Corrigendum: The Differences between NAD-ME and NADP-ME Subtypes of C4 Photosynthesis: More than Decarboxylating Enzymes
- 1BioDiscovery Institute and Department of Biological Sciences, University of North Texas, Denton, TX, USA
- 2BioEnergy Science Center, US Department of Energy, Oak Ridge, TN, USA
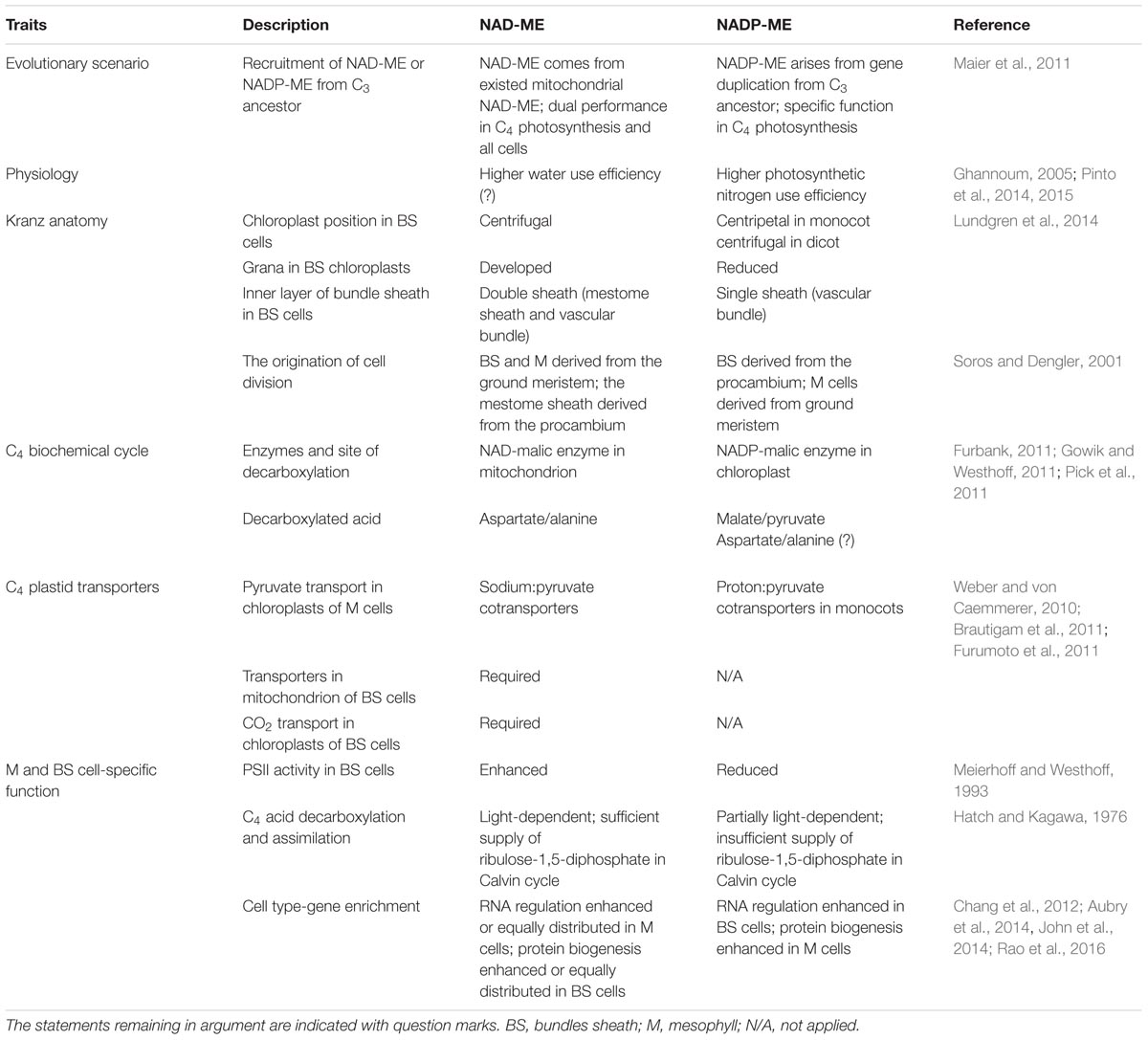
As an adaptation to changing climatic conditions that caused high rates of photorespiration, C4 plants have evolved to display higher photosynthetic efficiency than C3 plants under elevated temperature, high light intensities, and drought. The C4 plants independently evolved more than 60 times in 19 families of angiosperms to establish similar but not uniform C4 mechanisms to concentrate CO2 around the carboxylating enzyme Rubisco (ribulose bisphosphate carboxylase oxygenase). C4 photosynthesis is divided into at least two basic biochemical subtypes based on the primary decarboxylating enzymes, NAD-dependent malic enzyme (NAD-ME) and NADP-dependent malic enzyme (NADP-ME). The multiple polygenetic origins of these subtypes raise questions about the association of C4 variation between biochemical subtypes and diverse lineages. This review addresses the differences in evolutionary scenario, leaf anatomy, and especially C4 metabolic flow, C4 transporters, and cell-specific function deduced from recently reported cell-specific transcriptomic, proteomic, and metabolic analyses of NAD-ME and NADP-ME subtypes. Current omic analysis has revealed the extent to which component abundances differ between the two biochemical subtypes, leading to a better understanding of C4 photosynthetic mechanisms in NAD-ME and NADP-ME subtypes.
Introduction
In the warm-climate zones, C4 plants occupy nearly all grasslands and are a major component of the flora and biomass production through their improved photosynthetic, water and nutrient-use efficiencies (Sage, 2004; Gowik and Westhoff, 2011). C4 photosynthesis in C4 plants is not a single metabolic pathway. It has been established by a series of biochemical and morphological modifications to concentrate CO2 at the site of ribulose bisphosphate carboxylase oxygenase (Rubisco; Sage, 2004). In all C4 plants, CO2 is initially fixed by phosphoenolpyruvate (PEP) carboxylase. The resulting four-carbon acids are transported to an interior compartment where Rubisco is localized. Here, CO2 is released by a decarboxylating enzyme specific for the four carbon acid, and assimilated by Rubisco through the Calvin cycle. The decarboxylation reaction also produces a three-carbon acid, which diffuses back to the compartment where PEP carboxylase is located (Hatch, 1987; Sage, 2004; Sommer et al., 2012). Almost all C4 plants require the coordination of mesophyll (M) and bundle sheath (BS) cells (called Kranz anatomy) to separate primary and secondary carbon fixation reactions, while a few exceptions use internal subcellular compartmentalization within a single cell (Hatch, 1987; Offermann et al., 2015).
