- 1Department of Biochemistry and Biotechnology, Kenyatta University, Nairobi, Kenya
- 2Tropical Institute of Development Innovations, Kampala, Uganda
- 3Sudan Academy of Sciences, Khartoum, Sudan
- 4Agricultural Research Cooperation, Wad-Medani, Sudan
- 5International Crops Research Institute for the Semi-Arid Tropics, Nairobi, Kenya
- 6Commission for Biotechnology and Genetic Engineering, National Centre for Research, Khartoum, Sudan
- 7Department of Biology, University of Virginia, Charlottesville, VA, USA
Sorghum is a major food staple in sub-Saharan Africa (SSA), but its production is constrained by the parasitic plant Striga that attaches to the roots of many cereals crops and causes severe stunting and loss of yield. Away from cultivated farmland, wild sorghum accessions grow as weedy plants and have shown remarkable immunity to Striga. We sought to determine the extent of the resistance to Striga in wild sorghum plants. Our screening strategy involved controlled laboratory assays of rhizotrons, where we artificially infected sorghum with Striga, as well as field experiments at three sites, where we grew sorghum with a natural Striga infestation. We tested the resistance response of seven accessions of wild sorghum of the aethiopicum, drummondii, and arundinaceum races against N13, which is a cultivated Striga resistant landrace. The susceptible control was farmer-preferred variety, Ochuti. From the laboratory experiments, we found three wild sorghum accessions (WSA-1, WSE-1, and WSA-2) that had significantly higher resistance than N13. These accessions had the lowest Striga biomass and the fewest and smallest Striga attached to them. Further microscopic and histological analysis of attached Striga haustorium showed that wild sorghum accessions hindered the ingression of Striga haustorium into the host endodermis. In one of the resistant accessions (WSE-1), host and parasite interaction led to the accumulation of large amounts of secondary metabolites that formed a dark coloration at the interphase. Field experiments confirmed the laboratory screening experiments in that these same accessions were found to have resistance against Striga. In the field, wild sorghum had low Area under the Striga Number Progressive curve (AUSNPC), which measures emergence of Striga from a host over time. We concluded that wild sorghum accessions are an important reservoir for Striga resistance that could be used to expand the genetic basis of cultivated sorghum for resistance to the parasite.
Introduction
Domestication—the process of transforming wild species into elite cultivars—inevitably leads to decreased genetic diversity in the selected crops (Doebley et al., 2006). In some cases, the lost genetic diversity may represent the organism's capacity to adopt changes, such as pathogen resistance, in a dynamic ecosystem (Sakai and Itoh, 2010). Based on this possibility, many crop improvement programmes are now using genomics and molecular genetic technologies to reclaim lost genetic diversity by specifically targeting genes responsible for pathogen resistance (Zhu et al., 2012; Jones et al., 2014). Success of these programmes is based on well-documented evidence that wild relatives of crops are useful reservoirs of disease resistance genes (Brozynska et al., 2016). In this study, we explore the resistance interactions between wild sorghum accessions and the parasitic plant Striga hermonthica at their the parasite's center of origin in northeastern Africa.
Striga is also infamously known as witchweed and can destroy a crop with up to a 100% yield loss (Ejeta, 2007). It is estimated that over 60% of farmland under cultivation in sub-Saharan Africa (SSA) is infested with one or more species of Striga, which impacts over 300 million farmers in over 25 countries with yield losses of over seven billion dollars (Ejeta, 2007). Three species of Striga are particularly destructive—S. hermonthica (Del.) Benth. and S. asiatica (L) Kuntze, which attack cereals, and S. gesnerioides (Willd.) Vatke, which is parasitic to cowpea plants.
Such great losses indicate a successful parasite, and for Striga, this success can be attributed to two aspects of its lifecycle: elevated fecundity, each Striga flower spike can produce over 50,000 seeds that remain viable in the soil for up to 14 years (Yoder and Scholes, 2010), and its remarkable ability to intimately link its life cycle to that of a host, such as when germination of Striga seeds and attachment to the host only occur in response to chemical cues (strigolatone) contained in the host and in some cases, non-host root exudates (Bouwmeester et al., 2007).
To manage Striga, smallholder farmers in SSA have fought back with different control methods aimed at reducing Striga seed density in the soil. These methods include reducing the amount of Striga-seed-contaminated crop seed supplies, hand weeding, crop rotation, and the use of “trap crops” that induce germination of the parasite but are not hosts. Although these methods have been extensively encouraged for many years, crop losses and the host range of Striga has continued to increase, which underscores the need for a sustainable Striga management strategy (Cotter et al., 2012). The ideal strategy would be an integrated approach that greatly exploits natural resistance. However, sources of Striga resistance are limited and are often overcome by the parasite (Ejeta, 2007). Therefore, additional sources of Striga resistance need to be found for introduction to farming systems and to promote long-term resistance.
We hypothesized that Striga-resistant sorghum was likely to be found in northeastern Africa since this region harbors the greatest diversity of both wild and cultivated sorghum (Paterson et al., 2013) and because this area is the natural range of the Striga parasite (Musselman and Hepper, 1986). Sorghum in northeastern Africa is highly variable and complex, but most genotypes can be classified as wild, cultivated, or cultivated-wild crossbreeds—all species can be classified as subspecies that are completely inter-fertile (Harlan and de Wet, 1972). On one hand, cultivated types are classified as subsp. bicolor and further subspecies are classified into five different races based on grain shape, glume shape, and panicle type (Harlan and de Wet, 1972). The five basic races are bicolor, durra, kafir, caudatum, and guinea (Paterson et al., 2013). On the other hand, wild sorghums are classified as subsp. Verticilliflorum, which consists of the races arundinaceum, virgatum, aethiopicum, and verticilliflorum (Harlan and de Wet, 1972). In addition, crossbreeds between bicolor and wild sorghum occur and are classified as drummondii (Paterson et al., 2013).
Domesticated sorghum genotypes are to a great extent susceptible to Striga (Ejeta, 2007). In contrast, wild sorghum can be found on uncultivated land as weeds that are immune to Striga infestation, which suggests there is potential for wild sorghum to be used a source of Striga resistance.
Indeed, for cultivated sorghum, only a few races are known to harbor Striga resistance. Among these is landrace N13 (S. bicolor subspecies bicolor race durra), which has a resistance mechanism that was described in detail by Maiti et al. (1984). N13 has been known to resist Striga by cell wall-thickening as well as depositing silica. In addition, Striga can induce extra lignification in the pericycle cells to the point that the haustorium encounters the endodermis. The haustorium then becomes weak and is limited by xylem-xylem connections with the host (Maiti et al., 1984).
In this study, N13 was used as a resistance control alongside a susceptible control—Ochuti classified as S. bicolor, subspecies bicolor. Ochuti is a sorghum variety preferred by farmers and is a popular crop in Western Kenya (Ngugi et al., 2016).
With regard to the resistance of wild sorghum to Striga, several studies have previously described low germination stimulant production, germination inhibition, and low haustorial initiation production as a form of resistance (Rich et al., 2004) as well as hypersensitive reaction (HR) resistance against S. asiatica (Mohamed et al., 2003). Furthermore, HR has been observed as an incompatible response between the hypervirulent S. gesnerioides race SG3 and resistant b301 cowpea variety (Li and Timko, 2009).
There is another possible mechanism of resistance against Striga—physiological barriers could lead to deposits of material that obstruct haustorium penetration. This mechanism was demonstrated in resistance against S. gesnerioides (Okonkwo and Nwoke, 1978). Substances that are stained darkly by toluidine blue characterize this mechanism of resistance (Maiti et al., 1984). Although this resistance was not well characterized, it is believed that these substances soften and/or dissolve the cell wall of host tissues (Rogers and Nelson, 1962). Aside from enzymes that degrade host tissues, there are secondary metabolites whose role in defending hosts against pathogens is becoming apparent (van Dam and Bouwmeester, 2016). Although not characterized well in parasitic plants, their role in acquiring hosts is undeniable.
The wild sorghum accessions described in this paper include the S. bicolor subspecies verticilliflorum and the races arundinaceum, aethiopicum, and drummondii. These are part of a large collection maintained at the Agricultural Research Corporation (ARC) in Sudan. The collections were carried out in Striga-prone areas of Sudan.
To determine the resistance response of these wild sorghum accessions and their potential to function as donors of Striga resistance, we used laboratory and field screening assays. We showed remarkable Striga resistance in wild sorghum accessions of aethiopicum and arundinaceum races. We found this resistance was mediated by possible mechanical or biochemical barrier mechanisms. Our study thus provides the potential to increase the genetic basis of cultivated sorghum. These findings will have wide-reaching implications for Striga control because of the ability to pyramid (i.e., combine) multiple genes in a single variety so that resistance is durable and covers a broad spectrum.
