- 1State Key Laboratory of Crop Stress Biology for Arid Areas, College of Life Sciences, Northwest A&F University, Yangling, China
- 2State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, China
- 3Department of Plant Sciences and Plant Pathology, Montana State University, Bozeman, MT, USA
- 4China–Australia Joint Research Centre for Abiotic and Biotic Stress Management, Northwest A&F University, Yangling, China
RAR1 is a eukaryotic zinc-binding protein first identified as required for race-specific resistance to powdery mildew in barley. To study the function of TaRAR1 involvement in wheat (Triticum aestivum L.) defense against the infection of stripe rust pathogen Puccinia striiformis f. sp. tritici (Pst), we identified and cloned three wheat homeologous genes highly similar to the barley HvRar1, designated as TaRar1-2A, TaRar1-2B, and TaRar1-2D. The three TaRAR1 proteins all contain two conserved cysteine-and histidine-rich domains (CHORD-I and -II) shared by known RAR1-like proteins. Characterization of TaRar1 expression revealed that the expression was tissue-specific and up-regulated in wheat during stripe rust infection. Moreover, the transcription of TaRar1 was induced by methyl jasmonate, ethylene, and abscisic acid hormones. The same results were observed with drought and wound treatments. After TaRar1 was silenced in wheat cultivar Suwon11 containing the stripe rust resistance gene YrSu, the endogenous salicylic acid (SA) level, the hydrogen peroxide (H2O2) accumulation and the degree of hypersensitive response (HR) were significantly decreased, and the resistance to the avirulent pathotype of stripe rust was compromised. Meanwhile, the expression of catalase, an enzyme required for H2O2-scavenging, was up-regulated. Taken together, we concluded that TaRar1 is involved in wheat defense against stripe rust mediated by YrSu, and the defense was through SA to influence reactive oxygen species accumulation and HR.
Introduction
Plants have evolved sophisticated and effective mechanisms against most potential pathogens, such as non-host resistance, race specific and race non-specific resistance (Nürnberger and Lipka, 2005; Jones and Dangl, 2006). Non-host resistance is also called species-level resistance; it means that all genotypes of a given pathogen species can’t infect all genotypes of a plant species. Race-specific resistance is also known as gene-for-gene resistance (Flor, 1971), in which plant R genes can recognize cognate avirulence (Avr) genes from the pathogens to trigger defense responses. In most cases, R-gene-mediated resistance triggers a complex of signal transduction cascade leading to a local programmed cell death (PCD) namely the hypersensitive response (HR), and a systemic acquired resistance (SAR) (Heath, 2000; Durrant and Dong, 2004). HR occurs at the infection sites and immediate surrounding areas to restrict the pathogen growth. SAR is often induced along with the elevated pathogenesis-related (PR) gene expression through salicylic acid (SA) or jasmonic acid (JA) mediated signaling pathway to generate a global and a broad-spectrum resistance in plants (Durrant and Dong, 2004).
To date, the largest class of resistance genes cloned encodes proteins containing a nucleotide binding (NB) site and leucine-rich repeat (LRR) domains. In addition to R genes, many genetic components are also required in the regulation of R-gene-mediated defense signaling (Century et al., 1995; Parker et al., 1996; Shirasu et al., 1999; Austin et al., 2002). Among them, the gene Required for Mla12 Resistance1 in barley (HvRar1) was first identified as necessary for the function of multiple powdery mildew R genes (Jørgensen, 1994; Shirasu et al., 1999). Rar1 was also well documented to be required in R-gene-specific resistance in Arabidopsis and Nicotiana benthamiana (Liu et al., 2002; Tornero et al., 2002). In wheat, Rar1 was first reported to be involved in the Lr21-mediated resistance against leaf rust (Scofield et al., 2005), but was not required by the Sr33-mediated signaling pathway (Periyannan et al., 2013), suggesting Rar1 is not required by every R gene.
Barley Rar1 encodes a protein with two zinc-binding domains named as CHORD-I and -II (cysteine-and histidine-rich domain) (Azevedo et al., 2002). During initial yeast two-hybrid screening, an ubiquitin ligase protein containing a Skp1-cullin-F box, named SGT1, was identified as a RAR1-interacting partner (Shang et al., 2006; Tai, 2008; Cantu et al., 2013). Rar1 and Sgt1 are required by many but not all NB-LRR R genes to mediate resistances against viral, bacterial, oomycete, or fungal pathogens (Kitagawa et al., 1999; Austin et al., 2002; Liu et al., 2002; Tornero et al., 2002; Scofield et al., 2005; Periyannan et al., 2013). The molecular chaperone, RAR1 associated with SGT1 and HSP90, has been shown to regulate the correctly folded of R protein complexes and active the downstream signaling pathways (Shirasu and Schulze-Lefert, 2003).
Stripe rust disease is caused by Puccinia striiformis f. sp. tritici (Pst), which is one of the most common and destructive diseases of wheat (Triticum aestivum L.) in the world (Chen, 2005). To determine molecular mechanisms involved in wheat–Pst interaction, we isolated a gene highly upregulated from wheat cultivar Suwon11 infected with stripe rust fungus. The gene shares a high similarity with the barley HvRar1 gene and the wheat TaRAR1-1 gene, designated as TaRar1. The transcript abundance of TaRar1 was studied in Suwon11 seedlings inoculated with two different Pst pathotypes. Additionally, the expression patterns of TaRar1 were studied under different stresses and hormone treatments. Furthermore, the involvement of TaRar1 in defense against Pst was investigated using barley stripe mosaic virus induced gene silencing. The relationships between the TaRar1 silencing and the levels of SA accumulation and reactive oxygen species (ROS) accumulation were assayed; HR and pathogen growth were also studied. Our studies suggested that TaRar1 plays an important role in wheat defense against Pst pathogen through SA to modulate ROS accumulation and HR.
