- 1State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, China
- 2State Key Laboratory of Crop Stress Biology for Arid Areas, College of Life Science, Northwest A&F University, Yangling, China
- 3Shaanxi Rice Research Institute, Hanzhong Agricultural Science Institute, Hanzhong, China
Wheat stripe rust, caused by Puccinia striiformis f. sp. tritici (Pst), is one of the most devastating diseases of wheat in China. Rapid change to virulence following release of resistant cultivars necessitates ongoing discovery and exploitation of new resistance resources. Considerable effort has been directed at non-host resistance (NHR) which is believed to be durable. In the present study we identified rice mutant crr1 (compromised resistance to rust 1) that exhibited compromised NHR to Pst. Compared with wild type rice variety Nipponbare, crr1 mutant displayed a threefold increase in penetration rate by Pst, and enhanced hyphal growth. The pathogen also developed haustoria in crr1 mesophyll cells, but failed to sporulate. The response to the adapted rice pathogen Magnaporthe oryzae was unchanged in crr1 relative to the wild type. Several defense-related genes involved in the SA- and JA-mediated defense pathways response and in phytoalexin synthesis (such as OsPR1a, OsLOX1, and OsCPS4) were more rapidly and strongly induced in infected crr1 leaves than in the wild type, suggesting that other layers of defense are still in effect. Genetic analysis and mapping located the mutant loci at a region between markers ID14 and RM25792, which cover about 290 kb genome sequence on chromosome 10. Further fine mapping and cloning of the locus should provide further insights into NHR to rust fungi in rice, and may reveal new strategies for improving rust resistance in wheat.
Introduction
Wheat stripe rust, caused by Puccinia striiformis f. sp. tritici (Pst), is a devastating disease of wheat worldwide (Wellings, 2011). In China, the annual yield losses to stripe rust in wheat were estimated to be about 1 million metric tons (Chen et al., 2009). Cultivation of resistant varieties is the most effective, economical, and environmentally friendly way to control the disease. Although many resistance genes have been identified and utilized in wheat cultivars (Cheng et al., 2014; Lu et al., 2014; Han et al., 2015), the protection conferred has not been durable due to genetic variation in the pathogen population. One example is that of resistance gene Yr26 that was widely used in Chinese wheat breeding in recent years. New Yr26-virulent (CYR34, V26) races are now increasing and are causing unacceptable levels of disease on many of the cultivars with Yr26 (Han et al., 2015). Although resistances governed by quantitative trait loci (QTL) confer a broader-spectrum resistance, the level of resistance is not adequate to prevent significant crop losses, especially under severe epidemic conditions (Niks et al., 2015b). Thus, more durable control of stripe rust is urgently needed, and in addition to current exploration and identification of new resistance genes in wheat and its close relatives, a better understanding of non-host resistance (NHR) may offer opportunities in breeding for sustainable disease control.
In nature, a specific pathogen usually causes disease on a few plant species; that is, most plants are resistant to a wide range of phytopathogens. This form of disease resistance exhibited by all members of a plant species to all genetic variants of a non-adapted pathogen species [or possibly formae speciales (f. sp.)] is known as NHR. Due to its broad-spectrum effectiveness and durability NHR is of considerable interest for crop resistance improvement.
The genetic and molecular mechanisms underlying NHR remain largely unknown. Currently, the best-studied example of NHR is interaction between Arabidopsis and the non-adapted barley biotrophic fungal pathogen Blumeria graminis f. sp. hordei (Bgh), the causal agent of barley powdery mildew. Three NHR genes PEN1, PEN2, and PEN3 required for penetration resistance of Arabidopsis to Bgh have been isolated. Functional mutants of any one of the three PEN genes display increased penetration rates by Bgh. PEN1 (Collins et al., 2003; Lipka et al., 2005; Stein et al., 2006) encodes a membrane-associated syntaxin containing a SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) domain and is a member of a large family of proteins functioning in membrane fusion and secretion events. Cytological studies have demonstrated that PEN1 is involved in papilla formation. PEN2 encodes a glycoside hydrolase, which associates with the periphery of peroxisomes. PEN3 encode an ABC (ATP binding cassette) transporter that is localized to the plasma membrane. PEN2 and PEN3 may collaborate in transport of antimicrobial compounds to the apoplast. Importantly, the protein products of these PEN genes accumulate at sites of fungal penetration. Moreover, these genes also contribute to R gene-mediated resistance and cell death in response to both adapted and non-adapted pathogens (Johansson et al., 2014). While, for those plants that are evolutionary closely related to the natural host, the NHR is proposed to be predominantly mediated by multiple R genes that collectively confer resistance to all isolates of a pathogen species (Schulze-Lefert and Panstruga, 2011). For example, the NHR of Arabidopsis to non-adapted Albugo candida may result from recognition of pathogen effectors by multiple WRR4-like genes, which encode typical NB-LRR resistance proteins (Borhan et al., 2008).
There are many published reports on NHR to rust fungi. For example, NHR of Arabidopsis and Medicago to bean rust (Patto and Rubiales, 2014; Ishiga et al., 2015; Langenbach et al., 2016), barley and Brachypodium distachyon NHR to cereal rust pathogens (Zellerhoff et al., 2010; Dawson et al., 2015; Figueroa et al., 2015; Niks et al., 2015a). As the model of monocotyledonous plant, rice is unusual in not being affected by a rust pathogen (Ayliffe et al., 2011b). Several studies indicated that rust fungi have some potential to infect rice and trigger host defense responses such as production of reactive oxygen species and accumulation of Pathogenesis-related proteins (Ayliffe et al., 2011a; Li et al., 2012). Thus, it is of interest to characterize non-host interaction between rice and cereal rust pathogens and identify key genes involved in NHR. Proteomic studies revealed proteins that are involved in phytoalexin production, and glycerol-3-phosphate metabolism may have a role in rice NHR to P. triticina and Pst (Li et al., 2012; Zhao et al., 2014).
In the present study we identified rice mutant crr1 (compromised resistance to rust 1) that allowed a high level of penetration rates by Pst and enhanced hyphal growth. The fungus was able to develop haustoria in mesophyll cells of the mutant, but failed to sporulate. Histological analysis revealed that hydrogen peroxide (H2O2) production and callose deposition were not affected in the crr1 mutant. Furthermore, upon infected by Pst crr1 showed strikingly enhanced expression levels of defense-related genes involved in the SA-, and JA-mediated defense pathways as well as phytoalexin synthesis. These observations suggested different molecular mechanisms underlying NHR to Pst in rice compared to the host resistance. Genetic analysis demonstrated that the phenotype of crr1 was conditioned by a recessive gene between markers ID14 and RM25792 at the end of rice chromosome 10. Characterization and genetic study of crr1 would provide new insights into NHR, and assist in breeding wheat cultivars with durable resistance to stripe rust.
Results
crr1 Exhibited Compromised NHR to Pst
We screened 5,229 T2 rice mutant families and found nine putative mutants that allowed increased Pst growth in leaf tissue. These putative mutants were designated as Comprised resistance to rust fungus (crr1-9). Among them, crr1 showed the most Pst development in rice tissues. At 14 days after inoculation with Pst, wild type plants showed no visible symptoms, whereas brown flecks appeared on leaves of crr1 mutant plants, and these lesions were associated with hyphal colonization (Figures 1A,B). Microscopic observation revealed that most of the urediniospores germinated on the leaf surfaces of both wild type and crr1. However, on the wild type rice, only a few (0.2%) germinated urediniospores penetrated into stomates and successfully formed substomatal vesicles (ssv). In contrast, on crr1, the penetration rate was 0.6%, about three times higher than that on wild type plants (Figure 2A). Moreover, the rust fungus can produce haustoria in crr1 mesophyll cells, suggesting a defective of pre-invasion NHR in the mutant (Figures 1C,D). In addition, compared with the small colony (from 0 to 4,000 μm2 in area) developed in wild type rice, the clearly larger colonies were developed in crr1 with an average area over 10,000 μm2 (Figure 2B).