Materials and Methods
Plant Materials
Before this study was conducted, the Agricultural Research Cooperation (ARC) in Sudan conducted a countrywide collection of wild sorghum accessions and maintained them at a research station. The collection consisted of cultivated sorghum as well as wild sorghum. Genotypes collected from different locations were treated as different accessions. For this study, we selected a wild sorghum germplasm comprised of three wild races: aethiopicum (1 accession), arundinaceum (3 accessions), and drummondii (3 accessions). These accessions were referred to as WSE-1, WSA-1, WSA-2, WSA-3, WSD-1, WSD-2, and WSD-3, respectively. The initials “W,” “S,” “E,” “A,” and “D” denote Wild, Sudan, aEthiopicum, Arundinaceum, and Drummondii. The Striga-resistant landrace N13 was used as a resistant control for both laboratory and field experiments. In addition, Ochuti, which is sorghum cultivar that is popular among farmers in Kenya, was used for the susceptibility check. Both N13 and Ochuti were part of a sorghum collection maintained at Kenyatta University and originally obtained from the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) in Nairobi. S. hermonthica seeds were collected from sorghum growing in Striga-infested fields in Western Kenya at Kibos, Mbita, and Alupe.
Preconditioning of Striga Seeds
Prior to germination, Striga seeds were preconditioned as described in Gurney et al. (2003). Seeds (25 mg) were first surface sterilized in 10% (v/v) commercial bleach for 10 min with gentle agitation. The seeds were then rinsed three times with double distilled water and spread on a glass fiber filter paper (Whatman GFA) placed on sterile petri dishes. Sterile distilled water (5 ml) was added to Striga seeds followed by incubation at 29°C for 11 days for pre-conditioning. Preconditioned seeds were germinated by adding 3 ml of 0.1 ppm GR24 and incubated overnight at 29°C. Germinated Striga seedlings were analyzed for germination efficiency under a Leica MZ7F stereomicroscope fitted with a DFC320FX camera (Leica UK), and only plates showing >70% germination were used to infect sorghum roots.
Infection of Sorghum Roots with Striga
Sorghum seeds were germinated between moistened blocks of cotton wool lined with filter paper. After 7 days, each sorghum seedling was transferred to a root observation chamber (rhizotron)—25 × 25 × 5 cm Perspex plate—packed with vermiculite, as described by Gurney et al. (2006). The rhizotrons were covered with aluminum foil and supplied with 25 ml of 40% Long Ashton nutrient solution (Hudson, 1967) twice a day. The plants were maintained in a glasshouse for 11 days in a 12-h photoperiod. Day and night temperatures were set at 28 and 24°C, respectively, and the relative humidity was set at 60%.
After 11 days, sorghum seedlings with well-developed roots were infected with 25 mg of pre-germinated S. hermonthica seedlings by aligning them on sorghum roots using a soft paintbrush. Three different ecotypes of Striga (Kibos, Mbita, and Alupe) were used for infection. Five sorghum plants per accession were screened, and the experiment was replicated three times.
Macroscopic Screening of Sorghum for Striga Resistance
Rhizotrons containing sorghum roots infected with Striga were observed for resistance at 3, 9, and 21 days after infection (DAI). Observations at 3 and 9 DAI were done using a Leica MZ7F stereomicroscope fitted with a DFC320FX camera (Leica UK). At 21 DAI, rhizotrons were photographed using a Canon EOS600D camera.
To screen sorghum for post-attachment resistance and the effects of host plants on parasite development, Striga plants were harvested from the infected roots at 21 DAI. Harvested Striga seedlings from each host plant were placed in a 90 mm Petri dish and photographed using a digital camera. The number and length of Striga seedlings parasitizing each host plant was determined from the photographs using the image analysis software ImageJ, v. 1.45 (http://rsb.info.nih.gov/ij/). In addition, total Striga biomass was determined after extracting all Striga seedlings parasitizing each sorghum and drying the seedlings at 45°C for 2 days. The same metrics—Striga length, number, and biomass—were used to deduce the virulence of Striga ecotypes.
Microscopic Screening of Striga Resistance
To determine the extent of parasite development within the host root cortex, root tissue at the point of Striga haustoria attachment was dissected from host plants at 3 and 9 days (DAI) for sectioning. We used Technovit 7100 (embedding) and Technovit 3040 mounting kits (Haraeus Kulzer GmbH). The samples were fixed using Carnoy's fixative (4:1, ethanol:acetic acid) then dehydrated twice in 100% ethanol for 30 min. Next, the samples were pre-infiltrated in Ethanol-Technovit solution (1:1) for 1 h then in 100% Technovit solution for an additional 1 h. Fresh Technovit solution was added, and the samples were incubated for 3 days at 4°C.
To embed the tissues, the samples were placed into Eppendorf (1.5 ml) tube lid molds and Technovit solution:hardener 1 (1:15) was added. The molds were mounted onto wooden blocks using the Technovit 3040 kit according to the manufacturer's instructions. For sectioning, 5 micron-thick slices were cut using a Leica RM 2155 microtome (Leica instruments GmbH) and transferred to microscope slides. The sections were stained using 0.1% toluidine blue O (Sigma, USA) in 100 mM phosphate buffer at pH 7 for 2 min, then washed in distilled water and dried at 65°C for 30 min on a hot plate. The sections were then mounted onto glass slides with DePex (BDH, Poole, UK) then observed and photographed using a Zeiss microscope mounted with a Canon camera.
Field-Site Screening of Sorghum Accessions for Striga Resistance
The field evaluation of Striga resistance reported in this paper was part of a larger study involving 107 sorghum accessions that was conducted for a genome-wide association-mapping project. A sketch of the field layout is provided in Supplementary Figure 1. We planted sorghum in plots that are naturally infested with Striga in two field sites (Kumi and Bukedea) in eastern Uganda and one plot in Western Kenya (Alupe) for two seasons. The experiment was replicated three times in each of the locations using a completely randomized block design (CRBD). For control experiments, sorghum accessions were simultaneously planted in Striga-free plots within the same locations.
Each accession was planted in a sub-plot (size 3.2 × 2.5 m) separated by a one-meter footpath at a spacing of 80 cm between rows and 30 cm between plants (Supplementary Figure 1). To ensure uniform Striga infection in the infested fields, artificial inoculation with Striga seeds harvested from the same and adjacent fields was conducted. At planting, each hill was infested with approximately 3000 Striga seeds that were prepared by mixing 5 g of Striga seeds with 5 kg of washed sand and 1 tablespoon of inoculum and applied to each hill according to the methods of Jamil et al. (2013). Each accession was planted in five rows and five hills per row at a population density of 80,000 plants ha−1. In addition, 50 kg ha−1 of diammonium phosphate (DAP) fertilizer were applied. To ensure there were no gaps, three seeds were sown in each hole and thinned after 21 days, which left only one plant per hole. The first weeding was done 21 days after sowing using a hoe while subsequent weeding was conducted by hand pulling to avoid disturbing the emerging Striga plants.
Statistical Data Analysis
The generalized linear model (GLM) implemented in SAS version 9.1 was used for analysis of Striga resistance data from the laboratory experiments. Analysis of variance (ANOVA) was performed to compare the means for biomass, length and number of infecting Striga and to fit a factorial ANOVA for each replicate across the night accessions. In addition, Tukey's honest significant difference (HSD) test was performed to calculate mean separations. These data were presented as relative means ± SD in the form of graphs using Graph Pad Prism version 6 (http://www.graphpad.com). In addition, mean Striga attachments, length, and biomass data from sorghum accessions against the 3 Striga ecotypes were clustered and significant values (p ≤ 0.05) were visualized as a heat map using a custom hierarchical clustering R script. Finally, the median length data were used to generate a heat map as described above.
All nine accessions (the same ones used in laboratory screening) were evaluated for the number of Striga plants emerging 44, 58, 72, and 86 days after planting. Successive Striga counts were used to calculate the “Area under Striga number progression curve” (AUSNPC) as described by Rodenburg et al. (2005). The grain yield of sorghum in infected and non-infected fields from three randomly selected plants was weighed, and an average was obtained to provide the yield per plant in the nine sorghum accessions. This was then extrapolated to the yield in kg ha−1. The Percentage yield reduction was calculated as a function of the difference between the yield in Striga-free and Striga-infested fields. In addition, a t-test was used to determine the significant yield reduction as a result of Striga infestation. For AUSNPC and mean yields, ANOVA followed by Tukey's HSD test was performed to determine the mean separations.
Results
Resistance Response of Wild Sorghum Accessions under Controlled Laboratory Conditions
Striga infection in various sorghum hosts was variable with respect to the number, length, and biomass of Striga attachments. These three metrics were used to determine the resistance of wild sorghum. Susceptible sorghum had numerous Striga attachments. A representation of the resistance response of wild sorghum is provided in Figure 1.