Results
Sequence Analyses of TaRar1 cDNA and Protein
A 675-bp cDNA fragment was isolated due to its high expression level from the transcriptomes of wheat cultivar Suwon11 infected by avirulent Pst CYR23 at 1 day post inoculation (dpi). The cDNA shares a high homology with the Rar1 gene of barley HvRar1 (AF192261_1) and the wheat TaRAR1-1 or wRar1 amplified from WGRC7 (EF202841.1) (Tai, 2008; Cantu et al., 2013). The cDNA sequence was deposited in NCBI GenBank as TaRar1 (KX852426). A longer 696-bp cDNA sequence was amplified from Suwon11 mRNA using primers TaRar1-F/R (Table 1) designed based on TaRAR1-1. The deduced protein has 231 amino acids, with a predicted molecular weight of 25.29 kD and an isoelectric point (pI value) of 8.11. The protein contains two conserved regions highly similar to the CHORD domains of different RAR1 proteins identified (Figure 1). Search of the wheat genomic DNA sequence database at the International Wheat Genomic Sequence Consortium (IWGSC) for homologs of TaRAR1, found five wheat contigs each contains a homolog of TaRar1, contig64299699 (7,236 bp) and contig4730984 (4,311 bp) from 2AL, contig7991159 (6,558 bp) from 2BL, and contig9718756 (1,215 bp) and contig9716294 (1,350 bp) from 2DL. The contigs from 2A and 2D chromosomes contain only partial sequence of the homolog. The full-length sequence was obtained through the joint of the overlapped sequence from the two contigs. In total, three homeologs of TaRar1 have been identified in the wheat genomes, located on the long arms of chromosomes 2A, 2B, and 2D, thereafter referred to as TaRar1-2A. TaRar1-2B, and TaRar1-2D. The copy number and locations of the TaRar1 homologs were further confirmed by a Southern hybridization using Chinese Spring nulli-tetrasomic lines (Supplementary Figure S1). The three encoded proteins in Chinese Spring share about 95–99% similarity with each other (Supplementary Figure S2). The sequence of Suwon11 TaRar1 is the most similar to the nucleotides sequence of the Chinese Spring TaRar1-2B.
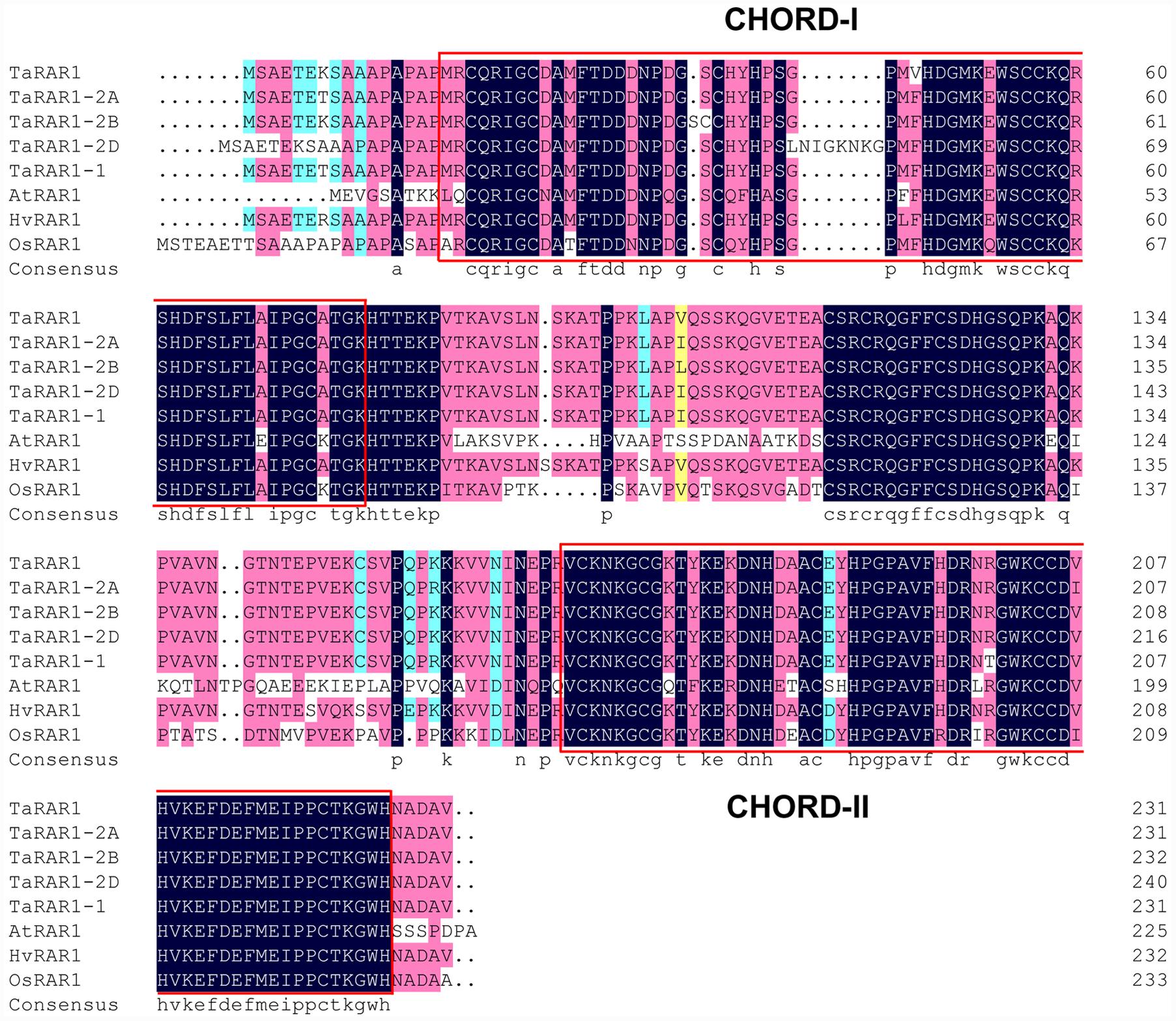
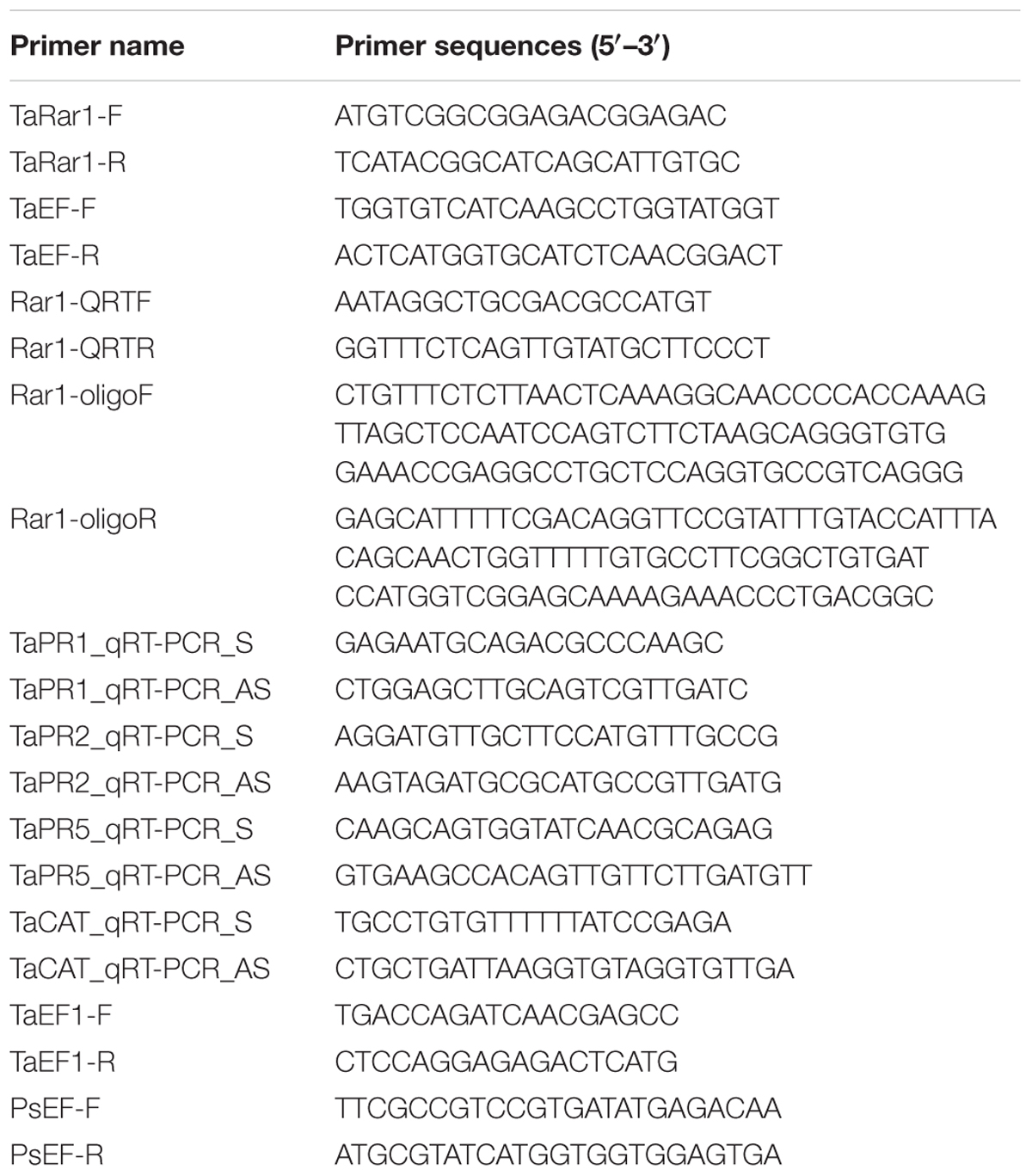
FIGURE 1. Alignment of RAR1 proteins from different species. Comparison of TaRAR1(KX852426), wheat alleles TaRAR1-2A, TaRAR1-2B, TaRAR1-2D, and the RAR1 protein(EF202841.1) from wheat cultivar WGRC7 with RAR1 proteins from rice OsRAR1 (XP_015623436.1), barley HvRAR1(AF192261_1) and Arabidopsis AtRAR1(BAB11239.1) revealed highly conserved sequences in the two conserved CHORD-I and -II domains indicated in two red boxes.
Alignments of the TaRAR1 with available RAR1 sequences of barley HvRAR1 (AF192261_1), rice OsRAR1 (XP_015623436.1), Arabidopsis AtRAR1 (BAB11239.1), and all the wheat alleles revealed highly conserved amino acid sequences at the two functional CHORD domains (Figure 1), implying RAR1 proteins from different plant species might have a similar function.
TaRar1 Transcriptional and SA Level Response to Pst
To study the expression profiles of the three TaRar1 homeologs during wheat–Pst interactions, we challenged Suwon11 with two Pst pathotypes, CYR23 and CYR31. Suwon11 is resistant to CYR23, forming an incompatible interaction with the pathogen; and susceptible to CYR31, forming a compatible interaction with the pathogen. The transcript abundances of the three homeologs can’t be assayed separately due to the high similarity at the RNA level. Therefore, the expression of all three alleles were measured together (Referred to as a group as TaRar1) using conserved primers via quantitative real-time PCR (qRT-PCR). Expression was measured in samples generated from leaf tissues collected at seven time points post inoculation (Figure 2A). In both interactions, the total of three TaRar1 homeologs expression was up-regulated starting from 6 h post inoculation (hpi), reached the highest level between 24 and 48 hpi and returned to the basal level (level of 0 hpi) at 120 hpi. The striking differences in the TaRar1 expression between the two interactions were the timing and gratitude; the highest TaRar1 level in the incompatible interaction was 24 h earlier and 13-fold stronger than that in the compatible interaction (Figure 2A). These results suggested that TaRar1 might play a role in defense response against Pst.
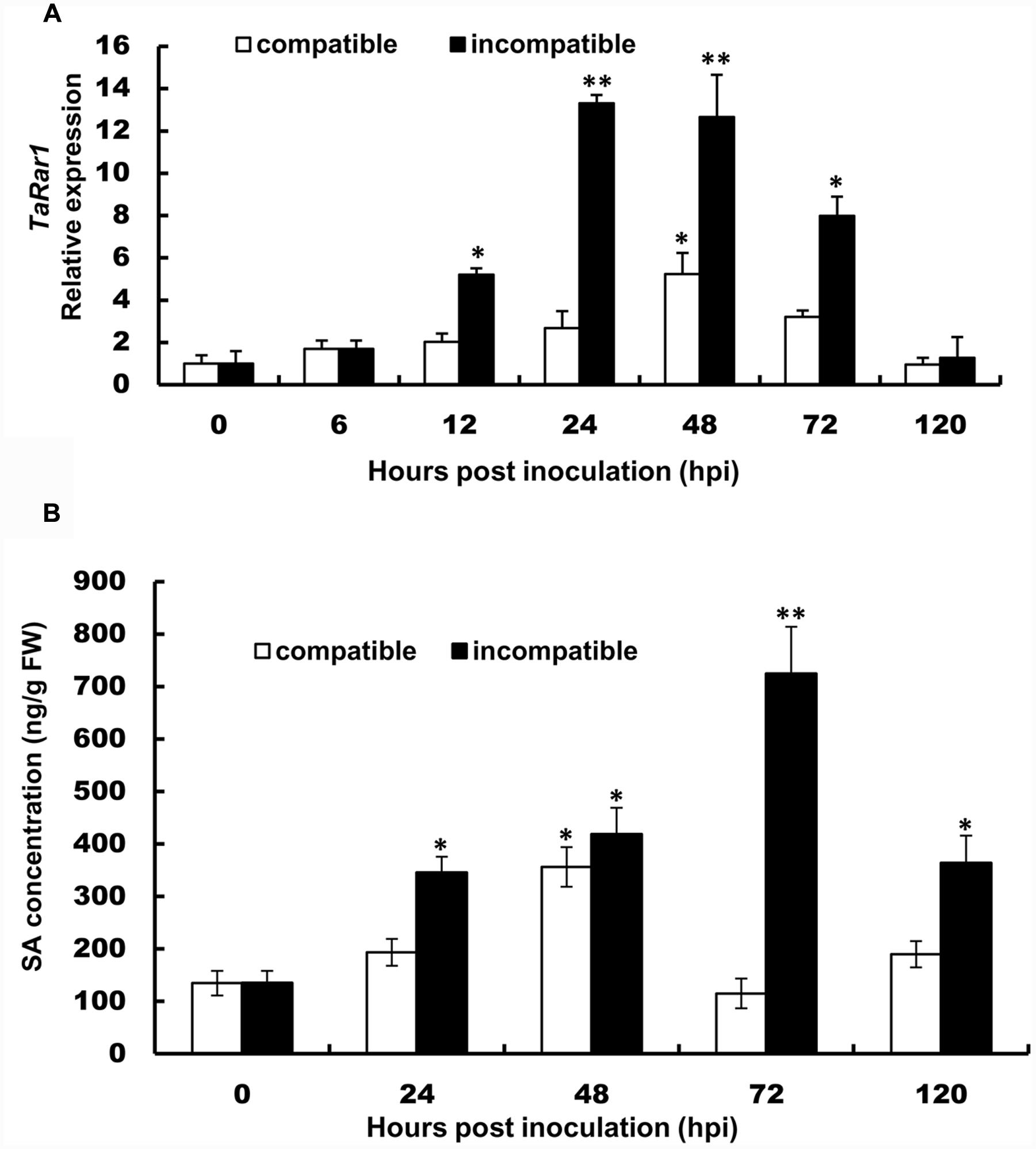
FIGURE 2. Transcript levels of TaRar1 (A) and salicylic acid (SA) concentration (B) in Suwon11 leaves after inoculation with CYR23 (incompatible interaction) and CYR31 (compatible interaction). Leaf tissues were sampled at 0, 6, 12, 24, 48, 72, and 120 hours post inoculation (hpi). Three independent biological replications were performed. Relative gene expression was calculated by the comparative ΔΔCt method and was relative to the mock at each corresponding time point using gene-specific oligonucleotide primers (Table 1). Transcript abundance was normalized to the reference gene TaEF-1a (wheat elongation factor) (GenBank accession Q03033). The mean expression values were calculated from three replications. Error bars represent standard deviation. (∗) and (∗∗) indicate a significant difference between a particular hpi and 0 hpi with a p-value < 0.05 and 0.01, respectively. Differences were assessed using Student’s t-tests.
The SA level was then measured in the two interactions at five time points (Figure 2B). SA level was significantly increased in both interactions during the course of pathogenesis. In the incompatible interaction, the earliest high SA level was detected at 24 hpi, peaked at 72 hpi and decreased at 120 hpi. The SA level was also increased significantly in the compatible interaction at 48 hpi, but was 12 h delayed than that in the incompatible interaction, similar to that observation in the TaRar1 expressions. In addition, the high SA level in the compatible interaction only lasted for a short time, by 72 hpi, the SA level has declined to the basal level at 0 hpi (Figure 2B).
