- 1Department of Plant Ecophysiology, Faculty of Biology, The Adam Mickiewicz University, Poznan, Poland
- 2Department of Plant Physiology, The University of Life Sciences in Poznan, Poznan, Poland
In the course of evolutionary changes pathogens have developed many invasion strategies, to which the host organisms responded with a broad range of defense reactions involving endogenous signaling molecules, such as nitric oxide (NO). There is evidence that pathogenic microorganisms, including two most important groups of eukaryotic plant pathogens, also acquired the ability to synthesize NO via non-unequivocally defined oxidative and/or reductive routes. Although the both kingdoms Chromista and Fungi are remarkably diverse, the experimental data clearly indicate that pathogen-derived NO is an important regulatory molecule controlling not only developmental processes, but also pathogen virulence and its survival in the host. An active control of mitigation or aggravation of nitrosative stress within host cells seems to be a key determinant for the successful invasion of plant pathogens representing different lifestyles and an effective mode of dispersion in various environmental niches.
Introduction
During plant–pathogen interactions a decisive role is played by the rate and intensity of reactions induced by both adversaries. In the initiation, coordination and transfer of information on the appearing threat a particularly significant role is exerted by endogenous signaling molecules, such as nitric oxide (NO). This free radical gas can diffuse rapidly through biological membranes and it is capable of acting as a transient, local, intra-, and intercellular signal within species from every biological system.
The signaling function of NO has been shown during both highly conserved pathogen-associated molecular pattern (PAMP) triggered immunity (PTI) and in a highly specific effector-triggered immunity (ETI), often accompanied by the hypersensitive response (HR) at the site of attempted host colonization (e.g., Mur et al., 2006; Floryszak-Wieczorek et al., 2007; Asai and Yoshioka, 2009; Schlicht and Kombrink, 2013). Importantly, the generation and potential functions of NO during these interactions have so far been analyzed solely from the point of view of the host plant. Many years ago Mur et al. (2006) highlighted that the role of NO within the pathogen was too often ignored when considering plant-pathogen interactions.
The plant–pathogen system is dynamic and in the course of evolutionary changes pathogens have developed numerous invasion strategies, to which the host organism has responded with an extensive range of defense traits known as “fight for their lives”. It is known from the scarce studies that also certain pathogenic microorganisms are capable of synthesizing NO, although the role of NO in this systematically heterogeneous group of organisms has not been specified. Definitely the greatest amount of experimental data, both in terms of the sources of synthesis and the potential function of NO in the host-pathogen interactions, has been supplied by studies on model bacterial and fungal human pathogens.
Since pathogens may use NO to its own benefit and concurrently, NO may prime the host organisms and activate their defense, this review focuses on the mode of NO action in two most important groups of eukaryotic plant pathogens, i.e., fungi and oomycetes.
The Origin of NO in Fungal and Oomycete Plant Pathogens
Pathogens are able to produce NO; however, the origin of NO seems to be as unclear as in plants (Arasimowicz-Jelonek and Floryszak-Wieczorek, 2014). In general, the biosynthetic pathways of NO in fungal and oomycete phytopathogens can be classified as either oxidative or reductive in operation.
The oxidative route involves NOS-like activity, which was evidenced in various groups of fungi. A pharmacological approach using mammalian NOS inhibitors, i.e., L-NAME and/or 1-[2-(Trifluoromethyl)phenyl]imidazole (TRIM), revealed a limited NO production including phytopathogenic fungi Macrophomina phaseolina, Blumeria graminis, and Colletotrichum coccodes, the mycoparasitic fungus Coniothyrium minitans and the aquatic fungus Blastocladiella emersonii (Wang and Higgins, 2005; Prats et al., 2008; Vieira et al., 2009; Li et al., 2010; Sarkar et al., 2014). Moreover, the NOS-like activity has been confirmed by measuring citrulline formation from 3H-labeled arginine in the endophytic and pathogenic fungus Neurospora crassa, the non-pathogenic Mucorales Phycomyces blakesleeanus and C. minitans (Ninnemann and Maier, 1996; Maier et al., 2001; Li et al., 2010). Importantly, some representatives of ascomycetes and zygomycetes possess tetrahydrobiopterin, a typical mammalian NOS cofactor essential for NO synthesis (Maier and Ninnemann, 1995). Additional experiments evidenced that the catalytic activity of the fungal NOS-like enzyme depends on NADPH and involves calcium ions (Vieira et al., 2009).
Based on genome analysis NOS-like sequences were found in genomes of fungal species from the genus Aspergillus (A. flavus, A. oryzae, and A. niger) as well as Glomerella graminicola, the teleomorph stage of the anthracnose pathogen Colletotrichum graminicola (Turrion-Gomez and Benito, 2011; Sarkar et al., 2014). More recently, NOS-like protein with conserved amino acid sequences was found in the genome of M. phaseolina, a necrotrophic fungus causing charcoal rot disease (Sarkar et al., 2014). What is more, multiple alignments of NOS sequences followed by motif enrichment analysis have generated two motifs, one in the oxygenase domain and the other in the flavodoxin/NO synthase domain presented in separate open reading frames (ORFs). As indicated by Sarkar et al. (2014), these motifs are conserved among the five necrotrophic plant pathogens including M. phaseolina, Neofusicoccum parvum, Colletotrichum gloeosporioides, G. graminicola, and A. oryzae.
Available data indicate that NOS-like activity in fungi could be greatly varied. For example, in mycelia of P. blakesleeanus it is 10 pmol/mg/min (Ninnemann and Maier, 1996) and in fruiting bodies of white-rot basidiomycete Flammulina velutipes it amounts to 500 pmol/mg/min (Song et al., 2000). In C. minitans the highest level of NOS-like activity, amounting to 20 pmol/min/mg, was recorded during conidiation (Li et al., 2010), suggesting that this route of NO synthesis is closely regulated depending on the species, developmental stage and environmental conditions.
