- 1U.S. Horticultural Research Laboratory, Agricultural Research Service, United States Department of Agriculture, Fort Pierce, FL, USA
- 2Institute of Food and Agricultural Sciences, Department of Plant Pathology, Indian River Research and Education Center, University of Florida, Fort Pierce, FL, USA
Candidatus Liberibacter asiaticus “Las” is a phloem-limited bacterial plant pathogen, and the most prevalent species of Liberibacter associated with citrus huanglongbing (HLB), a devastating disease of citrus worldwide. Although, the complete sequence of the Las genome provides the basis for studying functional genomics of Las and molecular mechanisms of Las-plant interactions, the functional characterization of Las effectors remains a slow process since remains to be cultured. Like other plant pathogens, Las may deliver effector proteins into host cells and modulate a variety of host cellular functions for their infection progression. In this study, we identified 16 putative Las effectors via bioinformatics, and transiently expressed them in Nicotiana benthamiana. Diverse subcellular localization with different shapes and aggregation patterns of the effector candidates were revealed by UV- microscopy after transient expression in leaf tissue. Intriguingly, one of the 16 candidates, Las5315mp (mature protein), was localized in the chloroplast and induced cell death at 3 days post inoculation (dpi) in N. benthamiana. Moreover, Las5315mp induced strong callose deposition in plant cells. This study provides new insights into the localizations and potential roles of these Las effectors in planta.
Introduction
Citrus huanglongbing (HLB), also known as citrus greening, is a devastating disease with high economical costs to the worldwide citrus industry (Hodges and Spreen, 2012). The disease is caused by three species of alpha-proteobacterium, “Candidatus Liberibacter asiaticus (Las),” “Ca. L. africanus,” and “Ca. L. americanus.” Las, the most widespread pathogen, is vectored by the Asian citrus psyllid (ACP) Diaphorina citri Kuwayama (Hemiptera: Psyllidae) (Jagoueix et al., 1996; Garnier et al., 2000; Bove, 2006; Hall et al., 2013). Las attacks all species and hybrids in the Citrus genus, and upon infection, Las resides in the phloem of the host, which causes a systemic disease and can eventually result in the death of the tree (Halbert and Manjunath, 2004; Bove, 2006; Gottwald, 2010). Once a tree is infected, it is extremely difficult to cure, and currently there is no adequate strategy for HLB management.
Host-pathogen relationships encompass a myriad of protein-protein interactions that include not only pathogen detection and removal by the host but also mechanisms to avoid such processes by the pathogen. Understanding this interplay would open the door to more successful HLB control methods, however, the corresponding mechanisms are currently largely unknown. Plants, including citrus, are able to detect conserved microbial molecular signatures termed pathogen-associated molecular patterns (PAMP), and initiate a process known as PAMP-triggered immunity (PTI) (Boller and Felix, 2009; Segonzac and Zipfel, 2011). Pathogens, in turn, can suppress the PTI response by deploying diverse effector proteins that modulate various host cellular functions in order to promote bacterial colonization and replication (Chisholm et al., 2006; Hogenhout et al., 2009; Dodds and Rathjen, 2010). Effectors that are deployed to suppress host defenses may be recognized by plant disease resistance (R) proteins in particular host genotypes, resulting in effector-triggered immunity (ETI) (Jones and Dangl, 2006; Boller and Felix, 2009). A common strategy for plant pathogens to avoid the immune responses is to either lose their effectors or mutate by developing new effectors that are once again able to suppress ETI (Stergiopoulos and De Wit, 2009). This is important because together PTI and/or ETI limit microbial entry, restrict pathogen propagation, or kill pathogens inside plant tissues (Dou and Zhou, 2012). Overall, an in-depth investigation into the roles of putative Las candidate effectors is key not only to understanding how Las manipulates host defense responses and results in HLB disease but to identifying pathogenicity mechanisms that can ultimately lead to the development of novel control methods.
In 2009, the complete genome sequence of Las was obtained (Duan et al., 2009), enabling heterologous expression of Las proteins to be performed. Expression studies have demonstrated that the Las bacterium can act as an “energy parasite” through its encoded ATP translocase, which allows for direct ATP/ADP importation from its host cells (Vahling et al., 2010), and that two novel autotransporter proteins encoded in the Las prophages target the mitochondria of plants (Hao et al., 2013). In addition, using Liberibacter crescens strain BT-1 as model system, a peroxidase of Las was confirmed to be involved in suppressing the plant innate immunity via detoxification of H2O2 (Jain et al., 2015). From the host plant aspect, Las infection is correlated with significant changes in protein regulation in Citrus paradisi, indicating that Las actively alters molecular processes in citrus (Nwugo et al., 2013).
Unfortunately, even with the examination of citrus transcriptomes in response to Las infection and comparative transcriptome analyses (Albrecht and Bowman, 2008, 2012; Kim et al., 2008; Rawat et al., 2015; Zhong et al., 2015), Las genes involved in psyllid or citrus colonization remain largely unknown. This lack of accomplishment is due in part to the limited success in culturing the bacterium (Sechler et al., 2009) and the large number of hypothetical proteins in the Las genome (Duan et al., 2009). Alternative tactics involving heterologous systems are necessary in order to overcome these hurdles and speed the process of identification and functional characterization of candidate Las effector proteins (Boller and Felix, 2009; Segonzac and Zipfel, 2011; Dou and Zhou, 2012). Genome sequencing together with computational methods have revealed a wide catalog of candidate effectors genes in filamentous plant pathogenic: fungi (Chaudhari et al., 2014; Petre and Kamoun, 2014), oomycetes (Ellis et al., 2009), nematodes (Davis et al., 2008), bacteria (Block et al., 2008; Zhou and Chai, 2008; Hogenhout et al., 2009), and insects such as aphids (Bos et al., 2010; Pitino and Hogenhout, 2013). To functionally characterize pathogen effectors and how these proteins manipulate host cells, it is crucial to identify the host compartments where the effectors localize (Alfano, 2009). Thus, subcellular localization is one of the first aspects to consider when assessing effector function.
Utilizing a pathogen secretome analysis we uncovered 16 putative effectors in the Las genome. Similar to other studies, in an attempt to characterize these candidate effector proteins, they were transiently expressed in N. benthamiana leaves to allow both the localization pattern as well as any overt phenotype to be observed (Vleeshouwers et al., 2008; Wang et al., 2011; Du et al., 2014; Petre et al., 2015). These findings not only help establish N. benthamiana as a valuable heterologous system for fast-forward effectoromic analysis of plant pathogens regardless of their host plant (Petre et al., 2016) but also represent the first attempt to characterize and locate putative Las effectors by combining a pathogen secretome analysis with an in planta transient expression assay. Knowledge obtained from this Las effector screen enhances our understanding of HLB and may aid in the development of new and more efficient management strategies.
Materials and Methods
Genomic DNA Extraction of Las Bacterial Pathogen
In addition to transmission by insects, Las can be experimentally transferred to non-rutaceous hosts such as periwinkle (Catharanthus roseus), which acts as a model organism for HLB because when infected via parasitic dodder (Cuscuta campestri) it allows Las to replicate to high titers (Garnier and Bové, 1983). Total genomic DNA was extracted from Las infected periwinkle leaves using the following protocol. Collected leaf tissue was chopped via razorblade and transferred to an autoclaved 2 ml screw-capped tube containing two 4 mm silicone-carbide sharp particles and four 2.3 mm chrome-steel beads in 800 μL of extraction buffer (100 mM HCl, 30 mM EDTA, 500 mM NaCl, 5% polyvinylpyrrolidone and 3% CTAB). Tissue was homogenized using a Fast Prep®-24 homogenizer at speed 6.0 m/s for 60 s and incubated at 65°C for 30 min after the addition of 80 μl 20% SDS. After adding one-third volume of 5 M potassium acetate, the tubes were incubated on ice for 5 min and centrifuged at 18,000 rpm (Eppendorf 5424, Sunnyvale, CA, USA) for 5 min to remove plant debris. The supernatants were removed and placed into a new tube and centrifuged for an additional 10 min at the same speed and then 800 μL of supernatant was transferred to a new 1.5 ml tube containing two-third volume of cold isopropanol. The sample was then placed in a Genesee filter column (Genesee Scientific, San Diego, CA), centrifuged 1 min at 10000 rpm and washed twice with 70% ethanol. Samples were then eluted with 100 μl nuclease-free water and analyzed using the Nanodrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE) with the concentrations being adjusted to 50 ng/μl DNA prior to PCR.
