- 1Graduate School of Bioagricultural Sciences, Nagoya University, Nagoya, Japan
- 2Department of Botany, University of Wisconsin–Madison, Madison, WI, USA
- 3Japan Science and Technology Agency, Precursory Research for Embryonic Science and Technology, Saitama, Japan
- 4Graduate School of Biological Sciences, Nara Institute of Science and Technology, Ikoma, Japan
- 5Graduate School of Life Sciences, Tohoku University, Sendai, Japan
- 6CREST, Japan Science and Technology Agency, Tokyo, Japan
Forward genetics is a powerful approach used to link genotypes and phenotypes, and mutant screening/analysis has provided deep insights into many aspects of plant physiology. Gravitropism is a tropistic response in plants, in which hypocotyls and stems sense the direction of gravity and grow upward. Previous studies of gravitropic mutants have suggested that shoot endodermal cells in Arabidopsis stems and hypocotyls are capable of sensing gravity (i.e., statocytes). In the present study, we report a new screening system using hypergravity conditions to isolate enhancers of gravitropism mutants, and we also describe a rapid and efficient genome mapping method, using next-generation sequencing (NGS) and single nucleotide polymorphism (SNP)-based markers. Using the endodermal-amyloplast less 1 (eal1) mutant, which exhibits defective development of endodermal cells and gravitropism, we found that hypergravity (10 g) restored the reduced gravity responsiveness in eal1 hypocotyls and could, therefore, be used to obtain mutants with further reduction in gravitropism in the eal1 background. Using the new screening system, we successfully isolated six ene (enhancer of eal1) mutants that exhibited little or no gravitropism under hypergravity conditions, and using NGS and map-based cloning with SNP markers, we narrowed down the potential causative genes, which revealed a new genetic network for shoot gravitropism in Arabidopsis.
Introduction
Plants alter the orientation of their growth in response to environmental stimuli, such as light, moisture, and gravity. Gravitropism is a well-known growth response in which shoots grow upward and roots grow downward, according to the direction of gravity, and the molecular mechanism of gravitropism has been investigated for decades, using physiological and genetic methods in Arabidopsis thaliana (Blancaflor and Masson, 2003; Morita, 2010). One method, forward genetics, is a powerful approach used to clarify relationships between phenotypes and genes and has contributed to a number of significant findings in plant physiology, including gravitropism. Null mutants of the GRAS family transcription factor SGR7/SHR, shoot gravitropism (sgr)7-1/short root (shr)-2, lack endodermal cells and fail to exhibit gravitropism in their hypocotyls and stems (Fukaki et al., 1998; Helariutta et al., 2000), and endodermal-amyloplast less 1 (eal1) mutants have a single amino acid deletion in SGR7/SHR that results in abnormal development of the endodermis and insufficient gravitropic responses (Fujihira et al., 2000; Morita et al., 2007). Shoot endodermal cells contain starch-filled plastids (amyloplasts) that distribute toward the gravity vector. The sgr2 and zig/sgr4 mutants have immobile amyloplasts in their endodermal cells, owing to abnormal vacuolar dynamics, and exhibit reduced shoot gravitropism (Saito et al., 2005; Toyota et al., 2013b). Amyloplasts in sgr9 mutants exhibit irregular interactions with actin filaments, which results in non-sedimentable amyloplasts and weak gravitropism (Nakamura et al., 2011). Comprehensive analyses of these gravitropic mutants support the idea that endodermal cells are gravity-sensing cells, i.e., statocytes, in Arabidopsis shoots and that the sedimentation of amyloplasts plays a crucial role in gravity perception. However, the main signaling pathway following amyloplast sedimentation remains largely unknown, since conventional screening has not identified such mutants/genes. Therefore, to isolate novel mutants/genes in gravity sensing/signaling, it is necessary to develop a new screening system under different gravitational conditions.
Hypergravity created by centrifugation provides a unique experimental environment that can be used to investigate the gravity responsiveness of plants living on Earth (1 g). Knight (1806) built a centrifuge (waterwheel) that was driven by the flow of a river and used it to determine that plants perceive gravitational and centrifugal acceleration and exhibit gravitropic responses. The starchless mutants, phosphoglucomutase (pgm), exhibit reduced gravity sensitivity in hypocotyls and roots at 1 g, which is restored by hypergravity, such as 10 g (Fitzelle and Kiss, 2001), and sgr2 mutants exhibit little gravitropism at 1 g, whereas hypergravity (10 and 30 g) moved the immobile amyloplasts of sgr2 mutants toward the gravity vector and induced obvious gravitropism in stems (Toyota et al., 2013b). However, sgr7-1/shr-2 mutants failed to exhibit gravitropism in stems, even at 30 g, which suggests that sgr7-1/shr-2 mutants completely lack gravity sensing machinery (Toyota et al., 2013b). Interestingly, the eal1 mutant, a weak allele of sgr7-1/shr-2, exhibits reduced but distinguishable gravitropism in hypocotyls at 1 g (Fujihira et al., 2000), which indicates that eal1 mutants still possess gravity sensing machinery in their abnormal endodermal cells.
In the present study, we designed a new screening system that used hypergravity conditions to search for enhancing mutations that further reduce the weak gravitropism of eal1 hypocotyls. We found that hypergravity intensified hypocotyl gravitropism in both wild-type and eal1 plants and that it enabled enhancer screening of the gravitropic mutant eal1. In addition, we also utilized whole genome sequencing and ethyl methanesulfonate (EMS)-derived single nucleotide polymorphism (SNP) markers to identify the causative mutations of the enhanced phenotypes (Hartwig et al., 2012). We report that six ENHANCER OF EAL1 (ENE) genes are related to gravitropism, two of which were novel gravitropic genes and the remaining were previously reported gravitropic genes. The identification of these genes provides deep insight into the molecular components of gravity sensing/signaling, and our screening system opens the possibility for new forward genetics in gravitropism.
Materials and Methods
Plant Materials and Growing Conditions
Arabidopsis seeds were sterilized, using 5% sodium hypochlorite and 1% Triton X-100, and then were sown on plates (AW2000; Eiken Chemical, Co., Ltd, Tokyo, Japan) with Murashige and Skoog (MS) medium that contained 1% [w/v] sucrose, 0.01% [w/v] myo-inositol, 0.05% MES-KOH (pH 5.7), and 0.05% [w/v] gellan gum. For the hypergravity condition, seeds were sown on plates (S01F04S; STEM, Co., Tokyo, Japan) with the MS medium that contained 0.03% [w/v] gelrite, instead of gellan gum. The seeds were vernalized at 4°C for 3 days, germinated at 23°C under continuous light, and then grown on the gel surface of vertical plates.
T-DNA Insertion Lines, Transgenic Lines, and ene Single Mutant Lines
abi4-1 and two T-DNA insertion lines, eif6b (SALK_139209) and atcambp25 (SALK_005722C), were obtained from Arabidopsis Biological Resource Center (ABRC).
For a transformation-rescue experiment, we generated the ARG1 construct (pARG1::ARG1), a KpnI-XhoI fragment of the genomic TAC K20N04 (∼8 kb), was cloned (Liu et al., 1999) and ligated into pENTR1A Gateway entry vector. The final construct was obtained by LR reaction in pFAST-R01 destination binary vector (Shimada et al., 2010). Transgenic seeds were generated by floral dip and were selected on MS medium supplied with hygromycin.
We crossed eal1 ene mutants with wild-type (Col) to generate ene single mutants. F2 seedlings were genotyped at two loci, eal1 and ene mutation site, using SNP markers (See below; Filtering SNPs and predicting causative genes). We considered individuals which are homozygous wild-type at the eal1 locus and homozygous mutant-type at the ene locus as ene single mutants.
Mutagenesis and Screening Using Hypergravity
Approximately, 7000 seeds were mutagenized using 0.4% [w/v] EMS for 10 h. The M1 seeds were divided into 144 batches (∼50 seeds per batch). The M2 seeds were sown on plates for hypergravity conditions as mentioned above, which were set in swinging buckets (TS-4TB; Tomy Digital Biology, Co., Ltd, Tokyo, Japan) in a centrifuge (LX-141; Tomy Digital Biology, Co., Ltd, Tokyo, Japan; Figure 1E) and then centrifuged at 10 g [200 revolution per minute (rpm)] at 23°C in darkness for 2 days. Hypocotyls that were inclined over ±30° toward the direction of the hypergravity 10 g (0°), were selected (first screening), and their offspring (M3) seed was obtained. Approximately, 50 M3 seedlings from each mutant candidate were examined, using the experimental conditions described above, and their agravitropic phenotypes were confirmed (second screening). Subsequently, M3 seedlings were grown in the light for 45 h and in the dark for 24 h, and after being exposed to unilateral irradiation with blue light for 29 h, their phototropism was investigated (third screening).