Evidence for the existence of an nitrate redctase (NR) dependent pathway of NO biosynthesis in A. nidulans was recently provided by Marcos et al. (2016). Although this reductive route was found to be functional during growth in both liquid and solid media, NR-dependent NO production was more abundant on solid media, where reproductive development occurred.
Fungi are able to produce NO as a result of denitrification processes, catalyzed by three enzymes: nitrate reductase (Nar), nitrite reductase (Nir), and NO reductase (Nor) (Ye et al., 1994; Morozkina and Kurakov, 2007). The denitrifying system coupled with the mitochondrial electron transport chain facilitates anaerobic respiration associated with ATP synthesis under hypoxia conditions. Nitrite reductases located in the intermembrane space of the fungal mitochondria have been shown to reduce NO2- to NO in a NADP-dependent manner (Röszer, 2012). A dissimilatory nitrite reductase was found in phytopathogens Fusarium oxysporum and Cylindrocarpon tonkinense as well as in a potentially pathogenic for humans yeast Fellomyces fuzhouensis (Abraham et al., 1993; Kobayashi and Shoun, 1995; Kobayashi et al., 1996; Uchimura et al., 2002). Moreover, using combination of the membrane inlet mass spectrometry (MIMS) and the restriction capillary inlet mass spectrometry (RIMS) techniques a nitrite-induced NO production has been demonstrated from cultures of plant pathogenic fungi Botrytis and Fusarium sp. and the oomycete Pythium (Conrath et al., 2004)
Interestingly, NO synthesis in the rice blast fungus Magnaporthe oryzae was associated neither with nitrite-dependent nor with the arginine-dependent pathway (Samalova et al., 2013). By creating mutants with a single and double knockout of genes potentially involved in NO synthesis, including NOS-like genes (NOL2 and NOL3), nitrate (NIA1) and nitrite reductase (NII1), the authors revealed that NO is not generated by the candidate proteins. A mammalian NOS inhibitor, L-NAME applied to a necrotroph Botrytis cinerea growing on a medium and in planta did not limit NO production either (Turrion-Gomez and Benito, 2011). The pathogen was also incubated with NO3- to identify the source of NO production via nitrate reductase, but NO signal was not detected implying that a physiological and genetic system other than NOS and NR is responsible for NO production in this plant pathogen (Turrion-Gomez and Benito, 2011). A plausible explanation for this elusive route of NO synthesis could involve an unidentified complex system of a fungus-type enzyme catalyzing NO production.
As suggested by Röszer (2012), the oxidative route of NO formation might be dominant in pathogen units under aerobic conditions and localized in the cytoplasm, whereas enzymes responsible for NO production under hypoxia or anoxia could involve mitochondrial cytochrome-c oxidase and mitochondrial nitrite reductase.
NO as a Developmental Signal in Fungal and Oomycete Pathogens
Recent studies have indicated that NO may play an important role in signaling networks in fungi. Based on the pharmacological approach (including also non-pathogenic fungi) it was shown that NO participates in the development of various fungal structures including sporangiophores, conidia, cleistothecia, pycnidia and appresoria (Maier et al., 2001; Wang and Higgins, 2005; Gong et al., 2007; Prats et al., 2008; Baidya et al., 2011). However, the intensity of NO generation and its location is strictly dependent on the developmental stage of the pathogen. Bio-imaging with fluorochrome DAF-FM DA showed NO presence in C. coccodes conidia, germ tubes, and immature appressoria, but the strongest NO-dependent fluorescence was observed in the cells with reduced cytoplasm in the conidium with a mature appressorium (Wang and Higgins, 2005). Also in the B. cinerea system the production of NO was detected in all fungal developmental stages, starting from spores up to mature mycelium (van Baarlen et al., 2004; Turrion-Gomez and Benito, 2011). A strong NO signal was found in young pycnidia of C. minitans and in the undifferentiated tissue of pycnidial primordia; in turn, weak fluorescence signals were observed in growing hyphal tips or hyphae, where pycnidia or primordia did not develop (Gong et al., 2007).
In general, both sexual and asexual reproduction in fungi was documented to be dependent on NO generation. What is more, NO could control a threshold to switch developmental phases. It was found that a NO-releasing compound (DETA NONOate) reduced asexual development in A. nidulans which has a limited, but significant, phytopathogenic potential (Dean and Timberlake, 1989). In turn, formation of sexual structures was increased after NO supplementation in several fungal species, including species from the genera Aspergillus and Neurospora, as well as the species F. velutipes (Song et al., 2000; Baidya et al., 2011). Alterations in A. nidulans conidiation induced by exogenous NO may be due to NO impairing the transcriptional activation of structural sporulation specific genes (Chiuchetta and Castro-Prado, 2005). NO levels influenced the balance between conidiation and sexual reproduction, since an artificial strong elevation of NO levels reduced conidiation and induced the formation of cleistothecia. As it was found by Marcos et al. (2016), different NO levels affected the expression of the regulator of sexual development nsdD and the regulator of conidiation brlA. Baidya et al. (2011) also showed that NO is involved in the switch of developmental phases. Deletion of fhbA, a gene encoding flavohaemoglobin (Fhb) protein involved in the reduction and detoxification of NO, resulted in increased Hülle cell production which nurse the young fruiting body during development (Baidya et al., 2011). This result suggests that fhbA is likely to suppress sexual development under stressful conditions accompanied by reactive nitrogen species (RNS) overproduction (Dyer and O’Gorman, 2012).
Nitric oxide also controls the development of sporangiophores in the zygomycete P. blakesleeanus (Maier et al., 2001). Exposure to light signals activated asexual reproduction concomitant with NO emission within developing cells of the fungus. Exogenous NO was able to mimic the light effect on sporangiophore formation indicating that NO could function as a light sensor molecule during light-mediated sporulation. The photoconidiation process in ascomycete N. crassa is also dependent on NO (Ninnemann and Maier, 1996). However, NO donor inhibited light-stimulated conidiation in N. crassa, whereas specific inhibitors of NOS activity enhanced conidiation in darkness and in the light. The role of NO in fungal physiology is related to spore germination as well. NO synthesis was found during germination of C. coccodes spores. Since NO trapping accelerated germination and exogenous NO delayed this phenomenon the authors suggested that a specific NO threshold could control the time of exit from spore dormancy (Wang and Higgins, 2005).
