- 1Marine Biotechnology and Ecology Division, CSIR-Central Salt and Marine Chemicals Research Institute, Bhavnagar, India
- 2Academy of Scientific and Innovative Research, Council of Scientific and Industrial Research, New Delhi, India
The universal stress protein (USP) is a ubiquitous protein and plays an indispensable role in plant abiotic stress tolerance. The genome of Salicornia brachiata contains two homologs of intron less SbUSP gene which encodes for salt and osmotic responsive USP. In vivo localization reveals that SbUSP is a membrane bound cytosolic protein. The role of the gene was functionally validated by developing transgenic tobacco and compared with control [wild-type (WT) and vector control (VC)] plants under different abiotic stress condition. Transgenic lines (T1) exhibited higher chlorophyll, relative water, proline, total sugar, reducing sugar, free amino acids, polyphenol contents, osmotic potential, membrane stability, and lower electrolyte leakage and lipid peroxidation (malondialdehyde content) under stress treatments than control (WT and VC) plants. Lower accumulation of H2O2 and radicals was also detected in transgenic lines compared to control plants under stress conditions. Present study confers that overexpression of the SbUSP gene enhances plant growth, alleviates ROS buildup, maintains ion homeostasis and improves the physiological status of the plant under salt and osmotic stresses. Principal component analysis exhibited a statistical distinction of plant response to salinity stress, and a significant response was observed for transgenic lines under stress, which provides stress endurance to the plant. A possible signaling role is proposed that some downstream genes may get activated by abiotic stress responsive cytosolic SbUSP, which leads to the protection of cell from oxidative damages. The study unveils that ectopic expression of the gene mitigates salt or osmotic stress by scavenging ROS and modulating the physiological process of the plant.
Introduction
Abiotic stresses, especially salinity and drought are an important limiting factor in plant growth due to increased salinization of soil and water as well as the global scarcity of water resources (Vinocur and Altman, 2005). Abiotic stresses (salinity, dehydration, extreme temperature, highlight intensity, etc.) pessimistically impinge on plant growth and development through biomass accumulation and grain yield. Abiotic stresses are multigenic in nature, and the cellular responses are often interconnected. Thus, abiotic stress tolerance in a plant is synchronized action of multiple stress responsive genes, which communicate in stress signal transduction pathways with other components (Tuteja, 2007). Salinity negatively affects plant growth in two distinct phases; initial osmotic phase and the late ionic phase (Munns and Tester, 2008). These phases can be experimentally distinguished by measuring effects in the short term (within minutes to a few days) upon addition of salt or after longer (several days to weeks) times (Yamamoto et al., 2015). The cellular and molecular responses include accumulation of stress proteins, up regulation of stress responsive genes for antioxidants, transcription factors, compatible solutes, transporters/channels, and phytohormones, etc. Molecular mechanism of stress responses plays a key role in devising a strategy to engineer stress tolerant plants (Zahur et al., 2009). It is observed that abscisic acid (ABA) accumulates under drought stress condition, which promotes expression of numerous ABA dependent drought and abiotic stress responsive genes. Though substantial researches have been carried on the abiotic stress response of plants, there is inadequate information on molecular regulatory mechanisms of environmental stress responsive genes and proteins (Isokpehi et al., 2011). There are a number of reports on stress associated genes and proteins, but they are still functionally unknown or poorly characterized. Functional validation of these stress responsive genes or proteins will unveil a new regulatory molecular mechanism, which may consider a promising approach for developing crop plants, tolerant to environmental stresses.
The universal stress proteins (Usp) were named initially as C13.5 protein on the basis of their migration on two-dimensional IEF-PAGE gel. Nevertheless, UspA quickly became extensively acknowledged, and the protein has specified its name to a rising orthologous group of proteins, the UspA superfamily (Kvint et al., 2003). The universal stress protein A (UspA) superfamily covers primordial and preserved group of proteins originated in bacteria, archaea, fungi, flies, and plants. Kerk et al. (2003) reported enigmatic functions of USPA containing protein kinases in Arabidopsis. Even though the regulation of the E. coli UspA gene has been known to some extent, but the precise role of Usp proteins and Usp domains stay unfathomable, they have been emerged to be linked for providing resistance to DNA damaging agents and respiratory uncouplers. In E. coli, the USPs have been grouped into four classes according to their structural analysis and amino acid sequence; Class-I: UspA, UspC, UspD; Class-II: UspF and UspG; Class-III and Class-IV: Two Usp domains of UspE (Isokpehi et al., 2011). There are six families of USP genes viz. UspA, UspC, UspD, UspE, UspF, and UspG, whereas, UspB is not considered as a bona fide Usp family protein (Kvint et al., 2003). The six USP genes of E. coli have different functions linked to motility, adhesion and oxidative stress resistance (Loukehaich et al., 2012). It is observed that the majority of Usp genes are monocistronically expressed from σ70 promoter, but high temperature, ethanol, and tetracycline stress reduce the expression level of Usp proteins (Kvint et al., 2003). This might be due to the inactivation of σ70 promoter at high temperature stress. USP homologs are ubiquitous in plants and encoded by gene families while the functions of USPs remain largely unknown (Loukehaich et al., 2012). It appears that USP boost up the rate of cell survival during prolonged exposure to stress agents, and it may endow plants with wide ranging stress endurance activity (Zahur et al., 2009). The UspA protects cells from detrimental effect of toxic chemicals and osmotic stress or UV light (Kerk et al., 2003). Few plant USP genes have been characterized viz. Gossypium arboreum, Astragalus sinicus, Vicia faba, Solanum pennellii, Barley, and Viridiplantae (Sauter et al., 2002; Chou et al., 2007; Merkouropoulos et al., 2008; Maqbool et al., 2009; Zahur et al., 2009; Li et al., 2010; Isokpehi et al., 2011; Loukehaich et al., 2012). USPs are divided into two categories; first group binds with ATP (UspF and UspG-type) and second do not, such as UspAs and UspA-like group (Tkaczuk et al., 2013). The number of copies of USPs varies amongst different organisms and majority of them have many paralogous of USPs. For instance, the known copies of genes encoding USPs in A. thaliana are four, Nitrosomonas europaea and Archaeoglobus fulgidus are six and eight, respectively (Tkaczuk et al., 2013). Kvint et al. (2003) proposed that the ATP-binding Usp domain is a member of a group of α/β domains called the HUP domain (HIGH-signature proteins, Usp-like domains, and PP-ATPases).
A halophyte Salicornia brachiata requires NaCl for tissue culture (Joshi et al., 2012), owns sulfur rich seed storage proteins (Jha et al., 2012), unique oligosaccharides (Mishra et al., 2013), and metabolites (Mishra et al., 2015). Several candidate genes from this halophyte extensively explored for abiotic stress responsive genes and promoters for developing abiotic stress tolerant transgenic plants (Jha et al., 2011; Chaturvedi et al., 2012, 2014; Joshi et al., 2013; Singh et al., 2014a,b; Tiwari et al., 2014, 2015, 2016; Udawat et al., 2014). The present study addresses first report on the functional characterization of the SbUSP gene by ectopic expression in a model plant and further analyses under varying abiotic stress condition. It was observed that overexpression of the SbUSP gene mitigates salt and osmotic stress and provides endurance to the transgenic plant by modulating physiological processes.
Materials and Methods
Genomic Organization of SbUSP Gene
Plant (S. brachiata) genomic DNA was extracted using modified CTAB (N-cetyl-N,N,N-trimethyl ammonium bromide) method (Bubner et al., 2004) with minor modifications, quantified using ND-1000 spectrophotometer and qualitatively analyzed by agarose gel electrophoresis. The SbUSP gene was amplified with primer pairs (F: 5′-ATGGCAATGTCTGATAAACC-3′ and R: 5′-TCAGTGCTTAATCTTAGGCTT-3′) using genomic DNA as template, cloned into the pGEM-T easy vector and sequenced (M/s Macrogen, S. Korea).
The copy number of SbUSP gene was determined using Southern blot analysis. Genomic DNA (2 μg) was digested with restriction enzymes (EcoR1 and BamH1) and transferred to Hybond N+ membrane (Amersham Pharmacia, UK) using alkaline transfer buffer (0.4 N NaOH along with 1 M NaCl) by upward capillary transfer. The blot was hybridized overnight at 42°C with PCR generated DIG-11-dUTP labeled SbUSP probe (F: 5′-ATGGCAATGTCTGATAAACC-3′ and R: 5′-AGGAGGACAATGCGTGCTT-3′) and signals were detected on X-ray film by using CDP-Star as a chemiluminescent substrate, as described by manufacturer instructions (Roche, Germany).
Subcellular Localization
The gateway technology was used to generate translational fusion cassette of SbUSP along with RFP (red fluorescent protein). The SbUSP CDS was PCR amplified using accuPrime Pfx DNA polymerase along with SbUSPF (5′-CACCATGGCAGAAGTTGATT-3′) and SbUSPR (5′-TCAGTGCTTAATCTTAGGCTT-3′) primers. The PCR product, amplified with blunt ends was cloned into entry vector (pENTER/D-TOPO, Invitrogen, USA) and sequenced. Subsequently, LR recombination reaction was executed between an attL-containing entry clone pENTER/D-TOPO-SbUSP vector and an attR-containing destination vector pSITE-4CA (RFP) using gateway LR Clonase II enzyme mix (Invitrogen, USA). Cloning was confirmed by PCR, and resulting expression cassette (RFP:SbUSP) and control vector (pSITE-4CA:RFP) were transformed to onion epidermal cells using particle bombardment with gene gun (PDS-1000/He Biolistic, Bio-Rad, USA). Transformants were observed after incubation in dark for 12 h for transient expression of RFP using an epifluorescence microscope (Axio Imager, Carl Zeiss AG, Germany).
Genetic Transformation to Tobacco Plants and Molecular Confirmation
The SbUSP CDS was amplified using proofreading enzyme accuPrime Pfx DNA polymerase along with primer pairs (F: 5′-CTCGAGATGGCAATGTCTGATAAACC-3′ and R: 5′-GGTACCTCAGTGCTTAATCTTAGGCTT-3′) having KpnI and XhoI sites, respectively. The SbUSP cDNA was cloned in pCAMBIA1301 via an intermediate pRT101 plant expression vector and recombinant pCAMBIA1301:35S:SbUSP vector was mobilized into Agrobacterium tumefaciens strain LBA4404. Tobacco (Nicotiana tabacum cv. Petit Havana) plants were transformed by leaf disk method (Horsch et al., 1985) and shoot buds directly generated from leaf disks were transferred to shoot elongation and subsequently to rooting media supplemented with hygromycin (20 mgL-1).
Putative transgenic lines (T0 and T1) were screened by growing on hygromycin (20 mgL-1), and the presence of transgenes was confirmed by PCR amplification of uidA (F: 5′-GATCGCGAAAACTGTGGAAT-3′ and R: 5′-TGAGCGTC GCAGAACATTAC-3′) and SbUSP genes. The activity of reporter gene β-glucuronidase in the leaf segment of transgenic (overexpressing SbUSP gene), VC (vector control; transgenic plant transformed with pCAMBIA1301 vector only) and WT (wild-type plant, non-transgenic) plants was visualized via GUS activity (Jefferson, 1987) using β-glucuronidase reporter gene staining kit (Sigma, USA). The integration of transgene and its copy number was determined by Southern blot analysis following manufacturer instructions (Roche, Germany).
Overexpression Analysis of SbUSP Gene
Total RNA was extracted from transgenic, VC and WT plants (abiotic stressed and unstressed), quantified and cDNA was prepared using Superscript II RT (Invitrogen, USA). The semi-quantitative reverse transcriptase PCR (RT-PCR) was performed thrice independently (each containing three plants) for the SbUSP gene and β-tubulin gene (F: 5′-GGAGTCACCGAGGCAGAG-3′ and R: 5′-ATCACATATCAGAAACCACAA-3′) was used as an internal reference. Similarly, quantitative Real Time PCR (qRT-PCR) was performed to study the relative fold expression of SbUSP gene in transgenic lines under salinity stress.
Analyses of T1 Transgenic Plants Exposed to Varying Abiotic Stresses
The T1 seeds (25 seeds from each plant) of SbUSP overexpressing transgenic lines (L7, L11, L14) along with VC and WT plants were germinated on MS media supplemented with NaCl (200 mM) or mannitol (300 mM) for three weeks under controlled conditions, and percentage seed germination was studied (Pandey et al., 2015). Percent germination was repeated three times and each contained three biological replicates. Equal size of hygromycin (20 mgL-1) positive T1 seedlings were transferred to MS media supplemented with NaCl (200 mM) or PEG (10%) and subsequently incubated for 21 days for root length, shoot length, and ion content analyses. Additionally, 45 days old plantlets (transgenic lines, VC and WT) were subjected to NaCl (200 mM), PEG (10%), ABA (20 μM) or SA (10 μM) stress for 24 h; thereafter samples were harvested. The function of SbUSP gene in transgenic plants was validated through morphological and physio-biochemical and molecular analyses viz. fresh weight, dry weight, osmotic potential, membrane stability index, electrolyte leakage, chlorophyll estimation, estimation, in vivo localization of H2O2 and radicals, etc.
Quantification of Osmotic Adjustment
The leaf segments of 45 days old stressed and unstressed tobacco plants were harvested and incubated at -20°C. After overnight incubation, plant samples were thawed and centrifuged at 13,000 rpm for 10 min. The cell sap extract was used to measure cellular osmotic potential using vapor pressure osmometer (VAPRO, Wescor Inc., USA). The experiment was carried out three times, and each contained three biological replicates.
Relative Water Content (RWC)
Fresh weight from control (WT and VC) and transgenic tobacco plants was determined, and leaf samples were floated overnight in deionised water. The turgid weight was determined, and all samples were dried at a constant temperature. Dry weight was measured, and percent relative water content (RWC, %) was calculated using following equation. RWC was measured three times, each containing three biological replicates.
Senescence Assay and Estimation of Chlorophyll Content
Leaf disks from 45 days old control and transgenic tobacco plants were punched out, and 11 leaf disks of similar size from each plant were floated in ½ MS (control) supplemented with NaCl (100, 150, or 200 mM) or PEG (10%) for 8 days. Effects of each treatment were gaged by visually analyzing phenotypic changes amongst leaf segments. Furthermore, total chlorophyll contents were isolated from leaf disks and determined as per gram fresh weight of tissue (Inskeep and Bloom, 1985). The experiment was repeated three times with three biological replicates each.
