- 1Institute of Biochemistry and Biology, University of Potsdam, Potsdam, Germany
- 2Max Planck Institute of Molecular Plant Physiology, Potsdam, Germany
The Arabidopsis thaliana NAC transcription factor JUNGBRUNNEN1 (AtJUB1) regulates growth by directly repressing GA3ox1 and DWF4, two key genes involved in gibberellin (GA) and brassinosteroid (BR) biosynthesis, respectively, leading to GA and BR deficiency phenotypes. AtJUB1 also reduces the expression of PIF4, a bHLH transcription factor that positively controls cell elongation, while it stimulates the expression of DELLA genes, which are important repressors of growth. Here, we extend our previous findings by demonstrating that AtJUB1 induces similar GA and BR deficiency phenotypes and changes in gene expression when overexpressed in tomato (Solanum lycopersicum). Importantly, and in accordance with the growth phenotypes observed, AtJUB1 inhibits the expression of growth-supporting genes, namely the tomato orthologs of GA3ox1, DWF4 and PIF4, but activates the expression of DELLA orthologs, by directly binding to their promoters. Overexpression of AtJUB1 in tomato delays fruit ripening, which is accompanied by reduced expression of several ripening-related genes, and leads to an increase in the levels of various amino acids (mostly proline, β-alanine, and phenylalanine), γ-aminobutyric acid (GABA), and major organic acids including glutamic acid and aspartic acid. The fact that AtJUB1 exerts an inhibitory effect on the GA/BR biosynthesis and PIF4 genes but acts as a direct activator of DELLA genes in both, Arabidopsis and tomato, strongly supports the model that the molecular constituents of the JUNGBRUNNEN1 growth control module are considerably conserved across species.
Introduction
Gibberellins (GAs) and brassinosteroids (BRs) play important roles in the regulation of plant growth and development. Because of their biological relevance, many key aspects of the metabolism and signaling of both phytohormones have been revealed in the past (Depuydt and Hardtke, 2011; Bai et al., 2012). GAs control processes like for example seed germination, leaf expansion, shoot elongation, flowering, fruit set and growth (Olszewski et al., 2002; Yamaguchi, 2008; Mutasa-Göttgens and Hedden, 2009; Seymour et al., 2013). Similarly, BRs function as growth-promoting hormones mediating many aspects of plant growth and development including the stimulation of growth through cell division and cell elongation, and the adaptation to environmental cues (Clouse, 2011). Mutants deficient in BR biosynthesis or perception exhibit various degrees of phenotypic similarity to GA-deficient mutants including dwarfism, short petioles and hypocotyls, curled dark-green leaves, and delayed flowering (Clouse and Sasse, 1998; Fleet and Sun, 2005).
With respect to the later steps of GA biosynthesis, GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox) are particularly important for controlling the levels of bioactive GAs (Phillips et al., 1995; Xu et al., 1995; Yamaguchi et al., 1998). The bioactive GAs produced by GA3ox enzymes are perceived by a soluble GA receptor, GIBBERELLIN-INSENSITIVE DWARF1 (GID1) (Ueguchi-Tanaka et al., 2005). The binding of GA to GID1 triggers the destruction of DELLA proteins by the ubiquitin-proteasome pathway, or their inactivation through a degradation-independent mechanism (Ariizumi et al., 2008, 2013; Ueguchi-Tanaka et al., 2008). DELLAs are major transcriptional growth repressors (Martí et al., 2007; Bai et al., 2012). In the absence of active GAs, DELLAs accumulate allowing them to interact with the transcription factors (TFs) PHYTOCHROME-INTERACTING FACTOR4 (PIF4) and BRASSINAZOLE-RESISTANT1 (BZR1) to inhibit their DNA-binding ability; this restricts cell elongation controlled by the two TFs thereby resulting in reduced plant growth (De Lucas et al., 2008; Bai et al., 2012).
Gibberellins and DELLAs have also been reported to control reproductive organ size and fruit development in several species (Martí et al., 2007; Dorcey et al., 2009; Fuentes et al., 2012). In tomato (Solanum lycopersicum), overexpression of SlDELLA (which is identical to SlGAI) suppresses fruit development by inhibiting cell expansion (Martí et al., 2007).
Brassinosteroids are synthesized from campesterol via two pathways, the early and the late C-6 oxidation pathways (Fujioka and Sakurai, 1997). DWARF4 (DWF4) encodes a cytochrome P450 protein that catalyzes sterol C-22 α-hydroxylation, a rate-limiting step in BR biosynthesis (Choe et al., 1998, 2001; Kim et al., 2006; Divi and Krishna, 2010). The BR signal is perceived by a membrane-bound receptor kinase, BRASSINOSTEROID INSENSITIVE1 (BRI1). BRs bind to BRI1 and initiate a signal transduction pathway which ultimately results in the dephosphorylation and thereby stabilization of BZR1 and BRI1-EMS SUPPRESSOR1 (BES1) (Wang et al., 2002; He et al., 2005; Yin et al., 2005), two TFs regulating BR-responsive genes to promote growth (Sun et al., 2010; Clouse, 2011; Yu et al., 2011). BR and GA signals are integrated through the direct interaction of BZR1 and BES1 with DELLAs, and they enhance each other’s signaling through inactivation and degradation of DELLA proteins, respectively (Bai et al., 2012; Gallego-Bartolomé et al., 2012; Li et al., 2012). In the presence of BRs, active forms of BZR1 and BES1 attenuate the transcriptional activity of DELLAs. On the other hand, in the absence of GA, accumulation of DELLAs and their binding to BZR1 or BES1 prevents their dephosphorylation and thus results in their inactivation. BR-deficient and -insensitive mutants have, e.g., been identified in Arabidopsis thaliana, pea (Pisum sativum), tomato (Lycopersicum solanum), and rice (Oryza sativa) (Clouse and Sasse, 1998; Bishop, 2003; Vert et al., 2005; Clouse, 2011). BR biosynthesis pathways appear to be conserved between Arabidopsis, tomato, and pea (Bancoş et al., 2002). Molecular genetic manipulation of BR biosynthesis and signaling has been used to improve crop yield and stress tolerance (Divi and Krishna, 2009). It has been shown that BR mutants in rice and barley have increased grain yield and productivity due to erect leaves and reduced plant height (Chono et al., 2003; Morinaka et al., 2006). Controlling plant architecture and height to obtain miniature ornamental plants has been achieved by co-suppression of genes controlling GA and BR biosynthesis pathways (Xie et al., 2014). Several studies have shown that BRs are required for normal fruit development. For example, BR levels are high in developing fruits of tomato (Montoya et al., 2005). Furthermore, tomato fruit ripening is accelerated by application of BRs, but is delayed by treatment with the BR biosynthesis inhibitor brassinazole (Vidya Vardhini and Rao, 2002; Lisso et al., 2006). In tomato, the BR-deficient dwarf mutant shows delayed flowering and fruit ripening. Dry mass content, starch and sugar levels are reduced in dwarf tomato fruits, but are rescued by BR application, indicating that BRs are required for normal fruit development in this species (Lisso et al., 2006).
We recently identified the Arabidopsis thaliana NAC transcription factor JUNGBRUNNEN1 (JUB1; in the following called AtJUB1) as a further element of the GA/BR control network. More specifically, AtJUB1 suppresses the expression of GA3ox1 and DWF4, two central genes of GA and BR biosynthesis, respectively, by directly binding to their promoters. Furthermore, AtJUB1 directly activates the expression of the DELLA encoding genes GAI and RGL1. The repression of the phytohormone biosynthesis genes and the activation of the DELLA genes leads to an accumulation of DELLA proteins in AtJUB1 overexpressors and growth characteristics typical of low-GA and low-BR mutants, mediated by the inhibition of cell elongation (Shahnejat-Bushehri et al., 2016). AtJUB1 further inhibits cell elongation by directly suppressing the expression of PIF4 (Shahnejat-Bushehri et al., 2016).
Here, we report that overexpression of AtJUB1 in tomato results in GA and BR deficiency phenotypes similar to those observed in Arabidopsis AtJUB1 overexpressors. Furthermore, we show that AtJUB1 directly regulates the expression of tomato genes orthologous to GA3ox1, DWF4, GAI, and PIF4 from Arabidopsis. Our study thus reveals considerable similarity of the GA/BR-related gene regulatory network controlled by JUB1 across species.
Materials and Methods
General
Tomato orthologs of Arabidopsis genes were identified using the PLAZA 3.0 database1 (Proost et al., 2015). Gene annotations were done using the PLAZA 3.0 and Sol Genomics2 database, and information taken from the literature. Quantitative real-time polymerase chain reaction (qRT-PCR) primers were designed using QuantPrime (Arvidsson et al., 2008); only primer pairs giving high PCR amplification efficiencies tested on at least 20 different RNAs isolated from diverse tomato tissues were chosen for the experiments performed here. Sequences of primers used for qRT-PCR and chromatin immunoprecipitation-polymerase chain reaction (ChIP-PCR) are given in Supplementary Table S1.
Plant Material and Growth Conditions
To generate AtJUB1-OX tomato plants, S. lycopersicum cv. Moneymaker was transformed with the 35S:AtJUB1-GFP construct previously reported (Wu et al., 2012) by Agrobacterium-mediated transformation. Seeds were germinated on full-strength Murashige-Skoog (MS) medium containing 2% (w/v) sucrose in a plant growth cabinet at 22°C, at a 16/8 h light (100–120 mmol photons m−2 s−1)/dark cycle. Two-week-old seedlings were transplanted to soil (potting and quartz sand; 2:1 [v/v]) and grown in the greenhouse at 25°C under a 16/8 h light (500 mmol photons m−2 s−1)/dark cycle, in individual pots (18 cm diameter). After transfer to soil, plants were fertilized with 5 g/pot Complesal Perfekt (15% N, 5% P2O5, 20% K2O, 2% MgO, 7% S; Manna, Duesseldorf, Germany); two further fertilizations (with each 4 g/pot) followed when the first flowers opened, and when the first stage B fruits appeared.
