- 1INRA, UMR BGPI INRA/CIRAD/SupAgro, Campus International de Baillarguet, Montpellier, France
- 2UMR – Interactions Plantes-Microorganismes-Environnement, IRD–Cirad–Université Montpellier, Institut de Recherche pour le Développement, Montpellier, France
Plants are constantly challenged by a wide range of pathogens and have therefore evolved an array of mechanisms to defend against them. In response to these defense systems, pathogens have evolved strategies to avoid recognition and suppress plant defenses (Brown and Tellier, 2011). Three recent reports dealing with the resistance of rice to Xanthomonas oryzae have added a new twist to our understanding of this fascinating co-evolutionary arms race (Ji et al., 2016; Read et al., 2016; Triplett et al., 2016). They show that pathogens also develop sophisticated effector mimics to trick recognition.
Transcription activator-like effectors (TALEs) are major virulence factors of Xanthomonas plant-pathogenic bacteria that modulate host transcription by trans-activating host susceptibility genes (Boch and Bonas, 2010). For this, TALEs possess a type III secretion signal mediating host cell translocation by the bacterial type III secretion system, nuclear localization signals (NLS) directing them into the plant nucleus and an activation domain that activates gene transcription in eukaryotic cells. In addition, TALEs possess a central repeat domain that mediates sequence specific DNA-binding. It is composed of variable numbers of a highly conserved 33–35 amino acid sequence in which residues 12 and 13, the so-called repeat variable di-residues (RVD), are hypervariable and determine the nucleotide binding specificity (Boch et al., 2009; Moscou and Bogdanove, 2009). Due to the discovery of the nucleotide binding specificity, a number of susceptibility genes targeted by Xanthomonas TALEs have been identified (Hutin et al., 2015a). Over the course of evolution, plants have acquired mutations in the promoter regions of susceptibility genes which abolish the binding and trans-activation by TALEs, hence leading to resistance by loss-of-susceptibility that is inherited in a recessive manner (Hutin et al., 2015b). Additionally, plants have evolved executor resistance genes whose transcription is induced by specific TALEs (upon binding to their promoters), resulting in a hypersensitive response, turning TALEs into avirulence (Avr) determinants (Zhang et al., 2015). The important role of the transcription machinery for TALE action in susceptibility and resistance is further highlighted by the recessive xa5 resistance gene, a natural allele of the gene for the transcription factor IIA gamma subunit 5 (TFIIA γ5). Direct interaction of TALEs with TFIIAγ5 from rice activates disease susceptibility genes (Yuan et al., 2016).
These TALE-based dominant or recessive resistances to xanthomonads differ profoundly from standard pathogen resistance in plants that relies on the recognition of patterns or effectors by immune receptors. The best studied case of pattern recognition is the rice Xa21 receptor kinase that recognizes by direct binding RaxX, a sulfated peptide widely present in Xanthomonas that can therefore be considered a microbial pattern (Pruitt et al., 2015). Other examples are the NLRs (Nucleotide-binding domain, leucine-rich repeat) Xa1, Bs2, and Bs4 that, respectively, confer resistance to specific Xanthomonas oryzae pv. oryzae (Xoo) and X. axonopodis pv. vesicatoria isolates (Yoshimura et al., 1998; Tai et al., 1999; Schornack et al., 2004). NLRs are immune receptors commonly found in plants that display a canonical multi-domain structure. At the N-terminal they have coiled-coil or TIR (Toll-Interleukin Receptor) domains, a central nucleotide-binding domain and a C-terminal leucine-rich repeat domain and which recognize cytoplasmic effectors in a direct or indirect manner (Ellis, 2016).
