- 1Graduate School of Natural Science and Technology, Okayama University, Okayama, Japan
- 2National Institute for Basic Biology, Okazaki, Japan
- 3Graduate School of Science and Technology, Kumamoto University, Kumamoto, Japan
- 4Department of Biology, Duke University, Durham, NC, USA
Polyamines are small basic compounds present in all living organisms and act in a variety of biological processes. However, the mechanism of polyamine sensing, signaling and response in relation to other metabolic pathways remains to be fully addressed in plant cells. As one approach, we isolated Arabidopsis mutants that show increased resistance to spermine in terms of chlorosis. We show here that two of the mutants have a point mutation in a nitrate transporter gene of the NRT1/PTR family (NPF), NRT1.3 (AtNPF6.4). These mutants also exhibit increased resistance to putrescine and spermidine while loss-of-function mutants of the two closest homologs of NRT1.3, root-specific NRT1.1 (AtNPF6.3) and petiole-specific NRT1.4 (AtNPF6.2), were shown to have a normal sensitivity to polyamines. When the GUS reporter gene was expressed under the control of the NRT1.3 promoter, GUS staining was observed in leaf mesophyll cells and stem cortex cells but not in the epidermis, suggesting that NRT1.3 specifically functions in parenchymal tissues. We further found that the aerial part of the mutant seedling has normal levels of polyamines but shows reduced uptake of norspermidine compared with the wild type. These results suggest that polyamine transport or metabolism is associated with nitrate transport in the parenchymal tissue of the shoot.
Introduction
Polyamines such as putrescine and spermidine are small basic compounds present in all living organisms and play a role in a variety of biological processes including DNA replication, protein synthesis, and ion channel modulation (Igarashi and Kashiwagi, 2010b; Pegg and Casero, 2011). The cellular content of polyamines is highly regulated by biosynthesis, oxidative degradation, transport, and, in plants, conjugation to hydroxycinnamic acids. Both in animals and plants, most of the mRNAs of S-adenosylmethionine decarboxylase (AdoMetDC), which is a key enzyme providing the aminopropyl group for the synthesis of spermidine and spermine, contain small upstream open reading frames (uORFs) that mediate polyamine-dependent repression of the main ORF translation (Raney et al., 2002; Hanfrey et al., 2005). Plants and some bacteria have a structural isomer of spermine, thermospermine (Takano et al., 2012). We have found that expression of ACAULIS5 (ACL5), which encodes thermospermine synthase in Arabidopsis, is under negative feedback control by thermospermine (Kakehi et al., 2008; Tong et al., 2014). In animals and fungi, degradation of ornithine decarboxylase (ODC), which catalyzes the conversion of ornithine into putrescine, a rate-limiting step in polyamine biosynthesis, is directed by ODC antizyme (OAZ) whose synthesis is induced by higher cellular polyamine levels (Matsufuji et al., 1995; Ivanov et al., 2000).
As for transport systems, antiporters of putrescine-ornithine and cadaverine-lysine as well as putrescine-specific and spermidine-preferential uptake carriers have been characterized in Escherichia coli (Kashiwagi and Igarashi, 2011). In yeast, GAP1 catalyzes the uptake of putrescine and spermidine together with the uptake of amino acids (Uemura et al., 2005). AGP2 acts as a high-affinity amino acid permease and selectively catalyzes the uptake of spermidine (Aouida et al., 2005). Three additional proteins, DUR3, SAM3, and tonoplast-localized UGA4 also function in polyamine uptake, while five excretion proteins, TPO1 to TPO5, have been identified (Igarashi and Kashiwagi, 2010a). In human, while multiple systems for uptake of polyamines have been identified, the organic cation transporter2 (OCT2) has been shown to function as a common uptake carrier (Higashi et al., 2014). In plants, earlier studies using petals of Saintpaulia ionantha revealed that putrescine uptake is dependent on the external pH (Bagni and Pistocchi, 1985). In carrot cell cultures, the uptake of putrescine and spermidine is remarkably rapid, reaching a maximum within only 1 min (Pistocchi et al., 1987; Antognoni et al., 1993). A genetic study of an Arabidopsis wild-type accession that is resistant to paraquat identified RMV1 as a polyamine uptake transporter (Fujita et al., 2012). In rice, OsPUT1 to OsPUT3 have been identified as spermidine-preferential transporters. AtPUT1 to AtPUT3 are the orthologous proteins in Arabidopsis and function as high affinity spermidine uptake transporters (Mulangi et al., 2012), among which AtPUT3 is identical to RMV1 (Fujita and Shinozaki, 2014).
Degradation of spermidine, spermine, and thermospermine in plants is catalyzed by polyamine oxidases (PAOs). PAOs in peroxisomes or the cytoplasm mediate back-conversion reaction with an end product of hydrogen peroxide and probably 3-aminopropanal, while extracellular PAOs catalyze terminal catabolic reactions to produce 1,3-diaminopropane and hydrogen peroxide (Moschou et al., 2008; Tavladoraki et al., 2012). Putrescine is catalyzed by copper-containing amine oxidases (CuAOs) to 4-aminobutanal along with ammonia and hydrogen peroxide (Angelini et al., 2010). Polyamine-derived hydrogen peroxide plays a critical role in biotic and abiotic stress responses and also in triggering secondary wall deposition (Cona et al., 2006; Moschou et al., 2012; Moschou and Roubelakis-Angelakis, 2014).
While cellular polyamine levels are maintained by the above-described regulatory systems, they may also be interrelated with other metabolic pathways. In poplar cell cultures, spermidine and spermine levels positively correlate with most amino acids (Mattoo et al., 2010). Increased polyamine biosynthesis may result in increased assimilation of both nitrogen and carbon by the cells (Majumdar et al., 2016). On the other hand, exogenous polyamines cause a rapid production of nitric oxide (NO) in Arabidopsis (Tun et al., 2006). A genetic screen of Arabidopsis mutants with increased tolerance to norspermidine identified a dominant mutant, par1-1D, with increased expression of QSO2 (Alejandro et al., 2007). QSO2 encodes a quiescin-sulfhydryl oxidase and may activate K+ efflux systems involved in xylem loading in roots thereby reducing the accumulation of toxic cations such as Na+ and polyamines. We are interested in how plant cells perceive and respond to external polyamines. To gain insight into sensing, signaling, and responsive mechanisms to polyamines, we isolated Arabidopsis mutants that show increased resistance to spermine. Among them, two mutants were found to be alleles of NRT1.3 encoding a member of the low-affinity nitrate transporter family.
Materials and Methods
Chemicals
The hydrochloride salts of putrescine, spermidine, spermine, and norspermidine were purchased from Sigma (MO, USA). Murashige and Skoog (MS) salts were purchased from Wako (Osaka, Japan).
Plant Materials
Arabidopsis thaliana accession Columbia-0 (Col-0) was used as the wide type. A T-DNA insertion allele of NRT1.3 (AtNPF6.4), which was obtained from the SALK collection (SALK_001553) and named here sper3-3, and a deletion allele of NRT1.1 (AtNPF6.3), chl1-5 (Tsay et al., 1993), were obtained from the Arabidopsis Biological Resource Center (ABRC) at Ohio State University (OH, USA). A T-DNA insertion allele of NRT1.4 (AtNPF6.2), nrt1.4-2 (Chiu et al., 2004), was a gift from Yi-Fang Tsay (Academia Sinica, Taiwan). A T-DNA insertion allele of NRT1.2 (AtNPF4.6), ait1-1 (SALK_146143; Kanno et al., 2012), and a paraquat-resistant accession Est-1 (Fujita et al., 2012) were also obtained from ABRC. spms-1 has been previously described (Imai et al., 2004). The wild-type accession Landsberg erecta (Ler) was used for mapping experiments.
Mutant Isolation, Mapping, and Next Generation Sequencing
For screening of mutants resistant to polyamines, 49,000 M2 seeds of ethyl methanesulfonate (EMS)-mutagenized Col-0 were sown on MS agar plates containing 3 mM spermine and 28 resistant plant lines were selected. Among them, three lines that reproducibly showed increased resistance to spermine in the progeny were named spermine resistant1 (sper1), sper2, and sper3, respectively.
