- 1Key Laboratory of Genetic Development and Germplasm Enhancement of Rare Plants in Three Gorges Area, College of Biology and Pharmacy, China Three Gorges University, Yichang, China
- 2The Genetic Engineering International Cooperation Base of Ministry of Science and Technology, Key Laboratory of Molecular Biophysics of Ministry of Education, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan, China
Environmental stresses frequently affect plant growth and development, and many genes have been found to be induced by unfavorable environmental conditions. Here, we reported the biological functions of TaNAC2D, a stress-related NAC (NAM, ATAF, and CUC) gene from wheat. TaNAC2D showed transcriptional activator activity in yeast. TaNAC2D-GFP fusion protein was localized in the nucleus of wheat mesophyll protoplasts. TaNAC2D transcript abundance was significantly induced by NaCl, PEG6000, and abscisic acid (ABA) at seedling stage, and repressed by NaCl and PEG6000 at mature plant stage. When TaNAC2D was introduced into Arabidopsis, the 35-day-old soil-grown TaNAC2D-overexpression (TaNAC2D-OX) plants displayed slower stomatal closure, higher water loss rate, and more sensitivity to salt and drought stresses compared with WT plants. In contrast, TaNAC2D-OX seedlings, grown on 1/2 MS medium supplemented with different concentrations of NaCl, Mannitol, and MV, had enhanced tolerances to salt, osmotic and oxidative stresses during seed germination and post-germination periods. The opposite stress-responsive phenotypes of transgenic Arabidopsis were consistent with the expression patterns of TaNAC2D in wheat. Moreover, under high salinity and dehydration conditions, three marker genes, including NCED3, RD29A, and RD29B, were down-regulated in 35-day-old TaNAC2D-OX plants grown in soil and up-regulated in 14-day-old TaNAC2D-OX seedlings grown on 1/2 MS medium. Our results suggest that the change in growth stages and environmental conditions may regulate TaNAC2D’s function.
Introduction
Environmental stresses may adversely affect plant development and yield. To protect plant against environmental stresses, many genes have been induced, and the products of these stress-inducible genes may provide a protection from stress damage. Stress-inducible genes are largely regulated by transcription factors (TFs). Transgenic plants overexpressing TF genes often show enhanced stress tolerance. For example, overexpression of TaASR1 in Nicotiana tabacum displays improved tolerance to dehydration and oxidative stresses (Hu et al., 2013). Arabidopsis plants overexpressing GsWRKY20 result in decreased water loss and increased drought tolerance (Luo et al., 2013). In rice (Oryza sativa), overexpression of OsMYB2 exhibits enhanced stress tolerance and increased sensitivity to exogenous ABA (Yang et al., 2012).
NAC TFs have been reported to participate in various plant developmental processes, including cell division (Kim et al., 2006), senescence (Yang et al., 2011; Wu et al., 2012; Kim et al., 2013), secondary cell wall formation (Zhong et al., 2010), hormone metabolism (Huh et al., 2012), light responses (Peng et al., 2015), and biotic/abiotic stress responses (Delessert et al., 2005; Wu et al., 2009). Genome-wide transcriptome analysis reveals that ∼20–25% NAC genes are involved in stress response (Puranik et al., 2012). Expression patterns of OsNAC family show that 140 putative OsNAC genes were predicted and at least 15% (21/140) OsNAC genes were induced by salt or drought (Fang et al., 2008). Recently, a new study has further revealed that ∼42% (63/149) OsNAC genes displayed overlapping expression patterns under biotic and abiotic stresses by microarray data analysis (Sun et al., 2015). Systematic sequence analysis of Brachypodium distachyon shows that 101 putative BdNAC genes were identified, 18 of which participated in stress responses (You et al., 2015). Moreover, to date many NAC TFs are responsive to abiotic stress in several plant species. Transgenic Arabidopsis plants overexpressing ATAF1/ANAC002, ANAC019, ANAC055, or ANAC072 have improved drought tolerance and hypersensitivity to exogenous ABA (Fujita et al., 2004; Tran et al., 2004; Wu et al., 2009). Overexpression of ANAC016 has low drought tolerance, while anac016 mutant plants exhibit high drought tolerance (Sakuraba et al., 2015). In rice, transgenic plants overexpressing OsNAC5, OsNAC9, or OsNAC10 show improved drought tolerance due to increased root diameter (Jeong et al., 2010, 2013; Redillas et al., 2012). SNAC1-overexpressing rice plants exhibit enhanced tolerance to abiotic stresses, increased ABA sensitivity, and decreased water loss rate through closing more stomatal pores (Hu et al., 2006). Compared with the model plant Arabidopsis and rice, to date only a few NAC TFs have been revealed to participate in the process of stress in bread wheat (Triticum aestivum). Plants overexpressing TaNAC2, TaNAC29, or TaNAC67 display improved tolerances to multiple abiotic stresses (Mao et al., 2012, 2014; Huang et al., 2015). Transgenic wheat overexpressing TaNAC69 shows a conspicuous drought-tolerant phenotype (Xue et al., 2011). Recently, a new study reveals that TaNAC2-5A-overexpressing wheat plants produce a higher grain yield through acquired more nitrogen from fertilizer, suggesting that TaNAC2-5A participates in the wheat nitrate-signaling pathway (He et al., 2015).
In this study, the full-length cDNA of TaNAC2D with high sequence identity to TaNAC2 was isolated from wheat. Gene expression patterns in response to salinity, dehydration, ABA, and oxidative stresses were examined by qRT-PCR in wheat. Functional analyses revealed that the 35-day-old soil-grown TaNAC2D-OX plants displayed reduced tolerance to salt and drought stresses, and slower ABA-mediated stomatal closure compared with WT plants. In contrast, TaNAC2D-OX seedlings, grown on 1/2 MS medium supplemented with 100 mM NaCl, 200 mM Mannitol, 1 μM MV, or 1 μM ABA, showed increased stress tolerance and reduced sensitivity to exogenous ABA during seed germination and post-germination periods. The opposite stress-responsive phenotypes of TaNAC2D-OX plants were caused by the change in growth stages and environmental conditions. Our results indicate that TaNAC2D may have a potential application value in genetic engineering of wheat.
Materials and Methods
Cloning and Bioinformatic Analysis of TaNAC2D
TaNAC2D (GenBank: GQ231954.1) was cloned from wheat (T. aestivum L. cv. Chinese spring) using gene-specific primers (Supplementary Table S1A). The NAC domain was searched using InterProScan tool1. Multiple sequence alignments were carried out by MEGA 5.0 software. The phylogenetic trees were generated using maximum likelihood method, and the bootstrap parameter was set at 1000 replicates. The ID regions of TaNAC2D were predicted using the program PONDR VL32.
Stress Treatments and qRT-PCR Analysis
Wheat seedlings were grown in the growth chamber at 22°C under a 12-h light/12-h dark cycle. The 10-day-old seedlings grown in petri dish with water were, respectively, exposed to different solutions, including 200 mM NaCl, 10 mM H2O2, 20% PEG6000, and 100 μM ABA. The 50-day-old mature plants grown in soil were uprooted and treated under the same stress conditions above. Mock-treated seedlings were used as control. Leaves were collected after 0, 1, 3, 6, 12, and 24 h post-treatment for gene expression analysis. Various organs of wheat were collected for organ-specific expression analysis. Total RNA were isolated from those collected materials, and then reverse-transcribed into cDNA using PrimeScript RT reagent Kit with gDNA Eraser (Tiangen, Beijing, China). The qRT-PCR analysis was performed using specific primers (Supplementary Table S1B) on a real time PCR machine (Bio-rad, Hercules, CA, USA). Each sample was analyzed using three biological replicates each with three technical replicates. The obtained values were calculated using the 2-ΔΔCT method (Livak and Schmittgen, 2001). TaActin (GenBank: AB181991.1) was used as an internal control.
Subcellular Localization of TaNAC2D
For subcellular localization assay, the recombinant construct of pMD18-35S-GFP (control vector) was firstly obtained as previously described (Huang et al., 2015). The TaNAC2D ORF, which is lacking the termination codon, was cloned into pMD18-35S-GFP control vector using specific primers (Supplementary Table S1C). The recombinant pMD18-35S-TaNAC2D-GFP vector and control vector were transformed into wheat mesophyll protoplasts according to previous described methods (Yoo et al., 2007). The GFP signal was visualized by fluorescence microscopy. The 4′,6-diamidino-2-phenylindole (DAPI) nuclear stain was used to determine the location of nucleus.
Transactivation Activity Analysis of TaNAC2D
The TaNAC2D ORF (TaNAC2D1-327) and two truncated versions (TaNAC2D1-172, and TaNAC2D173-327) were cloned into pGBKT7 vector (Clontech, USA) using specific primers (Supplementary Table S1D). Three constructed vectors and the pGBKT7 were separately transformed into AH109 (Clontech). The transactivation activity of TaNAC2D was evaluated based on the growth characteristic of transformants on SD medium (-Trp/-His/-Ade). X-α-Gal was used for the detection of α-galactosidase activity.