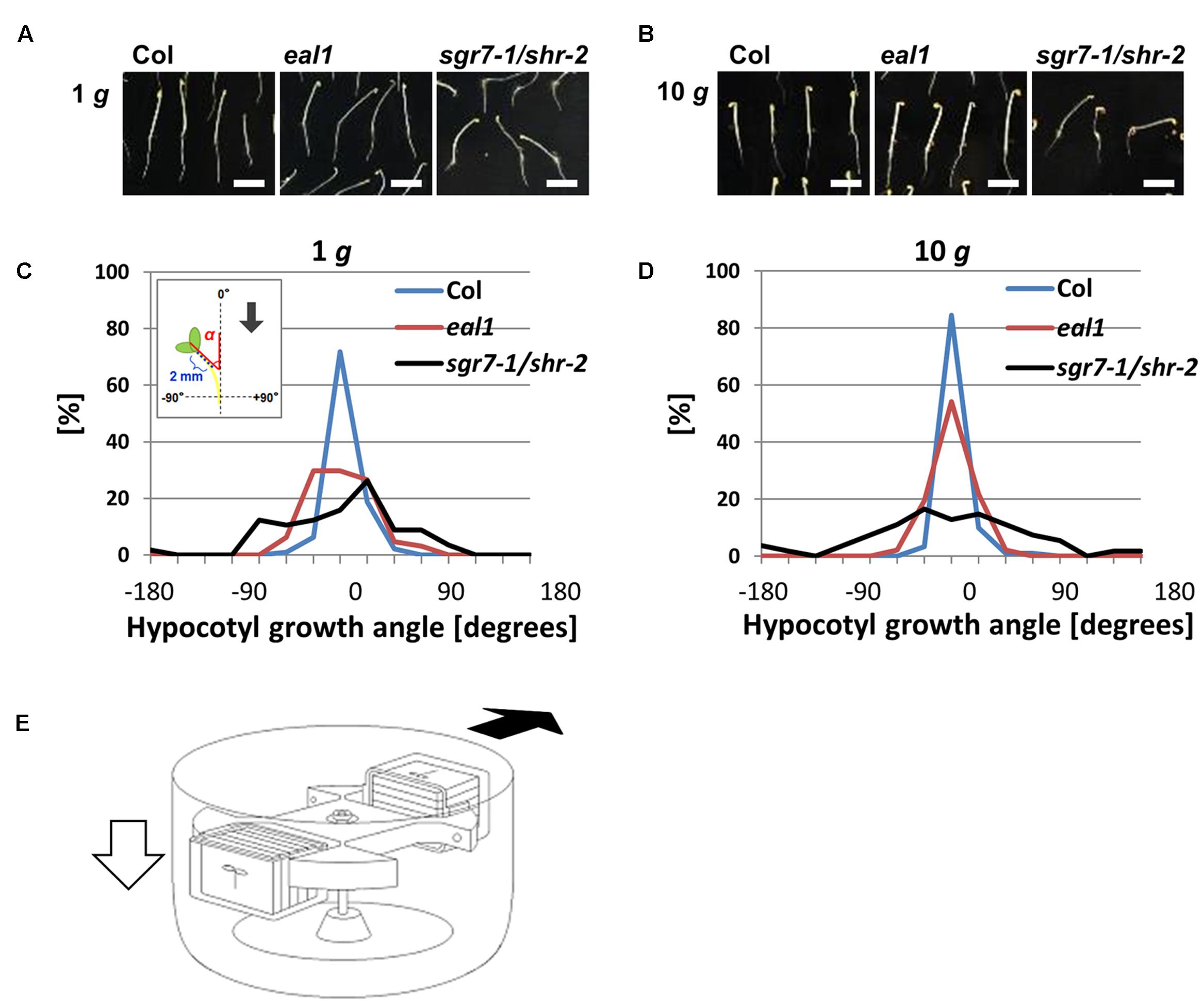
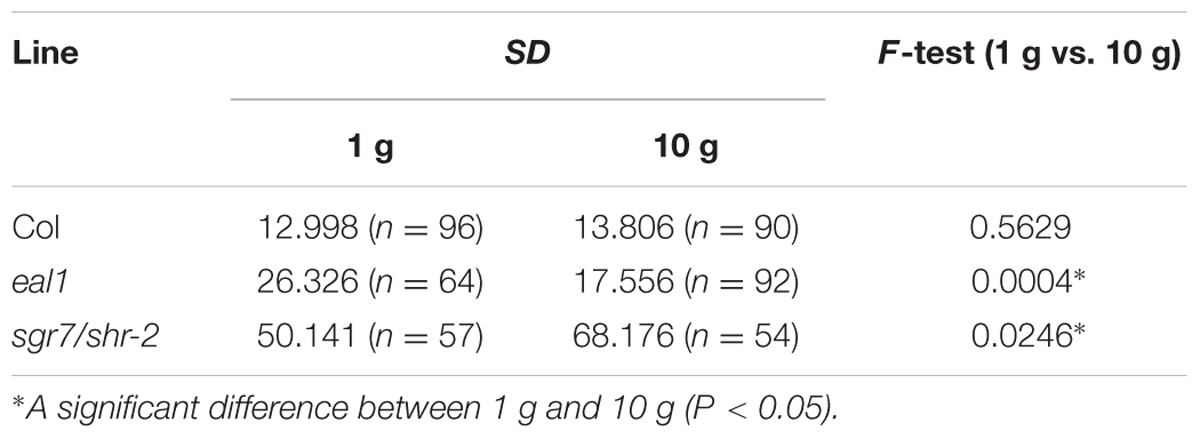
FIGURE 1. Phenotypes of eal1 and sgr7-1/shr-2 (A), (B) 2-day-old Col, eal1, and sgr7-1/shr-2. Etiolated hypocotyls were grown under 1 g (A) or 10 g (B) in the dark. All scale bars indicate 5 mm. (C,D) Distributions of angles in Col, eal1, and sgr7-1/shr-2 (X-axis, angle; Y-axis, percentage), under 1 g (C: n ≥ 57) and 10 g conditions (D: n ≥ 54). The inset denotes a schematic diagram of hypocotyl angles. Growth angle (α) was measured between hypocotyl tip (∼2 mm) and the vertical axis. Arrow indicates the gravity vector. (E) A schematic diagram of mutant screening using a centrifuge. White arrow indicates the gravity vector (1 g before starting rotation/centrifugation). Black arrow indicates the hypergravity vector (10 g after starting rotation/centrifugation).
Analysis of Gravitropism in Hypocotyls and Roots
All the gravitropic angles and growth rates (i.e., root elongation length) were analyzed using ImageJ1. In 3-day-old etiolated hypocotyls, we measured the gravitropic angle formed between the growing direction of 2-mm hypocotyl tip and the vertical axis (Figure 1C, inset; α: hypocotyl growth angle), whereas in 3-day-old etiolated roots, we rotated the roots 90°, kept them in the dark for 12 h, and then measured primary root tip angles and root length (Figure 3A; 𝜃: bending curvature after reorientation of primary roots).
Whole Genome Sequencing and SNP Detection
The eal1 ene1–6 plants were backcrossed to the parental eal1 plants, and the F2 population was screened under hypergravity conditions, as described above. Approximately, 80–300 individuals of the eal1 ene homozygous mutants and the eal1 control mutant were pooled and used to prepare DNA libraries, as reported previously (Uchida et al., 2011). The DNA libraries for all seven lines were sequenced, using Illumina Genome Analyzer IIx (75 bp single read). The resulting sequences were aligned, and SNPs were detected using Strand NGS ver. 2.1 (Strand Scientific Intelligence, Inc.2; Supplementary Table S1).
Filtering SNPs and Predicting Causative Genes
To identify line-specific SNPs, we compared the SNPs in each line to those detected in the other six lines and excluded a SNP when both the position and the base corresponded. Using the remaining SNPs, we searched for SNP-accumulating areas among the chromosomes (Supplementary Figure S2). These SNPs were also used as PCR markers (Supplementary Table S2) for mapping and confirmation of ene single mutants. Further, to predict the causative genes of the ene, synonymous SNPs and SNPs that were located in the intergenic regions or transposable elements were eliminated.
Results
New Screening System Using Hypergravity
When Arabidopsis seedlings are grown on the surface of a vertical agar plate, wild-type hypocotyls grow upward along the gravity vector, whereas the hypocotyls of gravitropic mutants grow in random directions. We measured angles (α) between the gravity vector (1 g) and growth direction in wild-type (Col), eal1, and sgr7-1/shr-2 seedlings (Figures 1A,C). Variance in growth angle was larger in sgr7-1/shr-2 and eal1 mutants than in wild-type plants, confirming that sgr7-1/shr-2 and eal1 were gravitropic mutants at 1 g (Table 1). To apply hypergravity to Arabidopsis seedlings, we set up a light-tight centrifuge in which 50–100 seedlings were continuously exposed for 2 days (Figures 1B,D,E; for detail, see Materials and Methods). Although, the variance of sgr7-1/shr-2 was not reduced at 10 g, likely owing to its lack of endodermis, that of eal1 was reduced (Table 1), which indicated that eal1 mutants retained gravity sensing machinery and that gravity sensitivity was restored. In the present study, we used the hypergravity condition (10 g) rather than the normal gravitational condition (1 g) to eliminate screening errors and to isolate eal1 enhancers.
Isolation of eal1 Enhancer Mutants
To find second mutations in different genes that attenuate gravity sensitivity in eal1, eal1 seeds were mutagenized with EMS. The resultant 57,600 M2 individuals were exposed to 10 g in the new screening system, and we selected 651 M2 candidates, hypocotyls of which were inclined over 30° to the gravity vector (Table 2; first screening). After obtaining the offspring of the M2 candidates, 50 individuals for each line were examined under the same screening condition and 181 M3 candidates were chosen (Table 2; second screening). Since this pool of M3 candidates should include mutants defective in not only gravitropism but also growth, we tested their phototropism as third screening. Finally, we isolated 12 candidates that exhibited normal phototropism (growth) but extremely reduced gravitropism at 10 g.