The sporulation of the blastocladiomycete B. emersonii was accompanied not only by an increased level of NO and NO-derived compounds but also by the expression of genes coding for guanylyl cyclase and cGMP phosphodiesterase. Using the pharmacological approach Vieira et al. (2009) demonstrated that the Ca2+-NO-cGMP signaling pathway facilitated control of zoospore biogenesis in the aquatic saprophytic fungus. Further support for the signaling role of NO during fungal development has been provided by Gong et al. (2007). The plant pathogen Cryphonectria parasitica failed to produce pycnidia in response to both L-NAME and the cGMP blocker (6-anilinoquinoline-5, 8-quinone), suggesting that the NO-mediated signal for conidiation may be common to multiple fungal genera, which produce pycnidia (Gong et al., 2007). Importantly, the dynamics of NO synthesis was closely related to changes in cGMP levels during pycnidial development.
NO Offensive and Necrotrophic Pathogens
Nitric oxide was found to be produced by various plant necrotrophs, including B. cinerea, A. nidulans, M. phaseolina, F. oxysporum, and C. coccodes (Conrath et al., 2004; Wang and Higgins, 2005; Floryszak-Wieczorek et al., 2007; Turrion-Gomez and Benito, 2011; Sarkar et al., 2014). Based on a model necrotroph B. cinerea NO generation was detected during both saprophytic growth and in planta (Conrath et al., 2004; Floryszak-Wieczorek et al., 2007). Based on DAF2-DA fluorochrome, the presence of NO was found in hyphae and spores of B. cinerea growing on a solid medium (Floryszak-Wieczorek et al., 2007). What is noteworthy, the necrotroph contact with pelargonium leaf tissue of the susceptible genotype resulted in the acquisition of the ability to generate much greater amounts of NO, favoring necrotic death of host cells and in consequence – disease development. The NO-dependent fluorescence surrounded germinating spores and mycelium of B. cinerea growing both on a complete medium and in planta, indicated the diffusion of NO produced inside the fungal cells (Turrion-Gomez and Benito, 2011). The observed NO spreading outside the fungal structures could have important physiological consequences in the establishment and progress of the disease, since pathogen-derived NO could reach plant cells and contribute to the hypersensitive cell death, facilitating subsequent tissue colonization (Turrion-Gomez and Benito, 2011; Arasimowicz-Jelonek and Floryszak-Wieczorek, 2014). Noteworthy, a compound required for full virulence of B. cinerea, the endopolygalactouronase 1 (BcPG1) was evidenced to trigger a phosphorylation-dependent NO production in grapevine cells (Vandelle et al., 2006). Thus, NO overaccumulation in planta induced by pathogen or originating both from the pathogen and the host plant might accelerate the spread of infection and constitute a significant element determining success of the necrotrophic aggressor. A strong accumulation of NO in host tissue correlated with disease development was also observed in the compatible lily–Botrytis elliptica interaction (van Baarlen et al., 2004). In the susceptible jute–M. phaseolina interaction NO overproduction followed 20 dpi and coincident with NO-derived compound accumulation localized in the vascular bundle region containing invaded mycelium and micro-sclerotia (Sarkar et al., 2014). It should be noted that the necrotroph-induced NO generation in plant cells could also correlate with enhanced disease resistance. An early and only transient NO burst synchronized with ROS generation was found to positively modulate resistance in various plants under attack by necrotrophic fungal pathogens, such as Botrytis cinerea or Sclerotinia sclerotiorum (Mur et al., 2006; Floryszak-Wieczorek et al., 2007; Asai and Yoshioka, 2009; Perchepied et al., 2010).
Production of NO could also have implications in virulence of fungal pathogens via regulation of mycotoxin biosynthesis. For example, the Fhb gene fhbA influences mycotoxin biosynthesis in necrotrophic A. nidulans ΔfhbA (or ΔfhbA ΔfhbB), since deletion mutants showed a reduction of sterigmatocystin production (Baidya et al., 2011). The diminished pool of mycotoxins coincided with a decrease in the expression of aflR, a transcription factor necessary for the activation of the sterigmatocystin gene cluster. The NO-releasing compound application to ΔfhbA strains resulted in an increase of aflR expression levels and in the recovery of mycotoxins near the wild-type levels (Baidya et al., 2011).
NO Offensive and Hemi-/Biotrophic Pathogens
Pathogens with the hemibiotrophic and biotrophic life strategies are able to produce NO. This RNS was found to be generated by mycelia of Oidium neolycopersici (Piterková et al., 2009), B. graminis f. sp. hordei (Prats et al., 2008), M. oryzae (Samalova et al., 2013) and by hyphae of the oomycete Bremia lactucae (Sedlárová et al., 2011).
In B. lactucae the presence of NO was observed in the infection structures of the pathogen grown both on susceptible and resistant lettuce genotypes; however, the plant genotype determined timing of the pathogen development. A strong NO signal was detected in the tip of the germ tube and appressorium, which is a prerequisite for tissue penetration. A weaker NO signal was detected in developing primary and secondary vesicles, intracellular hyphae and in haustoria on susceptible lettuce (Sedlárová et al., 2011). The genotype with an abundant HR showed NO generation frequently localized in penetrated cells undergoing HR before the occurrence of detectable necrosis (Sedlárová et al., 2011).