For chromosomal mapping of the mutant loci, F2 seeds from the cross between the mutants and Ler were sown on MS agar plates containing 3 mM spermine and the DNA was extracted from each individual that was resistant to spermine. Totally ca. 80–100 seedlings were selected for each mutant. PCR-based mapping was performed using simple sequence length polymorphism (SSLP) markers (Bell and Ecker, 1994) and cleaved amplified polymorphic sequence (CAPS) markers (Konieczny and Ausubel, 1993). Additional markers were designed according to the TAIR database1 and the primers used are shown in Supplementary Table S1.
Genome DNA sequences of the mutants were determined by next-generation sequencing with the SOLiD platform. The multiplex libraries were constructed using the SOLiD barcoding and sequenced on a single SOLiD slide (Tabata et al., 2013). The resulting sequence reads were aligned against the A. thaliana Col-0 genome reference TAIR9 (TAIR_chr_all.fas) using BioScope 1.3 software (Life Technologies) with default parameters. Single nucleotide polymorphisms (SNPs) were called using a diBayes SNP caller, a component of BioScope 1.3 and validated as described in Tabata et al. (2013).
Plasmid Construction and Plant Transformation
To generate the NRT1.3 promoter-GUS fusion gene, a 1.47-kb promoter region of NRT1.3 was amplified from Col-0 genomic DNA by PCR with primers SPER3pro-F, 5′-CACAG ACTCT TGGTT TCTAG A-3′ and SPER3pro-R, 5′-GGATC CTGAG AGAAG AAAGC AGAG-3′ and the PCR product was cloned into the pGEM-T easy vector (Promega), excised by restriction digestion with XbaI and BamHI, and cloned into the pBI101 Ti-plasmid vector (Clontech).
To construct the 35S promoter-driven NRT1.3-GFP fusion plasmid, a full-length cDNA of NRT1.3 was amplified by PCR with primers SPER3-F-ATG, 5′-CACCA TGGTT CATGT GTCAT CATCT CATG-3′ and SPER3-R-dSTOP, 5′-AGGAA TGTCT TAAGC TCAAA TTCG-3′ and the PCR product was cloned into the pENTR/D-TOPO vector (Invitrogen). Then, the fragment was transferred to the C-terminal GFP fusion vector pGWB5 (Nakagawa et al., 2007).
The constructs were introduced into Agrobacterium tumefaciens C58C1 by electroporation. Transformation of Arabidopsis with the GUS construct was performed by the floral dip method (Harrison et al., 2006) using 0.05% Silwet L-77 in the dipping solution.
Measurement of Chlorophyll
Chlorophyll was extracted from 50 mg of the aerial part of 10-day-old seedlings in 1 mL of N,N-dimethyl formamide at 4°C overnight in the dark and assayed as described (Porra et al., 1989).
GUS Staining
Samples were prefixed for 20 min in ice-cold 90% acetone (v/v), vacuum infiltrated for 20 min with the GUS staining solution containing 50 mM sodium phosphate buffer pH7.2, 2 mM potassium ferricyanide, 2 mM potassium ferrocyanide, 0.1% Triton X-100 (v/v) and 1 mM X-glucuronide, and incubated at 37°C overnight (Jefferson et al., 1987). The stained samples were cleared with 70% ethanol and observed under a light microscope. For sectioning, GUS-stained tissues were dehydrated in a graded ethanol series and embedded in Technovit 7100 (Heraeus Kulzer, Wehrheim, Germany) according to the manufacturer’s instructions. Sections were cut to 10 μm.
Transient Expression of GFP in Tobacco
Agrobacterium cells containing the NRT1.3-GFP construct were cultured in Luria broth (LB) with 50 μg/ml of kanamycin and hygromycin at 28°C overnight. Agrobacterium cells containing the p19 plasmid as a suppressor of silencing (Silhavy et al., 2002) were cultured in LB with 50 μg/ml kanamycin. Both cultures were centrifuged, dissolved in 10 ml infiltration buffer (10 mM MES, 10 mM MgCl2, 100 μM acetosyringone), and mixed together. The suspension was adjusted to a final OD600 of 0.8 for infiltration into leaves of 4-week-old Nicotiana benthamiana. At 2 days after infiltration, the GFP fluorescence signals were observed in the epidermal cells under confocal laser scanning microscopy (Eclipse C1, Nikon, Tokyo, Japan).
HPLC Analysis
Cellular polyamine levels were measured by HPLC. Acid-soluble free polyamines were extracted with perchloric acid from the aerial part of 10-day-old seedlings and benzoylated as described previously (Tong et al., 2014). The samples were analyzed using a reverse phase HPLC system equipped with TSKgel ODS-80Ts column (Toso, Tokyo, Japan).
Results
sper3-1 and sper3-2 Are Resistant to Polyamines
We identified three mutants that showed increased resistance to 3 mM spermine in an M2 population derived from EMS-mutagenized seeds of the accession Col-0 of A. thaliana and tentatively named sper1, sper2, and sper3. Because rough mapping data showed that sper1 is located on chromosome 4 while sper2 and sper3 are on chromosome 3 and, as shown below, sper3 and sper2 were found to be different alleles of the same gene, these alleles were designated sper3-1 and sper3-2, respectively. Under normal growth conditions, sper3-1 and sper3-2 have no obvious morphological abnormalities compared with the wild type (Figure 1A). However, when grown on MS agar plates containing different concentrations of spermine, sper3-1 and sper3-2 seedlings retained green leaves at 3 mM spermine while wild-type seedlings showed chlorosis at 1 mM spermine (Figure 1B). These mutant seedlings were also resistant to 25 mM putrescine and 5 mM spermidine while they were no more resistant to basic amino acids, lysine and arginine, than wild-type seedlings (Figure 1C and not shown). F1 seedlings from a backcross of sper3-1 to its wild-type parent Col-0 showed intermediate sensitivity to spermine in terms of the leaf development (Figure 2A) and the chlorophyll content (Figure 2B), indicating that sper3-1 represents a semi-dominant allele. Similarly, the seedlings heterozygous for sper3-2 showed intermediate sensitivity to spermine (Figure 2B), indicating that sper3-2 also has a semi-dominant effect.

FIGURE 1. Phenotypes of sper3-1 and sper3-2. (A) Gross morphology of 20-day-old wild-type (Col-0), sper3-1 and sper3-2 plants. (B) Phenotype of 10-day-old seedlings grown in MS agar plates with spermine (Spm). (C) Phenotype of 10-day-old seedlings grown in MS agar plates with spermidine (Spd), putrescine (Put), lysine (Lys), or arginine (Arg).