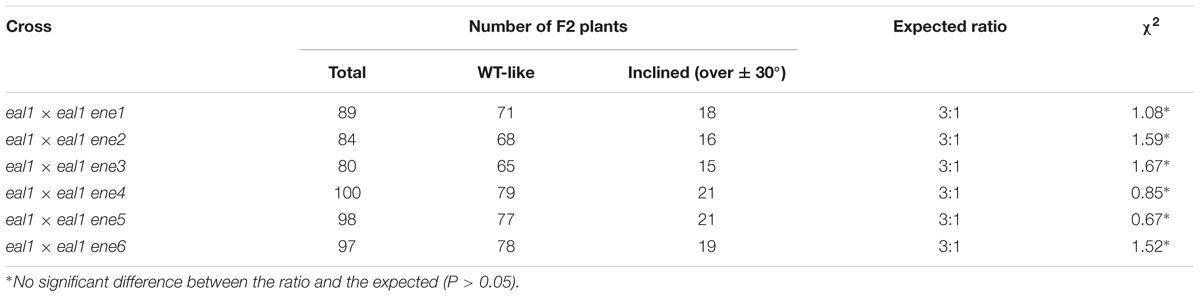
We named the eal1 enhancer lines eal1 ene (enhancer of eal1) and analyzed six of the 12 lines (eal1 ene1–6). Accordingly, eal1 ene1–6 were backcrossed with the parental eal1 line, and the gravitropism of the F2 population was investigated at 10 g. The segregation ratio of wild-type to mutant phenotypes was 3:1 (Table 3), which indicated that each eal1 ene was the result of a single recessive mutation.
Phenotypes of eal1 ene Mutants
We quantified the phenotype of eal1 ene hypocotyls under 1 g and hypergravity conditions (Figures 2A,C). The variance of growth angle was significantly higher in hypocotyls of the eal1 ene1–6 mutants than in those of eal1 at 10 g (Figure 2B), whereas no significant difference was observed at 1 g (Figure 2C).
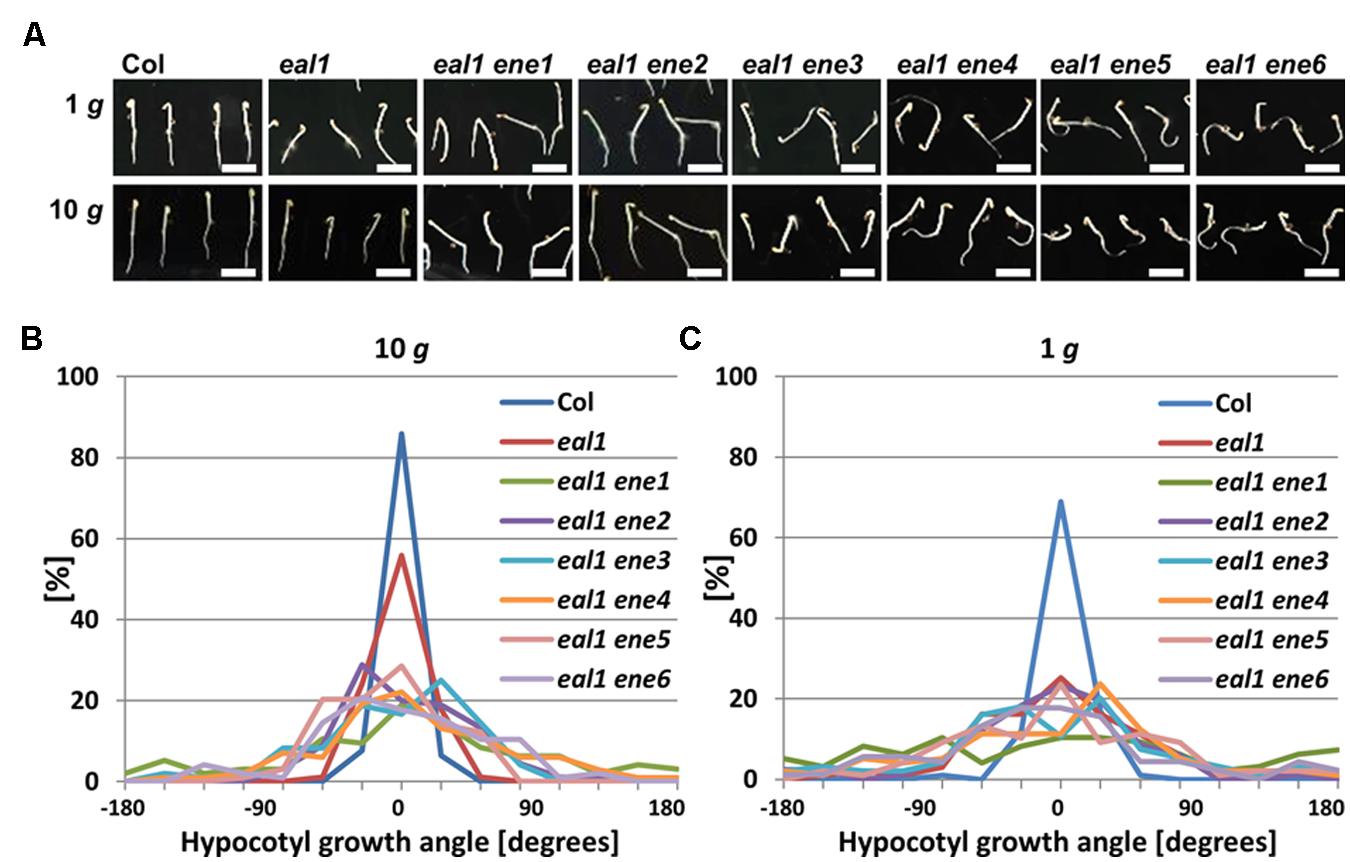
FIGURE 2. Quantification of hypocotyl growth angles in eal1 ene double mutants. (A) 3-day-old Col, eal1, and eal1 ene etiolated hypocotyls grown under 1 or 10 g. All scale bars indicate 5 mm. (B,C) Distributions of angles in Col, eal1, and eal1 ene hypocotyls grown under 10 (B: n ≥ 90) and 1 g conditions (C: n ≥ 82).
Since some eal1 ene mutants showed agravitropic responses in their roots (Figure 2A), we quantified growth rate and gravitropism in all the eal1 ene mutants. We found that root gravitropism was more random in eal1 ene3–6 roots than that in eal1 roots (Figure 3A). Meanwhile, eal1 ene2 plants exhibited slightly reduced gravitropism, and the phenotype of eal1 ene1 was identical to that of eal1 and wild-type plants. There was no statistical difference in the growth rate among wild-type, eal1, and eal1 ene1–6 plants (Figure 3B), and the reduced or agravitropic responses resulted from the ene2–6 mutations, since the parental eal1 mutants exhibited normal gravitropism in primary roots (Fujihira et al., 2000; Morita et al., 2007). Thus, ENE2–6 play a role in genetic pathways for both shoot and root gravitropism, whereas ENE1 is only involved in shoot gravitropism.
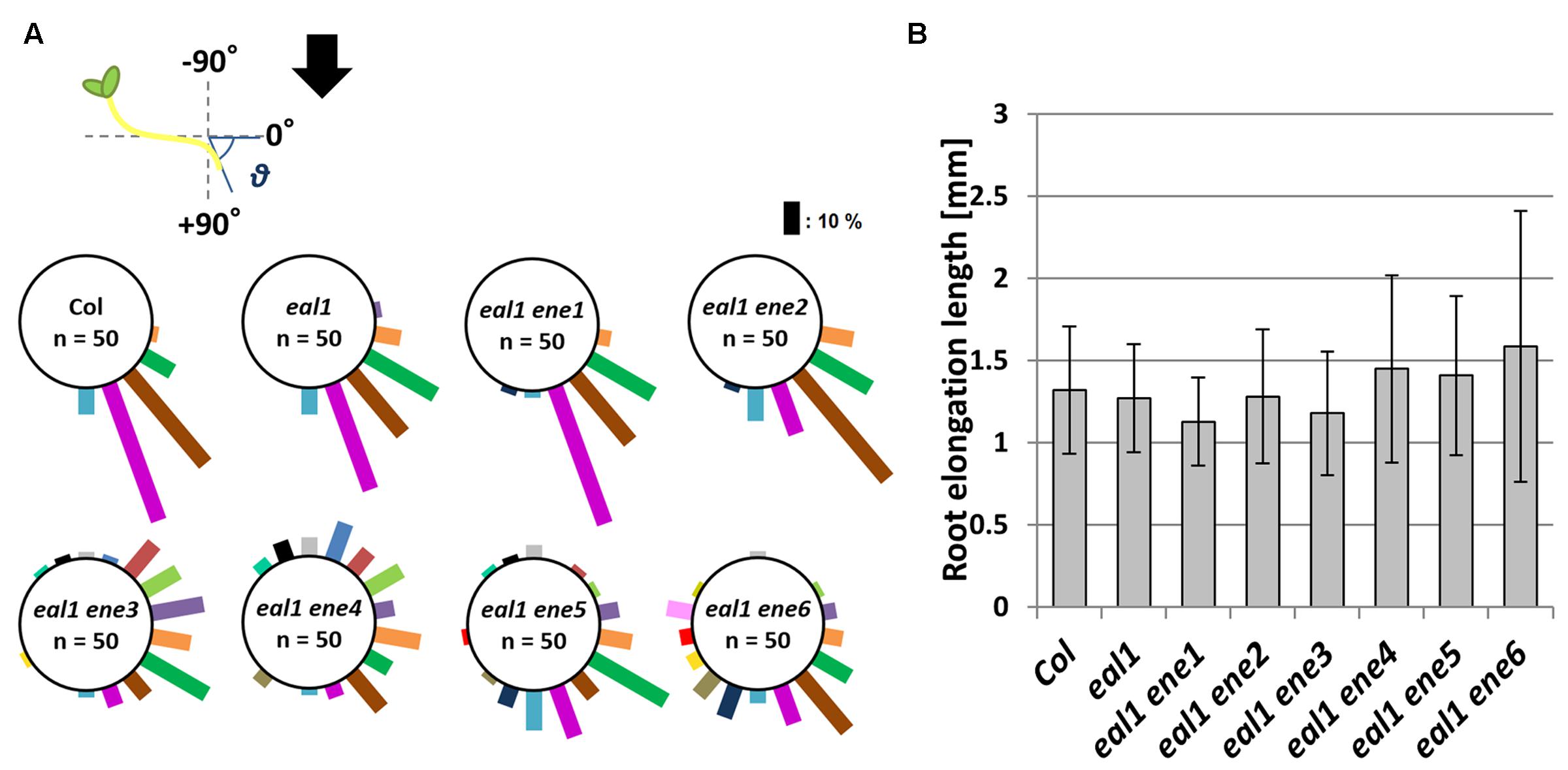
FIGURE 3. Root gravitropism in eal1 ene double mutants. (A) Measuring the angle (𝜃) from primary root tip to the horizontal axis. Arrow indicates the gravity vector. 3-day-old etiolated seedlings were used. The bar length is proportional to the number of plants observed in each 20° bin. (B) The root extension length in each line. Error bar shows standard deviation (SD). No significant difference was observed between eal1 and each eal1 ene (Student’s t-test, P < 0.01).