Expression of Las5315 Effector Gene from Infected and Uninfected Citrus by RT-PCR
Total RNA was extracted from Las infected and uninfected Citrus paradise (Grapefruit cultivar Duncan) and Citrus sinensis (Sweet orange cultivar Valencia) leaves using TRIzol reagent according to the manufacturer's protocol. DNA contaminations were removed by treating RNA extraction with RNase-free DNase (QIAGEN, West Sussex, UK) and subsequently purified with QIAamp columns (QIAGEN). First-strand cDNA was synthesized at 42°C from total RNA using M-MLV (Invitrogen) reverse transcriptase according to the manufacturer's instructions with the individual PCR reactions containing 1 μl of cDNA, 0.5 μl of each specific primer (10 pmol/μl) (Table S1), 10 μl of 2X Green GoTaq® Reaction Buffer (Promega) in a final volume of 20 μl. The following standard thermal profile was used for all amplifications: 95°C for 3 min followed by 30 cycles of 95°C for 30 s, 58°C for 30 s, and 70°C for 30 s.
Bioinformatic Identification of Las Secreted Effector Candidates
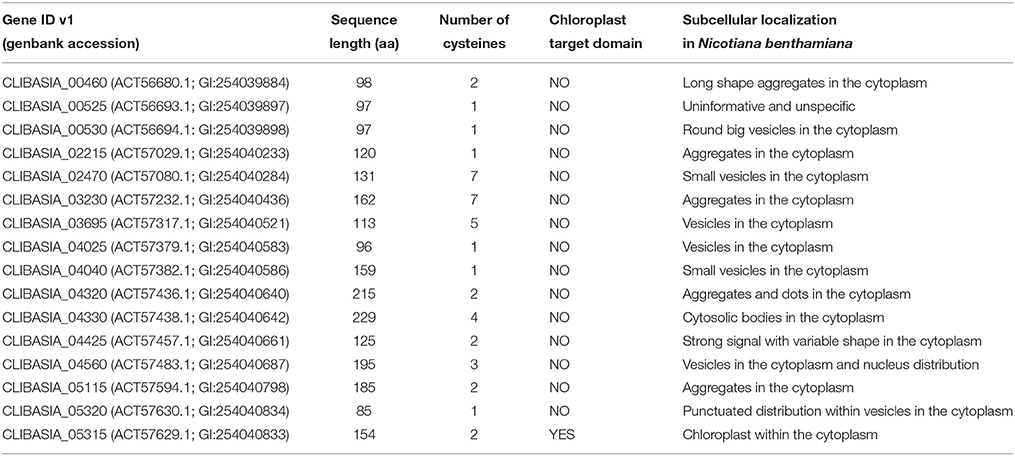
We used as input the genome version v1 from Las isolate psy62 (Duan et al., 2009) (genbank accession NC_012985.3), which is also deposited under accession 537021.9 at Pathosystems Resource Integration Center (PATRIC) (https://www.patricbrc.org/portal/) (Wattam et al., 2014). A protein was predicted to be secreted when a signal peptide was present using SignalP v2.0 and (Nielsen et al., 1997), signalP4.0 and no transmembrane domains were present using TMHMM v2 (Petersen et al., 2011). Then, putative Las secreted candidate effectors were screened for: (1) secreted proteins that were ≤250 amino acids and (2) secreted proteins with a previously uncharacterized function according to the NCBI BLAST database. The genbank “GI” accessions for all 16 secreted candidate effectors are shown in Table 1.
Amplification and Cloning of Las Secreted Effector Candidates
The mature form of each effector protein without its corresponding signal peptide was cloned to create a C-terminal translation fusion with green fluorescent protein (GFP) under the control of 35S promoter in the binary vector ImpGW405 (Nakagawa et al., 2007). Primers were designed to amplify the corresponding ORFs and contained the complete ATTB sites (Table S1). The sequences were amplified using Las infected periwinkle DNA and HIFI PCR mastermix (Clontech, Mountain View, CA, USA). The PCR fragments were first cloned into pDONRzeo by Gateway® BP Clonase® II (Invitrogen, Carlsbad, CA, USA) and then subcloned into the Gateway destination vectors ImpGWB405 (Nakagawa et al., 2007).
Transient Expression of Las Secreted Effectors in Nicotiana Benthamiana
Sequences encoding for mature protein form (lacking signal peptide) of a total of 16 effector candidates were cloned into the binary vector ImpGWB405 and transformed into Agrobacterium tumefaciens strain GV3101. Agrobacterium transformant cells were cultured overnight in LB medium with 50 μg ml−1 of rifampicin and 100 μg ml−1 spectinomycin and resuspended in 10 mM MgCl2. The culture was diluted to an optical density at 600 nm (OD600) of 0.5 and acetosyringone (final concentration of 100 μM) was added. For each construct, we infiltrated three leaves of young N. benthamiana plants with the A. tumefaciens suspension. Agroinfiltrated plants were kept in a greenhouse for the duration of the experiment. The agrobacteria-infiltrated leaves were observed for phenotypic changes and then detached from plants and analyzed under microscope (Olympus BX51-P, Center Valley, PA, USA) equipped with a UV light source at 3 dpi for protein localization.
Staining of Callose Deposition and Relative Callose Intensity Measurement
Aniline blue staining allowed visualization of callose through fluorescence microscopy. An aniline blue signal indicates deposited callose. Four N. benthamiana leaves were used after 1, 2, 3, and 4 dpi for Las5315mp and the GFP control. To distain the chlorophyll, ethanol (90% v/v) was added for 6 h at 37°C, followed by ethanol (50% v/v) for 1 h, ethanol (30% v/v) for 1 h, and finally dH2O for 2 h. The distained leaves were incubated for 2 h in 150 mM K2HPO4 and 0.01% aniline blue, samples were embedded in 50% glycerol before microscope analysis. Three leaves from 3 different plants were used in the analysis.
Callose was quantified from digital photographs by the number of white pixels (callose intensity) relative to the total number of pixels covering plant material, using Photoshop CS5 software (Adobe System, San Jose, CA, USA). Callose was selected automatically, using the “Color Range” tool. Data sets were analyzed for statistical differences by Student's t-tests (*P < 0.01).
H2O2 Production and Electrolyte Leakage Measurement
H2O2 detection was conducted as described in previous studies with minor modification (Zhang et al., 2001; Liao et al., 2013). Briefly, N. benthamiana leaf discs expressing Las5315mp were collected using a circular 4 mm diameter cork borer at 4 dpi. 6 leaf discs from three leaves were placed into a loading buffer with 50 mM Tris-KCl (pH 7.2) containing 100 μM of H2DCF-DA for 10 min. Fluorescence emission was measured by the LUMIstar microplate luminometer (excitation wavelength, 484 nm; emission wavelength, 525 nm). To quantify cell death, electrolyte leakage was performed according to (Huang et al., 2013). Five leaf discs (~5 mm in diameter) were collected from the Las5315mp and control Agrobacterium-infiltrated areas and immersed in 10 mL of non-ionic, double-distilled water. After incubation at room temperature for 1 h with shaking at 160 rpm, conductivity of the solution was measured using a conductivity meter (Horiba scientific, Edison, NJ, USA).
Results
Sixteen Putative Las Effectors were Selected
We analyzed 1,136 protein-encoding genes of Las Psy62 genome version 1 (genbank accession NC_012985.3) (Duan et al., 2009) for the presence of signal peptide and lack of transmembrane domain. To identify candidate effectors, we filtered the secretome for (1) proteins ≤250 amino acids in length and (2) previously uncharacterized proteins (Figure 1A). These 16 secreted effector proteins exhibit sequence lengths that varied from 85 to 229 amino acids. The mature proteins (Figure 1B), without their putative signal peptides, were transiently expressed in N. benthamiana in order to characterize the location of the putative effectors and analyze phenotypic changes produced by the proteins (Figure 1C).
