- 1Department of Biology, University of Florida, Gainesville, FL, USA
- 2Genetics Institute, University of Florida, Gainesville, FL, USA
- 3Department of Pharmacognosy, Faculty of Pharmacy, Zagazig University, Zagazig, Egypt
- 4Plant Molecular and Cellular Biology Program, University of Florida, Gainesville, FL, USA
- 5Thermo Fisher Scientific, West Palm Beach, FL, USA
- 6Interdisciplinary Center for Biotechnology Research, University of Florida, Gainesville, FL, USA
Glucosinolates (Gls) constitute a major group of natural metabolites represented by three major classes (aliphatic, indolic and aromatic) of more than 120 chemical structures. In our previous work, soluble proteins and metabolites in Arabidopsis mutants deficient of aliphatic (myb28/29) and indolic Gls (cyp79B2B3) were analyzed. Here we focus on investigating the changes at the level of membrane proteins in these mutants. Our LC/MS-MS analyses of tandem mass tag (TMT) labeled peptides derived from the cyp79B2/B3 and myb28/29 relative to wild type resulted in the identification of 4,673 proteins, from which 2,171 are membrane proteins. Fold changes and statistical analysis showed 64 increased and 74 decreased in cyp79B2/B3, while 28 increased and 17 decreased in myb28/29. As to the shared protein changes between the mutants, one protein was increased and eight were decreased. Bioinformatics analysis of the changed proteins led to the discovery of three cytochromes in glucosinolate molecular network (GMN): cytochrome P450 86A7 (At1g63710), cytochrome P450 71B26 (At3g26290), and probable cytochrome c (At1g22840). CYP86A7 and CYP71B26 may play a role in hydroxyl-indolic Gls production. In addition, flavone 3′-O-methyltransferase 1 represents an interesting finding as it is likely to participate in the methylation process of the hydroxyl-indolic Gls to form methoxy-indolic Gls. The analysis also revealed additional new nodes in the GMN related to stress and defense activity, transport, photosynthesis, and translation processes. Gene expression and protein levels were found to be correlated in the cyp79B2/B3, but not in the myb28/29.
Introduction
Glucosinolates (Gls) as natural anticancer compounds are represented by three major classes of chemical structures (aliphatic, indolic, and aromatic; Yan and Chen, 2007; Sønderby et al., 2010). In addition to their anti-carcinogenic activities, they have a distinct role in plant defense against herbivores (Halkier and Gershenzon, 2006; Yan and Chen, 2007) and pathogens (Kissen et al., 2009). The activities are attributed to their hydrolysis products, such as isothiocyanates, thiocyanates, and nitriles (Halkier and Gershenzon, 2006). Gls biosynthesis starts from methionine, tryptophan or phenylalanine to produce aliphatic, indolic, or aromatic Gls, respectively (Yan and Chen, 2007; Sønderby et al., 2010). Briefly, the substrate amino acid is converted to aldoxime, then to aci-nitro compounds, thiohydroximate, and desulfoglucosinolate. After sulfation, the core Gls structure is formed. In aliphatic Gls biosynthesis, the methionine chain-elongation and the core structure biosynthesis are under the control of three transcriptional factors MYB28, MYB29, and MYB76 (Yan and Chen, 2007; Frerigmann et al., 2012). In the core pathway, the formation of aldoximes is catalyzed by cytochrome P450s CYP79F1 and CYP79F2, and that of the aci-nitro compounds by CYP83A1 (Grubb and Abel, 2006). Then glutathione S-transferase U20 forms thiohydroximates, which are in turn rearranged to desulfoglucosinolate by UGT74B1 (Sønderby et al., 2010), followed by sulfation by SOT17 and SOT18 to produce intact Gls (Sønderby et al., 2010; Mostafa et al., 2016). Similar for indolic Gls, CYP79B2, CYP79B3, and CYP83B1 are responsible for aldoximes and aci-nitro compounds formation, followed by conversion to thiohydroximates by glutathione S-transferase F10, rearrangement to desulfoglucosinolates and sulfation to indolic Gls by SOT16 (Grubb and Abel, 2006; Mostafa et al., 2016). It is clear that the cytochrome P450s play a central role in the Gls biosynthesis, and these proteins are membrane localized (Neve and Ingelman-Sundberg, 2010).
