mM NaCl solution), and wounding for 2 h were immediately dipped into X-Gluc solution, as previously described (Jefferson et al., 1987). After overnight incubation at 37°C in the growth chamber, stained samples were bleached with 70% (v/v) ethanol, washed several times with ddH2O, and observed under a Zeiss Stemi 2000-C microscope, Germany.
A quantitative fluorometric GUS assay was conducted as described by Jefferson et al. (1987). The protein concentrations from a series of truncated constructs pGUSP1-P4 in transgenic soybean hairy roots were assessed by Bradford method, using bovine serum albumin (BSA) as a standard. GUS activity was normalized to the protein concentration of each sample and calculated as nmol of 4-MU per milligram of soluble protein per minute.
Generation of Transgenic Lotus japonicus Plants
The resulting pGUS-GmTIP2;3 promoter(p3):GUS plasmid was introduced into Agrobacterium rhizogenes strain K599 and used to transform small Lotus japonicus seedling to produce hairy roots, as in the soybean hairy root system. The hairy roots were transferred to MS medium with 0.5 mg/L 6-BA for 20 days to generate adventive buds, and 1–2 cm high regeneration seedlings without roots were transferred to 1/2 MS medium to produce roots. Finally, the whole seedlings were transferred to pots.
Yeast Transformation
The novel pYES2-GFP was reconstructed via the recombination of pYES2 and pJIT166-GFP using the same double-digestion by Hind III and EcoR I. The CDS with Hind III and BamH I digestion for the forward primer and reverse primer, respectively (Table 1), was inserted into the pYES2-GFP vector digested with the same enzymes.
The resulting pYES2-GmTIP2;3:GFP plasmid was introduced into S. cerevisiae INVSc1 strain cells using the lithium acetate method, with the empty vector pYES2-GFP as a control. S. cerevisiae INVSc1 strain cells transformed with the empty vector PYES2-GFP alone and with pYES2-GmTIP2;3:GFP were induced with galactose and spotted on the SC-Ura medium in 0, 10, 100, 1000, and 10,000-fold-dilutions, and the drought tolerance of yeast cells expressing GmTIP2;3 was tested by 30% PEG6000 treatment for 40 h. The GFP in yeast was observed using a fluorescence microscope (Olympus BX61). All experiments were repeated three times.
Accession Number
The accession numbers of proteins used for Multiple Sequence Alignment (MSA) and phylogenetic tree analysis are as follows: SsTIP1;1 (AJ242805.1) from Sporobolus stapfianus, PsTIP1;1 (AJ243309.1) from Pisum sativum, SoTIP2;1 (AJ245953.1) from Spinacia oleracea, PtTIP3;2 (XM_006372585.1) from Populus trichocarpa, and GmTIP2;3 (XM_006582773.1) from Glycine max were used for MSA. MtTIP2.1 (XP_003626979.1) from Medicago truncatula; AtTIP4.1 (NP_180152.1), AtTIP1.3 (NP_192056.1), AtTIP5.1 (NP_190328.1), AtTIP2.3 (NP_199556.1), AtTIP2.2 (NP_193465.1), AtTIP2.1 (NP_188245.1), AtTIP1.1 (NP_181221.1), AtTIP1.2 (NP_189283.1), AtTIP3.1 (NP_177462.1), and AtTIP3.2 (NP_173223.1) from Arabidopsis thaliana; OsTIP1.1 (P50156.1), OsTIP1.2 (NP_001045562.1) OsTIP2.1 (NP_001047632.1), OsTIP2.2 (Q5Z6F0.1), OsTIP3.1 (NP_001064933.1), OsTIP3.2 (NP_001053371.1), OsTIP4.1 (NP_001054979.1), OsTIP4.2 (BAA92993.1), OsTIP4.3 (NP_001042500.1), OsTIP5.1 (NP_001053493.1) from Oryza sativa; and ZmTIP1.1 (NP_001104896.1), ZmTIP1.2 (NP_001105029.1), ZmTIP2.1 (NP_001105030.1), ZmTIP2.2 (NP_001105031.1), ZmTIP2.3 (NP_001104907.1), ZmTIP3.1 (NP_001105032.1), ZmTIP4.1 (NP_001105033.1), ZmTIP4.2 (NP_001105034.1), ZmTIP4.3 (NP_001105035.1), ZmTIP4.4 (NP_001105641.1), and ZmTIP5.1 (NP_001105036.1) from Zea mays were used for phylogenetic tree analysis.
Statistical Analysis
The data were analyzed by ANOVA testing using the EXCEL software. Significant differences among means were determined by the LSD at P < 0.05, and a–f represent the different significant levels.
