- 1College of Horticulture, Sichuan Agricultural University, Chengdu, China
- 2Institute of Pomology and Olericulture, Sichuan Agricultural University, Chengdu, China
- 3Agricultural and Animal Husbandry College of Tibet University, Linzhi, China
Rubus L. is a large and taxonomically complex genus, species of which exhibit apomixis, polyploidy, and frequent hybridization. Most of Chinese Rubus are assigned in two major sections, Idaeobatus and Malachobatus. To explore the phylogenetic relationships within Chinese Rubus, inferences upon three chloroplast DNA (rbcL, rpl20-rps12, and trnG-trnS), nuclear ribosomal ITS, and two low-copy nuclear markers (GBSSI-2 and PEPC) were deduced in 142 Rubus taxa from 17 subsections in 6 sections. nrITS and GBSSI-2 were the most informative among the six DNA regions assessed. Phylogenetic relationships within Rubus were well-resolved by combined nuclear datasets rather than chloroplast markers. The phylogenetic inferences strongly supported that section Idaeobatus was a polyphyletic group with four distant clades. All samples of sect. Malachobatus formed a monophyletic clade, in which R. tsangorum and R. amphidasys of sect. Dalibardastrum, and R. peltatus from subsection Peltati of sect. Idaeobatus were always nested. Rubus pentagonus (2n = 2x = 14) from subsect. Alpestres of sect. Idaeobatus was a sister group to the polyploid sect. Malachobatus, as well as the polytomy of three sect. Cyalctis members. This suggests that some polyploids of Malachobatus might originate from common ancestors, via polyploidization of hybrids between R. pentagonus and sect. Cylactis species. They had experienced species explosion in a short time. Section Dalibardastrum species have potential parental lineages from subsects. Moluccani and Stipulosi of sect. Malachobatus. Based on molecular phylogenies, we also provided recommendations for the taxonomic treatments of four taxa. In addition, our results showed certain incongruence between chloroplast and nuclear markers, which might be due to hybridization and introgression.
Introduction
The genus Rubus L. comprises 900–1000 species and has a worldwide distribution (excluding Antarctica) (Focke, 1910, 1911, 1914). It has long been deemed taxonomically challenging due to its complexity for apomixis, polyploidy, lack of a universal species concept, and frequent hybridization (Thompson, 1997). The widely adopted taxonomic system built by Focke (1910, 1911, 1914) divided Rubus (Rubus hereafter) into 12 subgenera, with the three largest being Idaeobatus (117 species), Malachobatus (115 species), and Rubus (132 species, subg. Rubus hereafter). Among the remaining nine subgenera (Anoplobatus, Chamaebatus, Chamaemorus, Comaropsis, Cylactis, Dalibarda, Dalibardastrum, Lampobatus, and Orobatus), only three have more than six species. South-western China is thought to be a major center of origin for Rubus because of the rich genetic diversity, large number of subgenera, and extensive morphological variations found there (Lu, 1983). However, the taxonomic system, widely accepted in China, proposed that Chinese Rubus consists of eight sections (almost corresponding to the subgenera by Focke), in which 139 species are endemic (Yü et al., 1985; Lu and Boufford, 2003). Some representative sections and the species number are: Idaeobatus (88 spp.), Lampobatus (1 spp.), Rubus (1 spp.), Malachobatus (92 spp.), Dalibardastrum (11 spp.), Chamaebatus (5 spp.), Cylactis (9 spp.), and Chamaemorus (1 spp.) (Lu and Boufford, 2003). Among these sections, the two largest, Idaeobatus and Malachobatus are further classified into 11 and 13 subsections respectively (Yü et al., 1985). Both two systems are mainly based on morphological characters. However, the two classification systems are partially contradictory in placement for certain species (Table S1). It is hence a challenging task for researchers to classify Rubus species correctly, particularly when just based on morphological appearance.
Polyploidy and hybridization are prevalent in Rubus (Alice and Campbell, 1999). Species of sect. Idaeobatus are predominantly diploid, whereas sects. Malachobatus, Dalibardastrum, and Chamaebatus are exclusively polyploid (Thompson, 1997; Naruhashi et al., 2002; Wang et al., 2008). Hybridization in Rubus occurs commonly between closely related species (Bammi and Olmo, 1966; Iwatsubo and Naruhashi, 1991, 1992, 1993a, 1996, 1998; Randell et al., 2004; Mimura et al., 2014), but sometimes can occur between sections (Iwatsubo and Naruhashi, 1993b; Alice et al., 2001). For instance, Rubus parvifolius of sect. Idaeobatus can not only cross with R. coreanus (R. × hiraseanus, 2x and 3x) and R. phoenicolasius (R. × nikaii, 2x) from sect. Idaeobatus (Iwatsubo and Naruhashi, 1991, 1996, 1998), but also with R. sieboldii (R. × tawadanus, 3x) from sect. Malachobatus (Iwatsubo and Naruhashi, 1992, 1993b), resulting in ranges of morphological variations (Nybom and Schaal, 1990). Therefore, molecular data can be extremely useful in assessing the phylogenetic relationship among Rubus species to complement with morphological data.
A lot of studies have attempted to gain phylogenetic information within Rubus using either maternally inherited chloroplast markers or a bi-parentally inherited ribosomal DNA marker. Molecular data, such as ndhF (Howarth et al., 1997; Morden et al., 2003; Zhang et al., 2015), rbcL (Imanishi et al., 2008), rpl16 (Alice et al., 2008), trnG-trnS (Michael, 2006), trnL-trnF (Yang and Pak, 2006), and ITS (Alice and Campbell, 1999; Alice et al., 2001), have partially resolved some phylogenetic uncertainties of Rubus. However, most of these studies focused on European and American Rubus taxa (Alice and Campbell, 1999; Sochor et al., 2015). The phylogeny of Chinese Rubus, with a majority of endemic taxa remains unresolved to date. Research with a more extensive taxon sampling based on multiple chloroplast and nuclear regions is necessary to construct a comprehensive phylogeny within Chinese Rubus.
In terms of polyploidy, the use of low-copy nuclear genes (LCNGs) is particularly useful for reconstructing reticulate evolution (Zimmer and Wen, 2013). Previous phylogenetic studies have shown that granule-bound starch synthase I (GBSSI) exons and introns are useful in resolving relationships among closely-related species or genera (Rousseau-Gueutin et al., 2009), especially in detecting ancient hybridizations (Evans et al., 2000; Michael, 2006). Phosphoenolpyruvate carboxylase (PEPC) has also been reported to have one or few copies and to be phylogenetically informative in different flowering plant families (Lo et al., 2009). These LCNGs have been applied to reconstruct the phylogeny within the Rosaceae family (Evans et al., 2000; Lo et al., 2009; Rousseau-Gueutin et al., 2009), revealing promising prospects. Therefore, we also expected to find informative characters within Chinese Rubus by using these low copy nuclear genes.
In this study, we used three chloroplast (rbcL, rpl20-rps12, and trnG-trnS) and three nuclear (nrITS, GBSSI-2, and PEPC) genetic markers to reconstruct the phylogeny of Chinese Rubus. Our sampling covered 106 species from 17 out of 24 subsections (six out of eight sections) in Rubus. There are four specific objectives of this study: (1) to evaluate the phylogenetic information of the six markers at different taxonomic levels; (2) to obtain a well-resolved and thoroughly sampled phylogeny for Chinese Rubus; (3) to illustrate the evolutionary history for sects. Idaeobatus, Malachobatus, and Dalibardastrum; and (4) to provide recommendations for the taxonomic treatments of four taxa based on molecular phylogenies.
Materials and Methods
Taxon Sampling
In total, we sampled 142 Rubus individuals, of which 88 (representing 63 species) belong to sect. Idaeobatus, one belongs to sect. Rubus, 47 (representing 36 species) belong to sect. Malachobatus, two belong to sect. Dalibardastrum, one belongs to sect. Chamaebatus, and three belong to sect. Cylactis. This collection, with confirmed ploidy level, contains 70 diploids (2n = 2x = 14), one triploid (2n = 3x = 21), 38 tetraploids (2n = 4x = 28), three hexaploids (2n = 6x = 42), and one octoploid (2n = 8x = 56) (Table S1) (Thompson, 1997; Amsellem et al., 2001; Meng and Finn, 2002; Wang et al., 2008). Samples were collected in the wild field from Sichuan, Guizhou, Jiangxi, He'nan, Shaanxi, Gansu, and Tibet Province, China (Figure S1) and were all identified by at least three botanists. Voucher specimens were deposited in the herbarium for horticultural plants, Sichuan Agricultural University (These herbaria were not indexed in Index Herebariorum). The samples were classified according to Flora of China (Yü et al., 1985; Lu and Boufford, 2003) because of the endemicity of some species. Fragaria vesca L. and Rosa banksiae Ait. were chosen as outgroups based on a previous study (Morgan et al., 1994). Detailed information can be found in Table S1.