Rice blast fungus M. oryzae produced NO during germination and early development, and was critically required for the progress of appressorium formation. Importantly, the elimination of NO produced by M. oryzae significantly reduced the level of infection in a compatible interaction, confirming that the pathogen-derived NO is required for successful host colonization by the hemibiotrophic pathogen (Samalova et al., 2013). Most recently, Zhang et al. (2015) revealed that the M. oryzae gene MoSFA1 coding S-(hydroxymethyl)-glutathione dehydrogenase involved NO metabolism through the reduction of S-nitrosoglutathione (GSNO) contributing to full virulence in the pathogen. MoSFA1 mutants showed attenuated virulence on rice cultivar CO-39, as well as severe reduction of conidiation and appressorium turgor pressure. Importantly, the virulence of MoSFA mutants on wounded rice leaves was not affected, indicating that MoSFA1 significantly contributes to virulence during penetration or the biotrophic phase of the pathogen.
Differentiation of infection structures was also found to be determined by NO in the biotroph B. graminis f. sp. hordei. NO trapping as well as blocking of NOS-like activity significantly reduced the number of appressorial lobes, which in consequence affected host cell penetration (Prats et al., 2008).
In contrast to plant-necrotroph interactions, tomato powdery mildew development was not accompanied by NO over-accumulation in host cells, although pathogen-derived NO could favor infection. NO signal was detected in O. neolycopersici conidia, germ tubes and appressoria developed on various genotypes of S. lycopersicum at 24 and 48 hpi. At later stages of pathogenesis NO was absent in spreading structures of the pathogen in susceptible tomato and only attacked cells of moderately resistant and highly resistant plants showing a strictly localized NO generation (Piterková et al., 2011).
Since the functional role of NO in living organisms may be realized directly through the involvement of NO in the post-translational modification, NO is likely to have significant implications in pathogen virulence also via the nitration/S-nitrosylation phenomenon. The analysis of a crude extract of the plant biotrophic fungus Plasmopara halstedii revealed the presence of nitrated proteins; however, their role remains unknown (Chaki et al., 2009).
Pathogen Survival Under NO Stress
Colonization of the host tissues by pathogens often results in an over-production of NO and NO-derived molecules, which create boosted and pathophysiological levels of RNS, defined as nitrosative stress (Arasimowicz-Jelonek and Floryszak-Wieczorek, 2014). Therefore, the nitrosative stress response seems to be notably important during the early stages of infection when the fungus is battling with host defenses.
Although NO over-accumulation could favor necrotroph development and/or host colonization, most of the published data rather support an adverse effect of NO on fungal growth (Schlicht and Kombrink, 2013). It remains controversial whether NO itself can kill pathogens; however, the potential antimicrobial effect of NO on pathogens has been experimentally proven. Gaseous NO inhibited mycelial growth, spore germination and sporulation of three plant pathogenic fungi, including A. niger, Monilinia fructicola, and Penicillium italicum under in vitro conditions, indicating a direct effect of exogenous NO on fungal metabolism (Lazar et al., 2008). The inhibitory effect of NO on fungal growth and spore germination of the plant pathogen P. expansum was related to increased levels of intracellular ROS and elevated carbonylation damage. Simultaneously, the activities of SOD and CAT as well as ATP content were diminished in response to the NO modulator (Lai et al., 2011). Moreover, the 35S::nNOS Arabidopsis line, which contained constitutively enhanced NO levels due to the expression of the rat neuronal NOS, rendered the plants more resistant to infection by a biotrophic fungus Golovinomyces orontii (Schlicht and Kombrink, 2013). This transgenic Arabidopsis line displayed also enhanced resistance to a bacterial pathogen P. syringae and enhanced tolerance to salt and drought. Since the enhanced resistance was associated with SA accumulation and SA-responsive defense genes expression, the authors speculated that restriction of fungal growth was the result of diverse NO-mediated plant defense components; however, a direct effect of NO on biotroph development may not be excluded (Schlicht and Kombrink, 2013).
Phytopathogens cope both with innate NO and with plant-derived NO. In response to these huge amounts of NO pathogenic microorganisms have evolved constitutive and inducible mechanisms to prevent the adverse effects of NO, helping them to survive during the contact with host cells (Arasimowicz-Jelonek and Floryszak-Wieczorek, 2014). Thus, the pathogen metabolic equipment to counteract nitrosative stress might be implicated in its virulence. Among inducible mechanisms, the detoxification of NO governed by the evolutionarily ancient Fhb is the best described for model fungi and human fungal pathogens (Forrester and Foster, 2012). Genes coding Fhb-like proteins have also been identified in fungal phytopathogens, including B. cinerea, Cladosporium fulvum, F. oxysporum, Gibberella zeae, and N. crassa (Boccara et al., 2005); however, the in vivo ability to detoxify NO via NO-dioxygenase activity has only been confirmed for B. cinerea. Unlike human pathogens, deletion of Bcfhg1 did not affect pathogenicity of B. cinerea in relation to three different host species such as S. lycopersicum, A. thaliana, and P. vulgaris (Turrion-Gomez et al., 2010).
Another system contributing to nitrosative stress resistance employs S-nitrosoglutathione reductase (GSNOR). This GSH-dependent bi-functional enzyme is able to reduce GSNO to form GSSG plus NH3, as well as detoxify formaldehyde (Tillmann et al., 2015). In plants, GSNOR modulates the extent of cellular S-nitrosothiol (SNO) formation following nitrosative stress and is required for disease resistance (Feechan et al., 2005; Yu et al., 2012). As it was mentioned before, in M. oryze a MoSFA1 coding S-(hydroxymethyl)glutathione dehydrogenase belonging to a class III alcohol dehydrogenase was proven to be involved in NO metabolism via GSNO reduction (Zhang et al., 2015). As evidenced by Fernandez and Wilson (2014), GSH-recycling in the GSH-dependent antioxidant system is critical in the colonization phase of host cells by M. oryzae. Moreover, loss of MoSFA1 increases the level of SNOs creating indirectly NO over-accumulation and significantly attenuated the virulence on plant hosts (Zhang et al., 2015). It worth pointing that both enzymes counteracting nitrosative stress, i.e., Fhb and GSNOR, promoted virulence of the human fungal pathogen Cryptococcus neoformans (de Jesús-Berrɪos et al., 2003).