Results
The isolation and characterization of GmTIP2;3
The gene locus Glyma07 g02060 from the QTL region between the markers Satt590 and Satt567 on chromosome 7 in soybean (Specht et al., 2001) was selected as a candidate gene and further isolated by RT-PCR method. BLAST X showed that this locus encoded a protein with 89% identity to TIP2-1-like from Cicer arietinum. The phylogenetic trees were created using GmTIP and Arabidopsis thaliana TIPs (AtTIPs), Oryza saliva TIPs (OsTIPs), Zea mays TIPs (ZmTIPs), and Medicago sativa TIPs (MtTIPs). The phylogenetic tree showed that GmTIP had the highest similarity to TIP2-type proteins from other plants (Figure 1). Therefore, GmTIP was designated as GmTIP2;3. SMART software analysis showed that its protein sequence possessed two conserved NPA motifs and six transmembrane domains, indicating that it was a typical aquaporin protein (Figure 2).
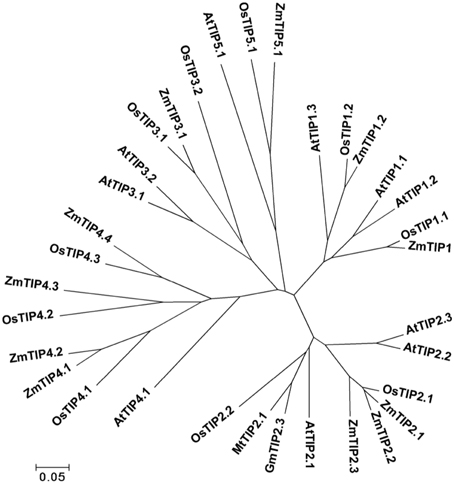
Figure 1. Phylogenetic tree of GmTIP2;3 and reported TIP proteins from Arabidopsis thaliana TIPs (AtTIPs), Oryza saliva TIPs (OsTIPs), Zea mays TIPs (ZmTIPs), and Medicago sativa TIPs (MtTIPs). The Glycine max TIP cloned in this paper showed the highest similarity to TIP2-Type proteins from other plants. Therefore, TIP was designated as GmTIP2;3.
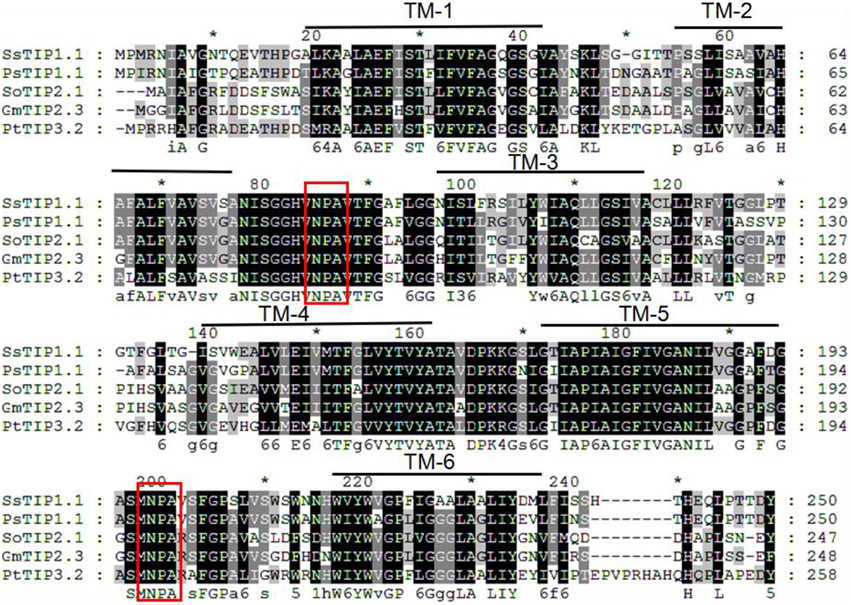
Figure 2. Multiple sequence alignment of GmTIP2;3 protein amino acid sequence with other species. TM1-6 represents the six membrane-spanning helices of GmTIP2;3. The two red boxes represent two conserved “NPA” motifs of the MIP superfamily proteins. *Indicates the consensus sequence at this site.
Expression Analysis of GmTIP2;3
The temporal and spatial expression patterns of GmTIP2;3 in various tissues/organs of soybean cv. Willimas 82 plants were examined using quantitative RT-PCR. GmTIP2;3 appears to be expressed in most parts of the plant, with the highest expression in the root, stem, and pod, Moreover, the expression patterns of GmTIP2;3 in different developmental stages of the same tissue, namely in different organs in the three-leaf, blooming, and podding stages, showed that the transcript abundance of GmTIP2;3 in the stem exhibited a slight increase at the blooming stage, then significantly decreased in both the root and stem at the podding stage except in the pod tissue (Figure 3A).