Quantitative Real-Time PCR
Total RNA from frozen powdered tissue (200 mg fruit, or 100 mg leaf) was isolated using Trizol reagent (Life Technologies). cDNA synthesis and qRT-PCR were performed as described (Caldana et al., 2007; Balazadeh et al., 2008). Prior to cDNA synthesis, RNA was treated with DNase (Ambion) to eliminate genomic DNA contamination. Purity of DNase-treated RNA was assessed with intron-specific primers on gene Solyc01g056340 using qRT-PCR. Prior to gene expression analysis, cDNA quality was determined using GAPDH-3′ and GAPDH-5′ (Solyc04g009030) primer pairs. Real-time qPCR was performed with SYBR Green (Applied Biosystems) and gene-specific primers, using an ABI PRISM 7900HT sequence detection system (Applied Biosystems). GAPDH served as reference gene for data analysis.
Chromatin Immunoprecipitation (ChIP)
The chromatin extract was isolated as previously described (Kaufmann et al., 2010) from leaves of mature AtJUB1-OX (35S:AtJUB1-GFP) plants, and wild type plants served as control. Anti-GFP antibody (Abcam) was used to immunoprecipitate protein–DNA complexes. After protein digestion, DNA was purified by QIAquick PCR Purification kit (Qiagen) and analyzed by qPCR using primers flanking AtJUB1 binding sites in tomato promoters. As a negative control for ChIP-qPCR data analysis, we used a primer pair amplifying the 5′ upstream regulatory region of Solyc01g090460, which lacks an AtJUB1 binding site. ChIP-qPCR data were analyzed as described (Wu et al., 2012).
Microarray Analysis
Total RNA was isolated from AtJUB1-OX and wild type fruits at MG and B+7 stages using Trizol reagent and quality-checked as for qRT-PCR experiments. Three μg of quality-checked RNA was processed for transcriptome profiling using GeneChip Tomato Genome Arrays (Affymetrix). Probe preparation and microarray hybridizations were performed by ATLAS Biolabs (Berlin, Germany). Data analysis was performed as in Balazadeh et al. (2010). Expression of differentially expressed genes (transgenic versus wild type) was reassessed by qRT-PCR using RNA isolated from plants of two independent biological experiments. Microarray expression data are available from the NCBI Gene Expression Omnibus (GEO) repository3 under accession number GSE87507.
Profiling of Primary Metabolites by GC–MS
Fruit materials were extracted in 100% methanol at 70°C for 15 min. After centrifugation, the resultant supernatant was dried under vacuum, and the residue was derivatized for 120 min at 37°C (in 50 μl of 20 mg ml-1 methoxyamine hydrochloride in pyridine) followed by a 30 min treatment at 37°C with 50 μl of N-methyl-N-(trimethylsilyl)-trifluoroacetamide (MSTFA). The gas chromatography–mass spectrometry (GC–MS) system used was a gas chromatograph coupled to a time-of-flight mass spectrometer (Pegasus III, Leco). An autosampler system (PAL) injected the samples. Helium was used as carrier gas at a constant flow rate of 2 ml s−1 and gas chromatography was performed on a 30 m DB-35 column. The injection temperature was 230°C and the transfer line and ion source were set to 250°C. The initial temperature of the oven (85°C) increased at a rate of 15°C min−1 up to a final temperature of 360°C. After a solvent delay of 180 s mass spectra were recorded at 20 scans s−1 with m/z 70–600 scanning range. Chromatograms and mass spectra were evaluated by using Chroma TOF 1.0 (Leco) and TagFinder 4.0 software (Roessner et al., 2001; Schauer et al., 2005).
Results
Tomato Plants Overexpressing AtJUB1 Exhibit Morphological Characteristics of GA- and BR-Deficient Mutants
To understand the potential role of the JUNGBRUNNEN1 TF in tomato, we generated transgenic plants (S. lycopersicum L. cv. Moneymaker) overexpressing AtJUB1-GFP (Wu et al., 2012) under the control of the Cauliflower Mosaic Virus (CaMV) 35S promoter. We previously showed that the JUB1-GFP fusion protein is fully functional when expressed in plants (Wu et al., 2012). We obtained more than 20 independent transgenic lines and determined the level of AtJUB1 expression in some of them by qRT-PCR in leaves (Figure 1A; lines OX1, OX2, and OX3) and fruits of different developmental stages (Figure 1B; OX2; thereafter called AtJUB1-OX). The formation of full-length AtJUB1 transcript in transgenic tomato plants was confirmed by PCR analysis of cDNA of leaves and fruits, using primers specific for AtJUB1 (Supplementary Figures S1A,B). Accumulation of AtJUB1-GFP fusion protein in nuclei of AtJUB1-OX tomato leaves was confirmed by confocal microscopy (Figure 1C). AtJUB1-OX lines displayed distinct morphological changes compared to the Moneymaker wild type (Figure 2). Strong AtJUB1 overexpressors (line OX2) had smaller shoots than wild type and developed smaller leaves (Figures 2A,B), while growth was not much reduced in plants moderately overexpressing AtJUB1, including OX1 and OX3 (not shown). Furthermore, AtJUB1-OX plants developed curly and dark-green leaves, a phenotype not observed in the wild type (Figure 2C). Additionally, overexpression of AtJUB1 in tomato delayed flowering, similar to Arabidopsis (Shahnejat-Bushehri et al., 2016). AtJUB1-OX tomato plants flowered when the main shoot had ∼13 leaves beneath the first inflorescence compared to ∼9 leaves in the wild type (Figure 2E). At the productive stage, AtJUB1-OX plants had smaller flowers and fruits than the wild type (Figures 2F,G), and fruit development and ripening were delayed. Fruits of AtJUB1-OX plants reached the mature green stage (MG) 4 days later than wild type tomatoes (measured as days after pollination) and ripening of AtJUB1-OX fruits was delayed by ∼6 days (Figure 2H). Upon visual inspection, AtJUB1-OX fruits showed reduced pigmentation at the breaker+7d (B+7) stage compared to wild type fruits at the same stage (not shown). Taken together, many of the growth phenotypes observed in AtJUB1-OX tomato plants were similar to those of Arabidopsis plants overexpressing AtJUB1, suggesting that JUB1 controls a similar set of growth-related genes in both species possibly including genes affecting hormone metabolism and signaling (Shahnejat-Bushehri et al., 2016).
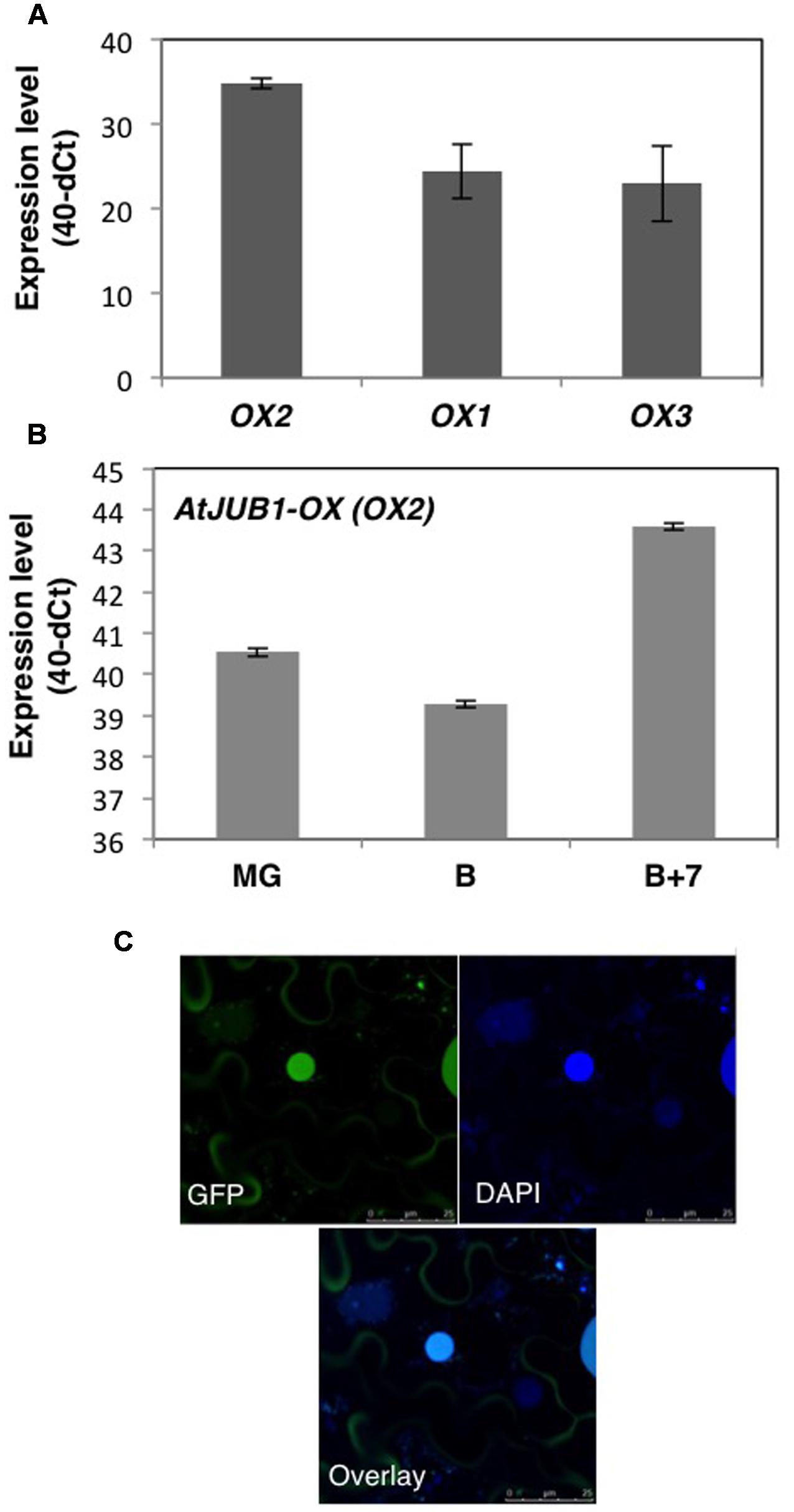
FIGURE 1. Analysis of AtJUB1-OX transgenic plants. (A) Expression level of AtJUB1 in leaves of transgenic lines OX1, OX2, and OX3 compared to wild type (WT). Mean ± SD (n = 3). (B) Expression of AtJUB1 in fruits of line OX2 at mature green (MG), breaker (B), and breaker+7d (B+7) stages. Mean ± SD (n = 3). For quantitative real-time polymerase chain reaction (qRT-PCR) analysis, values are expressed as the difference between an arbitrary value of 40 and dCt, so that high 40-dCt value indicates high gene expression level. (C) Nuclear localization of GFP-tagged AtJUB1 protein in leaves of transgenic tomato plants, imaged by confocal microscopy. Top left, fluorescence microscopy; top right, DAPI staining; bottom, overlay. Some AtJUB1-GFP protein appears to reside in the cytoplasm surrounding the central vacuole, likely due to incomplete nuclear import.