TaRar1 Transcript Level in Different Organs and Developmental Stages
TaRar1 expression was also analyzed in six different tissues (roots, culms, seedling leaves, flag leaves, floret, and spikelet) collected from plants grown under normal condition. The gene expression pattern appeared to be tissue-specific. If normalizing the transcript abundance in the seedling leaves as a control, roots, culms and spikelet had about 2.8-fold, 3.8-fold, and 3.5-fold higher expression than the control (Figure 3), respectively. Floret and flag leaves had about sixfold higher than the control (Figure 3), suggesting higher TaRar1 expression at the adult plant stage.
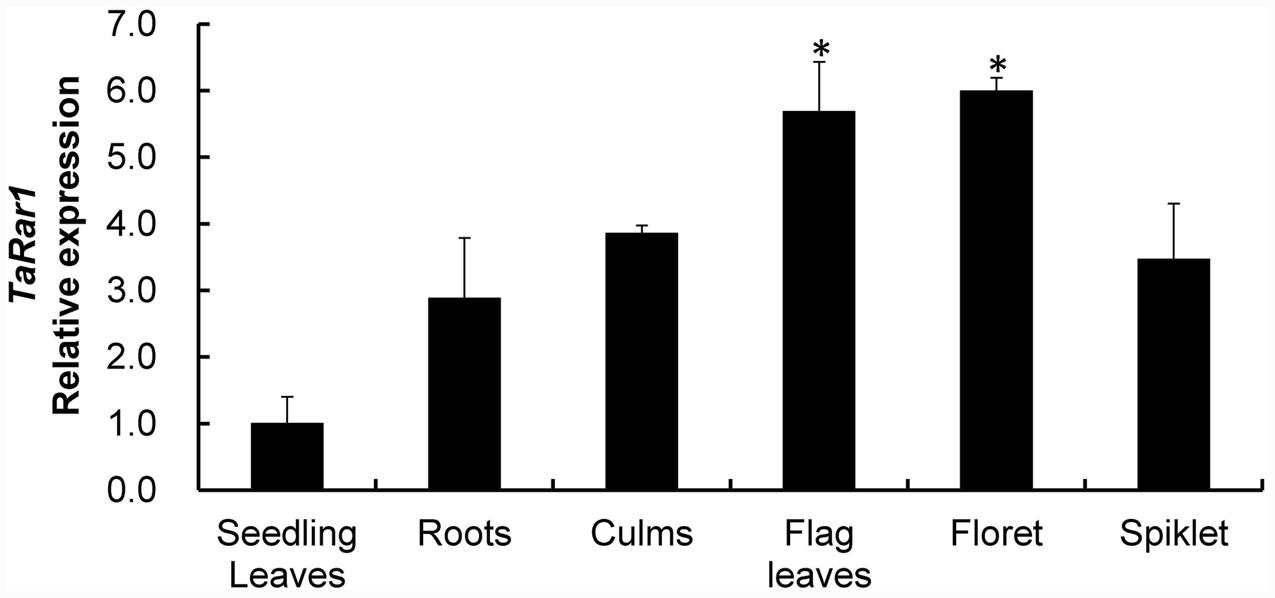
FIGURE 3. Relative transcript levels of TaRar1 in different wheat tissues. Samples were taken from seedling leaves, roots, culms, flag leaves, floret, and spikelet. Three independent biological replications were performed. Relative gene expression was calculated by the comparative ΔΔCt method. Transcript abundances were normalized to the reference gene TaEF-1a and relative to that in seedling leaves (normalized as 1). Error bars represent standard deviation among three biological replicates. (∗) indicate a significant difference between a particular tissue and seedling leaves with a p-value < 0.05. Differences were assessed using Student’s t-tests.
TaRar1 Expression in Response to Abiotic Stresses and Hormones
To study TaRar1 in response to abiotic stress, we treated plants with PEG6000 to induce drought stress and poked leaves by a sterilized scissors to cause wounding. TaRar1 transcription level was measured at five time points as shown in Figure 4. Real-time PCR revealed a rapidly induction of TaRar1 under drought stress as early as 6 h post PEG6000 treatment (hpt), and the highest fivefold increase at 12 hpt (Figure 4A), then restored to the 0-hpt control level at 24 hpt. Similarly, TaRar1 expression was also increased after wounding, but the induction was relatively slower compared to the PEG treatment, a significant threefold increase was detected at 24 hpt, and then backed to the control level by 48 hpt (Figure 4A).
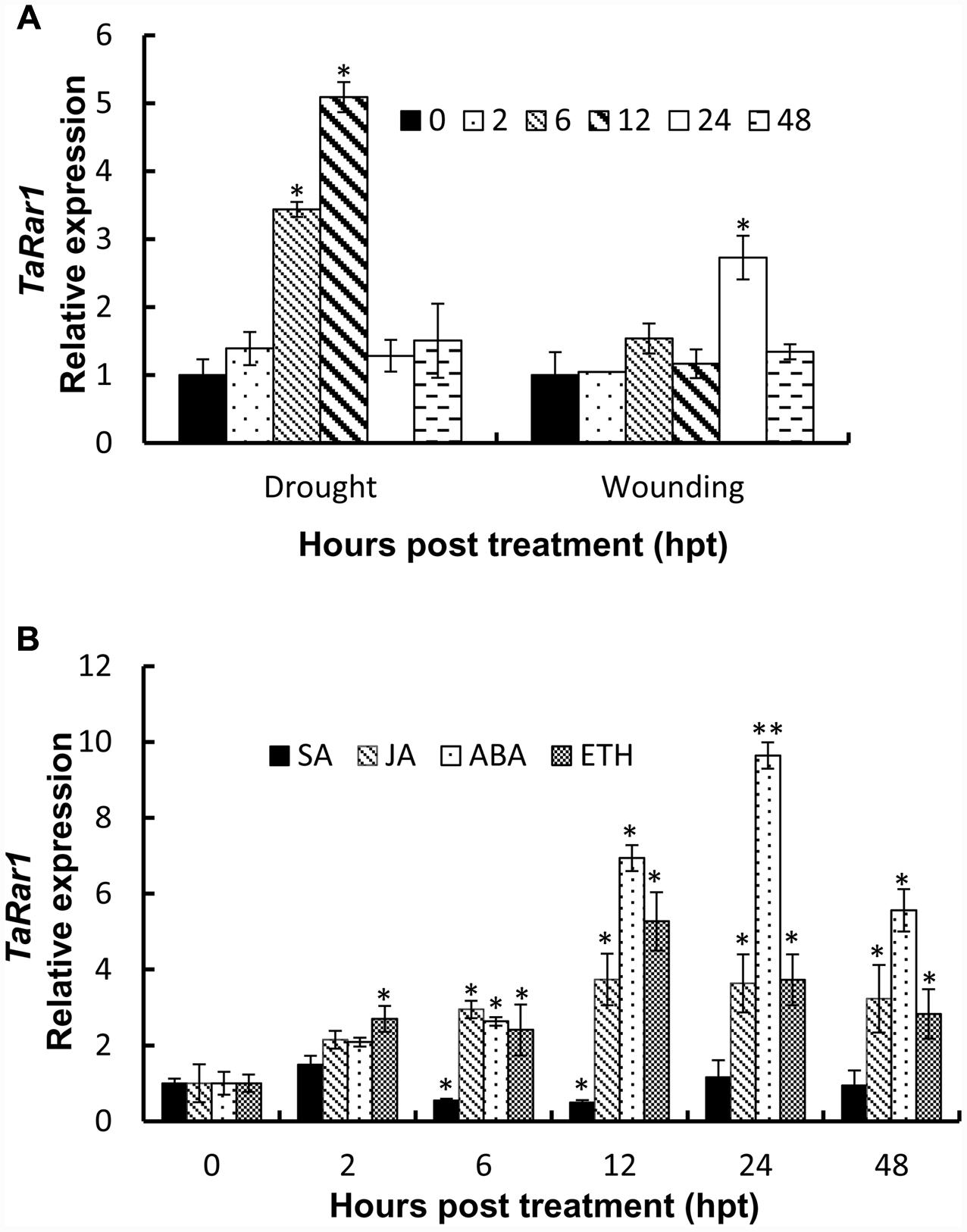
FIGURE 4. Expression profiles of TaRar1 in response to abiotic stresses (A) and exogenous hormones (B). Leaf tissues were sampled at 0, 2, 6, 12, 24, and 48 hours post treatment (hpt) both in wound and drought stresses (A). Transcript abundances were normalized to the wheat elongation factor TaEF-1a gene and relative to the level at 0 hpi. Three independent biological replications were performed. Error bars represent standard deviation among three biological replicates. The asterisks (∗) and (∗∗) indicate a significant differences between that time point and 0 hpt with a p-value < 0.05 and 0.01, respectively. Differences were assessed using Student’s t-tests. ABA, abscisic acid; SA, salicylic acid; ET, ethylene; MeJA, methyl jasmonate.
To understand the TaRar1 regulation by plant hormones, we treated leaves with four different plant hormones exogenously; including SA, JA, abscisic acid (ABA), and ethanol (ET) at the two leaves stage. Leaf tissues were collected at six different time points (Figure 4B). The 0 hpt level was normalized as 1 for each treatment. As shown in Figure 4B, when treated with SA, TaRar1 was either unchanged or significantly reduced during 6–12 hpt. In contrast, TaRar1 was significantly up-regulated after treating with JA, ABA, or ET. The highest 10-fold of that of the 0-hpt level TaRar1 expression was detected at 24 h post-ABA treatment. The TaRar1 induction by JA or ET treatment was not as high as that detected in the ABA treatment, but all was significantly higher than the control level at 0 hpt.
Down-Regulating TaRar1 Compromised Wheat Resistance to an Avirulent Strain of Stripe Rust
Because the higher TaRar1 expression was associated with the incompatible interaction, we investigated if down-regulating TaRar1 expression would compromise Suwon11 resistance to the avirulent Pst pathotype. We knocked down the endogenous TaRar1 transcripts in Suwon11 using a barley stripe mosaic virus induced gene silencing (BSMV-VIGS) assay targeting all three TaRar1 homeologs using a 190-bp highly conserved region (Supplementary Figure S3), the vector is designed as BSMV:Rar1. Two constructs carrying only the BSMV genomes and a 120-bp wheat phytoene desaturase (PDS) gene were included as controls in the study, named as BSMV:00 and BSMV:PDS. In addition, a mock control only inoculated with the FES buffer was also included.
Mild chlorotic mosaic symptoms were appeared after BSMV-inoculated plants at 5–8 dpi, and no distinct defects on newly emerged leaves were observed. At 9 dpi, photobleaching was displayed on the plants inoculated with BSMV:PDS (data not shown), indicating BSMV induced gene silencing was started. Two Pst pathotypes CYR23 and CYR31 were inoculated the TaRar1 silenced plants, respectively. Suwon11’s resistance level described as infection types (ITs) was scored at 15 dpi. As shown in Figure 5A, Suwon11 showed a resistant response after inoculated with CYR23 on the mock and BSMV:00 controls, characterized with a high necrosis areas at the infection sites. TaRar1 silenced leaves had more fungal sporulation than the mock and the BSMV:00 controls (Figure 5A, left), suggesting down-regulation of TaRar1 compromised the Suwon11 resistance to the avirulent pathotype CYR23. When inoculated with the virulent CYR31 pathotype, TaRar1 silenced leaves had the same IT as the controls (Figure 5A, right).
