- 1National Key Laboratory for Rice Biology, Institute of Biotechnology, Zhejiang University, Hangzhou, China
- 2National Navel Orange Engineering Research Center, College of Life and Environmental Sciences, Gannan Normal University, Ganzhou, China
The Ethylene-Responsive Factors (ERFs) comprise a large family of transcriptional factors that play critical roles in plant immunity. Gray mold disease caused by Botrytis cinerea, a typical necrotrophic fungal pathogen, is the serious disease that threatens tomato production worldwide. However, littler is known about the molecular mechanism regulating the immunity to B. cinerea in tomato. In the present study, virus-induced gene silencing (VIGS)-based functional analyses of 18 members of B3 group (also called Group IX) in tomato ERF family were performed to identify putative ERFs that are involved in disease resistance against B. cinerea. VIGS-based silencing of either SlERF.B1 or SlERF.C2 had lethal effect while silencing of SlERF.A3 (Pit4) significantly suppressed vegetative growth of tomato plants. Importantly, silencing of SlERF.A1, SlERF.A3, SlERF.B4, or SlERF.C3 resulted in increased susceptibility to B. cinerea, attenuated the B. cinerea-induced expression of jasmonic acid/ethylene-mediated signaling responsive defense genes and promoted the B. cinerea-induced H2O2 accumulation. However, silencing of SlERF.A3 also decreased the resistance against Pseudomonas syringae pv. tomato (Pst) DC3000 but silencing of SlERF.A1, SlERF.B4 or SlERF.C3 did not affect the resistance to this bacterial pathogen. Expression of SlERF.A1, SlERF.A3, SlERF.B4, or SlERF.C3 was induced by B. cinerea and by defense signaling hormones such as salicylic acid, methyl jasmonate, and 1-aminocyclopropane-1-carboxylic acid (an ethylene precursor). SlERF.A1, SlERF.B4, SlERF.C3, and SlERF.A3 proteins were found to localize in nucleus of cells and possess transactivation activity in yeasts. These data suggest that SlERF.A1, SlERF.B4, and SlERF.C3, three previously uncharacterized ERFs in B3 group, and SlERF.A3, a previously identified ERF with function in immunity to Pst DC3000, play important roles in resistance against B. cinerea in tomato.
Introduction
During last decade, extensive genetics and molecular studies revealed that higher plants have evolved to possess a sophisticated innate immunity system, which is similar to the innate immunity in animals (Dodds and Rathjen, 2010; Fu and Dong, 2013). The innate immunity system in plants consists of two layers of immune responses, called pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI), which are precisely regulated upon infection from different types of pathogens (Bernoux et al., 2011; Segonzac and Zipfel, 2011). Generally, both of PTI and ETI are required for resistance to biotrophic and hemibiotrophic pathogens while only PTI is believed to be active resistance response against necrotrophic pathogens (Mengiste, 2012). Upon sensing and recognition of the pathogen-derived PAMPs or effectors by surface/extracellular receptors such as receptor-like kinases and receptor-like proteins in plants (Osakabe et al., 2013), initiation of the innate immune response often requires transcriptional reprogramming to coordinately regulate expression of a large set of genes (Moore et al., 2011; Buscaill and Rivas, 2014). For example, approximately one-third of the Arabidopsis genome is differentially expressed during the early stage of infection by Botrytis cinerea (Windram et al., 2012). The dramatic transcription reprogramming is conferred by the concerted action of myriad transcription (co)factors (TFs) that function directly or indirectly to deploy their activity rapidly, transiently, spatially and hierarchically. In recent years, many TFs belonging to the AP2/ERF, NAC, MYB, WRKY, and bZIP (super)families have been identified to play important roles in regulating plant immune response against diverse pathogens (Eulgem and Somssich, 2007; Alves et al., 2013; Licausi et al., 2013; Nuruzzaman et al., 2013).
Botrytis cinerea is an airborne plant pathogen with a necrotrophic lifestyle attacking over 200 crop hosts worldwide (Williamson et al., 2007) and the interaction of tomato-B. cinerea has been developed as a useful pathosystem to study the molecular mechanism of plant immunity to necrotrophic fungal pathogens (Mengiste, 2012). Generally speaking, PTI but not ETI is effective in plant immunity to necrotrophic fungal pathogens such as B. cinerea (Mengiste, 2012). Several quantitative trait loci conferring resistance or susceptibility to B. cinerea have been identified and mapped in tomato (Finkers et al., 2007; Davis et al., 2009). Significant transcriptional reprogramming, metabolic and biochemical changes, and modification of signal pathways operated by different stress hormones such as ethylene (ET), salicylic acid (SA), jasmonic acid (JA), and abscisic acid (ABA) are involved in the response of tomato or its wild species to B. cinerea (Blanco-Ulate et al., 2013; Seifi et al., 2014; Smith et al., 2014; Camañes et al., 2015; Vega et al., 2015). During the infection process, B. cinerea can manipulate the antagonistic balance between the SA- and JA/ET-mediated signaling pathways and hijack the SA signaling pathway to accelerate disease development (El Oirdi et al., 2011; Rahman et al., 2012). Shortening in JA biosynthesis resulted in increased susceptibility to B. cinerea (Hind et al., 2011; Zhang S. et al., 2015), whereas ET-mediated signaling plays a positive role in immunity to B. cinerea (Francia et al., 2007; Lin et al., 2008; Nambeesan et al., 2012). It was shown that ABA regulates the immunity to B. cinerea in tomato through modulating the cuticle permeability and pectin composition in cell wall or suppressing the SA-mediated signaling pathway or the production of nitric oxide (Audenaert et al., 2002; Asselbergh et al., 2007; Curvers et al., 2010; Sivakumaran et al., 2016). A number of genes encoding receptor-like protein kinase TPK1b, transcriptional factors SHINE3, AIM1, SlDRW1, SlSRN1, SlSR1, and SlSR3L (Abuqamar et al., 2008, 2009; Buxdorf et al., 2014; Li et al., 2014a; Liu et al., 2014a,b), histone H2B monoubiquitination enzymes SlHUB1 and SlHUB2 (Zhang Y. et al., 2015), mitogen-activated protein kinase kinase SlMKK2 and SlMKK4 (Li et al., 2014b), phosphatidylinositol-phospholipase SlPLC2 (Gonorazky et al., 2016), NADPH oxidase SlRbohB (Li X. et al., 2015), 12-oxophytodienoate reductase SlOPR3 (Scalschi et al., 2015) and matrix metalloproteinase Sl3-MMP (Li D. et al., 2015) have been identified to play important roles in tomato immunity against B. cinerea. Enzymes involved in biosynthesis of vitamin B6 and trehalose-6-phosphate as well as concurrent over-activation of cytosolic glutamine synthetase and γ-aminobutyric acid shunt are also involved in tomato immune response to B. cinerea (Seifi et al., 2013; Zhang et al., 2014, 2016). Furthermore, simultaneous suppression of both polygalacturonase and expansin or accumulation of anthocyanin decreased the susceptibility of ripening fruits to B. cinerea (Cantu et al., 2008; Zhang et al., 2013). However, our knowledge on the molecular mechanism regulating the plant immunity to necrotrophic fungal pathogens is relatively lagging, as compared to the progress in plant immunity to (hemi)biotrophic pathogens.
The AP2/ERF superfamily is a large family with more than 100 members in plants [e.g., 147 in Arabidopsis (Nakano et al., 2006) and 139 in rice (Sharoni et al., 2011)] and represents a unique group of plant-specific TFs (Riechmann et al., 2000). A common structural feature of the proteins encoded by this superfamily is the presence of a highly conserved DNA-binding domain, called AP2 domain, containing 58 or 59 amino acids involved in the high-affinity binding to target DNA sequences (Ohme-Takagi and Shinshi, 1995). According to the numbers and characteristics of the AP2 domains, the AP2/ERF superfamily is basically divided into three families, e.g., AP2 family with two AP2 domains, ERF/DREB family with one AP2 domain and RAV family with a B3 DNA-binding domain (Riechmann et al., 2000; Nakano et al., 2014). The ERF/DREB family falls mainly into three subfamilies: ERFs (ethylene response factors), DREBs (dehydration-responsive element binding proteins) and the CBF (C-repeat binding factor) family (Nakano et al., 2014). Biochemical evidence indicates that the ERF proteins can specifically bind to a cis-element called GCC box (AGCCGCC) (Ohme-Takagi and Shinshi, 1995; Buttner and Singh, 1997), which is present in the promoter regions of many ethylene-regulated defense genes, and function as transcriptional activators or repressors (Fujimoto et al., 2000). It was also found that several members of the ERF family contain phosphorylation sites in C-terminals and need posttranslational modification for their biochemical activity and biological functions (Meng et al., 2013).
Extensive studies with loss-of-function and gain-of-function mutants in different plant species have demonstrated the importance of the ERF proteins during plant development and adaptation to abiotic stress conditions (Licausi et al., 2013). Meanwhile, a large number of functional investigations have also suggested that the ERF family play critical roles in plant response to biotic stresses (Licausi et al., 2013). Among the 12 major groups classified based on the type of AP2 domain (Nakano et al., 2006), the B3 group (also called as IX group) has been shown to play important roles in regulating defense responses in different plants against pathogens. For example, overexpression of Arabidopsis ERF1 or AtERF2, tobacco ERF5 or OPBP1, tomato Pti4 or Pti5, rice OsERF922 or OsBIERF3, wheat TaPIEP1 and Medicago truncatula MtERF1-1 in transgenic plants resulted in enhanced resistance against a variety of diseases caused by necrotrophic or biotriophic fungal and bacterial pathogens (Solano et al., 1998; He et al., 2001; Berrocal-Lobo et al., 2002; Gu et al., 2002; Fischer and Droge-Laser, 2004; Guo et al., 2004; Cao et al., 2006; Anderson et al., 2010; Dong et al., 2010; Son et al., 2011; Liu et al., 2012). The Arabidopsis ERF1, ERF5, ERF6, AtERF14, and ORA59 have been shown to act as regulators of the JA/ET signaling pathway that is required for resistance against necrotrophic fungal pathogens including B. cinerea (Lorenzo et al., 2003; Berrocal-Lobo and Molina, 2004; Oñate-Sánchez et al., 2007; Moffat et al., 2012). Together, at least 7 (ERF1, AtERF2, AtERF5, AtERF6, AtERF14, ORA59, and AtERF104) out of 17 members in the B3 group of the Arabidopsis ERF family have functions in regulating defense response against pathogen infections, demonstrating the importance of this group in plant disease resistance.
Recent genome-wide bioinformatics analyses identified a total of 146 members in the tomato AP2/ERF superfamily, of which 77 members belong to the ERF family (Sharma et al., 2010; Pirrello et al., 2012; Liu et al., 2016). Expression profiling analyses revealed that a large set of the tomato ERF genes are differentially induced by developmental cues, hormones and various stress factors (Gu et al., 2000; Tournier et al., 2003; Sharma et al., 2010; Pirrello et al., 2012). Functional studies have characterized a number of tomato ERF genes, e.g., SlERF1, Sl-ERF2, TSRF1, SlAP2a, Sl-ERF.B3, SlERF6, SlERF36, and SlERF52, that play important roles in regulating growth and developmental processes including seed germination, seedling development, stomatal density, fruit ripening, and flower pedicel abscission (Pirrello et al., 2006; Li et al., 2007; Zhang et al., 2008, 2009; Chung et al., 2010; Lee et al., 2012; Liu et al., 2013, 2016; Upadhyay et al., 2013; Liu M. et al., 2014; Nakano et al., 2014). Whereas TERF1, TERF2, TSRF1, JERF1, JERF3, LeERF3b, Sl-ERF.B.3, and ERF5 were found to be involved in regulating abiotic stress response (Huang et al., 2004; Wang et al., 2004; Zhang et al., 2004a,b, 2007, 2010; Chen et al., 2008; Wu et al., 2008; Quan et al., 2010; Zhang and Huang, 2010; Tian et al., 2011; Pan et al., 2012; Klay et al., 2014), Pti4, Pti5, Pti6, TSRF1, and SlERF3 have been demonstrated to play key roles in regulating defense responses against pathogens and insect pests (Zhou et al., 1997; He et al., 2001; Gu et al., 2002; Wu et al., 2002, 2015; Zhang et al., 2004b, 2007; Pan et al., 2010). Among the characterized tomato ERF genes, seven of them including Pti4, Pti5, LeERF1, LeERF4 (Sl-ERF.B3), TSRF1, TERF1, and ERF5 are members of the B3 group (Zhou et al., 1997; Tournier et al., 2003; Huang et al., 2004; Zhang et al., 2004a,b, 2007; Pan et al., 2012; Liu et al., 2013, 2016; Liu M. et al., 2014). Here, we performed virus-induced gene silencing (VIGS)-based functional analyses of 18 members in the B3 group members of the tomato ERF family to explore their possible involvement in disease resistance against B. cinerea. Our results data indicate that SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 positively regulate resistance of tomato plants against B. cinerea.
Materials and Methods
Plant Growth Condition and Treatments
Tomato (Solanum lycopersicum) cv. Suhong 2003 was used for all experiments. Tomato and Nicotiana benthamiana plants were grown in plastic pots containing compost soil mix (perlite: vermiculite: plant ash = 1: 6: 2) under 10 h fluorescent light (200 μE m2 s-1) at 22 ∼ 24°C with a 14 h light/10 h dark cycle. Generally, 2-week-old tomato seedlings were used for VIGS assays and 4-week-old plants were used for gene expression experiments. For analyses of gene expression in response to hormone treatments, tomato plants were treated by foliar spraying with 100 μM methyl jasmonate (MeJA), 1 mM SA, 100 μM 1-aminocyclopropane-1-carboxylic acid (ACC, a precursor of ethylene) in solutions containing 0.1% ethanol and 0.02% Tween-20. Plants treated with same volume of solution containing 0.1% ethanol and 0.02% Tween-20 were used as a control. For analyses of gene expression in response to B. cinerea, whole plant inoculation assays along with a corresponding mock-inoculation control were performed (see below for detail). Leaf samples were collected from hormone-treated or pathogen-inoculated plants at different time points as indicated after treatment or inoculation and stored at -80°C until use.
Vector Construction and VIGS Assays
According to the predicted cDNA and available full-length cDNAs, VIGS fragments of 250–330 bp (Supplementary File 1) were designed at the 5′ ends of the selected tomato B3 group ERF genes and amplified with gene-specific primers with different restriction enzyme sites (Supplementary Table S1) from cDNAs synthesized from total RNA prepared from tomato leaf samples. After cloning and sequencing, the VIGS fragments were cloned into pTRV2 vector (Liu et al., 2002), yielding pTRV2-SlERF-A, B and Cs, which were then introduced into Agrobacterium tumefaciens strain GV3101 by electroporation using GENE PULSER II Electroporation System (Bio-Rad Laboratories, Hercules, CA, USA). Agrobacteria carrying pTRV2-GUS (as a negative control) or pTRV2-SlERF-A, B and Cs were cultivated in YEP medium (10 g/l peptone, 10 g/l yeast extract, 5 g NaCl/l, 50 μg/ml rifampicin, 50 μg/ml kanamycin, and 25 μg/ml gentamicin) for 36 h with continuous shaking in a 28°C incubator. Cells were collected by centrifugation and resuspended in infiltration buffer (10 mM MgCl2, 150 μM acetosyringone, MES, pH5.7). The agrobacteria carrying pTRV2-GUS or pTRV2-SlERF-A, B and Cs were mixed with the agrobacteria carrying pTRV1 in a ratio of 1:1 and maintained at OD600 = 1.5 for 3 h at room temperature. The mixed agrobacteria suspension was infiltrated into the abaxial surface of the 2-week-old seedlings using 1 mL needleless syringes. Leaf samples were collected at 2 weeks after VIGS infiltration and used for analysis of the silencing efficiency and specificity by qRT-PCR using gene-specific primers (Supplementary Table S1).