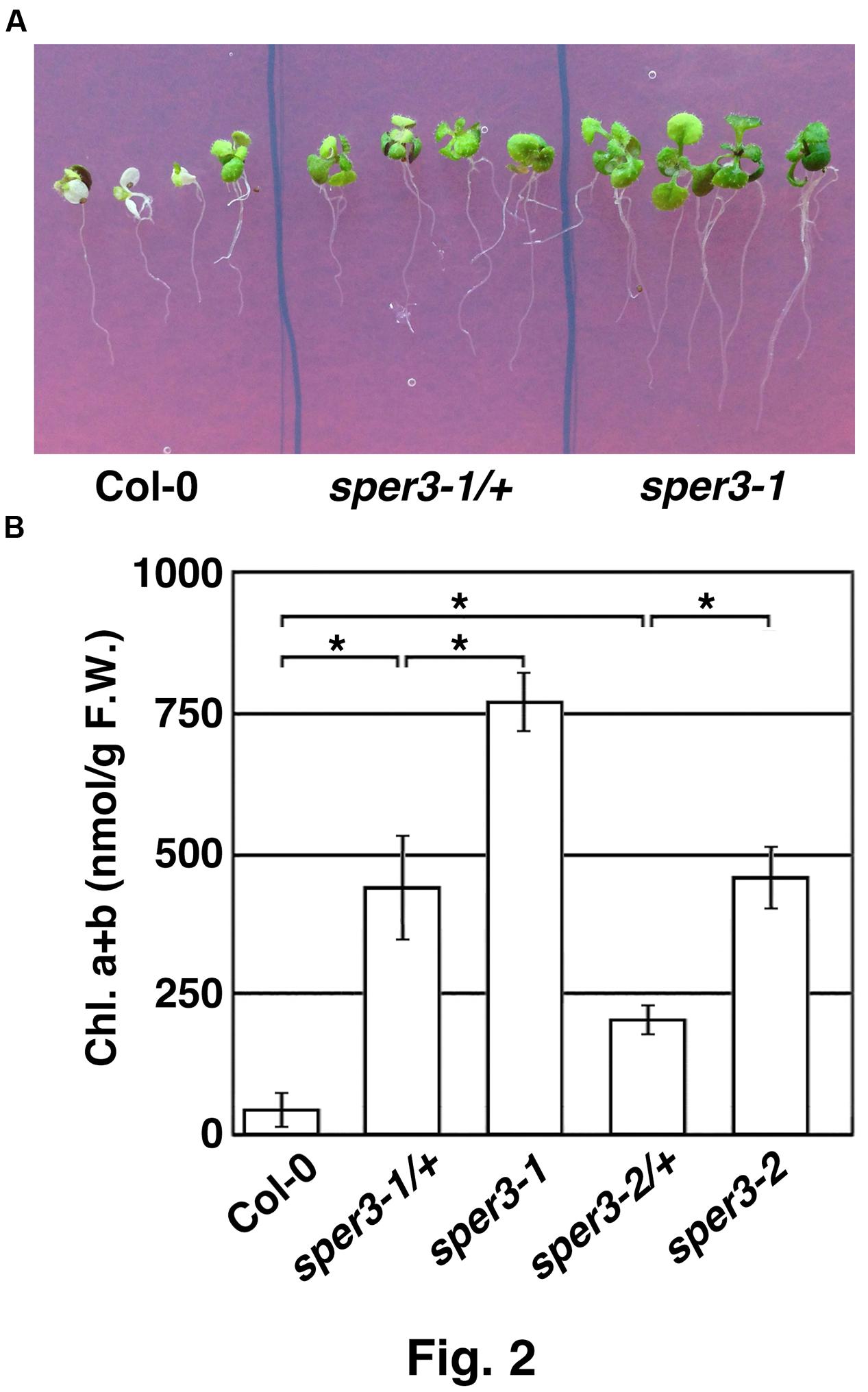
FIGURE 2. Semi-dominant traits of sper3-1 and sper3-2. (A) Phenotype of 10-day-old seedlings grown in MS agar plates with 2 mM spermine. (B) Chlorophyll content of 10-day-old seedlings grown in MS agar plates with 2 mM spermine. Error bars represent the SE (n = 3). Asterisks indicate statistically significant difference between groups assessed by ANOVA with Tukey-Kramer HSD (P < 0.05).
sper3-1 and sper3-2 Are Alleles of NRT1.3
Fine mapping using F2 plants from a cross between sper3-1 and the accession Ler delimited the locus to a 200-kb region on chromosome 3 (Figure 3A). Next-generation sequencing data of this region of the sper3-1 genomic DNA revealed a G-to-A point mutation in NRT1.3 (AtNPF6.4; Figure 3B), which belongs to the NRT1/PTR low-affinity nitrate transporter family (Tsay et al., 2007; Supplementary Figure S1). The G-to-A base substitution in sper3-1 changes glutamate 46, which is conserved in the family, to lysine (Figure 3C). On the other hand, sequencing of the sper3-2 genomic DNA revealed a C-to-T base substitution that changes alanine 433 of NRT1.3 (Figure 3B), which is also conserved in the family, to valine (Figure 3D).
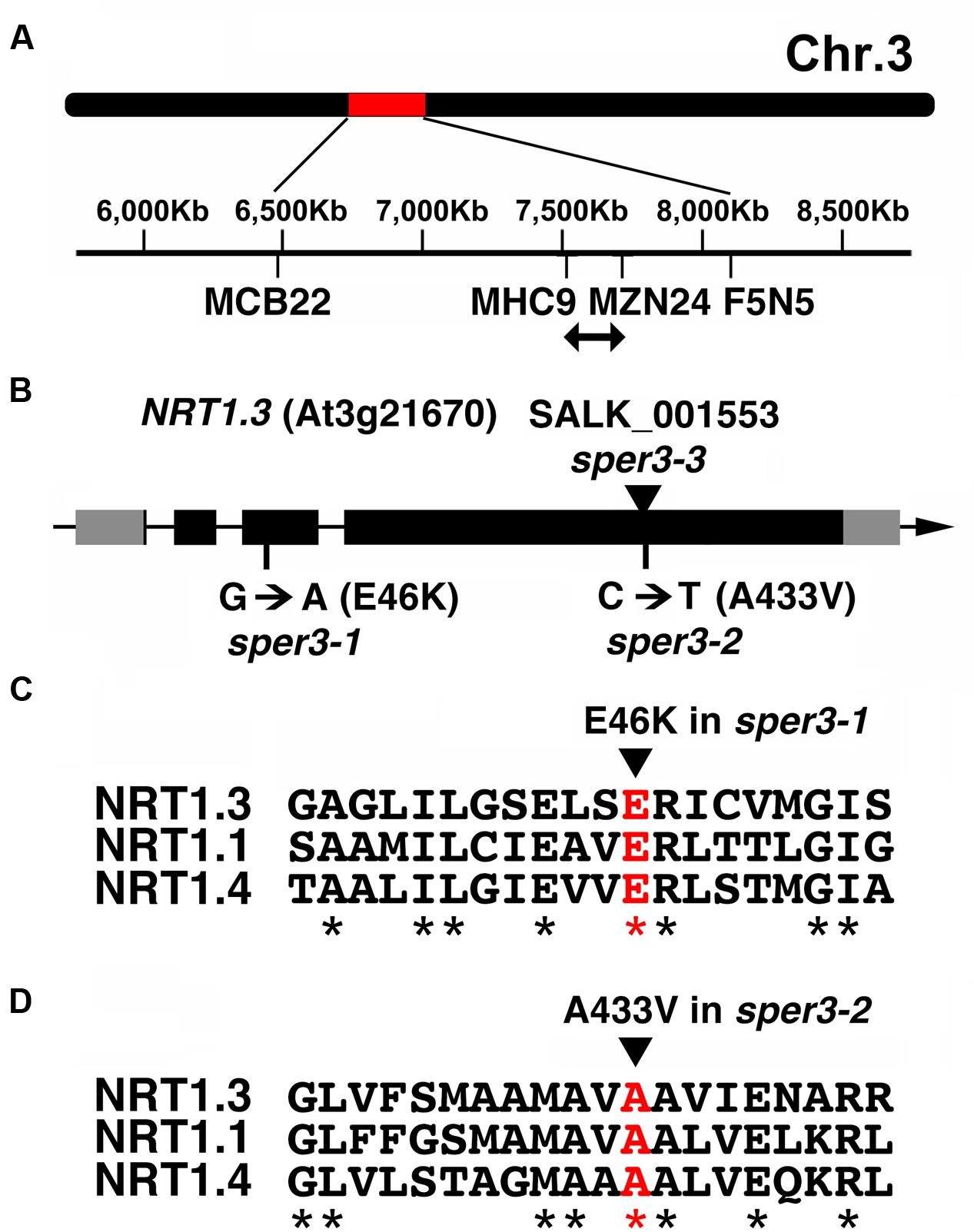
FIGURE 3. Gene structure of SPER3. (A) Chromosome location of sper3 alleles. (B) Genomic structure of NRT1.3 (AtNPF6.4, SPER3). Boxes indicate exons. Gray and black areas represent untranslated regions and protein coding regions, respectively. Base and amino acid substitutions in sper3-1 and sper3-2 are shown. The arrowhead indicates the position of T-DNA insertion in sper3-3. (C,D) Amino acid alignment of the region surrounding sper3-1 (C) and sper3-2 (D) mutated residues of NRT1.3 with corresponding regions of NRT1.1 (AtNPF6.3) and NRT1.4 (AtNPF6.2). Asterisks indicate conserved amino acids.
To confirm further that polyamine resistance is indeed caused by a mutation in NRT1.3, we examined whether a T-DNA insertion mutant of NRT1.3 shows increased resistance to spermine. The results revealed that the homozygous knockout allele designated sper3-3 (Figure 3B) had no detectable levels of full-length NRT1.3 mRNA (Supplementary Figure S2) and clearly showed increased resistance to 3 mM spermine (Figure 4A). Unlike sper3-1/+ and sper3-2/+, heterozygous sper3-3/+ seedlings showed no increased resistance to spermine as shown in the chlorophyll content (Figure 4B), indicating that sper3-3 is a recessive allele.