Historically, C4 photosynthesis in traditional text books has been classified into three subtypes based on the predominant decarboxylating enzymes of the four carbon acid, NAD-dependent malic enzyme (NAD-ME), NADP-dependent malic enzyme (NADP-ME), and PEP carboxykinase (PEPCK) (Hatch, 1987). However, multiple pieces of evidence challenge the establishment of the PEPCK subtype; no pure PEPCK-type C4 species has been discovered (Sage, 2004), and the robust model analysis of “pure PEPCK type” indicates the imbalance of energy requirements in BS and M cells (Wang et al., 2014). Therefore, currently NAD-ME and NADP-ME subtypes are suggested as distinct C4 biochemical pathways, both with or without the additional service of the PEPCK pathway. Many productive cereal, forage, and biofuel crops belong to either the NADP-ME C4 subtype, for example, maize (Zea mays), sugarcane (Saccharum spp.), and sorghum (Sorghum bicolor), or to the NAD-ME subtype, for example, switchgrass (Panicum virgatum L.), pearl millet [Pennisetum glaucum (L.) R. Br], and amaranth (Amaranthaceae) (Edwards and Walker, 1983).
During the past decade, high throughput tools have made it possible to quantify the transcriptome, proteome, and metabolome at the cell- or tissue-levels (Metzker, 2010). Such applications have expanded the borders and enhanced our knowledge of C4 photosynthesis, which was first reported in the 1950s. In this review, we focus on the differences associated with C4 photosynthesis in NAD-ME and NADP-ME subtypes in terms of genetic, physiological, cytological, biochemical, and molecular traits.
Evolutionary Scenarios of NAD-ME and NADP-ME Subtypes
The evolution of C4 photosynthesis has been achieved over 60 times through individually adaptive steps in 19 families of angiosperms (Sage, 2004), and was hypothetically triggered by the decrease of atmospheric CO2 concentration and plant hydraulics (Christin et al., 2008; Osborne and Sack, 2012). The NAD-ME and NADP-ME subtypes represent almost equal numbers of genera in the eudicots, and the NADP-ME subtype dominates in monocot families (Sage et al., 2011).
The distinct subtypes and lineages of C4 plants were hypothesized to have evolved in adaptation to selective pressure such as shortage of nitrogen and water (Liu and Osborne, 2015; Brautigam and Gowik, 2016). Global geographic surveys of C4 grasses have shown that the NAD-ME subtype occurs more in drier areas and the percentage of NADP-ME subtypes increases with annual precipitation (Vogel et al., 1978; Hattersley, 1992; Taub, 2000). Correspondingly, the largely NAD-ME grass lineage Chloridoideae exhibits a significantly greater enhancement of water use efficiency than NADP-ME grasses under drought condition, due to its leaf structure and faster leaf curling rates (Ghannoum et al., 2002; Liu and Osborne, 2015). High correlation was observed between photosynthetic nitrogen use efficiency and the NADP-ME subtype. Plants in the NADP-ME subtype (except those in the Aristidoideae tribe) tend to have higher photosynthetic nitrogen use efficiency compared with other C4 grasses under adequate or deficient nitrogen supply (Taub and Lerdau, 2000; Ghannoum, 2005; Pinto et al., 2014, 2015). A reduced content of nitrogen and faster Rubisco activity in leaves contribute to better nitrogen-use efficiency in NADP-ME grasses (Ghannoum, 2005). However, the association of C4 subtypes with particular physiological traits, and whether the optimization of nitrogen or water usage drives the evolution of at least some C4 lineages, remain to be determined.
Multiple origins of C4 photosynthesis have been suggested as C3 to C4 transitions and evolutionary conversions between two C4 subtypes (Grass Phylogeny Working Group II, 2012; Washburn et al., 2015). Figure 1 shows a simplified example of a phylogenetic tree with selected families of grasses and three origins (black squares in Figure 1) are considered as the evolutionary conversions between NAD-ME and NADP-ME subtypes (Grass Phylogeny Working Group II, 2012; Washburn et al., 2015). Three models have been proposed to describe the evolutionary divergence of C4 subtypes. One places the NAD-ME subtype as the ancestral C4 subtype, with the NADP-ME subtype evolving from it (Gutierrez et al., 1974; Washburn et al., 2015). The second model proposes that the NAD-ME and NADP-ME subtypes were shared at some level in a common C4 ancestor, then individually predominated in distinct lineages (Washburn et al., 2015). In the third model, NAD-ME and NADP-ME subtypes evolved independently from a C3–C4 intermediate as their most recent common ancestor (Washburn et al., 2015).
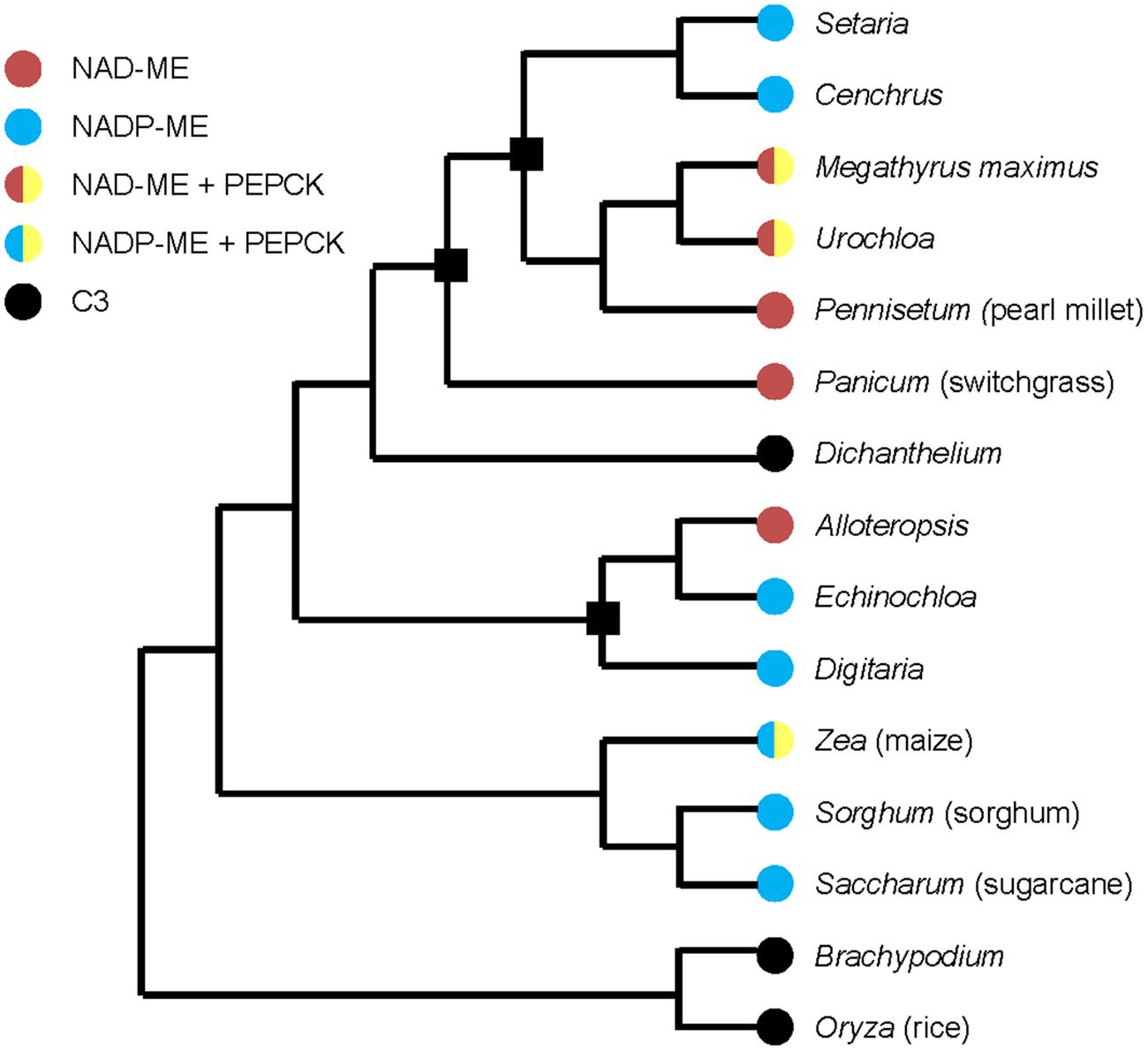
FIGURE 1. Simplified phylogenetic tree of grasses with selected C4 and C3 families, drawn based on Grass Phylogeny Working Group II (2012) and Washburn et al. (2015). Black square means the possible evolutionary conversions between NAD-ME and NADP-ME subtypes. Representative species of families are marked in parentheses.