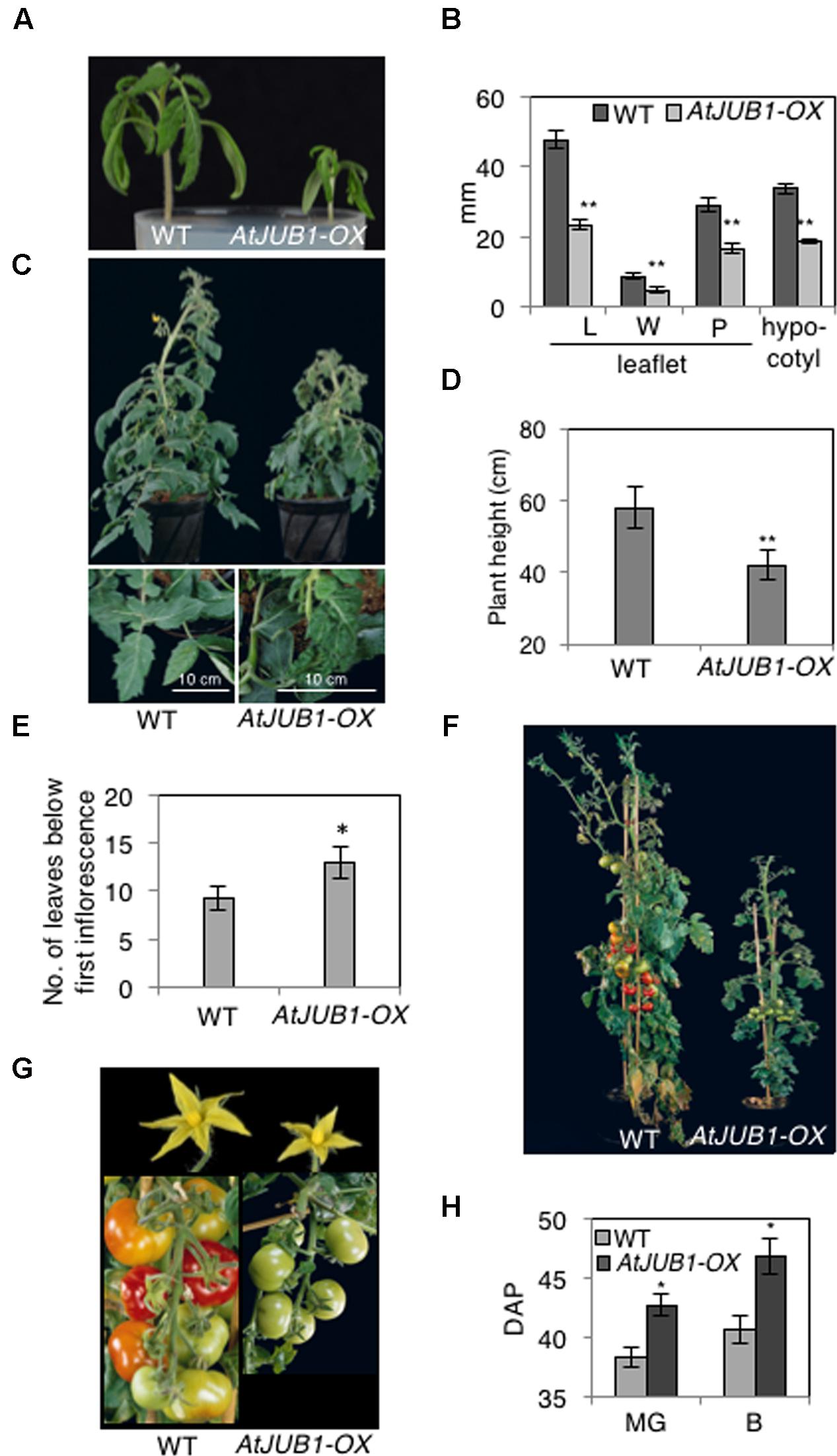
FIGURE 2. Phenotype of AtJUB1-OX tomato plants at different developmental stages. (A) Reduced growth in 3-week-old AtJUB1-OX transgenic plants strongly overexpressing AtJUB1, compared to wild type (WT). (B) Leaf parameters: length (L) and width (W) of leaflet blade; petiolule (P) length; hypocotyl length of 3-week-old AtJUB1-OX and WT tomato seedlings. Leaf parameters were determined for the two subterminal leaflets of leaf number 1. (C) Vegetative growth phenotype of 6-week-old plants and their leaves (lower panel). (D) Height of plants. (E) Flowering time of AtJUB1-OX and WT plants measured by counting the number of leaves on the date that the first inflorescence arose. (F) Phenotype of plants at reproductive stage. (G) Flowers and fruits of plants shown in (F). (H) Fruit ripening was scored in AtJUB1-OX plants compared to WT by counting the days after pollination (DAP) to reach the mature green and breaker stages. Data in graphs represent mean ± SD (n = 10) and ∗p < 0.05, ∗∗p < 0.01 by Student’s t-test. Asterisks denote significant differences to WT.
AtJUB1 Directly Regulates GA- and BR-Related Genes in Tomato
Previously, we reported that AtJUB1 extends longevity, dampens intracellular H2O2 level, and enhances tolerance to various abiotic stresses partly through direct transcriptional regulation of DREB2A, an important TF involved in the response to various abiotic stresses (Sakuma et al., 2006a,b; Wu et al., 2012). Furthermore, AtJUB1 restricts growth and primes plants for stress tolerance in a DELLA-dependent manner by regulating a complex transcriptional module composed of key components of GA and BR pathways. In Arabidopsis, AtJUB1 directly suppresses the expression of GA3ox1 (GA biosynthesis), DWF4 (BR biosynthesis), and PIF4 (regulation of cell elongation), but activates the expression of the GAI and RGA DELLA genes (Shahnejat-Bushehri et al., 2016). Considering the growth phenotypes observed for the AtJUB1-OX tomato plants (see above), we tested whether similar genes are targets of AtJUB1 in tomato. First, we checked the expression of tomato orthologs of Arabidopsis DWF4, GA3ox1, GAI and RGL1 in AtJUB1-OX tomato leaves and in fruits at the MG, B and B+7 stages. As shown in Figure 3A, expression of the AtDWF4 tomato orthologs Solyc02g085360 (denoted SlDWF4-1 in the following) and Solyc04g080650 (SlDWF4-2) was not altered in fruits of AtJUB1-OX fruits at all stages tested, but was strongly reduced in leaves of mature plants compared to wild type. Similarly, expression of the AtGA3ox1 tomato orthologs Solyc06g066820 (denoted SlGA3ox1-1 in the following) and Solyc03g119910 (SlGA3ox1-2) was significantly downregulated in leaves of mature AtJUB1-OX plants. Moreover, SlGA3ox1-1 was lower expressed in fruits of AtJUB1-OX plants at the breaker (B) stage, while expression of SlGA3ox1-2 was decreased in MG and B+7 fruits compared to wild type fruits of the same stage. Interestingly, expression of the tomato DELLA genes Solyc11g011260 (SlGAI) and Solyc01g009840 (SlGAI-like) was slightly and strongly upregulated, respectively, in MG, B and B+7 fruits of AtJUB1-OX plants, consistent with the reduced size of the fruits. Expression of SlGAI and SlGAI-like was not altered in leaves of AtJUB1-OX plants relative to wild type. Expression of Solyc07g043580 (SlPIF4), the positive regulator of cell elongation, was strongly suppressed by AtJUB1 in MG and B, but not affected in B+7 fruits or leaves of AtJUB1-OX plants.