Plant Inoculation and Disease Assays
Botrytis cinerea was cultivated on 2 × V8 agar (36% V8 juice, 0.2% CaCO3, 2% agar) at 22°C. Spores were collected from 10-day-old plates by rinsing the culture with 1% maltose buffer, filtered through bi-layered cheesecloth and adjusted to 2 × 105 spores/ml for inoculation (Li et al., 2014a). For gene expression analyses, 4-week-old plants were inoculated by foliar spraying with spore suspension or with same volume of 1% maltose buffer as a mock-inoculation control and leaf samples were collected at different time points after inoculation. For disease assays, simplified detached leaf inoculation assay and whole plant inoculation assay were performed. In the detached leaf inoculation assays, leaves were detached from at least 10 VIGS-infiltrated plants at 4 weeks after VIGS infiltration and inoculated by dropping 5 μl of spore suspension on leaf surface. In the whole plant inoculation assays, 6-week-old plants were inoculated by foliar spraying with spore suspension. The inoculated leaves and plants were kept at 22°C in sealed containers to retain the moist conditions favorable for disease development. Lesion sizes on inoculated detached leaves were measured. Fungal growth in planta was analyzed by amplification of the transcripts of B. cinerea Actin gene as a marker using a pair of primers BcActin-F and BcActin-R (Supplementary Table S1) (Li et al., 2014b). Relative fungal growth was expressed as folds of the transcript levels of BcActin vs the transcript levels of a tomato Actin gene.
Pseudomonas syringae pv. tomato DC3000 was grown overnight in King’s B (KB) liquid medium containing rifampicin at 50 μg/ml. Cells were collected and resuspended in 10 mM MgCl2 to OD600 = 0.0002 for plant inoculation. Plants were vacuum infiltrated with suspension of Pst DC3000 or with MgCl2 solution as a mock inoculation control at 4 weeks after VIGS treatment. The inoculated plants were kept in a sealed container to maintain high humidity (RH > 90%) and disease progress was observed daily. For measurement of in planta bacterial growth, disks (6 mm in diameter) from leaves of Pst DC3000-inoculated plants were surface sterilized in 75% ethanol for 10 s, homogenized in 200 μl of 10 mM MgCl2, diluted in 10 mM MgCl2, and plated on KB agar plates containing 50 μg/ml rifampicin. The plates were incubated at 28°C and the bacterial colonies were counted 3 days after incubation.
Subcellular Localization Assays
Coding sequences of SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 were amplified using gene-specific primers SlERF.A1-1F/1R, SlERF.A3-1F/1R, SlERF.B4-1F/1R, or SlERF.C3-12F/1R (Supplementary Table S1), respectively. After cloning and confirmation by sequencing, the coding sequences were released from recombinant plasmids and inserted into pFGC-Egfp vector, yielding pFGC-Egfp:SlERF.A1, pFGC-Egfp:SlERF.A3, pFGC-Egfp:SlERF.B4, and pFGC-Egfp:SlERF.C3. These sequence-verified constructs and the empty vector pFGC-Egfp were introduced into A. tumefaciens strain GV3101. Four-week-old N. benthamiana plants were infiltrated with agrobacteria carrying different constructs and allowed to grow at 25°C for 48 h. Fluorescence was excited at 488 nm and detected using a 500–530 nm emission filter preformed with confocal laser scanning microscope (Model LSM 510 META, Zeiss, Oberkochen, Germany).
Transactivation Activity Assays
The coding sequences of SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 were amplified using gene-specific primers SlERF.A1-2F/2R, SlERF.A1-2F/2R, SlERF.B4-2F/2R, and SlERF.C3-2F/2R (Supplementary Table S1), respectively, and cloned into pBD-GAL4Cam vector, yielding pBD-SlERF.A1, pBD-SlERF.A3, pBD-SlERF.B4, and pBD-SlERF.C3. The recombinant plasmids and the pBD empty vector (a negative control) were transformed into yeast strain AH109. The transformed yeasts were cultivated on the SD/Trp- and SD/Trp-His- medium for 3 days at 28°C, followed by addition of x-α-gal. Transactivation activity of the fused proteins was evaluated according to the growth situation and production of blue pigments after the addition of x-α-gal of the transformed yeast cells on the SD/Trp-/His- medium.
Analyses of Gene Expression
Gene-specific primers were designed using Primer Premier 6 and located near the 3′ ends of the target genes (Supplementary Table S1). Total RNA was extracted using TRIzol reagent (Invitrogen, Shanghai, China) and treated with RNase-free DNase (TaKaRa, Dalian, China). First-strand cDNA was synthesized from 1 μg total RNA using AMV reverse transcriptase (TaKaRa, Dalian, China). Each qRT-PCR reaction contained 12.5 μl SYBR Premix Ex Taq (TaKaRa, Dalian, China), 0.1 μg cDNA and 7.5 pmol of each gene-specific primer in a final volume of 25 μl, and performed in a CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA, USA). Three independent biological replicates were used for analyses and relative expression levels were calculated using the 2-ΔΔCT method. A tomato actin gene was used as an internal control and relative expression levels of the target genes were shown as folds of the expression level of the actin gene.
In situ Detection of H2O2
In situ detection of H2O2 in leaves of mock- and B. cinerea-inoculated plant was carried out by the 3, 3-diaminobenzidine (DAB) staining method (Thordal-Christensen et al., 1997). Accumulation of H2O2 in stained leaves was visualized by a digital camera.
Statistical Analysis
Experiments were repeated three times and three replicates were included in each experiment. Data obtained from three independent experiments were subjected to statistical analysis using the Student’s t-test and he probability values of p < 0.05 were considered as significant difference.
Results
The B3 Group of the Tomato ERF Family and VIGS-based Silencing of the Selected B3 Group ERF Genes
A total of 28 members of the B3 group in tomato ERF family were previously identified (Sharma et al., 2010; Pirrello et al., 2012; Liu et al., 2016). However, locus Solyc09g089910 corresponding to SlERF59 (Sharma et al., 2010) was not included as a B3 group member (Pirrello et al., 2012; Liu et al., 2016). Solyc09g089910 is phylogenetically related to subgroup IXc (Pirrello et al., 2012; Liu et al., 2016) and was named as SlERF.C11 (Liu et al., 2016). Thus, the B3 group of the tomato ERF family harbors 29 members in total, among which 5, 13, and 11 are assigned into IXa, IXb, and IXc subgroups, respectively.
To explore the possible involvement of the B3 group ERF genes in tomato resistance against B. cinerea, 18 members, among which 16 members had available full-length cDNAs and two members did not have full-length cDNAs, were selected for study. Fragments of 250–330 bp in size at the 5′ ends of the genes for each of these selected B3 group ERF genes were amplified and constructed into pTRV2 vectors (Supplementary File 1). Standard VIGS assays were performed on 2-week-old tomato seedlings (Liu et al., 2002). In our VIGS assays, efficiency of the VIGS protocol was >85%, estimated by the appearance of bleaching phenotype, which was resulted from silencing of a phytoene desaturase (PDS) gene as a positive control marker (Liu et al., 2002), in pTRV2-PDS-infiltrated plants. Silencing efficiency for each of the target genes selected under our experimental conditions was ∼60% (Figure 1A), as estimated by qRT-PCR analysis of the transcript levels for the target genes in pTRV2-SlERFs-infiltrated plants and compared with that in the pTRV2-GUS-infiltrated negative control plants. As SlERF.A1, SlERF.B4, and SlERF.C3 were later shown to be involved in resistance to B. cinerea (see below), silencing specificity of SlERF.A1, SlERF.B4, and SlERF.C3 genes in pTRV2-SlERF.A1-, pTRV2-SlERF.B4-, or pTRV2-SlERF.C3-infiltrated plants was further evaluated. In a phylogenetic tree constructed with cDNA sequences of the B3 group ERF genes, SlERF.A1 is closely related to SlERF.A4 and SlERF.A5, SlERF.B4 shows high levels of identity to SlERF.B2 and SlERF.B5, and SlERF.C3 is highly related to SlERF.C11 (Supplementary Figure S1). As shown in Figure 1B, the transcript levels of SlERF.A4 and SlERF.A5 in pTRV2-SlERF.A1-infiltrated plants, SlERF.B2 and SlERF.B5 in pTRV2-SlERF.B4-infiltrated plants and SlERF.C11 in pTRV2-SlERF.C3-infiltrated plants were comparable to those in pTRV2-GUS-infiltrated plants. These data indicate that silencing of SlERF.A1, SlERF.B4, or SlERF.C3 did not significantly affect the expression of their closely related genes.
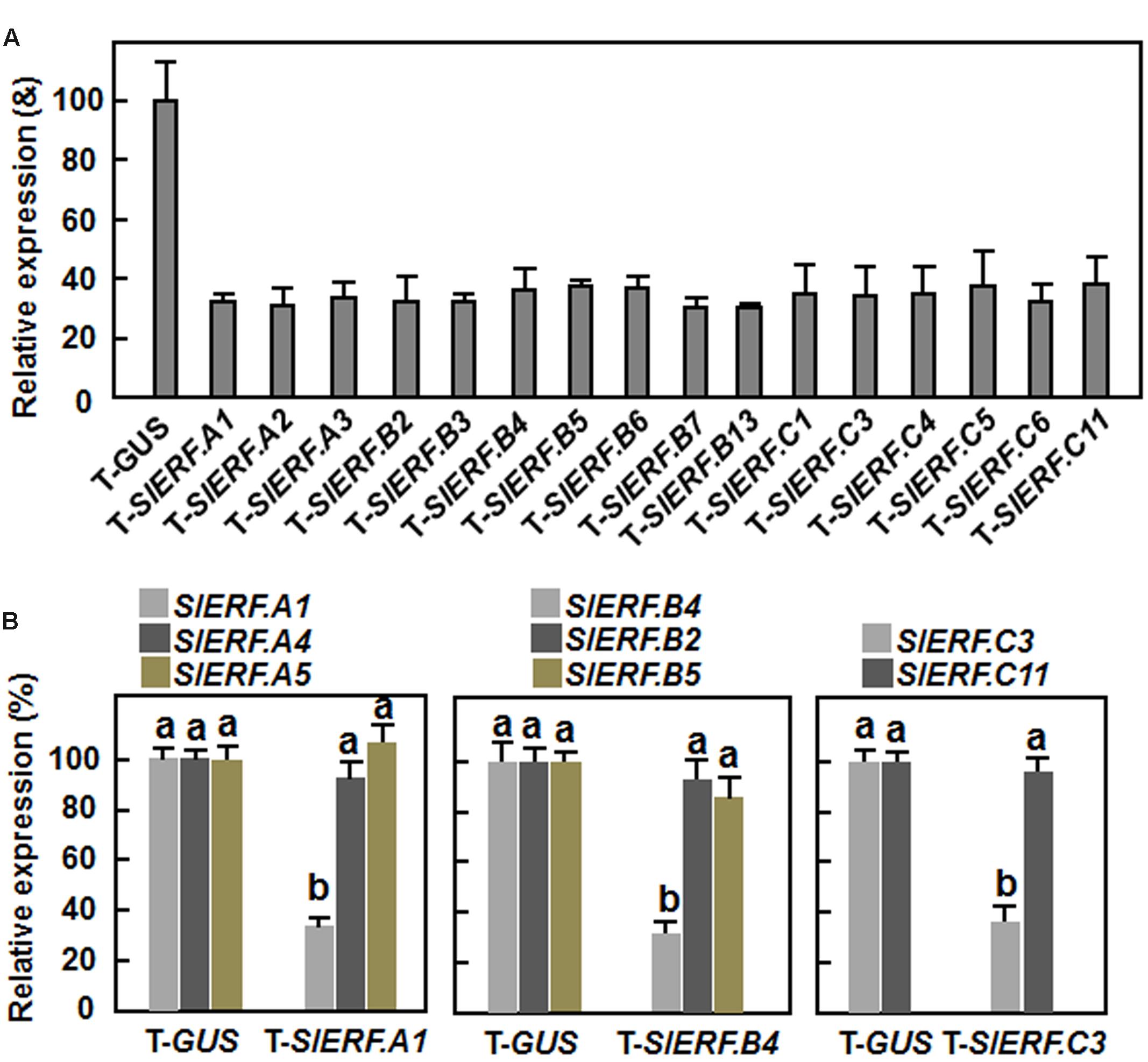
FIGURE 1. Silencing efficiency and specificity for target genes. Two-week-old seedlings were infiltrated with agrobacteria carrying different virus-induced gene silencing (VIGS) constructs and leaf samples were collected at 2 weeks after VIGS infiltration. (A) Silencing efficiency for target genes in VIGS-infiltrated plants. (B) Silencing specificity for target genes in VIGS-infiltrated plants. Transcript levels of the target genes were analyzed by qRT-PCR and data were normalized with a tomato actin gene. Silencing efficiency was calculated by comparing the transcript levels of each SlERF gene in corresponding pTRV2-SlERF-infiltrated plants to those in pTRV2-GUS-infiltrated plants. The transcript levels for each SlERF gene in the pTRV2-GUS-infiltrated plants were set as 100%. The transcript levels of each SlERF gene in VIGS-infiltrated plants were shown above the columns. Data are the mean ± SD from three independent experiments and different letters above the columns indicate significant difference at p < 0.05 level between pTRV2-SlERF- and pTRV2-GUS-infiltrated plants.
Silencing of SlERF.A3, SlERF.B1, or SlERF.C2 Affected Vegetative Growth
During our repeated experiments, the pTRV2-SlERF.B1- and pTRV2-SlERF.C2-infiltrated plants died within 7 days after VIGS infiltration while the pTRV2-GUS-infiltrated plants grew normally (Figure 2A), implying that silencing of either SlERF.B1 or SlERF.C2 had lethal effect on growth of tomato plants. Furthermore, silencing of SlERF.A3 (Pti4) suppressed significantly the growth of pTRV2-SlERF.A3-infiltrated plants (Figure 2B), as the heights of the pTRV2-SlERF.A3-infiltrated plants were approximately 55% of the pRTV2-GUS-infiltrated plants at 10 and 20 days after VIGS infiltration (Figure 2C), indicating that silencing of SlERF.A3 had an effect on the growth of tomato plants. During a period of 6 weeks, no significant defect in growth and development was observed for plants that were VIGS infiltrated and silenced for one of the other selected B3 group member ERF genes.