DNA Extraction, PCR Amplification, and Sequencing
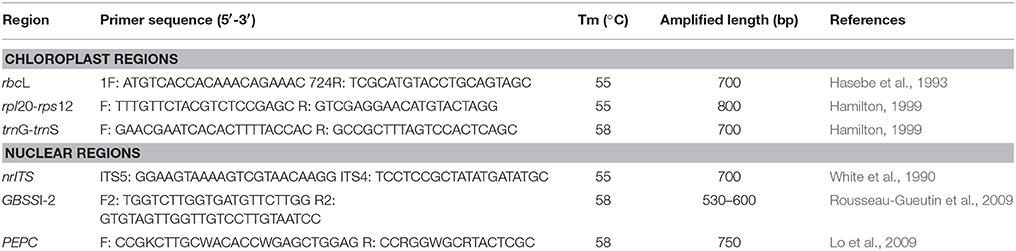
Total genomic DNA was isolated from silica-gel dried or frozen leaf tissues using a modified cetyltrimethyl ammonium bromide (CTAB) method (Zhou, 2005). Three chloroplast regions (rbcL, rpl20-rps12, and trnG-trnS), nuclear ribosomal internal transcribed spacers (nrITS), and two single copy nuclear genes coding for granule-bound starch synthase I (GBSSI), and phosphoenolpyruvate carboxylase (PEPC) were used in this study. The family Rosaceae has two copies of GBSSI: GBSSI-1 and GBSSI-2 (Evans et al., 2000). We selected GBSSI-2 gene with primers from strawberry (Rousseau-Gueutin et al., 2009) due to the observations of multiple copies of GBSSI-1 in Rubus polyploids during preliminary screening. Primers for above markers and the corresponding annealing temperature used in this study are listed in Table 1.
PCR amplification was performed in a 25 μL volume containing 20 ng of total DNA, 1.2 μL of MgCl2 (25 mmol·L−1), 1.4 μL of dNTP mix (10 mmol·L−1), 1 μL of each primer (5 μmol·L−1), 1.5 U of PfuDNA polymerase (Tiangen, Beijing), and 2.0 μL of 10 × PCR buffer (10 mmol·L−1 pH 8.0 Tris-HCl, 50 mmol·L−1 KCl, 1.5 mmol·L−1 EDTA). Conditions for amplification consisted of an initial denaturation at 94°C for 4 min, followed by 35 cycles at 94°C for 45 s, then at 55–58°C for 1 min and at 72°C for 1 min, with a final extension at 72°C for 10 min. Amplifications were conducted using a PTC-200 thermocycler (Bio-rad, Hercules, CA).
All reported genes in this study gave only one single band as determined by 1% agarose gel electrophosis. The products were purified using the UNIQ-10 Column MicroDNA Gel Extraction Kit (Sangon, Shanghai, China). Then they were sequenced directly in both directions using Big Dye Terminator Cycle Sequencing kit (version 2.0, Applied Biosystems, Foster City, CA, USA) on an ABI PRISM 3730 (Applied Biosystems, Foster City, CA, USA) automatic DNA sequencer (Beijing Genomics Institute (BGI), Shenzhen). All the sequences were deposited in the GenBank database with the following accession numbers: KU881049-KU881624, KU891076-KU891200, and KU926720-KU926855 (Table S1).
Sequence Alignment and Phylogenetic Analyses
Sequences of the six examined regions were edited and assembled using CLC Genomics Workbench (v7.5, CLC bio, Qiagen, Boston, MA). After manually editing, final datasets were aligned separately with Muscle (Edgar, 2004), and adjusted in the Molecular Evolutionary Genetics Analysis software (MEGA6) (Tamura et al., 2013) with gaps treated as missing data.
Before tree reconstruction, we performed partition homogeneity test (PHT) in PAUP v4.0 b10 (1000 replicates, invariable sites excluded) for the plastic and nuclear datasets (Swofford, 2002). According to the obtained PHT results, we applied partitioned phylogeny analyses by using maximum likelihood (ML) and bayesian inference (BI) methods for the combined cpDNA or nDNA datasets, with assignment that each partition had its own evolutionary rate. DNA substitution models were selected out from JModelTest v2.1.1 (Darriba et al., 2012) according to Akaike Information Criterion (AIC) (Akaike, 1974) for each gene.
We inferred the ML trees using the edge-linked partitioned phylogeny in IQ-TREE v1.4.2, which could implemented individual assigned substitution models for each partition (Nguyen et al., 2015; Chernomor et al., 2016). One thousand regular bootstrap replicates were performed to obtain confidence values for the branches. The values were considered to be low when strictly inferior to 65%, moderate between 65 and 80% and strong when superior to 80%. BI analyses were performed using MrBayes v3.2.1 with partitioned genes (Ronquist et al., 2012). The Markov chains Monte Carlo (MCMC) algorithm was run for 6,000,000 generations with one cold and three heated chains, starting with a random tree and sampling one tree every 1000 generations. The first 1,500,000 generations were treated as burn-in. An adequate burn-in value for each analysis was assessed using the software Tracer 1.5 (Rambaut and Drummond, 2009) and a 50% majority-rule consensus tree was then computed.
Phylogenetic network was constructed for combined nuclear datasets using SplitTree v4.14.2 (Huson and Bryant, 2006). Network analysis was performed using the NeighborNet algorithm with Kimura 2-parameter (K2P) distance and Ordinary Least Square Method implemented.
Results
Phylogeny of Combined Chloroplast Regions
After treating the gaps as missing characters, our aligned chloroplast rbcL, rpl20-rps12, and trnG-trnS DNA regions contained 668 base pairs (bp), 771 bp, and 694 bp in length, respectively (Table 2). The final combined cpDNA matrix consisted of 144 taxa and 2133 bp, of which 297 (13.92%) were variable. The selected best fit models for rbcL, rpl20-rps12 and trnG-trnS were TIM3+I+G, TVM+I+G, and GTR+I+G, respectively.
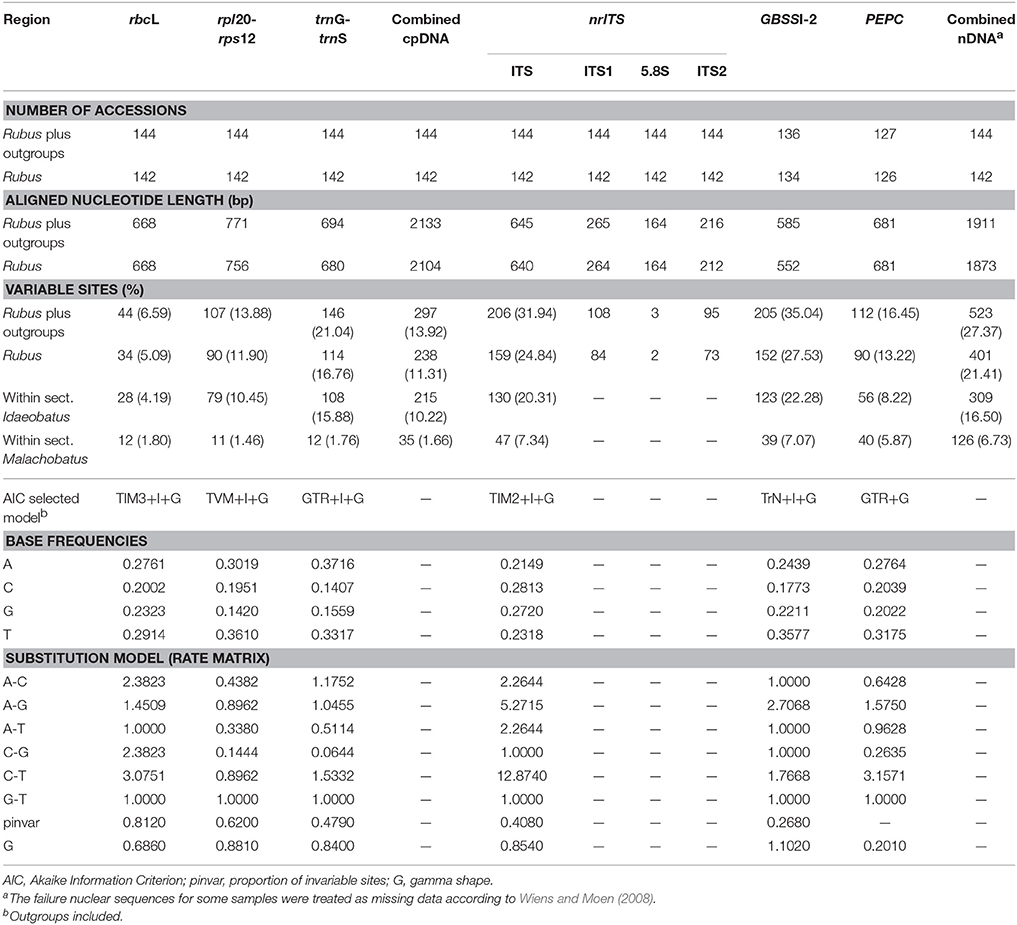
Table 2. Comparison of sequence variation and best-fitting models among different markers utilized in Rubus.