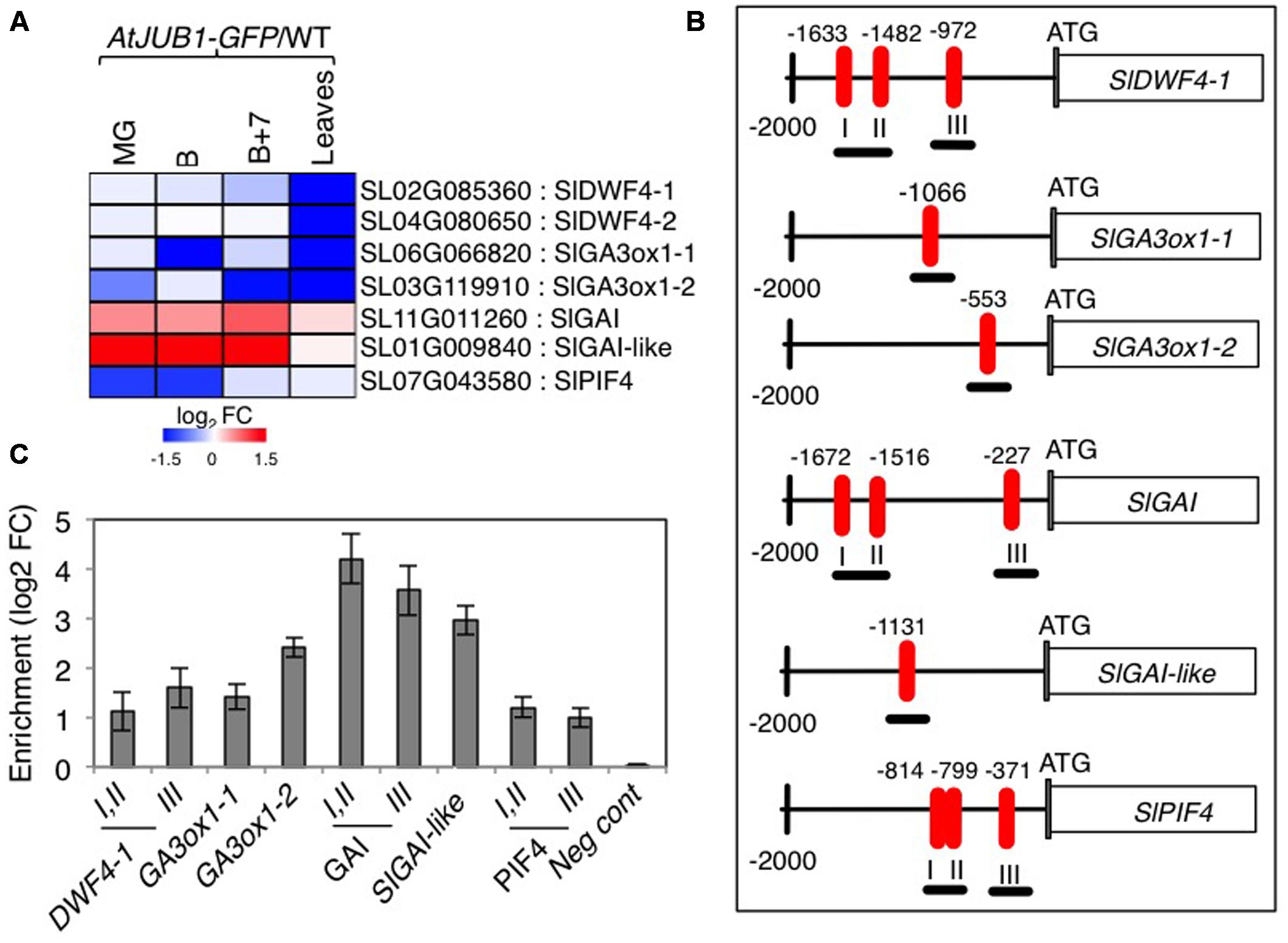
FIGURE 3. Direct regulation of GA- and BR-associated genes by AtJUB1. (A) Heat map showing the expression of orthologs of Arabidopsis DWF4, GA3ox1, GAI, RGL1, and PIF4 in tomato line OX2 at mature green (MG), breaker (B), breaker+7d (B+7) stages and leaves of mature plants compared to wild type (WT). The log2 fold change scale is indicated under the heat map. Complete data are given in Supplementary Table S2. (B) Schematic diagram showing positions of AtJUB1 binding sites (numbered I, II and III if three sites were present) in the promoters of genes orthologous to the AtJUB1 targets in Arabidopsis. ChIP amplicons are indicated as black thick underlines. (C) ChIP assays showing the direct binding of AtJUB1 to the promoters of SlDWF4-1, SlGA3ox1-1, SlGA3ox1-2, SlGAI, SlGAI-like and SlPIF4. Values were normalized to the values for Solyc01g090460 (promoter lacking AtJUB1 binding site), as a negative control. Error bars represent mean ± SD (two independent biological replicates, each with three technical replicates). FC, fold change.
The fact that JUB1 regulates orthologous genes in Arabidopsis and tomato when overexpressed indicates that this TF obeys a conserved function in plants as part of a complex regulatory network associated with GA and BR pathways. To consolidate this conclusion we next analyzed the promoters of selected GA- and BR-related genes whose expression is affected by AtJUB1 overexpression (shown in Figure 3A and Supplementary Table S2), and found that SlDWF4-1, SlGA3ox1-1, SlGA3ox1-2, SlGAI and SlGAI-like harbor the conserved AtJUB1 binding site (Wu et al., 2012) in their 5′ upstream regulatory regions (Figure 3B). An AtJUB1 binding site is also present in the promoter of SlPIF4 (Figure 3B). To test whether AtJUB1 is able to bind to the selected putative target genes in tomato, we performed chromatin immunoprecipitation followed by qPCR using leaves of mature AtJUB1-OX plants. Binding of AtJUB1 was observed for all promoters tested, while no binding was detected for a promoter region lacking an AtJUB1 binding site (negative control) (Figure 3C). Taken together, our data indicate a direct transcriptional regulation of SlDWF4-1, SlGA3ox1-1, SlGA3ox1-2, SlGAI, SlGAI-like and SlPIF4 by AtJUB1 in vivo.
Differentially Expressed Genes in AtJUB1 Overexpressing Tomato Fruits
To identify genes globally affected by AtJUB1 during fruit development and ripening, we compared the transcriptomes of AtJUB1-OX and wild type fruits at MG and B+7 stages, using microarray analysis. We selected 92 genes that were differentially expressed in MG and/or B+7 fruits of AtJUB1-OX compared to wild type (at a 1.5-fold cut-off in increase or decrease of gene expression) and re-analyzed their expression in additional biological replicates by qRT-PCR, which confirmed altered expression of 60 genes (Figure 4 and Supplementary Table S3). The AtJUB1-regulated genes fall into different functional categories including TFs, cell wall-modifying enzymes, antioxidant enzymes, enzymes involved in flavonoid biosynthesis, and hormone-related genes. Among the TFs, RIPENING INHIBITOR (RIN; Solyc05g012020), a MADS-box TF with a key function in fruit ripening, was significantly downregulated in AtJUB1-OX tomato fruits at MG stage. Similarly, ETHYLENE RESPONSE FACTOR H15 (SlERF.H15; Solyc06g050520), which shows increased expression at the onset of ripening and together with other ERFs was suggested to be involved in regulating the ripening process (Liu et al., 2016), was downregulated in AtJUB1-OX fruits at MG stage. Furthermore, expression of cell wall-related genes including a pectin esterase family protein (Solyc06g051960), a cellulase synthase (Solyc12g056580), and expansins (Solyc06g051800, Solyc02g088100, and Solyc05g007830) was reduced in AtJUB1-OX fruits. A group of genes associated with cellular oxidation and peroxidation processes, including several peroxidases (Solyc01g006300, Solyc03g080150, Solyc06g076630), were lower expressed in AtJUB1-OX fruits. Peroxidases are considered to affect fruit softening and ripening (Kumar et al., 2016). Expression of Solyc12g005350, encoding dihydroflavonol-4-reductase, a key gene in anthocyanin biosynthesis (Holton and Cornish, 1995), is slightly decreased in AtJUB1-OX. Among hormone-related genes, expression of 1-aminocyclopropane-1-carboxylate (ACC) synthase (ACS; Solyc08g081550) and ACC oxidases (ACO; Solyc02g036350, Solyc07g026650, Solyc11g072110) is either not altered or repressed in AtJUB1-OX fruits. These genes are known to be involved in ethylene biosynthesis and their expression is induced during fruit ripening (Barry et al., 2000). Moreover, Solyc02g037550 and Solyc02g082450, members of the auxin efflux carrier family, are significantly downregulated in AtJUB1-OX fruits at stages MG and B+7. These proteins are involved in the distribution of auxin, a growth hormone controlling many aspects of fruit development and ripening (Gillaspy et al., 1993; Srivastava and Handa, 2005). Next, we searched for the binding site of AtJUB1 in the promoters of the differentially expressed genes. The absence of the full AtJUB1 binding site in the 5′ upstream regulatory regions of the 60 genes differentially expressed between AtJUB1-OX and wild type suggests that AtJUB1 indirectly, e.g., in conjunction with hormonal networks, regulates their expression.