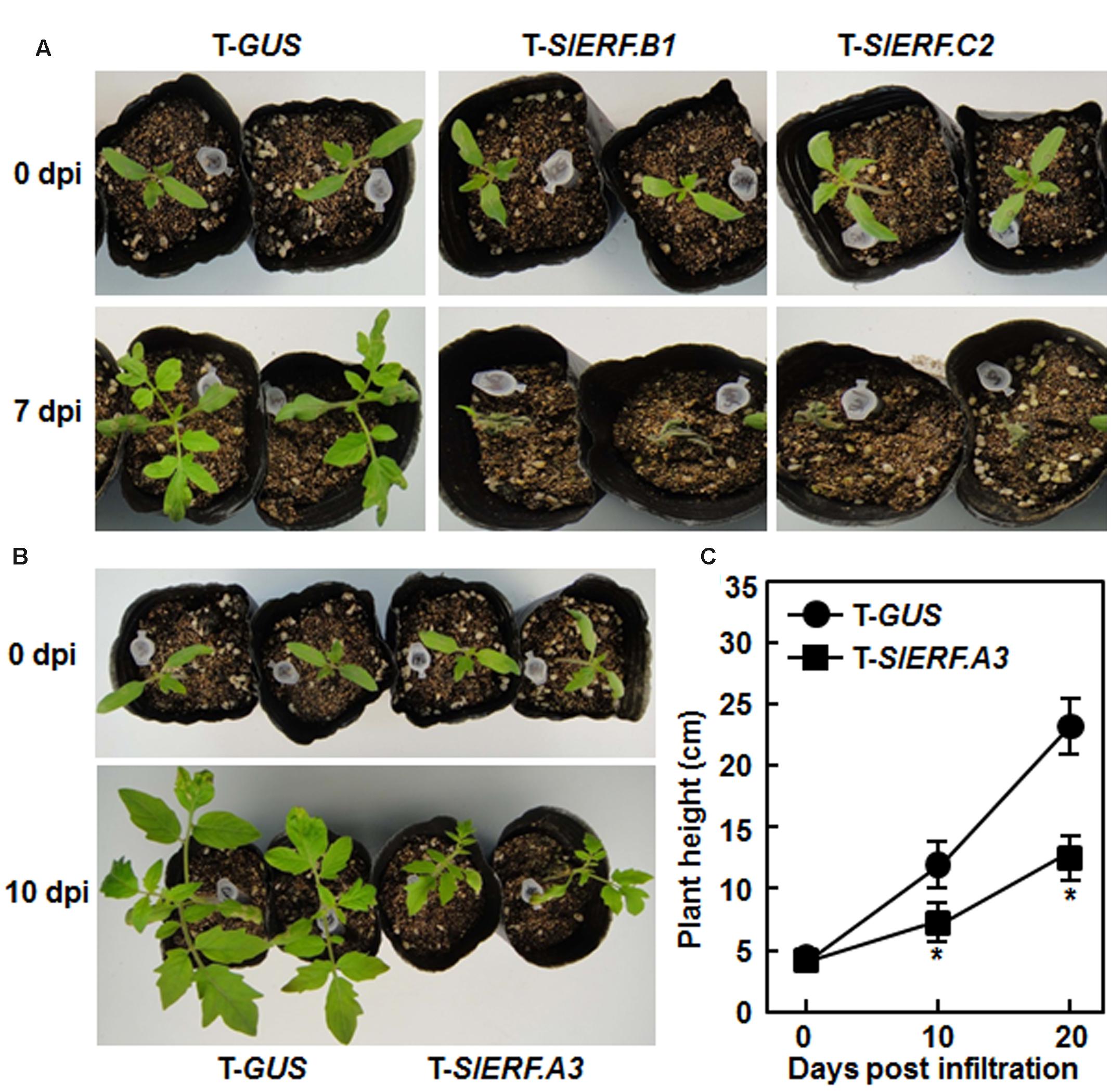
FIGURE 2. Silencing of SlERF.A3, SlERF.B1, and SlERF.C2 affected vegetative growth of VIGS-infiltrated plants. (A) Silencing of SlERF.B1 or SlERF.C2 led to death of the pTRV2-SlERF.B1- and pTRV2-SlERF.C2-infiltrated plants. (B) and (C) Silencing of SlERF.A3 suppressed vegetative growth of the pTRV2-SlERF.A3-infiltrated plants. Heights of the pTRV2-SlERF.A3- and pTRV2-GUS-infiltrated plants grown under same condition were measured at 10 and 20 days after VIGS infiltration. Data presented in (C) are the mean ± SD from three independent experiments. ∗Below the line in (C) indicates significant difference at p < 0.05 between the pTRV2-SlERF.A3- and pTRV2-GUS-infiltrated plants. dpi, days post-infiltration.
Silencing of SlERF.A1, SlERF.B4, SlERF.C3, or SlERF.A3 Led to Decreased Resistance to B. cinerea
Because of the lethality of the pTRV2-SlERF.B1- or pTRV2-SlERF.C2-infiltrated plants, a total of 16 B3 group ERF genes were further examined using VIGS-based phenotyping for their involvement in disease resistance against B. cinerea. In our experiments, B. cinerea-caused lesions on leaves from pTRV2-SlERF.A2-, pTRV2-SlERF.B2-, pTRV2-SlERF. B3-, pTRV2-SlERF.B5-, pTRV2-SlERF.B6-, pTRV2-SlERF.B7-, pTRV2-SlERF.B13-, pTRV2-SlERF.C1-, pTRV2-SlERF.C4-, pTRV2-SlERF.C5-, pTRV2-SlERF.C6-, and pTRV2-SlERF. C11-infiltrated plants were similar to that on leaves from pTRV2-GUS-infiltrated plants (Figure 3), implying that these B3 group ERF genes may not be involved in disease resistance against B. cinerea. However, B. cinerea-caused lesions caused on leaves from pTRV2-SlERF.A1-, pTRV2-SlERF.A3-, pTRV2-SlERF.B4-, and pTRV2-SlERF.C3-infiltrated plants were significantly larger than the lesions on leaves from the pTRV2-GUS-infiltrated plants (Figure 3A) at 3 days after inoculation (Figure 3B), indicating that SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 play roles in disease resistance against B. cinerea. Because the functions of SlERF.A1, SlERF.B4, and SlERF.C3 have not been characterized previously, we further analyzed and compared the B. cinerea-provoked disease progress and in planta fungal growth between the pTRV2-SlERF.A1-, pTRV2-SlERF.B4-, and pTRV2-SlERF.C3-infiltrated plants and the pTRV2-GUS-infiltrated plants. As shown in Figure 4A, separate B. cinerea-caused lesions were observed; however, sizes of lesions on leaves from pTRV2-SlERF.A1-, pTRV2-SlERF.A3-, pTRV2-SlERF.B4-, and pTRV2-SlERF.C3-infiltrated plants were larger than that on leaves from pTRV2-GUS-infiltrated plants at 2 day after inoculation. At 4 days after inoculation, the lesions merged into large necrotic area on leaves from pTRV2-SlERF.A1-, pTRV2-SlERF.A3-, pTRV2-SlERF.B4-, and pTRV2-SlERF.C3-infiltrated plants, whereas lesions on leaves from pTRV2-GUS-infiltrated plants remained separated (Figure 4A). The lesions on leaves from pTRV2-SlERF.A1-, pTRV2-SlERF.A3-, pTRV2-SlERF.B4-, and pTRV2-SlERF.C3-infiltrated plants showed 32, 39, 44, and 30% of increase over that in pTRV2-GUS-infiltrated plants (Figure 4B). In whole plant disease assays, the pTRV2-SlERF.A1-, pTRV2-SlERF.B4-, pTRV2-SlERF.C3-, and pTRV2-SlERF.A3-infiltrated plants showed severer disease than the pTRV2-GUS-infiltrated plants (Figure 4C). qRT-PCR analysis of the transcript levels of the B. cinerea actin gene BcActin, an indication of the rate of fungal growth in planta, in leaves of the inoculated whole plants, showed that the fungal growth in the pTRV2-SlERF.A1-, pTRV2-SlERF.B4-, pTRV2-SlERF.C3-, and pTRV2-SlERF.A3-infilrated plants were significantly increased, resulting in increases of 1.3, 1.9, 1.5, and 2.2-folds at 4 days after inoculation over that in the pTRV2-GUS-infiltrated plants (Figure 4D). These data demonstrate that silencing of SlERF.A1, SlERF.B4, SlERF.C3, or SlERF.A3 resulted in increased susceptibility to B. cinerea and thus these three B3 group ERF genes may function as positive regulators of disease resistance against B. cinerea.
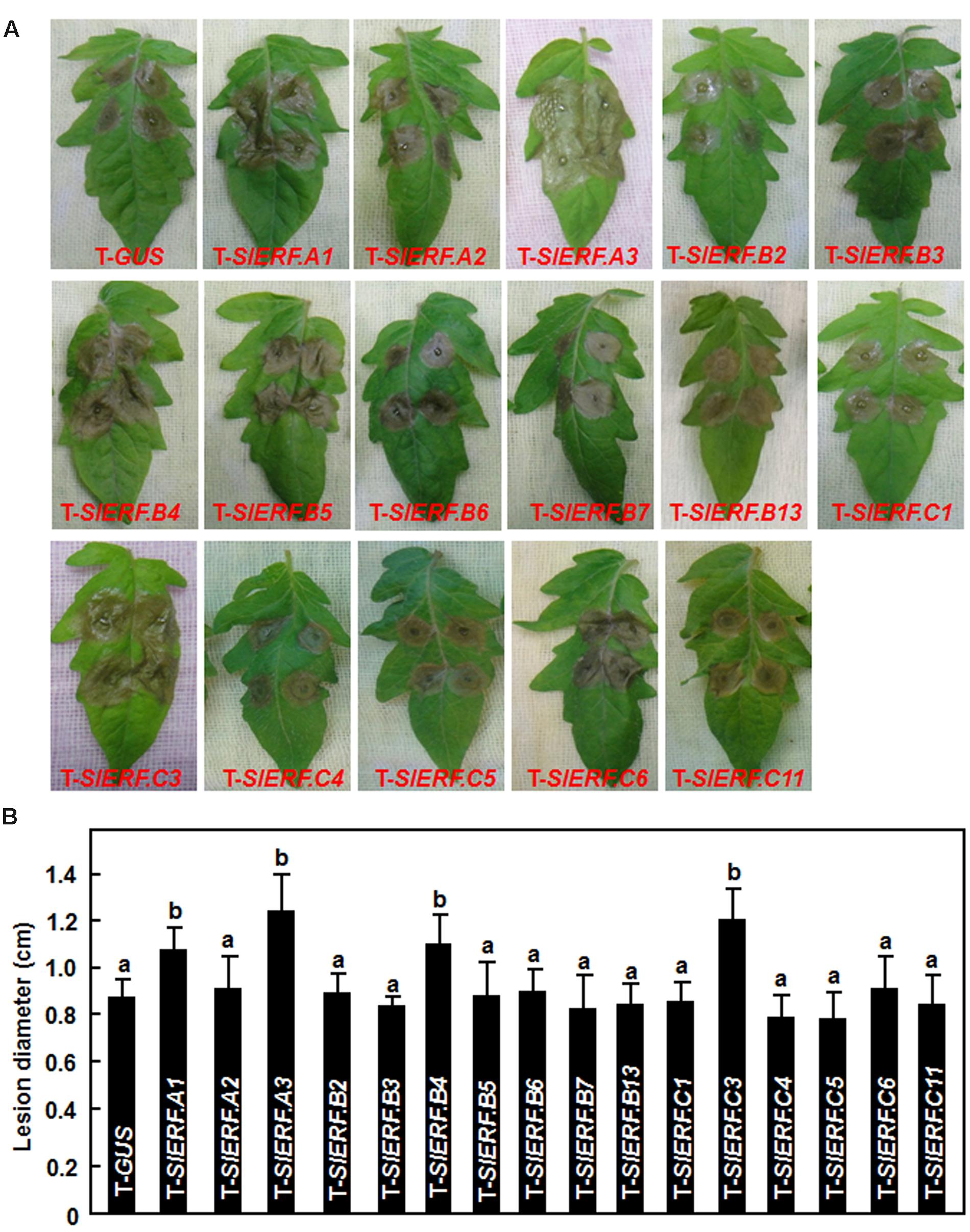
FIGURE 3. Botrytis cinerea-caused disease phenotype on leaves of VIGS-infiltrated plants with silencing of individual B3 group Ethylene-Responsive Factors (ERF) gene. Two-week-old tomato seedlings were infiltrated with agrobacteria carrying pTRV2-GUS or pTRV2-SlERFs constructs and fully expanded leaves were detached from pTRV2-GUS- or pTRV2-SlERF-infiltrated plants at 4 weeks after VIGS infiltration for disease assays. Inoculation with B. cinerea was done by dropping 5 μl of spore suspension (2 × 105 spores/ml). (A) Disease symptom on detached leaves at 3 days post inoculation (dpi). (B) Lesion sizes in leaves of the pTRV2-GUS- or pTRV2-SlERF-infiltrated plants at 3 days after inoculation. At least 10 leaves from ten individual plants were used for each experiment. Data presented (B) are the means ± SD from three independent experiments and different letters above the columns indicate significant differences at p < 0.05 level.