A recent report (Ji et al., 2016) demonstrates that several TALEs are recognized by rice Xa1 protein, a member of the NLR family. Remarkably, two other groups reported the recognition of several TALE effectors by the Xo1 locus a yet uncharacterized gene, which the authors argue to be likely a NLR protein (Read et al., 2016; Triplett et al., 2016). This rather non-specific recognition of TALEs does not lead to broad-spectrum resistance to Xoo and X. oryzae pv. oryzicola (Xoc) because TALE-derived (truncTALES and iTALES) effectors can suppress this resistance suggesting they might act as effector mimics. In the case of Xo1 this suppression is independent from DNA binding at least for the Tal2h truncTALE (Read et al., 2016). It will be interesting to determine whether Xa1 inhibition by iTALES is independent of DNA binding as well, or if its suppression is by a different mechanism. These findings provide an exciting novel insight into the evolutionary arms race between plants and pathogens and reveals new functions of TALEs. In these studies, the function of different combinations of TALEs was evaluated by reintroducing them into Xoo strains depleted for most TALEs (Ji et al., 2016; Triplett et al., 2016). The first discovery was that both Xa1 and the newly identified Xo1 locus trigger resistance by recognizing several unrelated X. oryzae TALEs that differ in their target sequence and their number of central repeats (Ji et al., 2016; Triplett et al., 2016). The structural motifs that are recognized and the mode of recognition, either direct or indirect, are not yet defined. However, it appears that at least 3.5 central repeats regardless of their RVDs are required to trigger both Xa1 and Xo1 resistance (Ji et al., 2016; Triplett et al., 2016). Additionally, in the case of Xo1, TALE recognition and activation of resistance does not require the activation domain and is independent of DNA-binding (Read et al., 2016; Triplett et al., 2016). As a consequence, Xa1 recognizes an entire effector family, and not as other NLRs, only individual effectors in a very specific manner. This is a new feature of NLR activity that has previously been rather associated with membrane bound receptor complexes which can in certain cases recognize entire, widely distributed effector families (Böhm et al., 2014).
The second major discovery of these studies is that truncated TALE gene variants, previously considered as pseudogenes, designated as truncTALEs (Read et al., 2016) can act as interfering TALEs (iTALEs; Ji et al., 2016). Thus, some truncTALEs can block Xa1- and Xo1-mediated recognition of full-length TALEs, hence acting as iTALEs and suppressing resistance. These iTALEs/truncTALEs are characterized by specific deletions in the conserved N- and C-terminal sequences, require at least 3.5 central repeats and do not depend on specific RVDs, suggesting that their activity does not rely on DNA-binding or the direct regulation of the transcription of target genes. As suggested by Read et al. (2016) at least in the case of Xa1 it is tempting to speculate that iTALEs compete with full-length TALEs for binding to the NLR receptor but, on the contrary of the genuine ligand, do not activate the immune receptor, thus acting as dominant suppressors. Suppressors of NLR-mediated resistance have been identified in various phytopathogenic organisms but in the cases where they have been molecularly identified they correspond to effectors that are unrelated to the recognized Avr effectors (Houterman et al., 2008; Bourras et al., 2015; Plissonneau et al., 2016). A completely new and extraordinary twist in microbial virulence comes from the discovery that the oomycete pathogen Phytophthora sojae deploys an effector mimic PsXLP1 which resembles the functional virulence protein PsXEG1 to disrupt plant defense (Ma et al., 2017). The discovery of iTALEs/truncTALEs suggests that Xoo and Xoc also deploy effectors that mimic other, recognized effectors. Similarly to PsXLP1/PsXEG1, iTALEs/truncTALEs can be viewed as effector mimics that the pathogen uses to interfere with recognition by the plant. These two examples open novel exciting dimensions in the understanding of plant–pathogen co-evolution. Whether other effectors act in a similar way, as suppressors of resistance, is an unexplored question and may force us to consider the large effector repertoires with a completely new perspective in which some effectors may in fact be effector mimics.