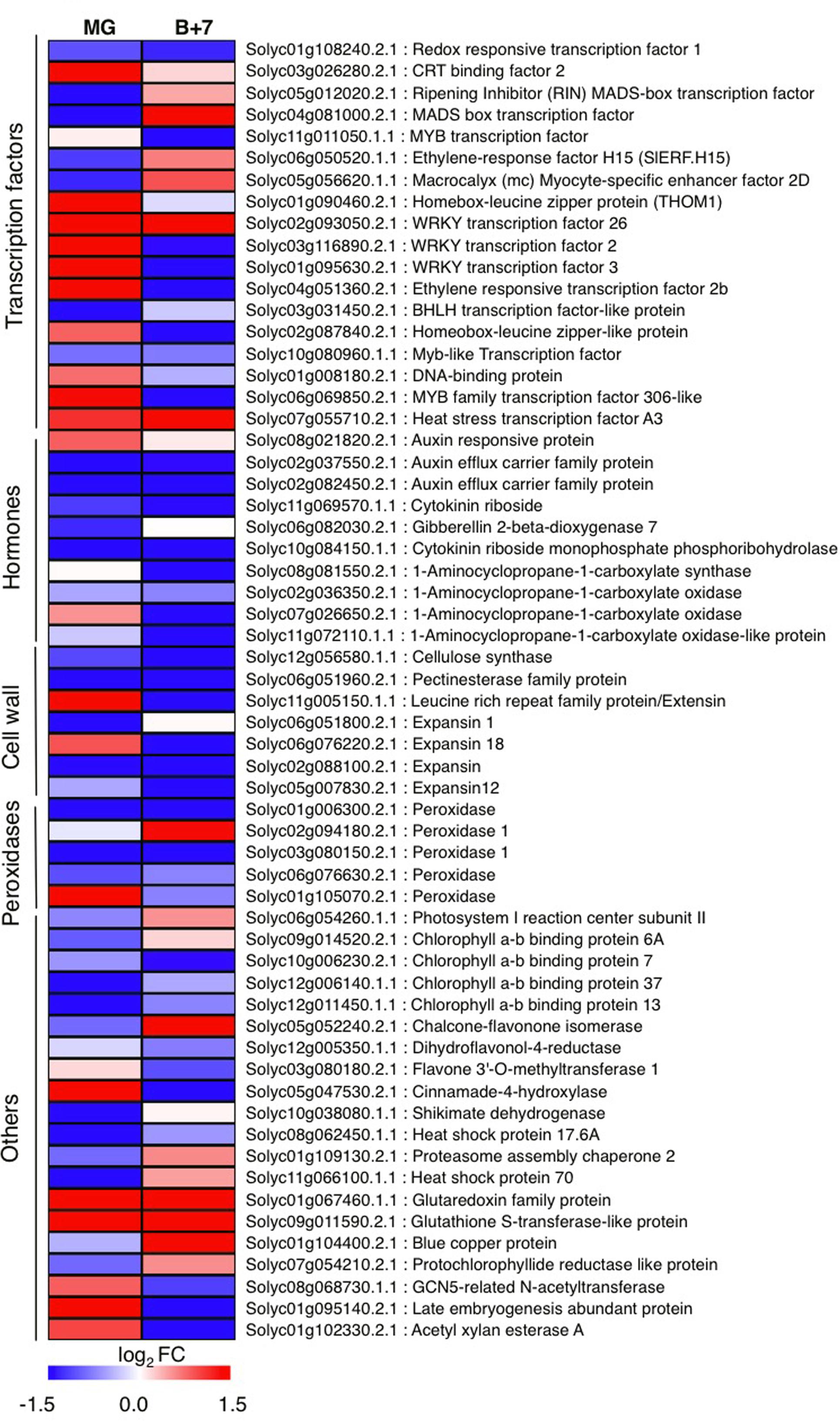
FIGURE 4. Quantitative real-time polymerase chain reaction analysis of AtJUB1-regulated genes in tomato fruits. Heat map showing differentially expressed genes in AtJUB1-OX fruits compared to wild type. Means of three experiments are shown; complete data are given in Supplementary Table S3. The log2 fold change scale is indicated under the heat map. Transcription factors, hormone metabolism-related, cell wall-related and peroxidase categories are shown.
AtJUB1 Affects Metabolite Profiles in Tomato Fruits
Metabolite profiling was performed on fruits of AtJUB1-OX and wild type plants at three independent ripening stages: mature green (MG), breaker (B) and red ripe fruit (B+7). Metabolites were separated and quantified by GC–MS. In total, 54 metabolites were quantified including amino acids, organic acids, sugars and a few miscellaneous compounds. Figure 5 shows the metabolic changes in the AtJUB1-OX line in comparison to wild type, where the relative changes are depicted in blue (decreased) or red (increased) (complete data are given in Supplementary Table S4). Thirty-four metabolites out of 54 (∼63%) were significantly increased in the overexpression line compared to the wild type, with similar trends being observed at all three ripening stages. In general, the content of amino acids increased in the overexpression line compared to the wild type. Among these the levels of proline, β-alanine and phenylalanine showed considerable increases being a maximum of 12-, 14- and 7-fold higher in the AtJUB1-OX line compared to wild type. However, the greatest change was observed in the gamma-aminobutyric acid (GABA) content which was approximately 34-fold higher in ripe fruit of the AtJUB1-OX line compared to wild type fruit at the same stage (B+7). In addition, a marked increase was observed in major organic acids, particularly for glutamic acid (12-fold), aspartic acid (4.8-fold) and pyroglutamic acid (3.6-fold). Among the detected disaccharides, the levels of trehalose and maltose were mostly changed. By contrast, relatively few metabolites decreased in the three ripening stages in the overexpression line compared to wild type with only succinic acid, galactinol and tyramine displaying this behavior, although urea, glycerol, raffinose, glyceric acid and quinic acids were less abundant in the overexpression line compared to wild type in red ripe fruit (B+7).
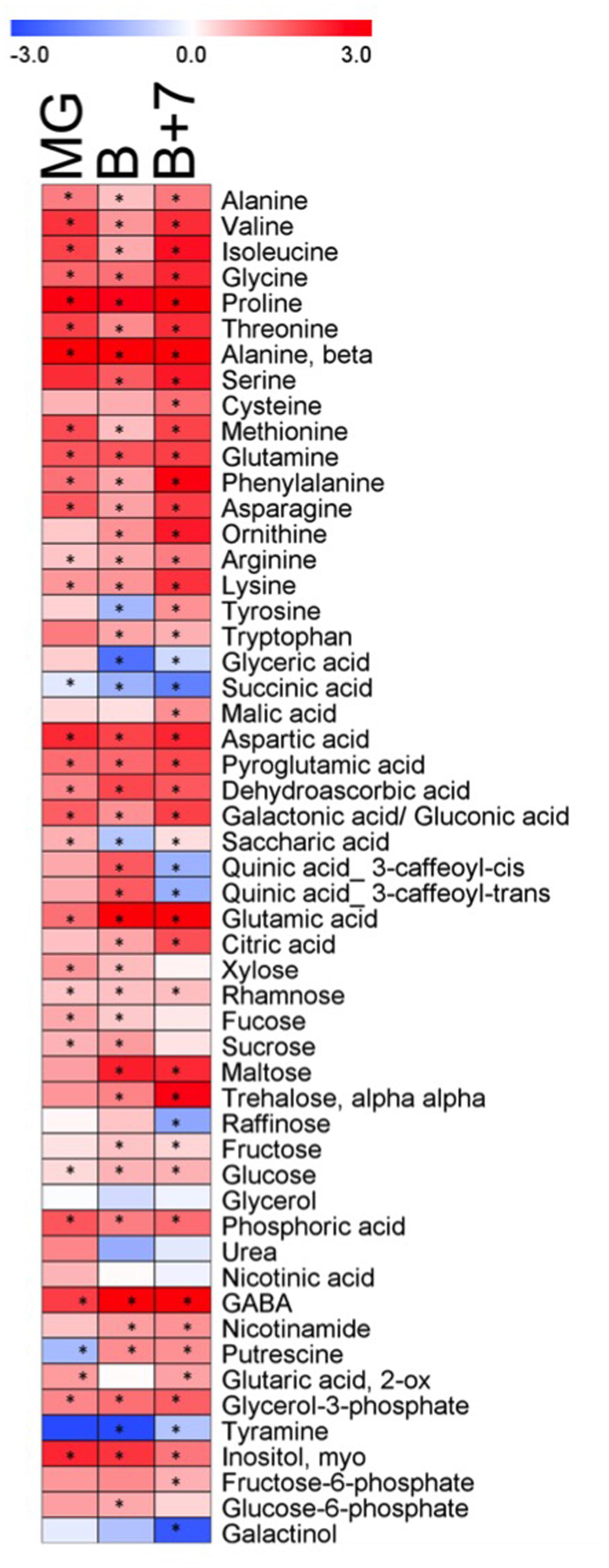
FIGURE 5. Heat map showing the metabolite changes in fruits of AtJUB1-OX plants in comparison with wild type at three ripening stages. Mature green fruit (MG), breaker stage (B), and red ripe fruit (B+7) were analyzed. Red or blue indicate that the metabolite content is increased or decreased, respectively. Asterisks denote significant difference (p ≤ 0.01, Student’s t-test) in AtJUB1-OX compared to wild type; complete data are given in Supplementary Table S4.
Discussion
In this study, we characterized the biological function exerted by the Arabidopsis NAC transcription factor AtJUB1 (JUNGBRUNNEN1) when expressed in the heterologous plant tomato (S. lycopersicum L. cv. Moneymaker). Similar to Arabidopsis, overexpression of AtJUB1 in tomato resulted in reduced plant size, dark-green curled leaves, a delay in flowering, and reduced reproductive organ size compared to wild type (Shahnejat-Bushehri et al., 2016); in addition, fruit growth and ripening was delayed in AtJUB1-OX tomatoes. In Arabidopsis, AtJUB1 regulates various growth-related genes by directly binding to their promoters: it represses expression of GA3ox1 and DWF4, which encode key genes of GA and BR biosynthesis, respectively, and of PIF4, a growth promoting TF of the bHLH family. In contrast, AtJUB1 activates the expression of the DELLA genes GAI and RGA, which encode important repressors of growth. Collectively, the transcriptional control exerted by AtJUB1 leads to the characteristic GA- and BR-deficiency phenotypes observed for AtJUB1 overexpressors, which is associated with a reduction of the levels of bioactive GAs and BRs and an accumulation of DELLA proteins (Shahnejat-Bushehri et al., 2016).
To gain insights into the mechanisms through which AtJUB1 affects growth in tomato, we analyzed genes orthologous to AtJUB1 targets in Arabidopsis (GA3ox1, DWF4, GAI, RGL1, and PIF4) in different tissues of AtJUB1-OX tomato plants and found that expression of all genes was altered, like in Arabidopsis itself.
In Arabidopsis, GA3ox1 encodes a key enzyme involved in the biosynthesis of bioactive GAs, and it is expressed at relatively high levels at all stages of development (Mitchum et al., 2006); ga3ox1 knockout mutants display a semi-dwarf phenotype due to reduced levels of bioactive GAs (Talon et al., 1990; Chiang et al., 1995). Here, we observed that expression of the GA3ox1 tomato orthologs, SlGA3ox1-1 and SlGA3ox1-2, was down-regulated in leaves as well as in three fruit developmental stages (MG, B and B+7) of AtJUB1-OX plants compared to wild type. The reduced expression of the rate-limiting GA biosynthetic enzymes may lead to lower levels of GA, thus contributing to GA-deficient phenotypes in AtJUB1-OX tomatoes similarly to Arabidopsis plants overexpressing AtJUB1 (Shahnejat-Bushehri et al., 2016).