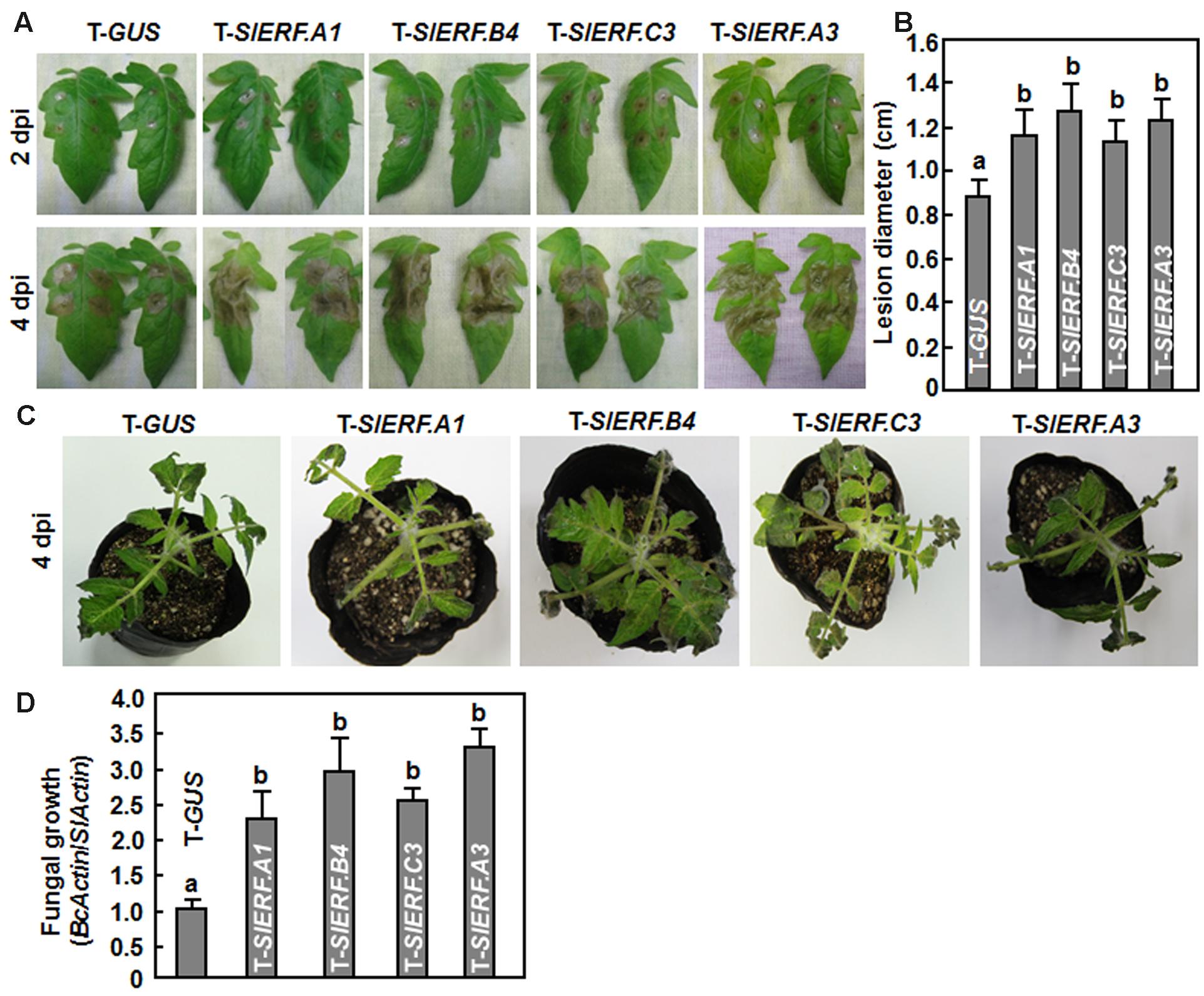
FIGURE 4. Silencing of SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 resulted in increased susceptibility to B. cinerea. Two-week-old seedlings were infiltrated with agrobacteria carrying pTRV2-SlERF.A1, pTRV2-SlERF.A3, pTRV2-SlERF.B4, pTRV2-SlERF.C3, or pTRV2-GUS and disease assays were performed on detached leaves and whole plants at 4 weeks after VIGS infiltration. Inoculation with B. cinerea was done by dropping 5 μl of spore suspension (2 × 105 spores/ml) on surface of the detached leaves or foliar spraying onto leaves of whole plants. (A) Disease symptom and progress on detached leaves of pTRV2-SlERF.A1-, pTRV2-SlERF.A3, pTRV2-SlERF.B4-, pTRV2-SlERF.C3-, or pTRV2-GUS-infiltrated plants at 2 and 4 days post inoculation (dpi). (B) Lesion sizes in leaves of the pTRV2-SlERF.A1-, pTRV2-SlERF.A3, pTRV2-SlERF.B4-, pTRV2-SlERF.C3-, or pTRV2-GUS-infiltrated plants at 4 days post inoculation. At least 10 leaves from ten individual plants were used for each experiment. (C) Disease phenotype on representative inoculated plants at 4 days post inoculation. (D) Growth of B. cinerea in inoculated leaves of pTRV2-SlERF.A1-, pTRV2-SlERF.A3, pTRV2-SlERF.B4-, pTRV2-SlERF.C3-, or pTRV2-GUS-infiltrated plants. Fungal growth in planta was assumed at 4 days after inoculation by qRT-PCR analyzing the transcript levels of B. cinerea BcActinA gene using SlActin gene. Relative fungal growth was shown as folds of transcript levels of BcActin compared to SlActin. Data presented in (B) and (C) are the means ± SD from three independent experiments and different letters above the columns indicate significant differences at p < 0.05 level.
Silencing of SlERF.A3 But Not SlERF.A1, SlERF.B4, or SlERF.C3 Decreased the Resistance to Pst DC3000
We further examined whether SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 were also involved in resistance against Pst DC3000. As shown in Figure 5A, the Pst DC3000-caused disease symptom on and the in planta bacterial growth in the inoculated leaves of the pTRV2-SlERF.A1-, pTRV2-SlERF.B4-, and pTRV2-SlERF.C3-infilrated plants were similar to those in leaves of the pTRV2-GUS-infiltrated plants at 4 days after inoculation (Figure 5B). However, disease symptom on the inoculated leaves of the pTRV2-SlERF.A3-infiltrated plants was severer and the bacterial growth in leaves of the pTRV2-SlERF.A3-infiltrated plants showed ∼100 times higher, as compared to those in leaves of the pTRV2-GUS-infiltrated plants (Figure 5). These results indicate that SlERF.A1, SlERF.B4, or SlERF.C3 may not have a function in tomato resistance against Pst DC3000 and that SlERF.A3 is required for the resistance to Pst DC3000.
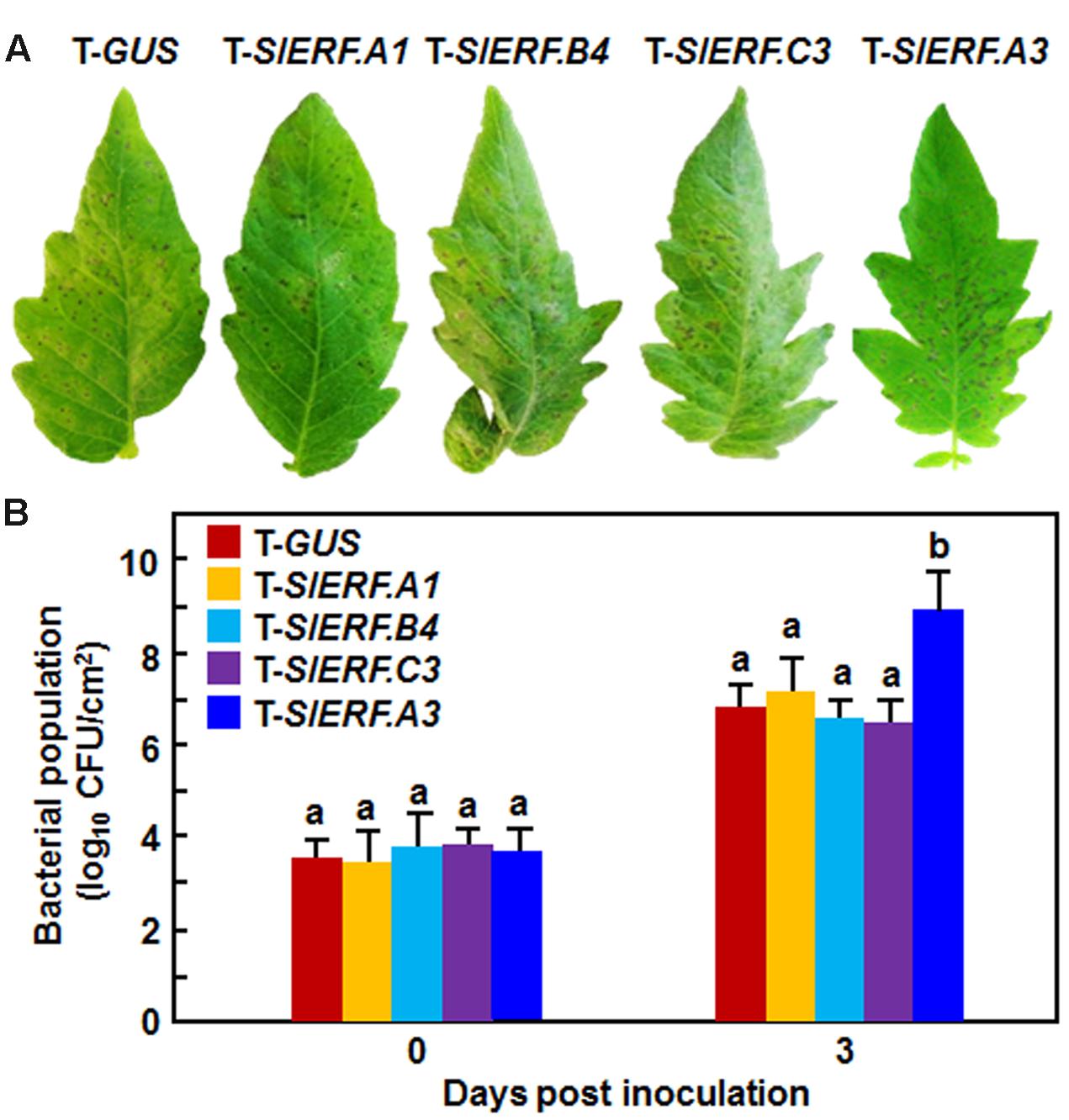
FIGURE 5. Silencing of SlERF.A3 but not SlERF.A1, SlERF.B4, and SlERF.C3 decreased the resistance against Pst DC3000. pTRV2-SlERF.A1-, pTRV2-SlERF.A3-, pTRV2-SlERF.B4-, pTRV2-SlERF.C3-, or pTRV2-GUS-infiltrated plants were inoculated with Pst DC3000 at 4 weeks after VIGS infiltration. Disease phenotype (A) and bacterial population (B) on leaves of SlERF.A1-, SlERF.A3-, SlERF.B4-, SlERF.C3-, and GUS-silenced plants were recorded and measured. Data presented (B) are the means ± SD from three independent experiments and different letters above the columns indicate significant differences at p < 0.05 level.
Silencing of SlERF.A1, SlERF.B4, SlERF.C3, or SlERF.A3 Attenuated Defense Response upon B. cinerea Infection
To explore whether silencing of SlERF.A1, SlERF.B4, and SlERF.C3 affected the defense response, we analyzed and compared the expression levels of some well-known defense-related genes between the pTRV2-SlERF.A1-, pTRV2-SlERF.B4-, and pTRV2-SlERF.C3-infilrated plants and pTRV-GUS-infiltrated plants with or without inoculation with B. cinerea. SlLapA1 and SlPin2 were considered to be regulated by the JA/ET signaling pathway while SlPR1a and SlPR-P2 were thought to be regulated by the SA-mediated signaling pathway (Kawazu et al., 2012). In mock-inoculation controls, the expression levels of these four selected defense-related genes were comparable between the pTRV2-SlERF.A1-, pTRV2-SlERF.B4-, pTRV2-SlERF.C3-, and pTRV2-SlERF.A3-infilrated plants and pTRV-GUS-infiltrated plants during a period of 48 h after mock-inoculation (Figure 6). However, the expression levels of SlLapA1 and SlPin2 in the pTRV2-SlERF.A1-, pTRV2-SlERF.B4-, pTRV2-SlERF.C3-, and pTRV2-SlERF.A3-infilrated plants were markedly reduced, leading to three–sixfolds of reduction as compared with those in the pTRV2-GUS-infiltrated plants at 12, 24, and 48 h after infection with B. cinerea (Figure 6, upper two rows). By contrast, the expression levels of SlPR1a and SlPR-P2 in the pTRV2-SlERF.A1-, pTRV2-SlERF.B4-, pTRV2-SlERF.C3-, and pTRV2-SlERF.A3-infilrated plants at 12, 24, and 48 h after inoculation were markedly induced by B. cinerea but showed comparable induction patterns to those in the pTRV2-GUS-infiltrated plants (Figure 6, lower two rows). These results suggest that silencing of SlERF.A1, SlERF.B4, SlERF.C3, or SlERF.A3 attenuated the B. cinerea-induced expression of the JA/ET signaling pathway-regulated defense-related genes but not affect the expression of the SA signaling pathway-regulated defense genes in tomato.
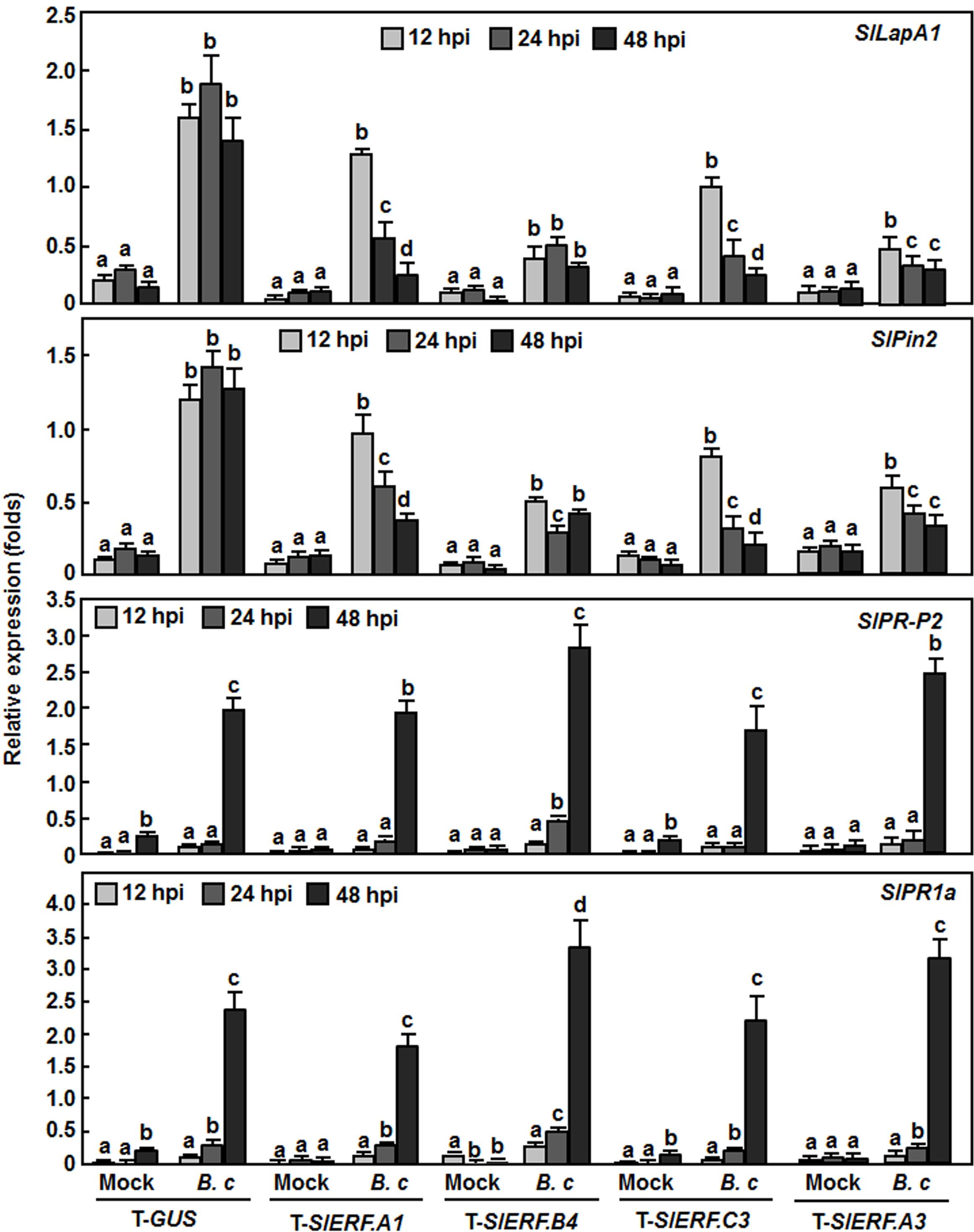
FIGURE 6. Silencing of SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 attenuated the B. cinerea-induced expression of JA/ET signaling pathway-regulated defense genes. Two-week-old seedlings were infiltrated with agrobacteria carrying pTRV2-SlERF.A1, pTRV2-SlERF.A3, pTRV2-SlERF.B4, pTRV2-SlERF.C3, or pTRV2-GUS and inoculated by spraying with spore suspension of B. cinerea or with 1% maltose buffer as a mock control at 4 weeks after VIGS infiltration. Leaf samples were collected at 12, 24, and 48 h post inoculation (hpi) for analyzing expression of selected defense-related genes by qRT-PCR. Relative expression folds were calculated after normalization with actin transcript values. Data presented are the means ± SD from three independent experiments and different letters above the columns indicate significant difference at p < 0.05 level between pTRV2-SlERF- and pTRV2-GUS-infiltrated plants.