Figure 1. Profile of differential Striga hermonthica attachments in wild sorghum accessions: WSD-1, Ochuti, WSA-1, N13, and WSE-1 21 days following infection of host roots with Striga seeds collected from Kibos. Red arrows indicate attachment points to the host by the parasite. Susceptible accessions represented here by WSD-1 and Ochuti showed a high number of Striga attachments compared to the control N13. Resistant accessions represented here by WSA-1 and WSE-1 showed few Striga attachments.
We observed a significantly higher level of resistance between three sorghum accessions (WSE-1, WSA-1, and WSA-2) and the positive control N13. These three accessions had the lowest number of Striga attachments (Figure 2A) and the lowest Striga biomass (Figure 2B). The low number of Striga attachments and biomass for WSE-1, WSA-1, and WSA-2 were consistently replicated for each Striga ecotype infecting the sorghum accessions (Figures 2A,B). Among the three most resistant accessions, there were significant differences in mean Striga attachments between WSE-1 and WSA-1 when only Striga from Kibos was used. However, there were significant differences in mean Striga biomass among the accessions (WSE-1, WSA-1, and WSA-2) when the sorghum accessions were infected with the Alupe Striga ecotype (Figures 2A,B).
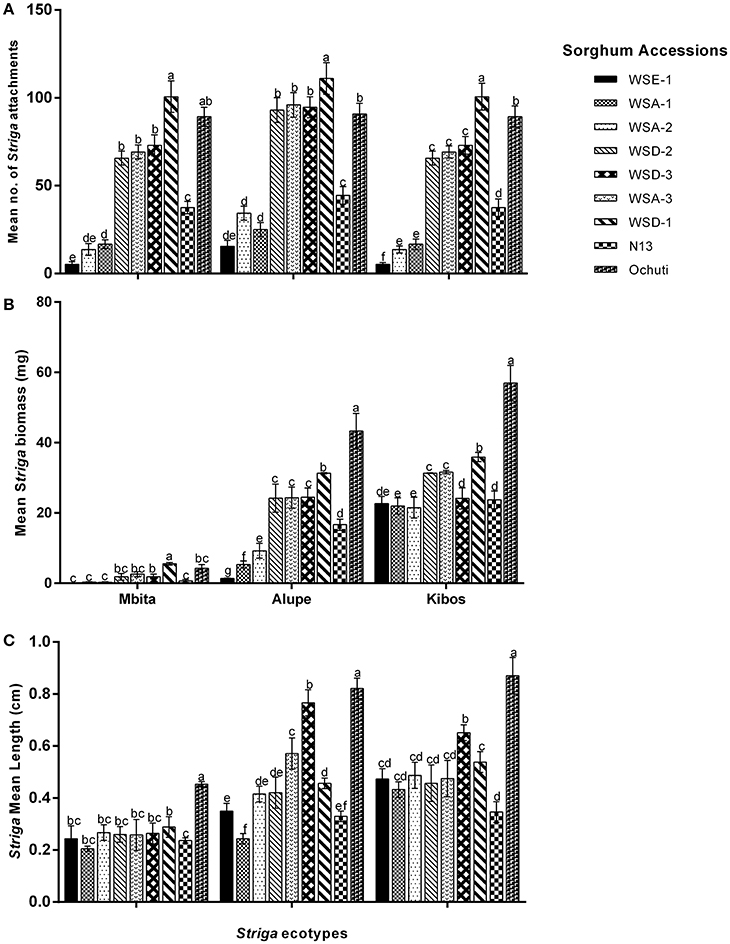
Figure 2. Resistance response of sorghum accessions to Striga hermonthica: (A) Mean Striga attachments; (B) Mean bioass of Striga plants; (C) mean Striga length on wild sorghum accessions using 3 ecotypes of Striga. The number of Striga plants was measured 21 days after infection of sorghum roots with S. hermonthica seeds. Vertical bars represent the mean ±SD while letters represent mean separations at p ≤ 0.05. The values given represent the mean for each plant.
Despite the variations observed with regard to Striga attachments and biomass among the resistant and susceptible sorghum, we did not observe those variations with regard to Striga length (Figure 2, Supplementary Figure 2). For example, in Mbita, there were only significant differences between the mean Striga length of Ochuti, WSD-1 and the rest of the accessions. Similarly, for Kibos, there were significant differences in Striga length between Ochuti—which had the highest length—and the accession with the next highest length, WSA-3. WSA-3 had a significantly higher Striga length than the rest of accessions. However, there were more significant variations with regard to length when Striga from Alupe was used. In this case we found that: (i) WSE-1 and WSA-1 had the shortest Striga seedlings attached to them; (ii) Striga seedlings attached to WSA-1 were significantly shorter than those attached to WSE-1 and; (iii) the length of Striga seedlings attached on N13 were statistically similar to those on WSA-1 and WSE-1. For Alupe, Ochuti had the highest mean length, which was significantly different compared to the accession that ranked second in Striga mean length, WSD-1. Remarkably, when we used median Striga length to rank and cluster resistance in sorghum, we obtained a heatmap that was strikingly similar to the one generated using biomass (Figure 3).
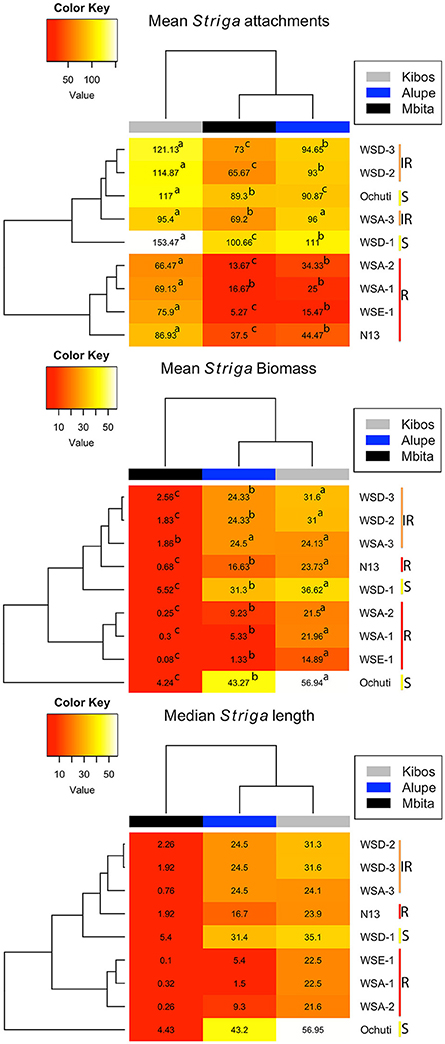
Figure 3. Heatmaps (generated using mean Striga attachments, biomass and median Striga length) showing levels of resistance of sorghum accessions infected with Striga seedlings. The yellow bar labeled “S” (Ochuti and WSD-1) represents the most susceptible accessions, and the orange bar labeled “IR” is a group consisting of WSA-3, WSD-2, and WSD-3 that had intermediate resistance, while the red bar labeled “R” represents the most resistant group, which consists of WSA-1, WSA-2 WSE-1, and N13. The heatmaps also provide an indication of the virulence of Striga ecotypes. More attachments, more biomass and longer Striga seedlings formed from Kibos and Alupe ecotypes compared to Mbita. The letters indicate significance differences between ecotypes on the same accession at p < 0.05.
We determined Striga resistance rankings based on mean Striga count and biomass because of the consistency of these metrics across the sorghum accessions and Striga ecotypes. In general, three groups of resistance with respect to N13 emerged: a highly resistant group comprised of WSE-1, WSA-1, and WSA-2; an intermediate resistance group comprised of WSA-3, WSD-2, and WSD-3, and a highly susceptible group with WSD-1 and Ochuti (Figures 2, 3).
Regarding Striga virulence, we found that the three Striga ecotypes—Kibos, Alupe, and Mbita—exhibited significant variations. There were significantly higher numbers of Striga attachments for all sorghum accessions except WSA-3 when they were infected with the Kibos ecotype. When we compared Mbita and Alupe ecotypes on how they infected sorghum, we found that the Alupe ecotype induced significantly more attachments in WSD-3, WSA-3, WSA-2, WSE-1, and N13 but not in Ochuti and WSD-2. For Striga biomass, all sorghum accessions had significantly more biomass when they were infected with all Striga ecotypes in the order of Kibos, Alupe, and Mbita. Similarly, the lengths of Striga seedlings attached to sorghum were ranked by virulence in the order of Kibos, Alupe, and Mbita. Six sorghum accessions (WSA-2, WSE-1, WSA-1, WSD-3, and WSD-1) showed significantly longer Striga when they were infected with Kibos seeds compared to Alupe. When the virulence of Alupe and Mbita were compared in sorghum with regard to length, all accessions had significantly longer Striga with regard to Alupe Striga.
Taken together, our results suggested that for the accessions that were screened, aethiopicum and the arundenaceum races of wild sorghum exhibited higher levels of resistance compared to the drummondii races. In particular, WSE-1, WSA-1, and WSA-2 could provide valuable resistance against S. hermonthica. Additionally, the resistance of these accessions was effective against three of the most common Striga ecotypes in Kenya, which indicated that the resistance had a broad spectrum.