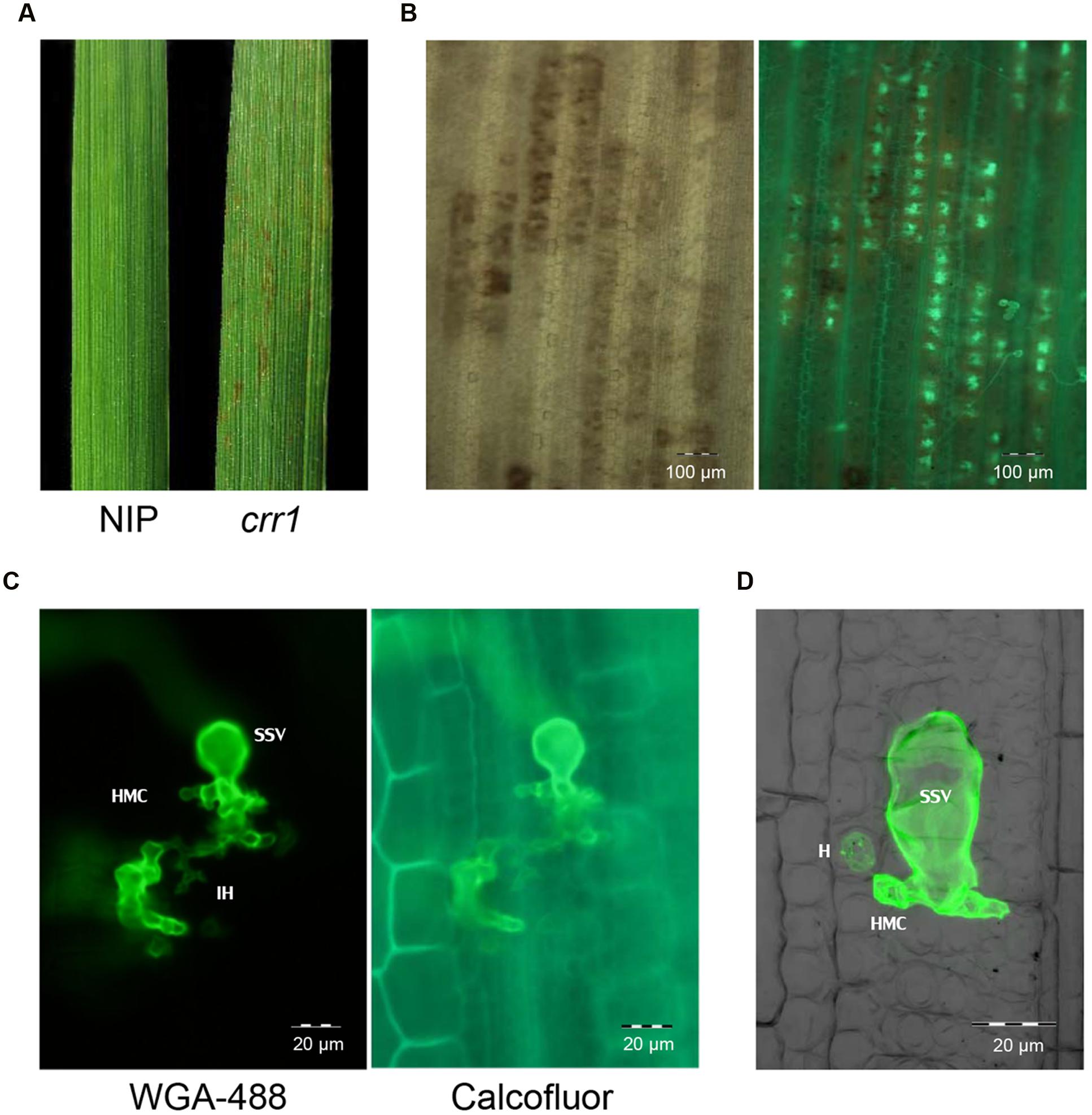
FIGURE 1. Infection of rice mutant crr1 and wild type Nipponbare with Puccinia Striiformis f.sp. tritici (Pst) race CYR32. (A) Macroscopic observation of Nipponbare and crr1 leaves infected with Pst. (B) Microscopic observation of Pst development in crr1 leaves using bright field (left) and fluorescence (right). (C) Pst infection site in crr1 showing formation of sub-stomatal vesicle (ssv), infection hyphae (IH) and haustoria mother cell (HMC). (D) A haustorium (H) in crr1 mesophyll cells. The image was composite of stacked fluorescent and blight field photographs by confocal microscopy.
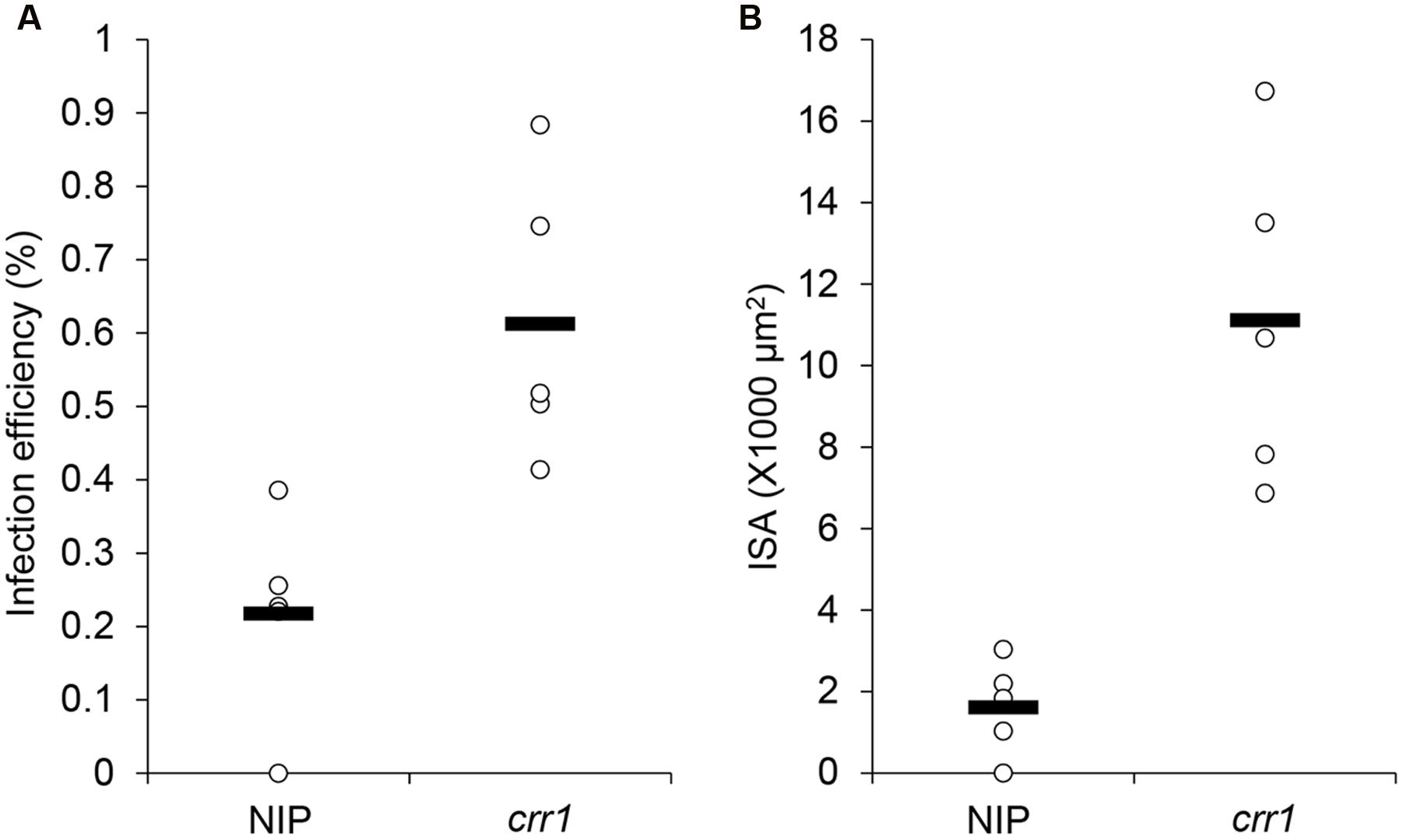
FIGURE 2. Penetration frequency and infection site area of Pst on Nipponbare and crr1. (A) Penetration frequency of Pst on Nipponbare and crr1. (B) Infection site area of Pst on Nipponbare and crr1. Open circles show measurements for each plants. Black lines show the medians of the data.