Superoxide Estimation and Cell Viability Test
Production of superoxide radicals under NaCl (200 mM) or PEG (10%) stress was quantified using 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) assay (Hema et al., 2014). Leaf segments (of equal size and weight) were incubated for 5 h in potassium phosphate buffer (20 mM, pH 6.0) containing 500 μM XTT and increase in absorbance was recorded at 470 nm.
Similarly, leaf segments (similar weight and size) of 45 days old stressed and unstressed plants were used for estimation of cell viability by 2,3,5-triphenyltetrazolium chloride (TTC) assay (Hema et al., 2014) in sodium phosphate buffer (pH 7.4). Leaf segments were thoroughly washed with sterile water and incubated in TTC solution at room temperature in the dark for 6 h. Samples were boiled in 5 ml of 2-methoxy ethanol till aridness to dig-out bound formazan. Subsequently, 5 ml of 2-methoxy ethanol was again added, and absorbance was recorded at 485 nm. Both experiments were carried out three times and each contained three biological replicates.
Membrane Stability Index (MSI) and Electrolyte Leakage (EL)
Healthy young leaves were collected from stressed and unstressed WT and transgenic tobacco plants, and MSI was calculated by the method described by Sairam (1994). Leaf segments were placed in sterile water, and one set (L1) was incubated at 40°C for 30 min whereas other set (L2) was incubated at 100°C for 10 min. Electrical conductivity of samples from both sets was measured, and MSI was calculated using following formula:
For electrolyte leakage, leaf samples were initially washed with sterile water to eliminate surface stacked electrolytes. Healthy young leaf samples were incubated in deionized water in closed vials and kept overnight on a rotary shaker. Thereafter, electrical conductivity was measured (Lt) using conductivity meter (SevenEasy, Mettler Toledo AG 8603, Switzerland). Subsequently, leaf samples were autoclaved at 120°C for 15 min, further, they were cooled to 25°C, and electrical conductivity (L0) was again determined (Lutts et al., 1996). The percent electrolyte leakage was calculated using following formula:
The MSI and EL experiments were repeated three times with three biological replicates each.
Quantification of Proline, H2O2, and Lipid Peroxidation
The free proline content and H2O2 level in the stressed and unstressed WT and transgenic tobacco plants were determined using methods described by Bates et al. (1973) and He et al. (2005), respectively. A standard curve was prepared using known amount of proline and H2O2, and absorbance was measured at 520 and 560 nm for determining free proline and H2O2 content, respectively. Lipid peroxidation was analyzed by quantifying the amount of MDA (malondialdehyde) generated by TBA (thiobarbituric acid) reaction (Draper and Hadley, 1990). All experiments were repeated three times with three biological replicates each.
Quantification of Total Soluble Sugar, Reducing Sugar, Starch, Total Amino Acid, and Polyphenol Content
The total soluble sugar, reducing sugar and starch contents were determined against a standard curve prepared with glucose (Sigma–Aldrich, USA). Stressed and unstressed control and transgenic leaf samples were homogenized in 85% ethanol and treated with anthrone reagent. Samples were boiled, cooled up to 25°C, absorbance was measured at 620 nm and total soluble sugar was calculated from the standard curve. Reducing sugar was quantified by a colorimetric test using DNS method (Yemm and Willis, 1954). An aliquot of glucose or leaf sample was mixed with 3 ml of DNS reagent, boiled, cooled to room temperature, and color intensity was measured at 540 nm. For starch estimation, homogenized samples were treated with 52% perchloric acid, centrifuged, the supernatant was treated with 4 ml of anthrone reagent and absorbance was measured at 630 nm (Ghose, 1987). Free amino acids and polyphenol contents were determined against a standard curve prepared with glycine and catechol solution, respectively. The ethanolic extract of samples was redissolved in ninhydrin reagent or folin–ciocalteu reagent, and absorbance (color intensity) was measured at 570 or 650 nm, respectively. All experiments were carried out three times with three biological replicates each.
In Vivo Localization of H2O2 and
Histochemical staining with 3,3-diaminobenzidine (DAB) or nitro-blue tetrazolium (NBT) was performed for in vivo analyses of H2O2 and radical production (Shi et al., 2010). A measure of 1 mg ml-1 each stain solution was prepared in phosphate buffer (10 mM pH 7.8 for NBT and pH 7.3 for DAB). Leaf samples were immersed in the respective staining solution in the dark for 2 h, thereafter exposed to white light until blue and brown spots appeared. Total chlorophyll content was bleached and leaf samples were documented.
Analyses of Ion Content
Seedlings from stressed and unstressed, control and transgenic plants were subjected to dryness at 70°C for 48 h and dry weight was determined. Samples were acid digested overnight in perchloric acid-nitric acid solution (3:1) heated to dryness and redissolved in sterile water. Ion content was measured using inductively coupled plasma optical emission spectrometer (Optima2000DV, PerkinElmer, Germany). ICP analysis was done three times with three biological replicates each.
Quantitative RT PCR Analysis
The cDNA of all control and treated plant samples (WT, VC, and transgenic lines) viz. NaCl, PEG, ABA, and SA was synthesized from 5 μg total RNA using Superscript RT III first strand cDNA synthesis kit (Invitrogen, USA). The resulting cDNAs were subjected to real time qRT PCR analyses for selected antioxidant enzyme encoding genes of host plant (NtSOD, F: 5′-AGCTACATGACGCCATTTCC-3′ and R: 5′-CCCTGTAAAGCAGCACCTTC-3′; NtAPX, F: 5′-CAAATGTAAGAGGAAACTCAGAGGA-3′ and R: 5′-CAGCCTTGAGCCTCATGGTACCG-3′; NtCAT, F: 5′-AGGTACCGCTCATTCACACC-3′ and R: 5′-AAGCAAGCTTTTGACCCAGA-3′) followed by melt curve analysis for the validation of specificity of reaction. The relative fold expression change was calculated using the CT method, and internal reference gene β-tubulin was used as a reference (Livak and Schmittgen, 2001).
Statistical Analysis
For each set of experiment, data from three replicates, each containing three biological replicates were documented. Data sets were articulated as mean ± SE and were used to determine the significance of difference by analysis of variance (ANOVA) amongst the means of WT, VC, and transgenic plants of every treatment set. For comparison of multiple means Tukey HSD was used, p < 0.05 was considered significantly different from each other and designated by different letters. All dataset was analyzed individually and in combination by principal component analysis (PCA) and respective heat maps were generated. PCA is a multivariate analysis method by which correlation between variables were studied among multidimensional datasets. Plants grown under varying stress were selected as observations, whereas different morphological and physio-biochemical measurements were taken as variables. Observation and variable data set were used to generate Pearson’s correlation matrix, and PCA was analyzed using different software SigmaPlot (version 12), SYSTAT (version 13) and Origin (version 15).
Results
In Vivo Localization and Genome Organization
The in vivo subcellular localization study was accomplished by transient expression analysis of translational fusion construct pSITE-4CA:SbUSP and vector pSITE-4CA:RFP (Figures 1A,B) expressing RFP only in onion epidermal cells. The expression with pSITE-4CA vector showed uniform red fluorescence, whereas RFP:SbUSP fusion construct revealed fluorescence localized to the cytosolic membrane (Figure 1B). DAPI (4′,6-diamidino-2-phenylindole) is a fluorescent stain, which binds to double strand DNA at A-T rich region. DAPI has an emission maximum at 461 nm, and in fluorescence microscopy, it is detected through a blue filter. In the in vivo localization study, blue spot of DAPI indicates the position of the nucleus (as it binds with dsDNA). Overlapping of DAPI blue spots was not observed with RFP spots in the cells, transformed with RFP:SbUSP fusion construct. In the merged image, SbUSP bound RFP shows red fluorescence in the cytosol, outside the nucleus and inner side of the membrane. The result suggested that SbUSP is a membrane bound cytosol-localized protein. The in silico analysis also confirmed that SbUSP is membrane bound cytosolic protein. The SbUSP ORF amplified using full length gene specific primers from cDNA, and genomic DNA showed similar size, 486 bp. The comparative sequence analysis of cDNA clone compared to genomic clone further reveals single exon structure and confirms intron-less genomic organization the gene (data not shown). Furthermore, Southern blot analysis was commenced to identify the number of SbUSP copies (homologs) in the genome of S. brachiata. The analysis revealed a double copy of the SbUSP gene as the probe hybridized with two fragments of genomic DNA (Figure 1C).
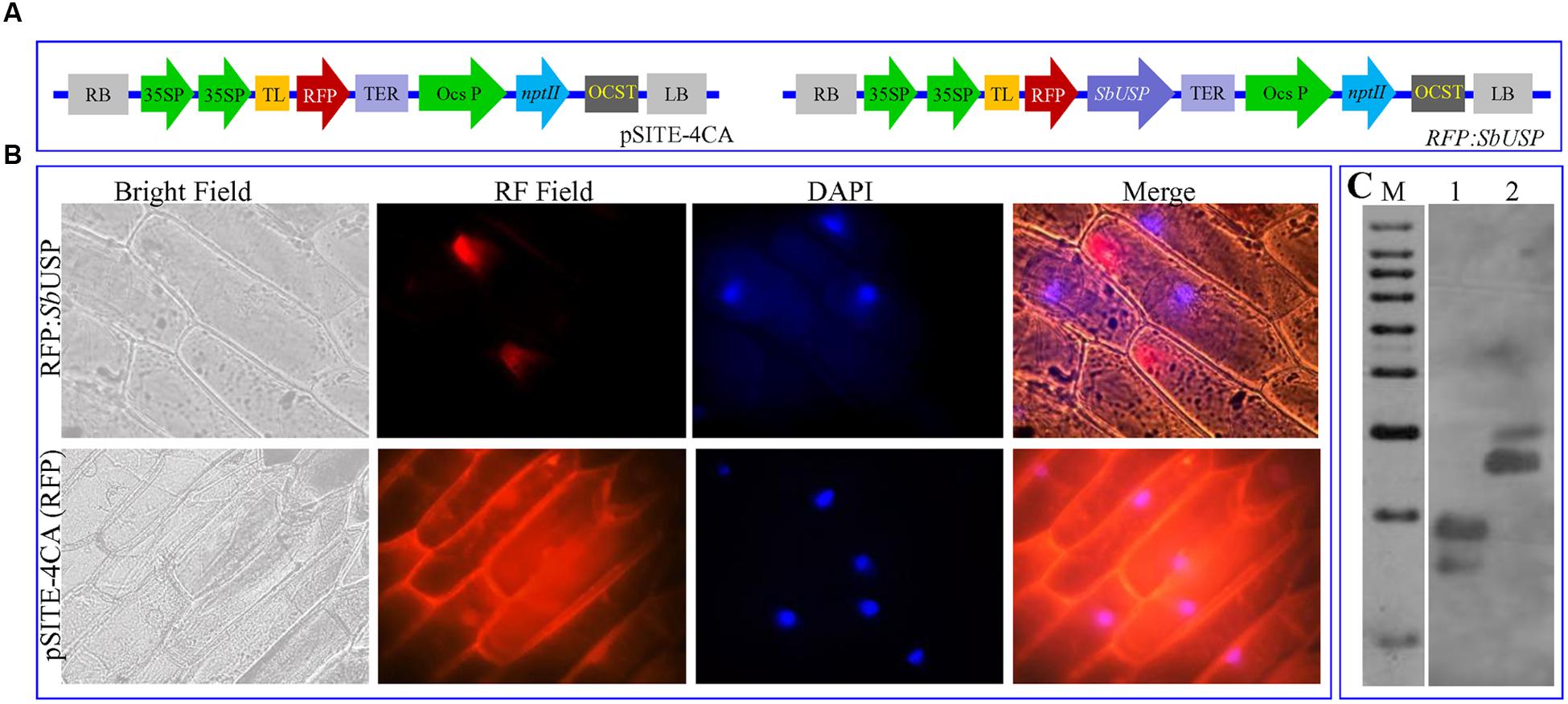
FIGURE 1. Sub-cellular localization of SbUSP protein and genome organization. (A) Vector pSITE-4CA(RFP) and SbUSP-1:RFP gene construct, (B) Transient expression of RFP alone and SbUSP:RFP translational fusion protein on transformed onion epidermal cells and (C) Southern hybridization of Salicornia brachiata genome to determine the copy number of SbUSP gene.
Molecular Analyses of Transgenic Tobacco Plants
Healthy leaves from the SbUSP gene (Figure 2A) overexpressing T1 lines (L1 to L17 lines), WT and VC plants were harvested and screened for the presence of transgenes (Supplementary Figure S1). Furthermore, Southern blot analysis confirmed presence of single copy transgene amongst L4, L7, L8, L11, L12, and L14 transgenic lines (Supplementary Figure S2A). The SbUSP gene overexpressing transgenic lines exhibited a high level of gus expression along with VC plants (Figure 2B). Comparative ectopic expression of the SbUSP gene was observed by semi-quantitative RT-PCR and quantitative Real Time PCR (qRT-PCR) for transgenic lines (Supplementary Figure S2) using β-tubulin as an internal reference gene. The analysis revealed high expression of the SbUSP gene in transgenic lines L7, L11, and L14, therefore, selected for further functional validation by morphological, physio-biochemical and molecular analyses.