We also observed morphology, such as leaf shape and lateral organs, in the eal1 enhancer lines at different ages. The morphologies of most of the aerial parts of 12-day-old eal1 ene seedlings were identical to those of the aerial parts of wild-type and eal1 seedlings (Supplementary Figure S1A), and the inflorescence stems of 4-week-old eal1 ene1–6 mutants exhibited gravitropism that was similar to that of eal1 seedlings at 1 g (Supplementary Figure S1B). Since the inflorescence stems in eal1 do not exhibit gravitropism at 1 g (Fujihira et al., 2000; Morita et al., 2007), we assumed that the phenotype was caused by the eal1 mutation. These data support the conclusion that ene1–6 have no obvious phenotypes, other than the gravitropism defect.
ENE3–6 were Revealed by Whole Genome Sequencing and SNP Analysis
Since EMS treatment causes thousands of SNPs in Arabidopsis chromosomes, we needed to eliminate irrelevant SNPs in order to identify ene mutations, using next-generation sequencing (NGS). Thus, we backcrossed eal1 ene1–6 with the parental eal1 line and used 80–300 F2 individuals that exhibited the agravitropic response at 10 g for NGS. Because the EMS-derived SNPs in ene lines are supposed to accumulate near ene mutations, rather than at other chromosomal sites, the frequency of SNPs was examined throughout the chromosomes of each eal1 ene mutant (Supplementary Figure S2).
Our NGS data revealed several regions with high SNP frequencies (Table 4; Supplementary Figure S2), so we targeted these regions to investigate SNPs and candidate genes. One SNP, in ene3, was located at a splice acceptor site in intron 10 of the ALTERED RESPONSE TO GRAVITY 1 (ARG1) gene (2255-1C > T), which is important for hypocotyl and root gravitropism (Fukaki et al., 1997; Harrison and Masson, 2008; Kumar et al., 2008). Therefore, the SNP in ene3 could affect the splicing process of ARG1 and gravitropism. In ene4, we found a SNP in exon 8 of the AUXIN RESISTANT 1 (AUX1) gene (Gly381Asp), which encodes an auxin influx carrier that is necessary for root gravitropism (Marchant et al., 1999). The SNP was located in a transmembrane domain, which is a new allele of AUX1 (Swarup et al., 2004). In ene5, we found a SNP in a lariat intron of the PIN-FORMED 2 (PIN2) gene (3128-17A > G), which encodes an auxin efflux carrier that is necessary for root gravitropism (Chen et al., 1998; Luschnig et al., 1998; Müller et al., 1998; Utsuno et al., 1998), suggesting that the mutation affects both PIN2 splicing and auxin distribution during gravitropism. In ene6, we found a SNP in exon 2 of the PIN2 gene (Pro111Leu). This amino acid substitution was in a transmembrane domain that could be critical for auxin transport. Taken together, our findings of these three genes (ARG1, AUX1, and PIN2) demonstrate the utility of our hypergravity screening system and SNP-based method for isolating and characterizing enhancer mutants in gravitropism, which we used to identify four novel alleles.
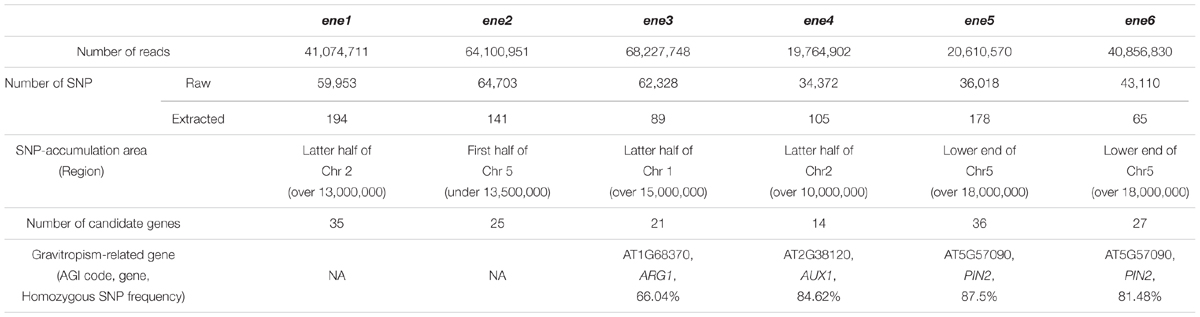
TABLE 4. Prediction of causative genes for ene using next-generation sequencing and SNP-based markers.
ENE3 is an Allele of ARG1
A SNP was located at a splice site in ARG1 in ene3 (Figure 4A). We crossed arg1–3, a line with a mutant allele of ARG1, with eal1 mutant to investigate whether arg1 mutation enhances eal1 phenotype in gravitropism (Harrison and Masson, 2008). The eal1 arg1–3 double mutant showed similar variance to that of eal1 ene3, whereas arg1–3 single mutant showed similar variance to that of eal1 (Figure 4B). This data indicates that arg1 mutation enhances eal1 gravitropism. Further, we obtained the ene3 single mutant and compared its gravitropism to the arg1–3 single mutant under 10 g. Since both single mutants showed eal1-like variances (Figure 4B), each allele has similar enhanced effects on eal1 gravitropism. We also generated transgenic lines carrying a pARG1::ARG1 in arg1–3 or eal1 ene3 background and conducted a transformation-rescue experiment. Although, single arg1–3 mutant showed large variance under 10 g condition, a transgenic line, arg1–3 ARG1#16-10, showed wild-type variance (Figure 4C) indicating that this construct functions properly. Two independent lines, eal1 ene3 ARG1#2-1 and #5-1, showed eal1-like variances in hypocotyl gravitropism (Figure 4C). Since ARG1 rescues the gravitropism defect in eal1 ene3, ene3 is a mutation in ARG1.
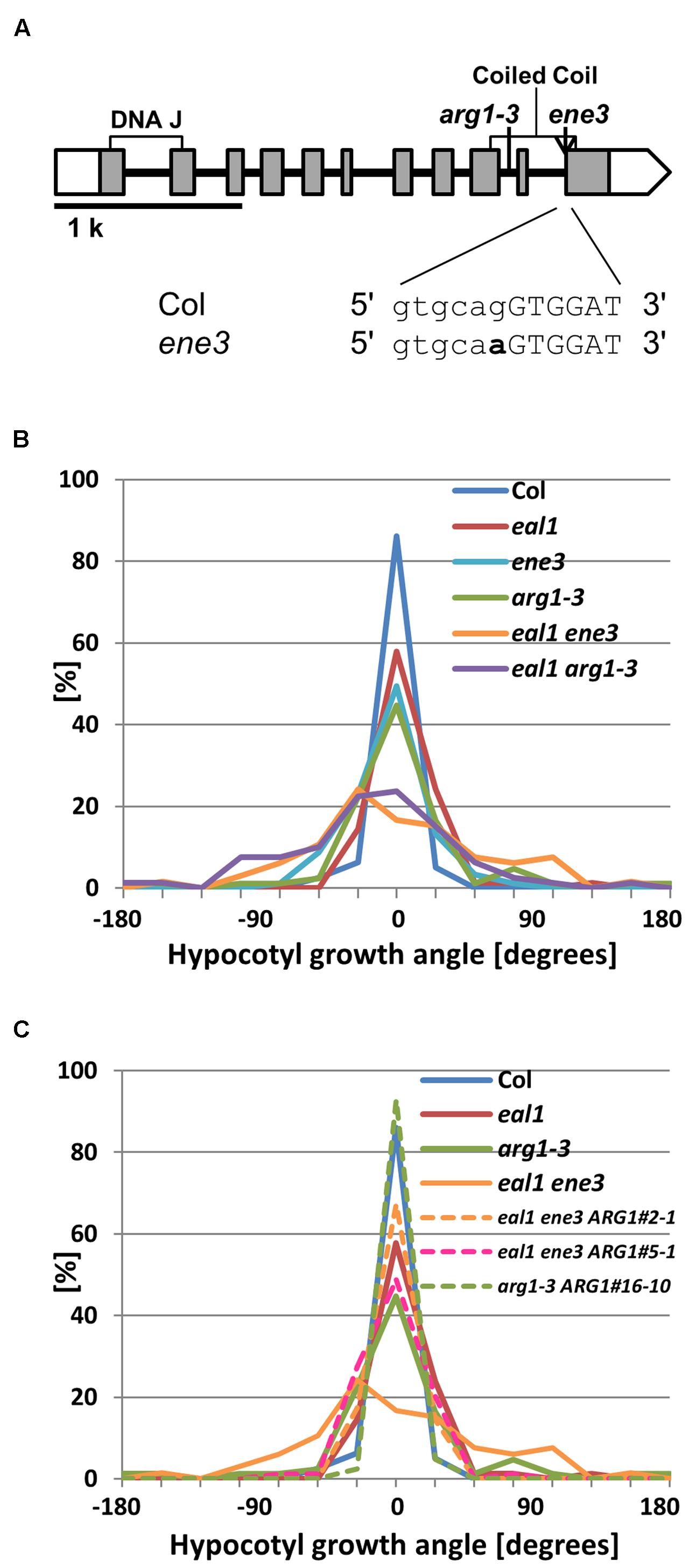
FIGURE 4. (A) ARG1 gene structure. White and shaded boxes indicate UTRs and coding regions, respectively. Black bold lines indicate introns. Two regions of the gene encoding DNA J and coiled coil domains within the protein are shown by open rectangles underneath the gene exons. At the bottom, bold letter indicates ene3 mutation. (B) Quantification of hypocotyl growth angles in ene3 single and the double mutants under 10 g (n ≥ 66). (C) Transformation-rescue experiment (n ≥ 66).