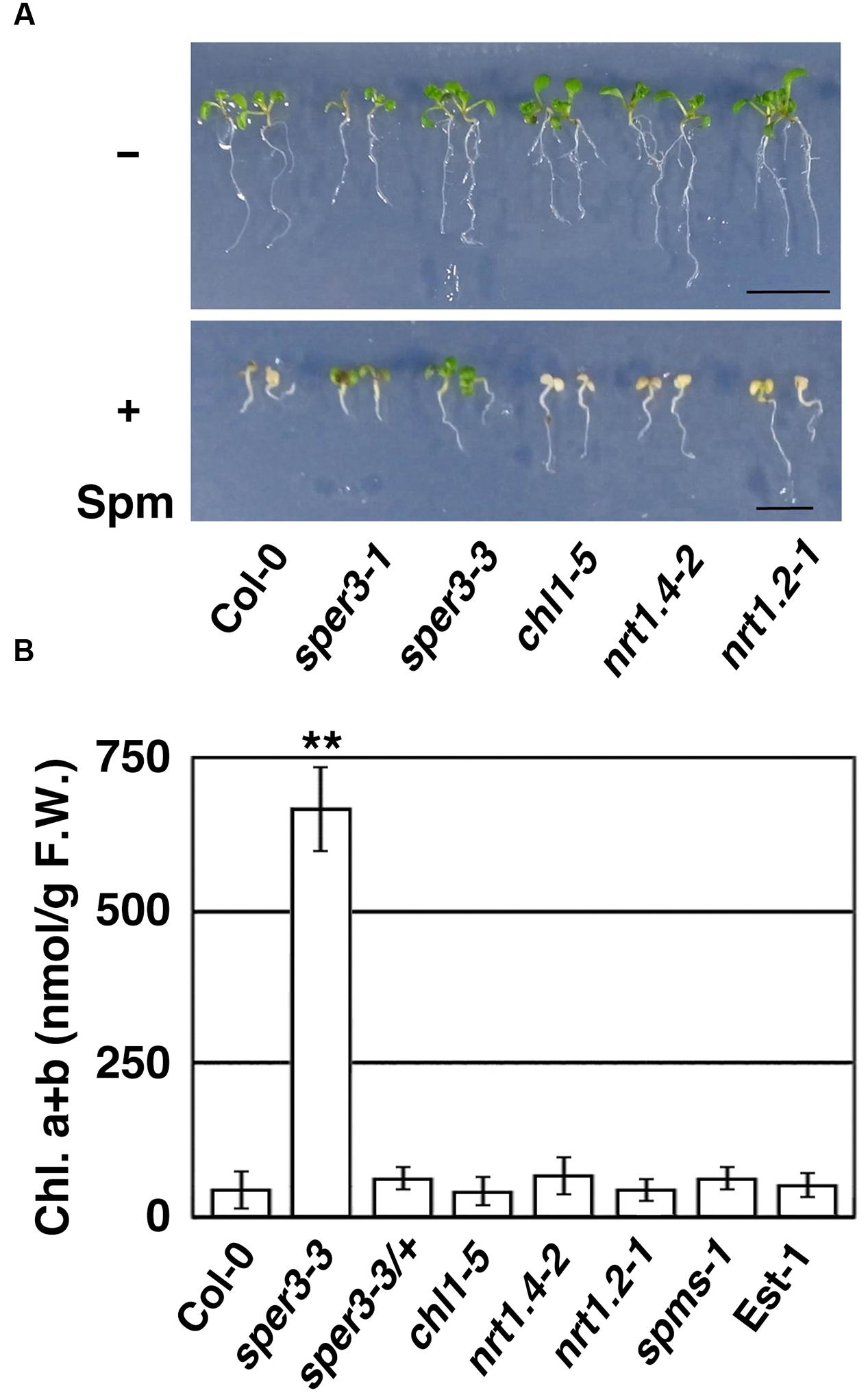
FIGURE 4. Growth response of sper3-3 and other knockout mutants of the AtNPF6 subfamily to spermine. (A) Phenotype of 7-day-old seedlings grown in MS agar plates with (+) or without (-) 2 mM spermine (Spm). chl1-5, nrt1.4-2, and nrt1.2-1 (ait1-1) represent loss-of-function mutants of AtNPF6.3, AtNPF6.2, and AtNPF4.6, respectively. (B) Chlorophyll content of 7-day-old seedlings grown in MS agar plates with 2 mM spermine. Error bars represent the SE (n = 3). Asterisks indicate statistically significant difference between the wild-type and mutants (∗∗P < 0.01 Student’s t-test).
NRT1.3 Is Preferentially Expressed in Parenchymal Tissues
NRT1.3 expression was detected by RT-PCR in all organs examined with the highest expression in flowers (Supplementary Figure S3A). When wild-type seedlings grown for 7 days on MS agar plates were incubated for 2 or 24 h in MS solutions with 100 μM spermine, no significant changes in the level of the NRT1.3 mRNA were detected (Supplementary Figure S3B). Although NRT1.3 expression is induced by NO3– in shoots (Okamoto et al., 2003), its tissue expression pattern has not been investigated in previous studies. Thus, we generated transgenic plants carrying the NRT1.3 promoter-GUS fusion gene and examined the GUS expression in more than eight independent plant lines. GUS staining was detected in the hypocotyl, cotyledons, and leaves but not in the root of seedlings (Figure 5A). In adult plants, inflorescence stems, pedicels, and sepals of flowers were strongly stained (Figures 5B,C). Tissue sections revealed that GUS expression was restricted to mesophyll cells of leaves and cortical cells of stems but not in epidermal cells (Figures 5D,E), suggesting that NRT1.3 specifically functions in parenchymal tissues.
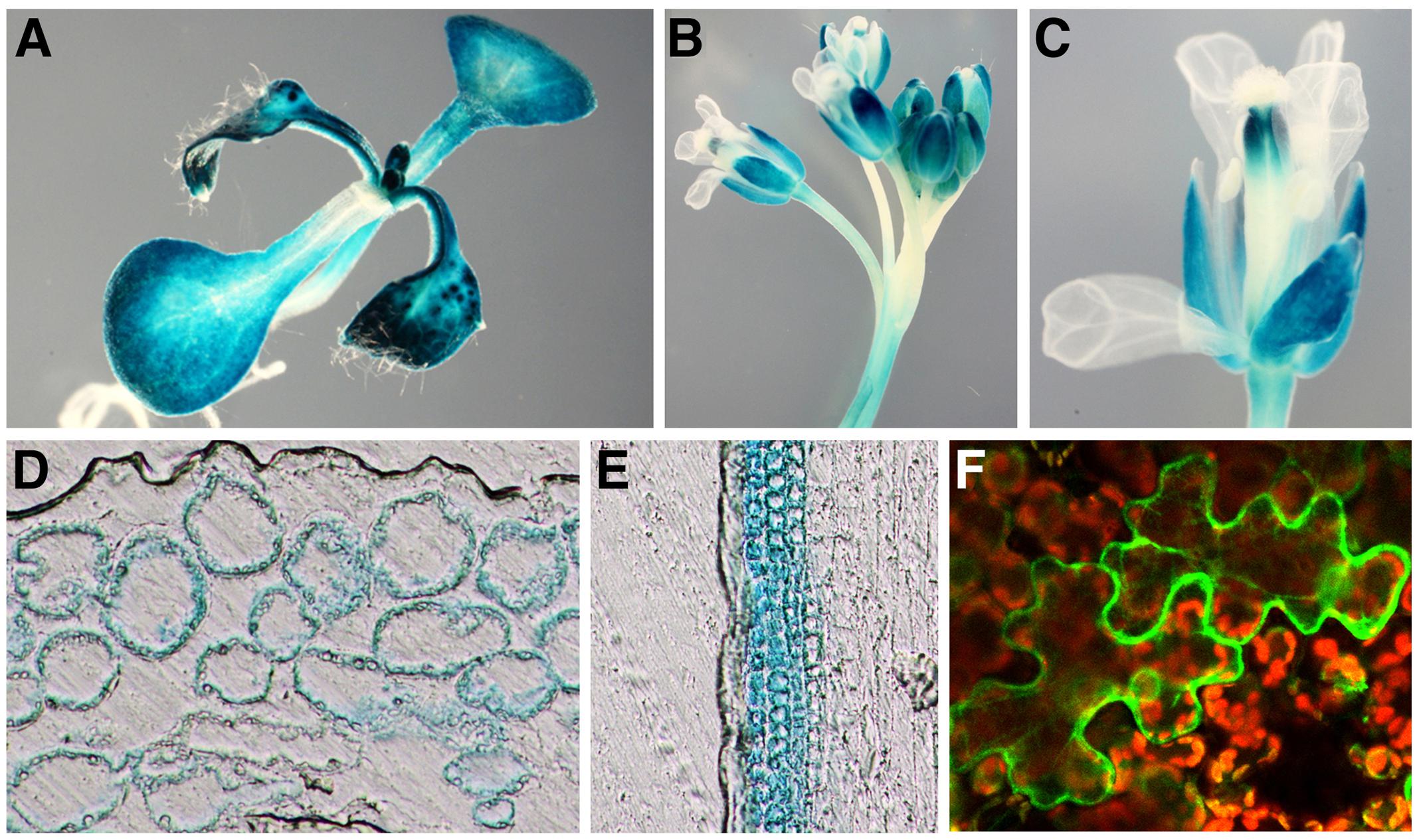
FIGURE 5. Expression of the AtNPF6.4 promoter-GUS gene and cellular localization of the AtNPF6.4-GFP fusion protein. (A–E) GUS staining pattern of a transgenic Arabidopsis plant carrying the AtNPF6.4 promoter-GUS fusion construct in 7-day-old seedlings (A), floral shoots (B), a single flower (C), a leaf section (D), and a stem section (E). (F) GFP fluorescence in a tobacco leaf epidermal cells carrying the AtNPF6.4-GFP fusion construct.