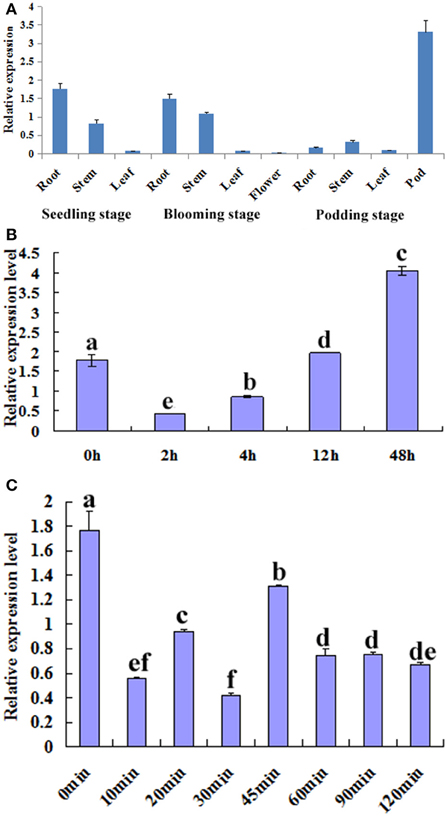
Figure 3. Expression analysis of GmTIP2;3. (A) The temporal and spatial expression patterns of GmTIP2;3 in different organs and at different stages. Root, stem, and leaf from three-leaf stage of young seedling; root, stem, leaf, and flower from blooming stage; and root, stem, leaf, and pod from podding stage. (B) The expression patterns of GmTIP2;3 gene in soybean roots under PEG6000 and 100 μM ABA treatments; a–f indicate the significant difference level at p < 0.05. (1) Expression patterns of GmTIP2;3 after PEG treatment for different time points. (2) Expression patterns of GmTIP2;3 after ABA treatment for different time points.
To test whether GmTIP2;3 responds to drought stress, soybean seedling roots were treated with PEG and ABA. Then, the expression of GmTIP2;3 was analyzed by quantitative real-time RT-PCR. The results indicated that the expression of GmTIP2;3 decreased within 2 h after PEG-6000 (20%) treatment, and then the mRNA level continuously increased from 4 to 12 h and reached a maximum at 12 h (Figure 3B-1) However, ABA treatment (100 μM) initially significantly suppressed GmTIP2;3 expression after 10 min treatment, reached its minimum at 30 min (p < 0.05), then increased from 30 to 45 min and continuously decreased from 45 to 120 min, followed by a stable expression level (Figure 3B-2).
The Promoter Activity Analysis of GmTIP2;3
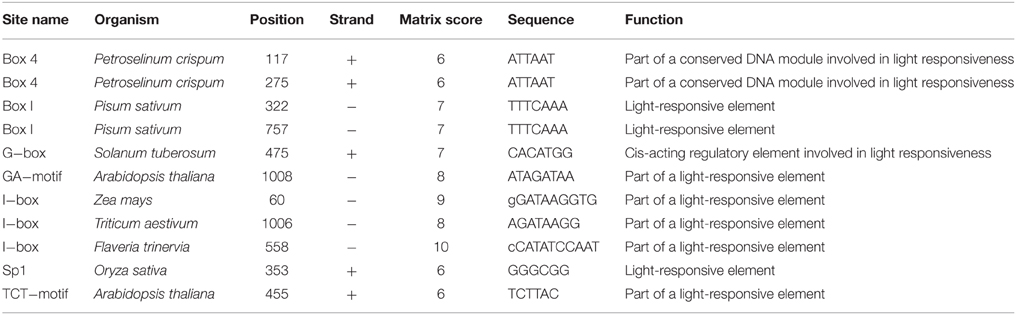
To analyze the elements contained in the promoter region and the promoter activities of GmTIP2;3, a more than 2 kb (2081 bp)-long promoter region located upstream of the ATG start codon was amplified and inserted into the pGUSP vector by the T/A cloning method. The resulting construct was transformed into Lotus japonicus, and the transgenic plant was successfully obtained (Figures 4A,B). The non-transgenic plant was used as a negative control (Figure 4C). GUS staining revealed that GmTIP2;3 was mainly expressed in the root (stele), stem (stele), and leaf (Figures 4D–F). Moreover, transgenic soybean hair roots using this construct also showed higher expression at the stele of the root (Figure 4G), which was consistent with its function as a water transporter. The GmTIP2;3 expression patterns in transgenic plants or hairy roots were identical to the patterns in different organs in the soybean plant. In addition, promoter sequence analysis using the PlantCARE online software indicated that it contained many light responsive elements, such as Box4, G-Box and I-Box, GATA-motif, MBS, and GARE-motif (Table 2). To further dissect the core region of the GmTIP2;3 promoter and explore the impact of external factors on its expression, a series of 8 truncated vectors were constructed, 2081, 1524, 1035, 935, 835, 735, 663, and 581 bp in length, named P1–P8, and transformed into Agrobacterium rhizogenes strain K599 to generate soybean hairy roots. The GUS staining and quantity assays demonstrated that only P1 and P3 exhibited GUS activities, and the activity of P3 was stronger than for P1 (Figure 5A). P5–P8 had no GUS signal, indicating that the core promoter region of GmTIP2;3 was 935 bp long from the ATG site. Interestingly, no GUS signal or GUS activity was detected for P2, implying that the inhibitor sequence occurred between P1 and P3, which also explained why the activity of P3 was stronger than for P1. Meanwhile, the expression of GmTIP2;3 was down-regulated under dark, drought (PEG and ABA), and salinity treatments for 2 h but showed no response to wounding treatment in transgenic lotus plants (Figure 5B). These results were consistent with the results of the expression patterns after treatments with ABA and PEG in soybean roots.