The evolutionary transition to C4 photosynthesis remains undetermined. Phylogenomics analysis has indicated the difference of recruitment of major decarboxylating enzymes into C4 photosynthesis in NAD-ME and NADP-ME subtypes (Maier et al., 2011; Christin et al., 2013). The non-photosynthetic NAD-ME in C3 plants is composed of α and β subunits, and functions as a homodimer and heterodimer in the respiration of malate in mitochondria of all cells (Tronconi et al., 2008). In leaves of the dicot C4 plant Cleome gynandra, transcripts corresponding to two genes encoding α and β subunits are abundant in BS cells (Brautigam et al., 2011; Aubry et al., 2014) and the formation of heterodimeric photosynthetic NAD-ME was found in leaves of Amaranthus hypochondriacus (Long et al., 1994), whereas in the monocot C4 plant switchgrass, only one gene encoding the NAD-ME β subunit is highly expressed in BS cells of leaves (Rao et al., 2016), and an octamer of only one type of subunit exists in Eleusine coracana and Panicum dichotomiflorum leaves (Murata et al., 1989). Phylogenetic analysis indicates that NAD-MEs in C4 plants evolved from the existing mitochondrial NAD-ME and may be acquired through changes in regulatory and kinetic properties, rather than gene duplication (Maier et al., 2011). In contrast, C4 NADP-ME, which is thought to derive from a C3 chloroplast-localized ancestor and rooted from an ancient cytosolic isoform, has specific function in C4 photosynthesis in BS cells of NADP-ME subtype plants (Maier et al., 2011).
The emergence of C4 NAD-ME and NADP-ME would include the steps of enriched expression in BS cells and optimization of enzymatic properties (Maier et al., 2011). However, the preferential expression of NAD-ME and NADP-ME may be not exclusively correlated with its corresponding subtype. Significant transcript levels of genes associated with NAD-ME subtype C4 photosynthesis and high NAD-ME activity have been observed in the C3–C4 intermediate species Flaveria ramosissima, which is close to the NADP-ME C4 Flaveria lineage (Gowik et al., 2011). Additionally, the NADP-ME ortholog was found to be preferentially accumulated in BS cells of NAD-ME subtype switchgrass, in which low NADP-ME activity was detected (Rao et al., 2016). These unpredicted transcript profiles may reflect the common C4 ancestor, within which NAD-ME and NADP-ME subtype C4 pathway are present together at some level (Gowik et al., 2011; Washburn et al., 2015).
Kranz Anatomy in Leaves of NAD-ME and NADP-ME Subtypes
In most C4 species, an altered arrangement of cells within the leaf known as Kranz anatomy facilitates the cellular compartmentation of carboxylation and decarboxylation (Nelson and Langdale, 1992; Heckmann, 2016). A typical Kranz anatomy includes an outer layer of chloroplast-containing M cells for initial carboxylation, and an inner layer of large, distinctive BS cells that surround the vascular bundle for carbon reduction (Sage, 2004).
Kranz form varies as a consequence of the distinct evolutionary origins of C4 plants (Fouracre et al., 2014). In the NADP-ME subtype, the layer of cells between the BS cells and the vascular bundle is absent, and suberin is deposited in the BS cell wall. BS chloroplasts with reduced grana are arranged centrifugally in monocotyledons and centripetally in dicotyledons (Gutierrez et al., 1974; Hattersley and Watson, 1976; Prendergast et al., 1987; Lundgren et al., 2014). Comparatively, the vasculature of the NAD-ME subtype is usually surrounded by a double sheath, consisting of the outer BS and the inner non-photosynthetic mestome sheath (Prendergast et al., 1987; Lundgren et al., 2014). Suberin ubiquitously deposits in the mestome sheath rather than in BS cells, and BS chloroplasts with developed grana are arranged centripetally (Hattersley and Watson, 1976; Nelson and Langdale, 1989; Fouracre et al., 2014; Mertz and Brutnell, 2014). Loss of one layer of mestome sheath cells in the NADP-ME type suggests differences in the origination of cell divisions. The single BS in C4 NADP-ME type grasses is derived from the procambium and M cells develop from the ground meristem. In the double-sheath species of the NAD-ME type, both the BS and M cells are derived from the ground meristem and the mestome sheath is derived from the procambium (Nelson and Langdale, 1989; Soros and Dengler, 2001).
Metabolite Flow of C4 Photosynthesis in NAD-ME and NADP-ME Subtypes
All C4 plants share a common enzymatic step, the initial carboxylation reaction catalyzed by PEPC to yield oxaloacetic acid (OAA) in M cells (Sage, 2004). Subsequent steps to concentrate CO2, the transported metabolites, and the subcellular localization of the decarboxylation reaction, differ between the different biochemical subtypes (Figure 2).