To determine whether disease response of crr1 was affected following infection by an adapted pathogen, we compared the responses of mutant and wild type plants to Pyricularia oryzae (strain Guy11), the causal agent of rice blast. As shown in Supplementary Figure S1, both Nipponbare and crr1 exhibited similar partial resistance responses to strain Guy11. These results suggesting that the host resistance is not affected by the mutation event in crr1.
Transmission Electronic Microscopy Observation of Pst Growth in crr1
To further understand the proliferation of Pst in crr1, we observed the colonization of Pst on crr1 leaves at 14 dpi using transmission electronic microscopy. Extensive growth was evident in crr1 leaf tissue (Figure 3A). More interesting, although at very low frequency, haustoria were observed in mesophyll cells (Figure 3D). Most of the haustoria were abnormal and were associated with host cell death. Host cells surrounding the hyphae remained living (Figure 3B) and there was no apparent cell wall thickening or papillae formation (Figure 3C). Some hyphae had begun to die and there was no evidence of sporulation (Figure 3D).
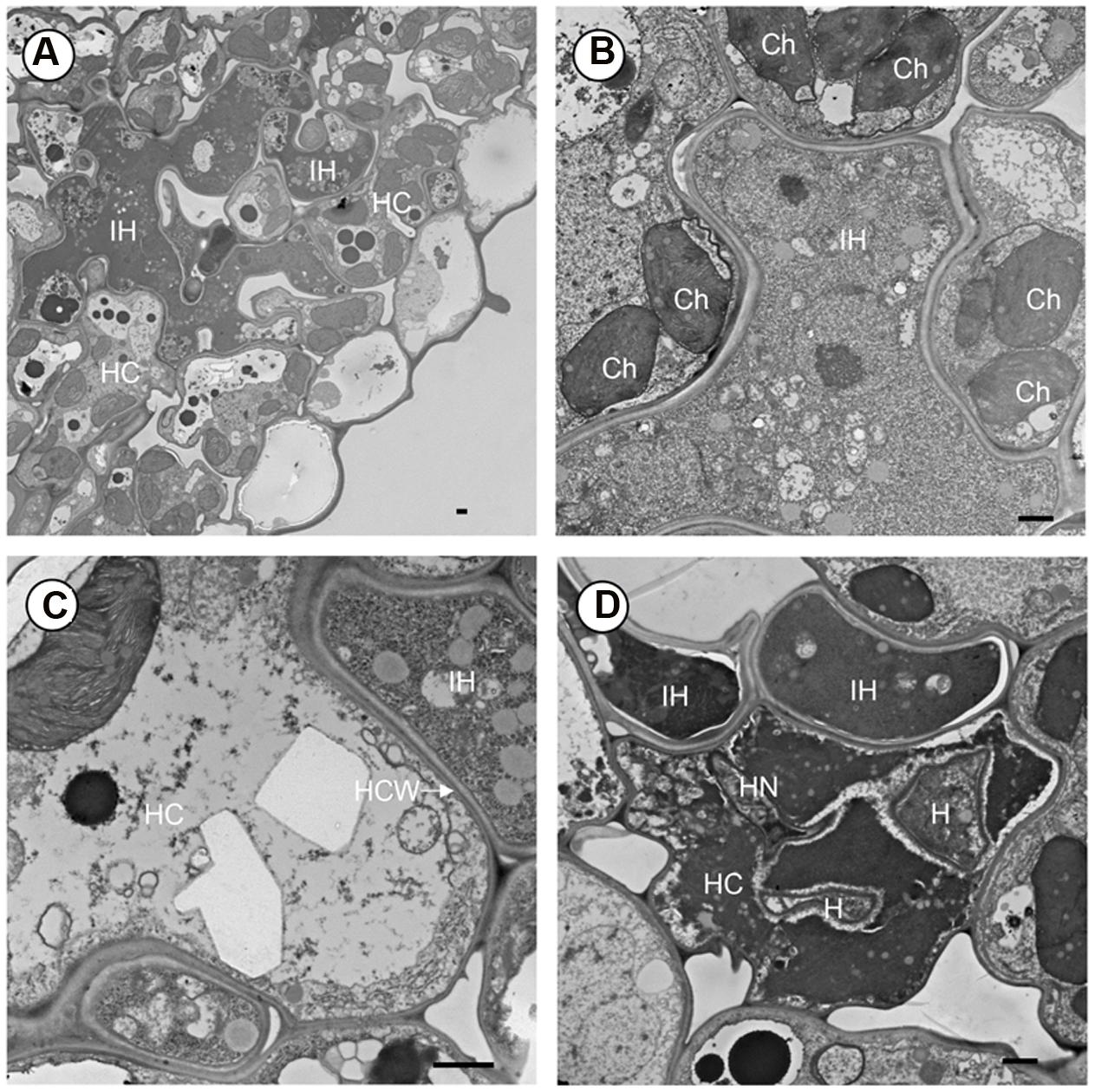
FIGURE 3. Transmission electron microscope (TEM) observation of Pst development in leaf tissue of rice mutant crr1. (A) Intercellular hyphae (IH) growing in the intercellular space and in the developing cavity between mesophyll and epidermis. (B) Intercellular hyphae (IH) surrounded by mesophyll cell. (C) Interface of host and fungus cell wall. (D) A haustorium in host cell and causing cell death. HC, host cell; IH, intercellular hyphae; Ch, chloroplast; HCW, host cell wall; H, haustorium; HN, haustoria neck. Scale bar: 1 μm.
H2O2 Production and Callose Deposition in crr1 Challenged by Pst
Hydrogen peroxide production plays a critical role in plant defense response. Previous studies have shown that H2O2 was induced in rice following rust infection (Ayliffe et al., 2011a; Yang et al., 2014). To examine whether the production of H2O2 was defective in crr1, we examined the H2O2 production at infection sites in mutant and wild type plants challenged by Pst. There was no apparent difference in H2O2 production between crr1 and wild type plants (Figure 4A). H2O2 was produced in the guard cells at 24 hpi in both crr1 and wild type plants, although some weak H2O2 signals were detected around the stomata. It is noteworthy that primary hyphae and ssv were formed in crr1 at 48 hpi despite of the production of H2O2 around the infection sites. We postulated that H2O2 mainly functions as a signal molecule in rice-Pst interaction and the weakened resistance of crr1 may result from lacking of some other downstream components. As another marker for plant defense response, callose deposition, was also compared between crr1 and wild type plants using the aniline blue staining method. More extensive callose deposition was observed with the growth of pathogen in crr1 (Figures 4B,C). These histological results suggested that the subdued NHR in crr1 should be independent of H2O2 production and callose deposition.