An unknown strategy to remove NO or suppress its excessive accumulation has been suggested for G. orontii. In Arabidopsis leaves infected with the host-adapted powdery mildew a localized and high peak of NO formation coincided in time with appressorium formation by G. orontii primary hyphae. Since the NO level rapidly declined to the background level after the initial burst, the biotrophic pathogen might actively modulate its amount by degradation/decomposition mechanisms and in consequence resume growth and colonize the whole leaf (Schlicht and Kombrink, 2013). In Candida albicans, a human fungal pathogen, RNS exerts fungistatic effects probably by causing damages through S-nitrosylation of proteins and glutathione. In response to this stress, a set of genes, including RNS detoxifying enzyme, is induced. Following RNS detoxification, C. albicans restored redox homeostatis and the resulted S-nitrosylated adducts could be repaired via denitrosylation, allowing the pathogen to resume growth (Brown et al., 2014).
It is worth pointing out that screening of a genomic DNA library makes it possible to identify a ntpA gene that conferred growth tolerance upon A. nidulans exposure to exogenous NO (Zhou et al., 2013). The ntpA disruption increased amounts of cellular SNO and provoked NO susceptibility. The gene coding a cysteine-rich 23-amino-acid peptide that reacts with NO and GSNO to generate an S-nitrosated peptide called inducible nitrosothionein (iNT). The NO scavenging role of iNT seems to be mediated by thioredoxin-dependent catalysis. Importantly, the authors highlighted that the ubiquitous distribution of iNT-like polypeptides constitutes a potent NO-detoxifying mechanism that is conserved among various organisms (Zhou et al., 2013).
Conclusion
Increasing knowledge from diverse systems indicates that NO plays a pivotal role in the immune response of plants attacked by pathogens with different lifestyle. However, despite extensive research on NO synthesis and signaling processes in plants interacting with biotic stressors, our understanding of NO in phytopathogens is very limited. What is more, a validity of the reports on the biological action of NO in pathogenic microorganisms should undergo some criticism due to the methods used for NO detection. For example, the specificity of widely used DAF-based dyes is questionable since under biological conditions the fluorescent triazole product can be formed from either nitrosative or oxidative chemistry (Bryan and Grisham, 2007). Also the specificity confirmation of NO-dependent fluorescence using the NO scavenger 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-l-oxyl-3-oxide (cPTIO) need to be used with caution because there is evidence that it can increase fluorescence (Arita et al., 2007). Finally, the use of NOS inhibitors in looking for NOS activity in fungal and oomycete pathogens does not provide an unequivocal confirmation for the enzyme presence, since these chemicals can inhibit other biosynthetic pathways as well (Planchet and Kaiser, 2006).
Fungi and oomycete pathogens have active sources of NO and pathways of its detoxication, which defend them against NO-induced damages and ensure the vital level required for the signaling function both in the pathogen physiological state and during host tissue colonization (Figure 1). Although pathogens with different lifestyles vary in their sensitivity to host-generated RNS, a flexible NO metabolism could constitute a good strategy to subdue the host plant.
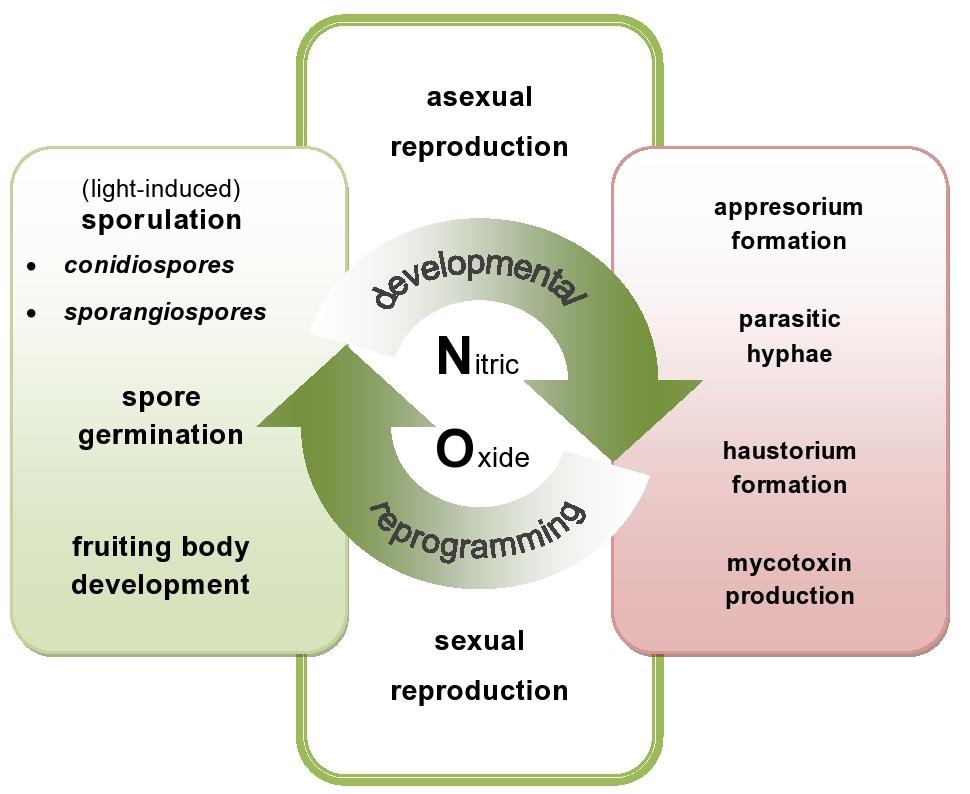
FIGURE 1. Nitric oxide (NO)-mediated events in plant pathogenic fungi and oomycete. NO might be directly or indirectly involved in the switch between developmental phases of the phytopathogens. On the one hand, endogenously produced NO participates in various developmental processes including sporulation, germination and fruiting bodies formation (green area – NO in developmental events). On the other hand, NO may be responsible for production of toxins and differentiation of various infection structures of the pathogens (red area – NO in offensive strategy).