Silencing of SlERF.A1, SlERF.A3, SlERF.B4 ,or SlERF.C3 Promoted H2O2 Accumulation upon B. cinerea Infection
It is generally accepted that pathogen-induced accumulation of reactive oxygen species (ROS) may benefit the infection of necrotrophic fungal pathogens including by B. cinerea (Mengiste, 2012). To examine whether silencing of SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 affected the accumulation of ROS, we analyzed and compared the accumulation of H2O2 between the pTRV2-SlERF.A1-, pTRV2-SlERF.A3-, pTRV2-SlERF.B4-, and pTRV2-SlERF.C3-infilrated plants and pTRV2-GUS-infiltrated plants with or without inoculation with B. cinerea. In mock-inoculated leaves, no significant accumulation of H2O2 was observed and no difference in H2O2 accumulation was seen between the pTRV2-SlERF.A1-, pTRV2-SlERF.A3-, pTRV2-SlERF.B4-, and pTRV2-SlERF.C3-infilrated plants and pTRV2-GUS-infiltrated plants (Figure 7). At 24 h after infection by B. cinerea, significant accumulation of H2O2 in inoculated leaves was detected in B. cinerea-infected leaves (Figure 7); however, more staining for H2O2 in B. cinerea-infected leaves of the pTRV2-SlERF.A1-, pTRV2-SlERF.A3-, pTRV2-SlERF.B4-, and pTRV2-SlERF.C3-infilrated plants was observed, as compared to that in B. cinerea-infected leaves of the pTRV2-GUS-infiltrated plants (Figure 7). These results indicate that silencing of SlERF.A1, SlERF.A3, SlERF.B4, or SlERF.C3 promoted the accumulation of H2O2 upon infection of B. cinerea.
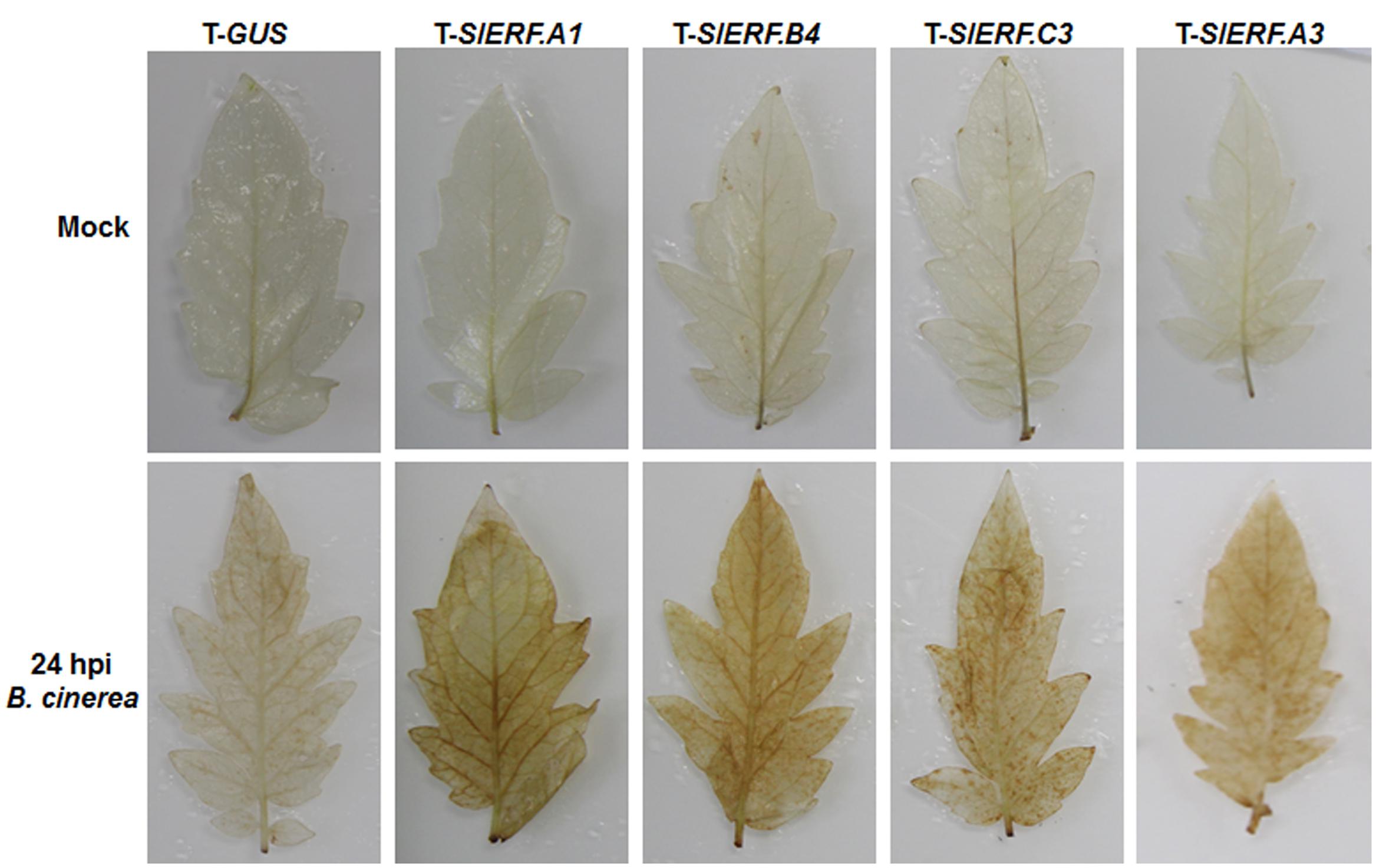
FIGURE 7. Silencing of SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 promoted the B. cinerea-induced H2O2 accumulation. Two-week-old seedlings were infiltrated with agrobacteria carrying pTRV2-SlERF.A1, pTRV2-SlERF.A3, pTRV2-SlERF.B4, pTRV2-SlERF.C3, or pTRV2-GUS and inoculated by spraying with spore suspension of B. cinerea or with 1% maltose buffer as a mock control at 4 weeks after VIGS infiltration. Leaf samples were collected at 24 hpi and accumulation of H2O2 was detected by DAB staining. Two independent experiments were performed with similar results.
Expression of SlERF.A1, SlERF.B4, SlERF.C3, and SlERF.A3 Was Induced by B. cinerea and Defense Signaling Hormones JA and ACC
To gain further insights into the involvement of SlERF.A1, SlERF.B4, SlERF.C3, and SlERF.A3 in disease resistance against B. cinerea, we analyzed their expression patterns in tomato plants after infection with B. cinerea or treatment with defense signaling hormones such as JA, SA, and ET-releasing precursor ACC. In mock-inoculated plants, the expression levels of SlERF.A1, SlERF.B4, SlERF.C3, and SlERF.A3 were not changed markedly during a 48 hr experimental period (Figure 8A). However, infection of B. cinerea significantly induced the expression of the SlERF.A1, SlERF.B4, SlERF.C3, and SlERF.A3 genes with similar patterns (Figure 8A). The expression levels in B. cinerea-inoculated plants showed 2.2–4.5, 4.7–6.7, 8.1–10.4, and 1.8–3.1-folds for SlERF.A1, SlERF.B4, SlERF.C3, and SlERF.A3, respectively, at 12, 24, and 48 h after inoculation, as compared with the levels in mock-inoculated plants (Figure 8A). Treatments with JA and ACC also induced the expression of SlERF.A1, SlERF.B4, SlERF.C3, and SlERF.A3, showing two–fourfolds of increases in JA- and ACC- treated plants, as compared with those in untreated control plants (Figure 8B). However, SA treatment did not affect the expression of SlERF.A1 but induced significantly the expression of SlERF.B4, SlERF.C3, and SlERF.A3 at 12 and/or 24 h after treatment (Figure 8B). These results indicate that expression of SlERF.A1, SlERF.B4, SlERF.C3, and SlERF.A3 can be induced by infection of B. cinerea and treatment with JA, SA, and ACC.
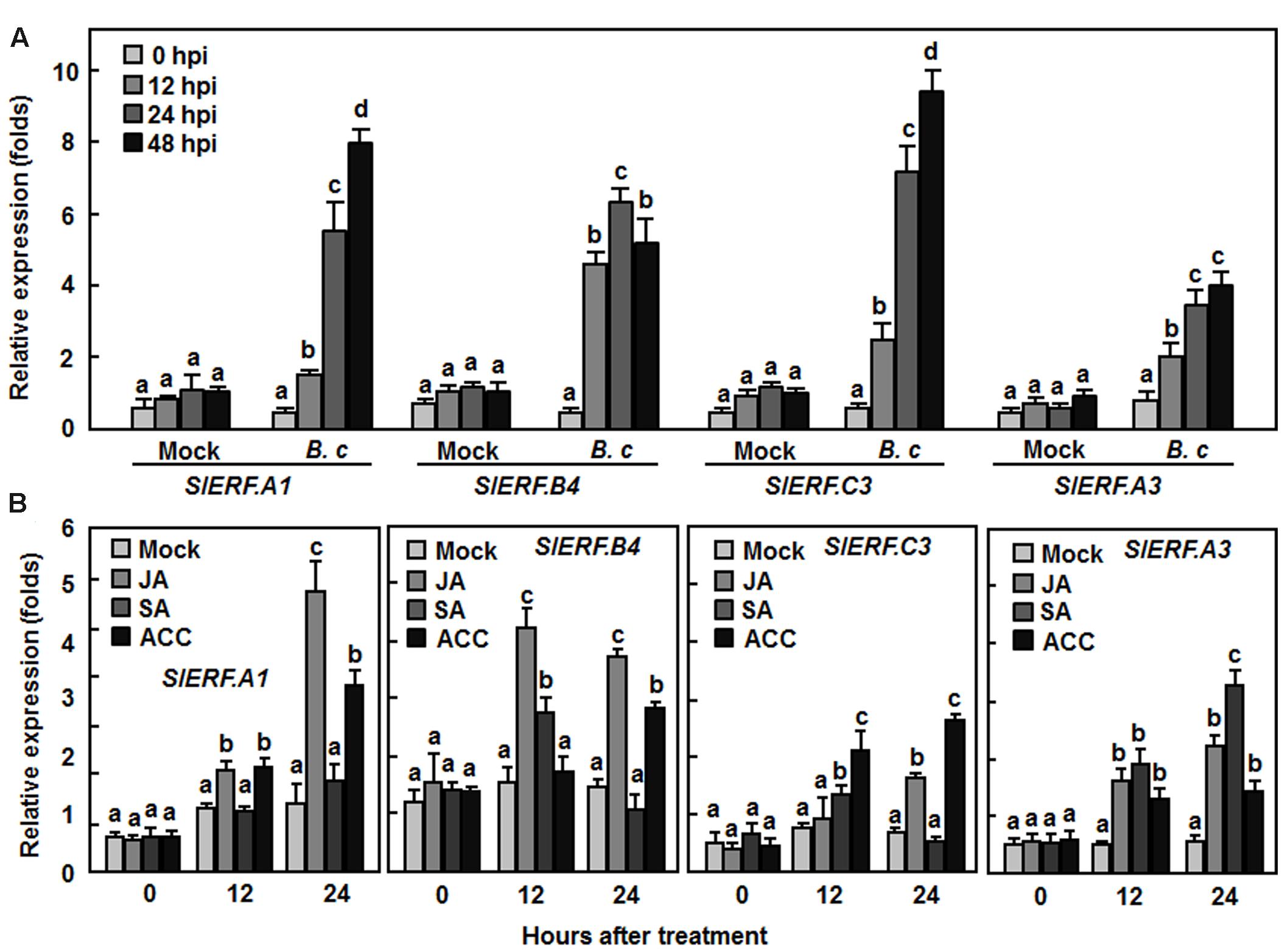
FIGURE 8. Expression of SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 induced by B. cinerea and by JA and ACC. (A) Expression of SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 in plants after inoculation with B. cinerea. Four-week-old plants were inoculated by foliar spraying with spore suspension of B. cinerea (B. c) or with 1% maltose buffer as a mock-inoculation control. (B) Expression of SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 in plants after treatment with MeJA, SA, and ACC. Four-week-old plants were treated by foliar spraying with 100 μM MeJA, 1 mM SA, 100 μM ACC solutions or sterilized distill water as a control. Leaf samples were collected at indicated time points after inoculation or treatment for analysis of gene expression by qRT-PCR. Relative expression folds were calculated after normalization with actin transcript values. Data presented are the means ± SD from three independent experiments and different letters above the columns indicate significant difference at p < 0.05 level between pTRV2-SlERF- and pTRV2-GUS-infiltrated plants.
SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 Were Localized in Nucleus and Had Transactivation Activity in Yeast
To examine the subcellular localization of SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 proteins, agrobacteria carrying pFGC-Egfp:SlERF.A1, pFGC-Egfp:SlERF.A3, pFGC-Egfp:SlERF.B4, pFGC-Egfp:SlERF.C3, and pFGC-Egfp (as a negative control) were infiltrated into leaves of 4-week-old N. benthamiana plants that expressed a red nuclear marker RFP–H2B protein (Chakrabarty et al., 2007). Confocal micrographs showed that SlERF.A1-GFP, SlERF.A3-GFP, SlERF.B4-GFP, and SlERF.C3-GFP were solely and clearly localized to the nucleus, co-localized with the known nucleus marker RFP–H2B protein (Figure 9A), whereas the GFP alone was detected in both the nucleus and cytoplasm (Figure 9A). These results demonstrate that the SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 proteins are localized to nucleus of the cells. Meanwhile, the transactivation activity of the SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 proteins were also determined using a yeast assay system. Yeast cells transformed with pBD-SlERF.A1, pBD-SlERF.A3, pBD-SlERF.B4, pBD-SlERF.C3 or pBD empty vector (as a negative control) grew well on SD/Trp- medium (Figure 9B). However, only the pBD-SlERF.A1-, pBD-SlERF.A3-, pBD-SlERF.B4-, or pBD-SlERF.C3-transformed yeast cells grew well on SD/His-/Trp- medium and produced blue pigments after addition of x-a-gal, whereas the pBD empty vector-transformed cells was unable to grow on SD/His-/Trp- medium (Figure 9B). These results suggest that SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 proteins have transactivation activity in yeasts.