The results of the ML and BI analyses were congruent on major lineages. ML bootstrap support (BS) and BI posterior probabilities (PP) are shown on the 50% majority-rule consensus tree from BI analysis (Figure 1). Four well-supported clades were recovered within Chinese Rubus. Clade A included “Arapaho” (Blackberry, Rubus spp.), belonging to sect. Rubus. Clade B could be divided into seven subclades. Subclade B1 contained “Chilcotin” (Raspberry, R. idaeus L.), R. pungens and its varieties from subsect. Pungentes of sect. Idaeobatus. All samples from sect. Malachobatus formed a monophyletic subclade (B2) with high support values (78% BS, 0.86 PP). Rubus tsangorum and R. amphidasys from sect. Dalibardastrum were nested within subclade B2. Rubus peltatus from subsect. Peltati of sect. Idaeobatus clustered with two subsect. Moluccani species of sect. Malachobatus with weak support (64% BS, 0.56 PP). Rubus fockeanus (B3), R. fragarioides var. pubescens (B4), and R. nyalamensis (B5) from sect. Cylactis, R. pentagonus and R. pentagonus var. modestus (B6) from subsect. Alpestres of sect. Idaeobatus were in a polytomy with subclade B2. Subclade B7 was represented by R. calycinus of sect. Chamaebatus and sister to the group of subclades B1-B6. The above mentioned subclades formed a well-supported (80% BS, 0.98 PP) clade B.
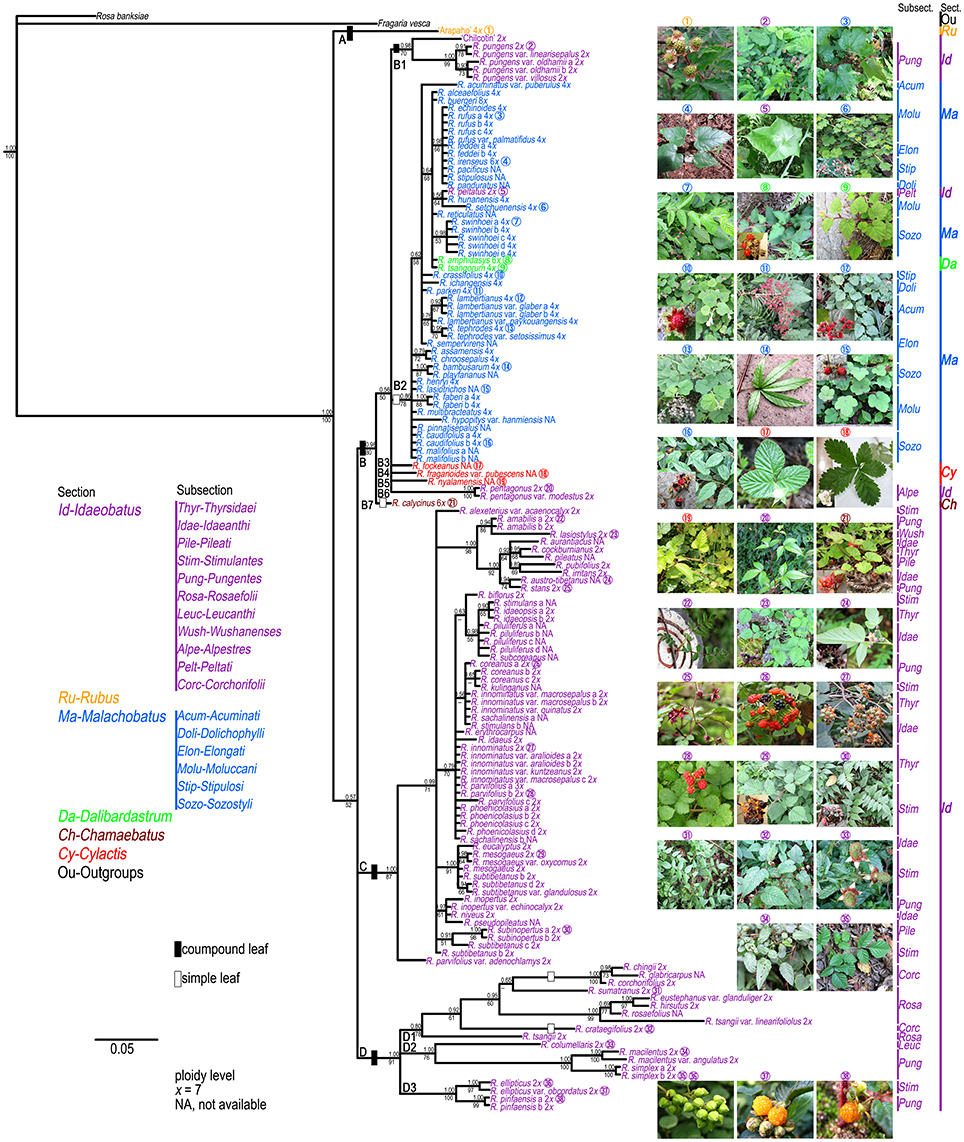
Figure 1. Bayesian Inference based phylogenetic tree through combined chloroplast datasets. Posterior probabilities from BI analysis and bootstrap values from ML analysis >50 are provided above and below the branches, respectively. Representative Rubus species with typical morphology for each section ( ) are shown in this figure.
) are shown in this figure.
The polyphyly of sect. Idaeobatus was strongly supported in the cpDNA tree (Figure 1). Clade C included all samples from subsections Thyrsidaei, Idaeanthi, Pileati, and Wushanenses, as well as most species from Stimulantes, and Pungentes of sect. Idaeobatus. The phylogenetic relationships among closely related species of clade C were not well-resolved. Clade D was composed of three subclades: a well-supported subclade D1 with samples from subsects. Rosaefolii and Corchorifolii; D2 with R. macilentus, R. macilentus var. angulatus, and R. simplex of subsect. Pungentes, as well as R. columellaris of subsect. Leucanthi; D3 with R. ellipticus and R. ellipticus var. obcordatus of subsect. Stimulantes, and two samples of R. pinfansis of subsect. Pungentes.
Phylogeny of Nuclear Markers
Via carefully checking the original tracing data, we detected few allelic like variations in ITS (3 out of 630 bp), GBSSI-2 (6 out of 491 bp), and PEPC datasets (2 out of 680 bp). Since each marker only gave single PCR product band, these variations seem more likely to be allelic sites, but might also be the consequences of PCR-generated mutations, which is inevitable with this methodological approach. All these sites that might interfere the accuracy of phylogeny were manually deleted as Brysting et al. (2011) did in their studies. We did not detect any other variations like insertion/deletion events of these three genes among the individuals of same species, even in the polyploids.
The ITS alignment of Rubus included 264 bp of ITS1, 164 bp of 5.8S rDNA, 212 bp of ITS2 (Table 2). Together with the outgroups, the final nrITS dataset contained 144 accessions and 645 aligned nucleotides, of which 206 (31.94%) were variable.
For the GBSSI-2 gene, we failed in getting the sequences for R. pileatus, R. kulinganus, R. simplex, R. acuminatus var. puberulus. Too much noise was found in the sequencing chromatograms (mostly after polyA segment) of these four taxa, which made it impossible to call out the base accurately, even after several runs of sequencing replication. While for other four samples from subsect. Corchorifolii, we only obtained partial sequence of GBSSI-1 using the same primers. We excluded these eight species in the following assay. Finally, the GBSSI-2 dataset included 136 accessions and 585 aligned nucleotides that contained 205 (35.04%) variable sites.
For PEPC, we failed to obtain sequences for 18 samples. Thirteen failed because of no PCR products, and other five samples failed due to too much noise in the tracing data. Among these, there are 16 diploids from subsects. Pungentes, Rosaefolii, Leucanthi, and Corchorifolii of sect. Idaeobatus, one tetraploid of sect. Malachobatus, and Rosa banksiae. The sequence for Fragaria vesca (XM011462481) was obtained from GenBank. Final PEPC dataset contained 127 accessions and 681 aligned nucleotides, which included 112 (16.45%) variable sites, much less than that of ITS and GBSSI-2 markers.
Based on the theoretic research by Wiens and Moen (2008), we treated those failure sequences as missing data for tree reconstruction when combined nuclear datasets. PHT indicated significant incongruence (P = 0.001) among the three nDNA markers. TIM2+I+G, TrN+I+G, and GTR+G models were then selected as best models for them (Table 2) when constructing partitioned phylogeny. Phylogenetic analyses of combined nDNA datasets using ML and BI methods showed similar topologies (Figure 2). Neither sect. Idaeobatus nor sect. Cylactis was recovered as monophyletic. In addition, those subclades, covering sects. Malachobatus plus Dalibardastrum and R. pelataus (B2), and Cylactis (B4–B6), as well as R. pentagonus (B3) identified in the cpDNA tree, were also supported here, while sect. Chamaebatus formed a separate clade C (Figure 2).