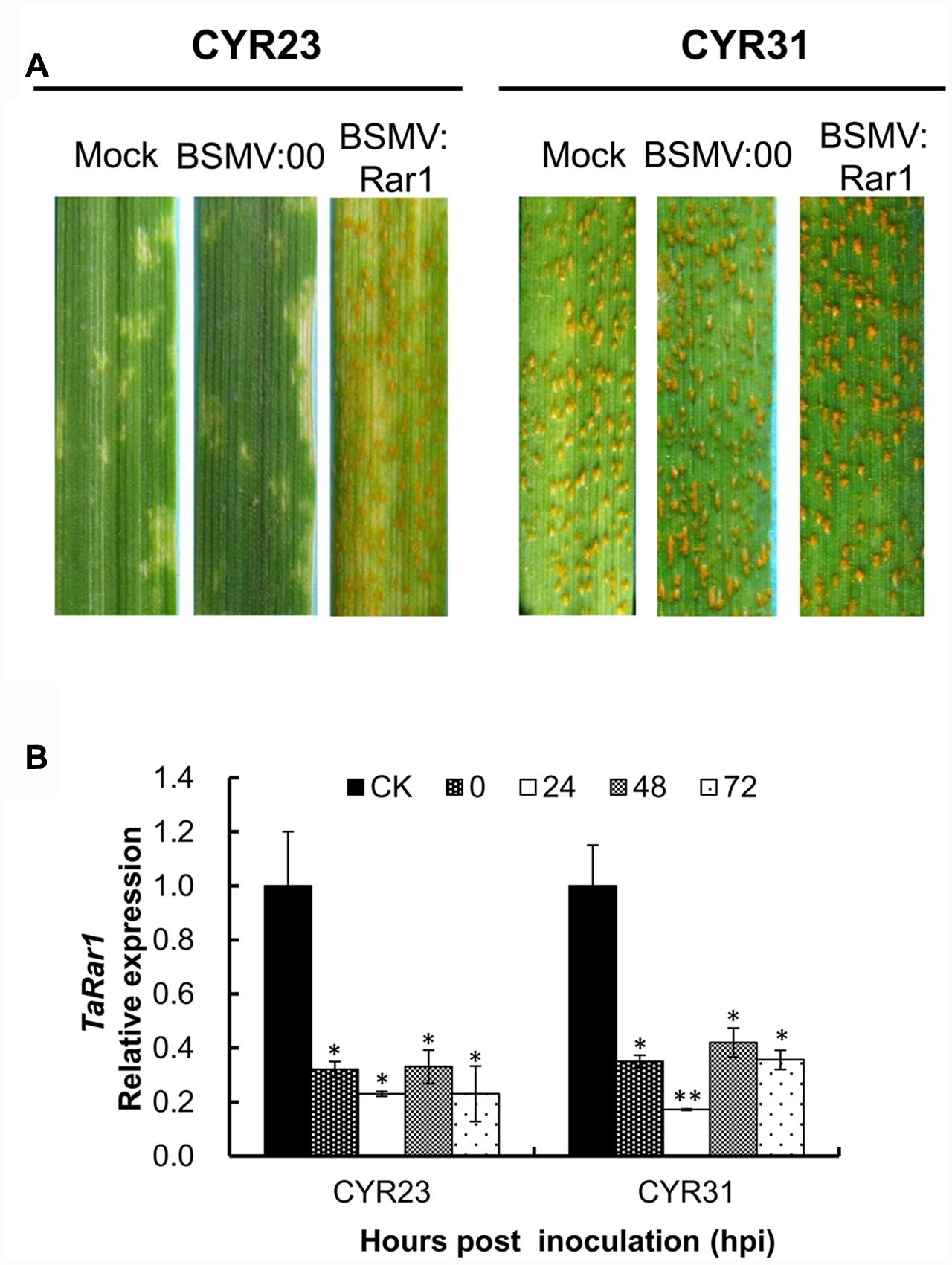
FIGURE 5. Functional analysis of the TaRar1 gene by the BSMV-induced gene silencing assay. (A) Infection type of Suwon11 to CYR23 (avirulent pathotype) and CY31 (virulent pathotype) after inoculated with BSMV:00 and BSMV:Rar1. Stripe rust was inoculated nine days post BSMV inoculation. Pictures were taken 15 days post rust inoculation. Mock: wheat leaves inoculated with FES buffer. BSMV:00 means only the BSMV genome; BSMV:Rar1 means a 190-bp fragment of TaRar1 was inserted into the BSMV gamma genome. (B) Transcript abundances of TaRar1 in Suwon11 during the course of rust infection. X-axis indicates the time post inoculations (hpi) by two Pst pathotypes at 0, 24, 48, and 72 h. CK: Suwon11 treated with BSMV:00. Different patterns of bars indicate different time points post rust inoculations. Relative expressions of TaRar1 were normalized to the CK at 0 hpi (as 1) in each of the rust inoculations. Error bars represent the variations among three independent replicates. (∗) and (∗∗) indicate a significant differences between that time point and CK with a p-value < 0.05 and 0.01, respectively.
Leaf tissues were collected to ensure the effective silencing assays from both BSMV:Rar1 and BSMV:00 plants right before Pst inoculations labeled as 0 hpi, and then 24 hpi, 48 hpi, and 72 hpi. The transcription level of TaRar1was knocked down by approximately about 70–80% in the BSMV:Rar1 silenced leaves compared to the leaves of the same growth stage from BSMV:00-infected plants, labeled as CK in Figure 5B. The results confirmed the silencing of TaRar1 before and during Pst infection.
Endogenous SA Level Decreased in TaRar1 Silenced Leaves
To analyze whether the endogenous concentration of SA was affected by the TaRar1 expression, we measured the SA level in BSMV:Rar1 silenced leaves and controls after the avirulent Pst inoculation at six time points (Figure 6A). In the BSMV:00 control plants, the SA levels were up-regulated and peaked at 48 hpi. However, the concentrations of SA were almost unchanged or reduced over the time course and had significantly lower expression than the control (BSMV:00) from 12 to 72 hpi (Figure 6A). In addition, three PR genes PR1. PR2, and PR5 were monitored during the same time course study. qRT-PCR revealed that all three PR genes had significantly low expressions when TaRar1 was silenced compared to the non-TaRar1 silenced control CK in Figure 6B.
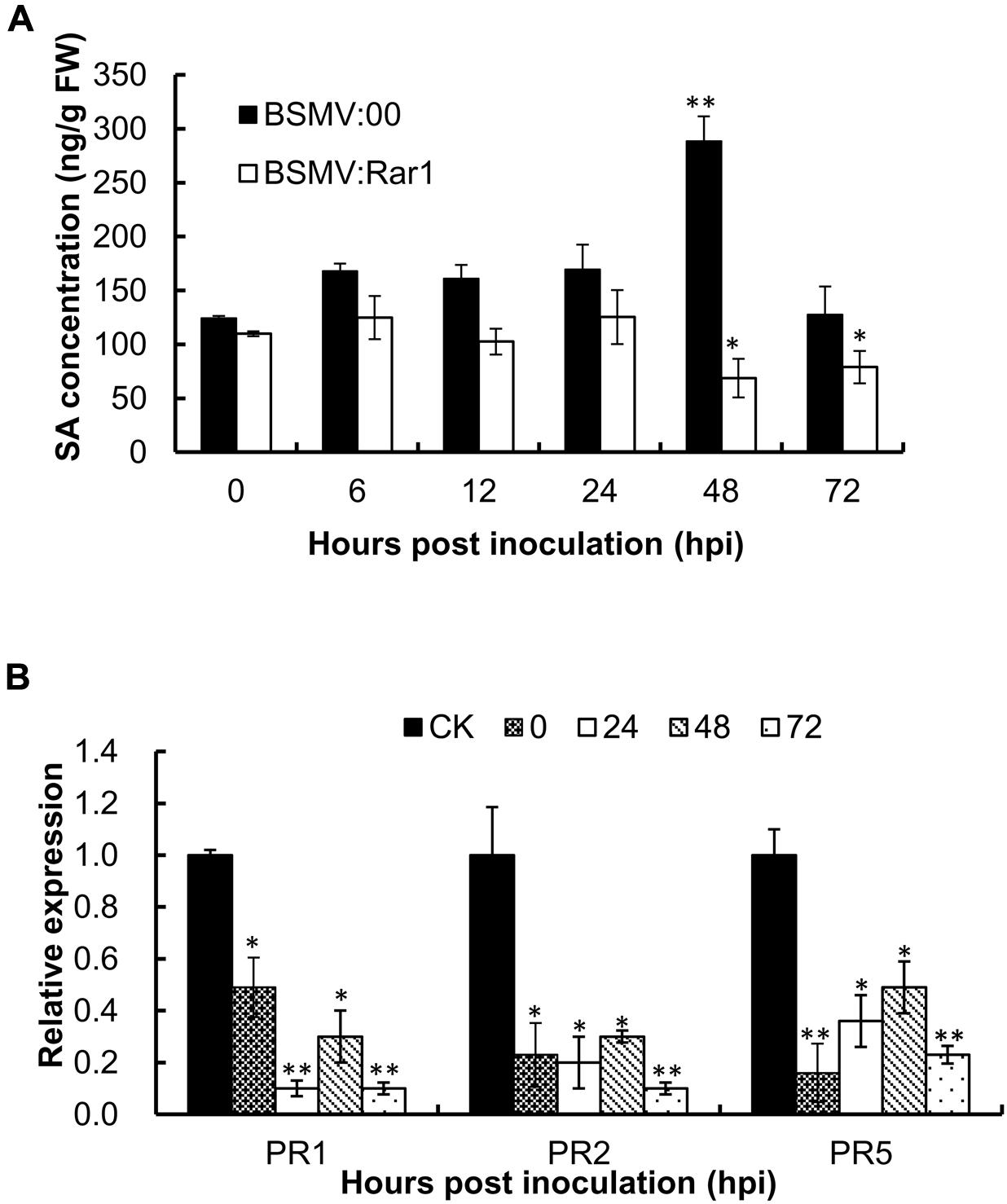
FIGURE 6. Salicylic acid concentration (A) and PR gene expression (B) in TaRar1 silenced leaves during the time course of stripe rust infection. Sample tissues were collected at 0, 6, 12, 24, 48, and 72 hhpi challenged with CYR23 Pst pathotype. Three independent biological replications were performed. Transcript abundance was normalized to the reference gene TaEF-1a (wheat elongation factor) (GenBank accession Q03033). The mean expression values were calculated from three replications. Error bars represent standard deviation among three biological replicates. (∗) and (∗∗) indicate a significant difference difference between a particular hpi and CK with a p-value < 0.05 and 0.01, respectively. Differences were assessed using Student’s t-tests.
Exogenous SA, JA, or ABA Treatment Enhanced Wheat Resistance to a Virulent Strain of Stripe Rust
Our studies have revealed that high TaRar1 expression after exogenous treatment of JA or ABA, and higher endogenous SA was associated with higher TaRar1 expression in the incompatible interaction. Therefore, we tested the level of endogenous SA and the wheat defense response after treatment with JA or ABA. As shown in Supplementary Figure S4A, endogenous SA concentration was increased as early as 12 h post ABA treatment (hpt) and with as much as eightfold increase at 24 hpt. A transient high SA level was detected post JA treatment at 24 hpt. In addition, Suwon11 showed less sporulation by the virulent CYR31 strain after exogenous SA, JA, and ABA treatments (Supplementary Figure S4B). The findings were confirmed by the less pustule counts per area (Supplementary Figure S4C) and relative lower fungal DNA concentration in the leaves pre-treated with a hormone compared to the control (Supplementary Figure S4D).