Generation of Transgenic Arabidopsis Plants
The TaNAC2D ORF containing the termination codon was cloned into the pBI121 using specific primers (Supplementary Table S1E). The pBI121-TaNAC2D plasmid and the pBI121 control vector were separately transformed into Arabidopsis plants by Agrobacterium-mediated floral dipping method (Clough and Bent, 1998). Seeds harvested from transgenic plants were screened on 1/2 MS medium supplemented with 50 mg/L kanamycin. The expression of TaNAC2D was detected by semi-quantitative analysis. Homozygous T3 progenies were used for further experiments.
Assessment of Stress Tolerance in TaNAC2D-OX Lines
For the analysis of germination and root growth, Arabidopsis seeds of WT, EV and TaNAC2D-OX lines were sown on 1/2 MS medium supplemented with 100 mM NaCl, 200 mM mannitol, 1 μM MV, and 1 μM ABA. After 3 days of cold treatment, the seeded plates were incubated in the growth chamber at 22°C with 16-h light/8-h dark cycle. Germination (seedlings with expanded cotyledons) and seedlings emergence (seedlings with green cotyledons) were examined 8 days. Root growth and plant survival of vertically grown seedlings were examined 8 or 21 days.
To assess stresses tolerance, WT and TaNAC2D-OX lines were grown in soil under non-stress conditions for 35 days. Subsequently, the 35-day-old plants were irrigated with 200 mM NaCl for 30 days after withholding of water for 2 weeks. For the drought tolerance assay, the 35-day-old plants were not irrigated for 20 days, and then re-watered for 7 days.
To further evaluate the function mechanism of TaNAC2D-OX lines to drought stress, the rosette leaves of 35-day-old soil-grown plants were sampled for the assays of water loss and stomatal movement. For assessing the degree of water loss, the detached leaves were placed on filter paper, and then dehydrated for 2 or 3 h at 25°C. Leaf shapes were photographed after dehydration treatment. Moreover, the rate of water loss was measured (n = 10). The assay of ABA-induced stomatal closure was further performed according to previous described methods (Osakabe et al., 2013; Ha et al., 2014). The leaf epidermal strips of 35-day-old soil-grown lines were soaked in buffer solution (0.2 mM CaCl2, 10 mM KCl, and 10 mM Mes-KOH, pH 6.15) for 6 h under light intensity of 300 μmol⋅m-2⋅s-1. Subsequently, the epidermal strips were exposed to 0 or 30 μM ABA solution for 1 h. The stomatal movement in guard cells was observed by light microcopy. The size of stomatal apertures was measured for WT and TaNAC2D-OX plants (n = 50).
Expression Analyses of NCED3, RD29A, and RD29B
To further investigate the molecular mechanism of stress tolerance, the expression levels of marker genes were detected in WT and TaNAC2D-OX plants. The 35-day-old soil-grown plants were submerged in 200 mM NaCl solution for 6 h or placed on dry filter paper, and then dehydrated for 3 h. The 14-day-old seedlings, grown on 1/2 MS medium, were incubated in 1/2 MS liquid medium containing 200 mM NaCl for 6 h or dehydrated for 1 h. The total RNAs were extracted from the whole plants. The qRT-PCR was performed using specific primers (Supplementary Table S1F) for the expression levels of marker genes, including NCED3 (nine-cis-epoxycarotenoid dioxygenase 3; At3g14440), RD29A (responsive-to-desiccation 29A; At5g52310), and RD29B (responsive-to-desiccation 29B; At5g52300). Actin2 (At3g18780) was used as a reference gene.
Measurement of H2O2 Content and Relative Electrolytic Leakage
The 35-day-old soil-grown plants were subjected to salt stress (200 mM NaCl) for 10 days or drought stress for 17 days, rosette leaves of WT and TaNAC2D-OX plants were collected to determine H2O2 content and REL. The same measured methods were used as previously described (Hu et al., 2012; Huang et al., 2015).
Results
TaNAC2D Encodes a Plant-Specific NAC Transcription Factor
TaNAC2D (GenBank: GQ231954.1) was cloned from bread wheat. The cDNA sequence of TaNAC2D contains a 984 bp ORF that encodes a protein of 327 amino acid residues. Sequence alignment and phylogenetic analyses (Supplementary Figures S1A and S2) revealed that TaNAC2D was highly homologous to HvSNAC1 (GenBank: AEG21060.1), OsSNAC1(GenBank: AIX03022.1), and almost identical to TaNAC2 (AAU08786.1), indicating that TaNAC2D, like these three NAC genes (Hu et al., 2006; Mao et al., 2012; Al Abdallat et al., 2014), participated in plant stress responses. The results of Blastn search in EnsemblPlants database showed that Traes_5BL_4497A137C.1 had a highest identity (96.8%) of nucleotide sequence with TaNAC2D, suggesting that TaNAC2D gene was located on chromosome 5BL of wheat.
TaNAC2D contains a NAM domain at N-terminus (amino acids 1∼172) with five subdomains (A–E) and a transcriptional regulatory domain at C-terminus (amino acids 173∼327; Supplementary Figure S1A). Transactivation assays demonstrated that TaNAC2D had transactivation activity, and the C-terminus region was enough to activate expression of reporter genes in yeast (Figure 1A). TaNAC2D-GFP fusion protein and DAPI were detected only in the nucleus of wheat mesophyll protoplasts (Figure 1B). These results indicate that TaNAC2D may function as a transcription factor. Additionally, the result of PONDR VL3 analysis showed that a largely ID region located in C-terminus of TaNAC2D (Supplementary Figure S1B), suggesting that the protein was a mostly non-folded conformation at its C-terminus.
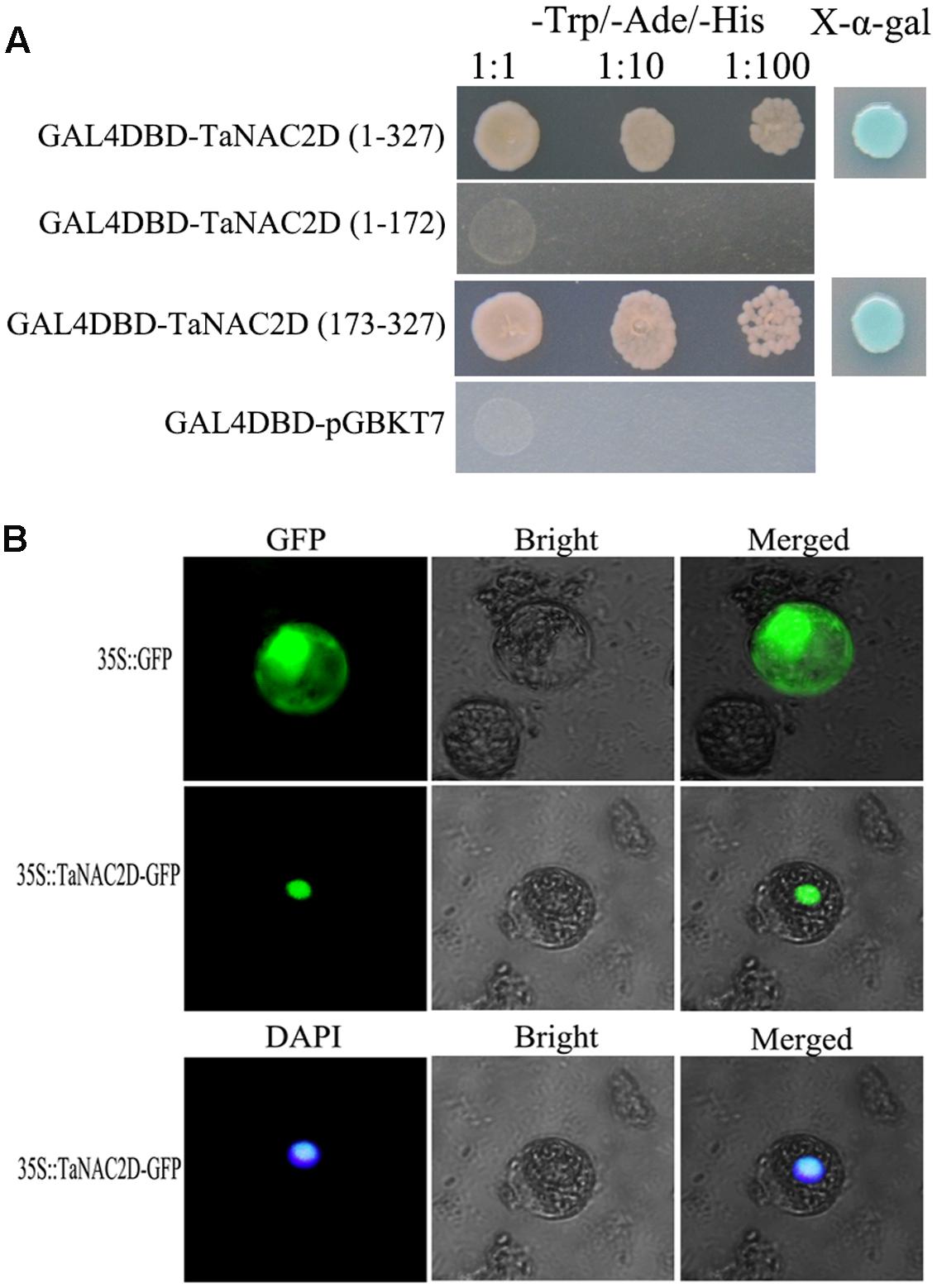
FIGURE 1. Transactivation activity and subcellular localization of TaNAC2D. (A) Transactivation activity of TaNAC2D in yeast strain AH109. Full-length and truncated versions of TaNAC2D were fused into pGBKT7 vector and the transformants were screened on the SD/-Trp-Ade-His plates with or without X-α-gal. (B) Subcellular localization of TaNAC2D. The fusion protein pMD18-35S-TaNAC2D-GFP and pMD18-35S-GFP (control) were transiently expressed in wheat mesophyll protoplasts and observed with fluorescence microscope.