Wild Sorghum Accessions Blocked Striga Penetration through Mechanical and Possible Biochemical Barriers
Striga infects its host by using a haustorium that connects the parasite with the host through xylem vessels (Dörr, 1997). To understand how wild sorghum accessions resisted Striga parasitism, we did microscopic observations of Striga haustorium at 3 and 9 DAI. At 3 DAI, Striga had successfully penetrated all accessions except for WSA-1, WSA-2, and WSE-1. In Figures 3, 4, we showed the resistance of WSA-1 and WSE-1 compared to the resistant control N13. We also showed a susceptible interaction represented by WSD-1.
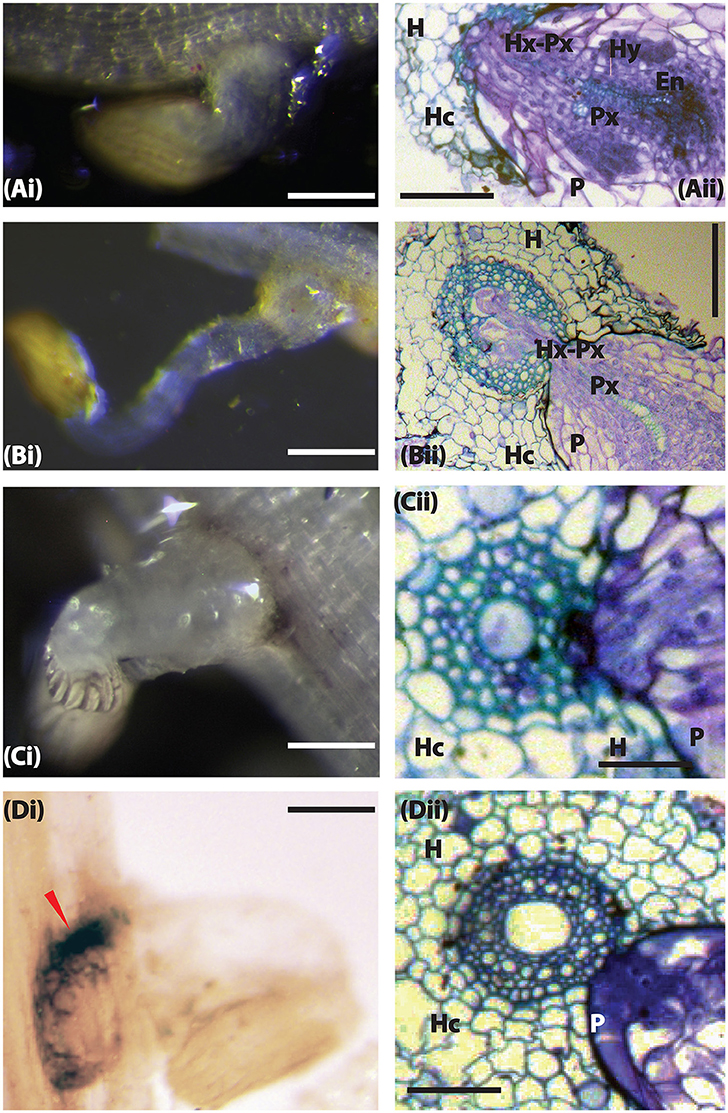
Figure 4. The host resistance mechanism of sorghum to Striga hermonthica 3 DAI. (Ai) Colonization of WSD-1 sorghum root by S. hermonthica (Kibos ecotype) showing a well-established haustorium (scale bar 1 mm). (Aii) Transverse section of an embedded root tissue of WSD-1 sorghum accession 3 days after infection with showing penetration of the host root cortex and endodermis as well as connections between the host and parasite xylem (Hx-Px). The scale bar is 0.1 mm. (Bi) Genotype N13. The haustorium is well-developed and shows swelling at the point of attachment. The scale bar is 1 mm. (Bii) Transverse section of embedded tissue from N13. By this time, the parasite had started developing vascular connections. The scale bar is 0.1 mm. (Ci) A resistant wild sorghum accession (WSA-1). The scale bar is 5 mm. (Cii) A transverse section of the resistant wild sorghum accession WSA-1. Striga was not able to penetrate the host endodermis to make vascular connections. The scale bar is 0.1 mm. (Di) Colonization of WSE-1 had more intense phenolic deposits (red arrow). The scale bar is 5 mm. (Dii) A transverse section through the haustorium of the resistant wild sorghum accession WSE-1.
In susceptible interactions, attachment, and subsequent formation of vascular connections was characterized by the swelling of the Striga radicle at the point of contact with the host roots (Figure 4Ai). Histological analysis of this section revealed a swollen haustorium with a well-differentiated Striga xylem that had already connected with the sorghum xylem (Figure 4Aii). As the infection progressed (at 9 DAI), the parasite's vegetative tissue grew vigorously, and the haustorium significantly expanded for interactions with susceptible sorghum and N13 (Figures 5Ai, Bi). A transverse section through a haustorium at 9 DAI showed that it was well-developed with a hyaline body (Hy), a vascular core (Vc) consisting of the xylem vessels, and an endophyte (En) that entered the host root cortex and endodermis (Figures 5Aii, Bii). This progression of infection and haustorium development was typical of most accessions. For the resistant control N13, we observed extra thickening on the pericycle (Figures 4Bii, 5Bii).
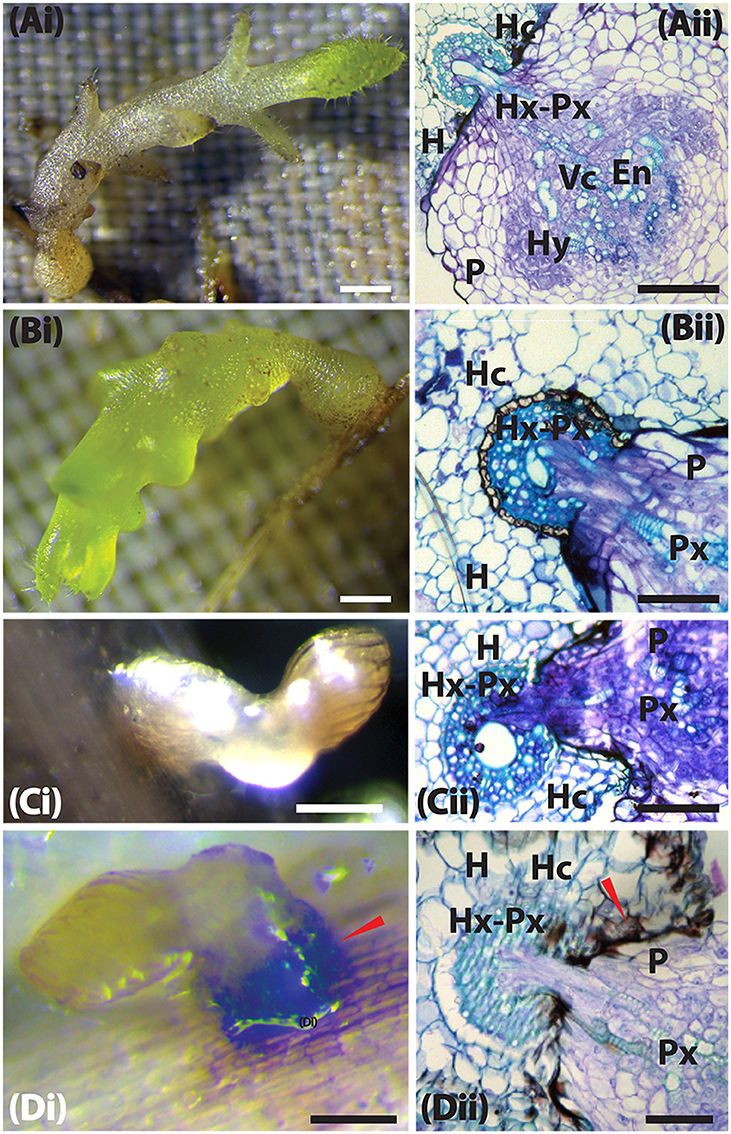
Figure 5. Host resistance mechanism of sorghum to Striga hermonthica 9 DAI. (Ai) Colonization of WSD-1 sorghum root by S. hermonthica (Kibos ecotype) 3 days after infection showing a well-established haustorium (the scale bar is 1 mm). (Aii) Transverse section of an embedded root tissue of WSD-1 sorghum accession showing penetration of the host root cortex and endodermis and vascular connections between the host and parasite xylem (Hx-Px). Scale bar is 0.1 mm. (Bi) Genotype N13 9 days after infection. The haustorium is well-developed and shows swelling at the point of infection. The scale bar is 1 mm. (Bii) Transverse section of embedded tissue of N13 9 DAI. By this time, the parasite had a well-developed endophyte (En) and hyaline body (Hy). The dark areas around the pericycle may indicate a resistance mechanism. The scale bar is 0.1 mm. (Ci) A resistant wild sorghum accession (WSA-1) infected with S. hermonthica. The parasite was weak and did not form much vegetative tissue. The scale bar is 5 mm. (Cii) A transverse section of a resistant wild sorghum accession (WSA-1). Only a small part of the parasite was able to penetrate the host vascular system. The scale bar is 0.1 mm. (Di) Colonization of WSE-1 9 DAI showing more intensive secondary metabolite deposits (red arrow). In most cases, the parasite died and did not persist past 14 DAI. The scale bar is 5 mm. (Dii) A transverse section through the haustorium of the resistant wild sorghum accession WSE-1. Only a small part of the parasite xylem was able to make vascular connections with the host. In addition, the hyaline body (Hy) as well as the endophyte (En) were much smaller compared to N13. The scale bar is 0.1 mm.