How TALEs are recognized by Xa1 and Xo1, directly or indirectly, and how truncTALEs/iTALEs interfere with recognition is unknown. However, for the case of Xa1, it is tempting to speculate that it might involve the BED-type zinc finger domain which is integrated in the N-terminal region of this NLR protein (Kroj et al., 2016). In fact, we and others showed recently that unconventional integrated domains in NLRs are involved in the detection of effectors either by direct binding or by posttranslational modifications; thus these integrated domains may mimic the true effector target proteins and therefore act as integrated decoys (Cesari et al., 2013, 2014; Le Roux et al., 2015; Maqbool et al., 2015; Sarris et al., 2015, 2016; Kroj et al., 2016). The BED domain has been shown to bind DNA and is present in transposases and transcription factors (Hayward et al., 2013). ZBED, a rice protein containing three BED domains was recently shown to be required for full resistance to the rice blast fungus (Kroj et al., 2016), suggesting a role of BED proteins in plant–pathogen interactions. It could therefore be that TALEs recruit BED proteins as co-factors for the trans-activation of target genes and are trapped by Xa1 according to the integrated decoy model (e.g., by binding directly the BED domain of Xa1). iTALEs may interfere with TALE recognition in a dominant-negative manner by occupying some binding sites of Xa1 for TALEs and by this outcompete TALEs for Xa1-binding without triggering receptor activation (Figure 1). This model is consistent with the finding of Ji et al. (2016) that Xa1-mediated TALE recognition and iTALE-mediated suppression of resistance require nuclear localization of TALEs and iTALEs. By contrast, Read et al. (2016) report that deleting the putative NLS in the iTALE Tal2h does not affect its ability to suppress Xo1. Yet, the localization of the Tal2h NLS mutant was not analyzed and we cannot rule out that Xo1-mediated TALE recognition and truncTALE suppression differs from Xa1 and can happen in the cytoplasm or in the nucleus in a NLS-independent manner. It will therefore be particularly interesting to investigate the location of Xa1 and Xo1, and to determine whether Xo1 is a NLR protein to gain a better insight into the potential similarities or differences of both.