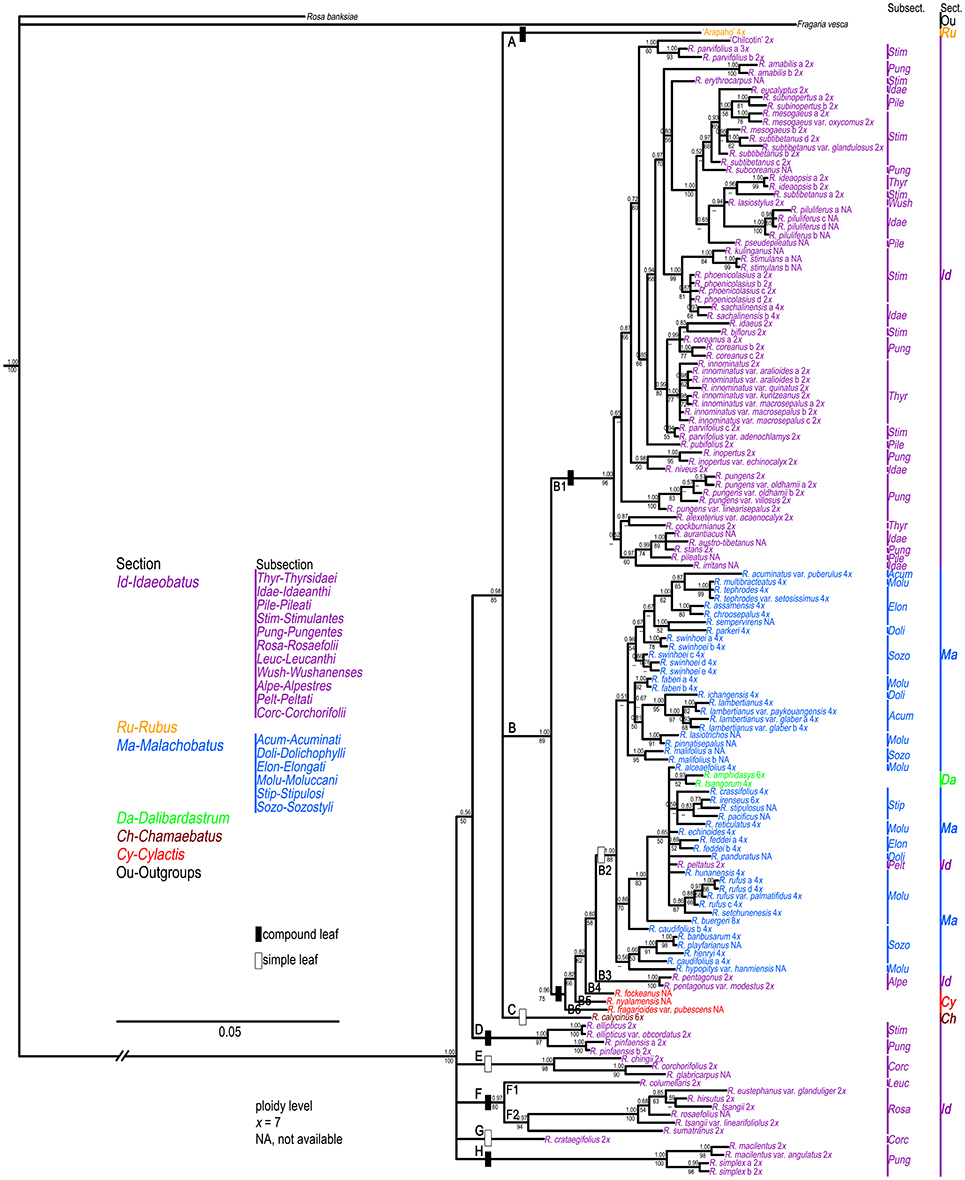
Figure 2. Bayesian Inference based phylogenetic tree through combined nuclear datasets. Double slashes on branches indicate branch lengths not in proportion. Posterior probabilities and bootstrap values >50 are provided above and below the branches, respectively.
Unlike cpDNA results, cultivar “Chilcotin,” and R. pungens and its derived varieties all clustered with most sect. Idaeobatus species (B1). These Rubus samples were nested in a well-supported (89% BS, 1.00 PP) clade B. The remaining five clades (D-H) included 19 samples of sect. Idaeobatus (Figure 2), which corresponded to clade D in the chloroplast tree (Figure 1). Clade D of R. ellipticus, R. ellipticus var. obcordatus, and R. pinfaensis was sister to the group (85% BS, 0.98 PP) composed of clades A, B and C. Subsections Rosaefolii (F2) and Leucanthi (F1) clustered together in clade F (94% BS, 0.97 PP), while four species of subsect. Corchorifolii formed clades E and G. Clade H was represented by four samples of subsect. Pungentes. Compared with cpDNA tree, the phylogeny within sects. Idaeobatus and Malachobatus was much better resolved in the nDNA tree.
Phylogenetic Network
Phylogenetic network of combined nuclear datasets (Figure 3) was similar to its corresponding phylogenetic tree (Figure 2), which revealed clearer backbones of genus Rubus. Section Idaeobatus was the most complicated section within Rubus, splitting into four distant clades in the network. Species of sect. Malachobatus formed a group, which contained R. tsangorum and R. amphidasys of sect. Dalibardastrum, as well as R. peltatus of sect. Idaeobatus. As predicted, three species of sect. Cylactis were split into different clades in nuclear network. Sections Rubus and Chamaebatus formed separate clades, respectively.
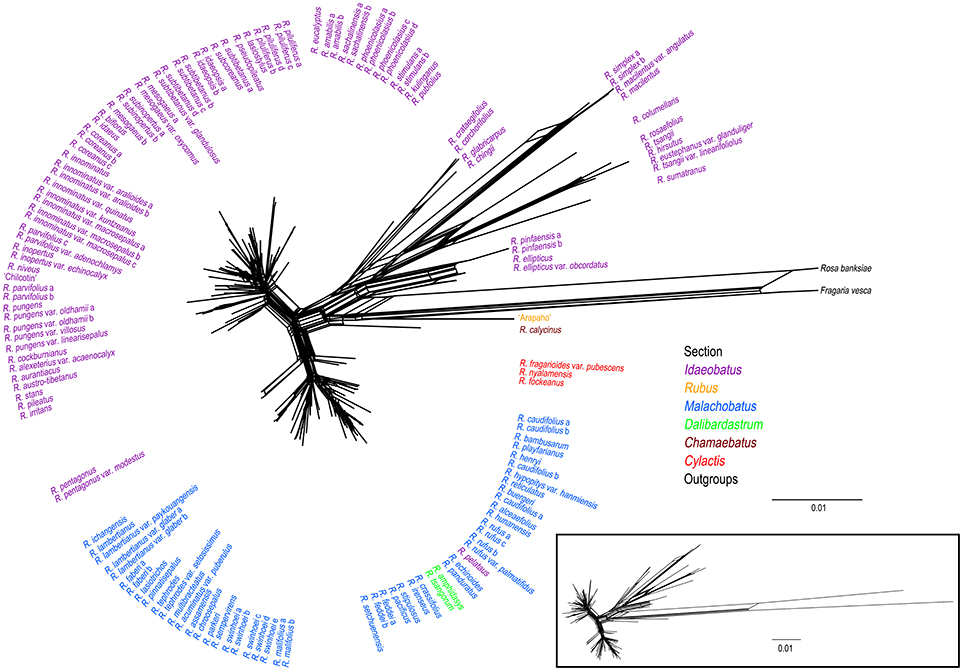
Figure 3. Phylogenetic network from NeighorNet analysis based on combined nuclear datasets. Different colors indicate sections in traditional taxonomy.
Discussion
Sequence Variation and Phylogenetic Incongruence
In this study, nrITS and GBSSI-2 datasets revealed much higher variation than cpDNA and PEPC regions within Rubus (Table 2), indicating their promising prospects in the genus. Our results provided insights into the evolutionary history of Rubus genus, although the phylogenetic relationships among closely related species were not fully resolved, especially within sects. Idaeobatus and Malachobatus. Low resolution in phylogeny can be caused by insufficient data, noisy sequences, rapid diversification, polyploidization, and reticulate evolution (Sochor et al., 2015; Spalink et al., 2016). The resolution was limited within sect. Malachobatus because of insufficient variation (Table 2). For sect. Idaeobatus, some reticulate evolution events were indicated in the nuclear network (Figure 3). When we combined the nuclear datasets (Figure 2), we obtained a much better resolved phylogeny of Rubus than each separate marker (Figures S2–4).
Phylogenetic incongruence (within subsects. Idaeanthi, Stimulantes, Pungentes, Moluccani, and Sozostyli etc.) was detected between the chloroplast and nuclear phylogenies (Figures 1, 2). For example, 19 species within sect. Idaeobatus formed a clade (D) in cpDNA tree (Figure 1), while scattered into five clades (D-H) in nDNA tree (Figure 2). Rubus calycinus of sect. Chamaebatus clustered with groups of sects. Malachobatus plus Dalibardastrum, Cylactis, as well as R. pentagonus in the cpDNA tree (Figure 1), whereas it formed a separate clade (Figure 2). Many studies suggested that convergent evolution, introgression following hybridization, incomplete lineage sorting, and horizontal gene transfer can cause phylogenetic incongruence (Cronn and Wendel, 2004; Linder and Rieseberg, 2004; Zou and Ge, 2008; Wang et al., 2016). Hybridization has occurred in the genus not only between closely related species but also between species from different sections (Bammi and Olmo, 1966; Iwatsubo and Naruhashi, 1991, 1992, 1993a,b, 1996, 1998; Thompson, 1997; Randell et al., 2004; Mimura et al., 2014). Some Rubus species were also proved to have undergone hybridization events along with asymmetric introgression (Mimura et al., 2014). Thus, incongruent phylogenetic relationships among our gene trees could be caused by frequent hybridization and genetic introgression.