Expression Patterns of TaNAC2D
Organ-specific analysis revealed that TaNAC2D transcripts were expressed in all organs except endosperm of wheat, and highly expressed in leaves (Supplementary Figure S3). Moreover, we further examined the expression levels of TaNAC2D under abiotic stress. The qRT-PCR results showed that TaNAC2D was significantly induced by NaCl, PEG, and ABA at seedling stage, and repressed by NaCl and PEG at mature plant stage (Figure 2). These results indicate that the expression of TaNAC2D is regulated at different stages under stress conditions.
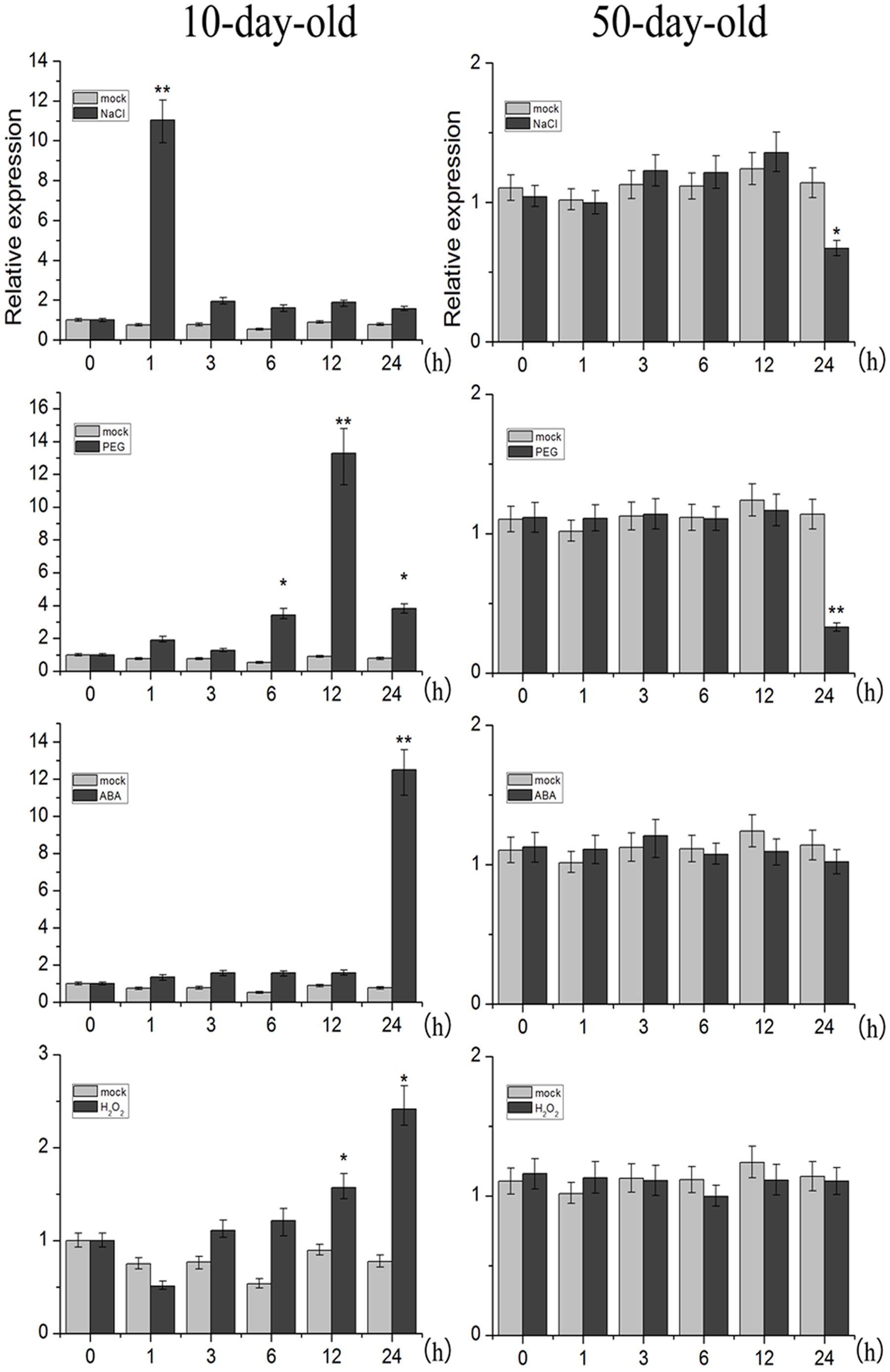
FIGURE 2. Expression patterns of TaNAC2D. The 10-day-old wheat seedlings and the 50-day-old mature wheat plants were treated with NaCl, PEG6000, ABA, and H2O2, transcript levels of TaNAC2D in wheat leaves were determined by qRT-PCR and normalized to TaActin. Data are means ± SE of three biological replicates. Asterisks indicate significant differences from mock (∗P < 0.05, ∗∗P < 0.01).
TaNAC2D-OX Plants Exhibit Hypersensitivity to Salt and Drought Stresses at Mature Plant Stage under Non-sterile Conditions
To study the function of gene, TaNAC2D was introduced into Arabidopsis. Eight transgenic lines (T3) were obtained through kanamycin screening. The TaNAC2D expression level in T3 lines was examined by semi-quantitative PCR analysis. Two TaNAC2D high expression lines, designated OX1 and OX8 (Supplementary Figure S4), were chosen for further experiments.
For the assessment of salt tolerance, when the 35-day-old soil-grown plants were subjected to 200 mM NaCl exposure for 14 days, the leaves of TaNAC2D-OX lines began to turn yellow, while the impact of salt stress on WT plants was slightly (Figure 3A). After 30 days of salt stress exposure, WT plants had a survival rate of over 60%, whereas OX1 and OX8 showed significantly lower survival rate than WT plants (26 and 22%, respectively; Figures 3A,C). This result indicated that TaNAC2D-OX plants showed more sensitivity to salt stress than that of WT.
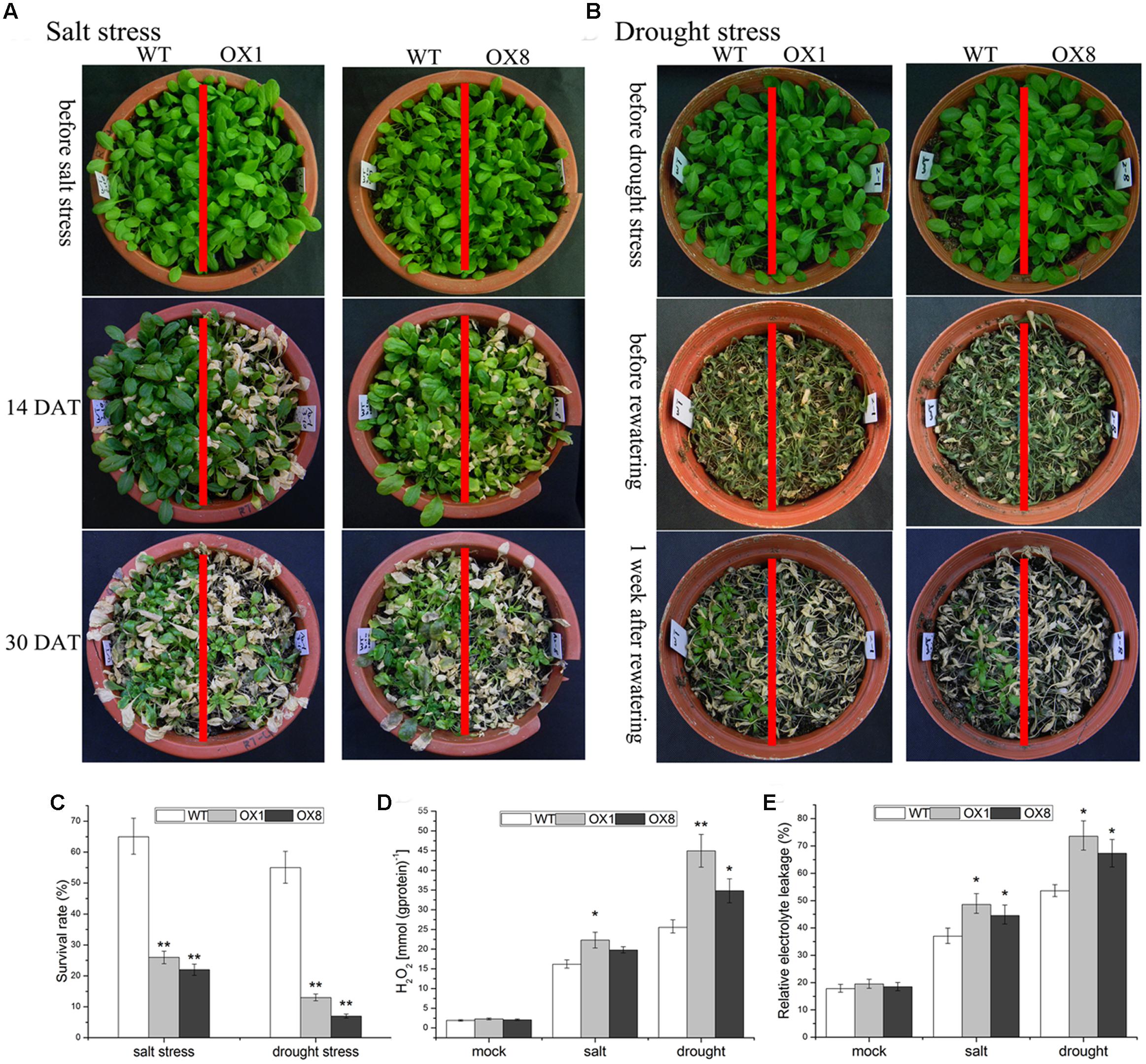
FIGURE 3. Hypersensitivity of TaNAC2D-OX plants to salt and drought stresses. (A) The 35-day-old WT and TaNAC2D-OX plants were irrigated with 200 mM NaCl for 30 days. (B) The 35-day-old WT and TaNAC2D-OX plants were subjected to drought stress for 20 days and then re-watered for 1 week. (C) Survival rate of WT and TaNAC2D-OX plants. (D,E) Plants were subjected to salt stress for 10 days and drought stress for 17 days, H2O2 content and relative electrolyte leakage were determined. Data in (C–E) are means ± SE of three biological replicates. Asterisks indicate significant differences from control (∗P < 0.05, ∗∗P < 0.01).