To examine cellular localization of NRT1.3, an NRT1.3-GFP translational fusion gene was constructed and transiently expressed under the CaMV 35S promoter in tobacco leaves by agro-infiltration. The NRT1.3-GFP fluorescence was detected mainly along the plasma membrane (Figure 5F).
Accumulation and Uptake of Polyamines in sper3-1
To examine whether or not sper3-1 affects polyamine transport or metabolism, cellular polyamine levels in the aerial part of sper3-1 seedlings were quantified by HPLC. The results revealed that the levels of all four polyamines, putrescine, spermidine, spermine, and thermospermine, were not affected by sper3-1 (Figure 6A). We next investigated polyamine uptake activity in sper3-1 by using norspermidine as a tracer because it is not present in wild-type plants. Seedlings grown for 7 days on MS agar plates were placed on the plate containing 100 μM norspermidine and the aerial part was subjected to polyamine extraction and HPLC. The results showed that the uptake level of norspermidine was initially indistinguishable between the wild type and sper3-1 but significantly declined after 24 h incubation in sper3-1 (Figure 6B).
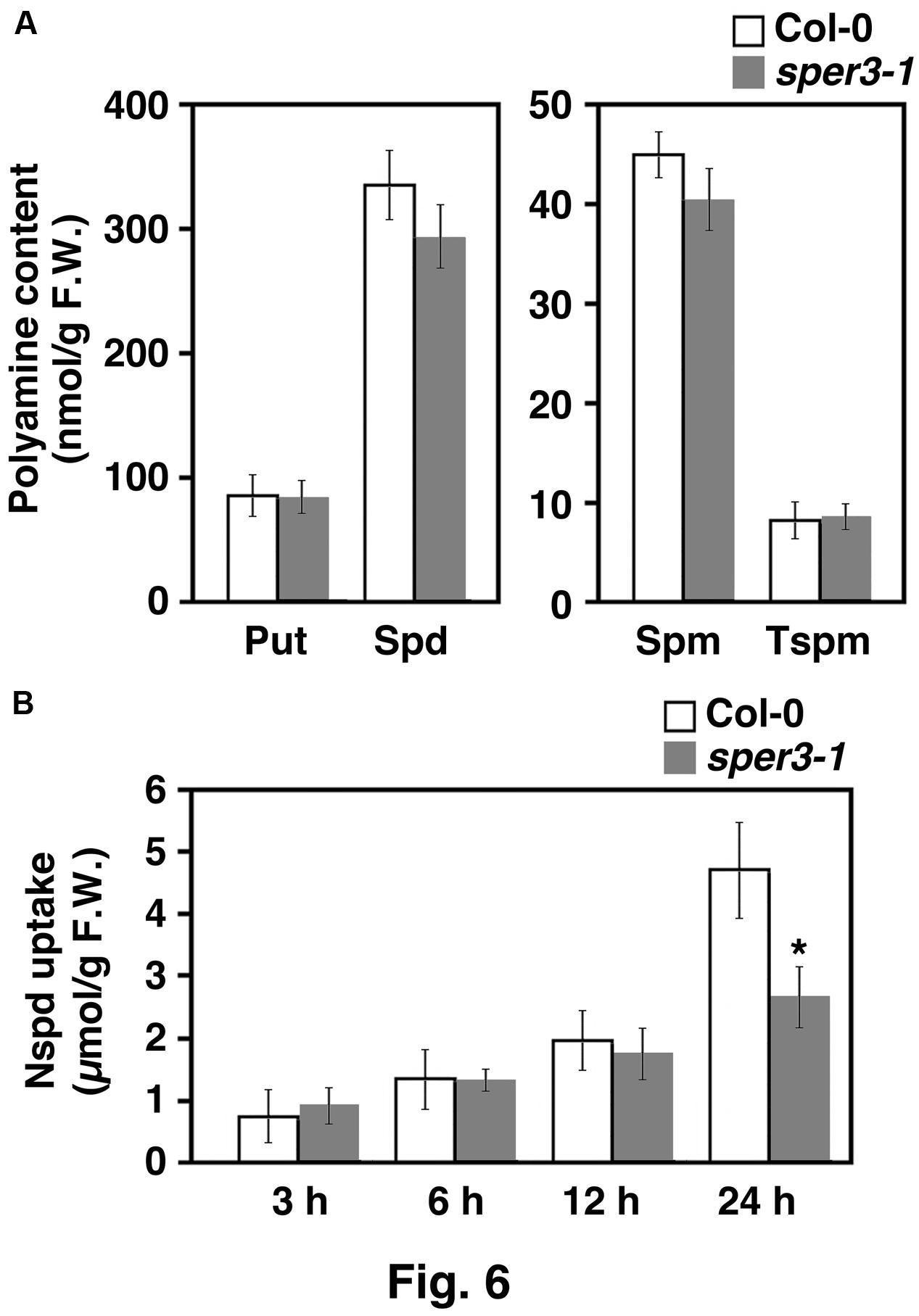
FIGURE 6. Polyamine content of wild-type and sper3-1 seedlings. (A) Level of putrescine (Put), spermidine (Spd), spermine (Spm), and thermospermine (Tspm) in grown in MS agar plates. Polyamines were extracted and analyzed by HPLC as described in section “Materials and Methods.” (B) Level of norspermidine (Nspd) in above-ground parts of seedlings. Wild-type and sper3-1 seedlings were grown for 7 days in MS agar plates, transferred to plates containing 100 μM norspermidine, and incubated for indicated hours. Error bars represent the SE (n = 3). An asterisk indicates statistically significant difference between wild-type and sper3-1 (∗P < 0.05 Student’s t-test).
Mutants of NRT1.1 and NRT1.4 Show no Increased Resistance to Spermine
NRT1.3 is classified into the same subfamily as root-specific NRT1.1 (AtNPF6.3) and leaf petiole-specific NRT1.4 (AtNPF6.2; Supplementary Figure S1; Wang et al., 2012). We thus examined whether these loss-of-function mutants and a mutant of vascular-specific NRT1.2 (AtNPF4.6; Kanno et al., 2012) also show increased resistance to spermine or not. chl1-5 is a deletion allele of NRT1.1 which shows increased resistance to chlorate (Tsay et al., 1993), while nrt1.4-2 and ait1-1 represent T-DNA insertion mutants of NRT1.4 and NRT1.2, respectively (Chiu et al., 2004; Kanno et al., 2012). When grown with 3 mM spermine, however, these mutant seedlings showed growth inhibition and chlorosis in a similar degree to wild-type seedlings (Figure 4A). Furthermore, neither spms-1, which is defective in the biosynthesis of spermine (Imai et al., 2004), nor the natural accession Est-1, which is defective in a plasma membrane-localized polyamine uptake transporter, RMV1/AtPUT3 (Fujita et al., 2012), showed increased resistance to spermine under our experimental condition (Figure 4B). We have also confirmed that, unlike the RMV1/AtPUT3-defective Est-1, which is resistant to paraquat, sper3-1 showed normal sensitivity to paraquat (Supplementary Figure S4).