Phylogeny of Chinese Rubus
All our phylogenetic results gave strong support to the monoplyly of genus Rubus, consistent with previous studies (Alice and Campbell, 1999; Morden et al., 2003; Yang and Pak, 2006). Based on morphological and chromosomal data, Lu (1983) suggested that evolution in Rubus proceeded from woody to herbaceous plants, and from compound to simple leaves. Alice and Campbell (1999) documented that primarily semi-herbaceous, simple-leaved species occupied basal positions in their trees, which was in disagreement with Lu's (1983) hypotheses. Sections Malachobatus, Dalibardastrum, Chamaebatus, and Cylactis might have common maternal diploid ancestors because they are all close to some sect. Idaeobatus species in the chloroplast tree (Figure 1). This also suggested that sect. Idaeobatus might be the most primitive group. Then species of different sections experienced distinct evolutionary history, along with various evolutionary rates. Specifically, all these datasets evolved three to six times within sect. Idaeobatus than that of sect. Malachobatus (Table 2).
Section Idaeobatus
Section Idaeobatus is one of the largest sections in Rubus, which contains predominantly diploid species (Thompson, 1997). These analyzed samples occurred in more than four clades within Chinese Idaeobatus inferred from different phylogenetic trees (Figures 1, 2). This demonstrated that sect. Idaeobatus was a polyphyletic group with at least four independent evolutionary routes. This is congruent with its morphological heterogeneity. Leaf type varies from pinnately compound in species of subsections 1 to 8 (Figure 1,  ), to palmately compound in subsect. Alpestres (Figure 1,
), to palmately compound in subsect. Alpestres (Figure 1,  ), to simple leaves in subsects. Peltati and Corchorifolii (Figure 1,
), to simple leaves in subsects. Peltati and Corchorifolii (Figure 1,  ,
,  ).
).
The first route was the “ancient” group, giving birth to species of subsects. Thyrsidaei, Idaeanthi, Pileati, Wushanenses, and a majority of Stimulantes and Pungentes species (Figures 1, 2). This group is composed of imparipinnately 3-11-foliolate species (Figure 1,  ), forming a mixed group without clear circumscription among subsections based on traditional taxonomy. For example, Rubus aurantiacus, R. austro-tibetanus, R. irritans, R. pileatus, and R. stans from three different subsections formed a well-supported clade (Figures 1, 2). They also share some similar morphological characters (Yü et al., 1985; Lu and Boufford, 2003). It has been documented that these Idaeobatus species exhibited both sexual and asexual reproduction, and some species could freely hybridize with each other and could produce fertile hybrids (Iwatsubo and Naruhashi, 1991, 1992, 1993a,b, 1996, 1998; Thompson and Zhao, 1993). This may partly cause their common morphology. Our molecular phylogenies also revealed the complex evolutionary history due to genetic introgression.
), forming a mixed group without clear circumscription among subsections based on traditional taxonomy. For example, Rubus aurantiacus, R. austro-tibetanus, R. irritans, R. pileatus, and R. stans from three different subsections formed a well-supported clade (Figures 1, 2). They also share some similar morphological characters (Yü et al., 1985; Lu and Boufford, 2003). It has been documented that these Idaeobatus species exhibited both sexual and asexual reproduction, and some species could freely hybridize with each other and could produce fertile hybrids (Iwatsubo and Naruhashi, 1991, 1992, 1993a,b, 1996, 1998; Thompson and Zhao, 1993). This may partly cause their common morphology. Our molecular phylogenies also revealed the complex evolutionary history due to genetic introgression.
As revealed from cpDNA tree (Figure 1), all samples from subsects. Rosaefolii, Leucanthi, Corchorifolii, and R. ellipticus and R. ellipticus var. obcordatus from subsect. Stimulantes, as well as R. pinfaensis, R. macilentus, R. macilentus var. angulatus, and R. simplex from subsect. Pungentes formed a well-supported clade. They not only show notable morphological differences with those species from other subsections, but also reveal high genetic divergence from them. It was evident that a second evolutionary pathway exists within sect. Idaeobatus. These species within the pathway are highly similar, with 3-7 leaflets per leaf (except subsect. Corchorifolii species with simple leaves, Figure 1,  ), with white flowers in cluster or in corymbs (Figure 1,
), with white flowers in cluster or in corymbs (Figure 1,  −
− ). However, these species formed five separate groups in the nDNA tree (Figure 2). The incongruence between chloroplast and nuclear phylogenies indicated that above mentioned species are probably involved in hybridization events. Additionally, subsection Pungentes was clearly demonstrated to be polyphyletic in this study. Taxonomic treatments of R. ellipticus, R. ellipticus var. obcordatus, and R. pinfaensis have been fraught with controversy for a long time (Van Thuan, 1968; Lauener, 1970; Yü et al., 1985; Li and He, 2001; Lu and Boufford, 2003; Wang et al., 2009). Rubus ellipticus and R. ellipticus var. obcordatus from subsect. Stimulantes, and R. pinfaensis from subsect. Pungentes formed a well-supported clade, which revealed obvious divergence with any other species from the two subsections by both cpDNA and nDNA trees (Figures 1, 2). Therefore, it seems more reasonable to assign them into another new subsection within sect. Idaeobatus.
). However, these species formed five separate groups in the nDNA tree (Figure 2). The incongruence between chloroplast and nuclear phylogenies indicated that above mentioned species are probably involved in hybridization events. Additionally, subsection Pungentes was clearly demonstrated to be polyphyletic in this study. Taxonomic treatments of R. ellipticus, R. ellipticus var. obcordatus, and R. pinfaensis have been fraught with controversy for a long time (Van Thuan, 1968; Lauener, 1970; Yü et al., 1985; Li and He, 2001; Lu and Boufford, 2003; Wang et al., 2009). Rubus ellipticus and R. ellipticus var. obcordatus from subsect. Stimulantes, and R. pinfaensis from subsect. Pungentes formed a well-supported clade, which revealed obvious divergence with any other species from the two subsections by both cpDNA and nDNA trees (Figures 1, 2). Therefore, it seems more reasonable to assign them into another new subsection within sect. Idaeobatus.
The third route corresponded to the ancestor of subsect. Alpestres (R. pentagonus, palmately 3-foliolate leaves, 2n = 2x = 14) (Figure 1,  ), sister to the clade of sect. Malachobatus species (Figures 1, 2). As a result, Rubus pentagonus and polyploid sect. Malachobatus might have a common origin from ancient diploid ancestors.
), sister to the clade of sect. Malachobatus species (Figures 1, 2). As a result, Rubus pentagonus and polyploid sect. Malachobatus might have a common origin from ancient diploid ancestors.
The fourth pathway produced the subsect. Peltati (R. peltatus, simple leaves, 2n = 2x = 14) (Figure 1,  ), always forming a clade with some species from subsect. Moluccani of sect. Malachobatus with moderate support. Rubus peltatus had common maternal ancestors with R. setchuenensis and R. hunanensis inferred from cpDNA tree (Figure 1). The species also shares some similar morphology, such as simple leaves and ovate stipules (Figure 1,
), always forming a clade with some species from subsect. Moluccani of sect. Malachobatus with moderate support. Rubus peltatus had common maternal ancestors with R. setchuenensis and R. hunanensis inferred from cpDNA tree (Figure 1). The species also shares some similar morphology, such as simple leaves and ovate stipules (Figure 1,  ), whereas cytogenetic research showed that it is diploid (Thompson and Zhao, 1993). Three suspected diploid species, Rubus fulvus, R. micropetalus, and R. paniculatus, have been reported to occur in this predominately polyploid section Malachobatus (Malik, 1965; Mehra and Dhawan, 1966; Subramanian, 1987). Should R. peltatus be moved from subsect. Peltati of sect. Idaeobatus into subsect. Moluccani of sect. Malachobatus? It is needed to be taken into consideration more cautiously.
), whereas cytogenetic research showed that it is diploid (Thompson and Zhao, 1993). Three suspected diploid species, Rubus fulvus, R. micropetalus, and R. paniculatus, have been reported to occur in this predominately polyploid section Malachobatus (Malik, 1965; Mehra and Dhawan, 1966; Subramanian, 1987). Should R. peltatus be moved from subsect. Peltati of sect. Idaeobatus into subsect. Moluccani of sect. Malachobatus? It is needed to be taken into consideration more cautiously.
Section Malachobatus
Section Malachobatus, as one of the largest groups in the genus, represents a polyploid complex with tetra-, hexa-, octo- or tetra-decaploids (Thompson, 1997; Naruhashi et al., 2002; Wang et al., 2008). Our samples of this section are all simple-leaved species (Figure 1,  ). Low variation and limited resolution were obvious within sect. Malachobaus (Table 2, Figure 1). A cytogenetic study showed species belonging to six subsections of sect. Malachobatus exhibited uniform intra-subsectional karyotypic features (Wang et al., 2008). Low resolution and uniform cytology indicated relatively narrow genetic background among sect. Malachobatus species. The taxonomy of subsect. Moluccani within Malachobatus was problematic (Nybom, 1986). Sixteen samples from this subsection scattered into several clusters in the phylogenetic trees. It seems that the result supported the taxonomy within Moluccani by Focke (1910) rather than Yü et al. (1985). Similar results were observed among subsects. Elongati and Sozostyli.