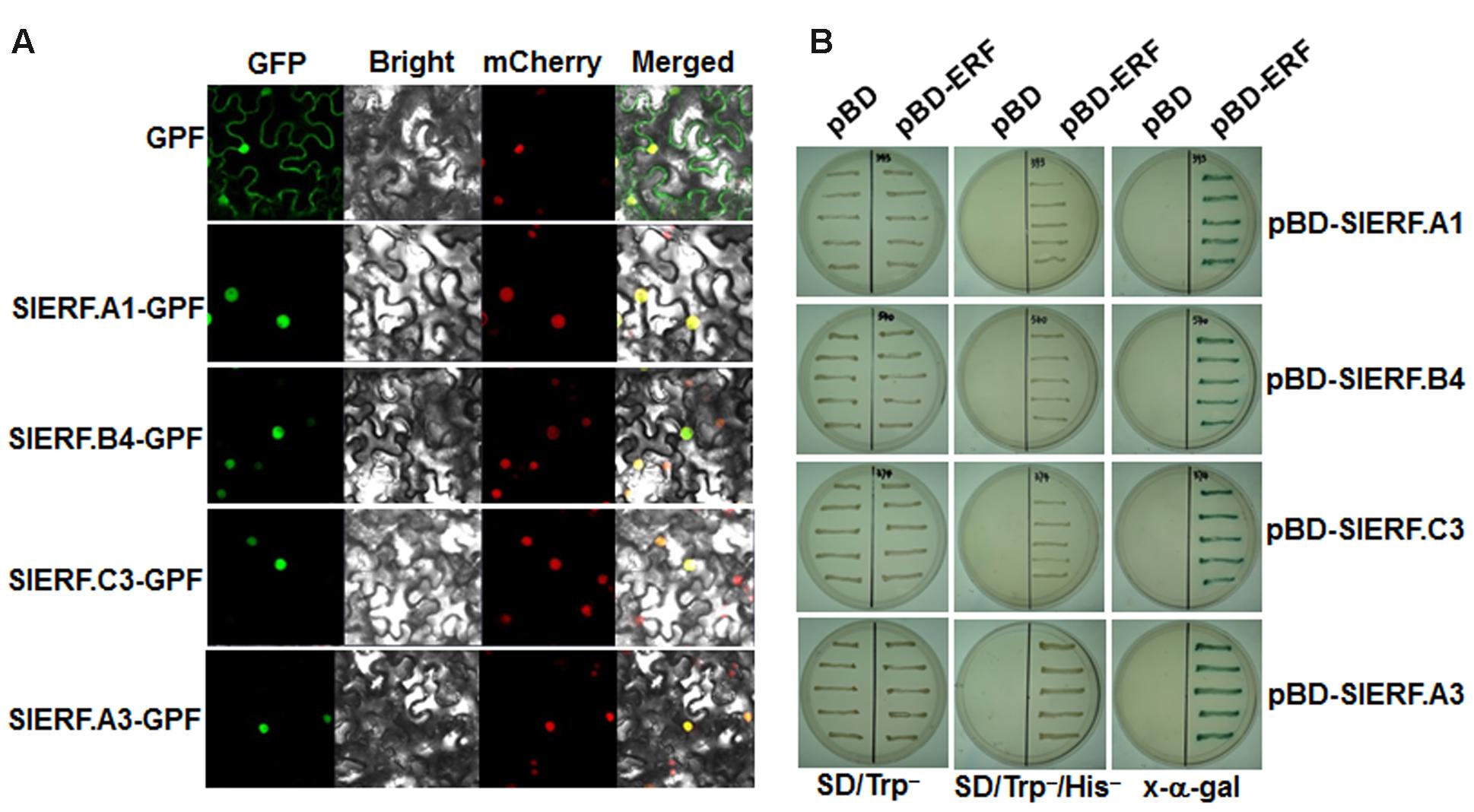
FIGURE 9. Subcellular localization and transactivation activity of SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 proteins. (A) Subcellular localization of SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 proteins. Agrobacteria carrying pFGC-Egfp-SlERF.A1, pFGC-Egfp-SlERF.A3, pFGC-Egfp-SlERF.B4, pFGC-Egfp-SlERF.C3, or pFGC-Egfp were infiltrated into leaves of N. benthamiana plants expressing a red nucleus marker RFP-H2B protein and the images were taken at 36 h after infiltration under dark field for green fluorescence (left) and red fluorescence (middle right), white field for cell morphology (middle left) and in combination (right), respectively. (B) Transactivation activity of SlERF.A1, SlERF.B4, and SlERF.C3 proteins in yeast. Yeast cells transformed with pBD-SlERF.A1, pBD-SlERF.A3, pBD-SlERF.B4, pBD-SlERF.C3 or pBD empty vector (as a negative control) were streaked on SD/Trp- plates (left) or SD/Trp-His- plates (middle) for 3 days at 28°C. The x-α-gal was added to the SD/Trp-His- plates and kept at 28°C for 6 hr (right).
Discussion
In the present study, VIGS-based functional analyses of 18 members in the B3 group of the tomato ERF family revealed that at least four members are required for disease resistance against B. cinerea and three members may have functions in regulating vegetative growth in tomato. Most importantly, our study presented evidence that SlERF.A1, SlERF.B4, and SlERF.C3, three previously uncharacterized members in B3 group of the ERF family, and SlERF.A3, a previously identified ERF with function in immunity to Pst DC3000 (Gu et al., 2002), positively regulate defense response against B. cinerea.
In Arabidopsis, several members in B3 group of the ERF family have been shown to function in defense response against B. cinerea, including ERF1, functionally redundant ERF5 and ERF6, and ORA59 (Berrocal-Lobo et al., 2002; Pré et al., 2008; Moffat et al., 2012). VIGS-based functional analyses of 16 members in B3 group of the tomato ERF family identified at least four members including SlERF.A1, SlERF.A3 (Pti4), SlERF.B4, and SlERF.C3 that are required for disease resistance against B. cinerea (Figures 3 and 4). The function of SlERF.A3 (Pti4) was previously reported to be associated with the resistance against Pst DC3000 in overexpression transgenic Arabidopsis plants (Gu et al., 2002) and regulate defense response through interaction with Pto and binding to GCC and non-GCC boxes in promoters of defense genes (Zhou et al., 1997; Wu et al., 2002; Chakravarthy et al., 2003). Our VIGS experiments further support that SlERF.A3 is required for the resistance of tomato plants against Pst DC3000 (Figure 5). However, the functions of the remaining three members SlERF.A1, SlERF.B4, and SlERF.C3 are previously uncharacterized and we demonstrated that these three members are required for disease resistance against B. cinerea. This hypothesis is supported by several lines of evidence. Firstly, silencing of SlERF.A1, SlERF.A3, SlERF.B4, or SlERF.C3 led to increased susceptibility to infection of B. cinerea, as evaluated by enhanced disease severity and in planta fungal growth in SlERF.A1-, SlERF.A3-, SlERF.B4-, and SlERF.C3-silenced plants (Figures 3 and 4). This is similar to the observation that the ORA59-silenced Arabidopsis plants showed increased susceptibility to B. cinerea (Pré et al., 2008). Secondly, expression of SlERF.A1, SlERF.A3, SlERF.B4 and SlERF.C3 was induced by infection of B. cinerea (Figure 8A), indicating that they are responsive to B. cinerea. Furthermore, the expression of SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 was induced by MeJA, SA, and ACC (Figure 8B). Previously, the expression of SlERF.A3 was shown to be induced by Pst DC3000 as well as by SA and ET (Thara et al., 1999; Gu et al., 2000). Recently, SlERF.B6, SlERF.B7, SlERF.B8, SlERF.B9, SlERF.B10, and SlERF.B11 were identified as jasmonate-responsive ERFs, which are involved in biosynthesis of steroidal glycoalkaloids (Thagun et al., 2016). Thirdly, the B. cinerea-induced expression of the JA/ET signaling pathway-regulated defense genes SlLapA1 and SlPin2 in SlERF.A1-, SlERF.A3-, SlERF.B4-, and SlERF.C3-silenced plants was attenuated significantly (Figure 6). These data indicate that SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 might function in disease resistance against B. cinerea through the JA/ET signaling pathway, which is considered to mediate defense response against necrotrophic fungi including B. cinerea (Grant and Jones, 2009; Mengiste, 2012). In Arabidopsis, ERF1, ERF5, ERF6, AtERF14, and ORA59 were shown to act as regulators of the JA/ET signaling pathway (Lorenzo et al., 2003; Berrocal-Lobo and Molina, 2004; Oñate-Sánchez et al., 2007; Pré et al., 2008; Zarei et al., 2011; Moffat et al., 2012). Notably, the B. cinerea-induced expression of SlLapA1 and SlPin2 in SlERF.A1-, SlERF.A3-, and SlERF.C3-silenced plants decreased gradually with the progress of disease development, whereas the expression of these two genes in SlERF.B4-silenced plants maintained at relatively low level without alteration over the time (Figure 6). This may imply that distinct mechanisms modulated by SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 are involved in regulating defense response against B. cinerea through the JA/ET signaling. Lastly, it is well known that pathogen-induced ROS play different roles in immune response against pathogens with different infection styles. Generally, pathogen-induced ROS plays a signaling role in immunity against Pst DC3000 while this pathogen-induced ROS may benefit the infection by necrotrophic fungi such as B. cinerea (Mengiste, 2012). The significant accumulation of H2O2 in leaves of the SlERF.A1-, SlERF.A3-, SlERF.B4-, and SlERF.C3-silenced plants after infection of B. cinerea (Figure 7) may suggest that the function of SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 in defense response against B. cinerea links to ROS generation. Similar correlation of enhanced ROS accumulation and increased B. cinerea susceptibility were also observed in SlSRN1-, SlMKK2-, or SlMKK4-silenced tomato plants (Li et al., 2014b; Liu et al., 2014b).
However, the expression of two SA signaling pathway-regulated defense genes SlPR1a and SlPR-P2 was not significantly suppressed in SlERF.A1-, SlERF.B4-, and SlERF.C3-silenced plants, as compared with their expressions in the control plants, after infection of B. cinerea (Figure 6), indicating that silencing of SlERF.A1, SlERF.B4, or SlERF.C3 may not affect the SA signaling pathway. This can be partially corroborated by the observation that silencing of SlERF.A1, SlERF.B4, or SlERF.C3 did not affect the phenotype of disease caused by Pst DC3000 (Figure 5). However, the involvement of SlERF.A1, SlERF.B4, and SlERF.C3 in SA signaling pathways cannot be excluded, as several members in B3 group of the Arabidopsis ERF family play roles in the modulating the balance between the JA/ET and SA signaling pathways to allow plants to mount an appropriate defense response against the attacking pathogen (Berrocal-Lobo et al., 2002; Moffat et al., 2012).
It is well known that some members of the AP2/ERF superfamily play important roles in regulating plant growth and development. However, evidence supporting the direct involvement of members in B3 group of the plant ERF family is limited. Recently, SlERF.A2 (LeERF1) and SlERF.B3 (Sl-ERF.B.3) were found to be involved in development and fruit ripening and softening in tomato (Li et al., 2007; Liu et al., 2013, 2016; Liu M. et al., 2014). In the present study, we found that silencing of SlERF.B1 or SlERF.C2 resulted in death of the pTRV2-SlERF.B1- and pTRV2-SlERF.C2-infiltrated plants within 7 days after VIGS infiltration (Figure 2A), even relative low concentrations of agrobacteria or relatively older plants were used for VIGS assays. Thus, it seems likely that both of SlERF.B1 and SlERF.C2 play roles in regulating vegetative growth of the tomato plants. On the other hand, it was previously reported that overexpression of Pti4 led to inhibition of growth in transgenic Arabidopsis, displaying dwarf phenotype (Zhou et al., 1997). The growth inhibition in Pti4-overexpressing Arabidopsis plants might be due to the constitutively upregulated expression of a set of GCC box-containing genes (Wu et al., 2002). In contrast, we found that silencing of SlERF.A3 (Pti4) significantly suppressed growth of the pTRV2-SlERF.A3-infitlrated plants (Figures 2B,C), suggesting that a proper expression level of SlERF.A3 (Pti4) is intrinsically required for at least vegetative growth in tomato.
It was demonstrated that plant ERF proteins could function as transcriptional activators or repressors (Fujimoto et al., 2000). However, several members of the B3 group of plant ERF family, including Arabidopsis ERF1 (AtERF92), AtERF15 (AtERF93), ORA59 (AtERF94), and ATERF14 (AtERF97) and tomato Pti4/5, TERF1, and TSRF1, have been shown to function as transcriptional activators (Zhou et al., 1997; Fujimoto et al., 2000; Gu et al., 2002; Wu et al., 2002; Huang et al., 2004; Zhang et al., 2007; Zarei et al., 2011). Recent biochemical studies showed that 12 members of the tomato B3 group, including SlERF.A1, SlERF.B4, and SlERF.C3, could bind to synthetic GCC promoter and acted as transcriptional activators, although the strength of transcriptional activity for members from different subgroups varied (Pirrello et al., 2012). In our study, we found that SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 were transcriptional activators and were localized in nucleus (Figure 9). It was found that, when overexpressed, several members in B3 group of the plant ERF family including tomato Pti4 and Arabidopsis ERF5 and ERF6 could upregulate expression of a large set of GCC box-containing genes (Wu et al., 2002; Moffat et al., 2012) and that the Arabidopsis ORA59 could directly bind to two functionally equivalent GCC boxes in the promoter of PDF1.2 to enable its responsiveness to activation of the JA/ET signaling pathway (Zarei et al., 2011).
In this study, we took advantage of the simply and fast VIGS approach (Liu et al., 2002) to knockdown endogenous expression of individual member in the B3 group of the tomato ERF family for investigating their involvement in disease resistance against B. cinerea. However, functional redundancy is a common phenomenon among plant ERF genes with high levels of sequence similarity/identity, leading to a difficulty to characterize the requirement of members of plant ERF family in disease resistance against pathogens when use knockout or knockdown mutants. For example, neither of erf5 nor erf6 mutant plants displayed altered resistance to B. cinerea, while the erf5 erf6 double mutant showed a significant increase in susceptibility to B. cinerea, demonstrating that ERF5 and ERF6, two members of IXb subgroup of B3 group of the Arabidopsis ERF family, play redundant roles in disease resistance against B. cinerea (Moffat et al., 2012). However, overexpression of either ERF5 or ERF6 led to increased disease resistance against B. cinerea (Moffat et al., 2012). In this regard, it is possible that we might miss discovery of other members in B3 group of the tomato ERF family that have function in disease resistance against B. cinerea. This is supported in part by the facts that silencing of SlERF.C4 (TSRF1) or SlERF.C6 (Pti5), which were previously shown to play important roles in disease resistance in overexpressing transgenic plants (He et al., 2001; Zhang et al., 2004b, 2007), did not affect disease resistance against B. cinerea (Figure 3). Although silencing of a specific ERF gene such as SlERF.A1, SlERF.B4, or SlERF.C3 did not affect the expression of closely related ERF genes (Figure 1B), further detailed studies with consideration of functional redundancy among different members should be helpful in understanding the biological function of the B3 group members in tomato ERF family.
Conclusion
Our VIGS-based functional analyses demonstrate that, SlERF.A1, SlERF.B4, and SlERF.C3, three previously uncharacterized members in the B3 group of the ERF family, and SlERF.A3, a previously identified ERF with function in immunity to Pst DC3000, are required for the resistance against B. cinerea. SlERF.A3 but not SlERF.A1, SlERF.B4 or SlERF.C3 is required for the resistance against Pst DC3000 in tomato. However, how SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 regulate tomato immunity against B. cinerea is still an open question. Currently, generation of stable transgenic tomato lines with overexpression and RNAi-mediated suppression of these defense-related ERF genes is undergoing in our lab. Once such transgenic lines are available, comparative RNA-seq and ChIP-seq analyses of transgenic plants with or without infection of B. cinerea will not only lead to the identification of SlERF.A1-, SlERF.A3-, SlERF.B4-, and SlERF.C3-dependent differentially expressed regulon, but also provide information on the SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 binding sites at genome-wide level. Data from these analyses will definitely be helpful to characterize the direct target genes, putative pathways and transcriptional network that are regulated by SlERF.A1, SlERF.A3, SlERF.B4, and SlERF.C3 during immune response against B. cinerea.