Among the three resistant wild sorghum accessions (WSE-1, WSA-1, and WSA-2), the haustorium did not make vascular connections by 3 DAI. These connections only occurred for a few attachments. The haustorium of the parasite invading a resistant accession did not display any swelling, and as histological analyses revealed, the haustorium did not penetrate the host endodermis to make vascular connections with the host (Figures 4Cii, Dii, 5Cii, Dii). This lack of connections was the case for WSA-1, WSA-2, and WSE-1. In WSA-1 and WSA-2, only a small section of the parasite haustorium had penetrated the host endodermis and made vascular connections (Figure 5Cii). We also observed deep blue staining at the point of contact between the Striga haustorium and the host endodermis for both WSA-1 and WSA-2 (Figures 4Cii, 5Cii). Additionally, WSE-1 showed deposits of secondary metabolites that caused an intense colouration at the site of parasite attachment (Figure 4Di) as early as 3 DAI. Most Striga attached to WSE-1 died within the first few days of attachment. Those attachments that persisted at 9 DAI showed intense coloration and a poorly developed Striga plant (Figures 5Ci,Di). A transverse section through a haustorium from this time point showed deposits of secondary metabolites characterized by increased colouration and limited vascular connections (Figure 5Dii).
These results suggest that the mechanical barriers that inhibited ingression of the haustorium into host tissue may be the cause of resistance in wild sorghum. The results from this experiment also suggested that the interactions between WSE-1 and S. hermonthica could be mediated by host-parasite biochemical integrations that lead to deposits of secondary metabolites.
Resistance Response of Wild Sorghum Accessions under Natural Striga Infestation
Under natural Striga infestation, we made the following general observations: (i) The three field sites (Kumi, Bukedea, and Alupe) had significant differences in their AUSNPC, which alluded to differences in Striga virulence at these sites; (ii) WSE-1 and WSA-2 were the most resistant sorghum accessions in both Kumi and Bukedea but not in Alupe and; (iii) Ochuti and WSD-1 were the least resistant accessions for all of the field sites. These results are presented in Figure 6.
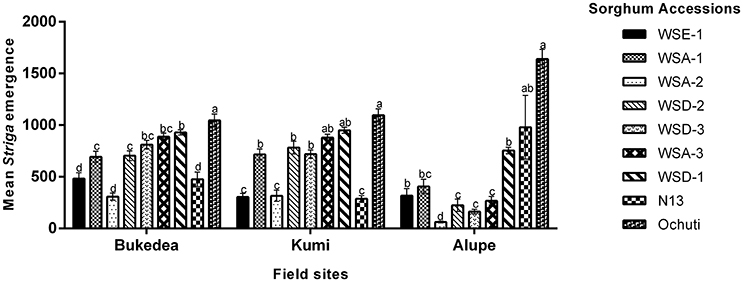
Figure 6. The resistance response of sorghum accessions to Striga hermonthica at the Kumi, Bukedea, and Alupe field sites. Mean Striga emergence was determined using AUSNPC. The vertical bars represent the mean ± SD while the letters (above each bar) indicate the mean separation and field sites, respectively, at p ≤ 0.05.
In Kumi, WSE-1 and WSA-2 had similar resistance to N13 (Figure 6). Similarly, in Bukedea, the most resistant accessions were WSA-2, WSE-1, and N13. The next four accessions: WSD-2, WSD-3, WSA-1, WSA-3, and Ochuti had similar resistance responses in Kumi. The same accessions (WSD-2. WSD-3, WSA-1, and WSA-3) had similar resistance in Bukedea. At this site, Ochuti was the most susceptible sorghum (Figure 6).
In Alupe—similar to Kumi and Bukedea—the most resistant accession was WSA-2. However, with an AUSPNC of 64.17 ± 17.83, this accession showed a significantly higher resistance compared to WSD-2 (AUSPNC, 182 ± 110.05) and WSD-3 (AUSPNC, 166.33 ± 53.79). The next cluster of accessions consisted of N13, WSE-1, WSA-1, and WSA-3 (Figure 6, Supplementary Table 1). In both Kumi and Bukedea, the least resistant accession was WSD-1 and the susceptible check Ochuti, although in Alupe N13 recorded a similar profile of emergence (Figure 6, Supplementary Table 1). Similar to Kumi and Bukedea, the most resistant sorghum accession was WSA-2. This resistance was significantly different from WSD-2, WSD-3, and WSA-3, which were the accessions ranked second, third, and fourth in terms of resistance at this field site. N13 had a significantly lower resistance than this group of accessions (Figure 6).
In summary, the field experiments confirmed the laboratory results that wild sorghum harbored resistance to Striga. In particular, aethiopicum (WSE-1) and arundinaceum (WSA-2) showed resistance comparable to the resistance of the control, N13, at all field sites except Alupe. In addition, the three field sites showed consistency in the resistance response of sorghum accessions to Striga.
Effects of Striga Infestation on Sorghum Yield
We assessed the yield performance of sorghum accessions for the reduction of yield due to Striga infestation and found the following: (i) Wild sorghum accessions yielded less compared to N13 in all field sites and (ii) Striga negatively affected the yields of all sorghum but more severely affected the susceptible accessions. These results are described in Figures 7, 8.
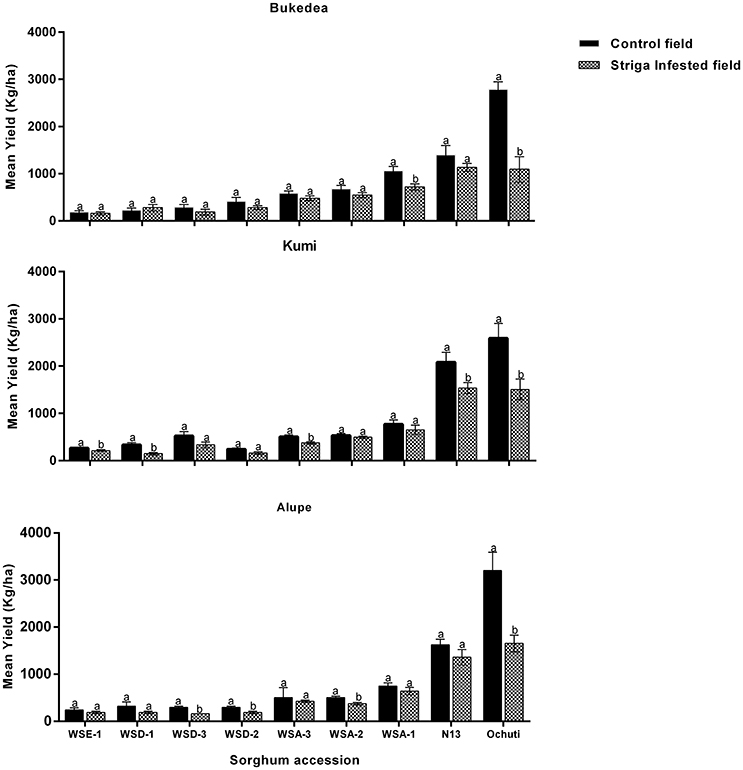
Figure 7. Yield performance of sorghum accessions at field sites under Striga and Striga-free (control) conditions. The vertical bars represent the mean ± SD (p ≤ 0.05) and different letters show presence of significant variations between the control and the Striga-infested fields.