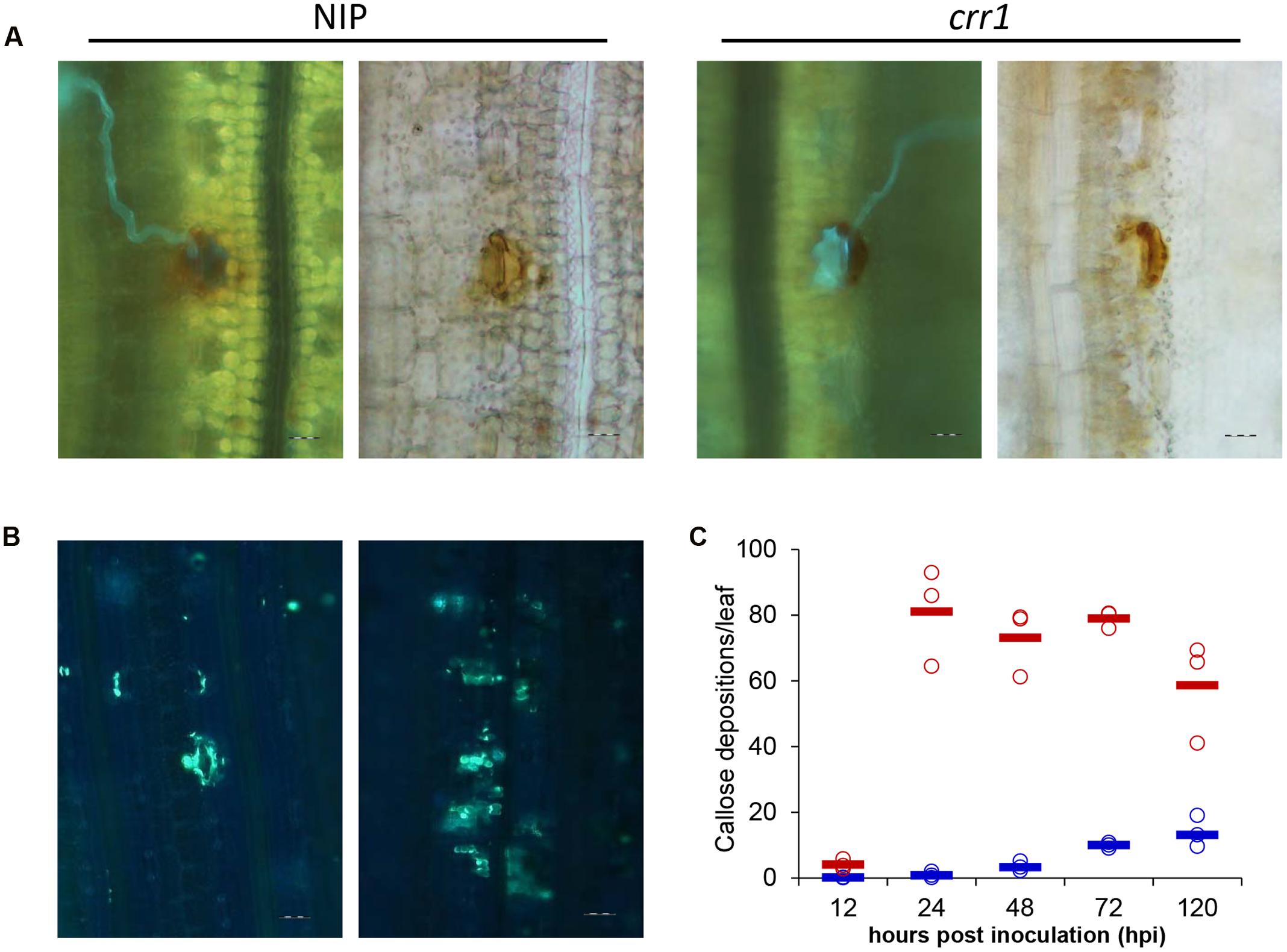
FIGURE 4. Histochemical examination of hydrogen peroxidase production and callose deposition at Pst infection sites in Nipponbare and crr1. (A) Fluorescence (left) and brightfield (right) images presenting hydrogen peroxidase production at infection sites in crr1 and wild type plants 48 hpi by Pst, Bar = 20 μm. (B) Callose deposition in crr1 and wild type plants at 120 hpi by Pst, Bar = 20 μm. (C) Quantification of callose deposition in crr1 and wild type plants at 12, 24, 48, 72, and 120 hpi by Pst. Open circles show measurements for each plants. Solid lines show the medians of the data. Blue and red symbols stand for data from Nipponbare and crr1, respectively.
Relative Expression Levels of Defense Related Genes
In plant–pathogen interactions many defense-related genes have been identified, and their analysis in crr1 could help in understanding factors involved in the NHR to Pst. Eight genes related to four functional categories were chosen to examine expression profiles. NPR1 and PR1 are marker genes for the salicylic acid-mediated defense pathway (Yuan et al., 2007; Yang et al., 2013). OsCATC and OsPOX encode catalase and peroxidase which function in ROS scavenging (Quan et al., 2008). OsAOC1 and OsLOX1 belong to the jasmonic acid pathway (Lyons et al., 2013; Yang et al., 2013). OsCPS4 and OsKSL8 represent genes involved in phytoalexin synthesis in rice (Hasegawa et al., 2010). Expression profiles of these genes were examined at four time points (0, 12, 24, and 72 hpi) in the mutant and wild type inoculated with Pst (Figure 5). The mRNA levels of NPR1 were increased at 12 and 24 hpi in Nipponbare and crr1, respectively, and then declined in Nipponbare but remained at a high level in crr1. PR1a was induced at 24 hpi in both Nipponbare and crr1. OsLOX1 was also induced at 24 hpi, while another gene OsAOC1 involved in the jasmonic acid pathway was slightly induced at 12 hpi only in Nipponbare. Both OsCPS4 and OsKSL8 were induced at 24 hpi in Nipponbare and crr1. OsCATC was induced in crr1 at 12 hpi, and at a considerably later stage (72 hpi) in Nipponbare. OsPOX was also induced at 12 hpi in crr1 where it kept increasing. In general, most of the defense-related genes were responsive to Pst infection in both wild type and crr1 mutant plants, but their induction was quicker and stronger in crr1 compared with that in wild type plants.
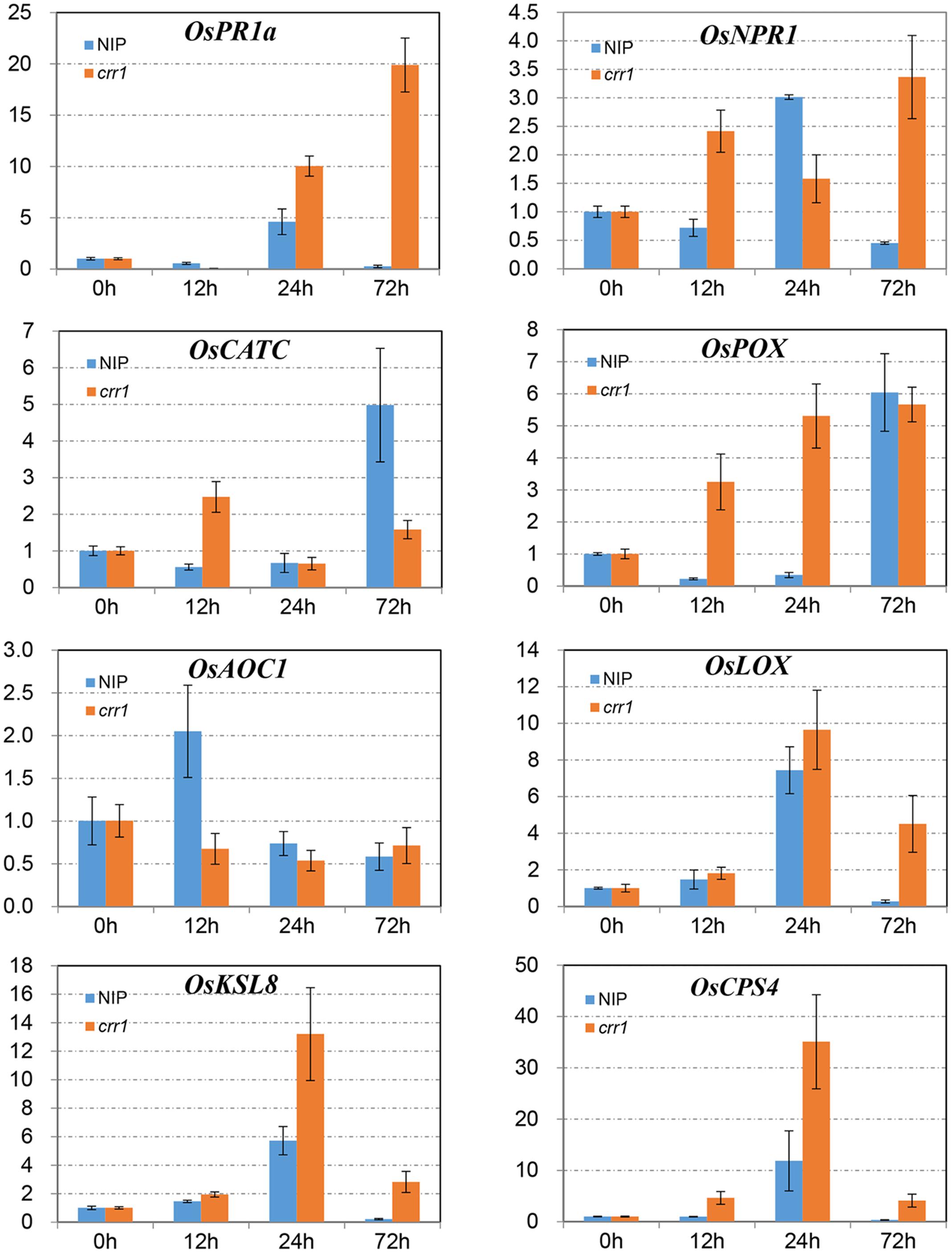
FIGURE 5. Relative expression patterns of defense-related genes in Nipponbare and crr1 infected with Pst. The mRNA levels of eight defense-related genes in Nipponbare and crr1 were examined at four time points (12, 24, 48, and 120 hpi). Relative expression levels are normalized to the values of the mock-inoculated plants.