Recognition of pathogen-derived NO during the battle between both adversaries will help us to resolve one of the central questions in plant pathology, namely what makes a pathogen successful and what makes a plant become a vulnerable host. To this end there are many important issues which need to be addressed. Firstly, research on additional genetic resources to unravel NO biosynthesis is needed. Secondly, the identification of cellular targets, degradation pathways and the mechanism/s of signal transduction need to be elucidated within different pathogen structures. Finally, fungal and oomycete pathogens, simultaneously exposed to combinations of different stimuli within the host cells, rather than to RNS alone, could activate specific NO sensing mechanisms relevant to the successful host colonization.
Author Contributions
MA-J and JF-W provided the idea, wrote the paper. All authors have read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported by the grant of National Science Centre, project NCN no. UMO-2014/13/B/NZ9/02177.
References
Abraham, L. D., Roth, A., Saddler, J. N., and Breuil, C. (1993). Growth, nutrition, and proteolytic activity of the sap-staining fungus Ophiostoma piceae. Can. J. Bot. 71, 1224–1230. doi: 10.1139/b93-144
Arasimowicz-Jelonek, M., and Floryszak-Wieczorek, J. (2014). Nitric oxide: an effective weapon of the plant or the pathogen? Mol. Plant Pathol. 15, 406–416. doi: 10.1111/mpp.12095
Arita, N. O., Cohen, M. F., Tokuda, G., and Yamasaki, H. (2007). “Fluorometric detection of nitric oxide with diaminofluoresceins (DAFs): applications and limitations for plant NO research,” in Nitric Oxide in Plant Growth, Development and Stress Physiology, eds L. Lamattina and J. C. Polacco (Berlin: Springer),268–280.
Asai, S., and Yoshioka, H. (2009). Nitric oxide as a partner of reactive oxygen species participates in disease resistance to necrotrophic pathogen Botrytis cinerea in Nicotiana benthamiana. Mol. Plant Microbe Interact. 22, 619–629. doi: 10.1094/MPMI-22-6-0619
Baidya, S., Cary, J. W., Grayburn, W. S., and Calvo, A. M. (2011). Role of nitric oxide and flavohemoglobin homolog genes in Aspergillus nidulans sexual development and mycotoxin production. Appl. Environ. Microbiol. 77, 5524–5528. doi: 10.1128/AEM.00638-11
Boccara, M., Mills, C. E., Zeier, J., Anzi, C., Lamb, C., Poole, R. K., et al. (2005). Flavohaemoglobin HmpX from Erwinia chrysanthemi confers nitrosative stress tolerance and affects the plant hypersensitive reaction by intercepting nitric oxide produced by the host. Plant J. 43, 226–237. doi: 10.1111/j.1365-313X.2005.02443.x
Brown, A. J. P., Budge, S., Kaloriti, D., Tillmann, A., Jacobsen, M. D., Yin, Z., et al. (2014). Stress adaptation in a pathogenic fungus. J. Exp. Biol. 217, 144–155. doi: 10.1242/jeb.088930
Bryan, N. S., and Grisham, M. B., (2007). Methods to detect nitric oxide and its metabolites in biological samples. Free Radic. Biol. Med. 43, 645–657. doi: 10.1016/j.freeradbiomed.2007.04.026
Chaki, M., Fernández-Ocaña, A. M., Valderrama, R., Carreras, A., Esteban, F. J., Luque, F., et al. (2009). Involvement of reactive nitrogen and oxygen species (RNS and ROS) in sunflower-mildew interaction. Plant Cell Physiol. 50, 265–279. doi: 10.1093/pcp/pcn196
Chiuchetta, S. J. R., and Castro-Prado, M. A. A. (2005). Genotoxic evaluation of sodium nitroprusside in Aspergillus nidulans. Genet. Mol. Biol. 28, 798–803. doi: 10.1590/S1415-47572005000500023
Conrath, U., Amoroso, G., Kohle, H., and Sultemeyer, D. F. (2004). Noninvasive online detection of nitric oxide from plants and some other organisms by mass spectrometry. Plant J. 38, 1015–1022. doi: 10.1111/j.1365-313X.2004.02096.x
de Jesús-BerrIos, M., Liu, L., Nussbaum, J. C., Cox, G. M., Stamler, J. S., and Heitman, J. (2003). Enzymes that counteract nitrosative stress promote fungal virulence. Curr. Biol. 13, 1963–1968. doi: 10.1016/j.cub.2003.10.029
Dean, R. A., and Timberlake, W. E. (1989). Production of cell wall-degrading enzymes by Aspergillus nidulans: a model system for fungal pathogenesis of plants. Plant Cell 1, 265–273. doi: 10.2307/3869006
Dyer, P. S., and O’Gorman, C. M. (2012). Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiol. Rev. 36, 165–192. doi: 10.1111/j.1574-6976.2011.00308.x
Feechan, A., Kwon, E., Yun, B. W., Wang, Y. Q., Pallas, J. A., and Loake, G. J. (2005). A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. U.S.A. 102, 8054–8059. doi: 10.1073/pnas.0501456102
Fernandez, J., and Wilson, R. A. (2014). Characterizing roles for the glutathione reductase, thioredoxin reductase and thioredoxin peroxidase-encoding genes of Magnaporthe oryzae during rice blast disease. PLoS ONE 9:e87300. doi: 10.1371/journal.pone.0087300