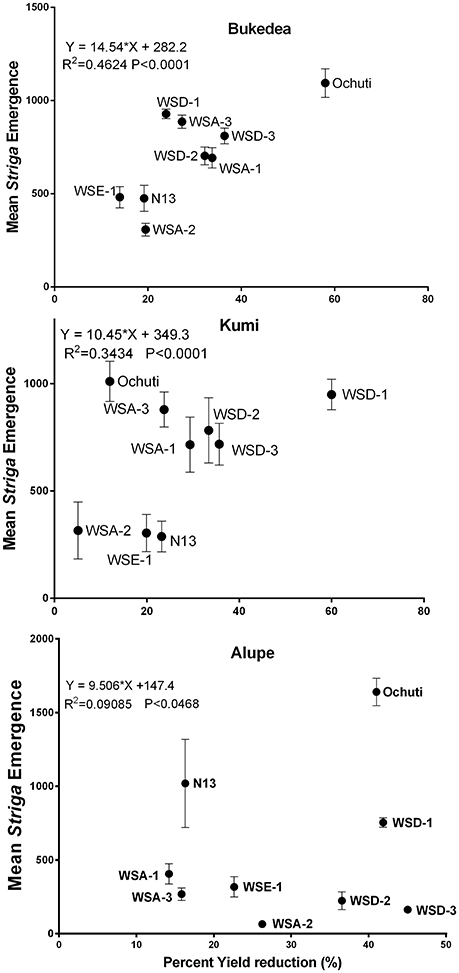
Figure 8. Correlation of Striga emergence to percentage yield loss at the three sites. The vertical axis represents mean emergence (AUSPNC) at 86 days after planting while the horizontal axis represents the percentage yield loss expressed as a yield loss between Striga-free fields and yield in Striga-infested fields as a function of yield in a Striga-free field. The data are presented as the means ± SD.
Yields of wild sorghum accessions in Striga-free plots varied significantly among one another with the average of the lowest yielding accessions being WSE-1 (230.9 kgha−1) compared to the highest Ochuti with an average yield of 2857.8 kgha−1 (Figure 7). Despite these variations, the yield of a particular accession did not vary with site. In other words, the yields of each accession were similar for all three field sites without statistical significance.
Striga had a negative effect on yield for all accessions. For some accessions, the effects of Striga infestation were more significant and depended on the field trial site. For example, Striga infestation only reduced the yields for N13 in Kumi (23.21 ± 2.00%) with statistical significance but not in Alupe or Bukedea (Figures 7, 8). For the most resistant wild accession, WSE-1, the percentage yield reduction was statistically significant only at the Kumi field site. Conversely, the yield reductions were more severe in the susceptible accessions (Figure 7). When considering the susceptible control (Ochuti), the percentage yield reduction was most dramatic in Alupe (48.33%), followed by Bukedea (41.75%) and Kumi (24.88%). These results suggest that Striga altered the yields of susceptible accessions more drastically compared to resistant accessions.
We further described the relationship between the percentage yield reduction and Striga infestation using regression analysis, and the results are shown in Figure 8. We observed a general positive relationship between Striga emergence and percentage yield reduction as well as a negative correlation between host resistance and percentage yield reduction.
The correlation between Striga emergence and yield reduction was more significant in Bukedea (R2 = 0.4624; p < 0.0001) and Kumi (R2 = 0.2136; p < 0.0001) compared to Alupe (R2 = 0.007; p = 0.047). In general, resistant accessions (WSE-1, WSA-2) had low Striga emergence and the lowest yield reduction, while for the susceptible accessions, including Ochuti, WSD-1, and WSD-3, the yield reduction became more severe with Striga emergence.
Discussion
Our goal was to determine to what extent wild sorghum could provide resistance to the parasitic weed Striga compared to the resistant cultivar N13. Successful parasitism by Striga in host roots was manifested by multiple, fast growing Striga attachments, i.e., high virulence. Accordingly, a few small Striga plants parasitizing host roots indicated a resistant host that did not support the parasite. We showed that: (i) Under laboratory conditions, wild sorghum accessions WSE-1, WSA-1, and WSA-2 have significantly higher resistance than the current Striga resistant control line—N13; (ii) Under field conditions, WSE-1 and WSA-2 have resistance comparable to that of N13 and; (iii) the resistance in wild sorghum occurred because Striga was unable to penetrate the host's endodermis and make vascular connections due to mechanical and/or biochemical barriers.
Pertaining to analyses of Striga resistance in the laboratory, we showed that wild sorghum accessions WSE-1, WSA-1, and WSA-2 were highly resistant against S. hermonthica compared to N13. The rankings for wild sorghum resistance had an overall similar pattern for all three post-attachment metrics of Striga resistance: Striga length, number and biomass. Previous studies using rhizotrons to determine Striga resistance in hosts have used these metrics with results comparable to ours (Gurney et al., 2006; Cissoko et al., 2011). For example Pescott (2013) found a mean of 75 attachments on the resistant sorghum genotype Brhan compared to our mean of 56 on N13. In rice, the most resistant rice cultivar (Nipponbare) averaged 30 attachments in a study conducted by Cissoko et al. (2011).
One result that was noteworthy was that although the mean number of attachments and mean biomass gave the same level/pattern of resistance and virulence, it was not possible to determine the resistance of sorghum using the mean length of Striga attachments. This outcome likely occurred because of the variability in Striga sizes. For example, a sorghum accession could have multiple small-sized attachments and one or two long attachments. This variation likely skewed the data and gave the impression that the accession was resistant. For this reason, mean Striga biomass seemed to be a good measure of resistance because it took both the number of attached parasites as well as their size into account. Interestingly, when we used median length to rank the resistance of sorghum accessions and presented the results in a heat map, we got the same pattern as we got for biomass. This result suggested that median length rather than mean length could be a better method to rank resistance if the Striga seedling length was highly variable.
With regard to mechanism of resistance in the wild sorghum accessions, we found that Striga had delayed penetration into the host endodermis for WSE-1, WSA-1, and WSA-2. In addition, we found that for WSE-1 Striga parasitism induced secretion of large amounts of secondary metabolites, which were probably phenolic compounds. These resistance mechanisms have important implications as platforms for further genetic improvement of cultivated sorghum for Striga resistance.
We described the resistance mechanisms of WSE-1, WSA-1, and WSA-2 as qualitative, and the mechanisms were similar to N13. Like N13, these accessions slowed the ability of Striga to penetrate host tissue and make vascular connections. For Striga, successful parasitism must involve overcoming mechanical barriers, such as the host's cell wall. This resistance—which varied from host to host—could be a manifestation of physiological and/or the biochemical incompatibility of the parasite growth on its host (Yoshida and Shirasu, 2009). The biological and genetic mechanisms underpinning this form of resistance in Striga are not completely understood but it is plausible that the effect was the result of multiple genes acting to fortify the host against invasion by the parasite. Such mechanisms include thickening of cell walls in the pericycle, lignification and silica deposition, which was suggested in Maiti et al. (1984). This form of resistance, which is exemplified by N13, has led to identification of several Quantitative Trail Loci (QTL) by mapping populations of resistant N13 and susceptible E103 (Haussmann et al., 2004). These QTL have been integrated into breeding programmes in Africa (Masiga et al., 2014; Mohamed et al., 2014; Yohannes et al., 2015). Therefore, for quantitative Striga resistance comparable to N13, WSE-1, WSA-1, and WSA-2 are good candidate accessions that could be integrated into varieties preferred by farmers. Additionally, these accessions could be used in the identification of Striga resistance loci by exploiting new and high throughput genotyping technologies, such as high-throughput sequencing. Therefore, unique markers could be identified, which would facilitate marker-assisted breeding of Striga resistance in sorghum.
Additionally, we singled out the resistance shown by WSE-1 as being mediated by biochemical reactions elicited by the parasite. Although this mechanism is not well-characterized in parasitic plants, this defense mechanism is reminiscent of studies that have shown chemical molecules could be produced as needed during pathogen attack (inducible) or starting with a small amount of preformed metabolites (Lanoue et al., 2010; Wurst et al., 2010). Some studies have further demonstrated the role of root secondary metabolites in induced plant defenses. For example, Ocimum basilicum secretes rosmarinic acid upon attack by the pathogenic fungus Pythium ultimum (Bais et al., 2002) and induction of iridoid glycosides in root exudates of Plantago lanceolata in the presence of nematodes was reported in Wurst et al. (2010). The induction of biochemical compounds—which was characterized by deep colorations at the host-parasite interphase in the resistant sorghum accession WSE-1 could be suggestive of a resistance mediated by these molecules. Further biochemical studies—such as metabolomics in a recent review (van Dam and Bouwmeester, 2016)—will be required to further identify the role of metabolites in Striga-host interactions.
Laboratory analysis provided the unique opportunity to minimize environmental variations and ensure that the resistance rankings that were observed could be confidently associated with the genotype. However, these data did not always translate to the field because other factors may influence resistance through genetic χ environment interactions, which have been observed before for Striga. These interactions were demonstrated in a study (Cissoko et al., 2011), who found an upland rice variety NERICA4 that was resistant using rhizotron assays. NERICA4 was, however, found to be susceptible under field conditions (Atera et al., 2012).
We therefore complemented our laboratory observations with field trial data. From the wide array of tools that are traditionally used to measure Striga virulence and the host resistance (Omanya et al., 2004; Rodenburg et al., 2005) at the field level, we determined AUSPNC, which was derived by integrating the curve of Striga emergence over time.