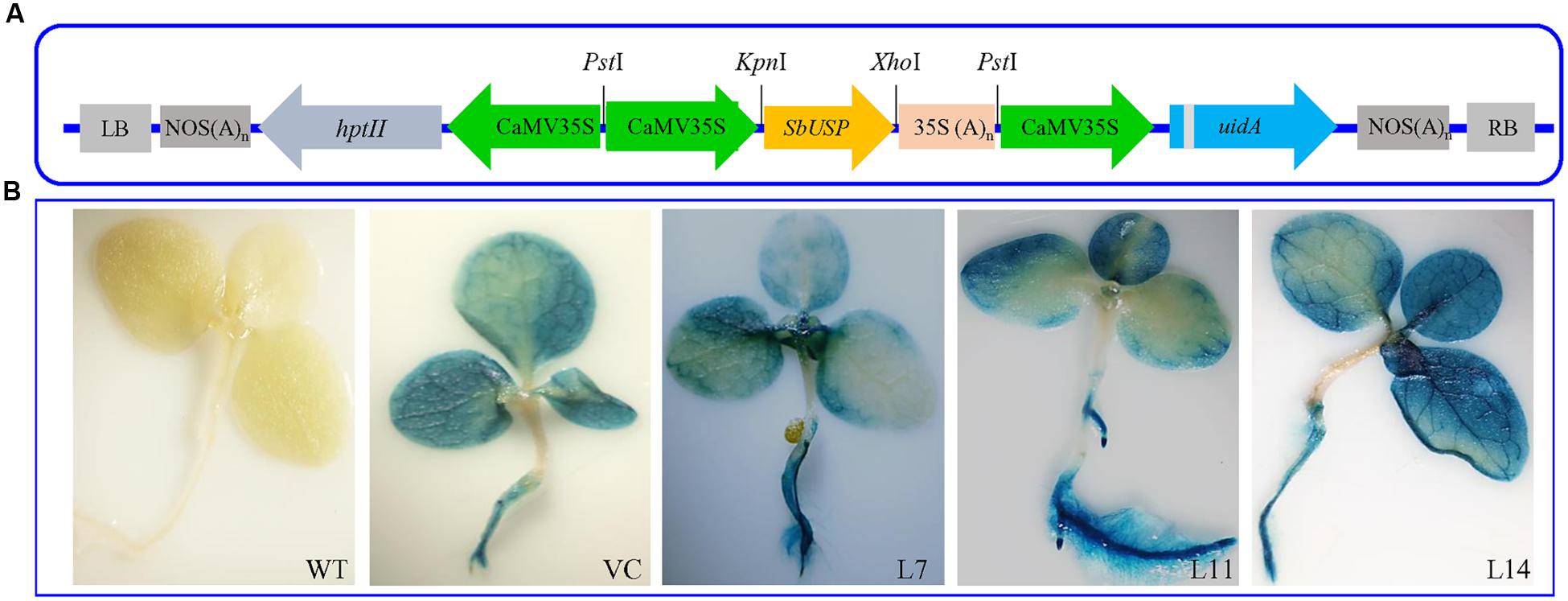
FIGURE 2. Genetic transformation and Histochemical GUS assay of transgenic tobacco plants. (A) Schematic representation of SbUSP-pCAMBIA1301 plant transformation vector construct and (B) Histochemical GUS assay of transgenic lines (L7, L11, and L14) along with control plants (WT and VC).
Overexpression of the SbUSP Gene Enhances Plant Growth under Salt and Osmotic Stress
Percentage seed germination was determined under control, NaCl, and osmotic stress condition (Figure 3). It was observed that transgenic lines (L7, L11, and L14) showed significantly (p < 0.05) higher percent seed germination compared to control plants (WT and VC) under the stressed condition. Similarly, growth parameters, including fresh weight, dry weight, shoot and root length were significantly (p < 0.05) improved in selected transgenic lines (L7, L11, and L14) compared to control plants (WT and VC) under stress conditions (Figure 4). Leaf segments reduce their viability and undergo senescence under salt and osmotic stress (Figure 5A). Salt and osmotic stress induced damages were lower in SbUSP overexpressing transgenic lines compared to their counterparts (WT and VC). Transgenic lines exhibited reduced rate of necrosis and therefore, significantly (p < 0.05) higher amount of chlorophyll content was observed in transgenic plants related to WT and VC plants. A degradation in cellular chlorophyll content was observed in WT and VC plants compared to transgenic lines (Figure 5B). Under control condition, chlorophyll content of WT, VC, and transgenic plants was comparable, whereas under increasing salt and osmotic stress, significantly higher amount of chlorophyll was found in transgenic lines. Additionally, transgenic lines showed significantly (p < 0.05) enhanced cell viability compared to WT and VC plants under stress condition (Figure 5C).
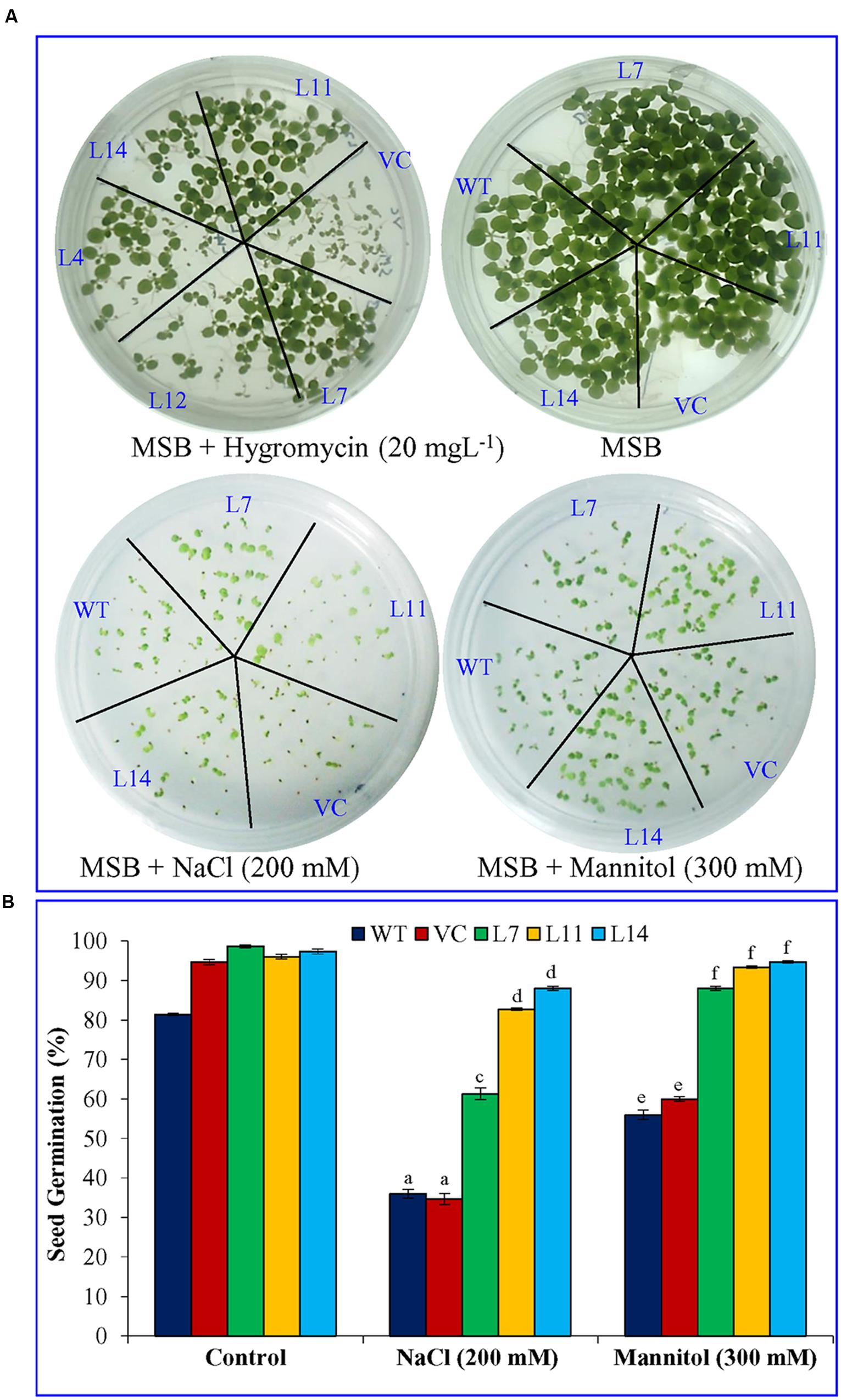
FIGURE 3. Seed germination analysis of transgenic tobacco plants under abiotic stress. (A) Morphology and (B) percent seed germination analysis of transgenic lines (L7, L11, and L14), WT and VC plants under salinity and osmotic stress condition. Bars represent means ± SE and values with different letters are significant at P < 0.05.
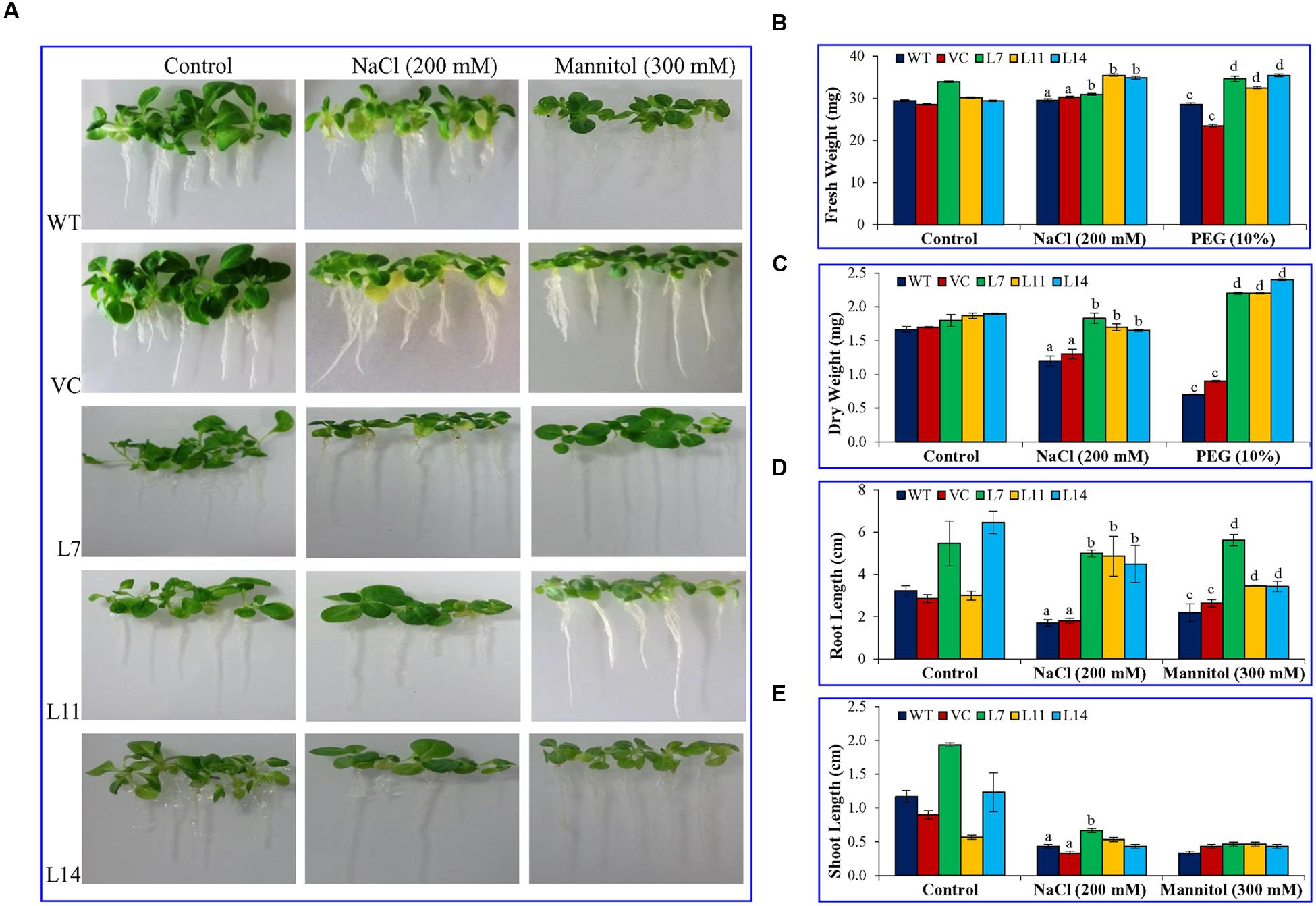
FIGURE 4. Plant growth analyses under salt and osmotic stress. Comparative study of (A) morphology, (B) fresh weight, (C) dry weight, (D) root length, and (E) shoot length of transgenic lines (L7, L11, and L14) and control plants (WT and VC) under salt and osmotic stress. Bars represent means ± SE and values with different letters are significant at P < 0.05.
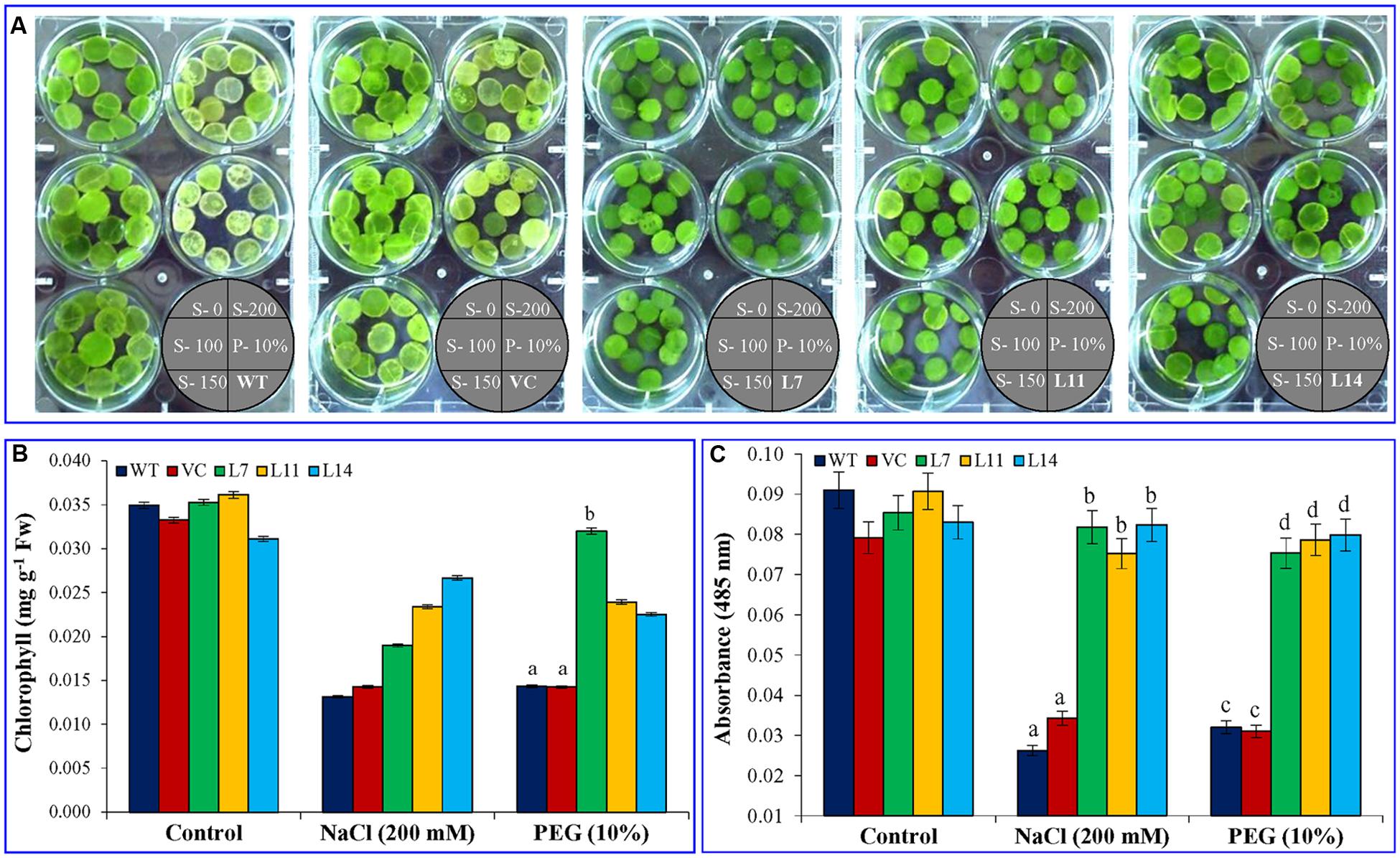
FIGURE 5. Leaf disk assay, estimation of chlorophyll content and cell viability. (A) Leaf disk assay, (B) total chlorophyll content, and (C) cell viability of transgenic (L7, L11, and L14) and control plants (WT and VC) under salt and osmotic stress condition. Bars represent means ± SE and values with different letters are significant at P < 0.05.