To investigate the impact of drought stress on TaNAC2D-OX lines, the 35-day-old soil-grown plants were not irrigated for 20 days, and then re-watered for 7 days. Approximately 90% of TaNAC2D-OX lines were dead, whereas over 50% of WT plants still survived (Figures 3B,C). This observation indicated that overexpression of TaNAC2D resulted in reduced tolerance to drought stress.
To further understand the physiological mechanisms underlying TaNAC2D functions that lead to reduced tolerance, H2O2 content and REL were measured. Under salinity and drought conditions, TaNAC2D-OX plants exhibited higher levels of H2O2 content and REL compared with those of WT plants (Figures 3D,E), indicating increased ROS production and greater membrane damage in transgenic plants. These results demonstrated that the TaNAC2D-OX plants exhibit hypersensitivity to salt and drought stresses, consistent with the suppression of TaNAC2D expression at mature stage of wheat (Figure 2).
TaNAC2D-OX Plants Show Reduced Sensitivity to Exogenous ABA
ABA signal is largely regulated by abiotic stress in plants (Yoshida et al., 2014). To investigate if ABA participates in transgenic plants responses to abiotic stresses, we examined responsiveness of TaNAC2D-OX plants to exogenous ABA during germination and post-germinative growth periods. The observation showed that exogenous ABA inhibited germination, seedling emergence (seedling with green cotyledon), and root growth in control plants more severely than observed in TaNAC2D-OX plants (Figure 4; Supplementary Figure S5), indicating that TaNAC2D-OX plants displayed reduced sensitivity to exogenous ABA than control plants. These results suggested that TaNAC2D might be regulated in part by ABA signal during abiotic stress responses in plants.
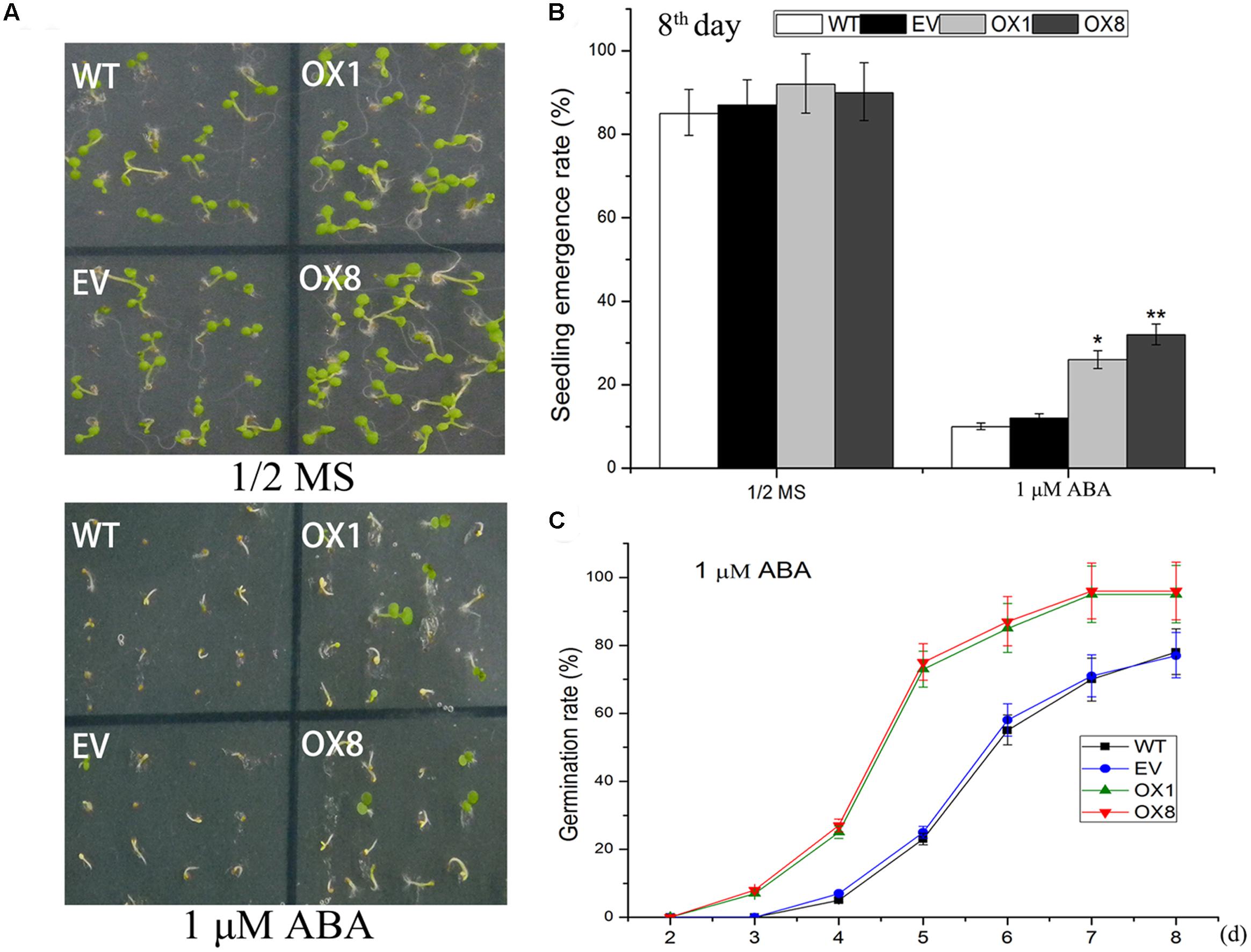
FIGURE 4. Response of TaNAC2D-OX plants to exogenous ABA. (A) Germination performance of WT, EV, and OXs grown for 8 days on 1/2 MS medium containing 1 μM ABA. (B) Seedlings emergence rate of WT, EV, and OXs. (C) Percent germination of WT, EV, and OXs. Data in (B,C) are means ± SE of three biological replicates (n = 40 to 50 seeds per genotype per experiment). Asterisks indicate significant differences from WT (∗P < 0.05, ∗∗P < 0.01).
Comparison of Water Loss and Stomatal Movement in Control and TaNAC2D-OX Plants
To further investigate the drought-sensitive phenotype of transgenic lines, detached rosette leaves of the 35-day-old soil-grown plants were sampled for dehydration and ABA-mediated stomatal closure. After 2 or 3 h of air drying, the leaves of OX1 and OX8 were severely curled, whereas the effect of dehydration process on WT and EV plants was slightly (Figure 5A). Additionally, TaNAC2D-OX lines showed higher water loss rate compared with control plants (Figure 5B), indicating that overexpression of TaNAC2D in Arabidopsis had a decreased water retention capacity.