Mapping of the Crr1 Gene
Sixty-six F2 individuals from the cross between crr1 and Nipponbare were evaluated for response to Pst infection. Fifty-two plants exhibited compromised resistance similar to crr1 and 14 plants displayed complete immune similar to Nipponbare. The phenotypic segregation fitted well to the expected 1:3 ratio indicative of a single gene model with homozygosity of the recessive allele leading to increased hyphal development at infection sites (χ2 = 0.81, P = 0.48). This observation suggested that the phenotype of crr1 resulted from a single locus mutation. Southern-blotting results indicated that crr1 possessed a single T-DNA insertion. The flanking sequence of the T-DNA was isolated by inverse PCR (Zhang et al., 2007) and the insertion site was located at the CDS of Os07g40020 (a GRAS family protein) but cosegregation analysis showed that the T-DNA insertion was independent of the locus segregating for the altered host response.
In order to map the gene, we developed F2 populations from crosses of crr1 with Zhonghua 11 and Mudanjiang 8. Using bulked segregant analysis (BSA) (Michelmore et al., 1991) the Crr1 locus was putatively mapped to the long arm of chromosome 10 and was linked to SSR marker RM25761. Twelve SSR and indel markers from this region were developed to generate a higher density map of the candidate region (Supplementary Table S1). Composite interval mapping (CIM) placed Crr1 between markers Id14 and RM25792 (Figure 6) with a logarithm of odds (LOD) score of 11.6. Variation of infection site area (ISA) and NIS at the locus accounted for 29.8% and 39.9% of the total variation, respectively (Table 1). This candidate region encompasses about 290 kb of genome sequence and contained 40 annotated genes.
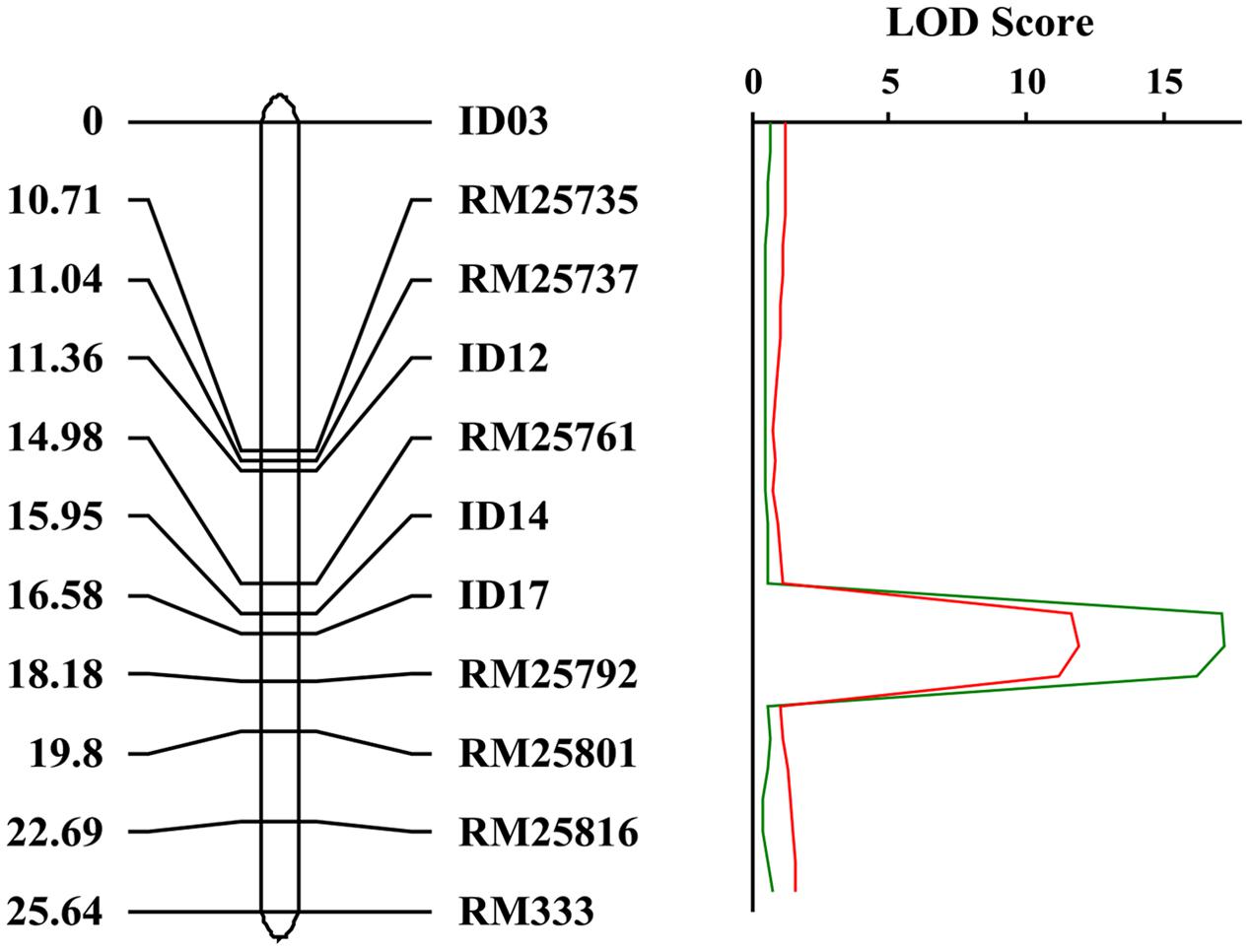
FIGURE 6. Genetic map of the crr1 locus based on the crr1/Zhonghua 11 F2 mapping population. The candidate crr1 gene was mapped to the region between two markers ID14 and RM25792 which located at the end of long arm of rice chromosome 10.

TABLE 1. Locus identified from composite interval mapping based on the infection site area (ISA) and number of infection sites (NIS) of the crr1/Zhonghua 11 F2 population inoculated with Pst.
Discussion
Non-host resistance has potential to provide non-specific, durable resistance to diseases, and to offer alternative possibilities to traditional R gene-mediated resistance. Unfortunately, no rice gene underlying NHR to rust has been identified so far. In this study, we characterized and identified a gene conferring NHR to Pst using a rice mutant crr1. This study may contribute to our understanding of rice NHR to Pst and exploiting rice NHR to develop wheat lines with more durable resistance to Pst.
According to the two-layered paradigm of plant active immune system, there are two types of NHR: pre-invasion defense mediated by PAMP-triggered immunity (PTI) and post-invasion defense mediated by effectors-triggered immunity (ETI) (Gill et al., 2015). Our data showed that the frequency of stomatal penetration of Pst urediniospores on wild type rice (0.2%) was significantly reduced than that on wheat (over 25% in previous study) (Cheng et al., 2013). Similar studies demonstrated that 0.5% urediniospores of P. triticina formed ssv on Arabidopsis leaves (Shafiei et al., 2007). These results suggested that rice NHR to Pst might largely act at the pre-invasion stage. Histologic studies revealed H2O2 production and callose accumulation concentrated in and around guard cells following Pst inoculation, but no hypersensitive cell death was detected. Therefore, PTI mediated defense responses that precede host mesophyll cell invasion appear to play the predominant roles in blocking attempted Pst. Compared with intermediate hosts of Pst, such as barley and Brachypodium, rice is more distantly related to wheat and resistant to all rusts (Vogel et al., 2010; Mayer et al., 2011). Thus our results fit well with the molecular evolution model that PTI play a key role when pathogens attempt to infect more distantly related non-host species (Schulze-Lefert and Panstruga, 2011). Increased frequency of stomatal penetration and larger infection sites in crr1 may result from a disrupted defense step preceding mesophyll cell invasion.