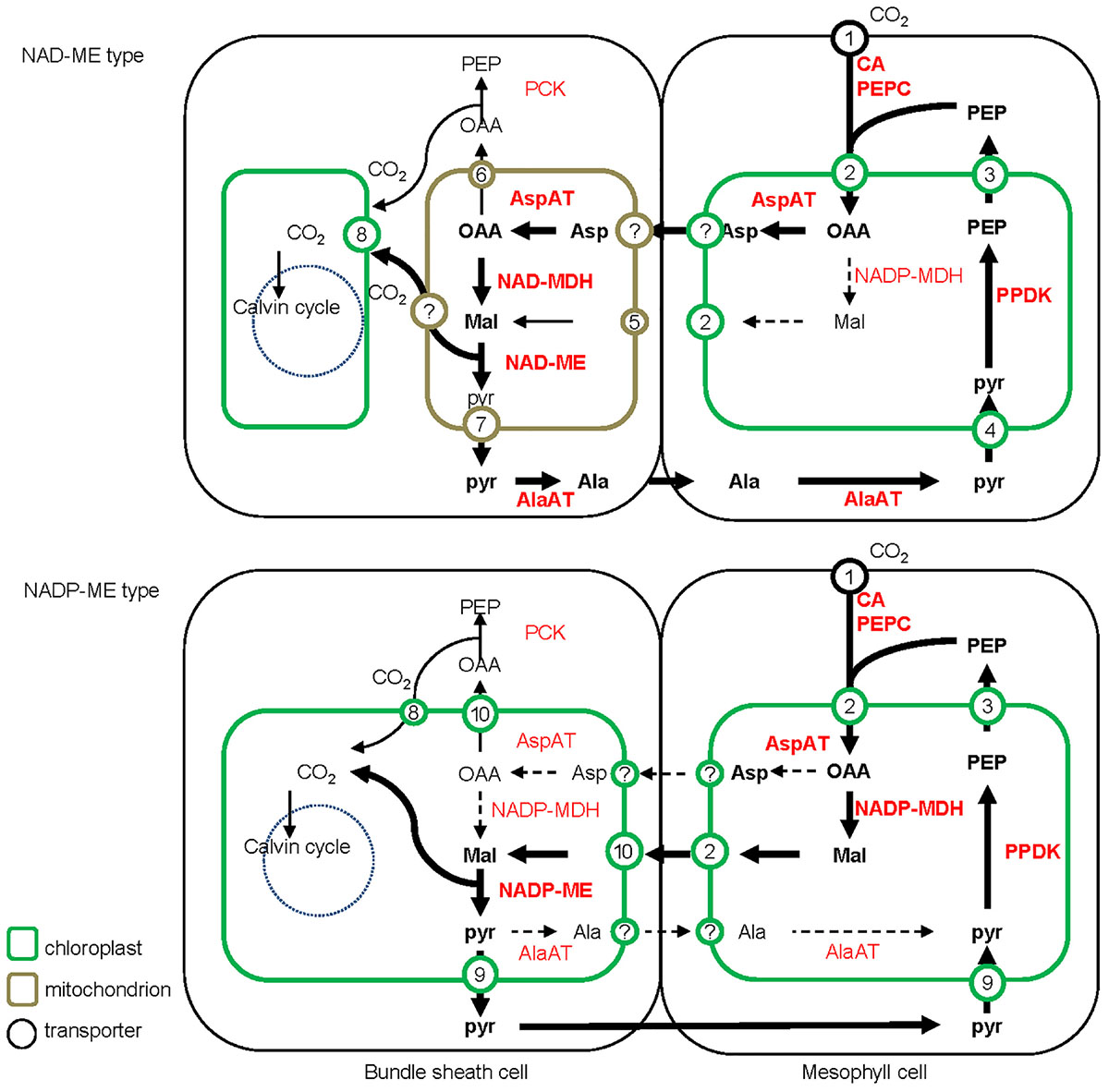
FIGURE 2. Detailed schematic of the C4 photosynthesis pathway of NAD-ME and NADP-ME subtypes. The major C4 biochemical pathway, the additional PEPCK pathway and the possible alternative pathway are indicated with bold, narrow, and dashed lines, respectively. The abundances of 4-carbon acids (metabolite level) and transporters (transcript level) are indicated with font style (with bold representing more abundant) and font/circle size (with larger representing more abundant), respectively. Ala, alanine; Asp, aspartate; Mal, malate; Pyr, pyruvate; OAA, oxaloacetate; PEP, phosphoenolpyruvate; CA, carbonic anhydrase; PEPC, phosphoenolpyruvate carboxylase; PPDK, pyruvate/orthophosphate dikinase; AspAT, aspartate aminotransferase; AlaAT, alanine aminotransferase; NADP-MDH, NADP-dependent malate dehydrogenase; NADP-ME, NADP-dependent malic enzyme; NAD-MDH, NAD-dependent malate dehydrogenase; NAD-ME, NAD-dependent malic enzyme; PCK, phosphoenolpyruvate carboxykinase. 1, Plasma membrane intrinsic protein (PIP); 2, dicarboxylate transporter 1 (DiT1, OMT1); 3, phosphate/phosphoenolpyruvate translocator (PPT); 4, sodium bile acid symporter 2 (BASS2) and sodium:hydrogen antiporter (NHD); 5, malate phosphate antiport 1 (DIC1) and phosphate proton symport (PIC); 6, mitochondrial carrier (DTC); 7, mitochondrial pyruvate carrier (MPC); 8, plasma membrane intrinsic protein (PIP) of chloroplast; 9, proton:pyruvate cotransporter (MEP); 10, dicarboxylate transport 2 (DiT2, DCT2).
The traditional biochemical view of the NADP-ME subtype places malate, derived from OAA, as the dominant transported metabolite to diffuse to the BS cells. Pyruvate is formed during the decarboxylation reaction, and returns to the M cells to be phosphorylated back to PEP. The synthesis of malate occurs in the M chloroplasts and the decarboxylation by NADP-ME in the BS chloroplasts. In contrast, NAD-ME plants use aspartate as the major transport metabolite, which is formed by transamination of OAA. After transfer to the BS cells, aspartate is converted to malate by a reductive deamination reaction. Pyruvate is also formed during the NAD-ME decarboxylation reaction, but is partially transported back to the M cells in the form of alanine to maintain the ammonia balance between the two cell types. Alanine in the M is converted through several steps into PEP, which provides the precursor for a new round of carboxylation and decarboxylation.
In the NAD-ME subtype, aspartate is synthesized in the M cytosol, while malate formation and decarboxylation by NAD-ME occur in the BS mitochondria (Hatch, 1971; Edwards and Walker, 1983; Hatch, 1987; Weber and von Caemmerer, 2010; Figure 2). In addition, the activity of PEPCK enzyme was detected at different levels in multiple lineages of NAD-ME and NADP-ME subtypes, which can decarboxylate OAA to PEP for CO2 release in the cytosol of BS cells (Pick et al., 2011; Sage et al., 2011; Figure 2). The supplementary utilization of the PEPCK pathway in some C4 plants is considered to enhance plant adaption to various environmental conditions (Wang et al., 2014; Furbank, 2016).
However, flexibility in the NADP-ME type C4 carbon fixation mechanism has been observed. Early C14 labeling experiments in maize (NADP-ME type C4 monocot) showed that label was incorporated into both malate and aspartate, with the latter occupying a minor but significant proportion (approximately 25%) of the active C4 acid pool (Hatch, 1971). Subsequent experiments showed that, in the presence of 2-oxoglutarate, aspartate can be decarboxylated by isolated BS cells of maize at lower rates (Chapman and Hatch, 1981). A similar study in Flaveria bidentis (NADP-ME type C4 dicotyledon) revealed that aspartate and malate contributed equally to transfer CO2 to the BS cells (Meister et al., 1996). High levels of transcripts encoding the major isoforms of aspartate aminotransferase (AspAT) and alanine aminotransferase (AlaAT) were detected in maize leaves, and enzymatic assays further confirmed the sufficiency of aminotransferase activity to carry out the carboxylation and decarboxylation reaction (Pick et al., 2011). Interestingly, cell-specific transcriptome analysis has revealed that AspAT and AlaAT are preferentially expressed at high levels in M and BS cells, respectively, in both NADP-ME type maize and Setaria viridis (Chang et al., 2012; John et al., 2014). The preferential accumulation of AspAT and AlaAT proteins was confirmed by proteomics analysis in isolated M (Majeran et al., 2005) and BS (Manandhar-Shrestha et al., 2013) cells of maize.