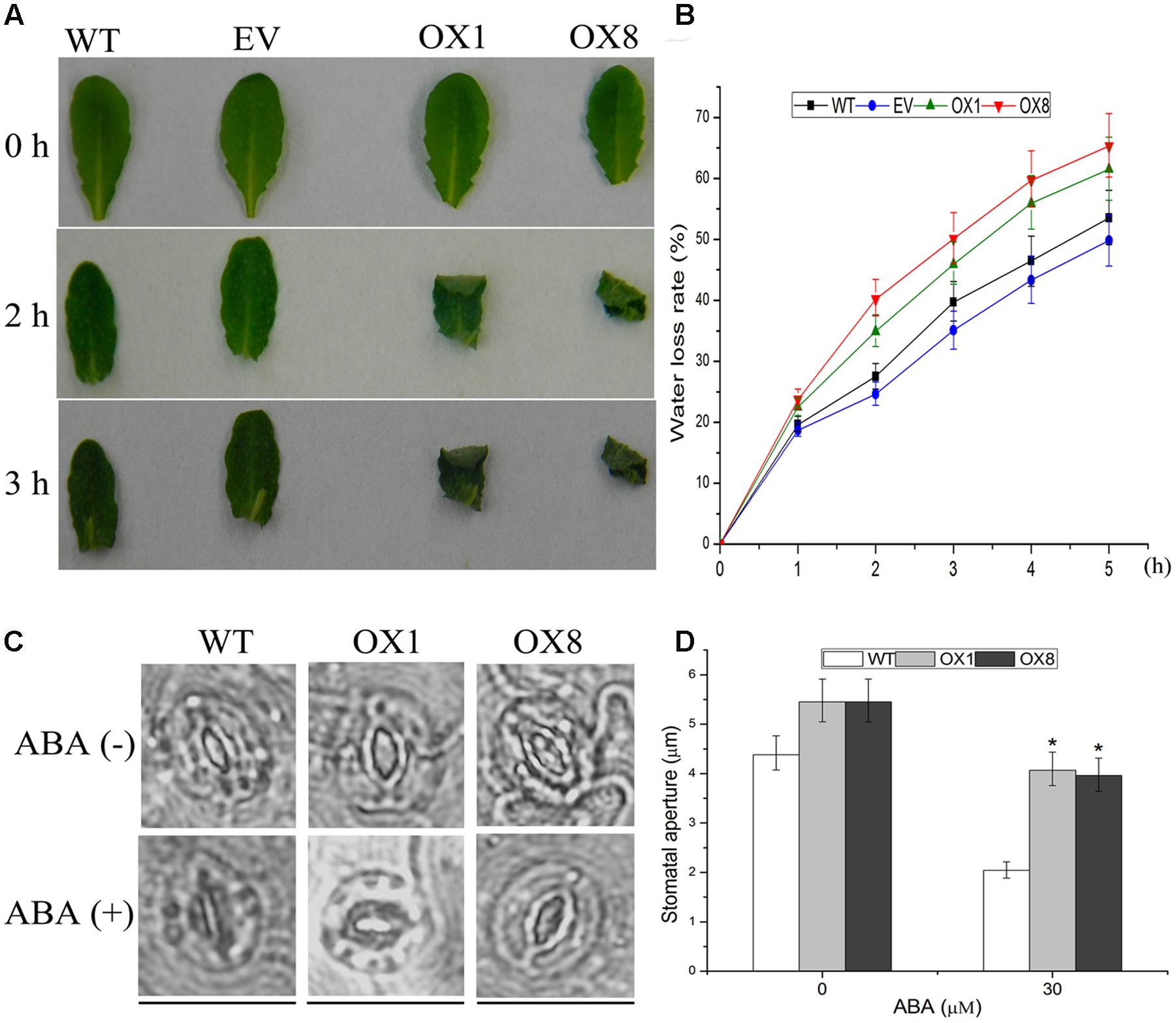
FIGURE 5. Response of TaNAC2D-OX plants to dehydration and ABA-mediated stomatal closure. (A) Phenotypes of leaves under dehydration stress for 0, 2, and 3 h. (B) Water loss rate of WT, EV, and OXs. Data are means ± SE of three independent experiments (n = 10). (C) Guard cells of 35-day-old soil-grown WT and OXs treated with or without 30 μM ABA for 1 h. Scale bars = 30 μm. (D) Quantitative analysis of the size of stomatal aperture. Data are means ± SE (n = 50). Asterisks indicate significant differences from WT (∗P < 0.05).
It is reported that ABA-mediated stomatal movement can affect transpiration rates during dehydration process (Osakabe et al., 2013; Ha et al., 2014). Because TaNAC2D-OX plants showed reduced sensitivity to exogenous ABA compared with WT plants (Figure 4; Supplementary Figure S5), ABA-mediated stomatal movement was further examined. The results revealed that the stomata of TaNAC2D-OX plants closed more slowly compared with that of WT plants (Figures 5C,D), indicating that the slower stomatal closure observed in TaNAC2D-OX plants was directly responsible for greater water loss and increased drought sensitivity of transgenic plants.
TaNAC2D-OX Plants Show Increased Tolerance to Salt, Osmotic and Oxidative Stresses at Seedling Stage under Sterile Conditions
Several studies reveal that stress tolerance of plants is associated with germination and root growth of seedlings (Ren et al., 2010; Zhu et al., 2010). To further investigate mechanisms of hypersensitivity to abiotic stress in TaNAC2D-OX plants, we examined germination, primary root length, and plant survive in control and TaNAC2D-OX lines under salt, osmotic and oxidative stresses. The statistical analysis of seed germination showed that TaNAC2D-OX lines grown on 1/2 MS medium supplemented with 100 mM NaCl, 200 mM mannitol or 1 μM MV, had higher germination rate than control plants (Figure 6). Root growth also exhibited that TaNAC2D-OX lines had longer primary root compared with control plants under salt and osmotic stresses (Supplementary Figure S6). Moreover, root growth in the control and TaNAC2D-OX lines was inhibited to similar extents by MV treatment, whereas TaNAC2D-OX lines had a higher survival rate than control plants under oxidative stress (Supplementary Figure S6). These results suggested that TaNAC2D-OX plants had improved tolerance to salt, osmotic and oxidative stresses during seed germination and post-germinative growth periods, consistent with the induced expression of TaNAC2D at seedling stage of wheat (Figure 2).
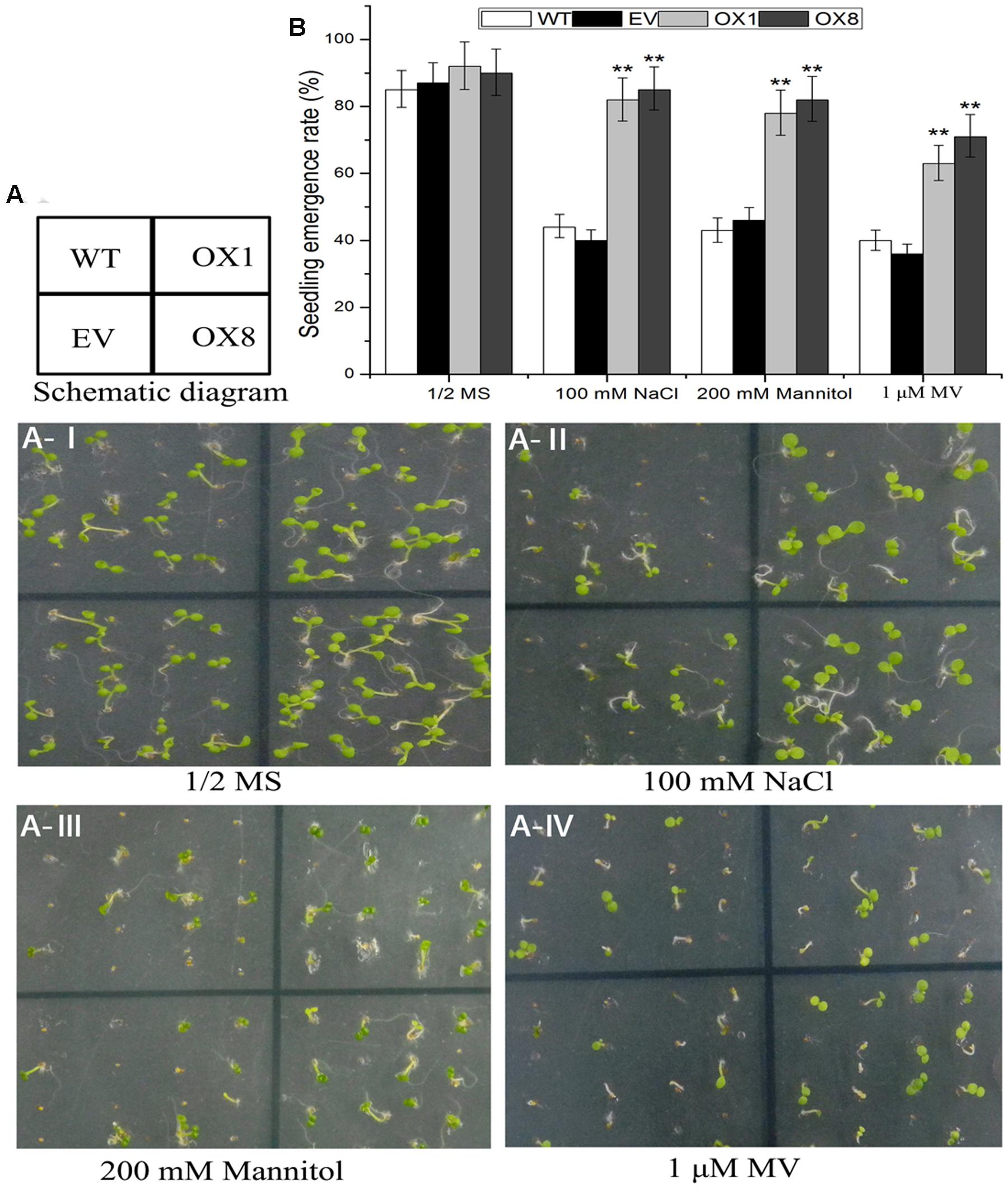
FIGURE 6. Seedling emergence assays of WT, EV and TaNAC2D-OX plants. (A) Seedling emergence performance of WT, EV, and OXs grown for 8 days on 1/2 MS medium (control) or 1/2 MS medium containing 100 mM NaCl, 200 mM Mannitol, and 1 μM MV. (B) Seedlings emergence rate of WT, EV, and OXs. Data are means ± SE of three biological replicates (n = 40 to 50 seeds per genotype per experiment). Asterisks indicate significant differences from WT (∗∗P < 0.01).