Author Contributions
FS and ZO designed the experiments. ZO, SL, LhH, YH, XL, LH, YZ, HZ, and DL carried out most of the experiments. FS and ZO drafted and revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation (No. 31272028), the Fundamental Research Fund for the Central Universities (2016FZA6014), and the Natural Science Foundation of Jiangxi Province of China (No. 20151BAB214017).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We are grateful to Dr. Michael Goodin (Department of Plant Pathology, University of Kentucky, USA) for providing the H2B-RFP N. benthamiana line.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01964/full#supplementary-material
References
Abuqamar, S., Chai, M. F., Luo, H., Song, F., and Mengiste, T. (2008). Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. Plant Cell 20, 1964–1983. doi: 10.1105/tpc.108.059477
Abuqamar, S., Luo, H., Laluk, K., Mickelbart, M. V., and Mengiste, T. (2009). Crosstalk between biotic and abiotic stress responses in tomato is mediated by the AIM1 transcription factor. Plant J. 58, 347–360. doi: 10.1111/j.1365-313X.2008.03783.x
Alves, M. S., Dadalto, S. P., Gonçalves, A. B., De Souza, G. B., Barros, V. A., and Fietto, L. G. (2013). Plant bZIP transcription factors responsive to pathogens: a review. Int. J. Mol. Sci. 14, 7815–7828. doi: 10.3390/ijms14047815
Anderson, J. P., Lichtenzveig, J., Gleason, C., Oliver, R. P., and Singh, K. B. (2010). The B-3 ethylene response factor MtERF1-1 mediates resistance to a subset of root pathogens in Medicago truncatula without adversely affecting symbiosis with rhizobia. Plant Physiol. 154, 861–873. doi: 10.1104/pp.110.163949
Asselbergh, B., Curvers, K., Franca, S. C., Audenaert, K., Vuylsteke, M., Van Breusegem, F., et al. (2007). Resistance to Botrytis cinerea in sitiens, an abscisic acid-deficient tomato mutant, involves timely production of hydrogen peroxide and cell wall modifications in the epidermis. Plant Physiol. 144, 1863–1877. doi: 10.1104/pp.107.099226
Audenaert, K., De Meyer, G. B., and Höfte, M. M. (2002). Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 128, 491–501. doi: 10.1104/pp.010605
Bernoux, M., Ellis, J. G., and Dodds, P. N. (2011). New insights in plant immunity signaling activation. Curr. Opin. Plant Biol. 14, 512–518. doi: 10.1016/j.pbi.2011.05.005
Berrocal-Lobo, M., and Molina, A. (2004). Ethylene response factor1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum. Mol. Plant Microbe Interact. 17, 763–770. doi: 10.1094/MPMI.2004.17.7.763
Berrocal-Lobo, M., Molina, A., and Solano, R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29, 23–32. doi: 10.1046/j.1365-313x.2002.01191.x
Blanco-Ulate, B., Vincenti, E., Powell, A. L., and Cantu, D. (2013). Tomato transcriptome and mutant analyses suggest a role for plant stress hormones in the interaction between fruit and Botrytis cinerea. Front. Plant Sci. 4:142. doi: 10.3389/fpls.2013.00142
Buscaill, P., and Rivas, S. (2014). Transcriptional control of plant defence responses. Curr. Opin. Plant Biol. 20, 35–46. doi: 10.1016/j.pbi.2014.04.004
Buttner, M., and Singh, K. B. (1997). Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. U.S.A. 94, 5961–5966. doi: 10.1073/pnas.94.11.5961
Buxdorf, K., Rubinsky, G., Barda, O., Burdman, S., Aharoni, A., and Levy, M. (2014). The transcription factor SlSHINE3 modulates defense responses in tomato plants. Plant Mol. Biol. 84, 37–47. doi: 10.1007/s11103-013-0117-1
Camañes, G., Scalschi, L., Vicedo, B., González-Bosch, C., and García-Agustín, P. (2015). An untargeted global metabolomic analysis reveals the biochemical changes underlying basal resistance and priming in Solanum lycopersicum, and identifies 1-methyltryptophan as a metabolite involved in plant responses to Botrytis cinerea and Pseudomonas syringae. Plant J. 84, 125–139. doi: 10.1111/tpj.12964
Cantu, D., Vicente, A. R., Greve, L. C., Dewey, F. M., Bennett, A. B., Labavitch, J. M., et al. (2008). The intersection between cell wall disassembly, ripening, and fruit susceptibility to Botrytis cinerea. Proc. Natl. Acad. Sci. U.S.A. 105, 859–864. doi: 10.1073/pnas.0709813105
Cao, Y., Wu, Y., Zheng, Z., and Song, F. (2006). Overexpression of the rice EREBP-like gene OsBIERF3 enhances disease resistance and salt tolerance in transgenic tobacco. Physiol. Mol. Plant Pathol. 67, 202–211. doi: 10.1016/j.pmpp.2006.01.004
Chakrabarty, R., Banerjee, R., Chung, S. M., Farman, M., Citovsky, V., Hogenhout, S. A., et al. (2007). pPSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana-virus interactions. Mol. Plant Microbe Interact. 20, 740–750. doi: 10.1094/MPMI-20-7-0740
Chakravarthy, S., Tuori, R. P., D’Ascenzo, M. D., Fobert, P. R., Despres, C., and Martin, G. B. (2003). The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 15, 3033–3050. doi: 10.1105/tpc.017574
Chen, G., Hu, Z., and Grierson, D. (2008). Differential regulation of tomato ethylene responsive factor LeERF3b, a putative repressor, and the activator Pti4 in ripening mutants and in response to environmental stresses. J. Plant Physiol. 165, 662–670. doi: 10.1016/j.jplph.2007.03.006
Chung, M. Y., Vrebalov, J., Alba, R., Lee, J., McQuinn, R., Chung, J. D., et al. (2010). A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 64, 936–947. doi: 10.1111/j.1365-313X.2010.04384.x
Curvers, K., Seifi, H., Mouille, G., de Rycke, R., Asselbergh, B., Van Hecke, A., et al. (2010). Abscisic acid deficiency causes changes in cuticle permeability and pectin composition that influence tomato resistance to Botrytis cinerea. Plant Physiol. 154, 847–860. doi: 10.1104/pp.110.158972
Davis, J., Yu, D., Evans, W., Gokirmak, T., Chetelat, R. T., and Stotz, H. U. (2009). Mapping of loci from Solanum lycopersicoides conferring resistance or susceptibility to Botrytis cinerea in tomato. Theor. Appl. Genet. 119, 305–314. doi: 10.1007/s00122-009-1039-9
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Dong, N., Liu, X., Lu, Y., Du, L., Xu, H., Liu, H., et al. (2010). Overexpression of TaPIEP1, a pathogen-induced ERF gene of wheat, confers host-enhanced resistance to fungal pathogen Bipolaris sorokiniana. Funct. Integr. Genomics 10, 215–226. doi: 10.1007/s10142-009-0157-4
El Oirdi, M., El Rahman, T. A., Rigano, L., El Hadrami, A., Rodriguez, M. C., Daayf, F., et al. (2011). Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 23, 2405–2421. doi: 10.1105/tpc.111.083394
Eulgem, T., and Somssich, I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. doi: 10.1016/j.pbi.2007.04.020
Finkers, R., van den Berg, P., van Berloo, R., ten Have, A., van Heusden, A. W., van Kan, J. A., et al. (2007). Three QTLs for Botrytis cinerea resistance in tomato. Theor. Appl. Genet. 114, 585–593. doi: 10.1007/s00122-006-0458-0
Fischer, U., and Droge-Laser, W. (2004). Overexpression of NtERF5, a new member of the tobacco ethylene response transcription factor family enhances resistance to tobacco mosaic virus. Mol. Plant Microbe Interact. 17, 1162–1171. doi: 10.1094/MPMI.2004.17.10.1162
Francia, D., Demaria, D., Calderini, O., Ferraris, L., Valentino, D., Arcioni, S., et al. (2007). Wounding induces resistance to pathogens with different lifestyles in tomato: role of ethylene in cross-protection. Plant Cell Environ. 30, 1357–1365. doi: 10.1111/j.1365-3040.2007.01709.x
Fu, Z. Q., and Dong, X. N. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Fujimoto, S. Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12, 393–404. doi: 10.1105/tpc.12.3.393
Gonorazky, G., Guzzo, M. C., Abd-El-Haliem, A. M., Joosten, M. H., and Laxalt, A. M. (2016). Silencing of the tomato phosphatidylinositol-phospholipase C2 (SlPLC2) reduces plant susceptibility to Botrytis cinerea. Mol. Plant Pathol. 17, 1354–1363. doi: 10.1111/mpp.12365
Grant, M. R., and Jones, J. D. (2009). Hormone (dis)harmony moulds plant health and disease. Science 324, 750–752. doi: 10.1126/science.1173771
Gu, Y. Q., Wildermuth, M. C., Chakravarthy, S., Loh, Y. T., Yang, C., He, X., et al. (2002). Tomato transcription factors Pti4, Pti5, and Pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 14, 817–831. doi: 10.1105/tpc.000794
Gu, Y. Q., Yang, C., Thara, V. K., Zhou, J., and Martin, G. B. (2000). Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 12, 771–786. doi: 10.2307/3871000
Guo, Z. J., Chen, X. J., Wu, X. L., Ling, J. Q., and Xu, P. (2004). Overexpression of the AP2/EREBP transcription factor OPBP1 enhances disease resistance and salt tolerance in tobacco. Plant Mol. Biol. 55, 607–618. doi: 10.1007/s11103-004-1521-3
He, P., Warren, R. F., Zhao, T., Shan, L., Zhu, L., Tang, X., et al. (2001). Overexpression of Pti5 in tomato potentiates pathogen-induced defense gene expression and enhances disease resistance to Pseudomonas syringae pv. tomato. Mol. Plant Microbe Interact. 14, 1453–1457. doi: 10.1094/MPMI.2001.14.12.1453
Hind, S. R., Pulliam, S. E., Veronese, P., Shantharaj, D., Nazir, A., Jacobs, N. S., et al. (2011). The COP9 signalosome controls jasmonic acid synthesis and plant responses to herbivory and pathogens. Plant J. 65, 480–491. doi: 10.1111/j.1365-313X.2010.04437.x
Huang, Z., Zhang, Z., Zhang, X., Zhang, H., Huang, D., and Huang, R. (2004). Tomato TERF1 modulates ethylene response and enhances osmotic stress tolerance by activating expression of downstream genes. FEBS Lett. 573, 110–116. doi: 10.1016/j.febslet.2004.07.064
Kawazu, K., Mochizuki, A., Sato, Y., Sugeno, W., Murata, M., Seo, S., et al. (2012). Different expression profiles of jasmonic acid and salicylic acid inducible genes in the tomato plant against herbivores with various feeding modes. Arthropod Plant Interact. 6, 221–230. doi: 10.1007/s11829-011-9174-z
Klay, I., Pirrello, J., Riahi, L., Bernadac, A., Cherif, A., Bouzayen, M., et al. (2014). Ethylene response factor Sl-ERF.B.3 is responsive to abiotic stresses and mediates salt and cold stress response regulation in tomato. Sci. World J. 2014:167681. doi: 10.1155/2014/167681
Lee, J. M., Joung, J. G., McQuinn, R., Chung, M. Y., Fei, Z., Tieman, D., et al. (2012). Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J. 70, 191–204. doi: 10.1111/j.1365-313X.2011.04863.x
Li, D., Zhang, H., Song, Q., Wang, L., Liu, S., Hong, Y., et al. (2015). Tomato Sl3-MMP, a member of the Matrix metalloproteinase family, is required for disease resistance against Botrytis cinerea and Pseudomonas syringae pv. tomato DC3000. BMC Plant Biol. 15:143. doi: 10.1186/s12870-015-0536-z
Li, X., Huang, L., Zhang, Y., Ouyang, Z., Hong, Y., Zhang, H., et al. (2014a). Tomato SR/CAMTA transcription factors SlSR1 and SlSR3L negatively regulate disease resistance response and SlSR1L positively modulates drought stress tolerance. BMC Plant Biol. 14:286. doi: 10.1186/s12870-014-0286-3
Li, X., Zhang, H., Tian, L., Huang, L., Liu, S., Li, D., et al. (2015). Tomato SlRbohB, a member of the NADPH oxidase family, is required for disease resistance against Botrytis cinerea and tolerance to drought stress. Front. Plant Sci. 6:463. doi: 10.3389/fpls.2015.00463
Li, X., Zhang, Y., Huang, L., Ouyang, Z., Hong, Y., Zhang, H., et al. (2014b). Tomato SlMKK2 and SlMKK4 contribute to disease resistance against Botrytis cinerea. BMC Plant Biol. 14:166. doi: 10.1186/1471-2229-14-166
Li, Y., Zhu, B., Xu, W., Zhu, H., Chen, A., Xie, Y., et al. (2007). LeERF1 positively modulated ethylene triple response on etiolated seedling, plant development and fruit ripening and softening in tomato. Plant Cell Rep. 26, 1999–2008. doi: 10.1007/s00299-007-0394-8
Licausi, F., Ohme-Takagi, M., and Perata, P. (2013). APETALA2/Ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. doi: 10.1111/nph.12291
Lin, Z., Alexander, L., Hackett, R., and Grierson, D. (2008). LeCTR2, a CTR1-like protein kinase from tomato, plays a role in ethylene signalling, development and defence. Plant J. 54, 1083–1093. doi: 10.1111/j.1365-313X.2008.03481.x
Liu, B., Hong, Y. B., Zhang, Y. F., Li, X. H., Huang, L., Zhang, H. J., et al. (2014a). Tomato WRKY transcriptional factor SlDRW1 is required for disease resistance against Botrytis cinerea and tolerance to oxidative stress. Plant Sci. 227, 145–156. doi: 10.1016/j.plantsci.2014.08.001
Liu, B., Ouyang, Z., Zhang, Y., Li, X., Hong, Y., Huang, L., et al. (2014b). Tomato NAC transcription factor SlSRN1 positively regulates defense response against biotic stress but negatively regulates abiotic stress response. PLoS ONE 9:e102067. doi: 10.1371/journal.pone.0102067
Liu, D., Chen, X., Liu, J., Ye, J., and Guo, Z. (2012). The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J. Exp. Bot. 63, 3899–3911. doi: 10.1093/jxb/ers079
Liu, M., Diretto, G., Pirrello, J., Roustan, J. P., Li, Z., Giuliano, G., et al. (2014). The chimeric repressor version of an Ethylene Response Factor (ERF) family member, Sl-ERF.B3, shows contrasting effects on tomato fruit ripening. New Phytol. 203, 206–218. doi: 10.1111/nph.12771
Liu, M., Gomes, B. L., Mila, I., Purgatto, E., Peres, L. E., Frasse, P., et al. (2016). Comprehensive profiling of ethylene response factor expression identifies ripening-associated ERF genes and their link to key regulators of fruit ripening in tomato. Plant Physiol. 170, 1732–1744. doi: 10.1104/pp.15.01859
Liu, M., Pirrello, J., Kesari, R., Mila, I., Roustan, J. P., Li, Z., et al. (2013). A dominant repressor version of the tomato Sl-ERF.B3 gene confers ethylene hypersensitivity via feedback regulation of ethylene signaling and response components. Plant J. 76, 406–419. doi: 10.1111/tpj.12305
Liu, Y., Schiff, M., and Dinesh-Kumar, S. P. (2002). Virus-induced gene silencing in tomato. Plant J. 31, 777–786. doi: 10.1046/j.1365-313X.2002.01394.x