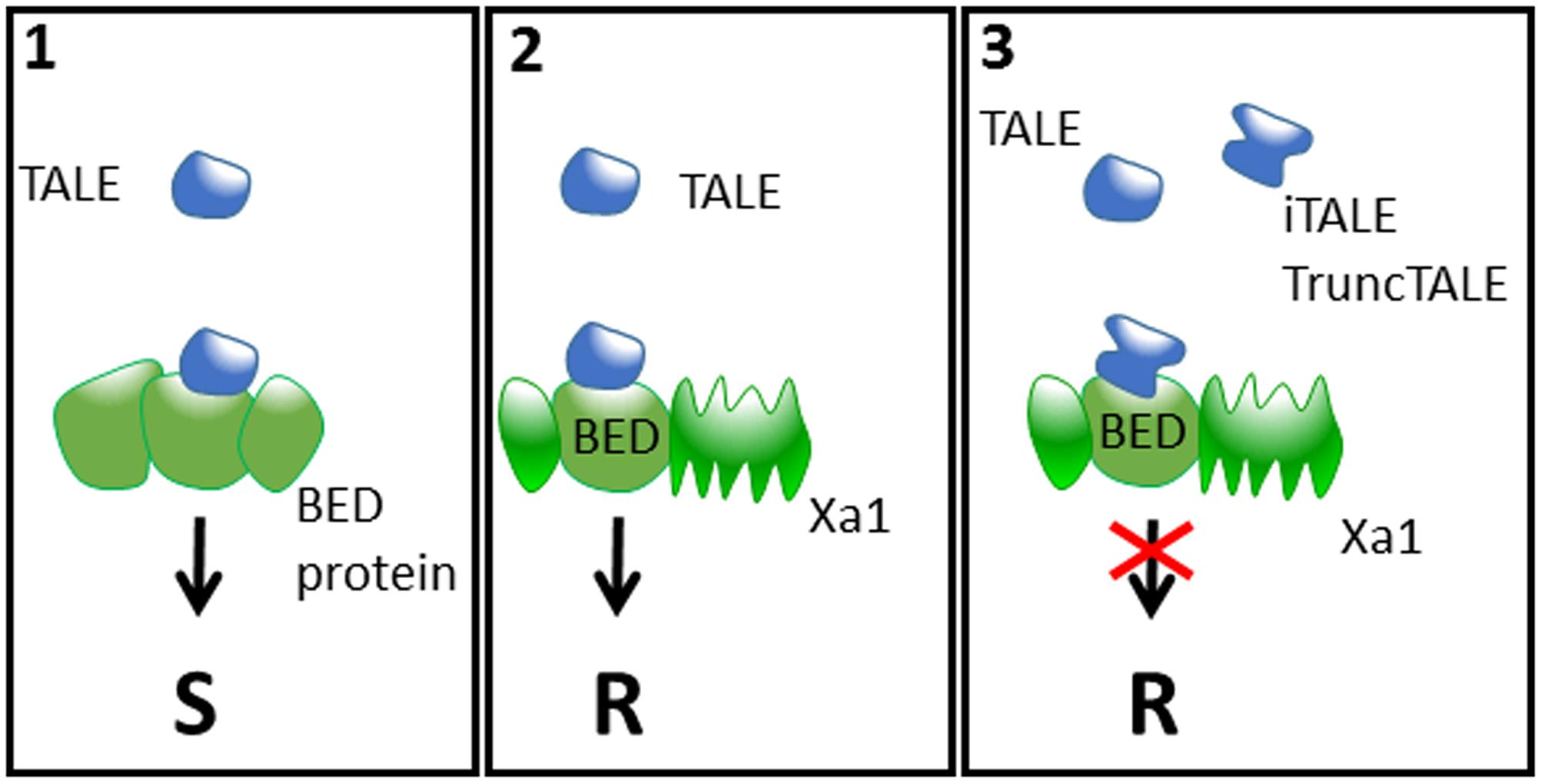
FIGURE 1. A speculative model for the arms race between plants and Xanthomonas. (1) Xanthomonas bacteria secrete TALEs into plant cells that presumably target protein(s) containing BED domain(s) to enhance susceptibility (S). (2) Plants have evolved immune receptors such as Xa1 that contain a BED domain which behaves as an integrated decoy, thus leading to TALE recognition and triggering resistance (R). (3) In turn, Xanthomonas evolved iTALEs and TruncTALEs, which are derivatives of full-length TALEs recognized by Xa1. These iTALEs and TruncTALEs interact directly or indirectly with Xa1 to inhibit its function, thus re-establishing susceptibility even in the presence of full-length TALEs.
The integrated decoy model for NLRs is a valuable concept to generate several hypotheses that can be challenged experimentally but that are entirely speculative at this point: Is the BED domain of Xa1 required for TALE recognition and iTALE-mediated suppression and if so, do these activities rely on direct binding between the BED domain and certain motifs in the TALE? Do TALEs interact with other Xa1 domains? Do TALEs interact directly or indirectly with other BED domain proteins and do such interactions contribute to target gene trans-activation? If yes, it is expected that mutants of these BED domain proteins are less susceptible to xanthomonads that rely on TALEs for virulence. Finally, a better molecular (structural) understanding of how TALEs and iTALEs interact with Xa1 may allow engineering NLR receptors that recognize full-length TALEs but not iTALEs and thus would truly confer broad-spectrum resistance against Xoo and Xoc.
Author Contributions
PZ, BS, RK, TK, and J-BM participated to the writing of this mini-review.