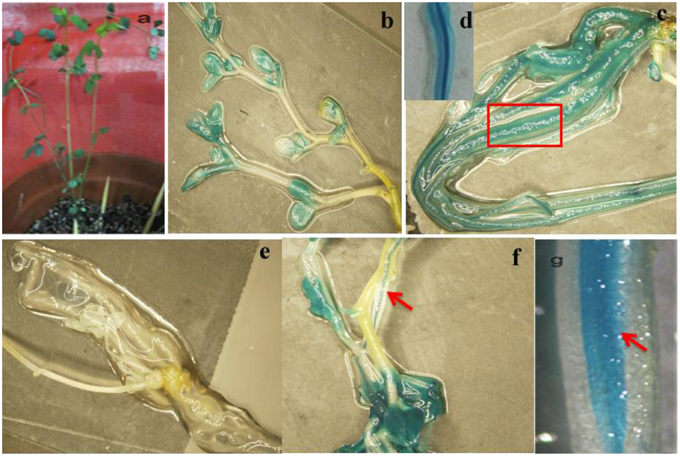
Figure 4. Promoter activity analysis of GmTIP2;3. (A) The positive transgenic lotus plant transformed with pGUSGmTIP2;3 promoter vector. (B) GUS staining of transgenic positive plants. (C) High expression in root of positive plants. (D) Special expression in stele of root, magnified from root (C). (E) GUS staining of negative plant. (F) High expression in leaf and stem stele of positive plant. (G) Special expression in stele of soybean hairy root. Red arrows indicate the stele of root and stem, and the red box indicates that this section was magnified into Panel (D).
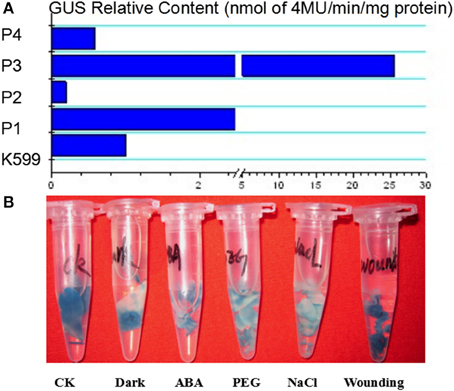
Figure 5. The relative content of GUS protein harboring differently truncated promoter regions of GmTIP2;3 and GUS expression patterns under different treatments using promoter 3.
Plasma Membrane Localization of GmTIP2;3
To examine the localization of the GmTIP2;3 protein, the coding sequences were fused in frame with the coding region of the N-terminal side of green fluorescent protein (GFP). The fusion genes were expressed under the control of the CaMV 35S promoter. GFP fluorescence was evident in the cell plasma membrane transformed with the GmTIP2;3::GFP fusion plasmid (Figure 6A), whereas GFP fluorescence (control) was detected throughout the cells transformed with GFP control plasmid (Figure 6B).
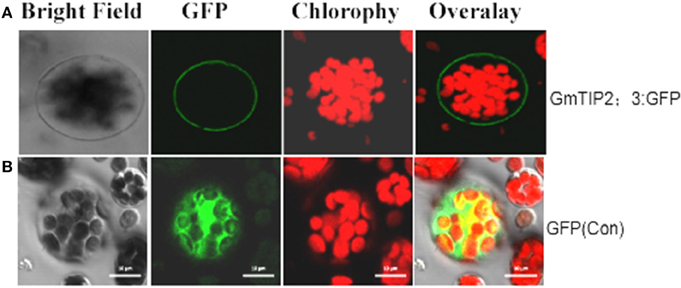
Figure 6. Subcellular localization of GmTIP2;3 protein in Arabidopsis protoplasts. The fusion construct of the GmTIP2;3-green fluorescent protein (p35S::GmTIP2;3-GFP) in the pJIT166-GFP vector without a termination codon to create an in-frame fusion between the CDS and GFP, and the GFP control plasmid (p35S::GFP), was transformed into Arabidopsis protoplasts by PEG4000-mediated method. The transformed Arabidopsis protoplasts were incubated for 18–24 h at room temperature and observed under a confocal fluorescence microscope. GmTIP2;3 was mainly located at the cell membrane (A). However, the GFP control was distributed throughout the whole cell (B). Scale bars = 10 μm.
Heterologous Expression GmTIP2;3 Improved Osmotic Stress Resistance in Yeast
Yeast cells carrying pYES2-GmTIP2;3:GFP or PYES2-GFP (control) were treated with PEG6000 for 40 h, and the survival state was detected. The results revealed that GmTIP2;3 was specifically expressed at the yeast cell membrane, and the heterologous expression of GmTIP2;3 in yeast cells could improve the survival efficiency under osmotic stress (Figure 7), indicating that GmTIP2;3 played an important role in osmotic tolerance in eukaryotes.