Silencing of TaRar1 Increased Fungal Growth and Reduced Cell Death
TaRar1 silenced Suwon11 leaves were examined microscopically after inoculation with the avirulent pathotype CYR23 to determine any histological changes associated with the enhanced susceptibility. Fungal development and host responses to CYR23 were similar to what have been described previously (Wang et al., 2008, 2013). At 48 hpi and 72 hpi after inoculated with CYR23, the fungal hyphae were significantly (∗P < 0.05, ∗∗P < 0.01) longer in the TaRar1 silenced leaves compared to that in the leaves of BSMV:00 inoculated control (Figure 7A). Meanwhile, the total infection area was significantly larger (∗∗P < 0.01) in the TaRar1-knockdown plants at 120 hpi relative to the control (Figure 7B).
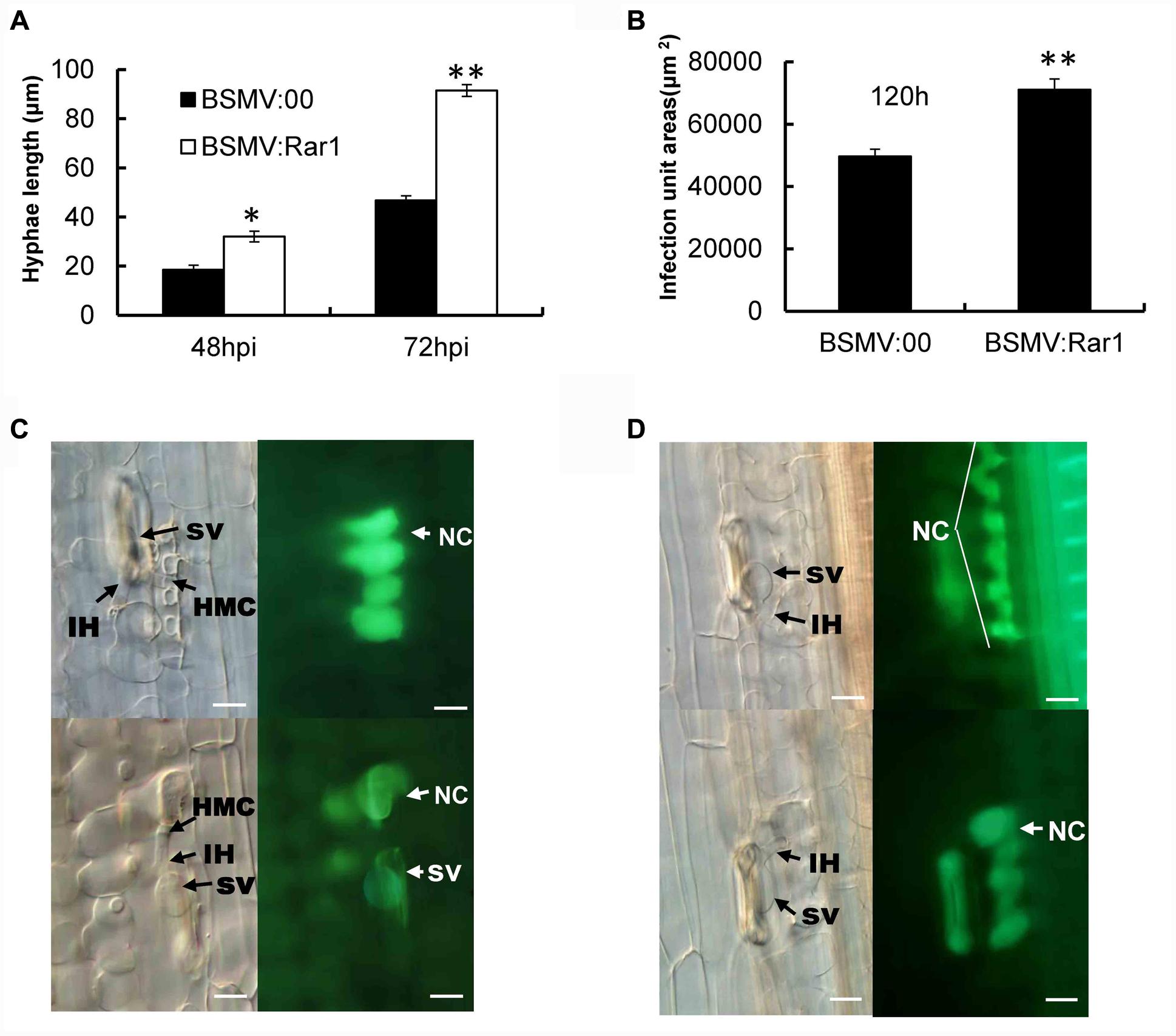
FIGURE 7. Pathotype CYR23 growth on Suwon11 measure by hyphae length (A), infection areas at 120 hpi (B) and cell death in Suwon11 leaves at 48 hpi (C) and 72 hpi (D). Suwon11 was inoculated with BSMV:00 or BSMV:Rar1 and then was inoculated with CYR23 nine days post inoculation. (A) Hyphal length was measured under a light microscope at 48 and 72 h post CYR23 inoculation in both TaRar1-silenced and non-silenced plants. The growth of hyphae was significantly increased in TaRar1-silenced Suwon11. (B) Infection areas were measured microscopically at 120 hpi in both TaRar1-silenced and non-silenced Suwon11. Infection area was significantly enlarged in TaRar1 silenced leaves. (C,D) Histological observations of cell death. Pictures were taken under an epifluorescence or light microscopy at 48 hpi (C) and 72 hpi (D). Significant reduced green florescence in the necrotic area per infection size in TaRar1-silenced plants. NC, necrotic cell; SV, substomatal vesicle; IH, initial hyphae; HMC, haustorial mother cell; Bars = 50 μm. Values represent mean ± standard errors of three independent samples. Differences were assessed using Student’s t-tests. (∗) and (∗∗) indicate a significant difference between a particular hpi and CK with a p-value < 0.05 and 0.01, respectively.
Hypersensitive cell death evidenced by the yellow auto-florescence under a florescence microscope was documented at 48 and 72 hpi and compared between TaRar1 silenced leaves and the control. The auto-fluorescence of attacked mesophyll cells were observed under an epifluorescence microscopy with excitation filter, 485 nm; dichromic mirror, 510 nm; and barrier filter, 520 nm. The yellow color is changed to green because of the settings of saturability and the contrast of light. The control leaves at the infection sites showed stronger auto-florescence compared to the TaRar1 silenced leaves at 48 hpi (Figure 7C) and 72 hpi (Figure 7D), suggesting that silencing of TaRar1 reduced the degree of cell death and resulted in more fungal growth.
TaRar1 Was Involved in Reactive Oxygen Accumulation Process
Because hydrogen peroxide (H2O2) has been associated with HR, we were interested to find out any changes of H2O2 accumulation in TaRar1 silenced leaves upon pathogen challenge. We used diaminobenzidine (DAB) polymerization to show H2O2 accumulation in infected leaf tissues in situ. Interestingly, similar to the control, BSMV: Rar1 inoculated plants had H2O2 accumulation in mesophyll cells at 24 hpi with the avirulent CYR23 pathotype (Figure 8A). However, the striking difference was that H2O2 accumulation was significantly (∗∗P < 0.01) reduced in the TaRar1 silenced leaves compared to the control (Figure 8B). In contrast, the expression of the wheat catalase gene (TaCAT), which is involved in ROS removal, was significantly increased (∗P < 0.05, ∗∗P < 0.01) in the TaRar1-knockdown leaves after infection with CYR23 (Figure 8C), implying that the reduction of H2O2 accumulation was the result of higher expression of TaCAT. Real-time PCR revealed little change in TaRar1 transcript level when treated wheat plants with exogenous H2O2 (100 mM) (Figure 8D). These results suggested that TaRar1 functions upstream of H2O2 accumulation in responses to Pst infection.
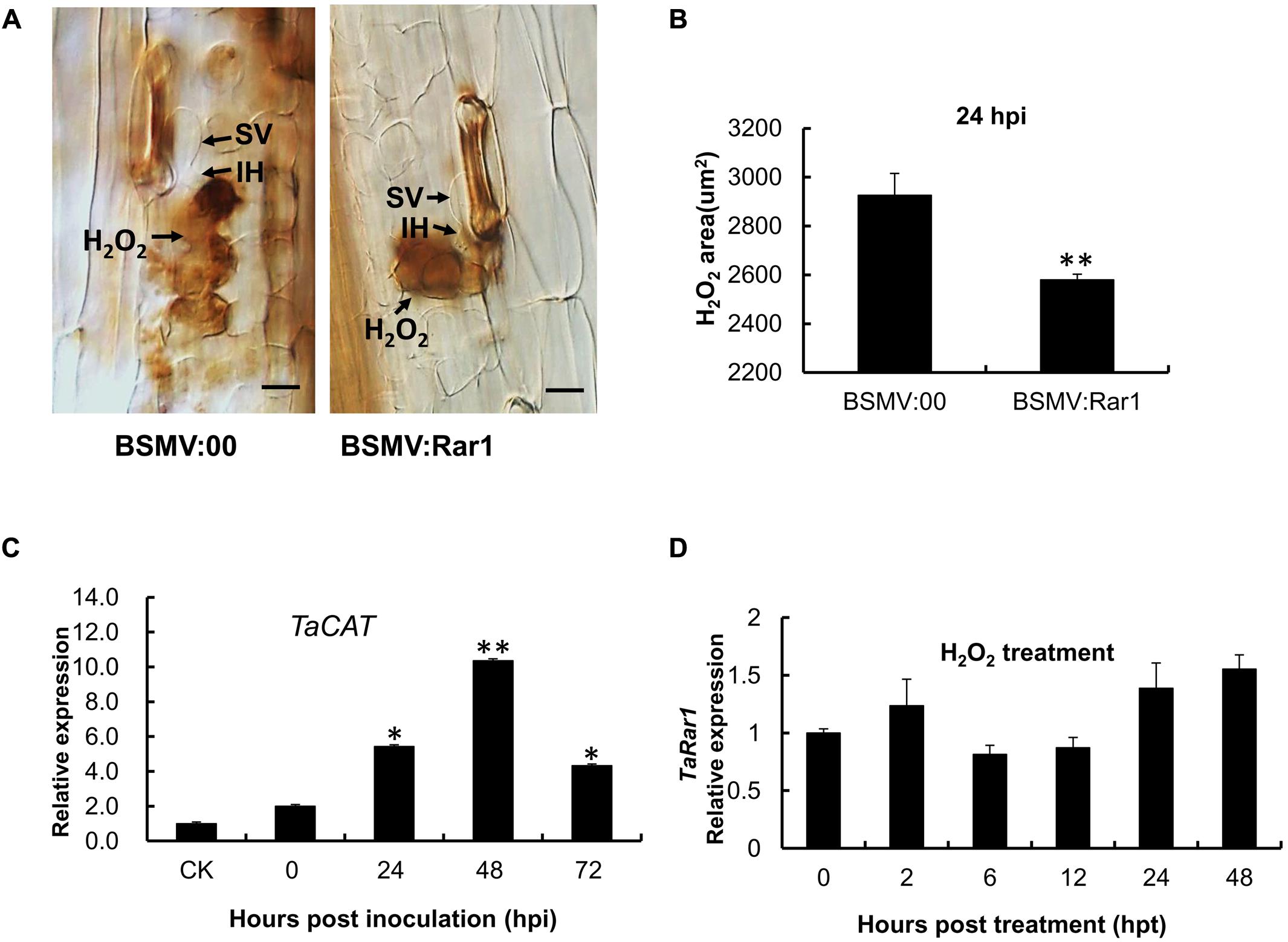
FIGURE 8. Reactive oxygen accumulation and related gene expression in the TaRar1 knockdown plants. (A) Histological observation of H2O2 accumulation (Brown color) by DAB stain in Suwon11 leaves 24 h post CYR23 inoculation in BSMV:00 and BSMV:Rar1 inoculations leaves. H2O2 accumulation was significantly reduced in TaRar1 silenced leaves compared to the control. SV, substomatal vesicle; IH, initial hyphae; Bars = 50 μm. (B) DAB stained areas measured microscopically at 24 hpi. (C) The expression level of TaCAT catalase (X94352) after BSMV:Rar1 inoculation at four time points relative to the expression in BSMV:00 (CK). (D) TaRar1 expression profile after exogenous H2O2 treatment. Transcript abundance of TaRar1 had no obvious change at different time points post treatment. (∗) and (∗∗) indicate a significant difference between a particular hpi and CK with a p-value < 0.05 and 0.01, respectively.