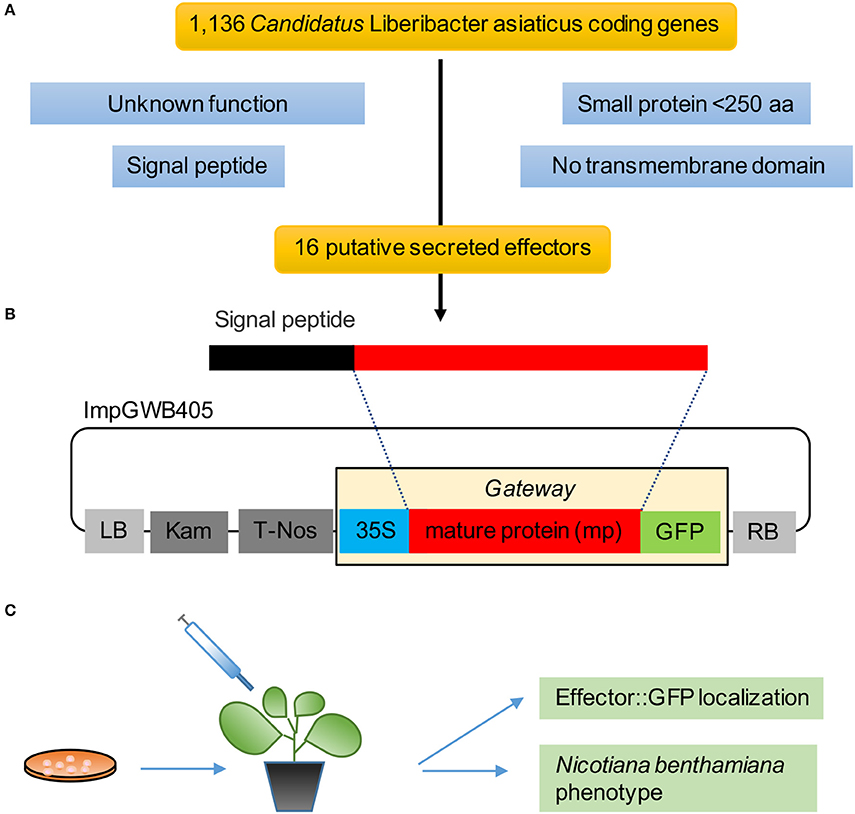
Figure 1. Pipeline to identify Las effectors. (A) Selection criteria for Las effectors yielded 16 putative genes from the 1,136 CDS in the Las genome. (B) Mature protein sequences were cloned in the gateway binary vector ImpGWB405. These clones were transformed into Agrobacterium tumefaciens strain GV3101 for transient expression in N. benthamiana leaves. (C) Functional analyses included GFP localization and in planta phenotype.
Las Candidate Effectors Exhibited Diverse Subcellular Localization Patterns in N. benthamiana
In order to determine the location of the 16 putative effectors in the plant cells, C-terminal translational fusions were produced using the mature form of each protein and the green-fluorescent protein (GFP) expressed from the 35S promoter in a gateway binary vector (Figure 1B). The leaf epidermal layer of the infiltrated zones for each of the 16 candidate effectors showed different localization and accumulation patterns in the plant cells (Figure 2, Figure S1) compared to the control GFP only (Figure 2S). In particular, Las460mp was distributed in the cytosol and accumulated in small vesicles and in elongated shapes. Las525mp was dispersed throughout the cytosol and the nucleus. Las530mp accumulated heavily in vesicle structures while Las2215mp accumulated in aggregated structures. Las2470mp had an intense signal with dot shape bodies covering the total cell area. Las3230mp formed small crystal shape aggregates, while Las3695mp showed an increased signal intensity and was localized in big vesicles, and Las4025mp accumulated in small vesicles and cytosol bodies. Las4040mp was similar to Las4025mp with small vesicles and punctate spots, Las4320mp was expressed evenly in the cell taking the shape of dots, Las4330mp small cytosolic bodies, Las4425mp accumulate in large cytosolic aggregates, Las4560mp was localized in large and small bodies and in the nucleus, Las5115mp accumulated mostly on the edge of the cell with large aggregates, and Las5320mp fluoresced strongly and accumulated in dots inside vesicles. The Las5315mp effector protein was localized in vesicles surrounding chloroplast (Figures 2P,Q), while the effector Las5315 that contained the signal peptide sequence was not present in chloroplast, but accumulated as crystal shape aggregates (Figure 2R).
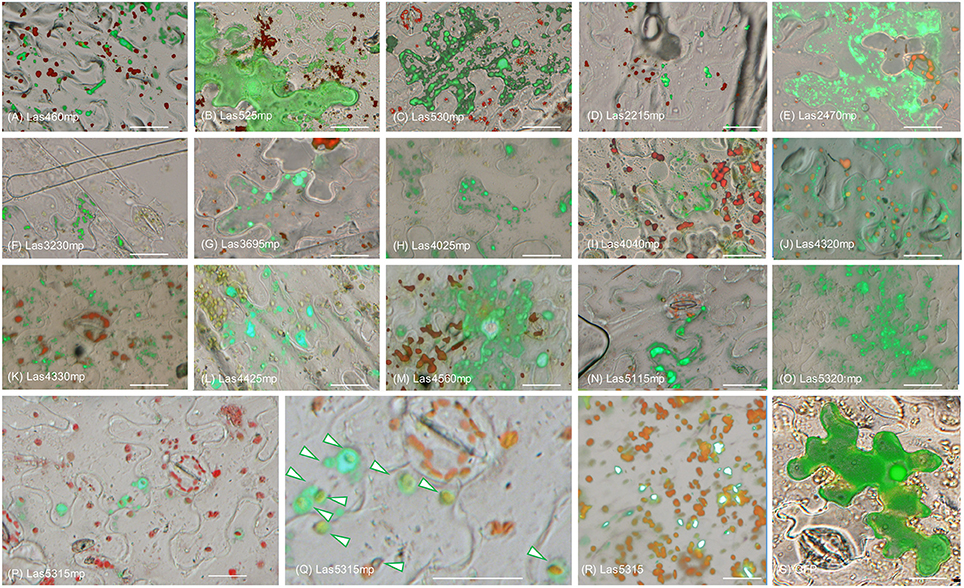
Figure 2. Subcellular localization of putative Las effector proteins in Nicotiana benthamiana. Epi-fluorescence micrograph overlaid with bright field micrograph of each of the 16 effector translation fusions expressed in N. benthamiana. Images depict protein effector: GFP (green) and chlorophyll autoflourescence (red). (A–Q) Micrograph of the localization of the 15 candidate effectors that did not elicit a phenotype in N. benthamiana. (P) Image shows localization of Las5315mp:GFP (green) surrounding chloroplasts (red). (Q) Enlarged image of panel (R) with arrows indicating Las5315mp localization surrounding chloroplasts. (R) Las5315::GFP accumulated in aggregate structures (S) GFP only. Scale bars are 20 μm.
Las5315mp Induced Cell Death and is Associated with H2O2 Accumulation, Electrolyte Leakage, and Callose Deposition in N. benthamiana
In an effort to further characterize these effectors and define a potential role regarding Las virulence, putative effector genes were transiently expressed in N. benthamiana and their phenotypes were observed. Of the 16 putative effectors only Las5315mp appeared to induce necrosis or cell death (Figure 3A and Figures S2A–C). Compared to the negative control at 3 dpi, cell death appeared in the N. benthamiana leaf infiltrated with the mature form of effector Las5315mp at three different concentrations: OD600 = 0.1, 0.3 and 0.5 (Figure S2A). The cell death phenotype was not observed in the premature form containing the signal peptide, Las5315, at the same three concentrations (Figure S2B). A side-by-side comparison on the same leaf demonstrated that chlorosis and increasing levels of cell death could be observed at 7 dpi in the infiltration zone expressing Las5315mp, while visually Las5315 remained unchanged (Figure S2C). Cell death is often associated with electrolyte leakage resulting from membrane damage (Bai et al., 2012). Ion leakage assays were performed to quantify the cell death induced by Las5315mp. As shown in (Figure 3B) significant amounts of ion leakage were detected 2, 3, and 4 days dpi after infiltration with the Agrobacterium harboring the Las5315mp. The cell death produced upon expression of Las5315mp is associated with H2O2 accumulation in the leaves (Figure 4) and electrolyte leakage (Figure 3B). Ion leakage was not induced by agroinfiltration of the control. These data confirm the cell death activity of Las5315mp.