Several hypotheses have been put forward to explain how aspartate contributes to carbon fixation in NADP-ME plants. Majeran et al. (2005) suggested that the abundance of AspAT in M chloroplasts serves as a metabolic link between amino acid synthesis and nitrogen assimilation to generate aspartate as the final step of incorporation of ammonia into amino acid. This view is consistent with the observation of accelerated turnover of aspartate in response to nitrogen deficiency in maize leaves (Khamis et al., 1992). The reduction of cellular aspartate may decrease the rate of protein synthesis, and transcriptome analysis has indeed revealed reduced expression of protein synthesis-related genes in some NADP-ME plants (Brautigam et al., 2011; Gowik et al., 2011). This likely causes the reduction of protein content and therefore higher nitrogen-use efficiency in NADP-ME plants, compared with that of their NAD-ME counterparts (Brautigam and Gowik, 2016).
The fate of aspartate after translocation from M to BS cells is unclear. One proposal is that it can serve as a C4 regulator by influencing the transport of malate or pyruvate across the BS chloroplast, rather than serving a metabolic role (Chapman and Hatch, 1979). Another proposal is that AspAT in BS cells converts aspartate into OAA. OAA can be directly decarboxylated in the cytosol by PEP-CK (route I), or re-reduced to malate and then decarboxylated by NADP-ME in the chloroplast (route II) (Furbank, 2011; Gowik and Westhoff, 2011; Pick et al., 2011). However, for route I, no or limited PEP-CK activity has been reported in some NADP-ME subtype plants such as S. bicolor and F. bidentis; for route II, the mixed model including four transfer acids (aspartate, malate, alanine, and pyruvate) of the NADP-ME subtype requires comparable amounts of AspAT and AlaAT in BS cells to those in M cells, which is not consistent with the finding of the unequal accumulation of aminotransferase in M and BS chloroplasts in maize (Majeran et al., 2005; Manandhar-Shrestha et al., 2013). Rigid definitions of decarboxylation pathways may be misleading, and variants of the C4 NADP-ME subtype may be considered (Wang et al., 2014). Aspartate may be transaminated and decarboxylated by PEP-CK in NADP-ME variants that present sufficient PEP-CK activity, such as maize, or be transformed into the donor of NADP-ME in NADP-ME variants such as F. bidentis, which has substantial PSII activity in BS to maintain redox balance during the reduction of aspartate (Meister et al., 1996).
Plastid Transporters Involved in C4 Photosynthesis in NAD-ME and NADP-ME Subtypes
The dispersed sub-localization of carboxylating, decarboxylating, and transaminase enzymes in M and BS cells of C4 plants requires the collaboration of multiple translocators to transfer reaction substrates and products across membranes. NAD-ME and NADP-ME subtype plants utilize different plastid transports to maintain this metabolite flux (Figure 2).
In all C4 versions, pyruvate is predominantly found in M cells, where it is converted to PEP as the precursor for fixing CO2. Pyruvate in M cells is compartmented in chloroplasts making its cytosolic concentration low (Ohnishi et al., 1990). Two different mechanisms for transport of pyruvate into M chloroplasts have been identified in a range of C4 species: proton-dependent and sodium-dependent (Aoki et al., 1992; Furumoto et al., 2011), with the assumption that NAD-ME and NADP-ME types might use sodium:pyruvate and proton:pyruvate cotransporters, respectively (Ohnishi et al., 1990; Weber and von Caemmerer, 2010). Recent comparative transcriptome analyses between NAD-ME and NADP-ME type C4 plants have supported this hypothesis; transcripts encoding sodium:pyruvate cotransporter were preferentially expressed in M cells of the NAD-ME-type plants switchgrass and C. gynandra, whereas transcripts encoding proton:pyruvate cotransporter were enriched in M cells of the NADP-ME-type plants S. viridis and maize (Chang et al., 2012; Aubry et al., 2014; John et al., 2014; Rao et al., 2016).
The decarboxylation and assimilation of CO2 both happen in BS chloroplasts of NADP-ME type plants, and metabolite transporters are required to transfer malate and pyruvate across the chloroplast envelope membrane. The major decarboxylating enzyme NAD-ME, NAD-MDH, and AspAT are restricted to mitochondria in NAD-ME subtypes. Compared with NADP-ME subtypes, additional mitochondrial transporters are required in NAD-ME subtypes, including those for imported OAA and glutamate, exported aspartate and 2-oxoglutarate for the AspAT processes and imported malate and exported pyruvate for the NAD-MDH and NAD-ME processes. These postulated carriers involved in the C4 biochemical pathways are indicated on Figure 2.
A high rate of CO2 diffusion across the plasma membrane of M cells is expected in all C4 versions. Compared with NADP-ME subtype plants, an additional transport process is required to facilitate the CO2 permeability of BS chloroplasts in NAD-ME subtype plants since CO2 is released outside of the chloroplast. The membrane channel aquaporins, PIPs (plasma membrane intrinsic protein), have been demonstrated to mediate M CO2 conductance in leaves of some C3 plants such as tobacco (NtAQP1; Uehlein et al., 2008), Arabidopsis (AtPIP1;2; Uehlein et al., 2012), and barley (PIP2 family; Mori et al., 2014). The role of PIPs in CO2 diffusion is still unclear in C4 plants. The diurnal expression of ZmPIPs in the M of maize leaves might suggest their possible roles as CO2 facilitators (Hachez et al., 2008), and CgPIP1B was suggested to be the candidate CO2 transporter across the M cell plasmalemma in Cleome (Brautigam et al., 2011). PIPs, especially the PIP2 subfamily, also show high water transport activity (Katsuhara and Hanba, 2008) and the activity of PIPs is dynamically controlled in BS cells in Arabidopsis as a response to hydraulic stress (Shatil-Cohen et al., 2011). The dual roles of PIPs in CO2 and water transport might be responsible for CO2 assimilation and water movement in C4 plants, also contributing to resistance of C4 plants to drought stress. Furthermore, it is worth considering whether the additional service of PIPs in BS cells of the NAD-ME subtype increases drought tolerance, at least in some lineages.
M and BS Cell-Specific Functions in NAD-ME and NADP-ME Subtypes
The spatial compartmentation of many metabolic pathways has been observed in M and BS cells of C4 plants (Majeran et al., 2005), and has generally been considered to be associated with the spatial separation of carboxylation and decarboxylation in the two cell types. There are both overlapping and differential cell-specific features of metabolic pathways in M and BS cells in NAD-ME and NADP-ME plants (Zhao et al., 2013; Koteyeva et al., 2014).