SbUSP Overexpression Alleviates ROS Buildup under Salinity and Oxidative Stress
The WT and VC plant leaves exhibited deeper NBT and DAB staining than transgenic lines exposed to salt and osmotic stress (Figure 6). Results confirmed that control plants (WT and VC) accumulate more and H2O2 compared to transgenic plants and thus validating the role of SbUSP gene in providing oxidative stress tolerance. Abiotic stress also leads to the generation of superoxide radicals. It was demonstrated by the reduction of tetrazolium salts XTT, resulting significant (p < 0.05) increase in absorbance (470 nm) in transgenic lines compared to control plants (WT and VC) under stress condition (Supplementary Figure S3).
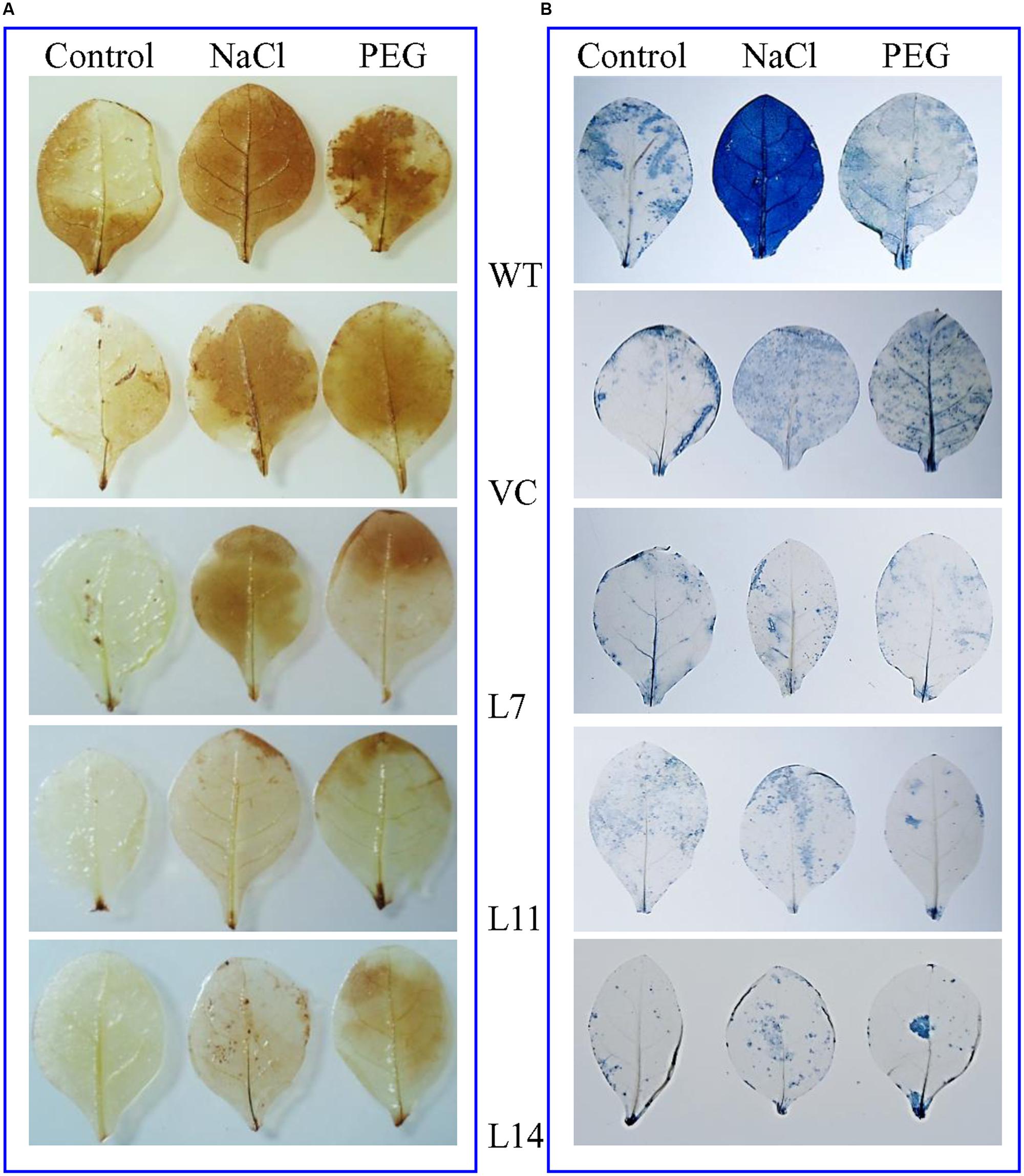
FIGURE 6. In vivo localization of peroxide and superoxide free radicals. (A) DAB and (B) NBT staining of transgenic (L7, L11, and L14) and control (WT and VC) leaves.
Proline and MDA contents were comparable in WT, VC, and transgenic lines under control condition (Figure 7). Transgenic lines accumulated significantly (p < 0.05) high proline compared to control plants under stress (Figure 7A). In contrast, elevated H2O2 and MDA contents (p < 0.05) were found in control plants under stress condition compared to transgenic lines (Figures 7B,C). Results further confirmed its (SbUSP) role in oxidative stress tolerance.
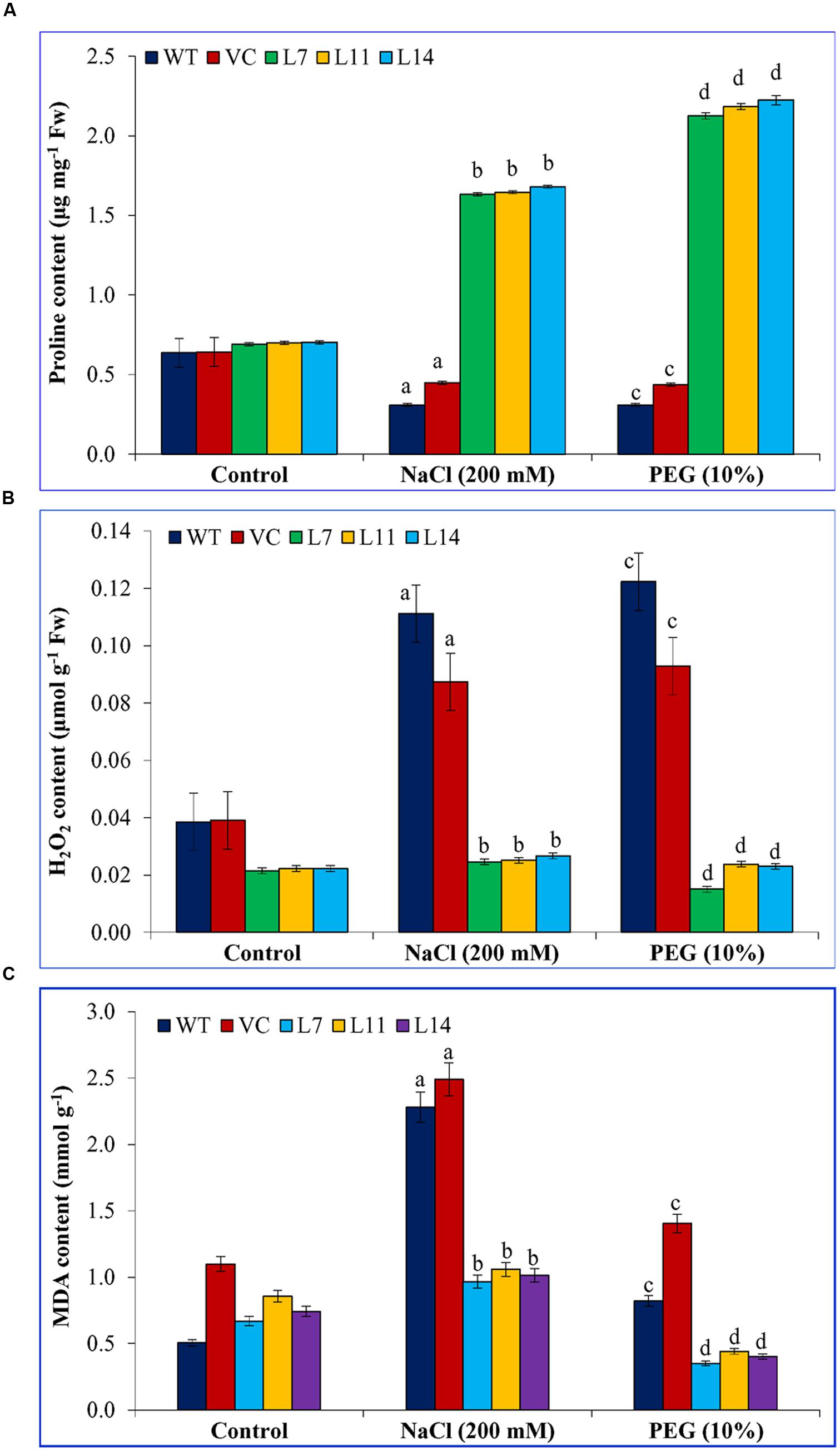
FIGURE 7. Biochemical analyses of transgenic tobacco plants under abiotic stress. Estimation of (A) proline, (B) H2O2, and (C) MDA contents in control (WT and VC) and transgenic plants (L7, L11, and L14) under salinity and osmotic stress condition. Bars represent means ± SE and values with different letters are significant at P < 0.05.
Overexpression of the SbUSP Gene Improves Physiological Status of Plant under Stress
Transgenic, WT and VC plants showed alike physiological status regarding RWC, EL, MSI, and OP under control condition (Figure 8). However, transgenic lines showed significantly (p < 0.05) higher RWC and MSI under stress condition compared to control plants. Furthermore, transgenic plants exhibited considerably (p < 0.05) reduced electrolyte leakage and osmotic potential under stress condition. Transgenic lines exhibited better osmotic adjustment and thus physiological status of transgenic plants improves.
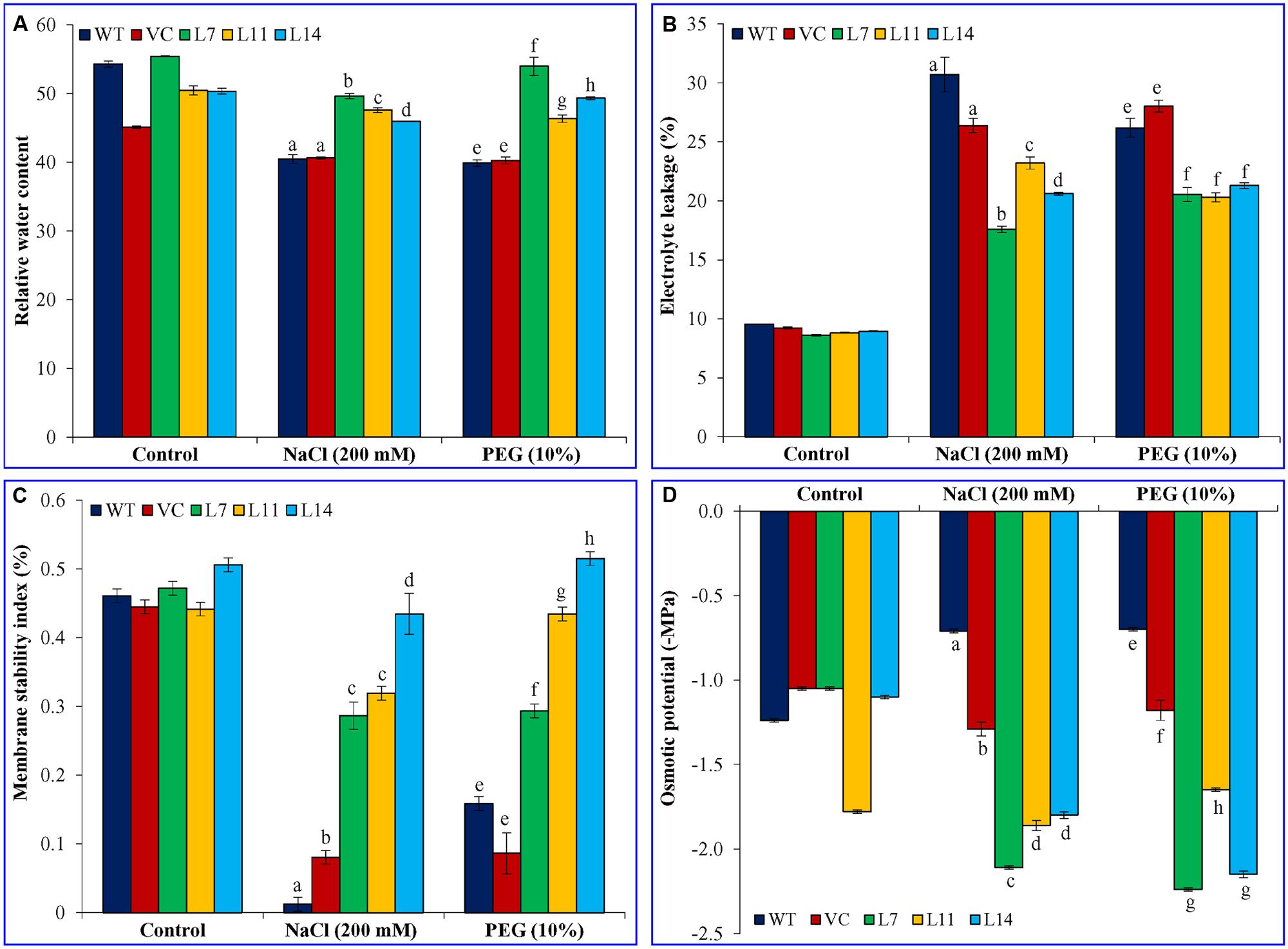
FIGURE 8. Physiological analyses of transgenic lines. Estimation of (A) RWC, (B) electrolyte leakage, (C) membrane stability index, and (D) osmotic potential from leaves of control plants (WT and VC) and transgenic lines (L7, L11, and L14) under control, salinity and osmotic stress conditions. Bars represent means ± SE and values with different letters are significant at P < 0.05.