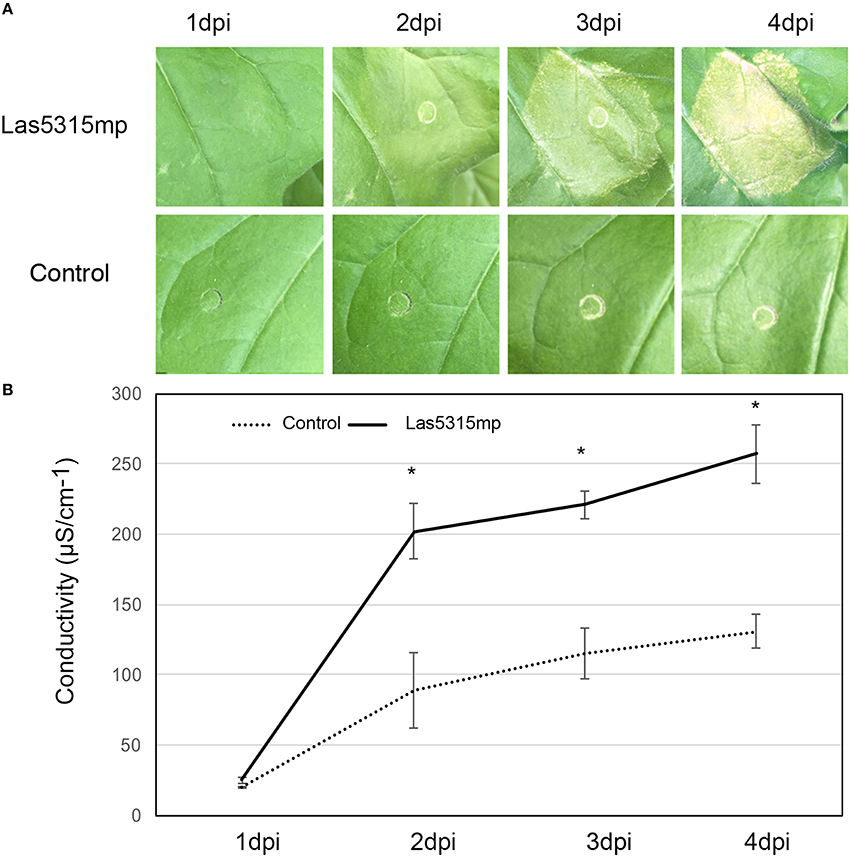
Figure 3. Cell death triggered by Las5315mp results in ion leakage. (A) Progression of cell death in N. benthamiana leaves infiltrated with Las5315mp. Example of leaves taken for electrolyte leakage. Photographs were taken at 1, 2, 3, and 4 dpi (dpi: days post infiltration). (B) Electrolyte leakage was measured in the infiltrated leaves at the indicated time points after agroinfiltration each data point represent the average + SD from 5 infiltrated leaves. The experiments were repeated three times with similar results. Asterisks signs indicate statistically significant differences from the control as calculated by Student's t-test (*P < 0.01).
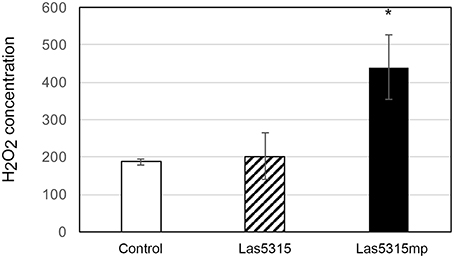
Figure 4. Cell death triggered by Las5315mp results in the accumulation of H2O2. The concentration of H2O2 was determined in leaves agro-infiltrated with the Las5315mp effector or the empty vector (EV) using H2DCFDA as an indicator at 4 dpi. Leaves were used as a control. Error bars represent the standard deviation (n = 8). Asterisks signs indicate statistically significant differences from the control as calculated by Student's t-test (*P < 0.01).
In addition to ion leakage and H2O2 accumulation, fluorescence microscopy with aniline blue staining revealed a strong callose deposition induced by Las5315mp from 1 dpi (Figure 5). This accumulation continued to increases both within the plant cells and inside the vascular tissue for the entire 4 day observation period.
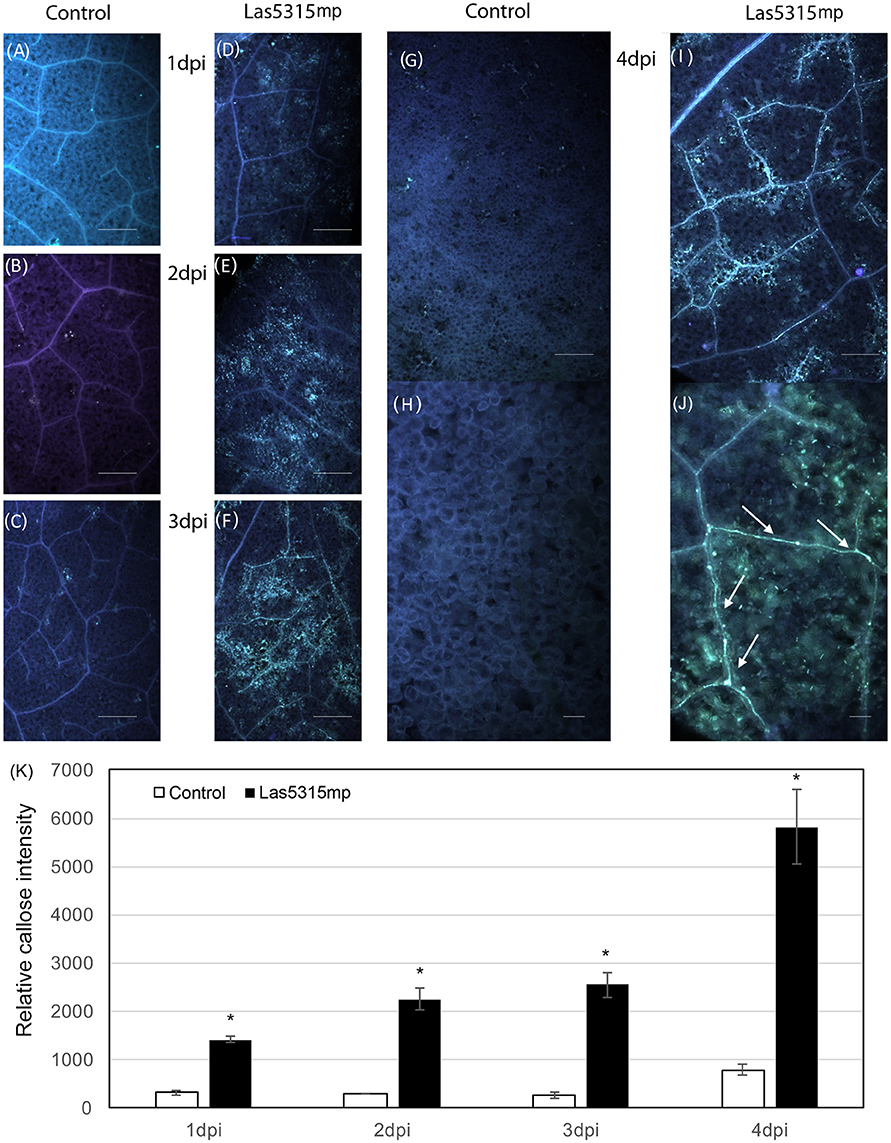
Figure 5. Callose deposition after Las5315mp agroinfiltration. N. benthamiana leaves agroinfiltrated with either the GFP control (A–C,G) or with Las5315mp (D–F,I) corresponding to 1, 2, 3, and 4 dpi respectively. Enlarged image of the leaf at 4 dpi infiltrated with either GFP (H) or Las5315mp (J) showing accumulation of callose in the vascular tissue (arrows). Leaves were stained with aniline blue and callose deposition was observed under a UV epifluorescence microscope. (K) Relative callose deposition was measured in the infiltrated leaves at the indicated time points after agroinfiltration (dpi: days post infiltration). Asterisks signs indicate statistically significant differences from the control as calculated by Student's t-test (*P < 0.01).
Las5315 Contained a Signal Peptide and a Chloroplast Targeting Domain
Sequence analysis of the Las5315mp revealed a 154 aa protein with a signal peptide predicted using SignalP v2.0 (Nielsen et al., 1997) and SignalP v4.0 (Petersen et al., 2011), a cleavage site between position 24 and 25 and no transmembrane domains using TMHMM v2 (Krogh et al., 2001) (Figure 6). Downstream of the N-terminal signal peptide sequence of Las5315 effector protein we found a chloroplast targeting peptide domain of 56 aa length (Figure 6) using ChloroP 1.1 (Emanuelsson et al., 1999) with a score of 0.562 and CS-score of 3.203.

Figure 6. Las5315mp secreted effector sequence and its predicted C-terminal chloroplast targeting domain. Amino acid sequence of Las5315mp with N-terminal signal peptide (highlighted in yellow) predicted using SignalP v4 (Petersen et al., 2011), the cleavage site between 24 and 25 amino acid positions (ALS-GS), and the chloroplast targeting (highlighted in green) predicted using ChloroP v1.1 (Emanuelsson et al., 1999).