The light-dependent reactions of photosynthesis are not equally distributed in M and BS cells of NADP-ME plants. There is a depletion of PSII activity and reduction of the associated development of grana generally present at various degrees in BS chloroplasts of NADP-ME type species such as maize, sorghum, and sugarcane (Chapman and Hatch, 1981; Meierhoff and Westhoff, 1993). In contrast, enhancement of PSII activity and grana development in BS chloroplasts is observed in NAD-ME plants (Edwards and Walker, 1983). This is because, in the NADP-ME subtype, the primary shuttle of malate from M cells to BS chloroplasts provides NADPH, the balance of which would be influenced by a high level of PSII activity, whereas the transferred C4 acid aspartate in NAD-ME subtype does not deliver NADPH as reductive power (Edwards and Walker, 1983; Koteyeva et al., 2014).
Biochemical studies have further revealed differences in metabolic control of the Calvin cycle in BS cells of NAD-ME and NADP-ME subtypes; the addition of ribose-5-phosphate significantly increased light-dependent CO2 fixation, and light is required in C4 acid decarboxylation and assimilation into the Calvin cycle in maize (NADP-ME type), but ribose-5-phosphate only partially or little affected light-dependent CO2 fixation in Atriplex spongiosa and Panicum miliaceum (NAD-ME type; Hatch and Kagawa, 1976). This suggests that there would be insufficient supply of ribulose 1,5-diphosphate in the Calvin cycle and the ratio of C4 assimilation into the cycle might be controlled in NADP-ME subtype, whereas the Calvin cycle functions independently in NAD-ME subtype plants (Hatch and Kagawa, 1976).
Recently, comparative transcriptome analysis has indicated differential enrichment of transcripts involved in RNA regulation and protein biogenesis/homeostasis in M and BS cells of two NAD-ME-type plants (switchgrass and Cleome) and two NADP-ME-type plants (maize and S. viridis) (Chang et al., 2012; Aubry et al., 2014; John et al., 2014; Rao et al., 2016). Transcripts involved in protein synthesis, folding, and assembly are more abundant in M cells in the two NADP-ME-type plants, but are preferentially or equally expressed in BS cells of the two NAD-ME-type plants. In contrast, transcripts involved in RNA regulation are enriched in BS cells of the two NADP-ME-type plants, but are more abundant in M cells of the NAD-ME-type plant switchgrass. The differentiation for transcriptional and post-transcriptional regulatory mechanisms in M and BS cells of NADP-ME and NAD-ME types might be associated with the unequal distribution of metabolites within the M and BS cells of these two subtypes (Rao et al., 2016).
Conclusion
A brief overview of the differences in features of NAD-ME and NADP-ME plants is shown in Table 1. C4 photosynthesis represents one of the most successfully evolutionary events in response to environmental change on the earth and can be divided into two broad biochemical groups, NAD-ME and NADP-ME. A clear statement of dichotomy in morphology and biochemistry can be made between the two C4 subtypes with some exceptions (Sage, 2004; Sage et al., 2011; Lundgren et al., 2014). The nature and commonality of C4 transporters and cell-type specific functional differentiation still remain to be determined beyond a few well-studied species, to explore whether these are common in most C4 plants or only within some C4 lineages. The diversification of physiological, biochemical, and molecular functions of the NAD-ME type and NADP-ME type might be a result of their distinct evolutionary pathways, and be associated with the accommodation of various environmental conditions.
Author Contributions
XR collected data from literature and wrote the manuscript. RD revised the article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The research from the authors’ laboratory was supported by grants to RD from both the US Department of Energy Advanced Research Projects Agency-Energy (ARPA-E) and the US Department of Energy Bioenergy Sciences Center (BESC, grant # BER DE-AC05-00OR2727), through the Office of Biological and Environmental Research in the DOE Office of Science.
References
Aoki, N., Ohnishi, J., and Kanai, R. (1992). Two different mechanisms for transport of pyruvate into mesophyll chloroplasts of C4 plants—a comparative study. Plant Cell Physiol. 33, 805–809.
Aubry, S., Kelly, S., Kumpers, B. M., Smith-Unna, R. D., and Hibberd, J. M. (2014). Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis. PLoS Gen. 10:e1004365. doi: 10.1371/journal.pgen.1004365
Brautigam, A., and Gowik, U. (2016). Photorespiration connects C3 and C4 photosynthesis. J. Exp. Bot. 67, 2953–2962. doi: 10.1093/jxb/erw056
Brautigam, A., Kajala, K., Wullenweber, J., Sommer, M., Gagneul, D., Weber, K. L., et al. (2011). An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiol. 155, 142–156. doi: 10.1104/pp.110.159442
Chang, Y. M., Liu, W. Y., Shih, A. C., Shen, M. N., Lu, C. H., Lu, M. Y., et al. (2012). Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiol. 160, 165–177. doi: 10.1104/pp.112.203810
Chapman, K., and Hatch, M. (1981). Aspartate decarboxylation in bundle sheath cells of Zea mays and its possible contribution to C3 photosynthesis. Funct. Plant Biol. 8, 237–248. doi: 10.1071/PP9810237
Chapman, K. S., and Hatch, M. D. (1979). Aspartate stimulation of malate decarboxylation in Zea mays bundle sheath cells: possible role in regulation of C4 photosynthesis. Biochem. Biophys. Res. Commun. 86, 1274–1280. doi: 10.1016/0006-291X(79)90254-7
Christin, P. A., Besnard, G., Samaritani, E., Duvall, M. R., Hodkinson, T. R., Savolainen, V., et al. (2008). Oligocene CO2 decline promoted C4 photosynthesis in grasses. Curr. Biol. 18, 37–43. doi: 10.1016/j.cub.2007.11.058
Christin, P. A., Boxall, S. F., Gregory, R., Edwards, E. J., Hartwell, J., and Osborne, C. P. (2013). Parallel recruitment of multiple genes into C4 photosynthesis. Genome Biol. Evol. 5, 2174–2187. doi: 10.1093/gbe/evt168
Edwards, G., and Walker, D. (1983). C3, C4: Mechanisms, and Cellular and Environmental Regulation, of Photosynthesis. Oxford: Blackwell Scientific Publications.