Furthermore, expression of SlDWF4-1 and SlDWF4-1, orthologs of the Arabidopsis DWF4 gene, was strongly reduced in leaves of AtJUB1-OX tomatoes, sharing this behavior with the repression of DWF4 by AtJUB1 in Arabidopsis (Shahnejat-Bushehri et al., 2016). DWF4 encodes a sterol C-22 α-hydroxylase that catalyzes a rate-limiting step in BR biosynthesis (Choe et al., 1998, 2001; Kim et al., 2006; Divi and Krishna, 2010); its reduced expression in tomato may contribute to the BR-deficiency growth characteristics observed.
Additionally, elevated expression was observed for the DELLA genes SlGAI and SlGAI-like in AtJUB1-OX fruits compared to wild type in the three developmental stages tested. While DELLA proteins are encoded by five genes in Arabidopsis (GAI, RGA, RGL1, RGL2, and RGL3) (Achard and Genschik, 2009), two DELLAs, namely SlGAI (identical to SlDELLA; Martí et al., 2007) and SlGAI-like, are encoded by the tomato genome (as deduced from the PLAZA 3.0 web database; Proost et al., 2015), and were analyzed here. GA promotes plant growth by removing DELLA proteins, whereas at low GA levels DELLAs accumulate and repress growth by directly inactivating several TFs including PIF4, a positive regulator of cell elongation (De Lucas et al., 2008; Feng et al., 2008; Arnaud et al., 2010; Leivar and Quail, 2011). In Arabidopsis, we found that AtJUB1 directly up-regulates DELLA genes thereby promoting their accumulation. Additionally, AtJUB1 represses the expression of PIF4, which further restricts cell elongation (Shahnejat-Bushehri et al., 2016). Similarly, reduced SlPIF4 expression was evident in MG and B stage fruits of AtJUB1-OX tomato plants compared to wild type. Activation of DELLAs and repression of SlPIF4 by AtJUB1 may contribute to the smaller fruit size of AtJUB1-OX plants.
In addition to the differential expression of GA-/BR-related genes in tomato upon AtJUB1 overexpression, the presence of the AtJUB1 binding site in their 5′ upstream regulatory regions and the presence of an AtJUB1 ortholog in the tomato genome and other plants (with in most case one predicted ortholog, see PLAZA 3.01) strongly argue for a JUB1 regulatory network that has a similar architecture and function in Arabidopsis and tomato, and likely other plants. In support of our hypothesis, ChIP-qPCR revealed a significant enrichment of promoter regions of all tomato GA- and BR-related gene orthologous to the AtJUB1 targets in Arabidopsis, while no enrichment was detected for a promoter region lacking an AtJUB1 binding site.
Additionally, we were interested in understanding the molecular mechanisms associated with the delay of fruit ripening in AtJUB1-OX plants. Toward this end, we performed transcriptome analysis of AtJUB1-OX and wild type fruits at MG and B+7 stages. Genes differentially expressed in AtJUB1-OX fruits compared to wild type encode TFs, genes involved in ethylene synthesis or signaling, cell wall modification, flavonoid biosynthesis, peroxidases, and others. Of note, expression of several ethylene-related ripening genes such as ACS (ACC synthase) and ACO (ACC oxidase), MADS-RIN, and SLERF.H15, among others, was repressed in AtJUB1-OX tomatoes. Tomato is a climacteric fruit that requires an ethylene burst for normal fruit ripening (Alexander and Grierson, 2002). In fruits of several species, including, e.g., tomato and melon, inhibition of key enzymes of ethylene biosynthesis (ACO and ACS) by antisense RNA leads to strong delay in ripening, demonstrating the requirement of ethylene in the process (Oeller et al., 1991; Picton et al., 1993; Ayub et al., 1996). With respect to TFs, the MADS-box protein RIPENING INHIBITOR (RIN) has been particularly well-characterized. It exerts its function in fruit ripening by directly regulating the expression of genes involved in ethylene production, lycopene accumulation, chlorophyll degradation, among several other physiological processes (Fujisawa et al., 2013). Ripening is completely abolished in the rin mutant (Vrebalov et al., 2002).
Apart from ACS and ACO, a PECTIN ESTERASE gene (Solyc06g051960) is repressed in AtJUB1-OX fruits compared to wild type; similarly, several pectin esterase encoding genes are repressed in the rin mutant during fruit ripening (Kumar et al., 2016). The absence of the AtJUB1 binding site in the promoters of the ripening-related genes, however, indicates indirect regulation by AtJUB1; the details of the mechanisms that underlie this regulation are currently unknown.
With respect to metabolites, in general, similar significant changes were found in AtJUB1 Arabidopsis and tomato overexpressors: 67 and 63% of the metabolites detected were significantly different between wild type and overexpression lines, respectively. However, here (in tomato AtJUB1 overexpressors) the relative changes were much higher than what we previously reported in Arabidopsis AtJUB1 overexpressors (Wu et al., 2012). In tomato, 44 out of 53 metabolites were significantly increased and nine decreased at least in one developmental stage. On the other hand, in Arabidopsis AtJUB1 overexpressors 21 metabolites were significantly increased and 17 decreased compared to wild type (Table 1). Comparing the metabolite changes indicates that 20% of the metabolites (nine increased and two decreased) overlapped and were significantly different in AtJUB1 overexpression plants compared to wild type, in both, tomato and Arabidopsis. On the other hand, 17 metabolites (34%) found to be significantly different between overexpression and wild type plants showed different behaviors in tomato and Arabidopsis.

TABLE 1. Summary comparing the number of metabolites in Arabidopsis and tomato AtJUB1 overexpression lines.
In tomato AtJUB1 overexpressor plants proline showed one of the most drastic increases in overexpression lines at all developmental stages, which increased up to 12-fold higher than in the wild type. We made a similar observation in Arabidopsis AtJUB1 overexpression plants where proline was increased up to twofold compared to wild type (Wu et al., 2012). Proline is known to be involved in the response to various environmental stresses. Among other metabolites that showed significant changes and similar trends to proline are trehalose, malic acid, and citric acid.
Another metabolite that had strongly increased levels in fruits of tomato AtJUB1 overexpressors compared to wild type fruits is GABA, which was 4-, 8-, and 34-fold higher in the MG, B and B+7 stages, respectively (Figure 5 and Supplementary Table S4). In wild type fruits GABA levels accumulate before the breaker stage, but shortly thereafter GABA is rapidly catabolized (Akihiro et al., 2008), most likely to support the high respiratory demand of later stages of fruit development. The fact that GABA content remains elevated in AtJUB1 overexpressors is in accordance with the delayed fruit ripening observed in these plants. The biological function of GABA in tomato fruits is not well-known, but may include the regulation of pH to prevent cellular acidification during fruit development when organic acids are synthesized, or help to maintain glutamate import from source organs by converting it to GABA (through the action of glutamate decarboxylase) (reviewed in Takayama and Ezura, 2015). The high accumulation of glutamate (9- and 12-fold in B and B+7 fruits, respectively; Supplementary Table S4) in AtJUB1 overexpressors compared to wild type supports this conclusion. Alternatively, GABA, which functions in defense against pests and pathogens might protect the immature seeds in developing fruits (Takayama and Ezura, 2015).
Also strongly elevated in AtJUB1 overexpressor fruits throughout all developmental stages is β-alanine (10- to 14-fold higher than in wild type; Figure 5 and Supplementary Table S4). Typically, β-alanine decreases during tomato fruit ripening (Carrari et al., 2006); the fact that its level remains high in AtJUB1 overexpressors even at later stages of fruit development is also in accordance with the delayed ripening observed in these plants.
In addition, among the few metabolites showing a significant decrease in overexpression lines in tomato AtJUB1 plants, raffinose (0.4-fold) and galactinol (0.2-fold), these metabolites decreased up to 0.6-fold and 0.2-fold in Arabidopsis overexpression plants compared to wild type, respectively. In contrast to Arabidopsis AtJUB1 overexpression lines, some organic acids, in particularly glyceric acid and succinic acid, showed a significant decrease in the tomato AtJUB1 plants (0.7- and 0.2-fold, respectively) compared to wild type. Succinic acid is a breakdown product of GABA and indeed is mobilized to augment flux through the TCA cycle (Studart-Guimarães et al., 2007); the low level of succinic acid in the presence of high GABA concentration (see above) indicates a reduced conversion of GABA to succinic acid in AtJUB1 overexpressor fruits compared to wild type fruits.
An accumulation of GABA levels was previously observed in fruits of transgenic tomato plants in which expression of GABA transaminase (GABA-T) was suppressed by RNA interference. GABA-T converts GABA to succinic semialdehyde which is then further metabolized to succinic acid by succinic semialdehyde dehydrogenase. Interestingly, inhibition of GABA-T led to plant dwarfism, concomitant with the over-accumulation of GABA (Koike et al., 2013), similar to the metabolic and growth phenotypes we observed here for AtJUB1 overexpressors. In addition, the excessive accumulation of GABA may negatively affect cell elongation (Renault et al., 2011), leading to the dwarfed phenotype of plants strongly overexpressing JUB1.
Taken together, we show here that the NAC TF JUNGBRUNNEN1, which we previously reported to control key elements of the GA/BR metabolism and signaling network in Arabidopsis thaliana (Shahnejat-Bushehri et al., 2016), exerts a similar control over orthologous genes in tomato. A key observation of our study is that in both plants, AtJUB1 negatively controls the expression of growth promoting genes (GA3ox1, DWF4, and PIF4), while it positively regulates the expression of growth repressing genes (DELLAs). This finding suggests that other, as yet unknown elements of the JUNGBRUNNEN1 growth control module are considerably conserved between species. Interestingly, JUB1 is a single-copy gene in most sequenced plant genomes, including dicots and monocots, with only few exceptions; e.g., two AtJUB1 orthologs are reported by PLAZA 3.0 for maize (Zea mays), poplar (Populus trichocarpa) and soybean (Glycine max), while three orthologs are reported for field mustard (Brassica rapa). The multiple regulatory roles that JUB1 exerts on central components of GA- and BR-related metabolism and signaling, which is key to the control of plant growth, suggests that JUB1 activity is finely tuned during growth and in different organs. Future research should focus on identifying further control elements of the JUB1 module in Arabidopsis and other species.