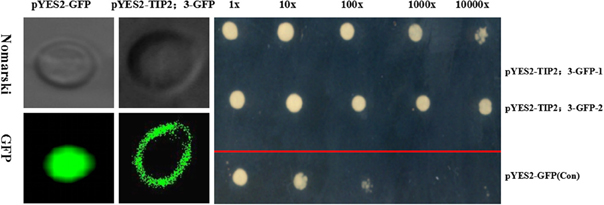
Figure 7. Osmotic response of yeast cells in heterogeneously expressing GmTIP2;3 S. cerevisiae INVSc1 strain cells transformed with the empty vector pYES2-GFP alone (Con) and pYES2-GmTIP2;3:GFP. GFP signal indicated that GmTIP2;3 was mainly located at the cell membrane, and the empty vector was distributed throughout the cells. S. cerevisiae INVSc1 strain cells transformed with the empty vector pYES2-GFP alone and pYES2-GmTIP2;3:GFP were spotted on SC-Ura medium in 0, 10, 100, 1000, and 10,000-fold dilutions, repeated twice for two different positive clones of pYES2-GmTIP2;3:GFP. The results showed the osmotic tolerance of yeast cells expressing GmTIP2;3. All experiments were repeated three times.
Discussion
In this study, we isolated and characterized GmTIP2;3, an MIP family protein showing the highest similarity to Arabidopsis, rice, and corn TIP5. SMART software showed that GmTIP2;3 contains six transmembrane domains, single “AEFH” and “NWIYWVGP” motifs, and two conserved NPA motifs. Fujiyoshi et al. (2002) reviewed the structure and function of water channels in mammalian aquaporins, reporting that the sequence alignment of aquaporins shows several highly conserved motifs including two “NPA” sequences and single “AEFL” and “HW[V/I][F/Y]WXGP” sequences. Here, we found that plant TIPs contain AEFI or AEFV/H, and TIPs from other plants do possess HW[V/I][F/Y]WXGP, but the soybean TIP5 had the motif NWIYWVGP, thereby implying the differences in function and localization between GmTIP2;3 and other plant TIPs.
Spatial and temporal expression analysis showed that GmTIP2;3 was constitutively expressed in all tested organs, with higher expression in the root and stem, indicating that it can absorb water from the soil through the root and then transport water through the stem to other organs, such as the leaf, flower, and pod. Tungngoen et al. (2009) cloned and characterized two aquaporins, HbPIP2;1 and HbTIP1;1, and induced expression analysis found that HbTIP1;1 was down-regulated in liber tissues but up-regulated in laticifers in response to bark Ethrel treatment. Regon et al. (2014) also analyzed the expression patterns of 100 TIP aquaporin genes from dicots and monocots and indicated that the expression of TIP genes varies during different developmental stages and under stressed conditions. da Silva et al. (2013) identified and analyzed the expression patterns of sugarcane aquaporin genes under water deficit, thereby finding the aquaporin transcription in sugarcane to be potentially genotype specific. These findings demonstrated that TIP expression was organ specific or genotype specific and performed different regulator roles in different tissues. Recently, Lee et al. (2015) showed that the expressions of barley HvTIP1;2 and HvTIP3;1 were regulated by gibberellic acid (GA) and ABA and that these two hormones were involved in the fusion of protein storage vacuoles in aleurone cells, indicating that TIP plays another role in vacuole formation and transportation. When subjected to drought stress (ABA and PEG), the expression of GmTIP2;3 showed a dynamic trend at different time points, with an increase after PEG and ABA treatments for 48 h and 45 min, respectively, indicating that the expression of GmTIP2;3 exhibited a response to osmotic stress.
In fact, GmTIP2;3 should be a plasma membrane intrinsic protein (PIP). It was predicted to be localized at the plasma membrane by the online software http://www.predictprotein.org/, and this subcellular localization was proven using Arabidopsis protoplasts, yeast cells, and onion epidermal cells (data not shown) harboring GFP. However, BLAST result at NCBI showed GmTIP2;3 to be a tonoplast intrinsic protein (TIP). Analysis of the promoter activity of GmTIP2;3 indicated that the activity of P3 (~1000 bp in length) was stronger than the activity of P1 (~2000 bp), implying that the inhibitor region occurred between these two regions, and P4 (~550 bp) exhibited no GUS activity. To further determine the core or minimum region for the GmTIP2;3 promoter, five truncated constructs at 100 bp intervals between P3–P4 were prepared, but no GUS signal was detected. Therefore, we concluded that the core promoter region for GmTIP2;3 was located +1000 bp upstream of the ATG start codon containing the 5′UTR region of the GmTIP2;3 gene. The promoter–GUS system was used to detect the GUS activity changes of transgenic Lotus leaf under different treatments, including ABA, Nacl, dark, wounding, and PEG for 2 h. The results showed that the expression of GmTIP2;3 decreased under all treatments except wounding. The plant CARE software revealed that the promoter region contains many light-responsive elements, so the down-regulated expression under dark conditions was reasonable. Moreover, GUS activity under drought treatment for 2 h was consistent with the expression patterns after ABA and PEG treatments for 2 h. Lee et al. (2015) detected the promoter activity of HvTIP3;1 in response to ABA and revealed that the ABA responsiveness of the HvTIP3;1 promoter is likely to occur via a unique regulatory system distinct from the one involving the ABA-response promoter complexes. Therefore, the mechanism of the ABA responsiveness of GmTIP2;3 should be further examined. Here, we can hypothesize that the plants first reduce the water hole number or close water channels to reduce the loss under stress by decreasing the transcription level of GmTIP2;3, and then when the plants have adapted to the stress environment, the expression of GmTIP2;3 recovers to its original level and continues to increase its transcript abundance to respond to stressed conditions.