Discussion
In this study, we found that alleles of NRT1.3 encoding a member of the NRT1/PTR family of nitrate transport proteins are responsible for the mutants with increased resistance to polyamines. The NRT1/PTR family in Arabidopsis consists of 53 members and includes both nitrate and oligopeptide transporters. The two closest homologs of NRT1.3, NRT1.4, and NRT1.1 share 51 and 47% amino acid sequence identities with NRT1.3, respectively. NRT1.1 is a dual-affinity transporter involved in both low- and high-affinity nitrate uptake, depending on cellular nitrate concentrations (Wang et al., 1998; Liu et al., 1999), and also facilitates the uptake of auxin in the absence of nitrate (Krouk et al., 2010). In contrast, NRT1.4 has been shown to be a low-affinity nitrate transporter (Chiu et al., 2004). Some other members are known to transport small organic anions as well as nitrate. NRT1.2 transports nitrate and abscisic acid (Huang et al., 1999; Kanno et al., 2012). NRT1.7, NRT1.9, and NRT1.10 (GTR2) play a role in the uptake of both nitrate and glucosinolates (Fan et al., 2009; Nour-Eldin et al., 2012). Although NRT1.3 remains to be characterized biochemically, its putative ortholog in Medicago truncatula, MtNRT1.3, has been shown to be a dual affinity transporter and also an ABA transporter (Morère-Le Paven et al., 2011; Pellizzaro et al., 2014). Given the cationic nature of polyamines, it is unlikely that NRT1.3 has the ability to transport polyamines, although its possibility cannot be excluded completely.
Nitrate is taken up from soil by the root tissue but its assimilation can take place in both the root and the shoot. NRT1.1 and NRT1.2 are involved in the uptake of nitrate from soil (Tsay et al., 1993; Wang et al., 1998; Huang et al., 1999; Liu et al., 1999). While NRT1.5 is expressed in root pericycle cells close to xylem and acts in loading nitrate into xylem vessels for root-to-shoot transport (Lin et al., 2008; Chen et al., 2012), NRT1.8, which is expressed in root xylem parenchyma cells, and NRT1.9, which is expressed in companion cells of the root phloem, are also involved in regulating root-to-shoot nitrate translocation (Li et al., 2010; Wang and Tsay, 2011). NRT1.7 is expressed in the phloem of the leaf vein, indicating that nitrate can be remobilized from older leaves to younger leaves (Fan et al., 2009). NRT1.4 is expressed in the leaf petiole and may be responsible for nitrate homeostasis in leaves (Chiu et al., 2004). NRT1.6 has been shown to function in delivering nitrate to the developing embryo in the seed (Almagro et al., 2008). We confirmed here that NRT1.3 is highly expressed in flowers, stems, and leaves. Our results of GUS expression (Figure 5) suggest that, complementarily to other members of the NRT1/PTR family, NRT1.3 may be expressed in mesophyll cells of the leaf and the cortex of the stem. However, because all sper3 alleles show wild-type phenotype under normal growth condition (Figure 1A), other nitrate transporters may function redundantly in these parenchymal tissues. Interestingly, the NRT1.3 promoter-GUS expression was not detected in epidermal cells. This might be related to the lack of chloroplasts in these cells. In shoot organs, nitrate assimilation is generally coupled to photosynthetic electron flow because the process of nitrite reduction occurs in the chloroplast stroma (Foyer and Noctor, 2002). The induction of NRT1.3 expression by NO3– is detected only in shoots (Okamoto et al., 2003). Furthermore, NRT1.3 expression is up-regulated by light while NRT1.1 and NRT1.5 by sugar (Lejay et al., 2008). Taken together, as a member of the NRT1/PTR family, NRT1.3 may play a role in the final step of nitrate supply to photosynthesizing cells. The strong fluorescence of NRT1.3-GFP at the cell periphery (Figure 5F) suggests that NRT1.3 is localized to the plasma membrane. However, its localization to endomembranes might also be possible. NRT2.7, one of the seven members of the NRT2 family, has been shown to be localized to the vacuolar membrane in seeds (Chopin et al., 2007).
We detected a reduced uptake of norspermidine in sper3-1 (Figure 6B). This raises a possibility that polyamine transport is associated or at least coincided with nitrate transport in parenchymal tissues and its defect may cause the reduced uptake of polyamines. sper3 mutants might have higher accumulation of nitrate in the root tissue and/or in the apoplast of shoot organs which could interfere with polyamine uptake. A recent study has revealed that mutants of a nitrate efflux channel, SLAH3, were more sensitive to ammonium, suggesting that the efflux of nitrate by SLAH3 alleviates ammonium toxicity in roots (Zheng et al., 2015). Although the sensitivity to polyamines in these mutants remains to be examined, SLAH3 might also serve to alleviate polyamine toxicity. On the other hand, considering that the initial uptake efficiency of norspermidine in sper3-1 was not different from that in the wild type (Figure 6B), it is also possible that the incorporated polyamines can be metabolized more rapidly under reduced availability of nitrate in shoot parenchymal tissues of sper3.
According to the crystal structure of NRT1.1, it forms a homodimer in the inward-facing conformation (Parker and Newstead, 2014; Sun et al., 2014). If this is also the case with NRT1.3, the semi-dominant trait of sper3-1 and sper3-2 might be due to a dominant negative effect by the presumed dimerization with the wild-type protein rather than a haplo-insufficiency. The NRT1/PTR family protein is also known as a proton-coupled transporter. In NRT1.1, H356 in the 7th transmembrane domain (TM7) is a binding site of nitrate, while the ExxER motif in TM1 plays a role in proton binding and co-transport (Steiner et al., 1995). This motif interacts with K164-E476 salt bridge involved in nitrate uptake (Doki et al., 2013; Parker and Newstead, 2014). The E46K amino acid substitution in sper3-1 occurs in the second E of the ExxER motif (Figure 3C), confirming the importance of this amino acid. Based on the phenotype of sper3-1 and sper3-2, the A433V amino acid substitution in sper3-2 may affect the function of NRT1.3 in a similar way to sper3-1. These alleles will provide useful tools for further biochemical study of this nitrate transporter.
Conclusion
This study suggests that nitrate transport in the parenchymal tissue of the shoot is closely interrelated with polyamine uptake or metabolism. Although mutants of NRT1.1, NRT1.2, and NRT1.4 showed normal sensitivity to spermine, involvement of some additional members in this interrelation may be possible, given the presence of multiple members of the NRT1/PTR family with various expression patterns. As a future study, it would be interesting to generate double and multiple mutants of these nitrate transporters and examine their sensitivity to polyamines.
Author Contributions
WT, AI, HM, and TT conceived and designed the experiments. WT, AI, RT, and TT performed the experiments. WT, HM, RT, and TT analyzed the data. SS, KY, MY, MH, and SS contributed materials and analysis tools. WT and TT wrote the paper.
Funding
This work was supported in part by the Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research on Innovative Areas No. 26113516 to TT, No. 25119715 to HM, and the National Institute for Basic Biology Cooperative Research Program (12-103) to SS, respectively.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We thank Yi-Fang Tsay for providing the nrt1.4-2 mutant strain.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00834
References
Alejandro, S., Rodríguez, P. L., Bellés, J. M., Yenush, L., García-Sanchez, M. J., Fernández, J. A., et al. (2007). An Arabidopsis quiescin-sulfhydryl oxidase regulates cation homeostasis at the root symplast-xylem interface. EMBO J. 26, 3203–3215. doi: 10.1038/sj.emboj.7601757
Almagro, A., Lin, S., and Tsay, Y. (2008). Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development. Plant Cell 20, 3289–3299. doi: 10.1105/tpc.107.056788
Angelini, R., Cona, A., Federico, R., Fincato, P., Tavladoraki, P., and Tisi, A. (2010). Plant amine oxidases ‘on the move’: an update. Plant Physiol. Biochem. 48, 560–564. doi: 10.1016/j.plaphy.2010.02.001
Antognoni, F., Pistocchi, R., and Bagni, N. (1993). Uptake competition between polyamines and analogues in carrot protoplasts. Plant Physiol. Biochem. 31, 693–698.