Funding
PZ is funded by the Agreenskills European program. Part of this research is funded by the French ANR project ImmuneReceptor ANR-15-CE20-0007 and the INRA-SPE project “Integrated decoys”. This work benefited from interactions promoted by COST Action FA 1208 (https://www.cost-sustain.org).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Boch, J., and Bonas, U. (2010). Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 48, 419–436. doi: 10.1146/annurev-phyto-080508-081936
Boch, J., Scholze, H., Schornack, S., Landgraf, A., Hahn, S., Kay, S., et al. (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512. doi: 10.1126/science.1178811
Böhm, H., Albert, I., Oome, S., Raaymakers, T. M., Van den Ackerveken, G., and Nürnberger, T. (2014). A conserved peptide pattern from a widespread microbial virulence factor triggers pattern-induced immunity in Arabidopsis. PLoS Pathog. 10:e1004491. doi: 10.1371/journal.ppat.1004491
Bourras, S., McNally, K. E., Ben-David, R., Parlange, F., Roffler, S., Praz, C. R., et al. (2015). Multiple avirulence loci and allele-specific effector recognition control the Pm3 race-specific resistance of wheat to powdery wildew. Plant Cell 27, 2991–3012. doi: 10.1105/tpc.15.00171
Brown, J. K., and Tellier, A. (2011). Plant-parasite coevolution: bridging the gap between genetics, and ecology. Annu. Rev. Phytopathol. 49, 345–367. doi: 10.1146/annurev-phyto-072910-095301
Cesari, S., Bernoux, M., Moncuquet, P., Kroj, T., and Dodds, P. N. (2014). A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy” hypothesis. Front. Plant Sci. 5:606. doi: 10.3389/fpls.2014.00606
Cesari, S., Thilliez, G., Ribot, C., Chalvon, V., Michel, C., Jauneau, A., et al. (2013). The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25, 1463–1481. doi: 10.1105/tpc.112.107201
Ellis, J. G. (2016). Integrated decoys and effector traps: how to catch a plant pathogen. BMC Biol. 14:13. doi: 10.1186/s12915-016-0235-8
Hayward, A., Ghazal, A., Andersson, G., Andersson, L., and Jern, P. (2013). ZBED evolution: repeated utilization of DNA transposons as regulators of diverse host functions. PLoS ONE 8:e59940. doi: 10.1371/journal.pone.0059940
Houterman, P. M., Cornelissen, B. J., and Rep, M. (2008). Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathog. 4:e1000061. doi: 10.1371/journal.ppat.1000061
Hutin, M., Perez-Quintero, A. L., Lopez, C., and Szurek, B. (2015a). MorTAL Kombat: the story of defense against TAL effectors through loss-of-susceptibility. Front. Plant Sci. 6:535. doi: 10.3389/fpls.2015.00535
Hutin, M., Sabot, F., Ghesquière, A., Koebnik, R., and Szurek, B. (2015b). A knowledge-based molecular screen uncovers a broad-spectrum OsSWEET14 resistance allele to bacterial blight from wild rice. Plant J. 84, 694–703. doi: 10.1111/tpj.13042
Ji, Z., Ji, C., Liu, B., Zou, L., Chen, G., and Yang, B. (2016). Interfering TAL effectors of Xanthomonas oryzae neutralize R-gene-mediated plant disease resistance. Nat. Commun. 7:13435. doi: 10.1038/ncomms13435
Kroj, T., Chanclud, E., Michel-Romiti, C., Grand, X., and Morel, J. B. (2016). Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 210, 618–626. doi: 10.1111/nph.13869
Le Roux, C., Huet, G., Jauneau, A., Camborde, L., Tremousaygue, D., Kraut, A., et al. (2015). A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 161, 1074–1088. doi: 10.1016/j.cell.2015.04.025
Ma, Z., Zhu, L., Song, T., Wang, Y., Zhang, Q., Xia, Y., et al. (2017). A paralogous decoy protects Phytophthora sojae apoplastic effector PsXEG1 from a host inhibitor. Science 17 355, 710–714. doi: 10.1126/science.aai7919