Materials and Methods
Plant Materials
Wheat (T. aestivum L.) cultivars Suwon11 (spring wheat) and two stripe rust Pst pathotypes CYR23 and CYR31 were the biological materials used in this study. Seedlings of Suwon11 was grown and maintained followed the procedure described by Kang and Li (1984). Suwon11 has been reported containing the stripe rust-resistance gene YrSu and highly resistant to CYR23 but highly susceptible to CYR31 (Stakman et al., 1962). Chinese Spring nulli-tetrasomic lines were kindly provided by the Wheat Genetics and Genomics Center at Kansas State University, Manhattan, KS, USA.
Cloning of TaRar1 and Sequence Analysis
To clone the TaRar1 gene, a pair of primers (forward and reverse) was designed using Primer 5.0 software. The primers were designed based on the cDNA sequence from our laboratory database, which was obtained from the interaction between the Suwon11 cultivar and CYR23 (avirulent) or CYR31 (virulent) (Table 1). The primers were used to amplify the open reading frame (ORF) of TaRar1. The template was a mixture of the first strand cDNA samples extracted from leaves of Suwon11 at 12, 24, 48, and 72 h post inoculated with CYR23 (an incompatible combination). The PCR products were cloned into the pGEM-T Easy Vector System (Promega, Madison, WI, USA) or the pMD18-T Simple Vector (TaKaRa Biotechnology)1 and sequenced using an ABI PRISM 3130XL Genetic Analyzer (Applied BioSystems). The amino acid sequence of TaRar1 was analyzed to determine the alignment of the deduced protein sequences using the DNAMAN (version 6) program (Lynnon Biosoft, Quebec, Canada).
Nulli-tetrasomic Analysis via Restriction Fragment Length Polymorphism (RFLP)
For RFLP, enzyme digestion, gel-electrophoresis, Southern blotting, probe, labeling, and hybridization were performed following the protocols described by Huang et al. (2003). A 200-bp DNA fragment from the CHORD-I region was used as a probe. Genomic DNA of CS nulli-tetrasomic lines were digested with EcoRV.
RNA Isolation and Real-Time PCR
Leaf tissues for time course study were collected at the corresponding time points from different plants. For tissue-specific expression analyses of TaRar1, intact seedling leaves were sampled at the two-leaf stage. The same plants then were used for root, stem, flag leaf, floret, and spikelet tissues at the adult stage at the same time. Sampled tissues were flash-frozen in liquid nitrogen and stored at -80°C before the isolation of total RNA. Each experiment has three independent biological replications included.
The RNA isolation was conducted with BiozolTM Reagent (BioFlux, Tokyo, Japan). The integrity and quality of the total RNA was characterized in a 1% agarose gel. Additionally, A NanoDropTM 2000 spectrophotometer (Thermo Fisher Scientific, USA) was used to estimate the RNA quantity. A Revert Aid First-strand cDNA synthesis kit from Fermentas2 was used to synthesize cDNA from RNA.
Real-time PCR reactions and primer design were performed as described by Wang et al. (2013). A CFX connectTM Real-Time PCR System was used to perform the quantitative real-time PCR (Applied Biosystems, Foster City, CA, USA). TaEF-1a gene (GenBank accession number Q03033), a wheat elongation factor was used as a housekeeping reference for real-time PCR analysis. An 85-bp TaEF-1a fragment and a 190-bp TaRar1 fragment as gene specific primers are synthesized to determine the gene expression, listed in Table 1 as TaEF-F/R and TaRar1-QRTF/QRTR. Specific amplification was contacted to ensure for each reaction according to a single peak dissociation curve. The comparative 2-ΔΔCT method was used to quantify relative gene expression according to the Threshold values (Ct) generated from Biosystems applied (Livak and Schmittgen, 2001).
Rust Inoculations and Chemical Treatments
Freshly stripe rust urediniospores were brushed to the surface of primary wheat leaves at 7-day-old seedlings. The control treatment was inoculated with sterile distilled water. Plants were incubated for 24 h in dark in a dew chamber with a temperature of 15°C and 100% humidity. Then the plants were subsequently transferred to a growth chamber with a temperature of 16°C and a 16 h photoperiod. Leaves were collected at 0, 6, 12, 24, 48, 72, and 120 hpi. These time points were related with a series of biological events in the interactions between Suwon11 and CYR23 or CYR31 (Wang et al., 2013).
Different chemical treatments were sprayed to the leaf surface of wheat seedlings with 20 mM SA, 2 mM methyl jasmonate (MeJA), 2 mM ABA, 2 mM ethylene (ET) and 100 mM H2O2, respectively (Wang et al., 2013). All chemicals were dissolved in 0.1 % (v/v) ET. The mock plants were sprayed with 0.1% (v/v) ET.
Analyses of the Expression of TaRar1 in Response to Different Abiotic Stress Treatments
To analyze the expression of TaRar1 under drought and wounded conditions, 30 wheat seedlings were prepared for each treatment. For the drought-stress treatment, roots of wheat seedlings were soaked in 200 mM NaCl or 20% PEG6000. The mock-treated seedlings were maintained in a growth chamber at a normal temperature with a 12 h photoperiod. The wound treatment was applied by cutting the wheat leaves using sterilized scissors. Leaf tissues were sampled at 0, 2, 6, 12, 24, and 48 hpt and rapidly frozen in liquid nitrogen and stored at -80°C. Three independent biological replicates were used for each time point and control.
BSMV-Mediated Gene Silencing
Method for constructing the silencing plasmids was conducted according by Holzberg et al. (2002). A 120-bp cDNA fragment was amplified from the wheat PDS gene TaPDS by reverse-transcriptase polymerase chain reaction (RT-PCR). BSMV:GFP (green fluorescent protein) cDNA as the starting material was used to create BSMV:PDS. A PDS fragment replaced the GFP coding sequence in BSMV:GFP resulting in BSMV:PDS. The same approach was constructed to get BSMV:Rar1, a 190-bp TaRar1 cDNA fragment amplified by the primers TaRar1-oligo-F/R(Table 1).
Tripartite BSMV genome was linearized and transcript to RNA in vitro (Holzberg et al., 2002) using the mMessage mMachine T7 in vitro transcription kit (Ambion, Austin, TX, USA). The BSMV inoculum was made by combining an equimolar ratio of α, β, and γ transcripts at a 1:1:1 ratio mixed with inoculation buffer (named as FES) containing a wounding agent. The inoculation was done on the second leaf of three-leaf stage seedling with BSMV RNA by gently rubbing the leaf surface with a gloved finger. Three independent sets of inoculations were performed, with a total of 72 seedlings inoculated for each of the three BSMV viruses (BSMV:00, BSMV:PDS, and BSMV:Rar1). Twenty-four seedlings inoculated with 1xFES buffer were included as a negative control. Post viral inoculation, wheat plants were maintained in a growth chamber at 25 ± 2°C, and examined for symptoms at regular intervals. Once photobleaching was observed, three independent sets of inoculations including CYR23, CYR31 and sterile water (as a mock), were performed. ITs of stripe rust were examined at 15 days post rust inoculation. The third leaves corresponding to the photobleached areas of BSMV: PDS infected plants were divided at 0, 24, 48, and 120 hpi for histological observation and real-time PCR assay. The primers used to assay the transcript abundances of the TaPR1. TaPR2. TaPR5, and TaCAT genes in the TaRar1-knockdown wheat seedlings are listed in Table 1.
SA Level Analysis with the HPLC-MS
Leaves were collected and immediately frozen in liquid nitrogen. The extraction of SA was according to Segarra et al.’s (2006) method and modified as followed. Frozen samples were then ground under liquid N2 with mortar and pestle. An amount of 200 mg of the resulting powder was extracted with 750 μl MeOH–H2O–HOAc (90:9:1, v/v/v) and centrifuged for 1 min at 10,000 rpm. The supernatant was collected and the extraction was repeated twice. Pooled supernatants were dried under N2, resuspended in 1000 μl of pure chromatographic grade MeOH, and finally filtered with a Millex-HV 0.22 μm filter from Millipore (Bedford, USA). Quantitation was done by the standard addition method by spiking control plant samples with SA solutions (ranging from 50 to 1000 ng ml-1), and extracting as described above.
Histological Observation of Fungal Growth and ROS Accumulation
Wheat leaves were sampled at 48 and 72 hpi with Pst after inoculated with BSMV and treated as Wang’s methods (Wang et al., 2013). Cleared leaf segments were observed using an Olympus BX-51 microscope (Olympus Corp., Tokyo) for infection areas and lengths of infection hyphae. Infection areas were the areas containing infection hypha at an infection unit. An infection unit is a site when intercellular fungal mycelium in the leaf mesophyll cell layer is formed by a germling that penetrated through a stoma. Auto-fluorescence was observed as a necrotic death area in infected mesophyll cells by epifluorescence microscopy. More than 30 infection sites were chosen to examine the auto-fluorescence on each of five randomly selected leaf segments per treatment. The infection sites were considered as successfully penetrated with fungal appressoria formation over stomata. The necrotic area was measured with a calibrated eyepiece micrometer and corresponding necrotic areas (square micrometers) calculated as Wang’s method (Wang et al., 2013). SPSS software was used to statistical analysis the standard deviations and Tukey’s test (SPSS, Inc. Chicago, IL, USA).
In order to study the host response, H2O2 accumulation was detected as described by Thordal-Christensen et al. (1997) in plant mesophyll cell. Cutting the inoculated wheat leaves and immersing the ends in a stained buffer containing 1 mg/ml 3,3′-DAB dissolved in HCl-acidified (pH 3.8) distilled water. Leaves were incubated for 8 h in the DAB buffer and transferred to the fixed buffer to terminate the reaction.
Relative Quantification of Pst in Inoculated Leaves
The single-copy target genes PsEF and TaEF1 were used to measure the relative quantification of Pst as carried out (Panwar et al., 2013). Standard curves were prepared with the genomic DNA of the wheat cultivar Suwon11 and the Pst pathotype CYR31 at seven serial dilutions, respectively. The correlation coefficients for the standard curves were above 0.99. The specific primers PsEF-F/R and TaEF1-F/R were used to do the quantification PCR listed in Table 1. The biology biomass of the real-time PCR products of PsEF and TaEF1 in infected sample leaves were calculated according to the gene-specific standard curves to analysis the quantification of Pst and wheat genome DNA.