ENE4 is an Allele of AUX1
In ene4, a SNP was found in a region of AUX1 that encodes a transmembrane domain within the corresponding proteins (Figure 5A). We generated eal1 aux1–7 double mutants to confirm whether a mutation in AUX1 can enhance defective gravitropism in eal1 (Pickett et al., 1990). Under 10 g, the variance of growth angle in eal1 aux1–7 was similar to that in eal1 ene4 (Figure 5B). The variance in aux1–7 was similar to that in eal1, indicating that aux1–7 mutation enhances eal1 gravitropism. Further, we obtained ene4 single mutant and compared it to aux1–7 single mutant. Single mutants of ene4 or aux1–7 show eal1-like variances, therefore these mutations have similar enhanced effects on eal1 gravitropism. We conducted an allelism test to determine whether ene4 and aux1–7 are allelic by crossing eal1 ene4 and eal1 aux1–7 (Figure 5B). The variance of F1 hypocotyls was identical to that in eal1 ene4 and eal1 aux1–7. These data demonstrate that ene4 is a mutation in AUX1 and is a new allele of AUX1.
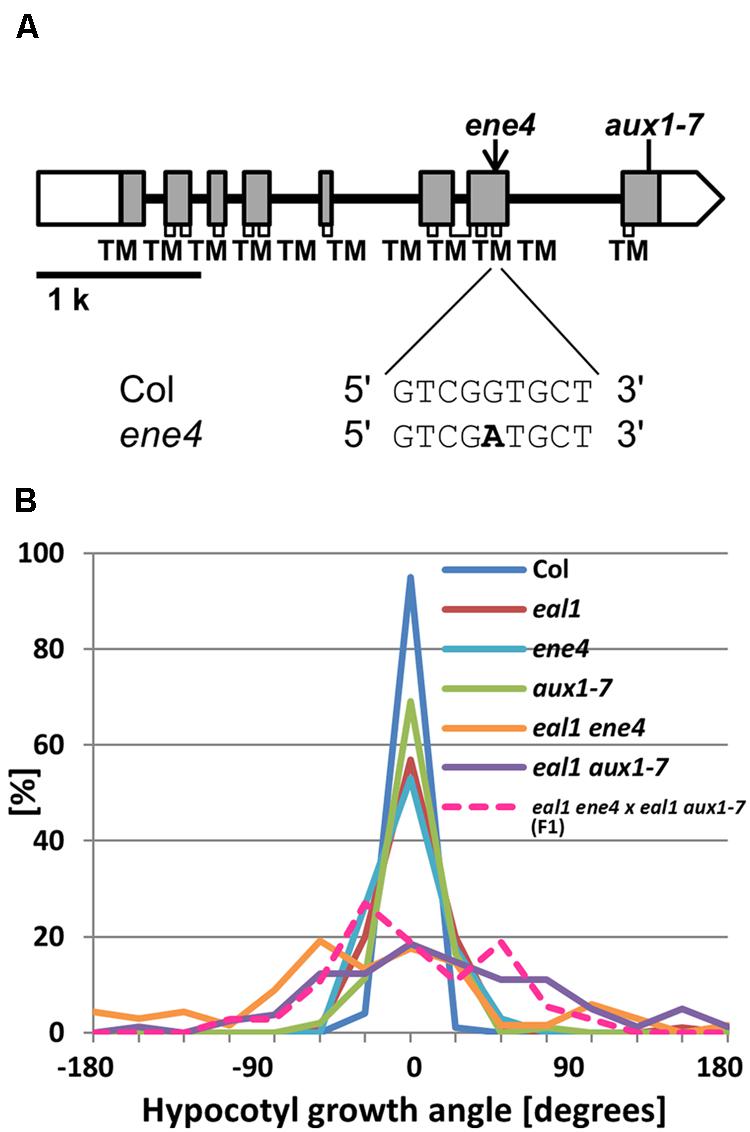
FIGURE 5. (A) AUX1 gene structure. White and shaded boxes indicate UTRs and coding regions, respectively. Black bold lines indicate introns. Regions of the gene encoding transmembrane domains (TMs) within the protein are shown by open rectangles underneath the gene exons. At the bottom, bold letter indicates ene4 mutation. (B) Quantification of hypocotyl growth angles in ene4 single and the double mutants under 10 g, along with F1 progeny from a cross between eal1 ene4 and eal1 aux1–7 plants (n ≥ 37).
ENE5 and ENE6 are Alleles of PIN2
Both lines ene5 and ene6 had a SNP located in PIN2 (Figure 6A). We generated eal1 eir1-1 double mutant and compared it to eal1 ene5 and eal1 ene6 under 10 g (Supplementary Figure S3). Since these variances were similar, ene5 and ene6 were suggested to be allele of PIN2. We obtained ene5 and ene6 single mutants and then compared to eir1-1 single mutant. All three showed wild-type variances. This indicates that these three mutations can enhance eal1 gravitropic defect. To determine whether ene5 and ene6 are in a same gene, we crossed eal1 ene5 to eal1 ene6 and then conducted an allelism test under 10 g (Supplementary Figure S3). The F1 mutants showed a variance similar to those of eal1 ene5 and eal1 ene6 therefore ene5 and ene6 are alleles of a same gene. We focused further on ene6 by crossing eal1 ene6 to eal1 eir1-1 for a second allelism test (Figure 6B). In F1 mutants, the variance was identical to that in parental eal1 ene6 and eal1 eir1-1 mutants. We revealed that both ene5 and ene6 are mutations in PIN2.
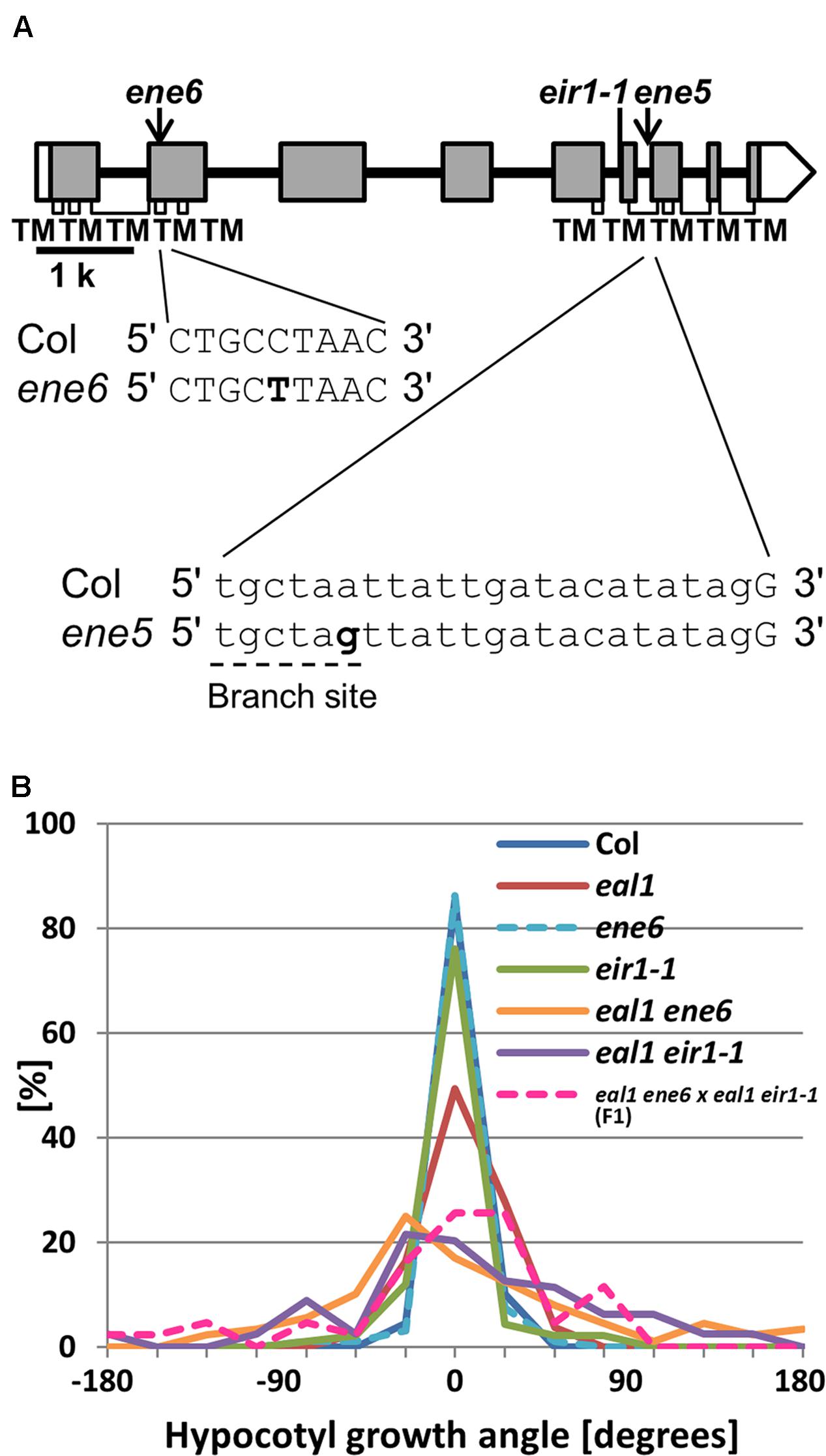
FIGURE 6. (A) PIN2 gene structure. White and shaded boxes indicate UTRs and coding regions, respectively. Black bold lines indicate introns. Regions of the gene encoding TMs with in the protein are shown by open rectangles underneath the gene exons. At the bottom, bold letters indicate ene5 and ene6 mutations. A dashed line indicates the branch site for lariat structure. (B) Quantification of hypocotyl growth angles in ene6 single and the double mutants under 10 g, along with F1 progeny from a cross between eal1 ene6 and eal1 eir1-1 plants (n ≥ 43).