Expression Analysis of NCED3, RD29A, and RD29B in WT and TaNAC2D-OX Plants under Salt and Dehydration Conditions
Many stress-inducible marker genes that are up-regulated in plants are expected to increase plant tolerance to abiotic stress (Cramer et al., 2011; Yoshida et al., 2014). To further understand the above-mentioned different stress-responsive phenotypes of TaNAC2D-OX plants, the expression analysis of marker genes, including NCED3, RD29A, and RD29B, was further performed. The 14-day-old seedlings, grown on 1/2 MS medium, were untreated (control) or treated with NaCl (200 mM) for 6 h or dehydration for 1 h, respectively, the transcript levels of NCED3, RD29A and RD29B were all significantly up-regulated in TaNAC2D-OX plants than in WT plants (Figures 7A–C); this was consistent with the stress-tolerant phenotypes of TaNAC2D-OX plants during germination and post-germinative growth periods (Figure 6; Supplementary Figure S6). In contrast, the 35-day-old soil-grown plants were incubated for 6 h in salt solution or dehydrated for 3 h, respectively, the transcript levels of NCED3, RD29A and RD29B were all significantly down-regulated in TaNAC2D-OX plants compared with in WT plants (Figures 7B,C); this was also in accordance with the stress-sensitive phenotypes of soil-grown TaNAC2D-OX plants (Figures 3A,B). These results suggested that the different growth stages and environmental conditions may regulate the transcriptional activity of TaNAC2D, leading to up-regulating or down-regulating of downstream genes, such as NCED3, RD29A, and RD29B.
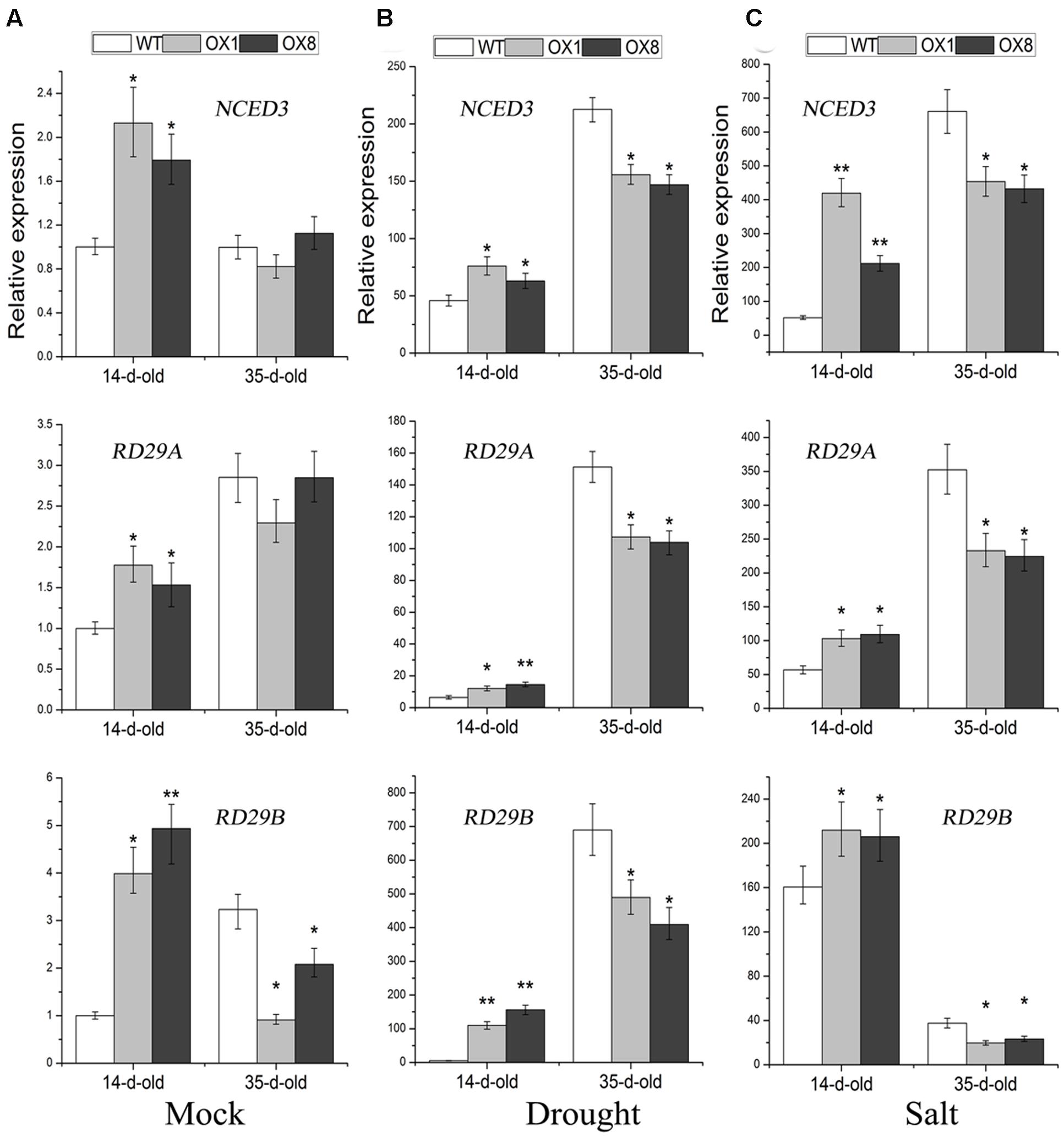
FIGURE 7. Expression pattern of NCED3, RD29A and RD29B in WT and TaNAC2D-OX plants. The 14-day-old seedlings grown on 1/2 MS medium were subsequently submerged in 1/2 MS liquid cultures supplemented with 200 mM NaCl for 6 h or placed on dry filter paper, and then dehydrated for 1 h. Moreover, the 35-day-old soil-grown plants were incubated for 6 h in 200 mM NaCl solution or dehydrated for 3 h. Transcript levels were determined by qRT-PCR and normalized to actin2. The values in 14-day-old untreated WT plants were set to 1. Data are means ± SE of three biological replicates. Asterisks indicate significant differences from WT (∗P < 0.05; ∗∗P < 0.01). (A) Effects of normal growth. (B) Effects of drought stress. (C) Effects of salt stress.
Discussion
TaNAC2D Is a NAC TFs that Participates in the Abiotic Stress Response
Many NAC TFs have been shown to participate in plant stress responses (Nakashima et al., 2012). In Arabidopsis, plant overexpressing ATAF1, ANAC019, ANAC055, or ANAC072 confers an improved ability to drought stress (Tran et al., 2004; Wu et al., 2009). Overexpression of VNI2/ANAC083 and JUB1/ANAC042 has enhanced tolerance to abiotic stress, meanwhile these two genes are also involved in senescence process (Yang et al., 2011; Wu et al., 2012). In rice (O. sativa), plant overexpressing either SNAC1, OsNAC5, OsNAC9, or OsNAC10 increases stress tolerance of transgenic rice (Hu et al., 2006; Jeong et al., 2010, 2013; Redillas et al., 2012). In wheat, overexpression of TaNAC2, TaNAC29, TaNAC67, and TaNAC69 exhibits stress-tolerant phenotypes of transgenic plants (Xue et al., 2011; Mao et al., 2012, 2014; Huang et al., 2015).
In this study, phylogenetic analysis of TaNAC2D showed a close relationship with TaNAC2, HvSNAC1 and OsSNAC1. The analysis of NCBI BLASTP revealed that TaNAC2D had 98% identity to TaNAC2 from T. aestivum, 95% identity to HvSNAC1 from Hordeum vulgare, 71% identity to OsSNAC1 from O. sativa. Transgenic Arabidopsis plants overexpressing TaNAC2 increase tolerance to salt, drought and cold stresses compared with WT plants (Mao et al., 2012). The present study showed that Arabidopsis plants overexpressing TaNAC2D displayed increased sensitivity to salt and drought stresses at mature stage, and only improved stress tolerance at seedling stage. Our results are only partly in agreement with the previous findings of TaNAC2. In addition, overexpression of HvSNAC1 in barley results in an increased resistance to drought and Ramularia leaf spot in the transgenic plants (Al Abdallat et al., 2014; McGrann et al., 2015). The transgenic barley plants overexpressing HvSNAC1 also delay dark-induced leaf senescence (McGrann et al., 2015). OsSNAC1 is the rice ortholog of TaNAC2D. Overexpression of OsSNAC1 in rice improves tolerance to drought and salt stresses at the vegetative stage (Hu et al., 2006). These previous findings together with our data imply that TaNAC2D may be involved in the stress response, pathogen defense, and senescence. In the present study, ectopic expression of TaNAC2D in Arabidopsis is a faster attempt to detect gene functions. However, the real role of gene needs to be further validated by overexpression and knock-down of TaNAC2D in wheat. We presume that TaNAC2D, like HvSNAC1 and OsSNAC1 (Hu et al., 2006; Al Abdallat et al., 2014; McGrann et al., 2015), is also involved in biotic and abiotic stress response in wheat.