Las5315 Gene is Expressed in Las-Infected Citrus
In order to determine if Las5315 is indeed expressed in citrus infected with Las, reverse transcription PCR was used to identify the level of expression of Las5315 in both Las infected and uninfected grapefruit and sweet orange. Amplicons produced via reverse transcription PCR demonstrate the expression of Las5315 both grapefruit and sweet orange but only when infected with Las.
Discussion
“Ca. Liberibacter asiaticus” is an insect-vectored obligate intracellular bacterium with a significantly reduced genome associated with HLB disease in citrus. Powerful functional genomic and genetic approaches integrated with comparative studies have contributed a great deal to the knowledge and identification of global gene expression changes during HLB disease development (Albrecht and Bowman, 2008, 2012; Fan et al., 2011; Martinelli et al., 2012, 2013; Aritua et al., 2013; Mafra et al., 2013; Rawat et al., 2015). Other studies have revealed the disruption in carbohydrate source and phloem plugging as main causes of HLB symptoms (Liao and Burns, 2012; Martinelli et al., 2013). However, knowing how effector proteins function in host plant cells is key to fully understanding the molecular basis behind the pathogen-plant interactions and for the development of sustainable management strategies.
Las, like spiroplasmas and phytoplasmas, is injected directly into the cytoplasm of phloem cells by its insect vector. It is the intracellular nature of these organisms that allows them to secrete proteins into the host cell cytoplasm despite their lack of type III secretion systems, permitting movement of these proteins to other plant cells via plasmodesmata (Hogenhout and Loria, 2008). Several examples of his include the plant gene-regulating effector, SAP11, which contains a nuclear localization signal that is functional in plant cells (Hogenhout et al., 2008) and SAP54, which manipulates its host to produce leaf-like flowers, making it more attractive for colonization by phytoplasma leafhopper vectors (Maclean et al., 2014). Las appears to contain all type I secretion system genes necessary for both multidrug efflux and toxin effector secretion. Moreover, all proteins required for the first step of the type II secretion system, the general secretory pathway (TC 3.A.5.) responsible for the export of proteins to the periplasm (Pugsley, 1993), were found in the “Ca. L. asiaticus” Psy62 genome (Duan et al., 2009). In addition, 10 putative proteins required for pilin secretion and assembly as part of the main terminal branch MTB (TC 3.A.15) were found, indicating the potential for a type IV pilus secretion assembly (Duan et al., 2009).
Therefore, in an attempt to identify novel proteins that function in the pathogenesis of HLB, we analyzed and selected the most promising candidate effector proteins from the current reference Las genome Psy62 (Duan et al., 2009). Based on the assumption that Las secretes proteins into the plant to manipulate host processes and facilitate colonization, proteins with a predicted signal peptide that did not contain transmembrane domains and had no predicted function were selected for further study. We obtained 16 candidates using this criteria and created C-terminal translation fusions for localization studies in plants by cloning the mature form of each candidate (sequence without the signal peptide regions) into the GFP vector ImPgwb405 (Figures 1A,B).
Pathogen effectors are known to target different parts of the host cell including nuclear components, membranes, and subcellular compartments such as the mitochondria or chloroplasts. This targeting of cellular components is a broadly conserved pathogenic strategy that controls the fate of cell organelles and regulates cellular activities (Deslandes and Rivas, 2012; Lindeberg et al., 2012). Mitochondria for instance play important roles in biological processes such as energy conversion for ATP synthesis, ion homeostasis, and calcium storage as well as in plant innate immunity signaling (Maxwell et al., 2002). Effector proteins produced by pathogens, such as HopG1 from Pseudomonas syringe, suppress the host's innate immunity by disrupting mitochondrial function (Block et al., 2010). The mitochondria is not the only system targeted by P. syringae, which produces the effectors HopI1, HopN1, HopK1, and AvrRps4 that localize to chloroplasts instead. Hopl1 and HopN1 are known to inhibit photosystem II (PSII) activity in chloroplast preparations by degrading PbsQ (Rodríguez-Herva et al., 2012) while HopK1 and AvrRps4 suppress plant immunity (Li et al., 2014).
Although only one effector displayed a clear cell death phenotype in N. benthamiana, all of the putative effectors tested showed unique subcellular accumulation patterns with the exception of Las525mp, which was diffused throughout the cytosol and the nucleus (Figures 2A–Q). The different accumulation and localization patterns may be indicative of a specific interaction between the Las protein and plant cell organelles and warrants further investigation in the future. It is important to point out that the Las5315mp effector protein (the mature protein) was localized in vesicles surrounding chloroplast (Figures 2P,Q), while the premature protein did not accumulate in the chloroplast (Figure 2R). These results are rationalized by the fact that the full length protein, Las5315, contains a signal peptide and a chloroplast targeting signal in succession (Figure 6). In an obligate intracellular bacterium such as Las, the signal peptide is assumed to be cleaved by the bacterium upon translocation; leaving only the mature Las5315mp effector protein in the host cell's cytosol. Here, the protein is no longer subjected to this cleavage reaction since it is being produced by the transformed plant not the bacterium and, thus, only the protein without the signal sequence is active.
Plant pathogens secrete effector proteins that are subsequently delivered to the inside of host cells and interfere with their molecular pathways, thereby promoting infection (Dangl and Jones, 2001; Nomura et al., 2005; Hann et al., 2010). These effectors often affect host immunity and cause phenotypes ranging from chlorosis to necrosis (Cunnac et al., 2009). The ability of protein Las5315mp to induce chlorosis and cell death at 3 dpi in N. benthamiana in the infiltration zone (Figure 3A) suggests that it may be affecting host immunity, an observation further supported by ion leakage assays (Figure 3B). Plant cell death is a common defense mechanism used by plants against invading pathogens to prevent the spread of the microbial pathogens and can occur in both resistant and susceptible plant–pathogen interactions. Cell death results when the host plant's receptors perceive the pathogen and trigger plant immunity reactions (Zipfel and Felix, 2005; Jones and Dangl, 2006; Takahashi et al., 2007; Coll et al., 2011). Interestingly, cell death has not been detected in citrus in response to Las infection comparable to that seen with N. benthamiana infiltrated with Las5315mp, suggesting that although N. benthamiana may possess resistance (R) genes, intracellular nucleotide-binding domains, or leucine-rich repeat containing (NB-LRR) immune receptors that specifically recognize Las5315mp effector protein, these NB-LRRs might not be conserved in citrus. Thus, only N. benthamiana can generate an effector triggered immunity response and induce cell death upon recognition of the pathogen effector Las5315mp. Indirect support of this is also demonstrated by the fact that previous attempts to infect Nicotiana tabacum with Las were unsuccessful (Duan, unpublished data), however Ca. L. americanus, which does not have a Las5315mp homolog, was able to infect N. tabacum L. cv Xanthi through Cuscuta spp (dodder) (Francischini et al., 2007).
Previously, NB-LRR genes have been identified and used to confer broad-spectrum resistance in different crops. Examples include the WRR4 gene from Arabidopsis that confers white rust resistance in transgenic oilseed brassica crops (Borhan et al., 2010); two other Arabidopsis genes (RFO1 and RPW8) that confer resistance against a diverse collection of Fusarium or powdery mildew fungi (Xiao et al., 2001; Diener and Ausubel, 2005); RCT1 from Medicago truncatula that confers resistance to races of the anthracnose fungi Colletotrichum trifolii and C. destructivum (Yang et al., 2008); the Mi-1.2 gene in tomato Solanum lycopersicum that confers resistance to the root-knot nematode Meloidogyne javanica in Solanum melongena (Goggin et al., 2006); and the RPS4/RRS1 genes from Arabidopsis that confer resistance to the fungal pathogen C. higginsianum to other brassicaceae plants including B. rapa and B. napus as well as in N. benthamiana and Solanum lycopersicum (Narusaka et al., 2013). In addition, RGA4 and RGA5 genes from rice mediate resistance to the fungal pathogen Magnaporthe oryzae in both rice protoplasts and N. benthamiana (Cesari et al., 2014). Therefore, it is quite plausible that identification of the N. benthamiana receptor of Las5315mp and its heterologous expression in citrus plants may trigger an innate immune response to the Las pathogen and confer resistance to HLB disease in citrus plants.