Lorenzo, O., Piqueras, R., Sánchez-Serrano, J. J., and Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178. doi: 10.1105/tpc.007468
Meng, X., Xu, J., He, Y., Yang, K. Y., Mordorski, B., Liu, Y., et al. (2013). Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25, 1126–1142. doi: 10.1105/tpc.112.109074
Mengiste, T. (2012). Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50, 267–294. doi: 10.1146/annurev-phyto-081211-172955
Moffat, C. S., Ingle, R. A., Wathugala, D. L., Saunders, N. J., Knight, H., and Knight, M. R. (2012). ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS ONE 7:e35995. doi: 10.1371/journal.pone.0035995
Moore, J. W., Loake, G. J., and Spoel, S. H. (2011). Transcription dynamics in plant immunity. Plant Cell 23, 2809–2820. doi: 10.1105/tpc.111.087346
Nakano, T., Fujisawa, M., Shima, Y., and Ito, Y. (2014). The AP2/ERF transcription factor SlERF52 functions in flower pedicel abscission in tomato. J. Exp. Bot. 65, 3111–3119. doi: 10.1093/jxb/eru154
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Nambeesan, S., AbuQamar, S., Laluk, K., Mattoo, A. K., Mickelbart, M. V., Ferruzzi, M. G., et al. (2012). Polyamines attenuate ethylene-mediated defense responses to abrogate resistance to Botrytis cinerea in tomato. Plant Physiol. 158, 1034–1045. doi: 10.1104/pp.111.188698
Nuruzzaman, M., Sharoni, A. M., and Kikuchi, S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4:248. doi: 10.3389/fmicb.2013.00248
Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182. doi: 10.1105/tpc.7.2.173
Oñate-Sánchez, L., Anderson, J. P., Young, J., and Singh, K. B. (2007). AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiol. 143, 400–409. doi: 10.1104/pp.106.086637
Osakabe, Y., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L. S. (2013). Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress. J. Exp. Bot. 64, 445–458. doi: 10.1093/jxb/ers354
Pan, I. C., Li, C. W., Su, R. C., Cheng, C. P., Lin, C. S., and Chan, M. T. (2010). Ectopic expression of an EAR motif deletion mutant of SlERF3 enhances tolerance to salt stress and Ralstonia solanacearum in tomato. Planta 232, 1075–1086. doi: 10.1007/s00425-010-1235-5
Pan, Y., Seymour, G. B., Lu, C., Hu, Z., Chen, X., and Chen, G. (2012). An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep. 31, 349–360. doi: 10.1007/s00299-011-1170-3
Pirrello, J., Jaimes-Miranda, F., Sanchez-Ballesta, M. T., Tournier, B., Khalil-Ahmad, Q., Regad, F., et al. (2006). Sl-ERF2, a tomato ethylene response factor involved in ethylene response and seed germination. Plant Cell Physiol. 47, 1195–1205. doi: 10.1093/pcp/pcj084
Pirrello, J., Prasad, N., Zhang, W. S., Chen, K. S., Mila, I., Zouine, M., et al. (2012). Functional analysis and binding affinity of tomato ethylene response factors provide insight on the molecular bases of plant differential responses to ethylene. BMC Plant Biol. 12:190. doi: 10.1186/1471-2229-12-190
Pré, M., Atallah, M., Champion, A., De Vos, M., Pieterse, C. M., and Memelink, J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147, 1347–1357. doi: 10.1104/pp.108.117523
Quan, R., Hu, S., Zhang, Z., Zhang, H., Zhang, Z., and Huang, R. (2010). Overexpression of an ERF transcription factor TSRF1 improves rice drought tolerance. Plant Biotechnol. J. 8, 476–488. doi: 10.1111/j.1467-7652.2009.00492.x
Rahman, T. A., Oirdi, M. E., Gonzalez-Lamothe, R., and Bouarab, K. (2012). Necrotrophic pathogens use the salicylic acid signaling pathway to promote disease development in tomato. Mol. Plant Microbe Interact. 25, 1584–1593. doi: 10.1094/MPMI-07-12-0187-R
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Scalschi, L., Sanmartín, M., Camañes, G., Troncho, P., Sánchez-Serrano, J. J., García-Agustín, P., et al. (2015). Silencing of OPR3 in tomato reveals the role of OPDA in callose deposition during the activation of defense responses against Botrytis cinerea. Plant J. 81, 304–315. doi: 10.1111/tpj.12728
Segonzac, C., and Zipfel, C. (2011). Activation of plant pattern-recognition receptors by bacteria. Curr. Opin. Microbiol. 14, 54–61. doi: 10.1016/j.mib.2010.12.005
Seifi, H., De Vleesschauwer, D., Aziz, A., and Höfte, M. (2014). Modulating plant primary amino acid metabolism as a necrotrophic virulence strategy: the immune-regulatory role of asparagine synthetase in Botrytis cinerea-tomato interaction. Plant Signal. Behav. 9, e27995. doi: 10.4161/psb.27995
Seifi, H. S., Curvers, K., De Vleesschauwer, D., Delaere, I., Aziz, A., and Höfte, M. (2013). Concurrent overactivation of the cytosolic glutamine synthetase and the GABA shunt in the ABA-deficient sitiens mutant of tomato leads to resistance against Botrytis cinerea. New Phytol. 199, 490–504. doi: 10.1111/nph.12283
Sharma, M. K., Kumar, R., Solanke, A. U., Sharma, R., Tyagi, A. K., and Sharma, A. K. (2010). Identification, phylogeny, and transcript profiling of ERF family genes during development and abiotic stress treatments in tomato. Mol. Genet. Genomics 284, 455–475. doi: 10.1007/s00438-010-0580-1
Sharoni, A. M., Nuruzzaman, M., Satoh, K., Shimizu, T., Kondoh, H., Sasaya, T., et al. (2011). Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol. 52, 344–360. doi: 10.1093/pcp/pcq196
Sivakumaran, A., Akinyemi, A., Mandon, J., Cristescu, S. M., Hall, M. A., Harren, F. J., et al. (2016). ABA suppresses Botrytis cinerea elicited NO production in tomato to influence H2O2 generation and increase host susceptibility. Front. Plant Sci. 7:709. doi: 10.3389/fpls.2016.00709
Smith, J. E., Mengesha, B., Tang, H., Mengiste, T., and Bluhm, B. H. (2014). Resistance to Botrytis cinerea in Solanum lycopersicoides involves widespread transcriptional reprogramming. BMC Genomics 15:334. doi: 10.1186/1471-2164-15-334
Solano, R., Stepanova, A., Chao, Q., and Ecker, J. R. (1998). Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714. doi: 10.1101/gad.12.23.3703
Son, G. H., Wan, J., Kim, H. J., Nguyen, X. C., Chung, W. S., Hong, J. C., et al. (2011). The ethylene responsive element binding factor 5, ERF5, is involved in the chitin-induced innate immunity response. Mol. Plant Microbe Interact. 25, 48–60. doi: 10.1094/MPMI-06-11-0165
Thagun, C., Imanishi, S., Kudo, T., Nakabayashi, R., Ohyama, K., Mori, T., et al. (2016). Jasmonate-responsive ERF transcription factors regulate steroidal glycoalkaloid biosynthesis in tomato. Plant Cell Physiol. 57, 961–975. doi: 10.1093/pcp/pcw067
Thara, V. K., Tang, X., Gu, Y. Q., Martin, G. B., and Zhou, J. M. (1999). Pseudomonas syringae pv tomato induces the expression of tomato EREBP-like genes pti4 and pti5 independent of ethylene, salicylate and jasmonate. Plant J. 20, 475–483. doi: 10.1046/j.1365-313x.1999.00619.x
Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D. B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x
Tian, Y., Zhang, H., Pan, X., Chen, X., Zhang, Z., Lu, X., et al. (2011). Overexpression of ethylene response factor TERF2 confers cold tolerance in rice seedlings. Transgenic Res. 20, 857–866. doi: 10.1007/s11248-010-9463-9
Tournier, B., Sanchez-Ballesta, M. T., Jones, B., Pesquet, E., Regad, F., Latché, A., et al. (2003). New members of the tomato ERF family show specific expression pattern and diverse DNA-binding capacity to the GCC box element. FEBS Lett. 550, 149–154. doi: 10.1016/S0014-5793(03)00757-9
Upadhyay, R. K., Soni, D. K., Singh, R., Dwivedi, U. N., Pathre, U. V., Nath, P., et al. (2013). SlERF36, an EAR-motif-containing ERF gene from tomato, alters stomatal density and modulates photosynthesis and growth. J. Exp. Bot. 64, 3237–3247. doi: 10.1093/jxb/ert162
Vega, A., Canessa, P., Hoppe, G., Retamal, I., Moyano, T. C., Canales, J., et al. (2015). Transcriptome analysis reveals regulatory networks underlying differential susceptibility to Botrytis cinerea in response to nitrogen availability in Solanum lycopersicum. Front. Plant Sci. 6:911. doi: 10.3389/fpls.2015.00911
Wang, H., Huang, Z., Chen, Q., Zhang, Z., Zhang, H., Wu, Y., et al. (2004). Ectopic overexpression of tomato JERF3 in tobacco activates downstream gene expression and enhances salt tolerance. Plant Mol. Biol. 55, 183–192. doi: 10.1007/s11103-004-0113-6
Williamson, B., Tudzynski, B., Tudzynski, P., and van Kan, J. A. (2007). Botrytis cinerea: the cause of grey mould disease. Mol. Plant Pathol. 8, 561–580. doi: 10.1111/j.1364-3703.2007.00417.x
Windram, O., Madhou, P., McHattie, S., Hill, C., Hickman, R., Cooke, E., et al. (2012). Arabidopsis defense against Botrytis cinerea: chronology and regulation deciphered by high-resolution temporal transcriptomic analysis. Plant Cell 24, 3530–3557. doi: 10.1105/tpc.112.102046
Wu, C., Avila, C. A., and Goggin, F. L. (2015). The ethylene response factor Pti5 contributes to potato aphid resistance in tomato independent of ethylene signalling. J. Exp. Bot. 66, 559–570. doi: 10.1093/jxb/eru472
Wu, K., Tian, L., Hollingworth, J., Brown, D. C., and Miki, B. (2002). Functional analysis of tomato Pti4 in Arabidopsis. Plant Physiol. 128, 30–37. doi: 10.1104/pp.010696
Wu, L., Zhang, Z., Zhang, H., Wang, X. C., and Huang, R. (2008). Transcriptional modulation of ethylene response factor protein JERF3 in the oxidative stress response enhances tolerance of tobacco seedlings to salt, drought, and freezing. Plant Physiol. 148, 1953–1963. doi: 10.1104/pp.108.126813
Zarei, A., Körbes, A. P., Younessi, P., Montiel, G., Champion, A., and Memelink, J. (2011). Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol. Biol. 75, 321–331. doi: 10.1007/s11103-010-9728-y
Zhang, H., Hong, Y., Huang, L., Liu, S., Tian, L., Dai, Y., et al. (2016). Virus-induced gene silencing-based functional analyses revealed the involvement of several putative trehalose-6-phosphate synthase/phosphatase genes in disease resistance against Botrytis cinerea and Pseudomonas syringae pv. tomato DC3000 in tomato. Front. Plant Sci. 7:1176. doi: 10.3389/fpls.2016.01176
Zhang, H., Huang, Z., Xie, B., Chen, Q., Tian, X., Zhang, X., et al. (2004a). The ethylene-, jasmonate-, abscisic acid- and NaCl-responsive tomato transcription factor JERF1 modulates expression of GCC box-containing genes and salt tolerance in tobacco. Planta 220, 262–270. doi: 10.1007/s00425-004-1347-x
Zhang, H., Li, W., Chen, J., Yang, Y., Zhang, Z., Zhang, H., et al. (2007). Transcriptional activator TSRF1 reversely regulates pathogen resistance and osmotic stress tolerance in tobacco. Plant Mol. Biol. 63, 63–71. doi: 10.1007/s11103-006-9072-4
Zhang, H., Yang, Y., Zhang, Z., Chen, J., Wang, X. C., and Huang, R. (2008). Expression of the ethylene response factor gene TSRF1 enhances abscisic acid responses during seedling development in tobacco. Planta 228, 777–787. doi: 10.1007/s00425-008-0779-0
Zhang, H., Zhang, D., Chen, J., Yang, Y., Huang, Z., Huang, D., et al. (2004b). Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum. Plant Mol. Biol. 55, 825–834. doi: 10.1007/s11103-005-2140-3
Zhang, S., Li, X., Sun, Z., Shao, S., Hu, L., Ye, M., et al. (2015). Antagonism between phytohormone signalling underlies the variation in disease susceptibility of tomato plants under elevated CO2. J. Exp. Bot. 66, 1951–1963. doi: 10.1093/jxb/eru538
Zhang, Y., Butelli, E., De Stefano, R., Schoonbeek, H. J., Magusin, A., Pagliarani, C., et al. (2013). Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr. Biol. 23, 1094–1100. doi: 10.1016/j.cub.2013.04.072
Zhang, Y., Li, D., Zhang, H., Hong, Y., Huang, L., Liu, S., et al. (2015). Tomato histone H2B monoubiquitination enzymes SlHUB1 and SlHUB2 contribute to disease resistance against Botrytis cinerea through modulating the balance between SA- and JA/ET-mediated signaling pathways. BMC Plant Biol. 15:252. doi: 10.1186/s12870-015-0614-2
Zhang, Y., Liu, B., Li, X., Ouyang, Z., Huang, L., Hong, Y., et al. (2014). The de novo biosynthesis of vitamin B6 is required for disease resistance against Botrytis cinerea in tomato. Mol. Plant Microbe Interact. 27, 688–699. doi: 10.1094/MPMI-01-14-0020-R
Zhang, Z., and Huang, R. (2010). Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol. Biol. 73, 241–249. doi: 10.1007/s11103-010-9609-4
Zhang, Z., Li, F., Li, D., Zhang, H., and Huang, R. (2010). Expression of ethylene response factor JERF1 in rice improves tolerance to drought. Planta 232, 765–774. doi: 10.1007/s00425-010-1208-8
Zhang, Z., Zhang, H., Quan, R., Wang, X. C., and Huang, R. (2009). Transcriptional regulation of the ethylene response factor LeERF2 in the expression of ethylene biosynthesis genes controls ethylene production in tomato and tobacco. Plant Physiol. 150, 365–377. doi: 10.1104/pp.109.135830
Keywords: tomato (Solanum lycopersicum), ethylene-responsive factor (ERF), B3 group, Botrytis cinerea, defense response
Citation: Ouyang Z, Liu S, Huang L, Hong Y, Li X, Huang L, Zhang Y, Zhang H, Li D and Song F (2016) Tomato SlERF.A1, SlERF.B4, SlERF.C3 and SlERF.A3, Members of B3 Group of ERF Family, Are Required for Resistance to Botrytis cinerea. Front. Plant Sci. 7:1964. doi: 10.3389/fpls.2016.01964
Received: 17 June 2016; Accepted: 12 December 2016;
Published: 27 December 2016.
Edited by:
Brigitte Mauch-Mani, University of Neuchâtel, SwitzerlandReviewed by:
Carmen González Bosch, University of Valencia, SpainMario Serrano, Universidad Nacional Autónoma de México, Mexico
Copyright © 2016 Ouyang, Liu, Huang, Hong, Li, Huang, Zhang, Zhang, Li and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fengming Song, fmsong@zju.edu.cn
 Zhigang Ouyang
Zhigang Ouyang Shixia Liu
Shixia Liu Lihong Huang
Lihong Huang Yongbo Hong
Yongbo Hong Xiaohui Li
Xiaohui Li Lei Huang
Lei Huang Yafen Zhang
Yafen Zhang Huijuan Zhang
Huijuan Zhang Dayong Li
Dayong Li Fengming Song
Fengming Song