Discussion
In this study, we cloned and characterized a TaRar1 gene from cultivar Suwon11. The gene has 14 polymorphisms at the nucleotide level but only three amino acid differences at the protein level (Figure 1 Supplementary Figure S5) compared to the TaRAR1-1 amplified from another wheat cultivar WGRC7 (Tai, 2008). Three TaRar1 homologs obtained from the wheat cultivar Chinese Spring genomic DNA sequence at the IWGSC share 94–99% similarity with each other (Supplementary Figure S3), and locate at wheat homeologous group 2 chromosomes 2A, 2B, and 2D (Supplementary Figure S1), therefore they were referred as homeologs. It is highly possible that three TaRar1 homeologs exist in Suwon11 as well. Since the three homeologs were knocked down simultaneously and measured together in this study, we drew the conclusion using TaRar1 to represent the function of the three homeologs.
RAR1 protein contains two CHORD domains rich in cysteine and histidine. The domains are involved in zinc-dependent protein–protein interactions. Comparison among RAR1 from different species or different cultivars of the same species revealed that the two functional domains of CHORD-I and CHORD-II are highly conserved (Figure 1; Supplementary Figure S2), suggesting an important and also similar function associated with the CHORD domains of the RAR1 protein from different species. It has been shown that the RAR1 CHORD-I interacts with the CS (CHORD-containing protein and SGT1) domain of SGT1 and CHORD-II interacts with M domain of HSP90 to form a complex. The protein complex functions as a chaperone complex for NLR immune sensors (Tornero et al., 2002; Bieri et al., 2004; Holt et al., 2005; Azevedo et al., 2006) to modulate downstream defense responses including HR and PR gene expressions. The function of RAR1 is conserved in different plant species, containing Arabidopsis, tobacco, and barley, and it is required by the subsets of both CC-NB-LRR and TIR-NB-LRR-type R proteins (Shirasu and Schulze-Lefert, 2003). In tobacco, silencing Rar1 by virus-induced gene silencing strongly reduced the resistance to tobacco mosaic virus mediated by the N gene (Liu et al., 2002). Similarly, silencing three TaRar1-1 homeologs in WGRC7 compromised the leaf resistance mediated by Lr21 (Scofield et al., 2005). HvRar1 was first identified as a requirement for Mla12 mediated resistance to powdery mildew. However, in the same species, mediating resistance to the same powdery mildew pathogen, but not all Mla genes need HvRar1 in the pathway (Azevedo et al., 2002). Similarly, leaf rust resistance gene Lr21 and stem rust resistance gene Sr33 are both CC-NBS-LRR type of R gene in wheat (Huang et al., 2003; Periyannan et al., 2013), TaRar1 is involved in the Lr21 (Scofield et al., 2005) but not in the Sr33-mediated resistance (Periyannan et al., 2013). Generalizing from the studies on Rar1 gene, it is clear that not all R genes with similar structures require Rar1 during defense response. This suggests a fine-tuning of defense response signaling mediated by each R gene. This also implies a different satiability of each R protein since a known function of RAR1-SGT1-HSP90 complex is protein chaperon. Comprehensive understanding of each R gene-mediated signaling pathway will provide a key to this puzzle. In our study, down-regulating three homeologs of TaRar1 in Suwon11 reduced the host resistance level to the avirulent Pst strain CYR23, suggesting this gene is required in the resistance to Pst infection mediated by YrSu in Suwon11.
In many cases, race-specific resistance is characterized by a rapid development of HR at the infection sites. Compared to the compatible interactions, we found the transcript level of TaRar1 was strikingly up-regulated at the early stages of the pathogen infection (Figure 2A) in incompatible interaction. When TaRar1 was silenced, the HR areas (Figures 7C,D) and production of H2O2 (Figures 8A,B) were significantly reduced, suggesting the TaRar1 gene was required for inducing HR and ROS. SA has been shown to be a signaling molecule involved in both local HR and production of ROS at the infection sites and SAR to further infection by broad range pathogens (Durner and Klessig, 1995). In the incompatible interaction, SA concentration in Suwon11 was significantly increased as early as 24 hpi (Figure 2B). Down-regulating TaRar1 reduced SA accumulation and HR compared to the control (Figure 6A), suggesting TaRar1 might modulate defense response through signaling molecule SA. This hypothesis was supported by the observation that the disease severity by the virulent strain on Suwon11 was reduced after exogenous SA treatment (Supplementary Figures S4B–D). However, interestingly, the expression of TaRar1 was reduced after exogenous SA treatment (Figure 4B). Notably, during the early infection course of pathogenesis, both TaRar1 and SA were up-regulated (Figures 2A,B), but the timing of the induction seemed to suggest a higher TaRar1 expression leaded to a higher SA accumulation. However, when SA level reached the peak at 72 hpi, TaRar1 level has dropped compared to its highest level at the same time point (Figures 2A,B). These results suggested a negative feedback regulation of SA to TaRar1 in this regulation, when SA accumulation exceeding a certain threshold level or exogenous high SA level was applied, TaRar1 expression started to reduce (Figure 4B). Because SA level started to drop once TaRar1 was reduced, suggesting TaRar1 functions upstream in the SA biosynthesis pathway. These results suggested TaRar1 involved in defense response against Pst through SA in the incompatible interaction (Figures 2A,B). Application of JA/ET and ABA also up-regulated TaRar1 and enhanced Suwon11 resistance to the virulent strain (Supplementary Figures S4A–D), suggesting TaRar1 involved in both race-specific defense and basal defense responses to Pst. These results are similar to those observed in the AtRar1 mediated pathogen-associated molecular pattern-trigged immunity (PTI) in the absence of the cognate resistance gene in Arabidopsis (Shang et al., 2006), and HvRar1-mediated basal resistance to Magnaporthe grisea in barley (Jarosch et al., 2005). The induction of PTI in Arabidopsis mediated by AtRAR1 was through JA signaling and the interaction with SGT1 (Shang et al., 2006).
Reactive oxygen species are thought to play key roles in defense responses, in which the most important component of ROS is H2O2 (Keppler et al., 1989; Hemetsberger et al., 2012). We previously studied the generation and accumulation of ROS in the interactions of Suwon11 and two races of Pst (avirulent and virulent). In the incompatible interaction, H2O2 was detected at 12 hpi and percentage of infection sites showing H2O2 accumulation further increased until 24 hpi, which coincided with primary haustoria formation in mesophyll cells (Wang et al., 2008). At these different time points, the transcript levels of TaRar1 were up-regulated compared with the control (Figure 2A). To analyze whether TaRar1 affects ROS accumulation in wheat, we measured the production of H2O2 after knocking down TaRar1 at 24 hpi. In control BSMV: 00 plants, abundant ROS accumulation were exhibited in a few mesophyll cells (Figure 8A, left). In contrast, mesophyll cells in contact with primary hyphae showed less ROS accumulation in BSMV: Rar1-silenced plants compared to the control (Figure 8A, right). The same event was observed in Mla12-triggered and Rar1-dependent oxidative burst, coinciding with fungal haustorium differentiation (Shirasu et al., 1999). SA radicals, which are generated from SA, binds and inhibits catalase which is function as a major H2O2-scavenging enzyme, thereby leading to an increase in the endogenous level of H2O2 (Durner and Klessig, 1995). We assayed the expression of catalase, which directly correlated with less SA accumulation, was upregulated after silencing TaRar1 at 48 hpi (Figure 8C). The results were consistent with the reduced H2O2 accumulation after knocking down TaRar1, suggesting TaRar1 activated the defense against Pst through modulating the H2O2 accumulation by SA signaling.
In addition to the involvement in defense against Pst, high level of TaRar1 transcript abundance was also detected in wheat tissues at the adult stage (Figure 3), under drought or wounding stresses (Figure 4A), and after treatment with three out of four tested hormones (Figure 4B). The result implied a cross-talk through TaRar1 among different signaling pathways modulating plant development and different stresses. The highest TaRar1 level was seen in response to exogenous ABA treatment. The role of ABA in plants is complicated and our knowledge on ABA regulation is incomplete yet. However, studies on this plant hormone have demonstrated that ABA plays an ambivalent role in plant defense response depending on the timing of the infection and the interaction of the host and pathogen pair. ABA is a global switch in modulation signaling pathways overlapped among plant development, biotic and abiotic stresses (Asselbergh et al., 2008). In our studies, among the four hormones tested, TaRar1 level was up-regulated after treatment with ABA, JA, or ET. An elevated SA level was seen after high level of TaRar1 (Figure 2A). The positive effect of JA and ABA on SA level was confirmed by the high SA concentration detected after exogenous JA or ABA treatment (Supplementary Figure S4A). Similarly, a cooperative or synergistic interaction between SA and JA/ET was reported in several other studies (Xu et al., 1994; Mur et al., 2006) although antagonistic interaction between JA/ET and SA was also well documented (Audenaert et al., 2002; Ellis and Turner, 2002; Spoel et al., 2003; Xu et al., 2013). Rar1 is a single copy gene in barley (Azevedo et al., 2002), A. thaliana (Muskett et al., 2002; Tornero et al., 2002), and potato (Pajerowska et al., 2005). However, in wheat, there are three highly conserved homeologs located on 2A, 2B, and 2D chromosomes, respectively (Supplementary Figure S3). With the VIGS strategy and real-time used in this study, we were unable to study the function and expression of each individual homeolog because of the highly similar DNA sequence among them. More investigation is needed to determine whether and how TaRar1 connects different signaling pathways, is a homeolog-dependent or a dosage-dependent. Knockout mutation on each individual homeolog via EMS or fast-neutron would make the functional study of each homeolog possible to answer the above questions.
Conclusion
We propose a working model, in which TaRar1 was placed in the upstream of defense response interacted directly or indirectly with the YrSu gene to mediate resistance, followed by the signaling molecule SA. When SA level was up-regulated, the H2O2 scavenging enzyme was activated and SA radicals generated and bound with TaCAT, ROS accumulated and HR appeared. Furthermore, PR genes expression increased.
Author Contributions
XgW, LH, and ZK conceived the study; XgW, XM, XL, XeW, LH, and ZK advised on the experimental design and drafted the manuscript; XgW, YW, PL, YD, MZ, and BH performed the experiments and did the data analysis. YW, YD, PL, MZ, BH, XM, XL, and XeW interpreted data. XgW, LH, and ZK wrote the manuscript and other authors reviewed and revised the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (No. 31501619), the National Key Research and Development Program of China (2016YFD0100602), the National Basic Research Program of China (Grant No. 2013CB127700), Natural Science Foundation Research Project of Shaanxi Province (2016JQ3032) , Dr. Startup Funds (2013BSJJ071) and Scientific and Technological Project of Yangling(2016NY-27).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank S. R Scofield for providing BSMV vectors.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00156/full#supplementary-material
FIGURE S1 | Nulli-tetrasomic analysis by RFLP revealed three homeologs of TaRar1 in bread wheat genome.
FIGURE S2 | Alignment of three homeologs RAR1 proteins from CS.
FIGURE S3 | Alignment of DNA sequence of the three TaRar1 homeologs. Yellow highlighted region was used for BSMV-VIGS.