The inhibition of photoassimilate export and the subsequent starch accumulation in the Las infected leaves indicates that Las infection perturbs normal carbon partitioning (Koh et al., 2012). Previous studies have demonstrated that Liberibacter infection is accompanied by callose deposition in sieve pores of the sieve tubes and it has been suggested this callose deposition is the main reason for the phloem plugging associated with starch accumulation and ultimately chlorosis of the leaf in infected plants (Masaoka et al., 2011). In addition, genes involved in callose deposition have been shown to be up-regulated in HLB-infected citrus (Koh et al., 2012; Zhong et al., 2015). In this work, we found that Las5315mp induces excessive callose in the infiltration zone starting at 1 dpi (Figure 5) and increases the accumulation of callose in the cells and inside the vascular tissue in a fashion similar to that seen in citrus plants infected by Las (Koh et al., 2012). Since Las5315 is also expressed in HLB infected citrus (Figure 7), it stands to reason that Las5315mp may be the protein associated with callose deposition in HLB affected citrus. Further functional characterization and identification of Las effectors associated with pathogenicity and virulence should lead to identification of potential targets for controlling Las infection and HLB disease progression.
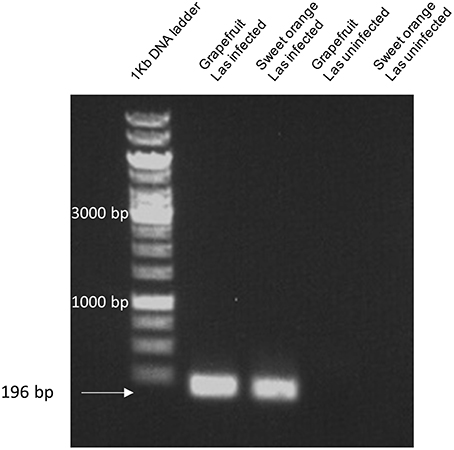
Figure 7. Las5315 expression in citrus using reverse transcription PCR. Amplicons corresponding to the expression levels of Las5315 in both Las infected and uninfected grapefruit and sweet orange were produced via reverse transcription PCR. Amplification products were subsequently run on a 1% agarose gel and visualized with ethidium bromide.
Author Contributions
Conceived and designed the experiments: MP, YD. Performed the experiments: MP. Analyzed the data: MP, CA, LC. Contributed reagents/materials/analysis tools: MP, CA, YD. Wrote the paper: MP, CA, YD.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer WU declared a shared affiliation, though no other collaboration, with several of the authors MP, CA, YD to the handling Editor, who ensured that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
We would like to thank Melissa Doud for her critical review, Carrie Vanderspool for the plants used in this study and Christina Latza for her excellent technical assistance. Funding was provided by the U. S. Department of Agriculture and Citrus Research and Development Foundation. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00982
Figure S1. Subcellular localization of putative Las effector proteins in Nicotiana benthamiana. Sixteen putative effector proteins expressed in leaves of N. benthamiana, visualized by epifluorescence microscopy 3 days post infiltration. (A–O) Micrograph of the localization of the 15 candidate effectors that did not elicit a phenotype in N. benthamiana. (P) Image shows localization of Las5315mp:GFP (green) surrounding chloroplasts (red). (Q) Enlarged image of panel (P) with arrows indicating Las5315mp localization surrounding chloroplasts. (R) Las5315::GFP accumulated in aggregate structures (S) GFP only. Scale bars are 20 μm.
Figure S2. Effector Las5315mp induces cell death in N. benthamiana. Phenotype of N. benthamiana leaves expressing (A) Las5315mp or (B) Las5315 at several different concentrations (OD600 = 0.1, 0.3, 0.5) compared to the empty vector control. (C) Overexpression of Las5315mp and Las5315 in the same leaf 7 days post infiltration (dpi) at an OD600 = 0.5.
Table S1. Primers used in this study.
References
Albrecht, U., and Bowman, K. D. (2008). Gene expression in Citrus sinensis (L.) Osbeck following infection with the bacterial pathogen Candidatus Liberibacter asiaticus causing Huanglongbing in Florida. Plant Sci. 175, 291–306. doi: 10.1016/j.plantsci.2008.05.001
Albrecht, U., and Bowman, K. D. (2012). Transcriptional response of susceptible and tolerant citrus to infection with Candidatus Liberibacter asiaticus. Plant Sci. 185, 118–130. doi: 10.1016/j.plantsci.2011.09.008
Alfano, J. R. (2009). Roadmap for future research on plant pathogen effectors. Mol. Plant Pathol. 10, 805–813. doi: 10.1111/j.1364-3703.2009.00588.x
Aritua, V., Achor, D., Gmitter, F. G., Albrigo, G., and Wang, N. (2013). Transcriptional and microscopic analyses of citrus stem and root responses to Candidatus Liberibacter asiaticus Infection. PLoS ONE 8:e73742. doi: 10.1371/journal.pone.0073742
Bai, S. W., Liu, J., Chang, C., Zhang, L., Maekawa, T., Wang, Q. Y., et al. (2012). Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog 8:e1002752. doi: 10.1371/journal.ppat.1002752
Block, A., Guo, M., Li, G. Y., Elowsky, C., Clemente, T. E., and Alfano, J. R. (2010). The Pseudomonas syringae type III effector HopG1 targets mitochondria, alters plant development and suppresses plant innate immunity. Cell. Microbiol. 12, 318–330. doi: 10.1111/j.1462-5822.2009.01396.x
Block, A., Li, G. Y., Fu, Z. Q., and Alfano, J. R. (2008). Phytopathogen type III effector weaponry and their plant targets. Curr. Opin. Plant Biol. 11, 396–403. doi: 10.1016/j.pbi.2008.06.007
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Borhan, M. H., Holub, E. B., Kindrachuk, C., Omidi, M., Bozorgmanesh-Frad, G., and Rimmer, S. R. (2010). WRR4, a broad-spectrum TIR-NB-LRR gene from Arabidopsis thaliana that confers white rust resistance in transgenic oilseed brassica crops. Mol. Plant Pathol. 11, 283–291. doi: 10.1111/j.1364-3703.2009.00599.x
Bos, J. I. B., Prince, D., Pitino, M., Maffei, M. E., Win, J., and Hogenhout, S. A. (2010). A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genet 6:e1001216. doi: 10.1371/journal.pgen.1001216
Bove, J. M. (2006). Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J. Plant Pathog. 88, 7–37. doi: 10.4454/jpp.v88i1.828
Cesari, S., Kanzaki, H., Fujiwara, T., Bernoux, M., Chalvon, V., Kawano, Y., et al. (2014). The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 33, 1941–1959. doi: 10.15252/embj.201487923
Chaudhari, P., Ahmed, B., Joly, D. L., and Germain, H. (2014). Effector biology during biotrophic invasion of plant cells. Virulence 5, 703–709. doi: 10.4161/viru.29652
Chisholm, S. T., Coaker, G., Day, B., and Staskawicz, B. J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814. doi: 10.1016/j.cell.2006.02.008
Coll, N. S., Epple, P., and Dangl, J. L. (2011). Programmed cell death in the plant immune system. Cell Death Differ. 18, 1247–1256. doi: 10.1038/cdd.2011.37
Cunnac, S., Lindeberg, M., and Collmer, A. (2009). Pseudomonas syringae type III secretion system effectors: repertoires in search of functions. Curr. Opin. Microbiol. 12, 53–60. doi: 10.1016/j.mib.2008.12.003
Dangl, J. L., and Jones, J. D. G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
Davis, E. L., Hussey, R. S., Mitchum, M. G., and Baum, T. J. (2008). Parasitism proteins in nematode-plant interactions. Curr. Opin. Plant Biol. 11, 360–366. doi: 10.1016/j.pbi.2008.04.003
Deslandes, L., and Rivas, S. (2012). Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci. 17, 644–655. doi: 10.1016/j.tplants.2012.06.011
Diener, A. C., and Ausubel, F. M. (2005). Resistance to Fusarium oxysporum 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171, 305–321. doi: 10.1534/genetics.105.042218
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Dou, D. L., and Zhou, J. M. (2012). Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12, 484–495. doi: 10.1016/j.chom.2012.09.003