Total Soluble Sugar, Reducing Sugar, Free Amino Acids, and Polyphenol Contents Increase in the SbUSP Overexpressing Lines
Total soluble sugar and reducing sugar were found almost similar in WT, VC, and transgenic lines under control conditions (Figure 9). A significant difference (p < 0.05) was observed amongst WT, VC, and transgenic plants exposed to salt and osmotic stress. Stress had no effect on total soluble sugar content in WT and VC plants, whereas transgenic lines accumulate significantly (p < 0.05) high content compared to control plants as well as the control condition (Figure 9A). The transgenic line L7 showed fivefold, whereas L11 and L14 showed a sixfold increase of soluble sugar content under salt stress. Similarly, under osmotic stress, line L7 showed more than sixfold increase and line L11 and L14 showed eightfold increases in total soluble sugar content. Reducing sugar, free amino acid and polyphenol content decreased under stress condition compared to the control condition, irrespective of plants. But, transgenic lines accumulated significantly (p < 0.05) higher content of reducing sugar, free amino acid and polyphenol compared to control plants (WT and VC) under stress condition (Figures 9B–D). Results showed the accumulation of primary metabolites/osmolytes in transgenic plants, which improve the performance of transgenic plants under stress condition.
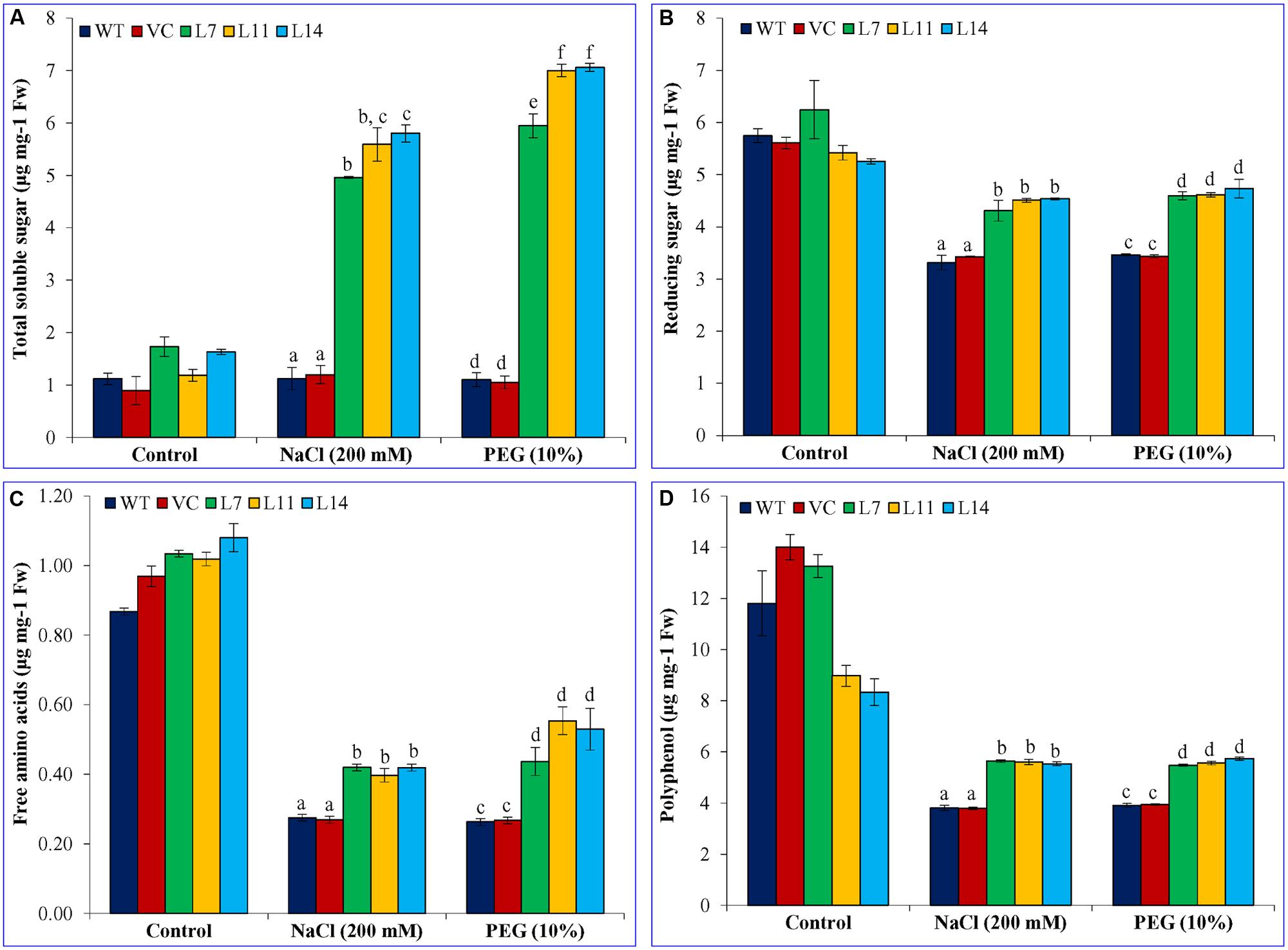
FIGURE 9. Estimation of sugar, free amino acids, and polyphenol contents. (A) Total soluble sugar, (B) reducing sugar, (C) free amino acids, and (D) polyphenol of transgenic lines (L7, L11, and L14) and control plants (WT and VC) under control and abiotic stress conditions. Bars represent means ± SE and values with different letters are significant at P < 0.05.
Overexpression of the SbUSP Maintains Ion Homeostasis
For analyses of ion content, the WT, VC, and SbUSP overexpressing transgenic seedlings were grown on MS media supplemented with NaCl (200 mM) and grown for 28 days. The Na+ and K+ content in whole seedlings were measured in WT, VC, and transgenic seedlings exposed to salt stress. Potassium ion content was increased in transgenic lines compared to control plants and control conditions (Supplementary Figure S4). In contrast, Na+ accumulation was observed in control plants (WT and VC) compared to transgenic lines under salt stress. The K+/Na+ ratio, an indicator of ion homeostasis was almost similar in all plants under control condition. The ratio decreased in control plants (WT and VC) under salt stress, whereas elevated ratio was detected in transgenic lines compared to control plants and control condition (Supplementary Figure S4). The high K+/Na+ ratio indicates that transgenic lines improved their ion homeostasis under salinity and thus performed better and showed stress endurance.
The SbUSP Gene May Also Involve in Transcriptional Regulation of Antioxidant Enzyme-Encoding Genes
Transcript expression of ROS scavenging enzyme genes viz. superoxide dismutase (NtSOD), catalase (NtCAT), and ascorbate peroxidase (NtAPX) were studied in transgenic lines under different abiotic stress condition, such as salt (200 mM), PEG (10%), heat (45°C), ABA (20 μM) and SA (10 μM), and compared with control plants and control condition (Figure 10). The transcript of NtSOD, NtCAT, and NtAPX genes was nearly steady in control plants (WT and VC) under every stress treatment compared to transgenic lines. A heat map showed the differential expression of antioxidant genes under varying stress condition (Supplementary Figure S5). The expression of NtSOD transcript under NaCl stress was increased up to twofold and sixfold in L7 and L14, respectively. Similarly, under osmotic stress, the transcript of lines L7 and L11 was up regulated by twofold. Exogenous application of ABA and SA increased the expression level of NtSOD transcript in L7 and L11. In addition, the expression of NtCAT was increased up to twofold and threefold in L7 and L14 under osmotic stress. Likewise, under salt stress, NtCAT transcript was accumulated up to twofold, whereas exogenous application of ABA and SA reduce the expression of NtCAT transcript. In contrast, ABA and SA treatments increased the expression of NtAPX transcript up to sixfold and ninefold, respectively, in L11. Similarly, the transcript of NtAPX accumulated by twofold and sevenfold in L7, L11, and L14 under salt stress. No significant difference was observed in NtAPX transcript level under osmotic stress. The transcripts of NtSOD, NtCAT, and NtAPX were down regulated under heat stress in all plants (WT, VC, and transgenic). However, down-regulation was significantly higher in transgenic lines compared to control plants. Transcriptional regulation of antioxidative genes in SbUSP overexpressing transgenic lines explicated its probable role as signaling molecule in salt and osmotic stress tolerance.
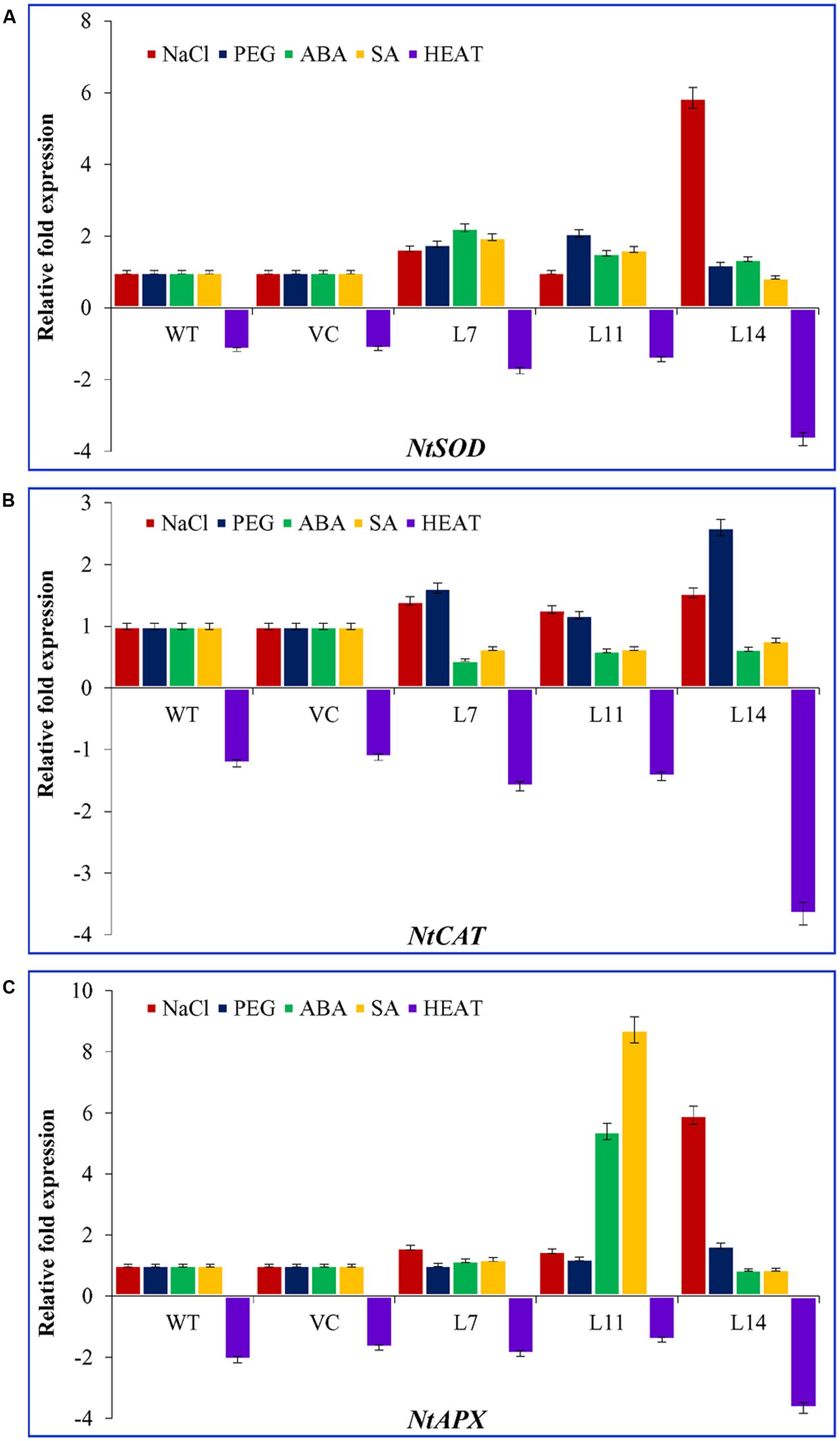
FIGURE 10. Transcript expression analysis of three antioxidant enzymes encoding genes. Comparative expression profile of (A) NtSOD, (B) NtCAT, and (C) NtAPX genes (encoding for superoxide dismutase, catalase, and ascorbate peroxidases enzymes, respectively) of transgenic lines (L7, L11, and L14) and control plants (WT and VC) under abiotic stress (salinity, osmotic, ABA, SA, and heat) conditions. Bars represent means ± SE and values with different letters are significant at P < 0.05.
Multivariate Data Analysis Distinguished Plant Responses under Stress Condition
Principal component analysis was performed for the comparative study of the morphological, biochemical, and physiological response of transgenic lines (L7, L11, and L14) and control plants (WT and VC) under normal and different stress (NaCl and osmotic) conditions. The integrated PCA showed the possible correlation of plant response to different variables and stress condition (Figure 11). Individually, the morphological, biochemical, and physiological response of plants explained 80.51, 87.46, and 91.25% of variations, respectively (Figure 12), whereas in-combination they explained 83.74% variations because of varying response as shown by heat map (Figure 11).