Aouida, M., Anick, L., Poulin, R., and Ramatar, D. (2005). AGP2 encodes the major permease for high affinity polyamine transport in Saccharomyces cerevisiae. J. Biol. Chem. 280, 24267–24276. doi: 10.1074/jbc.M503071200
Bagni, N., and Pistocchi, R. (1985). Putrescine uptake in Saintpaulia petals. Plant Physiol. 77, 398–402. doi: 10.1104/pp.77.2.398
Bell, C. J., and Ecker, J. R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. doi: 10.1006/geno.1994.1023
Chen, C. Z., Lv, X. F., Li, J. Y., Yi, H. Y., and Gong, J. M. (2012). Arabidopsis NRT1.5 is another essential component in the regulation of nitrate reallocation and stress tolerance. Plant Physiol. 159, 1582–1590. doi: 10.1104/pp.112.199257
Chiu, C. C., Lin, C. S., Hsia, A. P., Su, R. C., Lin, H. L., and Tsay, Y. F. (2004). Mutation of a nitrate transporter, AtNRT1.4, results in a reduced petiole nitrate content and altered leaf development. Plant Cell Physiol. 45, 1139–1148.
Chopin, F., Orsel, M., Dorbe, M. F., Chardon, F., Truong, H. N., Miller, A. J., et al. (2007). The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell 19, 1590–1602. doi: 10.1105/tpc.107.050542
Cona, A., Rea, G., Angelini, R., Federico, R., and Tavladoraki, P. (2006). Functions of amine oxidases in plant development and defence. Trends Plant Sci. 11, 80–88. doi: 10.1016/j.tplants.2005.12.009
Doki, S., Kato, H. E., Solcan, N., Iwaki, M., Koyama, M., Hattori, M., et al. (2013). Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT. Proc. Natl. Acad. Sci. U.S.A. 110, 11343–11348. doi: 10.1073/pnas.1301079110
Fan, S. C., Lin, C. S., Hsu, P. K., Lin, S. H., and Tsay, Y. F. (2009). The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 21, 2750–2761. doi: 10.1105/tpc.109.067603
Foyer, C. H., and Noctor, G. (2002). “Photosynthetic nitrogen assimilation: inter-pathway control and signalling,” in Photosynthetic Nitrogen Assimilation and Associated Carbon and Respiratory Metabolism. Advances in Photosynthesis Research Vol. 12, eds C. H. Foyer and G. Noctor (Amsterdam: Kluwer), 1–22.
Fujita, M., Fujita, Y., Iuchi, S., Yamada, K., Kobayashi, Y., Urano, K., et al. (2012). Natural variation in a polyamine transporter determines paraquat tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 6343–6347. doi: 10.1073/pnas.1121406109
Fujita, M., and Shinozaki, K. (2014). Identification of polyamine transporters in plants: paraquat transport provides crucial clues. Plant Cell Physiol. 55, 855–861. doi: 10.1093/pcp/pcu032
Hanfrey, C., Elliott, K. A., Franceschetti, M., Mayer, M. J., Illingworth, C., and Michael, A. J. (2005). A dual upstream open reading frame-based autoregulatory circuit controlling polyamine-responsive translation. J. Biol. Chem. 280, 39229–39237. doi: 10.1074/jbc.M509340200
Harrison, S. J., Mott, E. K., Parsley, K., Aspinall, S., Gray, J. C., and Cottage, A. (2006). A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 6:19. doi: 10.1186/1746-4811-2-19
Higashi, K., Imamura, M., Fudo, S., Uemura, T., Saiki, R., Hoshino, T., et al. (2014). Identification of functional amino acid residues involved in polyamine and agmatine transport by human organic cation transporter 2. PLoS ONE 9:e102234. doi: 10.1371/journal.pone.0102234
Huang, N. C., Liu, K. H., Lo, H. J., and Tsay, Y. F. (1999). Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11, 1381–1392. doi: 10.1105/tpc.11.8.1381
Igarashi, K., and Kashiwagi, K. (2010a). Characteristics of cellular polyamine transport in prokaryotes and eukaryotes. Plant Physiol. Biochem. 48, 506–512. doi: 10.1016/j.plaphy.2010.01.017
Igarashi, K., and Kashiwagi, K. (2010b). Modulation of cellular function by polyamines. Int. J. Biochem. Cell Biol. 42, 39–51. doi: 10.1016/j.biocel.2009.07.009
Imai, A., Akiyama, T., Kato, T., Sato, S., Tabata, S., Yamamoto, K. T., et al. (2004). Spermine is not essential for survival of Arabidopsis. FEBS Lett. 556, 148–152. doi: 10.1016/S0014-5793(03)01395-4
Ivanov, I. P., Matsufuji, S., Murakami, Y., Gesteland, R. F., and Atkins, J. F. (2000). Conservation of polyamine regulation by translational frameshifting from yeast to mammals. EMBO J. 19, 1907–1917. doi: 10.1093/emboj/19.8.1907
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Kakehi, J. I., Kuwashiro, Y., Niitsu, Y., and Takahashi, T. (2008). Thermospermine is required for stem elongation in Arabidopsis thaliana. Plant Cell Physiol. 49, 1342–1349. doi: 10.1093/pcp/pcn109
Kanno, Y., Hanada, A., Chiba, Y., Ichikawa, T., Nakazawa, M., Matsui, M., et al. (2012). Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. U.S.A. 109, 9653–9658. doi: 10.1073/pnas.1203567109
Kashiwagi, K., and Igarashi, K. (2011). Identification and assays of polyamine transport systems in Escherichia coli and Saccharomyces cerevisiae. Methods Mol. Biol. 720, 295–308. doi: 10.1007/978-1-61779-034-8_18
Konieczny, A., and Ausubel, F. M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR based markers. Plant J. 4, 403–410. doi: 10.1046/j.1365-313X.1993.04020403.x
Krouk, G., Lacombe, B., Bielach, A., Perrine-Walker, F., Malinska, K., Mounier, E., et al. (2010). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18, 927–937. doi: 10.1016/j.devcel.2010.05.008
Lejay, L., Wirth, J., Pervent, M., Cross, J. M., Tillard, P., and Gojon, A. (2008). Oxidative pentose phosphate pathway-dependent sugar sensing as a mechanism for regulation of root ion transporters by photosynthesis. Plant Physiol. 146, 2036–2053. doi: 10.1104/pp.107.114710
Li, J. Y., Fu, Y. L., Pike, S. M., Bao, J., Tian, W., Zhang, Y., et al. (2010). The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22, 1633–1646. doi: 10.1105/tpc.110.075242
Lin, S. H., Kuo, H. F., Canivenc, G., Lin, C. S., Lepetit, M., Hsu, P. K., et al. (2008). Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20, 2514–2528. doi: 10.1105/tpc.108.060244
Liu, K. H., Huang, C. Y., and Tsay, Y. F. (1999). CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11, 865–874. doi: 10.2307/3870820
Majumdar, R., Barchi, B., Turlapati, S. A., Gagne, M., Minocha, R., Long, S., et al. (2016). Glutamate, ornithine, arginine, proline, and polyamine metabolic interactions: the pathway is regulated at the post-transcriptional level. Front. Plant Sci. 7:78. doi: 10.3389/fpls.2016.00078
Matsufuji, S., Matsufuji, T., Miyazaki, Y., Murakami, Y., Atkins, J. F., Gesteland, R. F., et al. (1995). Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell 80, 51–60. doi: 10.1016/0092-8674(95)90450-6
Mattoo, A. K., Minocha, S. C., Minocha, R., and Handa, A. K. (2010). Polyamines and cellular metabolism in plants: transgenic approaches reveal different responses to diamine putrescine versus higher polyamines spermidine and spermine. Amino Acids 38, 405–413. doi: 10.1007/s00726-009-0399-4
Morère-Le Paven, M. C., Viau, L., Hamon, A., Vandecasteele, C., Pellizzaro, A., Bourdin, C., et al. (2011). Characterization of a dual-affinity nitrate transporter MtNRT1.3 in the model legume Medicago truncatula. J. Exp. Bot. 62, 5595–5605. doi: 10.1093/jxb/err243