Maqbool, A., Saitoh, H., Franceschetti, M., Stevenson, C. E. M., Uemura, A., Kanzaki, H., et al. (2015). Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. eLife 4:e08709. doi: 10.7554/eLife.08709.001
Moscou, M. J., and Bogdanove, A. J. (2009). A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501. doi: 10.1126/science.1178817
Plissonneau, C., Daverdin, G., Ollivier, B., Blaise, F., Degrave, A., Fudal, I., et al. (2016). A game of hide and seek between avirulence genes AvrLm4-7 and AvrLm3 in Leptosphaeria maculans. New Phytol. 209, 1613–1624. doi: 10.1111/nph.13736
Pruitt, R. N., Schwessinger, B., Joe, J., Thomas, N., Liu, F., Albert, M., et al. (2015). The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Sci. Adv. 1:e1500245. doi: 10.1126/sciadv.1500245
Read, A. C., Rinaldi, F. C., Hutin, M., He, Y. Q., Triplett, L. R., and Bogdanove, A. J. (2016). Suppression of Xo1-mediated disease resistance in rice by a truncated, non-DNA-binding TAL effector of Xanthomonas oryzae. Front. Plant Sci. 7:1516. doi: 10.3389/fpls.2016.01516
Sarris, P. F., Cevik, V., Dagdas, G., Jones, J. G. D., and Krasileva, K. V. (2016). Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 14:8. doi: 10.1186/s12915-016-0228-7
Sarris, P. F., Duxbury, Z., Huh, S. U., Ma, Y., Segonzac, C., Sklenar, J., et al. (2015). A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161, 1089–1100. doi: 10.1016/j.cell.2015.04.024
Schornack, S., Ballvora, A., Gürlebeck, D., Peart, J., Ganal, M., Baker, B., et al. (2004). The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J. 37, 46–60. doi: 10.1046/j.1365-313X.2003.01937.x
Tai, T. H., Dahlbeck, D., Clark, E. T., Gajiwala, P., Pasion, R., Whalen, M. C., et al. (1999). Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl. Acad. Sci. U.S.A. 96, 14153–14158. doi: 10.1073/pnas.96.24.14153
Triplett, L. R., Cohen, S. P., Heffelfinger, C., Schmidt, C. L., Huerta, A. I., Tekete, C., et al. (2016). A resistance locus in the American heirloom rice variety Carolina Gold Select is triggered by TAL effectors with diverse predicted targets and is effective against African strains of Xanthomonas oryzae pv. oryzicola. Plant J. 87, 472–483. doi: 10.1111/tpj.13212
Yoshimura, S., Yamanouchi, U., Katayose, Y., Toki, S., Wang, Z. X., Kono, I., et al. (1998). Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc. Natl. Acad. Sci. U.S.A. 95, 1663–1668. doi: 10.1073/pnas.95.4.1663
Yuan, M., Ke, Y., Huang, R., Ma, L., Yang, Z., Chu, Z., et al. (2016). A host basal transcription factor is a key component for infection of rice by TALE-carrying bacteria. eLife 5:e19605. doi: 10.7554/eLife.19605
Keywords: decoys, MIMICS, rice, Xanthomonas, Xa1, BED domain, Xo1, effector
Citation: Zuluaga P, Szurek B, Koebnik R, Kroj T, Morel J -B (2017) Effector Mimics and Integrated Decoys, the Never-Ending Arms Race between Rice and Xanthomonas oryzae. Front. Plant Sci. 8:431. doi: 10.3389/fpls.2017.00431
Received: 05 February 2017; Accepted: 14 March 2017;
Published: 28 March 2017.
Edited by:
Richard Berthomé, Centre Toulouse Midi-Pyrénées – Institut National de la Recherche Agronomique, FranceReviewed by:
Panagiotis F. Sarris, University of Exeter, UKMark James Banfield, John Innes Centre – Biotechnology and Biological Sciences Research Council, UK
Copyright © 2017 Zuluaga, Szurek, Koebnik, Kroj, Morel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jean-Benoit Morel, jean-benoit.morel@inra.fr
 Paola Zuluaga
Paola Zuluaga Boris Szurek
Boris Szurek Ralf Koebnik
Ralf Koebnik Thomas Kroj1
Thomas Kroj1 Jean-Benoit Morel
Jean-Benoit Morel