Floryszak-Wieczorek, J., Arasimowicz, M., Milczarek, G., Jelen, H., and Jackowiak, H. (2007). Only an early nitric oxide burst and the following wave of secondary nitric oxide generation enhanced effective defence responses of pelargonium to a necrotrophic pathogen. New Phytol. 175, 718–730. doi: 10.1111/j.1469-8137.2007.02142.x
Forrester, M. T., and Foster, M. W. (2012). Protection from nitrosative stress: a central role for microbial flavohemoglobin. Free Radic. Biol. Med. 52, 1620–1633. doi: 10.1016/j.freeradbiomed.2012.01.028
Gong, X. Y., Fu, Y. P., Jiang, D. H., Li, G. Q., Yi, X. H., and Peng, Y. L. (2007). L-arginine is essential for conidiation in the filamentous fungus Coniothyrium minitans. Fungal Genet. Biol. 44, 1368–1379. doi: 10.1016/j.fgb.2007.07.007
Kobayashi, M., Matsuo, Y., Takimoto, A., Suzuki, S., Maruo, F., and Shoun, H. (1996). Denitrification, a novel type of respiratory metabolism in fungal mitochondrion. J. Biol. Chem. 271, 16263–16267. doi: 10.1074/jbc.271.27.16263
Kobayashi, M., and Shoun, H. (1995). The copper-containing dissimilatory nitrite reductase involved in the denitrifying system of the fungus Fusarium oxysporum. J. Biol. Chem. 270, 4146–4151. doi: 10.1074/jbc.270.8.4146
Lai, T., Li, B., Qin, G., and Tian, S. (2011). Oxidative damage Involves in the Inhibitory effect of nitric oxide on spore germination of Penicillium expansum. Curr. Microbiol. 62, 229–234. doi: 10.1007/s00284-010-9695-1
Lazar, E. E., Wills, R. B. H., Ho, B. T., Harris, A. M., and Spohr, L. J. (2008). Antifungal effect of gaseous nitric oxide on mycelium growth, sporulation and spore germination of the postharvest horticulture pathogens, Aspergillus niger, Monilinia fructicola and Penicillium italicum. Lett. Appl. Microbiol. 46, 688–692. doi: 10.1111/j.1472-765X.2008.02373.x
Li, B., Fu, Y., Jiang, D., Xie, J., Cheng, J., Li, G., et al. (2010). Cyclic GMP as a second messenger in the nitric oxide-mediated conidiation of the mycoparasite Coniothyrium minitans. Appl. Environ. Microbiol. 76, 2830–2836. doi: 10.1128/AEM.02214-09
Maier, J., Hecker, R., Rockel, P., and Ninnemann, H. (2001). Role of nitric oxide synthase in the light-induced development of sporangiophores in Phycomyces blakesleeanus. Plant Physiol. 126, 1323–1330. doi: 10.1104/pp.126.3.1323
Maier, J., and Ninnemann, H. (1995). Biosynthesis of pteridines in Neurospora crassa, Phycomyces blakesleeanus and Euglena gracilis: detection and characterization of biosynthetic enzymes. Photochem. Photobiol. 61, 43–53. doi: 10.1111/j.1751-1097.1995.tb09241.x
Marcos, A. T., Ramos, M. S., Marcos, J. F., Carmona, L., Strauss, J., and Cánovas, D. (2016). Nitric oxide synthesis by nitrate reductase is regulated during development in Aspergillus. Mol. Microbiol. 99, 15–33. doi: 10.1111/mmi.13211
Morozkina, E. V., and Kurakov, A. V. (2007). Dissimilatory nitrate reduction in fungi under conditions of hypoxia and anoxia: a review. Appl. Biochem. Microbiol. 43, 544–549. doi: 10.1134/S0003683807050079
Mur, L. A., Carver, T. L., and Prats, E. (2006). NO way to live; the various roles of nitric oxide in plant–pathogen interactions. J. Exp. Bot. 57, 489–505. doi: 10.1093/jxb/erj052
Ninnemann, H., and Maier, J. (1996). Indications for the occurrence of nitric oxide synthases in fungi and plants and the involvement in photoconidiation of Neurospora crassa. Photochem. Photobiol. 64, 393–398. doi: 10.1111/j.1751-1097.1996.tb02477.x
Perchepied, L., Balagué, C., Riou, C., Claudel-Renard, C., Rivière, N., Grezes-Besset, B., et al. (2010). Nitric oxide participates in the complex interplay of defense-related signaling pathways controlling disease resistance to Sclerotinia sclerotiorumin Arabidopsis thaliana. Mol. Plant Microbe Interact. 23, 846–860. doi: 10.1094/MPMI-23-7-0846
Piterková, J., Hofman, J., Mieslerová, B., Sedlárová, M., Luhová, L., Lebeda, A., et al. (2011). Dual role of nitric oxide in Solanum spp.–Oidium neolycopersici interactions. Environ. Exp. Bot. 74, 37–44. doi: 10.1016/j.envexpbot.2011.04.016
Piterková, J., Petřivalský, M., Luhová, L., Mieslerová, B., Sedlářová, M., and Lebeda, A. (2009). Local and systemic production of nitric oxide in tomato responses to powdery mildew infection. Mol. Plant Pathol. 10, 501–513. doi: 10.1111/j.1364-3703.2009.00551.x
Planchet, E., and Kaiser, W. M. (2006). Nitric oxide (NO) detection by DAF fluorescence and chemiluminescence: a comparison using abiotic and biotic NO sources. J. Exp. Bot. 57, 3043–3055. doi: 10.1093/jxb/erl070
Prats, E., Carver, T. L. W., and Mur, L. A. J. (2008). Pathogen-derived nitric oxide influences formation of the appressorium infection structure in the phytopathogenic fungus Blumeria graminis. Res. Microbiol. 159, 476–480. doi: 10.1016/j.resmic.2008.04.001
Röszer, T. (2012). “NO synthesis in subcellular compartments of fungi,” in The Biology of Subcellular nitric Oxide, ed. T. Röszer (New York: Springer Science Business Media B.V), 83–102.