). Low variation and limited resolution were obvious within sect. Malachobaus (Table 2, Figure 1). A cytogenetic study showed species belonging to six subsections of sect. Malachobatus exhibited uniform intra-subsectional karyotypic features (Wang et al., 2008). Low resolution and uniform cytology indicated relatively narrow genetic background among sect. Malachobatus species. The taxonomy of subsect. Moluccani within Malachobatus was problematic (Nybom, 1986). Sixteen samples from this subsection scattered into several clusters in the phylogenetic trees. It seems that the result supported the taxonomy within Moluccani by Focke (1910) rather than Yü et al. (1985). Similar results were observed among subsects. Elongati and Sozostyli.
All samples of sect. Malachobatus clustered together in our chloroplast (Figure 1) and nuclear analyses (Figure 2), supporting its monophyly. Interestingly, this group was sister to R. pentagonus of sect. Idaeobatus and three sect. Cylactis members in the cpDNA tree (Figure 1), whereas it formed a polytomy composed of these four species in the nDNA tree (Figures 2). It has been documented that R. pentagonus formed a clade with R. calophyllus and R. lineatus from subsect. Lineati of sect. Malachobatus (Alice et al., 2008). They also share some morphological similarities, such as unarmed stems, abaxial leaf surfaces, and palmately 3- or 5-foliate leaves (Lu and Boufford, 2003). Rubus pentagonus is diploid (2n = 2x = 14) (Thompson, 1997), while ploidy levels for the three sect. Cylactis species have not been reported yet. Moreover, polyploid sect. Malachobatus was demonstrated to be of allopolyploid origin (Bammi, 1965; Wang et al., 2015). Therefore, we suggest that some sect. Malachobatus polyploids probably originate from common ancestors, via polyploidization of hybrids between R. pentagonus and sect. Cylactis species.
Section Dalibardastrum
Section Dalibardastrum, represented by R. tsangorum and R. amphidasys, was nested within sect. Malachobatus (Figures 1, 2). This was consistent with a previous study by Alice et al. (2008). The two species share some morphological similarities, such as simple leaves and weak, bristly, prostrate stems (Figure 1,  ). Rubus tsangorum was reported as a tetraploid (2n = 4x = 28), whereas R. amphidasys as a hexaploid (2n = 6x = 42) (Thompson, 1997). Therefore, it is likely that the two polyploids have at least one parental lineage from sect. Malachobatus. Their parental ancestors might be from subsects. Moluccani and Stipulosi based on phylogenetic trees (Figures 1, 2).
). Rubus tsangorum was reported as a tetraploid (2n = 4x = 28), whereas R. amphidasys as a hexaploid (2n = 6x = 42) (Thompson, 1997). Therefore, it is likely that the two polyploids have at least one parental lineage from sect. Malachobatus. Their parental ancestors might be from subsects. Moluccani and Stipulosi based on phylogenetic trees (Figures 1, 2).
Section Chamaebatus
In this study, we have collected just one hexaploid species (R. calycinus) within sect. Chamaebatus (Thompson, 1997), which has simple leaves and sparse prickles or unarmed stems (Figure 1,  ). Monophyly of sect. Chamaebatus has been demonstrated by Alice and Campbell (1999). Rubus calycinus was sister to a large group of sects. Malachobatus plus Dalibardastrum, and Cylactis, as well as R. pentagenus of sect. Idaeobatus in the chloroplast tree (Figure 1), while it formed a separate clade in the nDNA tree (Figure 2). A possible explanation was that the maternal ancestors of sect. Idaeobatus contributed to the formation of R. calycinus.
). Monophyly of sect. Chamaebatus has been demonstrated by Alice and Campbell (1999). Rubus calycinus was sister to a large group of sects. Malachobatus plus Dalibardastrum, and Cylactis, as well as R. pentagenus of sect. Idaeobatus in the chloroplast tree (Figure 1), while it formed a separate clade in the nDNA tree (Figure 2). A possible explanation was that the maternal ancestors of sect. Idaeobatus contributed to the formation of R. calycinus.
Section Cylactis
Three examined species of sect. Cylactis formed a clearly polyphyletic group (Figures 1, 2). This was also in agreement with its morphological heterogeneity, confirming their different origin patterns by cytological study. Leaf type varies from ternate in R. fockeanus (Figure 1,  ), and R. nyalamensis (Figure 1,
), and R. nyalamensis (Figure 1,  ), to palmately compound in R. fragarioides (Figure 1,
), to palmately compound in R. fragarioides (Figure 1,  ), and R. clivicola. Stem armature ranges from prickly in R. fockeanus, R. nyalamensis, to unarmed in R. fragarioides. There are three ploidy levels within this section, diploid (R. pedatus), tetraploid (R. saxatilis), and mixoploid (R. arcticus, 2x and 3x; R. humulifolius, 2x and 4x) (Thompson, 1997). Rubus pentagonus was also demonstrated to be closely related to sect. Cylactis (Figures 1, 2). Further research with more samples is needed to illustrate the phylogenetic relationships within sect. Cylactis.
), and R. clivicola. Stem armature ranges from prickly in R. fockeanus, R. nyalamensis, to unarmed in R. fragarioides. There are three ploidy levels within this section, diploid (R. pedatus), tetraploid (R. saxatilis), and mixoploid (R. arcticus, 2x and 3x; R. humulifolius, 2x and 4x) (Thompson, 1997). Rubus pentagonus was also demonstrated to be closely related to sect. Cylactis (Figures 1, 2). Further research with more samples is needed to illustrate the phylogenetic relationships within sect. Cylactis.
Origin and Evolution of Polyploids within Rubus
Polyploidy has long been considered a major driving force in plant evolution (Soltis and Soltis, 1999). Genus Rubus has a large proportion (60%) of polyploidy (Vamosi and Dickinson, 2006), often observed in predominately woody clades (sect. Malachobatus). Soltis and Soltis (2009) pointed out that formation of a new species is more likely via allopolyploidy rather than homoploid hybridization. The polyploids within sects. Malachobatus and Chamaemorus have been demonstrated to be of allopolyploidy origin by Bammi (1965), Michael (2006), and Wang et al. (2015). As predicted, many speciation events in Rubus are associated with a change in ploidy levels. Polyploidy in Rubus has arisen multiple times, found in subgenera Rubus, Lampobatus, and Malachobatus (Alice and Campbell, 1999). Multiple origins of polyploid species might also be the main contributor of the morphological complexity in Rubus (Rozanova, 1938, see Mavrodiev and Soltis, 2001). Moreover, recurrent hybridization events have been documented occurring over relatively short time spans and geographic distances (Soltis and Soltis, 1999). Ploidy in Rubus ranges from diploid (2n = 2x = 14) to dodecaploid (2n = 12x = 84), and the most frequent ploidy level is tetraploid (2n = 4x = 28) (Thompson, 1997). Polyploids show abundant morphological variation, while the genetic background was evident to be narrow in this study. We propose that sect. Malachobatus probably experience explosion of species from diploid ancestors to form polyploids within a short time, of which the allotetraploids are the most stable type. The production and fusion of unreduced reproductive cells is likely to be the most predominant pathway leading to allotetraploids (Wang et al., 2010, 2015; Satter et al., 2016).
Conclusions and Perspectives
This study presented phylogenies of genus Rubus based on three different kinds of genetic markers (chloroplast DNA, nuclear ribosomal DNA, and single copy nuclear DNA) with a comprehensive taxon sampling. Chloroplast and nuclear markers provided useful information for Rubus phylogenies, but general low resolution among closely related species because of insufficient variation and complex evolutionary history. The combined nuclear tree provided the most comprehensive knowledge of the reticulate evolution of Rubus. Pervasive incongruence was observed between chloroplast and nuclear trees, which might be due to frequent hybridization and genetic introgression. However, we confirmed that both sects. Idaeobatus and Cylactis are clearly polyphyletic with species present in several different clades. Section Malachobatus was monophyletic, probably originated via polyploidization of hybrids between R. pentagenus and sect. Cylactis species. Two sect. Dalibardastrum species probably have at least one parental lineage from sect. Malachobatus.
The molecular phylogenies presented here suggest the need for moderate taxonomic revisions of Rubus using modern approaches. Further studies including faster evolving low copy nuclear genes are indispensable and effective to better explore the complex evolutionary history of Rubus, especially polyploids. Recently, we have found another low copy nuclear gene, GBSSI-1, which showed only one copy in diploids, while exhibited two or three copies in polyploids (unpublished data). It will probably make it possible to obtain a more robust phylogeny, and even illustrate the origin and evolution of the polyploids.