Although crr1 plants exhibited more susceptibility to Pst, the H2O2 production was not affected relative to wild type. In addition, callose deposition increased as hyphal growth expanded in leaf tissues of the crr1 mutant. Similarly, the induction of defense related genes, such as genes involved in SA/JA mediated defense pathway and phytoalexin synthesis, were quicker and stronger in crr1 than that in wild type plants. Collectively, these findings suggest that increased development of Pst in apoplasts of crr1 plants triggers a more extensive defense response. Counter-intuitively, these defense responses seemed to have limited effects on preventing the growth of Pst. Similar findings were reported for non-host interaction between pen mutants and the barley mildew pathogen (Collins et al., 2003; Lipka et al., 2005; Stein et al., 2006). A natural variant of Arabidopsis accession Wa-1 which has compromised resistance to non-adapted wheat leaf rust pathogen exhibits increased SA and PR1 expression following P. triticina challenge (Shafiei et al., 2007). In these cases, there seems to be a “true” NHR which spanning multiple layers of defense. Crr1 mutants are indeed compromised for part of the defense (mainly related to ingress). However, the penetration rate is still very low and most defense responses still seem to be intact and in fact the pathogen cannot really proliferate/sporulate either.
Haustoria are special structures of biotrophic fungus that suppress host immune system and take up nutrients from host cells. Formation of haustoria is usually regarded as the symbol of parasitism establishment (Yi and Valent, 2013). Haustoria have been observed by fluorescence microscopy in several studies concerning rice-rust fungus interactions, while the details of haustoria is not clear yet (Ayliffe et al., 2011a; Yang et al., 2014). In the present study, we observed haustoria formation in mesophyll cells in rice mutant by TEM. This finding suggested that rust fungi have the potential to absorb nutrients and infect rice. However, only a limited number of haustoria were observed. Although urediniospores have stored nutrients for germination and initial infection, it was unbelievable that such a few hautoria can support the considerable colonization that encompassed hundreds of host cells in crr1. This suggests the possibility that hyphae are able to take up nutrients from the intercellular spaces. Although it is generally believed that the apoplast of plants leaves is a relatively nutrient-poor environment, a considerable number of microbes do derive nutrients from it. In a study of interaction between barley and P. hordei, sucrose and glucose were found in apoplast at much lower concentrations in infected than in healthy leaves, and uptake of hexoses by intercellular hyphae was suggested as the cause of the reduction (Tetlow and Farrar, 1993). A sucrose transporter SRT1 from Ustilago maydis was shown to take up sucrose from intercellular spaces of maize leaves allowing the hyphae to grow along the phloem (Wahl et al., 2010). Thus, we postulated that hyphae of Pst may obtain nutrients from the apoplast in leaves of the crr1 rice mutant. These nutrients absorbed from the intercellular spaces support limited hyphal growth, but they may be insufficient to support sporulation.
Using BSA and CIM strategies we located the crr1 locus between id14 and RM25792 at the end of chromosome 10. This region contains about 40 annotated genes, and there is no previously annotated host or non-host resistance gene in the interval. Recently, three genes involved in Brachypodium NHR to Pst were identified by flanking sequence isolation from mutant (An et al., 2016). Among them, Bradi5g17540 encodes a BAP28 domain containing protein, Bradi5g17540 encodes a MYB transcription factor and Bradi5g11590 encodes a lipoxygenase. However, none of them is present in the candidate region of crr1. Another gene conferring non-host resistance to P. striiformis was identified in barley, and designated as Rps6 (Andrew et al., 2016; Li et al., 2016). Rps6 was mapped on the long arm of barley chromosome 7H, which has no collinearity with rice chromosome10, suggesting that it is different with crr1 locus.
Several studies have demonstrated the feasibility of transferring single NHR-related genes across plant species to create durable, broad-spectrum resistance (Brutus and He, 2010; Wen et al., 2011; Schoonbeek et al., 2015; Langenbach et al., 2016). The mechanisms of rice resistance to Pst can be used to improve wheat resistance. Since it is not feasible to transfer gene through homoeologous recombination by hybridization, transgenic strategy might be a good alternative. Thus, further studies based on more comprehensive efforts will be needed to clone the corresponding rice gene conferring NHR to stripe rust fungus.
Materials and Methods
Plants and Growth Conditions
Rice mutant crr1 was identified from 5229 Nipponbare mutants made by T-DNA insertional mutagenesis at Huazhong Agricultural University, Wuhan, China (Wu et al., 2003). Two segregating F2 populations derived from crosses of crr1 with two highly resistant japonica varieties Mudanjiang 8 (MDJ8) and Zhonghua11 (ZH11) identified previously (Yang et al., 2014).
Maintenance and Inoculation of Pst
Pst isolate CYR32 (a predominant Pst race in China) were maintained on a susceptible wheat cultivar, Mingxian 169, following the procedures and conditions described by Zhang et al. (2011). For inoculation of rice, 3-week-old rice seedlings were pre-spayed with 0.1% Triton X-100, then fresh Pst urediniospores suspensions (50 mg urediniospores ml-1) were applied with a fine paintbrush onto the adaxial surface of the second leaf. Rice plants inoculated with sterilized distilled water were used as a negative control. The inoculated seedlings were kept in a dew chamber at 100% humidity for 36 h at 12°C in complete darkness to ensure the maximal rate of infection. Subsequently, the seedlings were transferred to a growth chamber at 16°C with a 16/8 h light/dark cycle. Leaf tissues were collected at specific time points for various analyses.
Histochemical Analysis of the Rice-Pst Interactions
At least six inoculated rice leaves were harvested at 10 days post inoculation (dpi) for histopathological analysis. Rice leaf segments of 4 cm were cut from the center of inoculated leaves. Leaf sections were fixed and decolorized in ethanol/trichloromethane (4:1, v/v) containing 0.15% (w/v) trichloroacetic acid for 2 days, and the fixation solution was replaced with fresh solution twice every other day. The specimens were cleared in saturated chloral hydrate until leaf tissues were translucent. For Calcofluor White (Sigma–Aldrich) staining, the method described by Zhang et al. (2011) was followed. For further visualization of internal infection structures, the wheat germ agglutinin (WGA) conjugated to the Fluorophore Alexa 488 (Invitrogen) staining was used (Ayliffe et al., 2011b). H2O2 was detected using the 3, 3-diaminobenzidine (DAB, Amresco, Solon, OH, USA) staining method (Thordal-Christensen et al., 1997) and observed under differential interference contrast (DIC) optics.
Data Collection and Analysis
For ISA and penetration frequency, rice leaves were harvested at 2 weeks post inoculation (dpi) and stained by Calcofluor for observation. Infection sites were photographed under 10× or 20× magnifications using a focal plane that maximized the area of each infection site. ISA was measured using Olympus CellSens® Digital Imaging Software (Version 1.5). At least 10 infection sites were measured for each individual F2 rice plant. Penetration frequencies were calculated using the number of infection sites developing ssv or hyphe divided by the number of all urediospores and 10 inoculated plants of Nipponbare and crr1 were scored per experiment.
For the quantification of callose deposition by Pst infection, rice leaves were harvested at 12, 24, 48, 72, and 120 hpi. A total of 30 infection sites were examined for each time point of Nipponbare or crr1 plants in one experiment and three independent experiments were performed.
For all data, means and standard errors were calculated from three independent biological replicates using Student’s t-tests.
Cytological Analysis of the Rice-Pst Interaction
Leaves were harvested from inoculated rice crr1 mutants at 2-week after inoculation and prepared for transmission electron microscope (TEM) examination according to procedures previously described (Zhang et al., 2011). The leaf samples were cut into small pieces and fixed with 3% (v/v) glutaraldehyde in 50 mmol/l phosphate buffer (pH 6.8) for 3–6 h at 4°C. After rinsing thoroughly with the same buffer and post-fixation with 1% (w/v) osmium tetroxide for 2 h at 4°C, the samples were dehydrated in a graded alcohol series, embedded in gelatin capsules filled with LR White resin (Sigma–Aldrich), and polymerized at 60°C for 48 h. For TEM observations, ultra-thin sections of the samples were cut with a diamond knife and collected on 200 mesh copper grids. After contrasting with uranyl acetate and lead citrate, the grids were examined with a JEM-1230 TEM (Jeol Co. Ltd, Tokyo, Japan) at 80 kV.
Genetic Mapping
Sequences of primers for SSR markers were obtained from The IRGSP (International Rice Genome Sequencing Project) (Matsumoto et al., 2005). For indel markers, polymorphisms markers primers were designed according to the DNA Polymorphisms information on NODAI Genome Research Centre1 (Arai-Kichise et al., 2011). The sequences of molecular markers were presented as Supplementary Table S1.