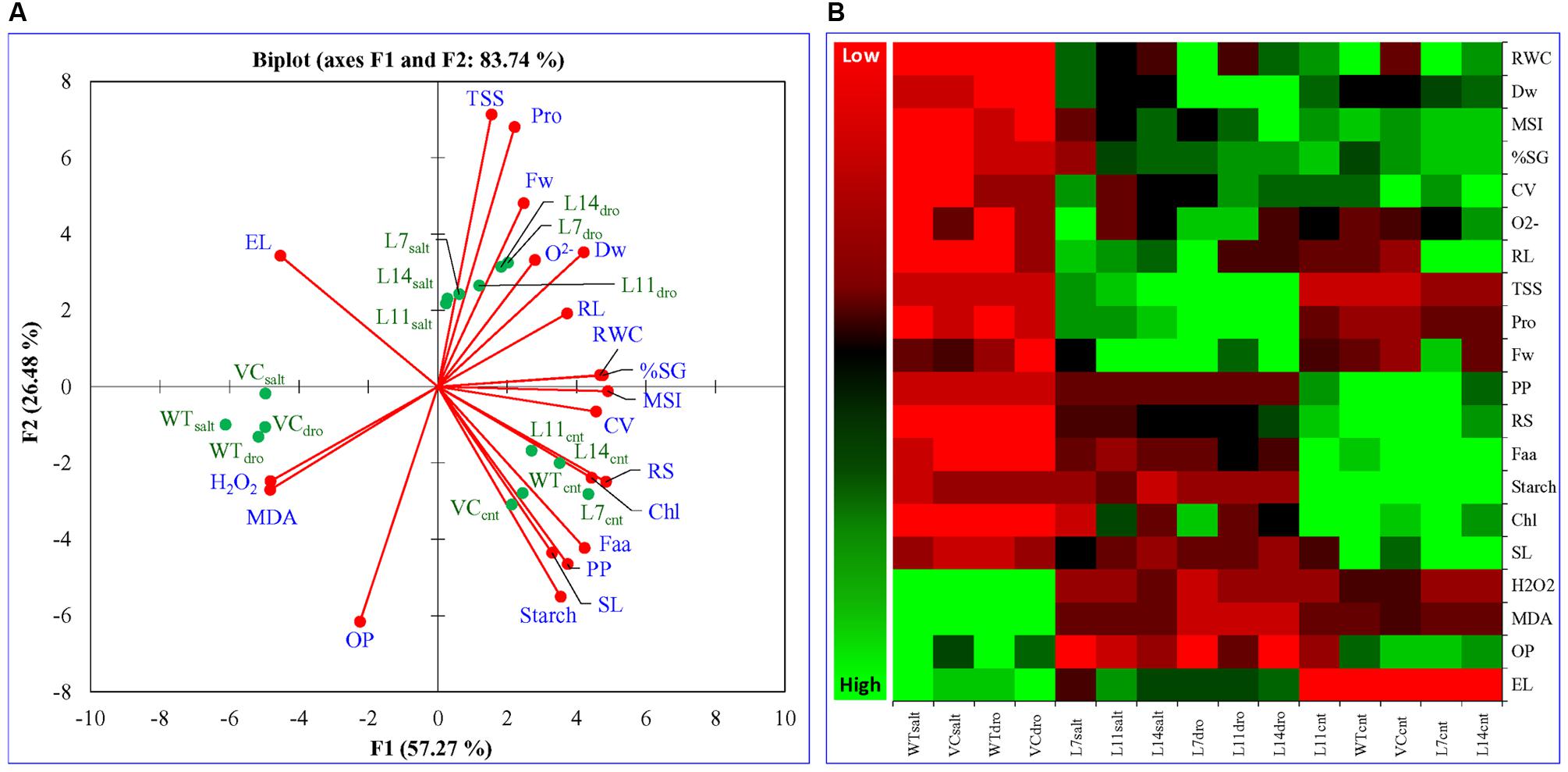
FIGURE 11. Multivariate data analysis. An integrated comparative (A) Bi-plot based principal component analysis (PCA) with first two principal components and (B) heat map showing the differential response of transgenic lines (L7, L11, and L14) and control plants (WT and VC) under normal and stress (NaCl and osmotic) condition. PCA is a multivariate analysis method by which correlation between variables was studied among multidimensional datasets. Plants grown under varying stress were selected as observations, whereas different morphological and physio-biochemical measurements were taken as variables. Observation and variable data set were used to generate Pearson’s correlation matrix, and PCA was analyzed.
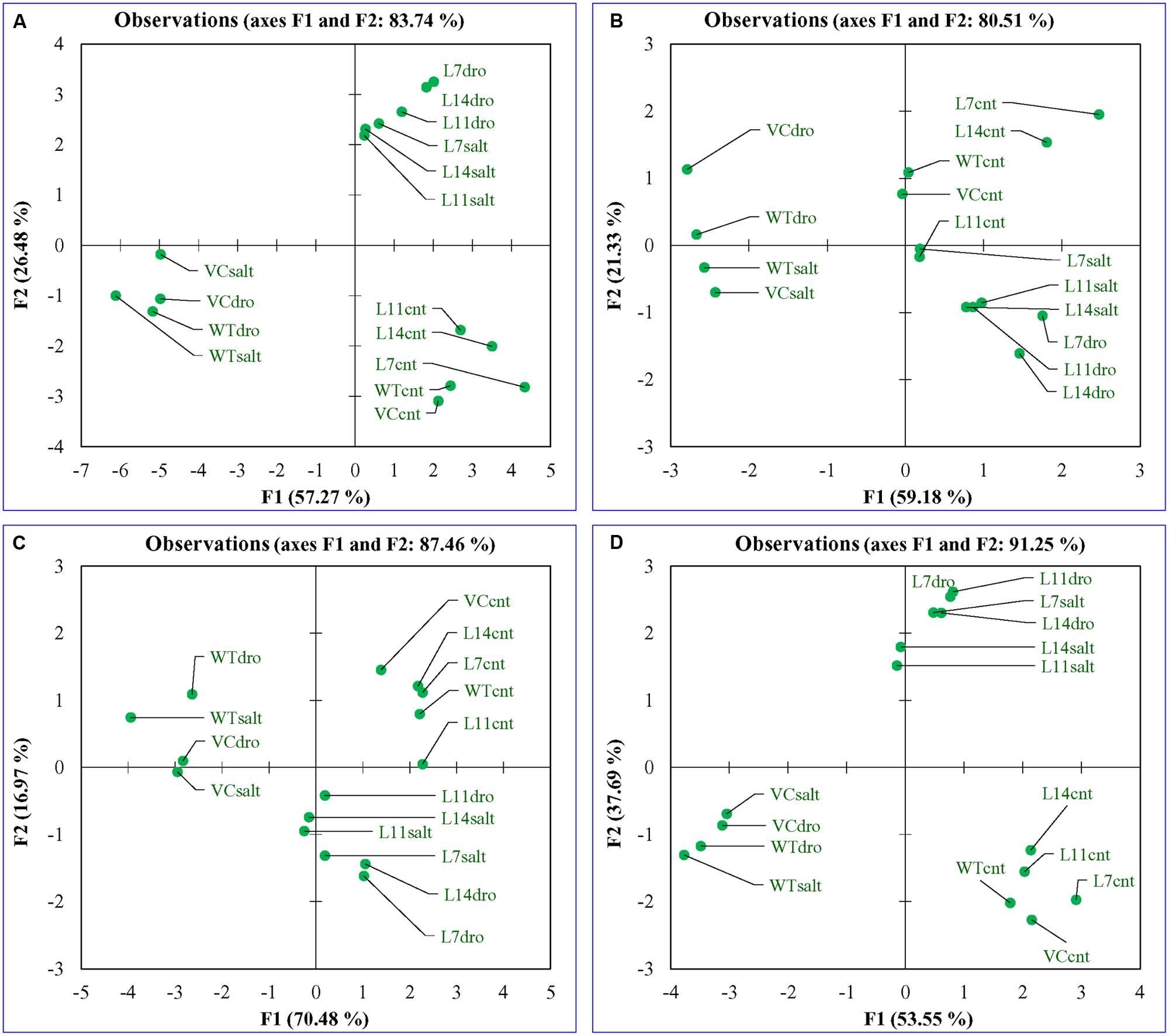
FIGURE 12. Principal component analysis. Comparative PCA of (A) combine, (B) morphological, (C) biochemical, and (D) physiological response of transgenic lines (L7, L11, and L14) and control plants (WT and VC) under control and stress (NaCl and osmotic) condition. Individually, the morphological, biochemical, and physiological response of plants explained 80.51, 87.46, and 91.25% of variations, respectively, whereas in-combination they explained 83.74% variations.
All plants (transgenic lines and control) showed comparable response under un-stress condition as revealed by the bi-plot analysis of individual and integrative analysis (Figures 11 and 12). Control plants (WT and VC) clustered together in all analysis (individual and integrated) and exhibited a similar response toward stress (salinity or drought) condition. Similarly, under stress condition, transgenic lines assembled at an axis and unveiled alike response to combat with environmental stress. A remarkable response was observed in the integrated PC analysis, which showed a significant correlation of plant response and the stress condition (Figure 13). Overall, the PCA exhibited a statistical distinction among morphological and physio-biochemical responses of control plants (WT and VC) and transgenic plants, also under control and stress conditions (salinity and drought).
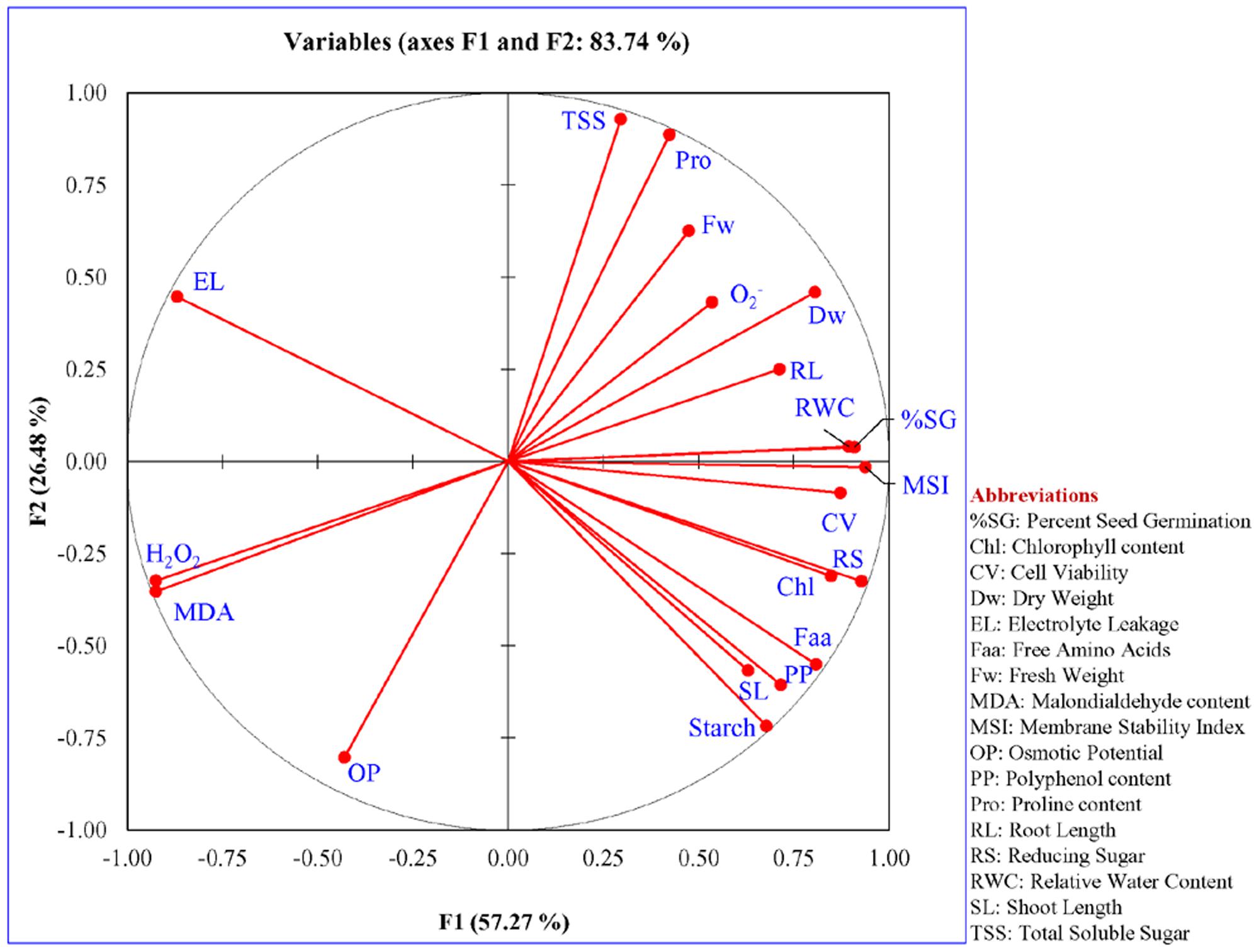
FIGURE 13. Statistical significant analysis of variables. The significance of parameters used to study the response of transgenic (L7, L11, and L14) and control (WT and VC) plants under different abiotic stress condition. A notable response was observed in the integrated PC analysis, showing a significant correlation of plant response and the stress condition.
Discussion
Due to sessile nature, plants have to widen their adaptive strategies to survive against continuous exposure to biotic and abiotic stresses. In a study of isolation and characterization of novel salt responsive genes, a novel gene encoding USP A-like from a euhalophyte S. brachiata (SbUSP A-like) has been isolated and functionally validated. The SbUSP gene encodes a protein that contains conserved 140–160 residues of a USP domain (Pfam Accession: PF00582) and has the ability to respond to a plethora of environmental stresses in bacteria, archaea, fungi, protozoa, and plants (Isokpehi et al., 2011). Some proteins of USP family in archaea, cyanobacteria, and plants are large proteins in which, the Usp domain is present together with other functional domains like Na+/H+ antiporter domains, Cl- voltage channels, amino acid permeases and protein kinase domains (Kvint et al., 2003). The present study explored the function of the SbUSP gene under different abiotic stresses in a model plant tobacco. In plants, very few homologs of the USP family have been isolated and characterized (Sauter et al., 2002; Chou et al., 2007; Merkouropoulos et al., 2008; Maqbool et al., 2009; Zahur et al., 2009; Li et al., 2010; Isokpehi et al., 2011; Loukehaich et al., 2012). The structure alignment and in silico analyses showed phosphorylation, glycosylation, and ATP binding sites (84% probability) of SbUSP and its interaction with adenosine monophosphate (Udawat et al., 2014). The SbUSP is a phosphoprotein that contains autophosphorylating serine and threonine residue and uses either GTP or ATP as phosphate donors (Kvint et al., 2003; Udawat et al., 2014). The multiple amino acid sequence alignment followed by phylogenetic analyses revealed that SbUSP clustered with RcUSP and generated a separate clad distinct from the USP of other plant species (Udawat et al., 2014). Based on pattern of conserved structural features, it was proposed that the USPA domain is a part of larger structural families, whose members has already diversified and present in the last universal common ancestor of all extant of life, furthermore it was suggested that ancestral function of the USPA domain was nucleotide binding and signal transduction (Kerk et al., 2003).