Author Contributions
YW, XW, and HT designed the research. YW carried out the experiments. YW, QC, and TC performed the data analyses. LL contributed to samples collection. YW wrote the manuscript. QC and XW made the revision of the manuscript. All authors approved the final revision to be published.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors sincerely thank Li Zhang and Yin Liu for their help in collecting samples. We are also grateful to Hongyan Yang for her help in experiments, and to Dr. Nian Wang (Shandong Agricultural University) for some useful suggestion and English editing on the manuscript. This study was supported by the National Natural Science Foundation of China (NSFC Grant Nos. 31272134, 31460206).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00968
Figure S1. Investigation sites of Chinese Rubus used for this study.
Figure S2. Bayesian Inference based phylogenetic tree through nrITS dataset. Double slashes on branches indicate branch lengths not in proportion. Posterior probabilities and bootstrap values >50 are provided above and below the branches, respectively.
Figure S3. Bayesian Inference based phylogenetic tree through GBSSI-2 dataset. Double slashes on branches indicate branch lengths not in proportion. Posterior probabilities and bootstrap values >50 are provided above and below the branches, respectively.
Figure S4. Bayesian Inference based phylogenetic tree through PEPC dataset. Posterior probabilities and bootstrap values >50 are provided above and below the branches, respectively..
Table S1. List of the studied Rubus taxa, their taxonomic position, herbarium voucher information, ploidy level, locality, and the GenBank accession numbers for this study
References
Akaike, H. (1974). A new look at the statistical model identification. IEEE Trans. Automat. Control 19, 716–723. doi: 10.1109/TAC.1974.1100705
Alice, L. A., and Campbell, C. S. (1999). Phylogeny of Rubus (Rosaceae) based on nuclear ribosomal DNA internal transcribed spacer region sequences. Am. J. Bot. 86, 81–97. doi: 10.2307/2656957
Alice, L. A., Dodson, T. M., and Sutherland, B. L. (2008). Diversity and relationships of Bhutanese Rubus (Rosaceae). Acta Hortic. 777, 63–69. doi: 10.17660/ActaHortic.2008.777.5
Alice, L. A., Eriksson, T., Eriksen, B., and Campbell, C. S. (2001). Hybridization and gene flow between distantly related species of Rubus (Rosaceae): evidence from nuclear ribosomal DNA internal transcribed spacer region sequences. Syst. Bot. 26, 769–778. doi: 10.1043/0363-6445-26.4.769
Amsellem, Noyer, J. L., and Hossaert-McKey, M. (2001). Evidence for a switch in the reproductive biology of Rubus alceifolius (Rosaceae) towards apomixis, between its native range and its area of introduction. Am. J. Bot. 88, 2243–2251. doi: 10.2307/3558386
Bammi, R. K. (1965). ‘Complement fractionation’ in a natural hybrid between Rubus procerus Muell. and R. laciniatus Willd. Nature 208, 608. doi: 10.1038/208608a0
Bammi, R. K., and Olmo, H. P. (1966). Cytogenetics of Rubus. V. Natural hybridization between R. procerus P. J. Muell. and R. laciniatus Willd. Evolution 20, 617–633. doi: 10.2307/2406596
Brysting, A. K., Mathiesen, C., and Marcussen, T. (2011). Challenges in polyploid phylogenetic reconstruction: a case story from the arctic-alpine Cerastium alpinum complex. Taxon, 60, 333–347. Available online at: http://www.jstor.org/stable/41317132
Chernomor, O. A., van Haeseler, A., and Minh, B. Q. (2016). Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. doi: 10.1093/sysbio/syw037. [Epub ahead of print].
Cronn, R., and Wendel, J. F. (2004). Cryptic trysts, genomic mergers, and plant speciation. New Phytol. 161, 133–142. doi: 10.1111/j.1469-8137.2004.00947.x
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). JModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772. doi: 10.1038/nmeth.2109
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl. Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Evans, R. C., Alice, L. A., Campbell, C. S., Kellogg, E. A., and Dickinsona, T. A. (2000). The granule-bound starch synthase (GBSSI) gene in the Rosaceae: multiple loci and phylogenetic utility. Mol. Phylogenet. Evol. 17, 388–400. doi: 10.1006/mpev.2000.0828
Focke, W. O. (1910). Species Ruborum. Monographiae Generis Rubi Prodromus part I. New York, NY: Stuttgart,E. Schweizerbart. 1–120.
Focke, W. O. (1911). Species Ruborum. Monographiae Generis Rubi Prodromus part II. New York, NY: Stuttgart,E. Schweizerbart. 121–223.
Focke, W. O. (1914). Species Ruborum. Monographiae Generis Rubi Prodromus part III. New York, NY: Stuttgart,E. Schweizerbart. 224–498.
Hamilton, M. B. (1999). Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Mol. Ecol. 8, 513–525. doi: 10.1046/j.1365-294x.1999.00510.x
Hasebe, M., Ito, M., Kofuji, R., Ueda, K., and Iwatsuki, K. (1993). Phylogenetic relationships of ferns deduced from rbcL gene sequence. J. Mol. Evol. 37, 476–482. doi: 10.1007/BF00160428
Howarth, D. G., Gardner, D. E., and Morden, C. W. (1997). Phylogeny of Rubus Subgenus Idaeobatus (Rosaeae) and its implications toward colonization of the Hawaiian Islands. Syst. Bot. 22, 433–441. doi: 10.2307/2419819
Huson, D. H., and Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267. doi: 10.1093/molbev/msj030
Imanishi, H., Nakahara, K., and Tsuyzaki, H. (2008). Genetic relationships among native and introduced Rubus species in Japan based on rbcL sequence. Acta Hortic. 769, 195–199. doi: 10.17660/ActaHortic.2008.769.26
Iwatsubo, Y., and Naruhashi, N. (1991). Karyomorphological and cytogenetical studies of Rubus parvifolius, R. coreanus and R. × hiraseanus (Rosaceae). Cytologia 56, 151–156. doi: 10.1508/cytologia.56.151
Iwatsubo, Y., and Naruhashi, N. (1992). Cytotaxonomical studies of Rubus (Rosaceae) I. Chromosome numbers of 20 species and 2 natural hybrids. J. Jpn. Bot. 67, 270–275.
Iwatsubo, Y., and Naruhashi, N. (1993a). Cytotaxonomical studies of Rubus (Rosaceae) II. Chromosome numbers of 21 species and 6 natural hybrids. J. Jpn. Bot. 68 (3), 159-165.
Iwatsubo, Y., and Naruhashi, N. (1993b). Cytogenetical study of Rubus × tawadanus (Rosaceae). Cytologia 58, 217–221. doi: 10.1508/cytologia.58.217
Iwatsubo, Y., and Naruhashi, N. (1996). A comparative chromosome study of Rubus × nikaii, R. parvifolius and R. phoenicolasius (Rosaceae). J. Jpn. Bot. 71, 333–337.
Iwatsubo, Y., and Naruhashi, N. (1998). Cytogenetic studies of natural hybrid, Rubus × hiraseanus, and artificial hybrid between R. coreanus and R. parvifolius (Rosaceae). Cytologia 63, 235–238. doi: 10.1508/cytologia.63.235
Lauener, L. A. (1970). Catalogue of the names published by Hector Leveille. VI. Notes Roy. Bot. Gard. Edinburgh 30, 239–294.
Li, W. L., and He, S. A. (2001). Taxonomic revision on several taxa in the genus Rubus (Rosaceae). Bull. Bot. Res. 21, 346–349. doi: 10.3969/j.issn.1673-5102.2001.03.007
Linder, C. R., and Rieseberg, L. H. (2004). Reconstructing patterns of reticulated evolution in plants. Am. J. Bot. 91, 1700–1708. doi: 10.3732/ajb.91.10.1700
Lo, E. Y. Y., Stefanovic, S., Christensen, K. I., and Dickinson, T. A. (2009). Evidence for genetic association between East Asian and western North American Crataegus, L. (Rosaceae) and rapid divergence of the eastern North American lineages based on multiple DNA sequences. Mol. Phylogenet. Evol. 51, 157–168. doi: 10.1016/j.ympev.2009.01.018
Lu, L. D., and Boufford, D. E. (2003). Rubus Linnaeus, Sp. P1. 1: 492. 1753. Flora China 9, 195–285.
Malik, C. P. (1965). Cytology of some Indian species of Rosaceae. Caryologia 18, 139–149. doi: 10.1080/00087114.1965.10796161
Mavrodiev, E. V., and Soltis, D. E. (2001). Recurring polyploid formation: an early account from the Russian literature. Taxon 50, 469–474. doi: 10.2307/1223892
Mehra, P. N., and Dhawan, H. (1966). Cytological investigations in the N. W. Himalayan Rosaceae. Proc. Indian Sci. Congr. Assoc. 53, 276–277.
Meng, R. G., and Finn, C. (2002). Determining ploidy level and nuclear DNA content in Rubus by flow cytometry. J. Am. Soc. Hortic. Sci. 127, 767–775.
Michael, K. (2006). Clarification of Basal Relationships in Rubus (Rosaceae) and the Origin of Rubus chamaemorus. Bowling Green: Western Kentucky University.