Du, J., Rietman, H., and Vleeshouwers, V. G. (2014). Agroinfiltration and PVX agroinfection in potato and Nicotiana benthamiana. J. Vis. Exp. 83:e50971. doi: 10.3791/50971
Duan, Y. P., Zhou, L. J., Hall, D. G., Li, W. B., Doddapaneni, H., Lin, H., et al. (2009). Complete genome sequence of Citrus Huanglongbing bacterium, ‘candidatus liberibacter asiaticus’ obtained through metagenomics. Mol. Plant Microbe Interact. 22, 1011–1020. doi: 10.1094/MPMI-22-8-1011
Ellis, J. G., Rafiqi, M., Gan, P., Chakrabarti, A., and Dodds, P. N. (2009). Recent progress in discovery and functional analysis of effector proteins of fungal and oomycete plant pathogens. Curr. Opin. Plant Biol. 12, 399–405. doi: 10.1016/j.pbi.2009.05.004
Emanuelsson, O., Nielsen, H., and Von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. doi: 10.1110/ps.8.5.978
Fan, J., Chen, C. X., Yu, Q. B., Brlansky, R. H., Li, Z. G., and Gmitter, F. G. (2011). Comparative iTRAQ proteome and transcriptome analyses of sweet orange infected by “Candidatus Liberibacter asiaticus”. Physiol. Plant. 143, 235–245. doi: 10.1111/j.1399-3054.2011.01502.x
Francischini, F. J. B., Oliveira, K. D. S., Astua-Monge, G., Novelli, A., Lorenzino, R., Matiolli, C., et al. (2007). First report on the transmission of 'Candidatus liberibacter americanus' from citrus to Nicotiana tabacum cv. Xanthi. Plant Dis. 91, 631–631. doi: 10.1094/PDIS-91-5-0631B
Garnier, M., and Bové, J. (1983). Transmission of the organism associated with citrus greening disease from sweet orange to periwinkle by dodder. Phytopathology 73, 1358–1363. doi: 10.1094/Phyto-73-1358
Garnier, M., Jagoueix-Eveillard, S., Cronje, P. R., Le Roux, H. F., and Bove, J. M. (2000). Genomic characterization of a liberibacter present in an ornamental rutaceous tree, Calodendrum capense, in the Western Cape province of South Africa. Proposal of 'Candidatus Liberibacter africanus subsp capensis'. Int. J. Syst. Evol. Microbiol. 50, 2119–2125. doi: 10.1099/00207713-50-6-2119
Goggin, F. L., Jia, L. L., Shah, G., Hebert, S., Williamson, V. M., and Ullman, D. E. (2006). Heterologous expression of the Mi-1.2 gene from tomato confers resistance against nematodes but not aphids in eggplant. Mol. Plant Microbe Interact. 19, 383–388. doi: 10.1094/MPMI-19-0383
Gottwald, T. R. (2010). Current epidemiological understanding of citrus huanglongbing. Annu. Rev. Phytopathol. 48, 119–139. doi: 10.1146/annurev-phyto-073009-114418
Halbert, S. E., and Manjunath, K. L. (2004). Asian citrus psyllids (Sternorrhyncha: Psyllidae) and greening disease of citrus: a literature review and assment of risk in Florida. Florida Entomol. 87, 330–353. doi: 10.1653/0015-4040(2004)087[0330:ACPSPA]2.0.CO;2
Hall, D. G., Richardson, M. L., Ammar, E. D., and Halbert, S. E. (2013). Asian citrus psyllid, Diaphorina citri, vector of citrus huanglongbing disease. Entomol. Exp. Appl. 146, 207–223. doi: 10.1111/eea.12025
Hann, D. R., Gimenez-Ibanez, S., and Rathjen, J. P. (2010). Bacterial virulence effectors and their activities. Curr. Opin. Plant Biol. 13, 388–393. doi: 10.1016/j.pbi.2010.04.003
Hao, G. X., Boyle, M., Zhou, L. J., and Duan, Y. P. (2013). The Intracellular Citrus Huanglongbing Bacterium, 'Candidatus Liberibacter asiaticus' encodes two novel autotransporters. PLoS ONE 8:e68921. doi: 10.1371/journal.pone.0068921
Hodges, A. W., and Spreen, T. H. (2012). “Economic impacts of citrus greening (HLB) in Florida, 2006/07-2010/11,” in Electronic Data Information Source (EDIS) FE903 (Gainseville, FL: University of Florida).
Hogenhout, S. A., and Loria, R. (2008). Virulence mechanisms of Gram-positive plant pathogenic bacteria. Curr. Opin. Plant Biol. 11, 449–456. doi: 10.1016/j.pbi.2008.05.007
Hogenhout, S. A., Oshima, K., Ammar El, D., Kakizawa, S., Kingdom, H. N., and Namba, S. (2008). Phytoplasmas: bacteria that manipulate plants and insects. Mol. Plant Pathol. 9, 403–423. doi: 10.1111/j.1364-3703.2008.00472.x
Hogenhout, S. A., Van Der Hoorn, R. A., Terauchi, R., and Kamoun, S. (2009). Emerging concepts in effector biology of plant-associated organisms. Mol. Plant Microbe Interact. 22, 115–122. doi: 10.1094/MPMI-22-2-0115
Huang, X. E., Liu, X. Y., Chen, X. H., Snyder, A., and Song, W. Y. (2013). Members of the XB3 family from diverse plant species induce programmed cell death in Nicotiana benthamiana. PLoS ONE 8:e63868. doi: 10.1371/journal.pone.0063868
Jagoueix, S., Bove, J. M., and Garnier, M. (1996). PCR detection of the two 'Candidatus' liberobacter species associated with greening disease of citrus. Mol. Cell. Probes 10, 43–50. doi: 10.1006/mcpr.1996.0006
Jain, M., Fleites, L. A., and Gabriel, D. W. (2015). Prophage-encoded peroxidase in 'Candidatus Liberibacter asiaticus' is a secreted effector that suppresses plant defenses. Mol. Plant Microbe Interact. 28, 1330–1337. doi: 10.1094/MPMI-07-15-0145-R
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kim, J., Sagaram, U. S., Burns, J. K., and Wang, N. (2008). Microscopy and microarray analyses of host response of sweet orange (Citrus sinensis) to Candidatus Liberibacter asiaticus infection. Phytopathology 98, S81–S81. doi: 10.1094/PHYTO-99-1-0050
Koh, E. J., Zhou, L. J., Williams, D. S., Park, J., Ding, N. Y., Duan, Y. P., et al. (2012). Callose deposition in the phloem plasmodesmata and inhibition of phloem transport in citrus leaves infected with “Candidatus Liberibacter asiaticus.” Protoplasma 249, 687–697. doi: 10.1007/s00709-011-0312-3
Krogh, A., Larsson, B., Von Heijne, G., and Sonnhammer, E. (2001). Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Li, G. Y., Froehlich, J. E., Elowsky, C., Msanne, J., Ostosh, A. C., Zhang, C., et al. (2014). Distinct Pseudomonas type-III effectors use a cleavable transit peptide to target chloroplasts. Plant J. 77, 310–321. doi: 10.1111/tpj.12396
Liao, H. L., and Burns, J. K. (2012). Gene expression in Citrus sinensis fruit tissues harvested from huanglongbing-infected trees: comparison with girdled fruit. J. Exp. Bot. 63, 3307–3319. doi: 10.1093/jxb/ers070
Liao, Y. W., Sun, Z. H., Zhou, Y. H., Shi, K., Li, X., Zhang, G. Q., et al. (2013). The role of hydrogen peroxide and nitric oxide in the induction of plant-encoded RNA-dependent RNA polymerase 1 in the basal defense against Tobacco mosaic virus. PLoS ONE 8:e76090. doi: 10.1371/journal.pone.0076090
Lindeberg, M., Cunnac, S., and Collmer, A. (2012). Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiol. 20, 199–208. doi: 10.1016/j.tim.2012.01.003
Maclean, A. M., Orlovskis, Z., Kowitwanich, K., Zdziarska, A. M., Angenent, G. C., Immink, R. G. H., et al. (2014). phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS-box proteins and promotes insect colonization in a RAD23-dependent manner. PLoS Biol. 12:e1001835. doi: 10.1371/journal.pbio.1001835
Mafra, V., Martins, P. K., Francisco, C. S., Ribeiro-Alves, M., Freitas-Astua, J., and Machado, M. A. (2013). Candidatus Liberibacter americanus induces significant reprogramming of the transcriptome of the susceptible citrus genotype. BMC Genomics 14:247. doi: 10.1186/1471-2164-14-247
Martinelli, F., Reagan, R. L., Uratsu, S. L., Phu, M. L., Albrecht, U., Zhao, W. X., et al. (2013). Gene regulatory networks elucidating huanglongbing disease mechanisms. PLoS ONE 8:e74256. doi: 10.1371/journal.pone.0074256
Martinelli, F., Uratsu, S. L., Albrecht, U., Reagan, R. L., Phu, M. L., Britton, M., et al. (2012). Transcriptome profiling of citrus fruit response to huanglongbing disease. PLoS ONE 7:e38039. doi: 10.1371/journal.pone.0038039