Mapping of ene1 Using SNP Markers
No known gravitropism-related genes were found in the targeted regions of the eal1 ene1 and eal1 ene2 mutants (Table 4). Therefore, the causative genes of ene1 and ene2 phenotypes should be novel. In eal1 ene2, we found SNP-accumulation areas at the center of chromosome 1 and in the first half of chromosome 5 (Supplementary Figure S2). Since all the SNPs in chromosome 1 were located in intergenic regions, we concluded that the causative gene of ene2 was located in the first half of chromosome 5 (Table 4). Meanwhile, we found 25 EMS-derived SNPs within 13.5 Mbp, in the first half of chromosome 5, which contains 25 genes (Table 4).
On the other hand, for ene1, we identified 35 SNPs within 6.2 Mbp, in the latter half of chromosome 2. To identify the causative gene of ene1, we designed new PCR markers that recognize the EMS-derived SNPs and performed map-based cloning (Figure 7A; Supplementary Table S2). Consequently, the responsible SNP was predicted to occur in a ∼650 kbp region between 16,502,619 (AT2G39550) and 17,153,976 (AT2G41150), in which there were five SNPs (Figure 7B). Two SNPs, 16,627,806 (AT2G39840) and 17,079,183 (AT2G40930), were located in introns and did not associate with any splice site or lariat formation. Two other SNPs, 16,619,455 (AT2G39820) and 17,114,526 (AT2G41010), caused amino acid substitutions. The former SNP (Asp203Asn) was located in exon 5 of EIF6B (AT2G39820), which encodes the eukaryotic initiation factor 6 domain (eIF6), whereas the latter SNP (Thr69Ile) was located in exon 1 of AtCaMBP25 (AT2G41010), which encodes a calmodulin binding protein. The last SNP creates a stop codon (Gln205X) at 16,796,973 (AT2G40220) in exon 1 of ABI4 (AT2G40220), which encodes a member of DREB subfamily A-3 of ERF/AP2 transcription factor family. We examined gravitropism in three single mutants of EIF6B, ABI4, and AtCaMBP25 (Figures 7C,D). Both SALK lines, eif6b and atcambp25, showed small variances, which are comparable to wild-type. However, the abi4-1 single mutant had a large variance as compared to wild-type suggesting that ENE1 is likely to be ABI4. This gene has not been reported in association with gravitropism so far. Identification of ENE1 and ENE2 will contribute to further understanding of the mechanism of gravity sensing in Arabidopsis hypocotyls.
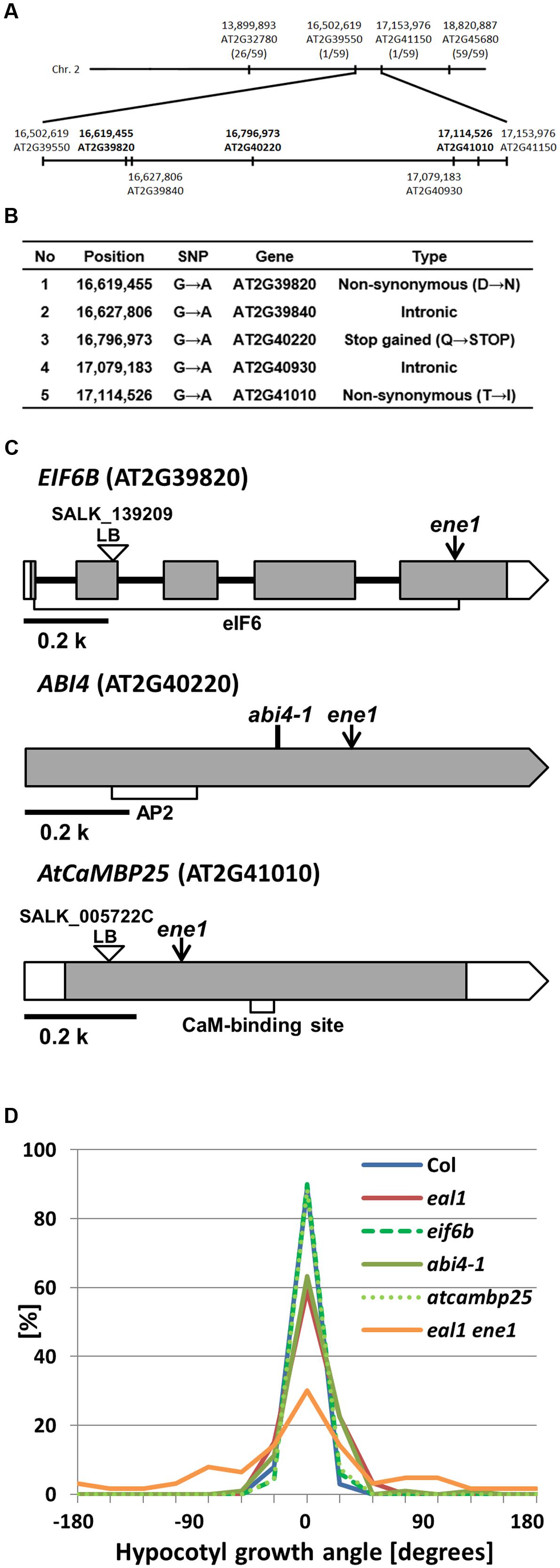
FIGURE 7. Mapping of eal1 ene1 using SNP-based markers. (A) Genomic locations of the PCR markers developed from the detected SNPs. Upper row shows chromosome 2. The positions of four PCR markers are indicated, with the number of recombinants. The ene1 mutation was (mapped on the latter half of chromosome 2, between markers AT2G39550 (16,502,619 bp) and AT2G41150 (17,153,976 bp). The candidate genes (five SNPs) for ene1 mutants are shown on the bottom. Bold letters indicate SNPs in the exon regions. (B) Annotation of five SNPs in the defined region. (C) EIF6B, ABI4, and AtCaMBP25 gene structures. White and shaded boxes indicate UTRs and coding regions, respectively. Black bold lines indicate introns. eIF6, APETALA 2 (AP2), and calmodulin binding domains (CaM-binding site) are shown by open rectangles under the corresponding gene exons in EIF6B, ABI4, and AtCaMBP25, respectively. Arrows indicate the position of SNPs identified in ene1. (D) Quantification of hypocotyl growth angles in single mutants under 10 g (n ≥ 63).)
Discussion
A variety of gravitropic mutants have been reported in Arabidopsis, but their enhancer/suppressor mutants have not been isolated under different gravitational conditions. Using our new screening system and NGS in combination with SNP-based markers, we isolated six eal1 ene mutants with enhanced gravitropism defect. Single and double mutant analyses revealed that three genes responsible are known to be related to gravitropism (ARG1, AUX1, and PIN2). The causative genes for ene3–6 phenotypes were successfully predicted by SNP analyses. We expect that remaining two genes are novel.
ENE3 is an Allele of ARG1
The gene ARG1 is recognized to be important for root and hypocotyl gravitropism (Sedbrook et al., 1999; Boonsirichai et al., 2003) and localization of the auxin efflux carrier PIN3 in columella cells, which are gravity-sensing cells in roots (Harrison and Masson, 2008). The SNP in ARG1 was found in a canonical splice site (GT-AG) in intron 10. Thus, the mutation likely causes abnormal splicing patterns, resulting in pre-mature stop codon in intron by no splicing or in the fifth amino acid in exon 11 by splicing with shifted one base toward the 5′. The ARG1 protein contains a DnaJ-like domain at its N terminus, a putative transmembrane region, and a coiled-coil domain at its C terminus (Sedbrook et al., 1999; Guan et al., 2003). The ene3 mutation located in the coiled-coil region may affect protein-protein interactions.
Single arg1 mutants exhibit decreased gravitropism in their hypocotyls and roots but normal phototropism in their hypocotyls (Sedbrook et al., 1999; Guan et al., 2003), which is similar to the eal1 ene3 phenotype (Figures 2 and 3A; Table 1; third screening), thus, supporting the idea that ene3 is a mutation in ARG1 and represents a new allele. Further, single and double mutant analysis and a transformation-rescue experiment revealed that the eal1 phenotypes are enhanced by the ene3 mutation in ARG1 (Figure 4B), which indicates that ARG1 and EAL1 regulate gravitropism independently. It has been reported that ARG1 proteins in endodermal cells function in hypocotyl gravitropism (Boonsirichai et al., 2003). In the eal1 mutant, a single amino acid deletion (E230X) occurs in the transcription factor SGR7/SHR (Morita et al., 2007). Therefore, in the endodermis, it is likely that the expressions of many gravitropism-related genes, such as SCARECROW (SCR), are downregulated. Nevertheless, because the ene3/arg1 mutation enhances the eal1 phenotypes, the expression of ARG1 may be regulated in a SHR-SCR independent manner.