Author Contributions
SB and BM-R conceived the idea for the study and supervised the work. SB and BM-R wrote the manuscript, with contributions from SS-B and AA. SS-B, AA, and NM performed the phenotype analysis; VT performed the confocal imaging; AA, NM, and VT performed the expression profiling; SS-B performed the ChIP-PCR experiments. SA and AF performed the metabolite profiling and analyzed the respective data.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
SB thanks the Deutsche Forschungsgemeinschaft (DFG) for funding (BA 4769/2-1). We thank Karin Koehl and her team (MPI of Molecular Plant Physiology) for plant care, and Eugenia Maximova (MPI of Molecular Plant Physiology) for help with microscopy. Financial support by the University of Potsdam and the MPI of Molecular Plant Physiology is gratefully acknowledged.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00214/full#supplementary-material
FIGURE S1 | AtJUB1 transcript in transgenic tomato plants. (A) Presence of full-length AtJUB1 transcript (arrow) in leaves of transgenic plants OX1, OX2 and OX3, confirmed by PCR using primers specific for AtJUB1. (B) Presence of AtJUB1 transcript in fruits and leaves of AtJUB1-OX (= OX2) tomato plants (arrow). Samples were taken from fruits of different developmental stages (MG, B, B+7). As expected, no AtJUB1 transcript is detected in wild type tomato plants (WT).
TABLE S1 | List of primers.
TABLE S2 | Expression of GA- and BR-associated genes.
TABLE S3 | Gene expression in tomato fruits.
TABLE S4 | Primary metabolites.
Footnotes
- ^ http://bioinformatics.psb.ugent.be/plaza/
- ^ https://www.solgenomics.net/
- ^ https://www.ncbi.nlm.nih.gov/geo/
References
Achard, P., and Genschik, P. (2009). Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J. Exp. Bot. 60, 1085–1092. doi: 10.1093/jxb/ern301
Akihiro, T., Koike, S., Tani, R., Tominaga, T., Watanabe, S., Iijima, Y., et al. (2008). Biochemical mechanism on GABA accumulation during fruit development in tomato. Plant Cell Physiol. 49, 1378–1389. doi: 10.1093/pcp/pcn113
Alexander, L., and Grierson, D. (2002). Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J. Exp. Bot. 53, 2039–2055.
Ariizumi, T., Hauvermale, A. L., Nelson, S. K., Hanada, A., Yamaguchi, S., and Steber, C. M. (2013). Lifting della repression of Arabidopsis seed germination by nonproteolytic gibberellin signaling. Plant Physiol. 162, 2125–2139. doi: 10.1104/pp.113.219451
Ariizumi, T., Murase, K., Sun, T.-P., and Steber, C. M. (2008). Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. Plant Cell 20, 2447–2459. doi: 10.1105/tpc.108.058487
Arnaud, N., Girin, T., Sorefan, K., Fuentes, S., Wood, T. A., Lawrenson, T., et al. (2010). Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 24, 2127–2132. doi: 10.1101/gad.593410
Arvidsson, S., Kwasniewski, M. M., Riano-Pachon, D., and Mueller-Roeber, B. (2008). QuantPrime–a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9:465. doi: 10.1186/1471-2105-9-465
Ayub, R., Guis, M., Ben Amor, M., Gillot, L., Roustan, J. P., Latché, A., et al. (1996). Expression of ACC oxidase antisense gene inhibits ripening of cantaloupe melon fruits. Nat. Biotechnol. 14, 862–866. doi: 10.1038/nbt0796-862
Bai, M.-Y., Shang, J.-X., Oh, E., Fan, M., Bai, Y., Zentella, R., et al. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14, 810–817. doi: 10.1038/ncb2546
Balazadeh, S., Riaño-Pachón, D. M., and Mueller-Roeber, B. (2008). Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol. (Stuttg). 10(Suppl. 1), 63–75. doi: 10.1111/j.1438-8677.2008.00088.x
Balazadeh, S., Siddiqui, H., Allu, A.-D., Matallana-Ramirez, L.-P., Caldana, C., Mehrnia, M., et al. (2010). A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 62, 250–264. doi: 10.1111/j.1365-313X.2010.04151.x
Bancoş, S., Nomura, T., Sato, T., Molnár, G., Bishop, G. J., Koncz, C., et al. (2002). Regulation of transcript levels of the Arabidopsis cytochrome P450 genes involved in brassinosteroid biosynthesis. Plant Physiol. 130, 504–513.
Barry, C. S., Llop-Tous, M. I., and Grierson, D. (2000). The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol. 123, 979–986. doi: 10.1104/pp.123.3.979
Bishop, G. J. (2003). Brassinosteroid mutants of crops. J. Plant Growth Regul. 22, 325–335. doi: 10.1007/s00344-003-0064-1
Caldana, C., Scheible, W.-R., Mueller-Roeber, B., and Ruzicic, S. (2007). A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods 3, 7. doi: 10.1186/1746-4811-3-7
Carrari, F., Baxter, C., Usadel, B., Urbanczyk-Wochniak, E., Zanor, M.-I., Nunes-Nesi, A., et al. (2006). Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol. 142, 1380–1396. doi: 10.1104/pp.106.088534
Chiang, H. H., Hwang, I., and Goodman, H. M. (1995). Isolation of the Arabidopsis thaliana GA4 locus. Plant Cell 7, 195–201. doi: 10.1105/tpc.7.2.195
Choe, S., Dilkes, B. P., Fujioka, S., Takatsuto, S., Sakurai, A., and Feldmann, K. A. (1998). The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22alphα-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10, 231–243. doi: 10.2307/3870701
Choe, S., Fujioka, S., Noguchi, T., Takatsuto, S., Yoshida, S., and Feldmann, K. A. (2001). Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 26, 573–582.
Chono, M., Honda, I., Zeniya, H., Yoneyama, K., Saisho, D., Takeda, K., et al. (2003). A semidwarf phenotype of barley uzu results from a nucleotide substitution in the gene encoding a putative brassinosteroid receptor. Plant Physiol. 133, 1209–1219.
Clouse, S. D. (2011). Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23, 1219–1230. doi: 10.1105/tpc.111.084475
Clouse, S. D., and Sasse, J. M. (1998). Brassinosteroids: essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 427–451.
De Lucas, M., Davière, J.-M., Rodríguez-Falcón, M., Pontin, M., Iglesias-Pedraz, J. M., Lorrain, S., et al. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484. doi: 10.1038/nature06520
Depuydt, S., and Hardtke, C. S. (2011). Hormone signalling crosstalk in plant growth regulation. Curr. Biol. 21, R365–R373. doi: 10.1016/j.cub.2011.03.013
Divi, U. K., and Krishna, P. (2009). Brassinosteroid: a biotechnological target for enhancing crop yield and stress tolerance. Nat. Biotechnol. 26, 131–136. doi: 10.1016/j.nbt.2009.07.006
Divi, U. K., and Krishna, P. (2010). Overexpression of the brassinosteroid biosynthetic gene AtDWF4 in Arabidopsis seeds overcomes abscisic acid-induced inhibition of germination and increases cold tolerance in transgenic seedlings. J. Plant Growth Regul. 29, 385–393.
Dorcey, E., Urbez, C., Blázquez, M. A., Carbonell, J., and Perez-Amador, M. A. (2009). Fertilization-dependent auxin response in ovules triggers fruit development through the modulation of gibberellin metabolism in Arabidopsis. Plant J. 58, 318–332. doi: 10.1111/j.1365-313X.2008.03781.x
Feng, S., Martinez, C., Gusmaroli, G., Wang, Y., Zhou, J., Wang, F., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479. doi: 10.1038/nature06448
Fleet, C. M., and Sun, T. (2005). A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 8, 77–85.
Fuentes, S., Ljung, K., Sorefan, K., Alvey, E., Harberd, N. P., and Østergaard, L. (2012). Fruit growth in Arabidopsis occurs via DELLA-dependent and DELLA-independent gibberellin responses. Plant Cell 24, 3982–3996. doi: 10.1105/tpc.112.103192
Fujioka, S., and Sakurai, A. (1997). Biosynthesis and metabolism of brassinosteroids. Physiol. Plant 100, 710–715.
Fujisawa, M., Nakano, T., Shima, Y., and Ito, Y. (2013). A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25, 371–386. doi: 10.1105/tpc.112.108118
Gallego-Bartolomé, J., Minguet, E. G., Grau-Enguix, F., Abbas, M., Locascio, A., Thomas, S. G., et al. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 13446–13451. doi: 10.1073/pnas.1119992109
Gillaspy, G., Ben-David, H., and Gruissem, W. (1993). Fruits: a developmental perspective. Plant Cell 5, 1439–1451.
He, J.-X., Gendron, J. M., Sun, Y., Gampala, S. S. L., Gendron, N., Sun, C. Q., et al. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307, 1634–1638. doi: 10.1126/science.1107580
Holton, T. A., and Cornish, E. C. (1995). Genetics and biochemisry of anthocyanin biosynthesis. Plant Cell 7, 1071–1083.