Moschou, P. N., and Roubelakis-Angelakis, K. A. (2014). Polyamines and programmed cell death. J. Exp. Bot. 65, 1285–1296. doi: 10.1093/jxb/ert373
Moschou, P. N., Sanmartin, M., Andriopoulou, A. H., Rojo, E., Sanchez-Serrano, J. J., and Roubelakis-Angelakis, K. A. (2008). Bridging the gap between plant and mammalian polyamine catabolism: a novel peroxisomal polyamine oxidase responsible for a full back-conversion pathway in Arabidopsis. Plant Physiol. 147, 1845–1857. doi: 10.1104/pp.108.123802
Moschou, P. N., Wu, J., Cona, A., Tavladoraki, P., Angelini, R., and Roubelakis-Angelakis, K. A. (2012). The polyamines and their catabolic products are significant players in the turnover of nitrogenous molecules in plants. J. Exp. Bot. 63, 5003–5015. doi: 10.1093/jxb/ers202
Mulangi, V., Chibucos, M. C., Phuntumart, V., Paul, F., and Morris, P. F. (2012). Kinetic and phylogenetic analysis of plant polyamine uptake transporters. Planta 236, 1261–1273. doi: 10.1007/s00425-012-1668-0
Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., et al. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41. doi: 10.1263/jbb.104.34
Nour-Eldin, H. H., Andersen, T. G., Burow, M., Madsen, S. R., Jørgensen, M. E., Olsen, C. E., et al. (2012). NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488, 531–534. doi: 10.1038/nature11285
Okamoto, M., Vidmar, J. J., and Glass, A. D. (2003). Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: responses to nitrate provision. Plant Cell Physiol. 44, 304–317. doi: 10.1093/pcp/pcg036
Parker, J. L., and Newstead, S. (2014). Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature 507, 68–72. doi: 10.1038/nature13116
Pegg, A. E., and Casero, R. A. Jr. (2011). Current status of the polyamine research field. Methods Mol. Biol. 720, 3–35. doi: 10.1007/978-1-61779-034-8_1
Pellizzaro, A., Clochard, T., Cukier, C., Bourdin, C., Juchaux, M., Montrichard, F., et al. (2014). The nitrate transporter MtNPF6.8 (MtNRT1.3) transports abscisic acid and mediates nitrate regulation of primary root growth in Medicago truncatula. Plant Physiol. 166, 2152–2165. doi: 10.1104/pp.114.250811
Pistocchi, R., Bangi, N., and Creus, J. A. (1987). Polyamine uptake in carrot cell cultures. Plant Physiol. 84, 374–380. doi: 10.1104/pp.84.2.374
Porra, R. J., Thompson, W. A., and Kriedemann, P. E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll II standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394. doi: 10.1016/S0005-2728(89)80347-0
Raney, A., Law, G. L., Mize, G. J., and Morris, D. R. (2002). Regulated translation termination at the upstream open reading frame in S-adenosylmethionine decarboxylase mRNA. J. Biol. Chem. 277, 5988–5994. doi: 10.1074/jbc.M108375200
Silhavy, D., Molnár, A., Lucioli, A., Szittya, G., Hornyik, C., Tavazza, M., et al. (2002). A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21, 3070–3080. doi: 10.1093/emboj/cdf312
Steiner, H. Y., Naider, F., and Becker, J. M. (1995). The PTR family: a new group of peptide transporters. Mol. Microbiol. 16, 825–834. doi: 10.1111/j.1365-2958.1995.tb02310.x
Sun, J., Bankston, J. R., Payandeh, J., Hinds, T. R., Zagotta, W. N., and Zheng, N. (2014). Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature 507, 73–77. doi: 10.1038/nature13074
Tabata, R., Kamiya, T., Shigenobu, S., Yamaguchi, K., Yamada, M., Hasebe, M., et al. (2013). Identification of an EMS-induced causal mutation in a gene required for boron-mediated root development by low-coverage genome re-sequencing in Arabidopsis. Plant Signal. Behav. 8:e22534. doi: 10.4161/psb.22534
Takano, A., Kakehi, J. I., and Takahashi, T. (2012). Thermospermine is not a minor polyamine in the plant kingdom. Plant Cell Physiol. 53, 606–616. doi: 10.1093/pcp/pcs019
Tavladoraki, P., Cona, A., Federico, R., Tempera, G., Viceconte, N., Saccoccio, S., et al. (2012). Polyamine catabolism: target for antiproliferative therapies in animals and stress tolerance strategies in plants. Amino Acids 42, 411–426. doi: 10.1007/s00726-011-1012-1
Tong, W., Yoshimoto, K., Kakehi, J. I., Motose, H., Niitsu, M., and Takahashi, T. (2014). Thermospermine modulates expression of auxin-related genes in Arabidopsis. Front. Plant Sci. 5:94. doi: 10.3389/fpls.2014.00094
Tsay, Y. F., Chiu, C. C., Tsai, C. B., Ho, C. H., and Hsu, P. K. (2007). Nitrate transporters and peptide transporters. FEBS Lett. 581, 2290–2300. doi: 10.1016/j.febslet.2007.04.047
Tsay, Y. F., Schroeder, J. I., Feldmann, K. A., and Crawford, N. M. (1993). The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72, 705–713. doi: 10.1016/0092-8674(93)90399-B
Tun, N. N., Santa-Catarina, C., Begum, T., Silveira, V., Handro, W., Floh, E. I., et al. (2006). Polyamines induce rapid biosynthesis of nitric oxide (NO) in Arabidopsis thaliana seedlings. Plant Cell Physiol. 47, 346–354. doi: 10.1093/pcp/pci252
Uemura, T., Tachihara, K., Tomitori, H., Kashiwagi, K., and Igarashi, K. (2005). Characteristics of the polyamine transporter TPO1 and regulation of its activity and cellular localization by phosphorylation. J. Biol. Chem. 280, 9646–9652. doi: 10.1074/jbc.M410274200
Wang, R., Liu, D., and Crawford, N. M. (1998). The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proc. Natl. Acad. Sci. U.S.A. 95, 15134–15139. doi: 10.1073/pnas.95.25.15134
Wang, Y. Y., Hsu, P. K., and Tsay, Y. F. (2012). Uptake, allocation and signaling of nitrate. Trends Plant Sci. 17, 458–467. doi: 10.1016/j.tplants.2012.04.006
Wang, Y. Y., and Tsay, Y. F. (2011). Arabidopsis nitrate transporter NRT1.9 is important in phloem nitrate transport. Plant Cell 23, 1945–1957. doi: 10.1105/tpc.111.083618
Keywords: Arabidopsis, nitrate transporter, NRT1/PTR family, parenchymal tissue, polyamines
Citation: Tong W, Imai A, Tabata R, Shigenobu S, Yamaguchi K, Yamada M, Hasebe M, Sawa S, Motose H and Takahashi T (2016) Polyamine Resistance Is Increased by Mutations in a Nitrate Transporter Gene NRT1.3 (AtNPF6.4) in Arabidopsis thaliana. Front. Plant Sci. 7:834. doi: 10.3389/fpls.2016.00834
Received: 29 February 2016; Accepted: 27 May 2016;
Published: 13 June 2016.
Edited by:
Wim Van den Ende, Katholieke Universiteit Leuven, BelgiumReviewed by:
Rubén Alcázar, Universitat de Barcelona, SpainPanagiotis N. Moschou, Swedish University of Agricultural Sciences, Sweden
Anthoni Pellizzzaro, University of Angers, France
Copyright © 2016 Tong, Imai, Tabata, Shigenobu, Yamaguchi, Yamada, Hasebe, Sawa, Motose and Takahashi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Taku Takahashi, perfect@cc.okayama-u.ac.jp
†Present address: Ryo Tabata, Graduate School of Science, Nagoya University, Nagoya, 464-8602, Japan
 Wurina Tong
Wurina Tong Akihiro Imai2
Akihiro Imai2 Ryo Tabata3†
Ryo Tabata3† Masashi Yamada
Masashi Yamada Mitsuyasu Hasebe
Mitsuyasu Hasebe Shinichiro Sawa
Shinichiro Sawa Hiroyasu Motose
Hiroyasu Motose Taku Takahashi
Taku Takahashi