AUX1 Plays a Role in Hypocotyl Gravitropism
In the eal1 ene4 mutant, we found a mutation in AUX1, which encodes an auxin influx carrier protein that plays a role in differential growth during gravitropism through the modulation of polar auxin transport (Michniewicz et al., 2007). In AUX1, 11 transmembrane regions were predicted by the PRED-TMR2 software, and the SNP (Gly381Asp) was identified in the 10th transmembrane region.
Single aux1 mutants exhibit reduced gravitropism in primary roots (Marchant et al., 1999; Swarup et al., 2004), and normal phototropism in hypocotyls (Okada and Shimura, 1992; Watahiki et al., 1999). In eal1 ene4 mutants, primary root gravitropism and hypocotyl phototropism are similar to single aux1 mutants, suggesting that ENE4 is identical to AUX1. A previous study mentioned that AUX1 functions as a modulator of gravitropism defect in arf7 hypocotyls (data not shown in Stone et al., 2008). Here we explicitly confirm the contribution of AUX1 to hypocotyl gravitropism.
PIN2 Functions in Hypocotyl Gravitropism
PIN2 is an auxin efflux carrier that is critical for differential growth during gravitropism, through its modulation of polar auxin transport (Chen et al., 1998; Luschnig et al., 1998; Müller et al., 1998; Utsuno et al., 1998). In both eal1 ene5 and eal1 ene6, we identified different point mutations in PIN2 (Table 4). PIN2 has 10 transmembrane regions (Müller et al., 1998) and the SNP (3128-17A > G) is located at the branch site for lariat structure within a region of the gene that encodes the seventh transmembrane domain, which suggests that a stop codon is generated in the intron. For the ene6 mutant, amino acid substitution (Pro111Leu) occurs in the third transmembrane region and should affect the function of auxin transport.
According to previous reports, single pin2 mutants exhibit decreased gravitropism in roots (Chen et al., 1998; Luschnig et al., 1998; Müller et al., 1998; Utsuno et al., 1998) and normal phototropism in hypocotyls (Wan et al., 2012). Both eal1 ene5 and eal1 ene6 mutants exhibit pin2 mutant-like phenotypes regarding the gravitropism in primary roots and the phototropism in hypocotyls. Our double mutant analysis and allelism test revealed that ene5 and ene6 are different alleles of PIN2 (Supplementary Figure S3; Figure 6B). pin2 hypocotyls are supposed to show a transient defect in gravitropism, although the data is not shown (E. R. and Kenneth L. Poff, unpublished results in Chen et al., 1998). Further, in 5-day-old etiolated hypocotyls, PIN2 is expressed in roots and also in shoots including hypocotyls and cotyledons (Chen et al., 1998). These results indicate that PIN2 functions in hypocotyl gravitropism, owing to overlap with the eal1 background.
ENE1 is a Novel Gene Related to Gravitropism in Hypocotyls
In the present study, we identified five possible mutations in the ene1 mutant, in a genomic region without any previously reported gravitropism-related genes (Figure 7B). Thus, the gene affected by the ene1 mutation is a novel gravitropism-related gene. Among the genes found in the region, ABI4 is known to play a role in sensitivity to abscisic acid during seed germination (Finkelstein, 1994) and is highly expressed in seeds and weakly expressed in hypocotyls and shoots (Finkelstein et al., 1998; Söderman et al., 2000). Previous studies have also suggested that ABI4 functions as a transcription factor (Finkelstein et al., 1998; Gregorio et al., 2014). The eal1 ene1 mutant has a mutation in ABI4 that generates a stop codon (Gln205X) in a transcriptional activation region. The expression levels of the downstream factors of ABI4 may be altered, leading to reduced gravitropism in hypocotyls.
The other candidate mutations were found in AtCaMBP25 and EIF6B. AtCaMBP25 is a calmodulin binding protein that is induced in response to various environmental stresses, such as drought, low temperature, and salt stress (Perruc et al., 2004). Due to the mutation in AtCaMBP25, the amino acid at position 69 was changed from hydrophilic threonine to hydrophobic isoleucine. Since it is known that cytosolic calcium signal and calmodulin network is important for gravity sensing in hypocotyls (Toyota and Gilroy, 2013; Toyota et al., 2013a) and roots (Sinclair and Trewavas, 1997), AtCaMBP25 might be involved in a downstream signal in an early gravitropic response. Meanwhile, eIF6 is a eukaryotic initiation factor concerned with the formation of the ribosome complex, and has two homologs in Arabidopsis thaliana: EIF6A and EIF6B (Guo et al., 2011; Moore et al., 2016). The eIF6 domain is composed of 206 amino acids. In this mutation, residue 201 is changed from aspartic acid to asparagine due to the mutation. This single amino acid substitution could affect the function of EIF6B as a translation initiator.
Although, the other possible mutations are located in introns of TOPP4 (AT2G39840) and AtUBP5 (AT2G40930) in the targeted region of ene1, these genes are unlikely to be responsible because the mutations do not occur in the splice site or the lariat site. In addition, only abi4-1 mutants showed gravitropism defect under 10 g by the single mutant analysis (Figure 7D). We concluded that the causative gene of ene1 might be ABI4, the function of which has not been clarified in gravitropism.
Enhancer Mutations Can be Mapped Efficiently Using Hypergravity and Parental Crossing
The hypocotyls of Arabidopsis eal1 mutants exhibit exceptionally weak gravitropism at 1 g. Thus, it is extremely difficult to isolate an enhancer mutant showing further reduced gravitropism at 1 g. In the present study, we used hypergravity to enhance the gravitropic response in eal1, which increased the efficiency of isolating enhancer lines. After identifying enhancer candidates, responsible genes were investigated using NGS. However, using SNP analysis to predict candidate genes has limitations, despite the adequate number of genomes and coverage (James et al., 2013). In conventional mapping, the F2 population is crossed with a different accession in which there is an abundance of polymorphic markers. Therefore, if used conventional methods to map eal1 enhancer mutations, 1/16 of the F2 population would be homozygous for both eal1 and ene mutations. In the present study, by crossing to the parental eal1, we expected 1/4 of the F2 population to be double mutant, eal1 ene. We developed new PCR markers based on SNPs that were identified by NGS and drastically reduced the number of candidate genes. Indeed, the mapping of eal1 ene1 in 59 F2 mutants allowed us to narrow the region containing the causative ene1 gene from ∼6.2 Mbp to 650 kbp and reduced the number of candidate genes from 35 to 5. We can apply this strategy to identify the causative gene of ene3 as well. Our unique screening and mapping system enabled us to discover new enhancer lines and to provide insight into the gravity sensory machinery of plants.
Author Contributions
MTM and MTo conceived and designed the study. TK advised on the experimental design of NGS. AM, MTo, MS, and MM performed experiments, and AM and MS analyzed NGS data. MTM, MTo, AM, and MTa interpreted data. AM wrote the manuscript and all authors reviewed and edited the manuscript.
Funding
This work was supported by Program for Leading Graduate Schools “Integrative Graduate Education and Research in Green Natural Sciences,” MEXT, Japan and by Plant Global Education Project, Graduate School of Biological Sciences, Nara Institute of Science and Technology, Japan.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer PM declared a shared affiliation, though no other collaboration, with one of the authors MT to the handling Editor, who ensured that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
We would like to thank Dr. Kotaro Yamamoto (Hokkaido University, Japan) for eal1 seeds and Dr. Simon Gilroy (University of Wisconsin-Madison, USA) for helpful comments. arg1–3 (SALK_024542) was a kind gift from Dr. Patrick Masson (University of Wisconsin-Madison, USA). We also thank Kaori Kaminoyama for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01443
Footnotes
References
Blancaflor, E. B., and Masson, P. H. (2003). Plant gravitropism. Unraveling the ups and downs of a complex process. Plant Physiol. 133, 1677–1690. doi: 10.1104/pp.103.032169
Boonsirichai, K., Sedbrook, J. C., Chen, R., Gilroy, S., and Masson, P. H. (2003). ALTERED RESPONSE TO GRAVITY is a peripheral membrane protein that modulates gravity-induced cytoplasmic alkalinization and lateral auxin transport in plant statocytes. Plant Cell 15, 2612–2625. doi: 10.1105/tpc.015560
Chen, R., Hilson, P., Sedbrook, J., Rosen, E., Caspar, T., and Masson, P. H. (1998). The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc. Natl. Acad. Sci. U.S.A. 95, 15112–15117. doi: 10.1073/pnas.95.25.15112
Finkelstein, R. R. (1994). Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 5, 765–771. doi: 10.1046/j.1365-313X.1994.5060765.x
Finkelstein, R. R., Wang, M. L., Lynch, T. J., Rao, S., and Goodman, H. M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10, 1043–1054. doi: 10.1105/tpc.10.6.1043
Fitzelle, K. J., and Kiss, J. Z. (2001). Restoration of gravitropic sensitivity in starch-deficient mutants of Arabidopsis by hypergravity. J. Exp. Bot. 52, 265–275. doi: 10.1093/jexbot/52.355.265
Fujihira, K., Kurata, T., Watahiki, M. K., Karahara, I., and Yamamoto, K. T. (2000). An agravitropic mutant of Arabidopsis, endodermal-amyloplast less 1, that lacks amyloplasts in hypocotyl endodermal cell layer. Plant Cell Physiol. 41, 1193–1199. doi: 10.1093/pcp/pcd046
Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). The RHG gene is involved in root and hypocotyl gravitropism in Arabidopsis thaliana. Plant Cell Physiol. 38, 804–810. doi: 10.1093/oxfordjournals.pcp.a029238
Fukaki, H., Wysocka-Diller, J., Kato, K., Fujisawa, H., Benfey, P. N., and Tasaka, M. (1998). Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J. 14, 425–430. doi: 10.1046/j.1365-313X.1998.00137.x
Gregorio, J., Hernández-Bernal, A. F., Cordoba, E., and León, P. (2014). Characterization of evolutionarily conserved motifs involved in activity and regulation of the ABA- INSENSITIVE (ABI) 4 transcription factor. Mol. Plant. 7, 422–436. doi: 10.1093/mp/sst132
Guan, C., Rosen, E. S., Boonsirichai, K., Poff, K. L., and Masson, P. H. (2003). The ARG1-LIKE2 gene of Arabidopsis functions in a gravity signal transduction pathway that is genetically distinct from the PGM pathway. Plant Physiol. 133, 100–112. doi: 10.1104/pp.103.023358
Guo, J., Wang, S., Valerius, O., Hall, H., Zeng, Q., Li, J. F., et al. (2011). Involvement of Arabidopsis RACK1 in protein translation and its regulation by abscisic acid. Plant Physiol. 155, 370–383. doi: 10.1104/pp.110.160663
Harrison, B. R., and Masson, P. H. (2008). ARL2, ARG1 and PIN3 define a gravity signal transduction pathway in root statocytes. Plant J. 53, 380–392.