Samalova, M., Johnson, J., Illes, M., Kelly, S., Fricker, M., and Gurr, S. (2013). Nitric oxide generated by the rice blast fungus Magnaporthe oryzae drives plant infection. New Phytol. 197, 207–222. doi: 10.1111/j.1469-8137.2012.04368.x
Sarkar, T. S., Biswas, P., Ghosh, S. K., and Ghosh, S. (2014). Nitric oxide production by necrotrophic pathogen Macrophomina phaseolina and the host plant in charcoal rot disease of jute: complexity of the interplay between necrotroph–host plant interactions. PLoS ONE 9:e107348. doi: 10.1371/journal.pone.0107348
Schlicht, M., and Kombrink, E. (2013). The role of nitric oxide in the interaction of Arabidopsis thaliana with the biotrophic fungi, Golovinomyces orontii and Erysiphe pisi. Front. Plant Sci. 4:351. doi: 10.3389/fpls.2013.00351
Sedlářová, M., Petřivalský, M., Piterková, J., Luhová, L., Kočířová, J., and Lebeda, A. (2011). Influence of nitric oxide and reactive oxygen species on development of lettuce downy mildew in Lactuca spp. Eur. J. Plant Pathol. 129, 267–280. doi: 10.1007/s10658-010-9626-9
Song, N. K., Jeong, C. S., and Choi, H. S. (2000). Identification of nitric oxide synthase in Flammulina velutipes. Mycologia 92, 1027–1032. doi: 10.2307/3761467
Tillmann, A. T., Strijbis, K., Cameron, G., Radmaneshfar, E., Thiel, M., Munro, C. A., et al. (2015). Contribution of Fdh3 and Glr1 to glutathione redox state, stress adaptation and virulence in Candida albicans. PLoS ONE 10:e0126940. doi: 10.1371/journal.pone.0126940
Turrion-Gomez, J. L., and Benito, E. P. (2011). Flux of nitric oxide between the necrotrophic pathogen Botrytis cinerea and the host plant. Mol. Plant Pathol. 12, 606–616. doi: 10.1111/j.1364-3703.2010.00695.x
Turrion-Gomez, J. L., Eslava, A. P., and Benito, E. P. (2010). The flavohemoglobin BCFHG1 is the main NO detoxification system and confers protection against nitrosative conditions but is not a virulence factor in the fungal necrotroph Botrytis cinerea. Fungal Genet. Biol. 47, 484–496. doi: 10.1016/j.fgb.2010.03.001
Uchimura, H., Enjoji, H., Seki, T., Taguchi, A., Takaya, N., and Shoun, H. (2002). Nitrate reductase-formate dehydrogenase couple involved in the fungal denitrification by Fusarium oxysporum. J. Biochem. 131, 579–586. doi: 10.1093/oxfordjournals.jbchem.a003137
van Baarlen, P., Staats, M., and van Kan, J. A. L. (2004). Induction of programmed cell death in lily by the fungal pathogen Botrytis elliptica. Mol. Plant Pathol. 5, 559–574. doi: 10.1111/j.1364-3703.2004.00253.x
Vandelle, E., Poinssot, B., Wendehenne, D., Bentéjac, M., and Alain, P. (2006). Integrated signaling network involving calcium, nitric oxide, and active oxygen species but not mitogen-activated protein kinases in BcPG1-elicited grapevine defenses. Mol. Plant Microbe Interact. 19, 429–440. doi: 10.1094/MPMI-19-0429
Vieira, A. L. G., Linares, E., Augusto, O., and Gomes, S. L. (2009). Evidence of a Ca2+-NO-cGMP signaling pathway controlling zoospore biogenesis in the aquatic fungus Blastocladiella emersonii. Fungal Genet. Biol. 46, 575–584. doi: 10.1016/j.fgb.2009.04.002
Wang, J., and Higgins, V. J. (2005). Nitric oxide has a regulatory effect in the germination of conidia of Colletotrichum coccodes. Fungal Genet. Biol. 42, 284–292. doi: 10.1016/j.fgb.2004.12.006
Ye, R. W., Averill, B. A., and Tiedje, J. M. (1994). Denitrification: production and consumption of nitric oxide. Appl. Environ. Microbiol. 60, 1053–1058.
Yu, M., Yun, B. W., Spoel, S. H., and Loake, G. J. (2012). A sleigh ride through the SNO: regulation of plant immune function by protein S-nitrosylation. Curr. Opin. Plant Biol. 15, 424–430. doi: 10.1016/j.pbi.2012.03.005
Zhang, Z., Wang, J., Chai, R., Qiu, H., Jiang, H., Mao, X., et al. (2015). An S-(Hydroxymethyl) Glutathione dehydrogenase is involved in conidiation and full virulence in the rice blast fungus Magnaporthe oryzae. PLoS ONE 10:e0120627. doi: 10.1371/journal.pone.0120627
Keywords: pathogen-derived nitric oxide, nitrosative stress resistance, defense response, biotrophic pathogens, necrotrophic pathogens
Citation: Arasimowicz-Jelonek M and Floryszak-Wieczorek J (2016) Nitric Oxide in the Offensive Strategy of Fungal and Oomycete Plant Pathogens. Front. Plant Sci. 7:252. doi: 10.3389/fpls.2016.00252
Received: 11 December 2015; Accepted: 15 February 2016;
Published: 04 March 2016.
Edited by:
Jeremy Astier, Helmholtz Zentrum München, GermanyReviewed by:
Marek Petrivalsky, Palacý University, Czech RepublicOlivier Lamotte, CNRS – UMR Agroécologie, France
Copyright © 2016 Arasimowicz-Jelonek and Floryszak-Wieczorek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Magdalena Arasimowicz-Jelonek, arasim@amu.edu.pl
 Magdalena Arasimowicz-Jelonek
Magdalena Arasimowicz-Jelonek Jolanta Floryszak-Wieczorek
Jolanta Floryszak-Wieczorek