Overexpression of TaNAC2D in Arabidopsis Reduces Drought Stress Tolerance at Mature Plant Stage
In the present study, leaves of 35-day-old soil-grown TaNAC2D-OX plants were found to be curled more severely and to lose water more rapidly compared with leaves of control plants when they were treated with dehydration, indicating that TaNAC2D-OX plants exhibited reduced drought tolerance than control plants due to the higher transpiration rate. The higher transpiration rate in TaNAC2D-OX plants was significantly correlation with the slower stomatal closure during dehydration. Additionally, TaNAC2D-OX plants displayed hyposensitivity to exogenous ABA during the germination and post-germinative growth periods. The slower ABA-mediated stomatal closure might be caused by ABA hyposensitivity, rendering more water loss and the increased drought sensitivity of TaNAC2D-OX plants. Several studies have reported that drought tolerance is associated with the transpiration rate of leaf and degree of stomatal closure. For example, max (more axillary growth) mutant plants exhibit hyposensitivity to exogenous ABA, the slower ABA-mediated stomatal closure, the higher transpiration rate, and the reduced drought tolerance compared with WT plants (Ha et al., 2014). KUP6, KUP8 and GORK mutant plants display decreased sensitivity to exogenous ABA, impaired stomatal closure, and reduced tolerance to drought stress (Osakabe et al., 2013). Consistent with the findings of previous studies, overexpression of TaNAC2D in Arabidopsis also exhibited the same characteristic.
Changes in Growth Stages and Environmental Conditions Alter Stress-Responsive Phenotypes of TaNAC2D-OX Plants
Phylogenetic analysis showed that TaNAC2D had close homology to ATAF1 and ATAF2 from Arabidopsis (Supplementary Figure S7), suggesting that TaNAC2D might have the similar function with them. Several studies have demonstrated the biological functions of ATAF1 and ATAF2. In one study, ATAF1-OX plants exhibit enhanced drought tolerance, and increased sensitivity to high salinity and oxidative stresses (Wu et al., 2009). Contrastingly, the earlier studies reveal that ataf1 mutant plants show improved drought tolerance (Lu et al., 2007; Jensen et al., 2008). Recently, two new studies show that ATAF1 directly interacts with the promoter of NCED3, which encodes a key enzyme in ABA biosynthesis pathway (Jensen et al., 2013; Garapati et al., 2015). Overexpression of ATAF1 improves the transcript level of NCED3, leading to an increased endogenous ABA level, while ataf1 mutant plants exhibit a significantly decreased NCED3 expression compared with WT plants (Jensen et al., 2013). The increased endogenous ABA level contributes to the enhanced tolerance of plants against abiotic stress (Jensen et al., 2013). Thus, the increased NCED3 expression level is responsible for the enhanced drought tolerance of ATAF1-OX plants observed by Wu et al. (2009), but cannot explain an increased drought tolerance of ataf1 mutants observed by Lu et al. (2007) and Jensen et al. (2008). These previous studies suggest that TaNAC2D may also be involved in regulating NCED3 expression because TaNAC2D had a close homology to ATAF1. Moreover, two abiotic stress-inducible marker genes, including RD29A and RD29B, are involved in ABA signaling pathway (Yamaguchi-Shinozaki and Shinozaki, 1993; Mizoguchi et al., 2010). ANAC083-OX seedlings displays enhanced salt tolerance, meanwhile the transcript levels of RD29A and RD29B are up-regulated in ANAC083-OX seedlings under salt stress (Yang et al., 2011). Thus, the comparative analysis of NCED3, RD29A and RD29B expression levels in WT and TaNAC2D-OX plants was conducted to detect the molecular mechanisms of TaNAC2D function. The results suggest that the down-regulation of NCED3, RD29A, and RD29B may be responsible for the reduced stress tolerance observed in the 35-day-old soil-grown TaNAC2D-OX plants. In contrast, the up-regulation of these three marker genes in 14-day-old TaNAC2D-OX seedling, grown on 1/2 MS medium, may contribute to the enhanced stress tolerance during germination and post-germinative growth periods. Our results indicate that the changes in growth stages and environmental conditions affect the expression levels of NCED3, RD29A and RD29B by regulating the transcriptional activity of TaNAC2D, leading to the variant stress-responsive phenotypes of TaNAC2D-OX plants. Several studies have demonstrated that the different growth stages and environmental conditions may regulate gene functions. For example, ATAF2 functions as a negative regulator of pathogenesis-related genes under sterile conditions (Delessert et al., 2005). In contrast, ATAF2 can increase the expression of pathogenesis-related genes under non-sterile growth conditions (Wang et al., 2009). Ha et al. (2014) revealed that the max mutant plants display increased sensitivity to salt and drought stresses compared with WT plants at mature plant stage, while primary root length is not significantly different between WT and max mutant plants at seedling stage under salt and osmotic stresses conditions. Based on previous findings and our own data, TaNAC2D’s function in the regulation of the stress response may vary depending on the growth stages and environmental conditions of plant.
Several NAC TFs act either as transcriptional repressors or activators to regulate the expression of downstream genes, leading to down-regulation or up-regulation of these target genes. VNI2 is a transcriptional repressor under normal growth conditions (Yamaguchi et al., 2010). However, the transcriptional activation activity of VNI2 was greatly elevated under salt-treatment conditions, resulting in up-regulation of COR and RD genes (Yang et al., 2011). Arabidopsis WUSCHEL acts as a repressor of transcription in shoot meristems, but it becomes an activator of transcription in floral patterning (Leibfried et al., 2005; Ikeda et al., 2009). These previous findings reveal that the transcriptional regulation activities are modulated by both intrinsic and environmental factors. Moreover, NAC TFs contain DNA-binding (DB) domain, NAC repression domain (NARD) and activation domain, which interact with each other to regulate transcriptional activities (Hao et al., 2010; Puranik et al., 2012). The D subdomain of NAC (Supplementary Figure S1A) contains NARD which acts to suppress transcriptional activity (Hao et al., 2010). If the function of NARD is stronger than that of activation domain, NAC TFs may be a transcriptional repressor. Conversely, NAC TFs act as a transcriptional activator. If the functions from NARD and activation domain are roughly equal, the transcriptional regulation activities may be dependent on the actual situation in plant (Hao et al., 2010). It is possible that the growth stages and environmental conditions may induce changes in the structure and/or activities of TaNAC2D. Based on previous findings and our data, we propose that TaNAC2D acts as a transcriptional activator at seedling stage under sterile conditions and as a transcriptional repressor at mature plant stage under non-sterile conditions.
Conclusion
Our results show that TaNAC2D encodes a typical NAC TFs. The 35-day-old soil-grown TaNAC2D-OX plants displayed greater water loss and significantly reduced salt and drought tolerance under non-sterile conditions, while TaNAC2D-OX seedlings had improved tolerance to salt, drought, and oxidative stresses at seedling stage under sterile conditions. The different growth stages and environmental conditions may induce change in the transcriptional activity of TaNAC2D, leading to up-regulation or down-regulation of downstream genes. The present study provides some insight into the complex mechanisms of NAC.
Author Contributions
QH conducted all experiments, analyzed the data, and drafted the manuscript. YW helped to conduct experiments. All authors read and approved the final manuscript.
Funding
This work was supported by Open Research Fund of Key Laboratory of Genetic Development and Germplasm Enhancement of Rare Plants in Three Gorges Area (2016KBC03); Research Fund for the Doctoral Program of China Three Gorges University.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We thank Dr. Jiapeng Han for general experimental technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01754/full#supplementary-material
Abbreviations
ABA, abscisic acid; EV, empty vector; ID, intrinsically disordered; MV, methylviologen; NAC, NAM, ATAF, and CUC; ORF, open reading frame; OX, overexpression; qRT-PCR, quantitative reverse transcription-PCR; REL, relative electrolyte leakage; WT, wild-type.