Hartwig, B., James, G. V., Konrad, K., Schneeberger, K., and Turck, F. (2012). Fast isogenic mapping-by-sequencing of ethyl methanesulfonate-induced mutant bulks. Plant Physiol. 160, 591–600. doi: 10.1104/pp.112.200311
Helariutta, Y., Fukaki, H., Wysocka-Diller, J., Nakajima, K., Jung, J., Sena, G., et al. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555–567. doi: 10.1016/S0092-8674(00)80865-X
James, G. V., Patel, V., Nordström, K. J., Klasen, J. R., Salomé, P. A., Weigel, D., et al. (2013). User guide for mapping-by-sequencing in Arabidopsis. Genome Biol. 14, R61. doi: 10.1186/gb-2013-14-6-r61
Knight, T. A. (1806). On the direction of the radicle and germen during the vegetation of seeds. Philos. Trans. R. Soc. Lond. 96, 99–108. doi: 10.1098/rstl.1806.0006
Kumar, N., Stevens, M. H., and Kiss, J. Z. (2008). Plastid movement in statocytes of the arg1 (altered response to gravity) mutant. Am. J. Bot. 95, 177–184. doi: 10.3732/ajb.95.2.177
Liu, Y. G., Shirano, Y., Fukaki, H., Yanai, Y., Tasaka, M., Tabata, S., et al. (1999). Complementation of plant mutants with large genomic DNA fragments by a transformation- competent artificial chromosome vector accelerates positional cloning. Proc. Natl. Acad. Sci. U.S.A. 96, 6535–6540. doi: 10.1073/pnas.96.11.6535
Luschnig, C., Gaxiola, R. A., Grisafi, P., and Fink, G. R. (1998). EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 12, 2175–2187. doi: 10.1101/gad.12.14.2175
Marchant, A., Kargul, J., May, S. T., Muller, P., Delbarre, A., Perrot-Rechenmann, C., et al. (1999). AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 18, 2066–2073. doi: 10.1093/emboj/18.8.2066
Michniewicz, M., Brewer, P. B., and Friml, J. (2007). Polar auxin transport and asymmetric auxin distribution. Arabidopsis Book 5:e0108. doi: 10.1199/tab.0108
Moore, M., Gossmann, N., and Dietz, K. J. (2016). Redox regulation of cytosolic translation in plants. Trends Plant Sci. 21, 388–397. doi: 10.1016/j.tplants.2015.11.004
Morita, T. M. (2010). Directional gravity sensing in gravitropism. Annu. Rev. Plant Biol. 61, 705–720. doi: 10.1146/annurev.arplant.043008.092042
Morita, T. M., Saito, C., Nakano, A., and Tasaka, M. (2007). endodermal-amyloplast less 1 is a novel allele of SHORT-ROOT. Adv. Space Res. 39, 1127–1133. doi: 10.1016/j.asr.2006.12.020
Müller, A., Guan, C., Gälweiler, L., Tänzler, P., Huijser, P., Marchant, A., et al. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17, 6903–6911. doi: 10.1093/emboj/17.23.6903
Nakamura, M., Toyota, M., Tasaka, M., and Morita, M. T. (2011). An Arabidopsis E3 ligase, SHOOT GRAVITROPISM9, modulates the interaction between statoliths and F-actin in gravity sensing. Plant Cell 23, 1830–1848. doi: 10.1105/tpc.110.079442
Okada, S., and Shimura, Y. (1992). Mutational analysis of root gravitropism and phototropism of Arabidopsis thaliana seedlings. Aust. J. Plant Physiol. 19, 439–448. doi: 10.1071/PP9920439
Perruc, E., Charpenteau, M., Ramirez, B. C., Jauneau, A., Galaud, J. P., Ranjeva, R., et al. (2004). A novel calmodulin-binding protein functions as a negative regulator of osmotic stress tolerance in Arabidopsis thaliana seedlings. Plant J. 38, 410–420. doi: 10.1111/j.1365-313X.2004.02062.x
Pickett, F. B., Wilson, A. K., and Estelle, M. (1990). The aux1 mutation of Arabidopsis confers both auxin and ethylene resistance. Plant Physiol. 94, 1462–1466. doi: 10.1104/pp.94.3.1462
Saito, C., Morita, T. M., Kato, T., and Tasaka, M. (2005). Amyloplasts and vacuolar membrane dynamics in the living graviperceptive cell of the Arabidopsis inflorescence stem. Plant Cell 17, 548–558. doi: 10.1105/tpc.104.026138
Sedbrook, J. C., Chen, R., and Masson, P. H. (1999). ARG1 (altered response to gravity) encodes a DnaJ-like protein that potentially interacts with the cytoskeleton. Proc. Natl. Acad. Sci. U.S.A. 96, 1140–1145. doi: 10.1073/pnas.96.3.1140
Shimada, T. L., Shimada, T., and Hara-Nishimura, I. (2010). A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 61, 519–528. doi: 10.1111/j.1365-313X.2009.04060.x
Sinclair, W., and Trewavas, A. J. (1997). Calcium in gravitropism. A re-examination. Planta 203, S85–S90.
Söderman, E. M., Brocard, I. M., Lynch, T. J., and Finkelstein, R. R. (2000). Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 124, 1752–1765. doi: 10.1104/pp.124.4.1752
Stone, B. B., Stowe-Evans, E. L., Harper, R. M., Celaya, R. B., Ljung, K., Sandberg, G., et al. (2008). Disruptions in AUX1-dependent auxin influx alter hypocotyl phototropism in Arabidopsis. Mol. Plant 1, 129–144. doi: 10.1093/mp/ssm013
Swarup, R., Kargul, J., Marchant, A., Zadik, D., Rahman, A., Mills, R., et al. (2004). Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 16, 3069–3083. doi: 10.1105/tpc.104.024737
Toyota, M., Furuichi, M., Sokabe, M., and Tatsumi, H. (2013a). Analyses of a gravistimulation- specific Ca2+ signature in Arabidopsis using parabolic flights. Plant Physiol. 163, 543–554. doi: 10.1104/pp.113.223313
Toyota, M., and Gilroy, S. (2013). Gravitropism and mechanical signaling in plants. Am. J. Bot. 100, 111–125. doi: 10.3732/ajb.1200408
Toyota, M., Ikeda, N., Sawai-Toyota, S., Kato, T., Gilroy, S., Tasaka, M., et al. (2013b). Amyloplast displacement is necessary for gravisensing in Arabidopsis shoots as revealed by a centrifuge microscope. Plant J. 76, 648–660. doi: 10.1111/tpj.12324
Uchida, N., Sakamoto, T., Kurata, T., and Tasaka, M. (2011). Identification of EMS-induced causal mutations in a non-reference Arabidopsis thaliana accession by whole genome sequencing. Plant Cell Physiol. 52, 716–722. doi: 10.1093/pcp/pcr029
Utsuno, K., Shikanai, T., Yamada, Y., and Hashimoto, T. (1998). AGR, an Agravitropic locus of Arabidopsis thaliana, encodes a novel membrane-protein family member. Plant Cell Physiol. 39, 1111–1118. doi: 10.1093/oxfordjournals.pcp.a029310
Wan, Y., Jasik, J., Wang, L., Hao, H., Volkmann, D., Menzel, D., et al. (2012). The signal transducer NPH3 integrates the phototropin1 photosensor with PIN2- based polar auxin transport in Arabidopsis root phototropism. Plant Cell. 24, 551–565. doi: 10.1105/tpc.111.094284
Keywords: gravitropism, enhancer mutant, hypergravity, whole genome sequencing, Arabidopsis thaliana
Citation: Mori A, Toyota M, Shimada M, Mekata M, Kurata T, Tasaka M and Morita MT (2016) Isolation of New Gravitropic Mutants under Hypergravity Conditions. Front. Plant Sci. 7:1443. doi: 10.3389/fpls.2016.01443
Received: 29 July 2016; Accepted: 09 September 2016;
Published: 29 September 2016.
Edited by:
Elison B. Blancaflor, Samuel Roberts Noble Foundation, USAReviewed by:
Enamul Huq, University of Texas at Austin, USAPatrick H. Masson, University of Wisconsin-Madison, USA
Copyright © 2016 Mori, Toyota, Shimada, Mekata, Kurata, Tasaka and Morita. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Miyo T. Morita, mimorita@agr.nagoya-u.ac.jp
 Akiko Mori
Akiko Mori Masatsugu Toyota
Masatsugu Toyota Masayoshi Shimada4
Masayoshi Shimada4 Tetsuya Kurata
Tetsuya Kurata Masao Tasaka
Masao Tasaka Miyo T. Morita
Miyo T. Morita