The Southern blot analysis identified two homologs of SbUSP in S. brachiata genome and the genome organization study revealed intronless genomic arrangement of the gene SbUSP (Figure 1). Subcellular localization, determined using 35S::SbUSP::RFP translational fusion construct, showed red fluorescence exclusively in the cytosol (Figure 1) and it is evident that SbUSP phosphoprotein is present in the cytosol as membrane bound form. The in silico study also predicted that SbUSP is membrane bound cytosolic protein (Udawat et al., 2014). Loukehaich et al. (2012) determined the subcellular localization of SpUSP using 35S::SpUSP::GFP translational fusion construct and detected green fluorescence exclusively in nucleus and cell membrane, whereas cells transformed with the vector containing GFP alone displayed fluorescence throughout the cells (Figure 1). To understand the role of SbUSP gene in abiotic stress tolerance, three homozygous transgenic tobacco lines (T1) L7, L11, and L14, containing single gene integration, overexpress high transgene(s) transcripts (Supplementary Figure S2) and high seed germination percentage under stress condition (Figure 3) were selected.
Transgenic lines showed improved plant biomass including, root length, shoot length, fresh weight (FW), and dry weight (DW) under different stress conditions compared to control plants (Figure 4), which reveal that the SbUSP gene reduces deleterious effects of salt and osmotic stresses. It is predicted that overexpression of SbUSP alters the development of the plant probably by changes in levels of phytohormones, like auxin and cytokinin. Furthermore, enhanced vegetative growth was observed in transgenic lines even under normal conditions, which is supported by the comparable chlorophyll content of transgenic lines (Figure 5). It is speculated that SbUSP gene regulates the expression of antioxidant encoding genes (Figure 10) and also induces genes of the abiotic stress defense system (Figure 14), which in turn provide stress endurance. It was established that total chlorophyll content, one of the markers of cellular stress decreased significantly under stress condition because of ROS generation in the chloroplast (Bartels and Sunkar, 2005). ROS inhibits the PSII repair system, which resulted in the degradation of chlorophyll (Bartels and Sunkar, 2005). In the present study, transgenic plants exhibited significantly higher chlorophyll content and also reduced leaf senescence under oxidative stress (Figure 5). Based on results, it is presumed that the SbUSP gene may be involved in the mechanism that protects chlorophyll from oxidative damage. Plants grown under control condition did not show the generation of peroxide and superoxide free radicals in a localization study (Figure 6). Control plants (WT and VC) showed brown and blue-colored precipitate after DAB and NBT staining, which indicates a higher level of free radicals compared to transgenic lines under stress condition. Similarly, negligible H2O2 content was measured in transgenic lines compared to control plants (WT and VC), which showed significant (p < 0.05) accumulation of H2O2 content under stress condition (Figure 7B). These observations indicate the role of SbUSP gene in alleviating oxidative stress by ROS scavenging activity. Recently, USP mediated ROS homeostasis was reported under anoxic conditions and oxygen sensing ability of USP followed by ROS signaling is proposed (Gonzali et al., 2015). The study enables an assumption that SbUSP may be involved in ROS scavenging activity.
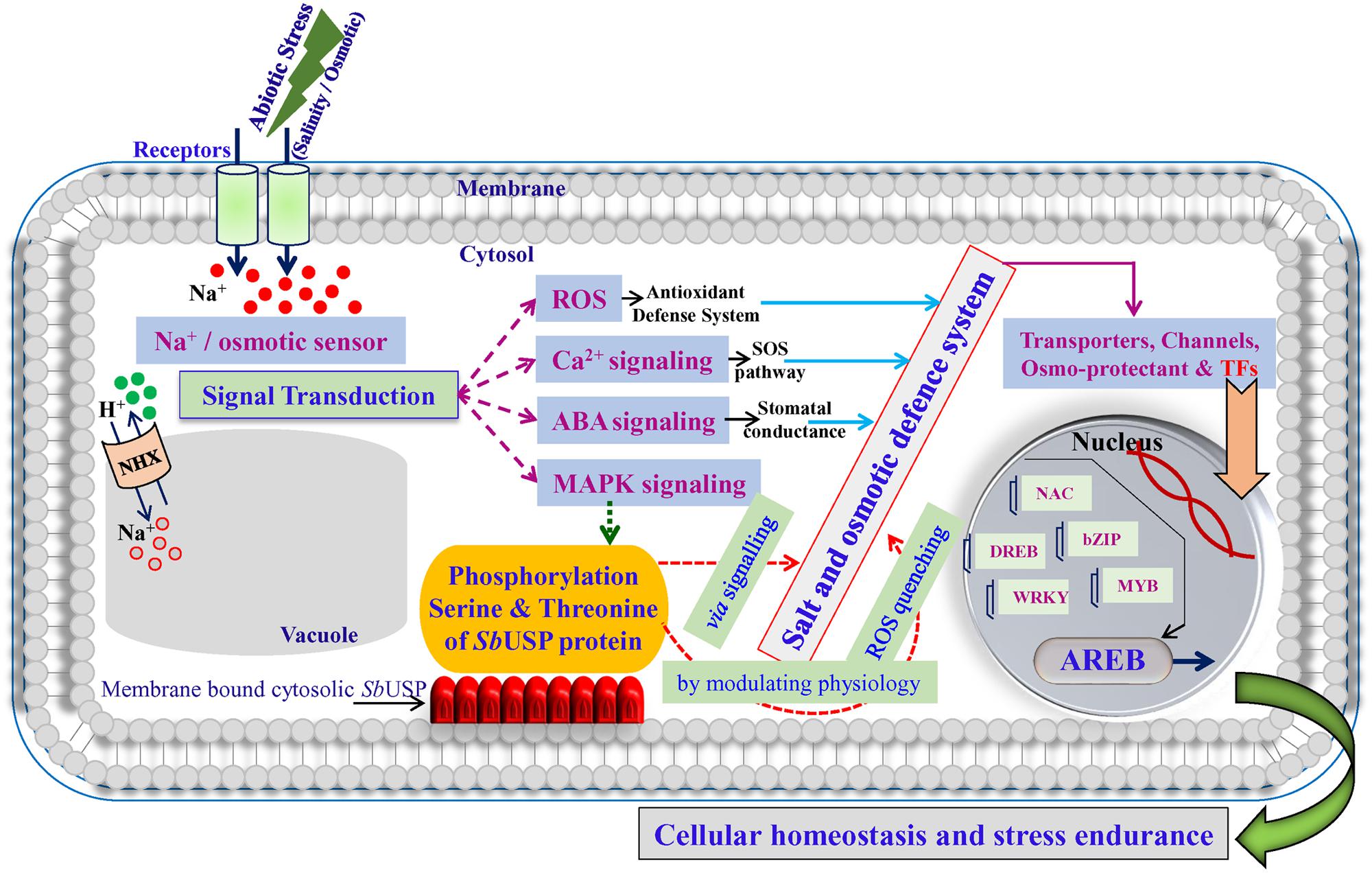
FIGURE 14. A hypothetical model for the role of membrane bound SbUSP in abiotic stress defense system.
The degree of injury caused by stress in a plant is commonly measured by the physiological status of the plant. It was generally studied by measuring RWC, EL, MSI, OP, proline, and MDA content under stress condition. Ectopic expression of the SbUSP gene maintains the physiology of plants and a significant (p < 0.05) increase in relative water content, membrane stability, osmotic potential, and proline content was observed compared to control plants (WT and VC) and control condition under salinity and osmotic stresses (Figures 7 and 8). Lipid peroxidation and electrolyte leakage are common stress markers, which decreased in transgenic lines, overexpressing SbUSP gene in the present study. Malondialdehyde is the decomposition product of polyunsaturated fatty acids of biomembranes, showed greater accumulation under salt stress, the degree of accumulation of MDA was higher in Guazuncho than in Pora, indicating a high rate of lipid peroxidation in Guazuncho due to salt stress (Meloni et al., 2003). It has been demonstrated that electrolyte leakage measurement may be correlated with several physiological and biochemical parameters conditioning the plant responses to environmental conditions, such as spectral reflectance, antioxidative enzyme synthesis, membrane acyl lipid concentrations, water use efficiency, transverse relaxation time of leaf water, stomatal resistance, osmotic potential, and leaf rolling index (Bajji et al., 2002).
Lipid peroxidation (measured by MDA content) and percent electrolyte leakage were decreased significantly in transgenic lines under stress condition. Overexpression of SpUSP induced stomatal closure and reduced water loss in tomato under osmotic stress and thus provides drought tolerance (Loukehaich et al., 2012). Salt and drought tolerant or resistant plant including halophytes are attributed by their capacity to adjust tissue water potential to a level that is more negative than that of the soil water potential of the habitat in which they are growing (Khan et al., 2000). In the present study, transgenic lines showed an improved osmotic potential and exhibiting endurance to the plants under stress condition. The stress tolerance of plants exposed to salinity was correlated with more negative solute potential (that is a greater concentration of total osmolytes), which permitted better water retention by desiccating plants, and with a larger ratio of K+ to Na+ (Pardo and Quintero, 2002).
Osmo-protectants are cellular stress markers, which accumulate to maintain the osmotic balance across the membrane under osmotic stress. Proline, total sugar, reducing sugars, polyphenols are considered as important biochemical markers to determine the plant response under oxidative condition. The SbUSP gene overexpressing transgenic plants accumulated a high level of proline, reducing sugar, total sugar, free amino acids, and polyphenol (Figures 7 and 9). In plants, polyphenol synthesis and/or accumulation is generally stimulated in response to biotic or abiotic stresses and polyphenolic compounds indeed participate in the defense against reactive oxygen species (Ksouri et al., 2007). There were a number of reports on transgenic plants, overexpressing abiotic stress responsive gene(s) cloned from S. brachiata, in which osmo-protectants increase under varying salt and osmotic stress (Jha et al., 2011; Joshi et al., 2012; Chaturvedi et al., 2014; Singh et al., 2014a,b; Patel et al., 2015; Tiwari et al., 2015). Although mechanisms involved in sugar signal transduction and sugar gene regulation in higher plants are entirely not clarified yet, important progresses have been made to obtain their understanding, principally, about signals that trigger these processes and how the regulation of photosynthetic carbon metabolism interacts with other processes during stress conditions (Rosa et al., 2009).
The SbUSP overexpressing transgenic lines showed high K+ content and K+/Na+ ratio and lower accumulation of Na+ (Supplementary Figure S4). Previously, it was observed that salt responsive genes, such as SbNHX1, SbpAPX1, SbASR-1 cloned from S. brachiata unveiled oxidative endurance to the model plant tobacco and crops, like jatropha, castor, and peanut (Jha et al., 2011; Joshi et al., 2012; Chaturvedi et al., 2014; Tiwari et al., 2014, 2015; Singh et al., 2014a,b; Patel et al., 2015). The present study unveiled the role of SbUSP gene in ROS scavenging mechanism directly as a non-enzymatic antioxidant or indirectly by enhancing the expression of antioxidative enzymes. To confirm its indirect role, transcript expression of NtSOD, NtCAT, and NtAPX genes were performed in transgenic lines and control plants subjected to control or stress (salinity and osmotic) condition (Figure 10).
The transcript of NtSOD, NtCAT, and NtAPX was induced by NaCl, desiccation and phytohormones, like ABA and SA in SbUSP overexpressing transgenic lines. Previously, the SpUSP transcript accumulated several folds under salt, drought, and ABA stress in tomato (Loukehaich et al., 2012). Similarly, a significant expression response (about eightfold) was observed for SbUSP under salt stress followed by drought, heat, and cold stress (Udawat et al., 2014). A high level of GUSP gene expression was detected in leaves, roots and stems of Gossypium arboreum under water stress (Maqbool et al., 2009). Li et al. (2010) reported putative barley (Steptoe) USP gene exhibited different expression at different time points analyzed under salt stress. Most of the drought responsive genes, studied till date, are induced by ABA and dehydration triggers the production of ABA, which in turn induces various genes (Zahur et al., 2009). Functional characterization of genes encoding ABA responsive cis- and trans-acting factors will provide useful insight to understand the ABA dependent signal transduction pathway. The ABA-responsive element (ABRE) was detected in the promoter of SbUSP gene. Nevertheless, a detailed involvement of ABA and SA in SbUSP regulation is further required to be explored. Even though, nearly 10% of the protein coding genes in Arabidopsis are likely to be regulated by ABA and these findings suggests that ABA-mediated gene expression plays a versatile and pivotal role in plants (Fujita et al., 2011). Although, heat stress down regulated NtSOD, NtAPX, and NtCAT transcript in SbUSP transgenic lines, in agreement to this, Kvint et al. (2003) reported that extreme temperature repressed the USP gene as σ70 gets inactivated at high temperature. In the coastal region the sea water is swamped twice in a month and remaining times the soil is almost dry, and the temperature is high.
Based on results of the study, it is hypothesized that abiotic stress responsive membrane bound cytosolic SbUSP protein may activate some downstream gene(s) (yet to be characterized; Figure 14), which lead to the accumulation of osmoprotectants to alleviate oxidative stress by scavenging ROS and thus provide salt or osmotic endurance by modulating physiological processes of the plant.
Conclusion
Results ascertain that overexpression of the SbUSP gene evidently enhances salt and osmotic tolerance by maintaining physiology of the plant. Functional characterization of an autophosphorylating USP was performed and it is expected that SbUSP may act as “molecular knob” by interacting with components of cellular signal transduction pathway (probably MAP Kinases). The expression data in this study designate that SbUSP act in ABA dependent manner and affects regulation of genes responsible for ROS quenching. Although additional studies are required to establish the exact mechanism of action of the USP gene in the plant under abiotic stress.