Strikingly, the resistance rankings of sorghum accessions screened in the laboratory were similar to those found with field experiments—for all field sites—with WSE-1, WSA-2 showing the most resistance. Additionally, the two least resistant accessions in all field sites were consistently WSD-1 and Ochuti. However, in Alupe N13 did not display the level of resistance observed in Kumi and Bukedea. In fact, the AUSPNC of N13 in Alupe was similar to that of Ochuti. Previous studies involving N13 in Kumi and Bukedea obtained an AUSPNC that averaged <500 (Olupot, 2011) which, is comparable to what we obtained—476 ± 70 and 288.17 ± 29.11 in Bukedea and Kumi respectively. This was lower than the AUSPNC of N13 in Alupe (978.83 ± 308.36). The variability in response to Striga infestation by site in our field experiments could be attributed to environmental effects such as different climatic conditions, differences in soil types or factors inherent to Striga characteristics such as seedbank or virulence.
With regard to the variability in Striga virulence, our rhizotron assays showed that the Striga ecotype from Alupe was significantly more virulent compared to Mbita but was less virulent than the ecotype from Kibos. This result showed there were variable differences in the virulence of Alupe seed ecotypes. In other words, even within the same Striga ecotype, variations in virulence occurred. This apparent lack of a clear association between virulence and Striga ecotypes supports the heterogeneity of S. hermonthica observed by Pescott (2013). Unlike S. asiatica, which has a highly inbreeding mating system (Gethi et al., 2005), and S. gesneroides, which has distinct race structures (Li et al., 2009), S. hermonthica has high outcrossing (Gethi et al., 2005), and that makes virulence grouping among ecotypes very nebulous. Therefore, it has been difficult to connect virulence to a specific eco-geographic region.
Another important consideration when comparing laboratory and field experiments for Striga resistance is the production of the germination stimulant strigolactone. Laboratory screens use artificially pre-germinated Striga seedlings and these screens forfeit the opportunity to test for pre-Striga germination resistance as well as resistance during other belowground stages (e.g., germination, attachment, below-ground development). In contrast, in field infestations, the host must produce enough strigolactone to allow Striga to germinate. Therefore, field screenings test for both pre- and post-Striga germination resistance. As such, it is important to establish the strigolactone profile of wild sorghum accessions to determine if they also exhibit low germination stimulant production as a resistance mechanism.
There is another important aspect of the Striga-host relationship, which is tolerance, i.e., the ability of a host to sustain a certain yield and experience less damage under Striga infestation (Parker and Riches, 1993; Rodenburg et al., 2006). Whereas, a resistant host “fights” the pathogen, a tolerant host “learns to live” with the pathogen thereby ameliorating the damage inflicted by the pathogen. As expected, Striga reduced the yields of all sorghum accessions, albeit the reductions in some accessions were not significant. N13, WSE-1, and WSA-1 tolerated Striga to the greatest extent in some field sites that did not have any significant yield reductions. In this study, the resistant accessions were also tolerant, which made it difficult to dissociate resistance and tolerance. Rodenburg et al. (2006) observed that tolerance was a complex trait that needs to take into account the resistance of the host as well as the biomass of the infecting Striga plants. The authors recommended that tolerance in resistant genotypes be quantified as a reduced yield loss per aboveground Striga plant and that the maximum relative yield loss could be used for susceptible genotypes. Further studies will be required to determine the correlation between resistance and tolerance in WSA-1 and WSE-1.
To summarize, our work revealed high Striga resistance in wild sorghum accessions. These accessions also had at least two Striga resistance mechanisms and therefore underscored the importance of wild sorghum as sources of resistance to Striga. Candidate wild sorghum accessions for Striga resistance have been identified and are available for multiplication and subsequent improvement. Our work thus sets a technology platform for future genetic improvement of cultivated sorghum. With modern techniques of genetics and genomics, it will soon be possible to “pinpoint” the genetic components that are responsible for resistance in wild sorghum. These insights will facilitate the stacking of appropriate resistance genes/loci in varieties preferred by farmers and Striga tolerant cultivars to enhance the durability and stability of defenses over the long-term.
Author Contributions
SR and MT conceived and designed the laboratory experiments. DM, EK, and JM performed laboratory experiments guided by SR and MW. Field experiments were designed and conceived by DO, AM, and NH, the performed by CM. SR, DM, CM, EK, and JM wrote the manuscript. All authors read and approved the final manuscript.
Funding
The National Academies of Science (NAS) supported this research project under the Partnerships Enhanced Engagement in Research (PEER) program (contract number NAS Sub-Grant Award Letter Agreement Number PGA-2000003439). The Sub-Grant Agreement was funded under Prime Agreement Number AID-OAA-A-11-00012, which was entered into by and between the NAS and the United States Agency for International Development (USAID). MPT was supported by grants from the National Science Foundation (DBI-0701748 and IBN-0322420). We further acknowledge the fieldwork support we received from the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00116/full#supplementary-material
Supplementary Figure 1. Experimental layout of the field sites. Each plot was 2.5 m by 3.2 m. Each plot was separated by a 1 m path and the spacing was 80 cm between rows and 30 cm between hills. The whole sorghum field was surrounded by susceptible sorghum (KARI Mtwapa).
Supplementary Figure 2. A heat map drawn using the Striga mean length showing levels of resistance of sorghum accessions infected with Striga seedlings. The yellow bar labeled “S” (Ochuti and WSD-1) represents the most susceptible accessions, the orange bar labeled “IR” is a group consisting of WSA-3, WSD-2 and WSD-3 that had intermediate resistance, while the red bar labeled “R” represents the most resistant group, which consists of WSA-1, WSA-2, WSE-1, and N13. The heat maps also provide an indication of the virulence of Striga ecotypes. Longer attachments formed from Kibos and Alupe ecotypes compared to Mbita ecotypes. The letters indicate significances differences between ecotypes for the same accession at p < 0.05.
Supplementary Table 1. Striga emergence values (Area under Striga Number Progressive Curve) of wild and cultivated sorghum accessions across three field sites.
References
Atera, E. A., Itoh, K., and Ishii, T. (2012). Response of NERICA Rice to Striga hermonthica Infections in Western Kenya. Int. J. Agric. 2, 271–275.
Bais, H. P., Walker, T. S., Schweizer, H. P., and Vivanco, J. M. (2002). Root specific elicitation and antimicrobial activity of rosmarinic acid in hairy root cultures of Ocimum basilicum. Plant Physiol. Biochem. 40, 983–995. doi: 10.1016/S0981-9428(02)01460–2
Bouwmeester, H. J., Roux, C., Lopez-Raez, J. A., and Bécard, G. (2007). Rhizosphere communication of plants, parasitic plants and AM fungi. Trends Plant Sci. 12, 224–230. doi: 10.1016/j.tplants.2007.03.009
Brozynska, M., Furtado, A., and Henry, R. J. (2016). Genomics of crop wild relatives: expanding the gene pool for crop improvement. Plant Biotech. J. 14, 1070–1085. doi: 10.1111/pbi.12454
Cissoko, M., Boisnard, A., Rodenburg, J., Press, M. C., and Scholes, J. D. (2011). New Rice for Africa (NERICA) cultivars exhibit different levels of post-attachment resistance against the parasitic weeds Striga hermonthica and Striga asiatica. New Phytol. 192, 952–963. doi: 10.1111/j.1469-8137.2011.03846.x
Cotter, M., de la Pena-Lavander, R., and Sauerborn, J. (2012). Understanding the present distribution of the parasitic weed Striga hermonthica and predicting its potential future geographic distribution in the light of climate change. Julius Kühn Arch. 0, 630. doi: 10.5073/jka.2012.434.082
Doebley, J. F., Gaut, B. S., and Smith, B. D. (2006). The molecular genetics of crop domestication. Cell 127, 1309–1321. doi: 10.1016/j.cell.2006.12.006
Dörr, I. (1997). How Striga parasitizes its host: a TEM and SEM Study. Ann. Bot. 79, 463–472. doi: 10.1006/anbo.1996.0385
Ejeta, G. (2007). “The Striga scourge in Africa: a growing pandemic,” in Integrating New Technologies for Striga Control, eds G. Ejeta and J. Gressel (Singapore: World Scientific Publishing Company), 3–16.