Mimura, M., Mishima, M., Lascoux, M., and Yahara, T. (2014). Range shift and introgression of the rear and leading populations in two ecologically distinct Rubus species. BMC Evol. Biol. 14:209. doi: 10.1186/s12862-014-0209-9
Morden, C. W., Gardner, D. E., and Weniger, D. A. (2003). Phylogeny and biogeography of Pacific Rubus subgenus Idaeobatus (Rosaceae) species: investigating the origin of the endemic Hawaiian raspberry R. macraei. Pac. Sci. 57, 181–197. doi: 10.1353/psc.2003.0018
Morgan, D. R., Soltis, D. E., and Robertson, K. R. (1994). Systematic and evolutionary implications of rbcL sequence variation in Rosaceae. Am. J. Bot. 81, 890–903. doi: 10.2307/2445770
Naruhashi, N., Iwatsubo, Y., and Peng, C. I. (2002). Chromosome numbers in Rubus (Rosaceae) of Taiwan. Bot. Bull. Acad. Sinica 43, 193–201.
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nybom, H. (1986). Chromosome numbers and reproduction in Rubus subgen. Malachobatus. Plant Syst. Evol. 152, 211–218. doi: 10.1007/BF00989428
Nybom, H., and Schaal, B. A. (1990). DNA fingerprints reveal genotypic distributions in natural populations of blackberries and raspberries (Rubus, Rosaceae). Am. J. Bot. 77, 883–888. doi: 10.2307/2444504
Rambaut, A., and Drummond, A. J. (2009). Tracer: MCMC Trace Analysis Tool. Version 1.5. Oxford: University of Oxford. Available online at: http://tree.bio.ed.ac.uk/software/tracer/
Randell, R. S., Howarth, D. G., and Morden, C. W. (2004). Genetic analysis of natural hybrids between endemic and alien Rubus (Rosaceae) species in Hawai'i. Conserv. Genet. 5, 217–230. doi: 10.1023/B:COGE.0000030006.30410.5d
Ronquist, F., Teslenko, M., Van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rousseau-Gueutin, M., Gaston, A., Aïnouche, A., Aïnouche, M. L., and Olbricht, K. (2009). Tracking the evolutionary history of polyploidy in Fragaria, L. (strawberry): new insights from phylogenetic analyses of low copy nuclear genes. Mol. Phylogenet. Evol. 51, 515–530. doi: 10.1016/j.ympev.2008.12.024
Rozanova, M. A. (1938). About the polymorphic origin of species. Rep. Soviet (U.S.S.R.) Acad. Sci. 18, 681–684.
Satter, M. C., Carvalho, C. R., and Clarindo, W. R. (2016). The polyploidy and its key role in plant breeding. Planta 243, 281–296. doi: 10.1007/s00425-015-2450-x
Sochor, M., Vašut, R. J., Sharbel, T. F., and Trávnícek, B. (2015). How just a few makes a lot: speciation via reticulation and apomixis on example of European brambles (Rubus subgen. Rubus, Rosaceae). Mol. Phylogenet. Evol. 89, 13–27. doi: 10.1016/j.ympev.2015.04.007
Soltis, P. S., and Soltis, D. E. (1999). Polyloidy: recurrent formation and genome evolution. Trends Eco. Evol. 14, 348–352. doi: 10.1016/S0169-5347(99)01638-9
Soltis, P. S., and Soltis, D. E. (2009). The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 60, 561–588. doi: 10.1146/annurev.arplant.043008.092039
Spalink, D., Drew, B. T., Pace, M. C., Zaborsky, J. G., Li, P., Cameron, K. M., et al. (2016). Evolution of geographical place and niche space: patterns of diversification in the North American sedge (Cyperaceae) flora. Mol. Phylogenet. Evol. 95, 183–195. doi: 10.1016/j.ympev.2015.09.028
Subramanian, D. (1987). Cytotaxonomic studies of South Indian Rosaceae. Cytologia 52, 395–403. doi: 10.1508/cytologia.52.395
Swofford, D. (2002). PAUP 4.0 b10: Phylogenetic Analysis using Parsimony. Sunderland, MA: Sinauer Associates.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Thompson, M. M. (1997). Survey of chromosome numbers in Rubus (Rosaceae: Rosoideae). Ann. Miss. Bot. Gard. 84, 128–164. doi: 10.2307/2399958
Thompson, M. M., and Zhao, C. M. (1993). Chromosome numbers of Rubus species in Southwest China. Acta Hortic. 352, 493–502. doi: 10.17660/ActaHortic.1993.352.71
Vamosi, J. C., and Dickinson, T. A. (2006). Polyploidy and diversification: a phylogenetic investigation in Rosaceae. Int. J. Plant Sci. 167, 349–358. doi: 10.1086/499251
Van Thuan, N. (1968). Flore du Cambodge, du Laos, et du Vietnam. Fascicule 7: Rosaceae II (Rubus). Paris: Museum National d'Histoire Naturelle.
Wang, N., McAllister, H. A., Bartlett, P. R., and Buggs, R. J. A. (2016). Molecular phylogeny and genome size evolution of the genus Betula (Betulaceae). Ann. Bot. 117, 1023–1035. doi: 10.1093/aob/mcw048
Wang, X. R., Liu, Y., Zhong, B. F., Dong, X. L., Chen, Q., Xia, W. F., et al. (2010). Cytological and RAPD data revealed genetic relationships among nine selected populations of the wild bramble species, Rubus parvifolius and R. coreanus (Rosaceae). Genet. Resour. Crop Evol. 57, 431–441. doi: 10.1007/s10722-009-9483-6
Wang, X. R., Tang, H. R., Duan, J., and Li, L. (2008). A comparative study on karyotypes of 28 taxa in Rubus sect. Idaeobatus and sect. Malachobatus (Rosaceae) from China. J. Syst. Evol. 46, 505–515. doi: 10.3724/SP.J.1002.2008.07079
Wang, X. R., Tang, H. R., Zhang, H. W., Zhong, B. F., Xia, W. F., and Liu, Y. (2009). Karyotypic, palynological, and RAPD study on 12 taxa from two subsections of section Idaeobatus in Rubus, L., and taxonomic treatment of R. ellipticus, R. pinfaensis, and R. ellipticus var. obcordatus. Plant Syst. Evol. 283, 9–18. doi: 10.1007/s00606-009-0190-8
Wang, Y., Wang, X. R., Chen, Q., Zhang, L., Tang, H. R., Luo, Y., et al. (2015). Phylogenetic insight into subgenera Idaeobatus and Malachobatus inferring from ISH analysis. Mol. Cytogenet. 8, 11. doi: 10.1186/s13039-015-0114-y
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky and T. J. White (New York, NY: Academic Press), 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Wiens, J. J., and Moen, D. S. (2008). Missing data and the accuracy of Bayesian phylogenetics. J. Syst. Evol. 46, 307–324. doi: 10.1080/10635150390218330
Yang, J. Y., and Pak, J. H. (2006). Phylogeny of Korean Rubus (Rosaceae) based on ITS (nrDNA) and trnL/F intergenic region (cpDNA). J. Plant Biol. 49, 44–54. doi: 10.1007/BF.03030787
Yü, D. J., Lu, L. D., Gu, C. Z., Guan, K. J., and Li, C. L. (1985). “Rubus, Rosaceae,” in Flora Reipublicae Popularis Sinicae, Vol. 37,” eds Chinese Academy of Sciences Editorial Board (Beijing: Science Press), 10–218.
Zhang, L., Wang, Y., Chen, Q., Luo, Y., Zhang, Y., Tang, H. R., et al. (2015). Phylogenetic utility of Chinese Rubus (Rosaceae) based on ndhF sequences. Acta Hortic. Sinica 42, 19–30. doi: 10.16420/j.issn.0513-353x.2014-0752
Zhou, Y. Q. (2005). “DNA extraction and detection in plants,” in Application on DNA Molecular Markers Technology in Plant Study, ed Y. Q. Zhou (Beijing: Chemical Industry Press), 9–34.
Zimmer, E. A., and Wen, J. (2013). Reprint of: using nuclear gene data for plant phylogenetics: progress and prospects. Mol. Phylogenet. Evol. 66, 539–550. doi: 10.1016/j.ympev.2013.01.005
Keywords: Chinese Rubus, phylogeny, section Idaeobatus, section Malachobatus, hybridization, polyploidy
Citation: Wang Y, Chen Q, Chen T, Tang H, Liu L and Wang X (2016) Phylogenetic Insights into Chinese Rubus (Rosaceae) from Multiple Chloroplast and Nuclear DNAs. Front. Plant Sci. 7:968. doi: 10.3389/fpls.2016.00968
Received: 06 April 2016; Accepted: 16 June 2016;
Published: 29 June 2016.
Edited by:
Federico Luebert, Universität Bonn, GermanyReviewed by:
Nakatada Wachi, JT Biohistory Research Hall, JapanXinfen Gao, Chengdu Institute of Biology, China
Copyright © 2016 Wang, Chen, Chen, Tang, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaorong Wang, wangxrtj@163.com
 Yan Wang
Yan Wang Qing Chen1
Qing Chen1 Tao Chen
Tao Chen Xiaorong Wang
Xiaorong Wang