FIGURE S4 | SA concentration (A) and infection type of Suwon11 to CYR31 (B), sporulation (C), and relative fungal DNA to wheat DNA (D) after exogenous application of hormones.
FIGURE S5 | Alignment of DNA sequence of between the TaRar1 from Suwon11 and the TaRar1-1 from WGRC7.
Footnotes
- ^http://www.takara.com.cn
- ^https://www.thermofisher.com/cn/zh/home/brands/thermo-scientific/molecular-biology/thermo-scientific-molecular-biology-products/fermentas.html?cid=fl-ts-fermentas
References
Asselbergh, B., De Vleesschauwer, D., and Höfte, M. (2008). Global switches and fine-tuning-ABA modulates plant pathogen defense. Mol. Plant Microbe Interact. 21, 709–719. doi: 10.1094/MPMI-21-6-0709
Audenaert, K., De Meyer, G. B., and Höfte, M. M. (2002). Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 128, 491–501. doi: 10.1104/pp.010605
Austin, M. J., Muskett, P., Kahn, K., Feys, B. J., Jones, J. D., and Parker, J. E. (2002). Regulatory role of SGT1 in early R gene-mediated plant defensesfenses. Science 295, 2077–2080. doi: 10.1126/science.1067747
Azevedo, C., Betsuyaku, S., Peart, J., Takahashi, A., Noel, L., Sadanandom, A., et al. (2006). Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 25, 2007–2016. doi: 10.1038/sj.emboj.7601084
Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295, 2073–2076. doi: 10.1126/science.1067554
Bieri, S., Mauch, S., Shen, Q.-H., Peart, J., Devoto, A., Casais, C., et al. (2004). RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell 16, 3480–3495. doi: 10.1105/tpc.104.026682
Cantu, D., Yang, B., Ruan, R., Li, K., Menzo, V., Fu, D., et al. (2013). Comparative analysis of protein-protein interactions in the defense response of rice and wheat. BMC Genomics 14:166. doi: 10.1186/1471-2164-14-166
Century, K., Holub, E. B., and Staskawicz, B. J. (1995). NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a funga pathogen. Proc. Natl. Acad. Sci. U.S.A. 92, 6597–6601. doi: 10.1073/pnas.92.14.6597
Chen, X. M. (2005). Epidemiology and control of stripe rust (Puccinia striiformis f. sp. Tritici) on wheat. Can. J. Plant Pathol. 27, 314–337. doi: 10.1080/07060660509507230
Durner, J., and Klessig, D. F. (1995). Inhibition of ascorbate peroxidase by salicylic acid and 2,6-dichloroisonicotinic acid, two inducers of plant defense responses. Proc. Natl. Acad. Sci. U.S.A. 92, 11312–11316. doi: 10.1073/pnas.92.24.11312
Durrant, W. E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. doi: 10.1146/annurev.phyto.42.040803.140421
Ellis, C., and Turner, J. G. (2002). A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215, 549–556. doi: 10.1007/s00425-002-0787-4
Flor, H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. doi: 10.1146/annurev-phyto-072910-095339
Heath, M. C. (2000). Hypersensitive response-related death. Plant Mol. Biol. 44, 321–334. doi: 10.1023/A:1026592509060
Hemetsberger, C., Herrberger, C., Zechmann, B., Hillmer, M., and Doehlemann, G. (2012). The Ustilago maydis effector Pep1 suppresses plant immunity by inhibition of host peroxidase activity. PLoS Pathog. 8:e1002684. doi: 10.1371/journal.ppat.1002684
Holt, B. F. III, Belkhadir, Y., and Dangl, J. L. (2005). Antagonistic control of disease resistance protein stability in the plant immune system. Science 309, 929–932. doi: 10.1126/science.1109977
Holzberg, S., Brosio, P., Gross, C., and Pogue, G. P. (2002). Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 30, 315–327. doi: 10.1046/j.1365-313X.2002.01291.x
Huang, L., Brooks, S. A., Li, W., Fellers, J. P., Trick, H. N., and Gill, B. S. (2003). Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploid genome of bread wheat. Genetics 164, 655–664.
Jarosch, B., Collins, N. C., Zellerhoff, N., and Schaffrath, U. (2005). RAR1, ROR1, and the actin cytoskeleton contribute to basal resistance to Magnaporthe grisea in barley. Mol. Plant Microbe Interact. 18, 397–404. doi: 10.1094/MPMI-18-0397
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jørgensen, J. H. (1994). Genetics of powdery mildew resistance in barley. Crit. Rev. Plant Sci. 13, 97–119. doi: 10.1080/713608055
Kang, Z. S., and Li, Z. Q. (1984). Discovery of a normal T type new pathogenic strain to Lovrin10. Acta Cllegii Septentrionali Occidentali Agric. 4, 18–28.
Keppler, L. D., Baker, C. J., and Atkinson, M. M. (1989). Active oxygen production during a bacteria-induced hypersensitive reaction in tobacco suspensio cells. Phytopathology 79, 974–978. doi: 10.1094/Phyto-79-974
Kitagawa, K., Skowyra, D., Elledge, S. J., Harper, J. W., and Hieter, P. (1999). SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell 4, 21–33. doi: 10.1016/S1097-2765(00)80184-7
Liu, Y. L., Schiff, M., Marathe, R., and Dinesh-Kumar, S. P. (2002). Tobacco Rar1, EDS1and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. doi: 10.1046/j.1365-313X.2002.01297.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mur, L. A. J., Kenton, P., Atzorn, R., Miersch, O., and Wasternack, C. (2006). The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140, 249–262. doi: 10.1104/pp.105.072348
Muskett, P. R., Kahn, K., Austin, M. J., Moisan, L. J., Sadanandom, A., Shirasu, K., et al. (2002). Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens. Plant Cell 14, 979–992. doi: 10.1105/tpc.001040
Nürnberger, T., and Lipka, V. (2005). Non-host resistance in plants: new insights into an old phenomenon. Mol. Plant Pathol. 6, 335–345. doi: 10.1111/j.1364-3703.2005.00279.x
Pajerowska, K. M., Parker, J. E., and Gebhardt, C. (2005). Potato homologs of Arabidopsis thaliana genes functional in defense signaling — Identification, genetic mapping, and molecular cloning. Mol. Plant Microbe Interact. 18, 1107–1119. doi: 10.1094/MPMI-18-1107
Panwar, V., McCallum, B., and Bakkeren, G. (2013). Endogenous silencing of Puccinia triticina pathogenicity genes through in planta-expressed sequences leads to the suppression of rust diseases on wheat. Plant J. 73, 521–532. doi: 10.1111/tpj.12047
Parker, J. E., Holub, E. B., Frost, L. N., Falk, A., Gunn, N. D., and Daniels, M. J. (1996). Characterization of edsl, a mutation in Arabidopsis suppressing resistance to Peronospora parasíiíca specified by severa1 different RPP genes. Plant Cell 8, 2033–2046. doi: 10.1105/tpc.8.11.2033
Periyannan, S., Moore, J., Ayliffe, M., Bansal, U., Wang, X., Huang, L., et al. (2013). The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science 341, 786–788. doi: 10.1126/science.1239028
Scofield, S. R., Huang, L., Brandt, A. S., and Gill, B. S. (2005). Development of a virus induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol. 138, 2165–2173.38. doi: 10.1104/pp.105.061861
Segarra, G., Jáuregui, O., Casanova, E., and Trillas, I. (2006). Simultaneous quantitative LC-ESI-MS/MS analyses of salicylic acid and jasmonic acid in crude extracts of Cucumis sativus under biotic stress. Phytochemistry 67, 395–401. doi: 10.1016/j.phytochem.2005.11.017
Shang, Y., Li, X., Cui, H., He, P., Thilmony, R., Chintamanani, S., et al. (2006). RAR1, a central player in plant immunity, is targeted by Pseudomonas syringae effector AvrB. Proc. Natl. Acad. Sci. U.S.A. 103, 19200–19205. doi: 10.1073/pnas.0607279103
Shirasu, K., Lahaye, T., Tan, M. W., Zhou, F. S., Azevedo, C., and Schulze-Lefert, P. (1999). A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99, 355–366. doi: 10.1016/S0092-8674(00)81522-6
Shirasu, K., and Schulze-Lefert, P. (2003). Complex formation, promiscuity and multi-functionality: protein interactions in disease resistance pathways. Trends Plant Sci. 8, 252–258. doi: 10.1016/S1360-1385(03)00104-3
Spoel, S. H., Koornneef, A., Claessens, S. M., Korzelius, J. P., Van Pelt, J. A., Mueller, M. J., et al. (2003). NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15, 760–770. doi: 10.1105/tpc.009159
Stakman, E. C., Stewart, D. M., and Loegering, W. Q. (1962). Identification of Physiological Races of Puccinia graminis var. tritici. Washington, DC: US Department of Agricultural Publication, E617.
Tai, Y. S. (2008). Interactome of signaling networks in wheat: the protein-protein interaction between TaRAR1 and TaSGT1. Mol. Biol. Rep. 35, 337–343. doi: 10.1007/s11033-007-9091-5
Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D. B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x
Tornero, P., Merritt, P., Sadanandom, A., Shirasu, K., Innes, R. W., and Dangl, J. L. (2002). RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell 14, 1005–1015. doi: 10.1105/tpc.001032
Wang, X., Wang, X., Duan, Y., Yin, S., Zhang, H., Huang, L., et al. (2013). TaAbc1, a member of Abc1-like family involved in hypersensitive response against the stripe rust fungal pathogen in wheat. PLoS ONE 8:e58969. doi: 10.1371/journal.pone.0058969
Wang, Y., Gao, M., Li, Q., Wang, L., Wang, J., Jeon, J. S., et al. (2008). OsRAR1 and OsSGT1 physically interact and function in rice basal disease resistance. Mol. Plant Microbe Int. 21, 294–303. doi: 10.1094/MPMI-21-3-0294
Xu, J., Audenaert, K., Hofte, M., and De Vleesschauwer, D. (2013). Abscisic acid promotes susceptibility to the rice leaf blight pathogen Xanthomonas oryzae pv oryzae by suppressing salicylic acid-mediated defenses. PLoS ONE 8:e67413. doi: 10.1371/journal.pone.0067413
Keywords: TaRar1, Puccinia striiformis f. sp. tritici, salicylic acid, reactive oxygen species, virus-induced gene silencing
Citation: Wang X, Wang Y, Liu P, Ding Y, Mu X, Liu X, Wang X, Zhao M, Huai B, Huang L and Kang Z (2017) TaRar1 Is Involved in Wheat Defense against Stripe Rust Pathogen Mediated by YrSu. Front. Plant Sci. 8:156. doi: 10.3389/fpls.2017.00156
Received: 18 September 2016; Accepted: 25 January 2017;
Published: 14 February 2017.
Edited by:
Brigitte Mauch-Mani, University of Neuchâtel, SwitzerlandReviewed by:
Ulrich Schaffrath, RWTH Aachen University, GermanyRoger Wise, Iowa State University, USA Antony Chapman contributed to the review of Roger Wise
Copyright © 2017 Wang, Wang, Liu, Ding, Mu, Liu, Wang, Zhao, Huai, Huang and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Huang, lhuang@montana.edu Zhensheng Kang, kangzs@nwsuaf.edu.cn
 Xiaojing Wang
Xiaojing Wang Yaru Wang
Yaru Wang Peng Liu2
Peng Liu2 Yan Ding
Yan Ding Xiaoqian Mu
Xiaoqian Mu Zhensheng Kang
Zhensheng Kang