Fouracre, J. P., Ando, S., and Langdale, J. A. (2014). Cracking the Kranz enigma with systems biology. J. Exp. Bot. 65, 3327–3339. doi: 10.1093/jxb/eru015
Furbank, R. T. (2011). Evolution of the C4 photosynthetic mechanism: are there really three C4 acid decarboxylation types? J. Exp. Bot. 62, 3103–3108. doi: 10.1093/jxb/err080
Furbank, R. T. (2016). Walking the C4 pathway: past, present, and future. J. Exp. Bot. 67, 4057–4066. doi: 10.1093/jxb/erw161
Furumoto, T., Yamaguchi, T., Ohshima-Ichie, Y., Nakamura, M., Tsuchida-Iwata, Y., Shimamura, M., et al. (2011). A plastidial sodium-dependent pyruvate transporter. Nature 476, 472–475. doi: 10.1038/nature10250
Ghannoum, O. (2005). Faster Rubisco is the key to superior nitrogen-use efficiency in NADP-malic enzyme relative to NAD-malic enzyme C4 grasses. Plant Physiol. 137, 638–650. doi: 10.1104/pp.104.054759
Ghannoum, O., von Caemmerer, S., and Conroy, J. P. (2002). The effect of drought on plant water use efficiency of nine NAD-ME and nine NADP-ME Australian C4 grasses. Funct. Plant Biol. 29, 1337–1348. doi: 10.1071/FP02056
Gowik, U., Brautigam, A., Weber, K. L., Weber, A. P., and Westhoff, P. (2011). Evolution of C4 photosynthesis in the genus Flaveria: how many and which genes does it take to make C4? Plant Cell 23, 2087–2105. doi: 10.1105/tpc.111.086264
Gowik, U., and Westhoff, P. (2011). The path from C3 to C4 photosynthesis. Plant Physiol. 155, 56–63. doi: 10.1104/pp.110.165308
Grass Phylogeny Working Group II (2012). New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytol. 193, 304–312. doi: 10.1111/j.1469-8137.2011.03972.x
Gutierrez, M., Gracen, V. E., and Edwards, G. E. (1974). Biochemical and cytological relationships in C4 plants. Planta 119, 279–300. doi: 10.1007/BF00388331
Hachez, C., Heinen, R. B., Draye, X., and Chaumont, F. (2008). The expression pattern of plasma membrane aquaporins in maize leaf highlights their role in hydraulic regulation. Plant Mol. Biol. 68, 337–353. doi: 10.1007/s11103-008-9373-x
Hatch, M. D. (1971). The C4 pathway of photosynthesis. Evidence for an intermediate pool of carbon dioxide and the identity of the donor C4 -dicarboxylic acid. Biochem. J. 125, 425–432.
Hatch, M. D. (1987). C4 photosynthesis: a unique elend of modified biochemistry, anatomy and ultrastructure. Biochim. Biophys. Acta 895, 81–106. doi: 10.1016/s0304-4173(87)80009-5
Hatch, M. D., and Kagawa, T. (1976). Photosynthetic activities of isolated bundle sheath cells in relation to differing mechanisms of C4 pathway photosynthesis. Arch. Biochem. Biophys. 175, 39–53. doi: 10.1016/0003-9861(76)90483-5
Hattersley, P. (1992). “C4 photosynthetic pathway variation in grasses (Poaceae): its significance for arid and semi-arid lands,” in Desertified Grasslands: Their Biology and Management, ed. G. P. Chapman (London: Academic Press), 181–212.
Hattersley, P. W., and Watson, L. (1976). C4 grasses: an anatomical criterion for distinguishing between NADP-malic enzyme species and PCK or NAD-malic enzyme species. Aust. J. Bot. 24, 297–308. doi: 10.1071/bt9760297
Heckmann, D. (2016). C4 photosynthesis evolution: the conditional Mt. Fuji. Curr. Opin. Plant Biol. 31, 149–154. doi: 10.1016/j.pbi.2016.04.008
John, C. R., Smith-Unna, R. D., Woodfield, H., Covshoff, S., and Hibberd, J. M. (2014). Evolutionary convergence of cell-specific gene expression in independent lineages of C4 grasses. Plant Physiol. 165, 62–75. doi: 10.1104/pp.114.238667
Katsuhara, M., and Hanba, Y. T. (2008). Barley plasma membrane intrinsic proteins (PIP Aquaporins) as water and CO2 transporters. Pflügers Arch. 456, 687–691. doi: 10.1007/s00424-007-0434-9
Khamis, S., Lamaze, T., and Farineau, J. (1992). Effect of nitrate limitation on the photosynthetically active pools of aspartate and malate in maize, a NADP malic enzyme C4 plant. Physiol. Plant. 85, 223–229. doi: 10.1111/j.1399-3054.1992.tb04726.x
Koteyeva, N. K., Voznesenskaya, E. V., Cousins, A. B., and Edwards, G. E. (2014). Differentiation of C4 photosynthesis along a leaf developmental gradient in two Cleome species having different forms of Kranz anatomy. J. Exp. Bot. 65, 3525–3541. doi: 10.1093/jxb/eru042
Liu, H., and Osborne, C. P. (2015). Water relations traits of C4 grasses depend on phylogenetic lineage, photosynthetic pathway, and habitat water availability. J. Exp. Bot. 66, 761–773. doi: 10.1093/jxb/eru430
Long, J. J., Wang, J. L., and Berry, J. O. (1994). Cloning and analysis of the C4 photosynthetic NAD-dependent malic enzyme of amaranth mitochondria. J. Biol. Chem. 269, 2827–2833.
Lundgren, M. R., Osborne, C. P., and Christin, P. A. (2014). Deconstructing Kranz anatomy to understand C4 evolution. J. Exp. Bot. 65, 3357–3369. doi: 10.1093/jxb/eru186
Maier, A., Zell, M. B., and Maurino, V. G. (2011). Malate decarboxylases: evolution and roles of NAD(P)-ME isoforms in species performing C4 and C3 photosynthesis. J. Exp. Bot. 62, 3061–3069. doi: 10.1093/jxb/err024
Majeran, W., Cai, Y., Sun, Q., and van Wijk, K. J. (2005). Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. Plant Cell 17, 3111–3140. doi: 10.1105/tpc.105.035519
Manandhar-Shrestha, K., Tamot, B., Pratt, E. P., Saitie, S., Brautigam, A., Weber, A. P., et al. (2013). Comparative proteomics of chloroplasts envelopes from bundle sheath and mesophyll chloroplasts reveals novel membrane proteins with a possible role in c4-related metabolite fluxes and development. Front. Plant Sci. 4:65. doi: 10.3389/fpls.2013.00065
Meierhoff, K., and Westhoff, P. (1993). Differential biogenesis of photosystem II in mesophyll and bundle-sheath cells of monocotyledonous NADP-malic enzyme-type C4 plants: the non-stoichiometric abundance of the subunits of photosystem II in the bundle-sheath chloroplasts and the translational activity of the plastome-encoded genes. Planta 191, 23–33.