The plant response to drought is dependent on the SPAC (Soil-Plant-Air-Continuum). Root absorption and soil play important roles in plant adaption to drought stress (Shao et al., 2009). Higher expression of aquaporin proteins in plants can allow them to effectively absorb water from the soil using the roots and then transport water by the stem to other organs, such as the leaf, flower, and seed, especially under osmotic stress (Devi et al., 2015; Ding et al., 2015; Miniussi et al., 2015; Olaetxea et al., 2015). Here, the higher expression of GmTIP2;3 in the steles of the root and stem might promote and speed up water transportation from the roots to other organs under osmotic stress, improving plant tolerance to osmotic stress.
Azad et al. (2009) analyzed water channels by yeast heterologous expression of tulip petal plasma membrane aquaporins from Pichia pastoris and monitored their water channel activity (WCA) by in vivo spheroplast-bursting and hypo-osmotic shock assays, suggesting that P. pastoris can be employed as a heterologous expression system to assay the WCA and to monitor the AQP-mediating channel gating mechanism of aquaporins. The yeast heterologous expression assay in this study showed that GmTIP2;3 could effectively improve the tolerance of yeast to drought stress. Previously, we performed this assay using salinity and drought treatments simultaneously, but the results indicated that yeast cells expressing GmTIP2;3 did not show improved survival rates under salinity stress, implying that GmTIP2;3 had the ability to transport water but not ions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was sponsored by the National Science Foundation of China (31101166), the Jiangsu Natural Science Foundation, China (BK20151364), the Open Foundation of the Jiangsu Key Laboratory for Bioresources of Saline Soils (JKLBS2014002), the One Hundred Talent Plan of Foreign Experts of Jiangsu Province (JSB2015005), the Project in the National Science and Technology Pillar Program during the Twelfth Five-year Plan Period (2011BAD35B06-4-3), and Jiangsu Autonomous Innovation of Agricultural Science and Technology [CX(15)1005].
Abbreviations
GFP, Green Fluorescent Protein; GUS, Glucuronidase; TIP, Tonoplast Intrinsic Protein; QRT-PCR, Quantitative reverse transcriptase Chain Reaction; CDS, Coding sequence; PEG, Polyethylene glycol; ABA, Abscisic acid.
References
Abel, S., and Theologis, A. (1994). Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. Plant J. 5, 421–427. doi: 10.1111/j.1365-313X.1994.00421.x
Azad, A. K., Sawa, Y., Ishikawa, T., and Shibata, H. (2009). Heterologous expression of tulip petal plasma membrane aquaporins in Pichia pastoris for water channel analysis. Appl. Environ. Microbiol. 75, 2792–2797. doi: 10.1128/AEM.02335-08
Boursiac, Y., Chen, S., Luu, D. T., Sorieul, M., van den Dries, N., and Maurel, C. (2005). Early effects of salinity on water transport in Arabidopsis roots. Molecular and cellular features of aquaporin expression. Plant Physiol. 139, 790–805. doi: 10.1104/pp.105.065029
Chaumont, F., Barrieu, F., Wojcik, E., Chrispeels, M. J., and Jung, R. (2001). Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol. 125, 1206–1215. doi: 10.1104/pp.125.3.1206
Danielson, J. A., and Johanson, U. (2008). Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biol. 8:45. doi: 10.1186/1471-2229-8-45
da Silva, M. D., Silva, R. L., Costa Ferreira Neto, J. R., Guimaraes, A. C., Veiga, D. T., Chabregas, S. M., et al. (2013). Expression analysis of sugarcane aquaporin genes under water deficit. J. Nucleic Acids 2013:763945. doi: 10.1155/2013/763945
Denker, B. M., Smith, B. L., Kuhajda, F. P., and Agre, P. (1988). Identification, purification, and partial characterization of a novel Mr 28,000 integral membrane protein from erythrocytes and renal tubules. J. Biol. Chem. 263, 15634–15642.