Gethi, J. G., Smith, M. E., Mitchell, S. E., and Kresovich, S. (2005). Genetic diversity of Striga hermonthica and Striga asiatica populations in Kenya. Weed Res. 45, 64–73. doi: 10.1111/j.1365-3180.2004.00432.x
Gurney, A. L., Grimanelli, D., Kanampiu, F., Hoisington, D., Scholes, J. D., and Press, M. C. (2003). Novel sources of resistance to Striga hermonthica in Tripsacum dactyloides, a wild relative of maize. New Phytol. 160, 557–568. doi: 10.1046/j.1469–8137.2003.00904.x
Gurney, A. L., Slate, J., Press, M. C., and Scholes, J. D. (2006). A novel form of resistance in rice to the angiosperm parasite Striga hermonthica. New Phytol. 169, 199–208. doi: 10.1111/j.1469-8137.2005.01560.x
Harlan, J. R., and de Wet, J. M. J. (1972). A simplified classification of cultivated sorghum. Crop Sci. 12, 172–176. doi: 10.2135/cropsci1972.0011183X001200020005x
Haussmann, B. I., Hess, D. E., Omanya, G. O., Folkertsma, R. T., Reddy, B. V., Kayentao, M., et al. (2004). Genomic regions influencing resistance to the parasitic weed Striga hermonthica in two recombinant inbred populations of sorghum. Theor. Appl. Genet. 109, 1005–1016. doi: 10.1007/s00122-004–1706-9
Hudson, J. P. (1967). Review of E. J. Hewitt 'sand and Water Culture Methods used in the Study of plant Nutrition. Kent, WA: Cambridge University Press
Jamil, M., Van Mourik, T. A., Charnikhova, T., and Bouwmeester, H. J. (2013). Effect of diammonium phosphate application on strigolactone production and Striga hermonthica infection in three sorghum cultivars. Weed Res. 53, 121–130. doi: 10.1111/wre.12003
Jones, J. D., Witek, K., Verweij, W., Jupe, F., Cooke, D., Dorling, S., et al. (2014). Elevating crop disease resistance with cloned genes. Philos. Trans. R. Soc. Lond. B 369, 20130087–20130087. doi: 10.1098/rstb.2013.0087
Lanoue, A., Burlat, V., Henkes, G. J., Koch, I., Schurr, U., and Röse, U. S. R. (2010). De novo biosynthesis of defense root exudates in response to Fusarium attack in barley. New Phytol. 185, 577–588. doi: 10.1111/j.1469-8137.2009.03066.x
Li, J., Lis, K. E., and Timko, M. P. (2009). Molecular genetics of race-specific resistance of cowpea to Striga gesnerioides (Willd.). Pest Manag. Sci. 65, 520–527. doi: 10.1002/ps.1722
Li, J., and Timko, M. P. (2009). Gene-for-gene resistance in Striga-cowpea associations. Science 325, 1094–1094. doi: 10.1126/science.1174754
Maiti, R. K., Ramaiah, K. V., Bisen, S. S., and Chidley, V. L. (1984). A comparative study of the haustorial development of Striga asiatica (L.) kuntze on sorghum cultivars. Ann. Bot. 54, 447–457.
Masiga, C. W., Mugoya, C., Ali, R., Mohamed, A., Osama, S., Ngugi, A., et al. (2014). “Enhanced utilization of biotechnology research and development innovations in eastern and central Africa for agro-ecological intensification,” in Challenges and Opportunities for Agricultural Intensification of the Humid Highland Systems of Sub-Saharan Africa (Cham: Springer International Publishing), 97–104.
Mohamed, A., Ali, R., Elhassan, O., Suliman, E., Mugoya, C., Masiga, C. W., et al. (2014). First products of DNA marker-assisted selection in sorghum released for cultivation by farmers in sub-saharan Africa. J. Plant Sci. Mol. Breed. 3:3. doi: 10.7243/2050-2389-3-3
Mohamed, A., Ellicott, A., Housley, T. L., and Ejeta, G. (2003). Hypersensitive Response to Infection in. Crop Sci. 43, 1320–1324. doi: 10.2135/cropsci2003.1320
Musselman, L. J., and Hepper, F. N. (1986). The witchweeds (Striga, Scrophulariaceae) of the Sudan Republic. Kew Bull. 41, 205. doi: 10.2307/4103043
Ngugi, K., Ngugi, A. J., Osama, S., and Mugoya, C. (2016). Combating Striga weed in sorghum by transferring resistance quantitative trait loci through molecular marker assisted introgression. J. Plant Breeding Crop Sci. 3, 67–76.
Okonkwo, S. N. C., and Nwoke, F. I. O. (1978). Initiation, development and structure of the primary haustorim in Striga gesnerioides (Scrophulariaceae). Ann. Bot. 42, 455–463.
Olupot, J., R. (2011). Genetic Analysis of Striga hermonthica in Sorghum (Sorghum Bicolor) Genotypes in Eastern Uganda. PhD thesis, Pietermaritzburg South Africa: University of KwaZulu-Natal.
Omanya, G. O., Haussmann, B. I. G., Hess, D. E., Reddy, B. V. S., Kayentao, M., Welz, H. G., et al. (2004). Utility of indirect and direct selection traits for improving Striga resistance in two sorghum recombinant inbred populations. Field Crop Res. 89, 237–252. doi: 10.1016/j.fcr.2004.02.003
Parker, C., and Riches, C. R. (1993). Parasitic Weeds of the World: Biology and Control. Oxfordshire: CAB International.
Paterson, A. H., Moore, P. H., and Tew, T. L. (2013). “The gene pool of Saccharum species and their improvement,” in Genomics of the Saccharinae, ed A. H. Paterson (New York, NY: Springer), 43–71. doi: 10.1007/978-1-4419-5947-8_3
Pescott, O. (2013). The Genetics of Host Adaptation in the Parasitic Plant Striga Hermonthica. PhD thesis, Sheffield: The University of Sheffield.
Rich, P. J., Grenier, C., and Ejeta, G. (2004). Striga resistance in the wild relatives of sorghum. Crop Sci. 44, 2221–2229. doi: 10.2135/cropsci2004.2221
Rodenburg, J., Bastiaans, L., and Kropff, M. J. (2006). Characterization of host tolerance to Striga hermonthica. Euphytica 147, 353–365. doi: 10.1007/s10681-005-9030-2
Rodenburg, J., Bastiaans, L., Weltzien, E., and Hess, D. E. (2005). How can field selection for Striga resistance and tolerance in sorghum be improved? Field Crop Res. 93, 34–50. doi: 10.1016/j.fcr.2004.09.004
Rogers, W. E., and Nelson, R. R. (1962). Penetration and nutrition of Striga asiatica. Phytopathology 52, 1064–1070
Sakai, H., and Itoh, T. (2010). Massive gene losses in Asian cultivated rice unveiled by comparative genome analysis. BMC Genomics 11:121. doi: 10.1186/1471-2164-11-121
van Dam, N. M., and Bouwmeester, H. J. (2016). Metabolomics in the rhizosphere: tapping into belowground chemical communication. Trends Plant Sci. 21, 256–265. doi: 10.1016/j.tplants.2016.01.008
Wurst, S., Wagenaar, R., Biere, A., and van der Putten, W. H. (2010). Microorganisms and nematodes increase levels of secondary metabolites in roots and root exudates of Plantago lanceolata. Plant Soil 329, 117–126. doi: 10.1007/s11104-009-0139-2
Yoder, J. I., and Scholes, J. D. (2010). Host plant resistance to parasitic weeds; recent progress and bottlenecks. Curr. Opi. Plant Sci. 13, 478–484. doi: 10.1016/j.pbi.2010.04.011
Yohannes, T., Abraha, T., Kiambi, D., Folkertsma, R., Tom Hash, C., Ngugi, K., et al. (2015). Marker-assisted introgression improves Striga resistance in an eritrean farmer-preferred sorghum variety. Field Crops Res. 173, 22–29. doi: 10.1016/j.fcr.2014.12.008
Yoshida, S., and Shirasu, K. (2009). Multiple layers of incompatibility to the parasitic witchweed, Striga hermonthica. New Phytol. 183, 180–189. doi: 10.1111/j.1469-8137.2009.02840.x
Keywords: Striga resistance, witchweed, sorghum, wild sorghum relatives, Sub-Saharan Africa
Nomenclature: Striga hermonthica benth., witchweed, sorghum bicolor
Citation: Mbuvi DA, Masiga CW, Kuria E, Masanga J, Wamalwa M, Mohamed A, Odeny DA, Hamza N, Timko MP and Runo S (2017) Novel Sources of Witchweed (Striga) Resistance from Wild Sorghum Accessions. Front. Plant Sci. 8:116. doi: 10.3389/fpls.2017.00116
Received: 15 August 2016; Accepted: 19 January 2017;
Published: 06 February 2017.
Edited by:
Hanan Eizenberg, Agricultural Research Organization, IsraelReviewed by:
Satoko Yoshida, Nara Institute of Science and Technology, JapanAlejandro Pérez-de-Luque, Andalusian Institute of Agricultural and Fisheries Research and Training, Spain
Copyright © 2017 Mbuvi, Masiga, Kuria, Masanga, Wamalwa, Mohamed, Odeny, Hamza, Timko and Runo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael P. Timko, mpt9g@virginia.edu
Steven Runo, runo.steve@ku.ac.ke
 Dorothy A. Mbuvi
Dorothy A. Mbuvi Clet W. Masiga
Clet W. Masiga Eric Kuria
Eric Kuria Joel Masanga
Joel Masanga Mark Wamalwa1
Mark Wamalwa1 Steven Runo
Steven Runo