Meister, M., Agostino, A., and Hatch, M. D. (1996). The roles of malate and aspartate in C4 photosynthetic metabolism of Flaveria bidentis (L.). Planta 199, 262–269. doi: 10.1007/bf00196567
Mertz, R. A., and Brutnell, T. P. (2014). Bundle sheath suberization in grass leaves: multiple barriers to characterization. J. Exp. Bot. 65, 3371–3380. doi: 10.1093/jxb/eru108
Metzker, M. L. (2010). Applications of next-generation sequencing technologies - the next generation. Nat. Rev. Genet. 11, 31–46. doi: 10.1038/Nrg2626
Mori, I. C., Rhee, J., Shibasaka, M., Sasano, S., Kaneko, T., Horie, T., et al. (2014). CO2 transport by PIP2 aquaporins of barley. Plant Cell Physiol. 55, 251–257. doi: 10.1093/pcp/pcu003
Murata, T., Ohsugi, R., Matsuoka, M., and Nakamoto, H. (1989). Purification and characterization of NAD malic enzyme from leaves of Eleusine coracana and Panicum dichotomiflorum. Plant Physiol. 89, 316–324. doi: 10.1104/pp.89.1.316
Nelson, T., and Langdale, J. A. (1989). Patterns of leaf development in C4 plants. Plant Cell 1, 3–13. doi: 10.1105/tpc.1.1.3
Nelson, T., and Langdale, J. A. (1992). Developmental genetics of C4 photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43, 25–47. doi: 10.1146/annurev.pp.43.060192.000325
Offermann, S., Friso, G., Doroshenk, K. A., Sun, Q., Sharpe, R. M., Okita, T. W., et al. (2015). Developmental and subcellular organization of single-cell C4 photosynthesis in Bienertia sinuspersici determined by large-scale proteomics and cDNA assembly from 454 DNA sequencing. J. Proteome Res. 14, 2090–2108. doi: 10.1021/pr5011907
Ohnishi, J., Flugge, U. I., Heldt, H. W., and Kanai, R. (1990). Involvement of Na+ in active uptake of pyruvate in mesophyll chloroplasts of some C4 plants : Na+ pyruvate cotransport. Plant Physiol. 94, 950–959. doi: 10.1104/pp.94.3.950
Osborne, C. P., and Sack, L. (2012). Evolution of C4 plants: a new hypothesis for an interaction of CO2 and water relations mediated by plant hydraulics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 583–600. doi: 10.1098/rstb.2011.0261
Pick, T. R., Brautigam, A., Schluter, U., Denton, A. K., Colmsee, C., Scholz, U., et al. (2011). Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. Plant Cell 23, 4208–4220. doi: 10.1105/tpc.111.090324
Pinto, H., Powell, J. R., Sharwood, R. E., Tissue, D. T., and Ghannoum, O. (2015). Variations in nitrogen use efficiency reflect the biochemical subtype while variations in water use efficiency reflect the evolutionary lineage of C4grasses at inter-glacial CO2. Plant Cell Environ. 39, 514–526. doi: 10.1111/pce.12636
Pinto, H., Sharwood, R. E., Tissue, D. T., and Ghannoum, O. (2014). Photosynthesis of C3, C3-C4, and C4 grasses at glacial CO2. J. Exp. Bot. 65, 3669–3681. doi: 10.1093/jxb/eru155
Prendergast, H. D. V., Hattersley, P. W., and Stone, N. E. (1987). New structural/biochemical associations in leaf blades of C4 grasses (Poaceae). Aust. J. Plant Physiol. 14, 403–420. doi: 10.1071/pp9870403
Rao, X., Lu, N., Li, G., Nakashima, J., Tang, Y., and Dixon, R. A. (2016). Comparative cell-specific transcriptomics reveals differentiation of C4 photosynthesis pathways in switchgrass and other C4 lineages. J. Exp. Bot. 67, 1649–1662. doi: 10.1093/jxb/erv553
Sage, R. F. (2004). The evolution of C4 photosynthesis. New Phytol. 161, 341–370. doi: 10.1111/j.1469-8137.2004.00974.x
Sage, R. F., Christin, P. A., and Edwards, E. J. (2011). The C4 plant lineages of planet Earth. J. Exp. Bot. 62, 3155–3169. doi: 10.1093/jxb/err048
Shatil-Cohen, A., Attia, Z., and Moshelion, M. (2011). Bundle-sheath cell regulation of xylem-mesophyll water transport via aquaporins under drought stress: a target of xylem-borne ABA? Plant J. 67, 72–80. doi: 10.1111/j.1365-313X.2011.04576.x
Sommer, M., Brautigam, A., and Weber, A. P. (2012). The dicotyledonous NAD malic enzyme C4 plant Cleome gynandra displays age-dependent plasticity of C4 decarboxylation biochemistry. Plant Biol. 14, 621–629. doi: 10.1111/j.1438-8677.2011.00539.x
Soros, C. L., and Dengler, N. G. (2001). Ontogenetic derivation and cell differentiation in photosynthetic tissues of C3 and C4 Cyperaceae. Am. J. Bot. 88, 992–1005. doi: 10.2307/2657080
Taub, D. R. (2000). Climate and the U.S. distribution of C4 grass subfamilies and decarboxylation variants of C4 photosynthesis. Am. J. Bot. 87, 1211–1215. doi: 10.2307/2656659
Taub, D. R., and Lerdau, M. T. (2000). Relationship between leaf nitrogen and photosynthetic rate for three NAD-ME and three NADP-ME C4 grasses. Am. J. Bot. 87, 412–417. doi: 10.2307/2656637
Tronconi, M. A., Fahnenstich, H., Weehler, M. C. G., Andreo, C. S., Flugge, U. I., Drincovich, M. F., et al. (2008). Arabidopsis NAD-malic enzyme functions as a homodimer and heterodimer and has a major impact on nocturnal metabolism. Plant Physiol. 146, 1540–1552. doi: 10.1104/pp.107.114975
Uehlein, N., Otto, B., Hanson, D. T., Fischer, M., McDowell, N., and Kaldenhoff, R. (2008). Function of Nicotiana tabacum aquaporins as chloroplast gas pores challenges the concept of membrane CO2 permeability. Plant Cell 20, 648–657. doi: 10.1105/tpc.107.054023
Uehlein, N., Sperling, H., Heckwolf, M., and Kaldenhoff, R. (2012). The Arabidopsis aquaporin PIP1;2 rules cellular CO2 uptake. Plant Cell Environ. 35, 1077–1083. doi: 10.1111/j.1365-3040.2011.02473.x
Vogel, J. C., Fuls, A., and Ellis, R. P. (1978). Geographical distribution of Kranz grasses in South Africa. S. Afr. J. Sci. 74, 209–215.
Wang, Y., Brautigam, A., Weber, A. P., and Zhu, X. G. (2014). Three distinct biochemical subtypes of C4 photosynthesis? A modelling analysis. J. Exp. Bot. 65, 3567–3578. doi: 10.1093/jxb/eru058
Washburn, J. D., Schnable, J. C., Davidse, G., and Pires, J. C. (2015). Phylogeny and photosynthesis of the grass tribe Paniceae. Am. J. Bot. 102, 1493–1505. doi: 10.3732/ajb.1500222
Weber, A. P., and von Caemmerer, S. (2010). Plastid transport and metabolism of C3 and C4 plants–comparative analysis and possible biotechnological exploitation. Curr. Opin. Plant Biol. 13, 257–265. doi: 10.1016/j.pbi.2010.01.007
Keywords: C4 photosynthesis, C4 plants, NAD-ME subtype, NADP-ME subtype, comparative transcriptome analysis
Citation: Rao X and Dixon RA (2016) The Differences between NAD-ME and NADP-ME Subtypes of C4 Photosynthesis: More than Decarboxylating Enzymes. Front. Plant Sci. 7:1525. doi: 10.3389/fpls.2016.01525
Received: 22 June 2016; Accepted: 28 September 2016;
Published: 13 October 2016.
Edited by:
Iker Aranjuelo, Agrobiotechnology Institute–Spanish National Research Council–University of Navarra, SpainReviewed by:
Masanori Arita, National Institute of Genetics, JapanVeronica Graciela Maurino, University of Düsseldorf, Germany
Copyright © 2016 Rao and Dixon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaolan Rao, xiaolan.rao@unt.edu Richard A. Dixon, richard.dixon@unt.edu
 Xiaolan Rao
Xiaolan Rao Richard A. Dixon1,2*
Richard A. Dixon1,2*