Kaufmann, K., Muiño, J. M., Østerås, M., Farinelli, L., Krajewski, P., and Angenent, G. C. (2010). Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 5, 457–472. doi: 10.1038/nprot.2009.244
Kim, H. B., Kwon, M., Ryu, H., Fujioka, S., Takatsuto, S., Yoshida, S., et al. (2006). The regulation of DWARF4 expression is likely a critical mechanism in maintaining the homeostasis of bioactive brassinosteroids in Arabidopsis. Plant Physiol. 140, 548–557. doi: 10.1104/pp.105.067918
Koike, S., Matsukura, C., Takayama, M., Asamizu, E., and Ezura, H. (2013). Suppression of G-aminobutyric acid (GABA) transaminases induces prominent GABA accumulation, dwarfism and infertility in the tomato (Solanum lycopersicum L.). Plant Cell. Physiol. 54, 793–807. doi: 10.1093/pcp/pct035
Kumar, V., Irfan, M., Ghosh, S., Chakraborty, N., Chakraborty, S., and Datta, A. (2016). Fruit ripening mutants reveal cell metabolism and redox state during ripening. Protoplasma 253, 581–594. doi: 10.1007/s00709-015-0836-z
Leivar, P., and Quail, P. H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28. doi: 10.1016/j.tplants.2010.08.003
Li, Q.-F., Wang, C., Jiang, L., Li, S., Sun, S. S. M., and He, J.-X. (2012). An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci. Signal. 5:ra72. doi: 10.1126/scisignal.2002908
Lisso, J., Altmann, T., and Müssig, C. (2006). Metabolic changes in fruits of the tomato dx mutant. Phytochemistry 67, 2232–2238. doi: 10.1016/j.phytochem.2006.07.008
Liu, M., Gomes, B. L., Mila, I., Purgatto, E., Peres, L. E., Frasse, P., et al. (2016). Comprehensive profiling of ethylene response factor expression identifies ripening-associated ERF genes and their link to key regulators of fruit ripening in tomato. Plant Physiol. 170, 1732–1744. doi: 10.1104/pp.15.01859
Martí, C., Orzáez, D., Ellul, P., Moreno, V., Carbonell, J., and Granell, A. (2007). Silencing of DELLA induces facultative parthenocarpy in tomato fruits. Plant J. 52, 865–876. doi: 10.1111/j.1365-313X.2007.03282.x
Mitchum, M. G., Yamaguchi, S., Hanada, A., Kuwahara, A., Yoshioka, Y., Kato, T., et al. (2006). Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 45, 804–818. doi: 10.1111/j.1365-313X.2005.02642.x
Montoya, T., Nomura, T., Yokota, T., Farrar, K., Harrison, K., Jones, J. D. G., et al. (2005). Patterns of Dwarf expression and brassinosteroid accumulation in tomato reveal the importance of brassinosteroid synthesis during fruit development. Plant J. 42, 262–269. doi: 10.1111/j.1365-313X.2005.02376.x
Morinaka, Y., Sakamoto, T., Inukai, Y., Agetsuma, M., Kitano, H., Ashikari, M., et al. (2006). Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol. 141, 924–931. doi: 10.1104/pp.106.077081
Mutasa-Göttgens, E., and Hedden, P. (2009). Gibberellin as a factor in floral regulatory networks. J. Exp. Bot. 60, 1979–1989. doi: 10.1093/jxb/erp040
Oeller, P., Lu, M., Taylor, L., Pike, D., and Theologis, A. (1991). Reversible inhibition of tomato fruit senescence by antisense RNA. Science 254, 437–439.
Olszewski, N., Sun, T.-P., and Gubler, F. (2002). Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14, S61–S80.
Phillips, A. L., Ward, D. A., Uknes, S., Appleford, N. E., Lange, T., Huttly, A. K., et al. (1995). Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 108, 1049–1057.
Picton, S., Barton, S. L., Bouzayen, M., Hamilton, A. J., and Grierson, D. (1993). Altered fruit ripening and leaf senescence in tomatoes expressing an antisense ethylene-forming enzyme transgene. Plant J. 3, 469–481. doi: 10.1111/j.1365-313X.1993.tb00167.x
Proost, S., Van Bel, M., Vaneechoutte, D., Van de Peer, Y., Inzé, D., Mueller-Roeber, B., et al. (2015). PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res. 43, D974–D981. doi: 10.1093/nar/gku986
Renault, H., El Amrani, A., Palanivelu, R., Updegraff, E. P., Yu, A., Renou, J. P., et al. (2011). GABA accumulation causes cell elongation defects and a decrease in expression of genes encoding secreted and cell wall-related proteins in Arabidopsis thaliana. Plant Cell Physiol. 52, 894–908. doi: 10.1093/pcp/pcr041
Roessner, U., Luedemann, A., Brust, D., Fiehn, O., Linke, T., Willmitzer, L., et al. (2001). Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13, 11–29. doi: 10.1105/tpc.13.1.11
Sakuma, Y., Maruyama, K., Osakabe, Y., Qin, F., Seki, M., Shinozaki, K., et al. (2006a). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18, 1292–1309.
Sakuma, Y., Maruyama, K., Qin, F., Osakabe, Y., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2006b). Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. U.S.A. 103, 18822–18827.
Schauer, N., Steinhauser, D., Strelkov, S., Schomburg, D., Allison, G., Moritz, T., et al. (2005). GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 579, 1332–1337.
Seymour, G. B., Østergaard, L., Chapman, N. H., Knapp, S., and Martin, C. (2013). Fruit development and ripening. Annu. Rev. Plant Biol. 64, 219–241. doi: 10.1146/annurev-arplant-050312-120057
Shahnejat-Bushehri, S., Tarkowska, D., Sakuraba, S., and Balazadeh, S. (2016). Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signalling. Nat. Plants 2, 16013. doi: 10.1038/nplants.2016.13
Srivastava, A., and Handa, A. K. (2005). Hormonal regulation of tomato fruit development: a molecular perspective. J. Plant Growth Regul. 24, 67–82.
Studart-Guimarães, C., Fait, A., Nunes-Nesi, A., Carrari, F., Usadel, B., and Fernie, A. R. (2007). Reduced expression of succinyl-coenzyme A ligase can be compensated for by up-regulation of the gamma-aminobutyrate shunt in illuminated tomato leaves. Plant Physiol. 145, 626–639.
Sun, Y., Fan, X. Y., Cao, D. M., Tang, W., He, K., Zhu, J. Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19, 765–777. doi: 10.1016/j.devcel.2010.10.010
Takayama, M., and Ezura, H. (2015). How and why does tomato accumulate a large amount of GABA in the fruit? Front. Plant Sci. 6:612. doi: 10.3389/fpls.2015.00612
Talon, M., Koornneef, M., and Zeevaart, J. A. (1990). Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of the semidwarf ga4 and ga5 mutants. Proc. Natl. Acad. Sci. U.S.A. 87, 7983–7987.
Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., et al. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698.
Ueguchi-Tanaka, M., Hirano, K., Hasegawa, Y., Kitano, H., and Matsuoka, M. (2008). Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant. Plant Cell 20, 2437–2446. doi: 10.1105/tpc.108.061648
Vert, G., Nemhauser, J. L., Geldner, N., Hong, F., and Chory, J. (2005). Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 21, 177–201.
Vidya Vardhini, B., and Rao, S. S. R. (2002). Acceleration of ripening of tomato pericarp discs by brassinosteroids. Phytochemistry 61, 843–847.
Vrebalov, J., Ruezinsky, D., Padmanabhan, V., White, R., Medrano, D., Drake, R., et al. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296, 343–346.
Wang, Z. Y., Nakano, T., Gendron, J., He, J., Chen, M., Vafeados, D., et al. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2, 505–513.
Wu, A., Allu, A. D., Garapati, P., Siddiqui, H., Dortay, H., Zanor, M.-I., et al. (2012). JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24, 482–506. doi: 10.1105/tpc.111.090894
Xie, Q., Chen, G., Chen, X., Deng, L., Liu, Q., Zhang, Y., et al. (2014). Jointly silencing BoDWARF, BoGA20ox and BoSP (SELF-PRUNING) produces a novel miniature ornamental Brassica oleracea var. acephala f. tricolor variety. Mol. Breed. 34, 99–113.
Xu, Y. L., Li, L., Wu, K., Peeters, A. J., Gage, D. A., and Zeevaart, J. A. (1995). The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc. Natl. Acad. Sci. U.S.A. 92, 6640–6644.
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804
Yamaguchi, S., Smith, M. W., Brown, R. G., Kamiya, Y., and Sun, T. (1998). Phytochrome regulation and differential expression of gibberellin 3beta-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10, 2115–2126.
Yin, Y., Vafeados, D., Tao, Y., Yoshida, S., Asami, T., and Chory, J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120, 249–259. doi: 10.1016/j.cell.2004.11.044
Keywords: Arabidopsis, tomato, fruit, growth, transcription factor, gibberellic acid, brassinosteroid, DELLA proteins
Citation: Shahnejat-Bushehri S, Allu AD, Mehterov N, Thirumalaikumar VP, Alseekh S, Fernie AR, Mueller-Roeber B and Balazadeh S (2017) Arabidopsis NAC Transcription Factor JUNGBRUNNEN1 Exerts Conserved Control Over Gibberellin and Brassinosteroid Metabolism and Signaling Genes in Tomato. Front. Plant Sci. 8:214. doi: 10.3389/fpls.2017.00214
Received: 20 October 2016; Accepted: 06 February 2017;
Published: 07 March 2017.
Edited by:
Zuhua He, Shanghai Institute for Biological Sciences (CAS), ChinaReviewed by:
Xiaoquan Qi, Institute of Botany (CAS), ChinaJeffrey Charles Waller, Mount Allison University, Canada
Copyright © 2017 Shahnejat-Bushehri, Allu, Mehterov, Thirumalaikumar, Alseekh, Fernie, Mueller-Roeber and Balazadeh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Salma Balazadeh, balazadeh@mpimp-golm.mpg.de Bernd Mueller-Roeber, bmr@uni-potsdam.de
†Present address: Annapurna D. Allu, The Faculty of Veterinary and Agricultural Sciences, The University of Melbourne, Melbourne, VIC, Australia; Nikolay Mehterov, Department of Medical Biology, Medical University of Plovdiv, Plovdiv, Bulgaria
‡These authors are joint first authors.
 Sara Shahnejat-Bushehri
Sara Shahnejat-Bushehri Annapurna D. Allu
Annapurna D. Allu Nikolay Mehterov
Nikolay Mehterov Venkatesh P. Thirumalaikumar
Venkatesh P. Thirumalaikumar Saleh Alseekh
Saleh Alseekh Alisdair R. Fernie
Alisdair R. Fernie Bernd Mueller-Roeber
Bernd Mueller-Roeber Salma Balazadeh
Salma Balazadeh