For bulk segregant analysis (BSA), genomic DNA was extracted from leaves of the parents and 163 F2 plants of crr1/Zhonghua 11 and 86 F2 plants of crr1/Mudanjiang 8. According to the data of the number of infection sites and ISA, five most susceptible and resistant F2 plants were selected and equal amounts of their DNA were mixed to form the susceptible bulk (SB) resistant bulk (RB). Genotypes for 96 polymorphism markers evenly distributed in rice genome were analyzed among SB, RB together with the parents. Genotypes from the parents and the bulks were used to identify molecular markers linked to the target loci.
Quantitative trait loci mapping for crr1 locus was conducted based on the number of infection sites and ISA of each F2 plants. The inclusive composite interval mapping (ICIM) analysis was performed using the software QTL IciMapping V3.3. In the first step of the stepwise regression of ICIM, probabilities for including and excluding marker variables were set at 0.01 and 0.02, respectively. In the second step of interval mapping of ICIM, the threshold LOD score was set at 2.5 to declare significant QTL for all phenotyping methods.
Author Contributions
ZK and JZ conceived and designed research. JZ and YY performed the genetic mapping and analyzed the data. JZ and DY performed the histological observation and gene expression analysis. GZ, HZ, and YC screen and identified the mutant. JW and KZ developed genetic population and molecular marker. MJ performed the TEM experiment. LH and ZK contribute comments during manuscript preparation. JZ wrote the manuscript and ZK revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was financially supported by the National Basic Research Program of China (2013CB127700), the Modern Agro-industry Technology Research System in China (CARS-3-1-11) and the 111 Project from the Ministry of Education of China (B07049).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Shiping Wang (Huazhong Agricultural University, Wuhan) for providing the 5229 rice T-DNA insertion mutants. We are grateful to the review of this manuscript by Prof. R. A. McIntosh, Plant Breeding Institute, University of Sydney, Australia.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01822/full#supplementary-material
TABLE S1 | Primer sequences of molecular markers for crr1 genetic mapping.
Footnotes
References
An, T. Y., Cai, Y. L., Zhao, S. Z., Zhou, J. H., Song, B., Bux, H., et al. (2016). Brachypodium distachyon T-DNA insertion lines: a model pathosystem to study nonhost resistance to wheat stripe rust. Sci. Rep. 6:25510. doi: 10.1038/srep25510
Andrew, M. D., John, N. F., Matthew, G., Phon, G., Amelia, H., and Matthew, J. M. (2016). Isolation and fine mapping of Rps6: an intermediate host resistance gene in barley to wheat stripe rust. Theor. Appl. Genet. 129, 831–843. doi: 10.1007/s00122-015-2659-x
Arai-Kichise, Y., Shiwa, Y., Nagasaki, H., Ebana, K., Yoshikawa, H., Yano, M., et al. (2011). Discovery of genome-wide DNA polymorphisms in a landrace cultivar of japonica rice by whole-genome sequencing. Plant Cell Physiol. 52, 274–282. doi: 10.1093/pcp/pcr003
Ayliffe, M., Devilla, R., Mago, R., White, R., Talbot, M., Pryor, A., et al. (2011a). Nonhost resistance of rice to rust pathogens. Mol. Plant Microbe Interact. 24, 1143–1155. doi: 10.1094/mpmi-04-11-0100
Ayliffe, M., Jin, Y., Kang, Z., Persson, M., Steffenson, B., Wang, S., et al. (2011b). Determining the basis of nonhost resistance in rice to cereal rusts. Euphytica 179, 33–40. doi: 10.1007/s10681-010-0280-2
Borhan, M. H., Gunn, N., Cooper, A., Gulden, S., Tör, M., Rimmer, S. R., et al. (2008). WRR4 encodes a TIR-NB-LRR protein that confers broad-spectrum white rust resistance in Arabidopsis thaliana to four physiological races of Albugo candida. Mol. Plant Microbe Interact. 21, 757–768. doi: 10.1094/mpmi-21-6-0757
Brutus, A., and He, S. Y. (2010). Broad-spectrum defense against plant pathogens. Nat. Biotechnol. 28, 330–331. doi: 10.1038/nbt0410-330
Chen, W. Q., Wu, L. R., Liu, T. G., Xu, S. C., Jin, S. L., Peng, Y. L., et al. (2009). Race dynamics, diversity, and virulence evolution in Puccinia striiformis f. sp tritici, the causal agent of wheat stripe rust in China from 2003 to 2007. Plant Dis. 93, 1093–1101. doi: 10.1094/Pdis-93-11-1093
Cheng, P., Xu, L. S., Wang, M. N., See, D. R., and Chen, X. M. (2014). Molecular mapping of genes Yr64 and Yr65 for stripe rust resistance in hexaploid derivatives of durum wheat accessions PI 331260 and PI 480016. Theor. Appl. Genet. 127, 2267–2277. doi: 10.1007/s00122-014-2378-8
Cheng, Y. L., Zhang, H. C., Yao, J. N., Han, Q. M., Wang, X. J., Huang, L. L., et al. (2013). Cytological and molecular characterization of non-host resistance in Arabidopsis thaliana against wheat stripe rust. Plant Physiol. Biochem. 62, 11–18. doi: 10.1016/j.plaphy.2012.10.014
Collins, N. C., Thordal-Christensen, H., Lipka, V., Bau, S., Kombrink, E., Qiu, J. L., et al. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425, 973–977. doi: 10.1038/nature02076
Dawson, A. M., Bettgenhaeuser, J., Gardiner, M., Green, P., Hernandez-Pinzon, I., Hubbard, A., et al. (2015). The development of quick, robust, quantitative phenotypic assays for describing the host-nonhost landscape to stripe rust. Front. Plant Sci. 6:876. doi: 10.3389/fpls.2015.00876
Figueroa, M., Castell-Miller, C. V., Li, F., Hulbert, S. H., and Bradeen, J. M. (2015). Pushing the boundaries of resistance: insights from Brachypodium-rust interactions. Front. Plant Sci. 6:558. doi: 10.3389/fpls.2015.00558
Gill, U. S., Lee, S., and Mysore, K. S. (2015). Host versus nonhost resistance: distinct wars with similar arsenals. Phytopathology 105, 580–587. doi: 10.1094/phyto-11-14-0298-rvw
Han, D. J., Wang, Q. L., Chen, X. M., Zeng, Q. D., Wu, J. H., Xue, W. B., et al. (2015). Emerging Yr26-virulent races of Puccinia striiformis f. tritici are threatening wheat production in the Sichuan basin, China. Plant Dis. 99, 754–760. doi: 10.1094/Pdis-08-14-0865-Re
Hasegawa, M., Mitsuhara, I., Seo, S., Imai, T., Koga, J., Okada, K., et al. (2010). Phytoalexin accumulation in the interaction between rice and the blast fungus. Mol. Plant Microbe Interact. 23, 1000–1011. doi: 10.1094/Mpmi-23-8-1000
Ishiga, Y., Uppalapati, S. R., Gill, U. S., Huhman, D., Tang, Y., and Mysore, K. S. (2015). Transcriptomic and metabolomic analyses identify a role for chlorophyll catabolism and phytoalexin during Medicago nonhost resistance against Asian soybean rust. Sci. Rep. 5:13061. doi: 10.1038/srep13061
Johansson, O. N., Fantozzi, E., Fahlberg, P., Nilsson, A. K., Buhot, N., Tor, M., et al. (2014). Role of the penetration-resistance genes PEN1, PEN2 and PEN3 in the hypersensitive response and race-specific resistance in Arabidopsis thaliana. Plant J. 79, 466–476. doi: 10.1111/tpj.12571
Langenbach, C., Schultheiss, H., Rosendahl, M., Tresch, N., Conrath, U., and Goellner, K. (2016). Interspecies gene transfer provides soybean resistance to a fungal pathogen. Plant Biotechnol. J. 14, 699–708. doi: 10.1111/pbi.12418
Li, H., Goodwin, P. H., Han, Q., Huang, L., and Kang, Z. (2012). Microscopy and proteomic analysis of the non-host resistance of Oryza sativa to the wheat leaf rust fungus, Puccinia triticina f. sp tritici. Plant Cell Rep. 31, 637–650. doi: 10.1007/s00299-011-1181-0