Devi, M. J., Sinclair, T. R., Jain, M., and Gallo, M. (2015). Leaf aquaporin transcript abundance in peanut genotypes diverging in expression of the limited-transpiration trait when subjected to differing vapor pressure deficits and aquaporin inhibitors. Physiol. Plant. doi: 10.1111/ppl.12378. [Epub ahead of print].
Ding, L., Gao, C., Li, Y., Li, Y., Zhu, Y., Xu, G., et al. (2015). The enhanced drought tolerance of rice plants under ammonium is related to aquaporin (AQP). Plant Sci. 234, 14–21. doi: 10.1016/j.plantsci.2015.01.016
Fujiyoshi, Y., Mitsuoka, K., de Groot, B. L., Philippsen, A., Grubmüller, H., Agre, P., et al. (2002). Structure and function of water channels. Curr. Opin. Struct. Biol. 12, 509–515. doi: 10.1016/S0959-440X(02)00355-X
Gao, Z., He, X., Zhao, B., Zhou, C., Liang, Y., Ge, R., et al. (2010). Overexpressing a putative aquaporin gene from wheat, TaNIP, enhances salt tolerance in transgenic Arabidopsis. Plant Cell Physiol. 51, 767–775. doi: 10.1093/pcp/pcq036
Guenther, J. F., and Roberts, D. M. (2000). Water-selective and multifunctional aquaporins from Lotus japonicus nodules. Planta 210, 741–748. doi: 10.1007/s004250050675
Guo, L., Wang, Z. Y., Lin, H., Cui, W. E., Chen, J., Liu, M., et al. (2006). Expression and functional analysis of the rice plasma-membrane intrinsic protein gene family. Cell Res. 16, 277–286. doi: 10.1038/sj.cr.7310035
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Kaldenhoff, R., and Fischer, M. (2006). Functional aquaporin diversity in plants. Biochim. Biophys. Acta 1758, 1134–1141. doi: 10.1016/j.bbamem.2006.03.012
Lee, S. E., Yim, H. K., Lim, M. N., Yoon, I. S., Kim, J. H., and Hwang, Y. S. (2015). Abscisic acid prevents the coalescence of protein storage vacuoles by upregulating expression of a tonoplast intrinsic protein gene in barley aleurone. J. Exp. Bot. 66, 1191–1203. doi: 10.1093/jxb/eru467
Ligaba, A., Katsuhara, M., Shibasaka, M., and Djira, G. (2011). Abiotic stresses modulate expression of major intrinsic proteins in barley (Hordeum vulgare). C. R. Biol. 334, 127–139. doi: 10.1016/j.crvi.2010.11.005
Miniussi, M., Del Terra, L., Savi, T., Pallavicini, A., and Nardini, A. (2015). Aquaporins in Coffea arabica L.: identification, expression, and impacts on plant water relations and hydraulics. Plant Physiol. Biochem. 95, 92–102. doi: 10.1016/j.plaphy.2015.07.024
Olaetxea, M., Mora, V., Bacaicoa, E., Garnica, M., Fuentes, M., Casanova, E., et al. (2015). Abscisic acid regulation of root hydraulic conductivity and aquaporin gene expression is crucial to the plant shoot rise growth enhancement caused by rhizosphere humic acids. Plant Physiol. 169, 2587–2596. doi: 10.1104/pp.15.00596
Park, W., Scheffler, B. E., Bauer, P. J., and Campbell, B. T. (2010). Identification of the family of aquaporin genes and their expression in upland cotton (Gossypium hirsutum L.). BMC Plant Biol. 10:142. doi: 10.1186/1471-2229-10-142
Quigley, F., Rosenberg, J. M., Shachar-Hill, Y., and Bohnert, H. J. (2002). From genome to function: the Arabidopsis aquaporins. Genome Biol. 3, 1–17.