Author Contributions
Conceived and designed the experiments: AM and BJ. Performed the experiments: PU, RJ, and DS. Analyzed the data: PU and AM. Wrote the paper: PU and AM.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
CSIR-CSMCRI Communication No.: PRIS-199/2015. This study was supported by the Council of Scientific and Industrial Research (CSIR; www.csir.res.in), Government of India, New Delhi [BSC0109-SIMPLE; Junior and Senior Research Fellowship to PU]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00518
References
Bajji, M., Kinet, J. M., and Lutts, S. (2002). The use of the electrolyte leakage method for assessing cell membrane stability as a water stress tolerance test in durum wheat. Plant Growth Regul. 36, 61–70. doi: 10.1023/A:1014732714549
Bartels, D., and Sunkar, R. (2005). Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24, 23–58. doi: 10.1080/07352680590910410
Bates, L. S., Waldern, R., and Teare, I. D. (1973). Rapid determination of free proline for water stress studies. Plant Soil 39, 205–207. doi: 10.1016/j.dental.2010.07.006
Bubner, B., Gase, K., and Baldwin, I. T. (2004). Two fold differences are the detection limit for determining transgene copy numbers in plants by real time PCR. BMC Biotechnol. 4:14. doi: 10.1186/1472-6750-4-14
Chaturvedi, A. K., Mishra, A., Tiwari, V., and Jha, B. (2012). Cloning and transcript analyses of type 2 Metallothionein gene (SbMT-2) from extreme halophyte Salicornia brachiata and its heterologous expression in E. coli. Gene 499, 280–287. doi: 10.1016/j.gene.2012.03.001
Chaturvedi, A. K., Patel, M. K., Mishra, A., Tiwari, V., and Jha, B. (2014). The SbMT-2 gene from a halophyte confers abiotic stress tolerance and modulates ROS scavenging in transgenic tobacco. PLoS ONE 9:e111379. doi: 10.1371/journal.pone.0111379
Chou, M. X., Wei, X. Y., Chen, D. S., and Zhou, J. C. (2007). A novel nodule enhanced gene encoding a putative universal stress protein from Astragalus sinicus. J. Plant Physiol. 164, 764–772. doi: 10.1016/j.jplph.2006.05.009
Draper, H. H., and Hadley, M. (1990). Malondialdehyde determination as index of lipid peroxidation. Methods Enzymol. 186, 421–431. doi: 10.1016/0076-6879(90)86135-I
Fujita, Y., Fujita, M., Shinozaki, K., and YamaguchiShinozaki, K. (2011). ABA mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–525. doi: 10.1007/s10265-011-0412-3
Ghose, T. K. (1987). Measurements of cellulase activities. Pure Appl. Chem. 59, 257–268. doi: 10.1351/pac198759020257
Gonzali, S., Loreti, E., Cardarelli, F., Novi, G., Parlanti, S., Pucciariello, C., et al. (2015). Universal stress protein HRU1 mediates ROS homeostasis under anoxia. Nat. Plants 1:15151. doi: 10.1038/nplants.2015.151
He, J., Xu, H., She, X., Song, X., and Zhao, W. (2005). The role and the interrelationship of hydrogen peroxide and nitric oxide in the UV-B-induced stomatal closure in broad bean. Funct. Plant Biol. 32, 237–247. doi: 10.1071/FP04185
Hema, R., Vemanna, R., Sreeramulu, S., Reddy, C., Senthil-Kumar, M., and Udayakumar, M. (2014). Stable expression of mtlD gene imparts multiple stress tolerance in Finger millet. PLoS ONE 9:e99110. doi: 10.1371/journal.pone.0099110
Horsch, R. B., Fry, J. E., Hoffmann, N. L., Eichholtz, D., Rogers, S. G., and Farley, R. T. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1231. doi: 10.1126/science.227.4691.1229
Inskeep, W. P., and Bloom, P. R. (1985). Extinction coefficients of chlorophyll a and b in N, N-dimethylformamide and 80% Acetone. Plant Physiol. 77, 483–485. doi: 10.1104/pp.77.2.483
Isokpehi, R. D., Simmons, S. S., Cohly, H., Ekunwe, S., Begonia, G. B., and Ayensu, W. K. (2011). Identification of drought-responsive universal stress proteins in Viridiplantae. Bioinformat. Biol. Insights 5, 41–58. doi: 10.4137/BBI.S6061
Jefferson, R. A. (1987). Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. doi: 10.1007/BF02667740
Jha, B., Sharma, A., and Mishra, A. (2011). Expression of SbGSTU (tau class glutathione S-transferase) gene isolated from Salicornia brachiata in tobacco for salt tolerance. Mol. Biol. Rep. 38, 4823–4832. doi: 10.1007/s11033-010-0625-x
Jha, B., Singh, N. P., and Mishra, A. (2012). Proteome profiling of seed storage proteins reveals the nutritional potential of Salicornia brachiata Roxb., an extreme halophyte. J. Agric. Food Chem. 60, 4320–4326. doi: 10.1021/jf203632v
Joshi, M., Jha, A., Mishra, A., and Jha, B. (2013). Developing transgenic Jatropha using the SbNHX1 gene from an extreme halophyte for cultivation in saline wasteland. PLoS ONE 8:e71136. doi: 10.1371/journal.pone.0071136
Joshi, M., Mishra, A., and Jha, B. (2012). NaCl plays a key role for in vitro micropropagation of Salicornia brachiata, an extreme halophyte. Ind. Crops Prod. 35, 313–316. doi: 10.1016/j.indcrop.2011.06.024
Kerk, D., Bulgrien, J., Smith, D. W., and Gribskov, M. (2003). Arabidopsis Proteins containing similarity to the Universal Stress stress Protein protein domain of bacteria. Plant Physiol. 131, 1209–1219. doi: 10.1104/pp.102.016006
Khan, M. A., Ungar, I. A., and Showalter, A. M. (2000). Effects of salinity on growth, water relations and ion accumulation of the subtropical perennial halophyte. Atriplex griffithii var. stocksii. Annu. Bot. 85, 225–232.
Ksouri, R., Megdiche, W., Debez, A., Falleh, H., Grignon, C., and Abdelly, C. (2007). Salinity effects on polyphenol content and antioxidant activities in leaves of the halophyte Cakile maritime. Plant Physiol. Biochem. 45, 244–249. doi: 10.1016/j.plaphy.2007.02.001
Kvint, K., Nachin, L., Diez, A., and Nystrom, T. (2003). The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6, 140–145. doi: 10.1016/S1369-5274(03)00025-0
Li, W. T., Wei, Y. M., Wang, J. R., Liu, C. J., Lan, X. J., Jiang, Q. T., et al. (2010). Identification, localization and characterization of putative USP genes in barley. Theor. Appl. Genet. 121, 907–917. doi: 10.1007/s00122-010-1359-9
Livak, K. J., and Schmittgen, T. D. (2001). Analyses of relative gene expression data using real time quantitative PCR and the 2(-Delta Delta C (T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Loukehaich, R., Wang, T., Ouyang, B., Ziaf, K., Li, H., Zhang, J., et al. (2012). SpUSP, an annexin-interacting universal stress protein, enhances drought tolerance in tomato. J. Exp. Bot. 63, 5593–5606. doi: 10.1093/jxb/ers220
Lutts, S., Kinet, J. M., and Bouharmont, J. (1996). NaCl-induced senescence in leaves of rice (Oryza sativa L.) cultivars differing in salinity resistance. Ann. Bot. 78, 389–398. doi: 10.1006/anbo.1996.0134
Maqbool, A., Zahur, M., Husnain, T., and Riazuddin, S. (2009). GUSP1 and GUSP2, two drought-responsive genes in Gossypium arboreum have homology to Universal Stress Proteins. Plant Mol. Biol. Rep. 27, 109–114. doi: 10.1007/s11105-008-0049-0
Meloni, D. A., Oliva, M. A., Martinez, C. A., and Cambraia, J. (2003). Photosynthesis and activity of superoxide dismutase, peroxidase and glutathione reductase in cotton under salt stress. Environ. Exp. Bot. 49, 69–76. doi: 10.1016/S0098-8472(02)00058-8
Merkouropoulos, G., Andreasson, E., Hess, D., Boller, T., and Peck, S. C. (2008). An Arabidopsis protein phosphorylated in response to microbial elicitation, AtPHOS32, is a Substrate substrate of MAP Kinases 3 and 6. J. Biol. Cem. 283, 10493–10499. doi: 10.1074/jbc.M800735200
Mishra, A., Joshi, M., and Jha, B. (2013). Oligosaccharide mass profiling of nutritionally important Salicornia brachiata, an extreme halophyte. Carbohyd. Polym. 92, 1942–1945. doi: 10.1016/j.carbpol.2012.11.055
Mishra, A., Patel, M. K., and Jha, B. (2015). Non-targeted metabolomics and scavenging activity of reactive oxygen species reveal the potential of Salicornia brachiata as a functional food. J. Funct. Foods 13, 21–31. doi: 10.1016/j.jff.2014.12.027
Munns, R., and Tester, M. (2008). Mechanism of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Pandey, S., Patel, M. K., Mishra, A., and Jha, B. (2015). Physio-biochemical composition and untargeted metabolomics of cumin (Cuminum cyminum L.) make it promising functional food and help in mitigating salinity stress. PLoS ONE. 10:e0144469. doi: 10.1371/journal.pone.0144469
Pardo, J. M., and Quintero, F. J. (2002). Plants and sodium ions: keeping company with the enemy. Genome Biol. 3, 1017.1–1017.4. doi: 10.1186/gb-2002-3-6-reviews1017
Patel, M., Joshi, M., Mishra, A., and Jha, B. (2015). Ectopic expression of SbNHX1 gene in transgenic castor (Ricinus communis L.) enhances salt stress by modulating physiological process. Plant Cell Tissue Organ. Cult. 122, 477–490. doi: 10.1007/s11240-015-0785-4
Rosa, M., Prado, C., Podazza, G., Interdonato, R., Gonzalez, J. A., Hilal, M., et al. (2009). Soluble sugars-Metabolism, sensing and abiotic stress- a complex network in the life of plants. Plant Signal. Behav. 4, 388–393. doi: 10.4161/psb.4.5.8294
Sairam, R. K. (1994). Effect of homobrassinolide application on metabolism and grain yield under irrigated and moisture stress conditions of two wheat varieties. J. Plant Growth Regul. 14, 173–181. doi: 10.1007/BF00025220
Sauter, M., Rzewuski, G., Marwedel, T., and Lorbiecke, R. A. (2002). The novel ethylene-regulated gene OsUsp1 from rice encodes a member of a plant protein family related to prokaryotic universal stress proteins. J. Exp. Bot. 53, 2325–2331. doi: 10.1093/jxb/erf096
Shi, J., Fu, X. Z., Peng, T., Huang, X. S., Fan, Q. J., and Liu, J. H. (2010). Spermine pretreatment confers dehydration tolerance of citrus in vitro plants via modulation of antioxidative capacity and stomatal response. Tree Physiol. 30, 914–922. doi: 10.1093/treephys/tpq030
Singh, N., Mishra, A., and Jha, B. (2014a). Ectopic over-expression of peroxisomal ascorbate paroxidase (SbpAPX) gene confers salt tolerance in transgenic peanut (Arachis hypogea). Gene 547, 119–125. doi: 10.1016/j.gene.2014.06.037
Singh, N., Mishra, A., and Jha, B. (2014b). Over-expression of the peroxisomal ascorbate paroxidase (SbpAPX) gene cloned from halophyte Salicornia brachiata confers salt and drought stress tolerance in transgenic tobacco. Mar. Biotechnol. 16, 321–332. doi: 10.1007/s10126-013-9548-6
Tiwari, V., Chaturvedi, A. K., Mishra, A., and Jha, B. (2014). The transcriptional regulatory mechanism of the peroxisomal ascorbate peroxidase (pAPX) gene cloned from an extreme halophyte, Salicornia brachiata. Plant Cell Physiol. 55, 1774–1871. doi: 10.1093/pcp/pct172
Tiwari, V., Chaturvedi, K. A., Mishra, A., and Jha, B. (2015). Introgression of the SbASR-1 gene cloned from a halophyte Salicornia brachiata enhances salinity and drought endurance in transgenic groundnut (Arachis hypogea) and act as a transcription factor. PLoS ONE 10:e0131567. doi: 10.1371/journal.pone.0131567
Tiwari, V., Patel, M. K., Chaturvedi, A. K., Mishra, A., and Jha, B. (2016). Functional Characterization of the Tau class glutathione-s-transferases gene (SbGSTU) promoter of Salicornia brachiata under salinity and osmotic stress. PLoS ONE 11:e0148494. doi: 10.1371/journal.pone.0148494
Tkaczuk, L. K., Shumilin, A., Igor Chruszcz, M., Evdokimova, E., Savchenko, E., Minor, W., et al. (2013). Structural and functional insight into the universal stress protein family. Evol. Appl. 6, 434–449. doi: 10.1111/eva.12057
Udawat, P., Mishra, A., and Jha, B. (2014). Heterologous expression of an uncharacterized universal stress protein gene (SbUSP) from the extreme halophyte, Salicornia brachiata, which confers salt and osmotic tolerance to E. coli. Gene 536, 163–170. doi: 10.1016/j.gene.2013.11.020
Vinocur, B., and Altman, A. (2005). Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr. Opin. Biotechnol. 16, 123–132. doi: 10.1016/j.copbio.2005.02.001
Yamamoto, N., Takano, T., Tanaka, K., Ishige, T., Terashima, S., Endo, C., et al. (2015). Comprehensive analysis of transcriptome response to salinity stress in the halophytic turfgrass Sporobolus virginicus. Front. Plant Sci. 6:241. doi: 10.3389/fpls.2015.00241
Yemm, E. W., and Willis, A. J. (1954). The estimation of carbohydrates in plant extracts by Anthrone. Biochem. J. 57, 508–514. doi: 10.1042/bj0570508
Keywords: abiotic stress, drought, halophyte, salinity, transgenic
Citation: Udawat P, Jha RK, Sinha D, Mishra A and Jha B (2016) Overexpression of a Cytosolic Abiotic Stress Responsive Universal Stress Protein (SbUSP) Mitigates Salt and Osmotic Stress in Transgenic Tobacco Plants. Front. Plant Sci. 7:518. doi: 10.3389/fpls.2016.00518
Received: 14 December 2015; Accepted: 01 April 2016;
Published: 21 April 2016.
Edited by:
Shabir Hussain Wani, Sher-e-Kashmir University of Agricultural Sciences and Technology, IndiaReviewed by:
Anil Kumar Singh, Indian Council of Agricultural Research – Indian Institute of Agricultural Biotechnology, IndiaMaria Azucena Ortega-Amaro, Instituto Potosino de Investigación Científica y Tecnológica, Mexico
Copyright © 2016 Udawat, Jha, Sinha, Mishra and Jha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Avinash Mishra, avinash@csmcri.org; Bhavanath Jha, bjha@csmcri.org
 Pushpika Udawat
Pushpika Udawat Rajesh K. Jha
Rajesh K. Jha Dinkar Sinha
Dinkar Sinha Avinash Mishra
Avinash Mishra Bhavanath Jha
Bhavanath Jha