Footnotes
References
Al Abdallat, A. M., Ayad, J. Y., Abu Elenein, J. M., Al Ajlouni, Z., and Harwood, W. A. (2014). Overexpression of the transcription factor HvSNAC1 improves drought tolerance in barley (Hordeum vulgare L.). Mol. Breed. 33, 401–414. doi: 10.1007/s11032-013-9958-1
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cramer, G. R., Urano, K., Delrot, S., Pezzotti, M., and Shinozaki, K. (2011). Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 11:163. doi: 10.1186/1471-2229-11-163
Delessert, C., Kazan, K., Wilson, I. W., Van Der Straeten, D., Manners, J., Dennis, E. S., et al. (2005). The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J. 43, 745–757. doi: 10.1111/j.1365-313X.2005.02488.x
Fang, Y., You, J., Xie, K., Xie, W., and Xiong, L. (2008). Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genomics 280, 547–563. doi: 10.1007/s00438-008-0386-6
Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., et al. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876. doi: 10.1111/j.1365-313X.2004.02171.x
Garapati, P., Xue, G. P., Munne-Bosch, S., and Balazadeh, S. (2015). Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol. 168, 1122–1139. doi: 10.1104/pp.15.00567
Ha, C. V., Leyva-Gonzalez, M. A., Osakabe, Y., Tran, U. T., Nishiyama, R., Watanabe, Y., et al. (2014). Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc. Natl. Acad. Sci. U.S.A. 111, 851–856. doi: 10.1073/pnas.1322135111
Hao, Y. J., Song, Q. X., Chen, H. W., Zou, H. F., Wei, W., Kang, X. S., et al. (2010). Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta 232, 1033–1043. doi: 10.1007/s00425-010-1238-2
He, X., Qu, B., Li, W., Zhao, X., Teng, W., Ma, W., et al. (2015). The nitrate-inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol. 169, 1991–2005. doi: 10.1104/pp.15.00568
Hu, H., Dai, M., Yao, J., Xiao, B., Li, X., Zhang, Q., et al. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. U.S.A. 103, 12987–12992. doi: 10.1073/pnas.0604882103
Hu, W., Huang, C., Deng, X., Zhou, S., Chen, L., Li, Y., et al. (2013). TaASR1, a transcription factor gene in wheat, confers drought stress tolerance in transgenic tobacco. Plant Cell Environ. 36, 1449–1464. doi: 10.1111/pce.12074
Hu, W., Yuan, Q., Wang, Y., Cai, R., Deng, X., Wang, J., et al. (2012). Overexpression of a wheat aquaporin gene, TaAQP8, enhances salt stress tolerance in transgenic tobacco. Plant Cell Physiol. 53, 2127–2141. doi: 10.1093/pcp/pcs154
Huang, Q., Wang, Y., Li, B., Chang, J., Chen, M., Li, K., et al. (2015). TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 15:268. doi: 10.1186/s12870-015-0644-9
Huh, S. U., Lee, S. B., Kim, H. H., and Paek, K. H. (2012). ATAF2, a NAC transcription factor, binds to the promoter and regulates NIT2 gene expression involved in auxin biosynthesis. Mol. Cells 34, 305–313. doi: 10.1007/s10059-012-0122-2
Ikeda, M., Mitsuda, N., and Ohme-Takagi, M. (2009). Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21, 3493–3505. doi: 10.1105/tpc.109.069997
Jensen, M. K., Hagedorn, P. H., de Torres-Zabala, M., Grant, M. R., Rung, J. H., Collinge, D. B., et al. (2008). Transcriptional regulation by an NAC (NAM-ATAF1,2-CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. sp. hordei in Arabidopsis. Plant J. 56, 867–880. doi: 10.1111/j.1365-313X.2008.03646.x
Jensen, M. K., Lindemose, S., de Masi, F., Reimer, J. J., Nielsen, M., Perera, V., et al. (2013). ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio 3, 321–327. doi: 10.1016/j.fob.2013.07.006
Jeong, J. S., Kim, Y. S., Baek, K. H., Jung, H., Ha, S. H., Do Choi, Y., et al. (2010). Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 153, 185–197. doi: 10.1104/pp.110.154773
Jeong, J. S., Kim, Y. S., Redillas, M. C., Jang, G., Jung, H., Bang, S. W., et al. (2013). OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 11, 101–114. doi: 10.1111/pbi.12011
Kim, Y. S., Kim, S. G., Park, J. E., Park, H. Y., Lim, M. H., Chua, N. H., et al. (2006). A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18, 3132–3144. doi: 10.1105/tpc.106.043018
Kim, Y. S., Sakuraba, Y., Han, S. H., Yoo, S. C., and Paek, N. C. (2013). Mutation of the Arabidopsis NAC016 transcription factor delays leaf senescence. Plant Cell Physiol. 54, 1660–1672. doi: 10.1093/pcp/pct113
Leibfried, A., To, J. P., Busch, W., Stehling, S., Kehle, A., Demar, M., et al. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438, 1172–1175. doi: 10.1038/nature04270
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, P. L., Chen, N. Z., An, R., Su, Z., Qi, B. S., Ren, F., et al. (2007). A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol. Biol. 63, 289–305. doi: 10.1007/s11103-006-9089-8
Luo, X., Bai, X., Sun, X., Zhu, D., Liu, B., Ji, W., et al. (2013). Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J. Exp. Bot. 64, 2155–2169. doi: 10.1093/jxb/ert073
Mao, X., Chen, S., Li, A., Zhai, C., and Jing, R. (2014). Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in Arabidopsis. PLoS ONE 9:e84359. doi: 10.1371/journal.pone.0084359
Mao, X., Zhang, H., Qian, X., Li, A., Zhao, G., and Jing, R. (2012). TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 63, 2933–2946. doi: 10.1093/jxb/err462
McGrann, G. R., Steed, A., Burt, C., Goddard, R., Lachaux, C., Bansal, A., et al. (2015). Contribution of the drought tolerance-related stress-responsive NAC1 transcription factor to resistance of barley to Ramularia leaf spot. Mol. Plant Pathol. 16, 201–209. doi: 10.1111/mpp.12173
Mizoguchi, M., Umezawa, T., Nakashima, K., Kidokoro, S., Takasaki, H., Fujita, Y., et al. (2010). Two closely related subclass II SnRK2 protein kinases cooperatively regulate drought-inducible gene expression. Plant Cell Physiol. 51, 842–847. doi: 10.1093/pcp/pcq041
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Osakabe, Y., Arinaga, N., Umezawa, T., Katsura, S., Nagamachi, K., Tanaka, H., et al. (2013). Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 25, 609–624. doi: 10.1105/tpc.112.105700
Peng, H., Zhao, J., and Neff, M. M. (2015). ATAF2 integrates Arabidopsis brassinosteroid inactivation and seedling photomorphogenesis. Development 142, 4129–4138. doi: 10.1242/dev.124347
Puranik, S., Sahu, P. P., Srivastava, P. S., and Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. doi: 10.1016/j.tplants.2012.02.004
Redillas, M. C., Jeong, J. S., Kim, Y. S., Jung, H., Bang, S. W., Choi, Y. D., et al. (2012). The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol. J. 10, 792–805. doi: 10.1111/j.1467-7652.2012.00697.x
Ren, Z., Zheng, Z., Chinnusamy, V., Zhu, J., Cui, X., Iida, K., et al. (2010). RAS1, a quantitative trait locus for salt tolerance and ABA sensitivity in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107, 5669–5674. doi: 10.1073/pnas.0910798107
Sakuraba, Y., Kim, Y. S., Han, S. H., Lee, B. D., and Paek, N. C. (2015). The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant Cell 27, 1771–1787. doi: 10.1105/tpc.15.00222
Sun, L., Huang, L., Hong, Y., Zhang, H., Song, F., and Li, D. (2015). Comprehensive analysis suggests overlapping expression of rice ONAC transcription factors in abiotic and biotic stress responses. Int. J. Mol. Sci. 16, 4306–4326. doi: 10.3390/ijms16024306
Tran, L. S., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. doi: 10.1105/tpc.104.022699
Wang, X., Goregaoker, S. P., and Culver, J. N. (2009). Interaction of the Tobacco mosaic virus replicase protein with a NAC domain transcription factor is associated with the suppression of systemic host defenses. J. Virol. 83, 9720–9730. doi: 10.1128/JVI.00941-09
Wu, A., Allu, A. D., Garapati, P., Siddiqui, H., Dortay, H., Zanor, M. I., et al. (2012). JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24, 482–506. doi: 10.1105/tpc.111.090894
Wu, Y., Deng, Z., Lai, J., Zhang, Y., Yang, C., Yin, B., et al. (2009). Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 19, 1279–1290. doi: 10.1038/cr.2009.108
Xue, G. P., Way, H. M., Richardson, T., Drenth, J., Joyce, P. A., and McIntyre, C. L. (2011). Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol. Plant 4, 697–712. doi: 10.1093/mp/ssr013
Yamaguchi, M., Ohtani, M., Mitsuda, N., Kubo, M., Ohme-Takagi, M., Fukuda, H., et al. (2010). VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22, 1249–1263. doi: 10.1105/tpc.108.064048
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1993). Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol. Gen. Genet. 236, 331–340. doi: 10.1007/BF00277130
Yang, A., Dai, X., and Zhang, W. H. (2012). A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 63, 2541–2556. doi: 10.1093/jxb/err431
Yang, S. D., Seo, P. J., Yoon, H. K., and Park, C. M. (2011). The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23, 2155–2168. doi: 10.1105/tpc.111.084913
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Yoshida, T., Mogami, J., and Yamaguchi-Shinozaki, K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. doi: 10.1016/j.pbi.2014.07.009
You, J., Zhang, L., Song, B., Qi, X., and Chan, Z. (2015). Systematic analysis and identification of stress-responsive genes of the NAC gene family in Brachypodium distachyon. PLoS ONE 10:e0122027. doi: 10.1371/journal.pone.0122027
Zhong, R., Lee, C., and Ye, Z. H. (2010). Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol. Plant 3, 1087–1103. doi: 10.1093/mp/ssq062
Keywords: wheat, Arabidopsis, NAC, TaNAC2D, stress
Citation: Huang Q and Wang Y (2016) Overexpression of TaNAC2D Displays Opposite Responses to Abiotic Stresses between Seedling and Mature Stage of Transgenic Arabidopsis. Front. Plant Sci. 7:1754. doi: 10.3389/fpls.2016.01754
Received: 07 August 2016; Accepted: 07 November 2016;
Published: 23 November 2016.
Edited by:
Sheo Mohan Prasad, Allahabad University, IndiaReviewed by:
Anil Kumar Singh, Indian Institute of Agricultural Biotechnology (ICAR), IndiaHao Peng, Washington State University, USA
Copyright © 2016 Huang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quanjun Huang, 405478393@qq.com
 Quanjun Huang
Quanjun Huang Yan Wang2
Yan Wang2