Regon, P., Panda, P., Kshetrimayum, E., and Panda, S. K. (2014). Genome-wide comparative analysis of tonoplast intrinsic protein (TIP) genes in plants. Funct. Integr. Genomics 14, 617–629. doi: 10.1007/s10142-014-0389-9
Reuscher, S., Akiyama, M., Mori, C., Aoki, K., Shibata, D., and Shiratake, K. (2013). Genome-wide identification and expression analysis of aquaporins in tomato. PLoS ONE 8:e79052. doi: 10.1371/journal.pone.0079052
Rodrigues, M. I., Bravo, J. P., Sassaki, F. T., Severino, F. E., and Maia, I. G. (2013). The tonoplast intrinsic aquaporin (TIP) subfamily of Eucalyptus grandis: characterization of EgTIP2, a root-specific and osmotic stress-responsive gene. Plant Sci. 213, 106–113. doi: 10.1016/j.plantsci.2013.09.005
Sade, N., Vinocur, B. J., Diber, A., Shatil, A., Ronen, G., Nissan, H., et al. (2009). Improving plant stress tolerance and yield production: is the tonoplast aquaporin SlTIP2;2 a key to isohydric to anisohydric conversion? New Phytol. 181, 651–661. doi: 10.1111/j.1469-8137.2008.02689.x
Sakurai, J., Ishikawa, F., Yamaguchi, T., Uemura, M., and Maeshima, M. (2005). Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol. 46, 1568–1577. doi: 10.1093/pcp/pci172
Shao, H. B., Chu, L. Y., Jaleel, C. A., Manivannan, P., Panneerselvam, R., and Shao, M. A. (2009). Understanding water deficit stress-induced changes in the basic metabolism of higher plants—biotechnologically and sustainably improving agriculture and the ecoenvironment in arid regions of the globe. Crit. Rev. Biotechnol. 29, 131–151. doi: 10.1080/07388550902869792
Specht, J. E., Chase, K., Macrander, M., Graef, G. L., Chung, J., Markwell, J. P., et al. (2001). Soybean response to water: a QTL analysis of drought tolerance. Crop Sci. 41, 493–509. doi: 10.2135/cropsci2001.412493x
Srivastava, A. K., Penna, S., Nguyen, D. V., and Tran, L. P. (2015). Multifaceted roles of aquaporins as molecular conduits in plant responses to abiotic stresses. Crit. Rev. Biotechnol. doi: 10.3109/07388551.2014.973367. [Epub ahead of print].
Subramanian, S., Graham, M. Y., Yu, O., and Graham, T. L. (2005). RNA interference of soybean isoflavone synthase genes leads to silencing in tissues distal to the transformation site and to enhanced susceptibility to Phytophthora sojae. Plant Physiol. 137, 1345–1353. doi: 10.1104/pp.104.057257
Tao, P., Zhong, X., Li, B., Wang, W., Yue, Z., Lei, J., et al. (2014). Genome-wide identification and characterization of aquaporin genes (AQPs) in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol. Genet. Genomics MGG289, 1131–1145. doi: 10.1007/s00438-014-0874-9
Tungngoen, K., Kongsawadworakul, P., Viboonjun, U., Katsuhara, M., Brunel, N., Sakr, S., et al. (2009). Involvement of HbPIP2;1 and HbTIP1;1 aquaporins in ethylene stimulation of latex yield through regulation of water exchanges between inner liber and latex cells in Hevea brasiliensis. Plant Physiol. 151, 843–856. doi: 10.1104/pp.109.140228
Wang, L. L., Chen, A. P., Zhong, N. Q., Liu, N., Wu, X. M., Wang, F., et al. (2014). The Thellungiella salsuginea tonoplast aquaporin TsTIP1;2 functions in protection against multiple abiotic stresses. Plant Cell Physiol. 55, 148–161. doi: 10.1093/pcp/pct166
Wang, X., Li, Y., Ji, W., Bai, X., Cai, H., Zhu, D., et al. (2011). A novel Glycine soja tonoplast intrinsic protein gene responds to abiotic stress and depresses salt and dehydration tolerance in transgenic Arabidopsis thaliana. J. Plant Physiol. 168, 1241–1248. doi: 10.1016/j.jplph.2011.01.016
Xin, S., Yu, G., Sun, L., Qiang, X., Xu, N., and Cheng, X. (2014). Expression of tomato SlTIP2;2 enhances the tolerance to salt stress in the transgenic Arabidopsis and interacts with target proteins. J. Plant Res. 127, 695–708. doi: 10.1007/s10265-014-0658-7
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zelazny, E., Borst, J. W., Muylaert, M., Batoko, H., Hemminga, M. A., and Chaumont, F. (2007). FRET imaging in living maize cells reveals that plasma membrane aquaporins interact to regulate their subcellular localization. Proc. Natl. Acad. Sci. U.S.A. 104, 12359–12364. doi: 10.1073/pnas.0701180104
Keywords: soybean, GmTIP2;3, heterologous expression, promoter, osmotic stress
Citation: Zhang D, Tong J, He X, Xu Z, Xu L, Wei P, Huang Y, Brestic M, Ma H and Shao H (2016) A Novel Soybean Intrinsic Protein Gene, GmTIP2;3, Involved in Responding to Osmotic Stress. Front. Plant Sci. 6:1237. doi: 10.3389/fpls.2015.01237
Received: 14 August 2015; Accepted: 20 December 2015;
Published: 08 January 2016.
Edited by:
Manoj Prasad, National Institute of Plant Genome Research, IndiaReviewed by:
Xuebin Zhang, Brookhaven National Laboratory, USASatish Kumar Guttikonda, Dow AgroSciences, USA
Copyright © 2016 Zhang, Tong, He, Xu, Xu, Wei, Huang, Brestic, Ma and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dayong Zhang, cotton.z@126.com;
Hongbo Shao, shaohongbochu@126.com
†These authors have contributed equally to this work.
 Dayong Zhang
Dayong Zhang Jinfeng Tong2†
Jinfeng Tong2† Marian Brestic
Marian Brestic Hongxiang Ma
Hongxiang Ma Hongbo Shao
Hongbo Shao