Masaoka, Y., Pustika, A., Subandiyah, S., Okada, A., Hanundin, E., Purwanto, B., et al. (2011). Lower concentrations of microelements in leaves of citrus infected with 'candidatus Liberibacter asiaticus'. Jpn. Agric. Res. Q. 45, 269–275. doi: 10.6090/jarq.45.269
Maxwell, D. P., Nickels, R., and Mcintosh, L. (2002). Evidence of mitochondrial involvement in the transduction of signals required for the induction of genes associated with pathogen attack and senescence. Plant J. 29, 269–279. doi: 10.1046/j.1365-313X.2002.01216.x
Nakagawa, T., Suzuki, T., Murata, S., Nakamura, S., Hino, T., Maeo, K., et al. (2007). Improved gateway binary vectors: high-performance vectors for creation of fusion constructs in Transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71, 2095–2100. doi: 10.1271/bbb.70216
Narusaka, M., Kubo, Y., Hatakeyama, K., Imamura, J., Ezura, H., Nanasato, Y., et al. (2013). Interfamily transfer of dual NB-LRR genes confers resistance to multiple pathogens. PLoS ONE 8:e55954 doi: 10.1371/journal.pone.0055954
Nielsen, H., Engelbrecht, J., Brunak, S., and Von Heijne, G. (1997). A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 8, 581–599. doi: 10.1142/S0129065797000537
Nomura, K., Melotto, M., and He, S. Y. (2005). Suppression of host defense in compatible plant- Pseudomonas syringae interactions. Curr. Opin. Plant Biol. 8, 361–368. doi: 10.1016/j.pbi.2005.05.005
Nwugo, C. C., Lin, H., Duan, Y. P., and Civerolo, E. L. (2013). The effect of 'Candidatus Liberibacter asiaticus' infection on the proteomic profiles and nutritional status of pre-symptomatic and symptomatic grapefruit (Citrus paradisi) plants. BMC Plant Biol. 13:59 doi: 10.1186/1471-2229-13-59
Petersen, T. N., Brunak, S., Von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Petre, B., and Kamoun, S. (2014). How do filamentous pathogens deliver effector proteins into plant cells? PLoS Biol. 12:e1001801. doi: 10.1371/journal.pbio.1001801
Petre, B., Saunders, D. G. O., Sklenar, J., Lorrain, C., Krasileva, K. V., Win, J., et al. (2016). Heterologous expression screens in Nicotiana benthamiana identify a candidate effector of the wheat yellow rust pathogen that associates with processing bodies. PLoS ONE 11:e0149035. doi: 10.1371/journal.pone.0149035
Petre, B., Saunders, D. G. O., Sklenar, J., Lorrain, C., Win, J., Duplessis, S., et al. (2015). Candidate effector proteins of the rust pathogen Melampsora larici-populina target diverse plant cell compartments. Mol. Plant Microbe Interact. 28, 689–700. doi: 10.1094/MPMI-01-15-0003-R
Pitino, M., and Hogenhout, S. A. (2013). Aphid protein effectors promote aphid colonization in a plant species-specific manner. Mol. Plant Microbe Interact. 26, 130–139. doi: 10.1094/MPMI-07-12-0172-FI
Pugsley, A. P. (1993). The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57, 50–108.
Rawat, N., Kiran, S. P., Du, D. L., Gmitter, F. G., and Deng, Z. A. (2015). Comprehensive meta-analysis, co-expression, and miRNA nested network analysis identifies gene candidates in citrus against Huanglongbing disease. BMC Plant Biol. 15:184. doi: 10.1186/s12870-015-0568-4
Rodríguez-Herva, J. J., González-Melendi, P., Cuartas-Lanza, R., Antúnez-Lamas, M., Río-Alvarez, I., Li, Z., et al. (2012). A bacterial cysteine protease effector protein interferes with photosynthesis to suppress plant innate immune responses. Cell. Microbiol. 14, 669–681. doi: 10.1111/j.1462-5822.2012.01749.x
Sechler, A., Schuenzel, E. L., Cooke, P., Donnua, S., Thaveechai, N., Postnikova, E., et al. (2009). Cultivation of 'Candidatus Liberibacter asiaticus', 'Ca. L. africanus', and 'Ca. L. americanus' associated with Huanglongbing. Phytopathology 99, 480–486. doi: 10.1094/PHYTO-99-5-0480
Segonzac, C., and Zipfel, C. (2011). Activation of plant pattern-recognition receptors by bacteria. Curr. Opin. Microbiol. 14, 54–61. doi: 10.1016/j.mib.2010.12.005
Stergiopoulos, I., and De Wit, P. J. G. M. (2009). Fungal effector proteins. Annu. Rev. Phytopathol. 47, 233–263. doi: 10.1146/annurev.phyto.112408.132637
Takahashi, Y., Bin Nasir, K. H., Ito, A., Kanzaki, H., Matsumura, H., Saitoh, H., et al. (2007). A high-throughput screen of cell-death-inducing factors in Nicotiana benthamiana identifies a novel MAPKK that mediates INF1-induced cell death signaling and non-host resistance to Pseudomonas cichorii. Plant J. 49, 1030–1040. doi: 10.1111/j.1365-313X.2006.03022.x
Vahling, C. M., Duan, Y. P., and Lin, H. (2010). Characterization of an ATP translocase identified in the destructive plant pathogen “Candidatus Liberibacter asiaticus”. J. Bacteriol. 192, 834–840. doi: 10.1128/JB.01279-09
Vleeshouwers, V. G., Rietman, H., Krenek, P., Champouret, N., Young, C., Oh, S. K., et al. (2008). Effector genomics accelerates discovery and functional profiling of potato disease resistance and phytophthora infestans avirulence genes. PLoS ONE 3:e2875. doi: 10.1371/journal.pone.0002875
Wang, Q. Q., Han, C. Z., Ferreira, A. O., Yu, X. L., Ye, W. W., Tripathy, S., et al. (2011). Transcriptional programming and functional interactions within the Phytophthora sojae RXLR Effector Repertoire. Plant Cell 23, 2064–2086. doi: 10.1105/tpc.111.086082
Wattam, A. R., Abraham, D., Dalay, O., Disz, T. L., Driscoll, T., Gabbard, J. L., et al. (2014). PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 42, D581–D591. doi: 10.1093/nar/gkt1099
Xiao, S. Y., Ellwood, S., Calis, O., Patrick, E., Li, T. X., Coleman, M., et al. (2001). Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291, 118–120. doi: 10.1126/science.291.5501.118
Yang, S. M., Gao, M. Q., Xu, C. W., Gao, J. C., Deshpande, S., Lin, S. P., et al. (2008). Alfalfa benefits from Medicago truncatula: the RCT1 gene from M-truncatula confers broad-spectrum resistance to anthracnose in alfalfa. Proc. Natl. Acad. Sci. U.S.A. 105, 12164–12169. doi: 10.1073/pnas.0802518105
Zhang, X., Zhang, L., Dong, F. C., Gao, J. F., Galbraith, D. W., and Song, C. P. (2001). Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol. 126, 1438–1448. doi: 10.1104/pp.126.4.1438
Zhong, Y., Cheng, C. Z., Jiang, N. H., Jiang, B., Zhang, Y. Y., Wu, B., et al. (2015). Comparative transcriptome and iTRAQ proteome analyses of citrus root responses to Candidatus Liberibacter asiaticus Infection. PLoS ONE 10:e0126973. doi: 10.1371/journal.pone.0126973
Zhou, J. M., and Chai, J. (2008). Plant pathogenic bacterial type III effectors subdue host responses. Curr. Opin. Microbiol. 11, 179–185. doi: 10.1016/j.mib.2008.02.004
Keywords: huanglongbing, Candidatus Liberibacter asiaticus, bacterial effectors, cell death, Nicotiana benthamiana, callose deposition, chloroplast localization
Citation: Pitino M, Armstrong CM, Cano LM and Duan Y (2016) Transient Expression of Candidatus Liberibacter Asiaticus Effector Induces Cell Death in Nicotiana benthamiana. Front. Plant Sci. 7:982. doi: 10.3389/fpls.2016.00982
Received: 14 April 2016; Accepted: 21 June 2016;
Published: 06 July 2016.
Edited by:
Pietro Daniele Spanu, Imperial College London, UKReviewed by:
William Underwood, Agricultural Research Service, United States Department of Agriculture, USADaolong Dou, Nanjing Agricultural University, China
Copyright © 2016 Pitino, Armstrong, Cano and Duan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongping Duan, yongping.duan@aus.usda.gov
 Marco Pitino
Marco Pitino Cheryl M. Armstrong
Cheryl M. Armstrong Liliana M. Cano
Liliana M. Cano Yongping Duan
Yongping Duan