Li, K., Hegarty, J., Zhang, C. Z., Wan, A. M., Wu, J. J., Guedira, G., et al. (2016). Fine mapping of barley locus Rps6 conferring resistance to wheat stripe rust. Theor. Appl. Genet. 129, 845–859. doi: 10.1007/s00122-015-2663-1
Lipka, V., Dittgen, J., Bednarek, P., Bhat, R., Wiermer, M., Stein, M., et al. (2005). Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310, 1180–1183. doi: 10.1126/science.1119409
Lu, Y., Wang, M. N., Chen, X. M., See, D., Chao, S. M., and Jing, J. X. (2014). Mapping of Yr62 and a small-effect QTL for high-temperature adult-plant resistance to stripe rust in spring wheat PI 192252. Theor. Appl. Genet. 127, 1449–1459. doi: 10.1007/s00122-014-2312-0
Lyons, R., Manners, J. M., and Kazan, K. (2013). Jasmonate biosynthesis and signaling in monocots: a comparative overview. Plant Cell Rep. 32, 815–827. doi: 10.1007/s00299-013-1400-y
Matsumoto, T., Wu, J. Z., Kanamori, H., Katayose, Y., Fujisawa, M., Namiki, N., et al. (2005). The map-based sequence of the rice genome. Nature 436, 793–800. doi: 10.1038/nature03895
Mayer, K. F. X., Martis, M., Hedley, P. E., Simkova, H., Liu, H., Morris, J. A., et al. (2011). Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell 23, 1249–1263. doi: 10.1105/tpc.110.082537
Michelmore, R. W., Paran, I., and Kesseli, R. V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U.S.A. 88, 9828–9832. doi: 10.1073/pnas.88.21.9828
Niks, R. E., Alemu, S. K., Marcel, T. C., and van Heyzen, S. (2015a). Mapping genes in barley for resistance to Puccinia coronata from couch grass and to P. striiformis from brome, wheat and barley. Euphytica 206, 487–499. doi: 10.1007/s10681-015-1516-y
Niks, R. E., Qi, X. Q., and Marcel, T. C. (2015b). Quantitative resistance to biotrophic filamentous plant pathogens: concepts, misconceptions, and mechanisms. Annu. Rev. Phytopathol. 53, 445–470. doi: 10.1146/annurev-phyto-080614-115928
Patto, M. C. V., and Rubiales, D. (2014). Unveiling common responses of Medicago truncatula to appropriate and inappropriate rust species. Front. Plant Sci. 5:618. doi: 10.3389/fpls.2014.00618
Quan, L. J., Zhang, B., Shi, W. W., and Li, H. Y. (2008). Hydrogen peroxide in plants: a versatile molecule of the reactive oxygen species network. J. Integr. Plant Biol. 50, 2–18. doi: 10.1111/j.1744-7909.2007.00599.x
Schoonbeek, H.-J., Wang, H.-H., Stefanato, F. L., Craze, M., Bowden, S., Wallington, E., et al. (2015). Arabidopsis EF-Tu receptor enhances bacterial disease resistance in transgenic wheat. New Phytol. 206, 606–613. doi: 10.1111/nph.13356
Schulze-Lefert, P., and Panstruga, R. (2011). A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci. 16, 117–125. doi: 10.1016/j.tplants.2011.01.001
Shafiei, R., Hang, C., Kang, J. G., and Loake, G. J. (2007). Identification of loci controlling non-host disease resistance in Arabidopsis against the leaf rust pathogen Puccinia triticina. Mol. Plant Pathol. 8, 773–784. doi: 10.1111/J.1364-3703.2007.00431.X
Stein, M., Dittgen, J., Sanchez-Rodriguez, C., Hou, B. H., Molina, A., Schulze-Lefert, P., et al. (2006). Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18, 731–746. doi: 10.1105/tpc.105.038372
Tetlow, I. J., and Farrar, J. F. (1993). Apoplastic sugar concentration and pH in barley leaves infected with brown rust. J. Exp. Bot. 44, 929–936. doi: 10.1093/Jxb/44.5.929
Thordal-Christensen, H., Zhang, Z. G., Wei, Y. D., and Collinge, D. B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x
Vogel, J. P., Garvin, D. F., Mockler, T. C., Schmutz, J., Rokhsar, D., Bevan, M. W., et al. (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768. doi: 10.1038/nature08747
Wahl, R., Wippel, K., Goos, S., Kamper, J., and Sauer, N. (2010). A novel high-affinity sucrose transporter is required for virulence of the plant pathogen Ustilago maydis. PLoS Biol. 8:e1000303. doi: 10.1371/journal.pbio.1000303
Wellings, C. R. (2011). Global status of stripe rust: a review of historical and current threats. Euphytica 179, 129–141. doi: 10.1007/s10681-011-0360-y
Wen, Y. Q., Wang, W. M., Feng, J. Y., Luo, M. C., Tsuda, K., Katagiri, F., et al. (2011). Identification and utilization of a sow thistle powdery mildew as a poorly adapted pathogen to dissect post-invasion non-host resistance mechanisms in Arabidopsis. J. Exp. Bot. 62, 2117–2129. doi: 10.1093/jxb/erq406
Wu, C. Y., Li, X. J., Yuan, W. Y., Chen, G. X., Kilian, A., Li, J., et al. (2003). Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 35, 418–427. doi: 10.1046/j.1365-313X.2003.01808.x
Yang, D. L., Yang, Y. N., and He, Z. H. (2013). Roles of plant hormones and their interplay in rice immunity. Mol. Plant 6, 675–685. doi: 10.1093/mp/sst056
Yang, Y. H., Zhao, J., Xing, H. J., Wang, J. Y., Zhou, K., Zhan, G. M., et al. (2014). Different non-host resistance responses of two rice subspecies, japonica and indica, to Puccinia striiformis f. sp tritici. Plant Cell Rep. 33, 423–433. doi: 10.1007/s00299-013-1542-y
Yi, M., and Valent, B. (2013). Communication between filamentous pathogens and plants at the biotrophic interface. Annu. Rev. Phytopathol. 51, 587–611. doi: 10.1146/annurev-phyto-081211-172916
Yuan, Y. X., Zhong, S. H., Li, Q., Zhu, Z. R., Lou, Y. G., Wang, L. Y., et al. (2007). Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol. J. 5, 313–324. doi: 10.1111/j.1467-7652.2007.00243.x
Zellerhoff, N., Himmelbach, A., Dong, W., Bieri, S., Schaffrath, U., and Schweizer, P. (2010). Nonhost resistance of barley to different fungal pathogens is associated with largely distinct, quantitative transcriptional responses. Plant Physiol. 152, 2053–2066. doi: 10.1104/pp.109.151829
Zhang, H. C., Wang, C. F., Cheng, Y. L., Wang, X. J., Li, F., Han, Q. M., et al. (2011). Histological and molecular studies of the non-host interaction between wheat and Uromyces fabae. Planta 234, 979–991. doi: 10.1007/s00425-011-1453-5
Zhang, J., Guo, D., Chang, Y. X., You, C. J., Li, X. W., Dai, X. X., et al. (2007). Non-random distribution of T-DNA insertions at various levels of the genome hierarchy as revealed by analyzing 13 804 T-DNA flanking sequences from an enhancer-trap mutant library. Plant J. 49, 947–959. doi: 10.1111/j.1365-313X.2006.03001.x
Keywords: non-host resistance, rice mutant, wheat stripe rust, defense-related genes, genetic mapping
Citation: Zhao J, Yang Y, Yang D, Cheng Y, Jiao M, Zhan G, Zhang H, Wang J, Zhou K, Huang L and Kang Z (2016) Characterization and Genetic Analysis of Rice Mutant crr1 Exhibiting Compromised Non-host Resistance to Puccinia striiformis f. sp. tritici (Pst). Front. Plant Sci. 7:1822. doi: 10.3389/fpls.2016.01822
Received: 03 September 2016; Accepted: 18 November 2016;
Published: 30 November 2016.
Edited by:
Benjamin Schwessinger, Australian National University, AustraliaReviewed by:
Roger W. Innes, Indiana University Bloomington, USARemco Stam, Technische Universität München, Germany
Copyright © 2016 Zhao, Yang, Yang, Cheng, Jiao, Zhan, Zhang, Wang, Zhou, Huang and Kang This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhensheng Kang, kangzs@nwsuaf.edu.cn
 Jing Zhao
Jing Zhao Yuheng Yang
Yuheng Yang Donghe Yang
Donghe Yang Yulin Cheng1
Yulin Cheng1 Hongchang Zhang
Hongchang Zhang Lili Huang
Lili Huang Zhensheng Kang
Zhensheng Kang