Several studies have reported the relationship between the Gls biosynthetic pathway and other biological pathways in plants, e.g., amino acid and carbohydrate pathways using CYP79F1 RNAi lines (Chen et al., 2012), auxin biosynthesis using cyp79B2/B3 mutant (Zhao et al., 2002) and stress response pathways through environmental perturbation (Martínez-Ballesta et al., 2013). In our previous work, we used Arabidopsis double mutants (cyp79B2/B3 deficient in indolic Gls production and myb28/29 deficient in aliphatic Gls production), and discovered new nodes in the glucosinolate molecular network (GMN) that include stress and defense related proteins like glucan endo-1,3-beta-glucosidase, glutathione S-transferase F7 and glutathione S-transferase F2 and the electron carriers cytochrome B5 isoform C and cytochrome c oxidase subunit 5b-2 (Mostafa et al., 2016). To date, no studies have reported the glucosinolate molecular networks in the membrane proteome context.
Since many known glucosinolate proteins such as the cytochrome P450s are membrane or membrane associated proteins, here we investigated how perturbation of Gls metabolism using the aforementioned mutants affects the Arabidopsis membrane proteome using Tandem Mass Tag (TMT) labeling LC-MS/MS based quantitative proteomics. Analyses of protein interaction networks using STRING and functional enrichment of the identified proteins using agriGO allowed us to discover new nodes and edges in the GMN. With qRT-PCR, we were able to determine the correlation between gene transcripts and membrane proteins in the two mutants. Together with our published soluble proteomics work (Mostafa et al., 2016), this study enables a comprehensive understanding of the Arabidopsis GMNs.
Materials and Methods
Plant Genotyping, Growth, and Sample Collection
Arabidopsis thaliana (L.) Heynh ecotype Columbia (Col-0) seeds were obtained from the Arabidopsis Biological Resource Center (Columbus, OH, USA). The seeds of cyp79B2/B3 and myb28/29 were kindly provided by Dr. John Celenza (Boston University, Boston, MA, USA) and Dr. Masami Hirai (RIKEN Plant Science Center, Yokohama, Japan), respectively. The mutant genotyping and chemotyping were reported in our previous study (Mostafa et al., 2016). Seed germination and seedling growth were conducted as previously described (Mostafa et al., 2016). Leaves from 5-week old wild type (WT), cyp79B2/B3 and myb28/29 were collected, frozen in liquid nitrogen and stored at −80°C. Four replicates were included per genotype, and each replicate contains 2 g leaves pooled from 12 plants.
Protein Extraction and Peptide TMT Labeling
Protein was extracted according to Pang et al. (2010) by grinding the leaf tissues in liquid nitrogen and then homogenizing on ice in 10 mM Tris-HCl (pH 7.4), 10 mM KCl, 1.5 mM MgCl2, 10 mM dithiothritol (DTT), 0.5 M sucrose, and 10 mM phenylmethylsulfonyl fluoride (PMSF). The protein extracts were filtered through cheesecloth and centrifuged at 800 g for 10 min at 4°C. The supernatant was transferred to ultracentrifuge tubes and centrifuged again at 100,000 g for 1.5 h at 4°C. The formed microsomal membrane was washed with 100 mM sodium carbonate using a glass dounce homogenizer, followed by centrifugation at 100,000 g for 1.5 h at 4°C. The microsome pellets were rinsed with 500 μl resuspension buffer containing 100 mM HEPES (pH 7), 1% triton X-100 and 0.5 M sucrose, and centrifuged at 800 g for 10 min at 4°C. Protein was precipitated using 5 volumes ice cold 90% acetone overnight at −20°C, followed by washing the pellets once with ice cold 90% acetone and twice with ice cold acetone before solubilizing in 7 M urea, 2 M thiourea, 4% CHAPS, and 0.25% Triton X-100. The protein amount was assayed using an EZQ assay kit (Invitrogen Inc., Eugene, OR, USA).
A total of 50 μg protein from each replicate was precipitated with ice cold 90% acetone at −20°C overnight, followed by 20,000 g centrifugation at 4°C for 15 min. After washing with ice cold 90% acetone, the pellets were solubilized, reduced, alkylated and digested with modified trypsin (Promega, Madison, WI, USA) at a 1:25 (w/w) ratio for 16 h at 37°C, followed by TMT labeling according to the TMT 6-plex kit manual (Thermo Scientific Inc., San Jose, CA, USA). The WT replicates were labeled with 126 and 127 tags, cyp79B2/B3 replicates with 128 and 129 tags and myb28/29 replicates with 130 and 131 tags at room temperature for 2 h. After quenching with 8 μl 5% hydroxylamine for 30 min, the labeled samples were combined and lyophilized. Two independent experiments and four biological replicates each sample were performed.
Peptide Desalting, Strong Cation Exchange Fractionation, and LC-MS/MS Analysis
The TMT labeled peptides were desalted on Macrospin C-18 reverse phase mini-column (The Nestgroup Inc., Southborough, MA, USA) and fractionated using an Agilent HPLC 1260 strong cation exchange system as previously described (Mostafa et al., 2016). A total of 12 fractions were collected from each experiment. Each fraction was lyophilized, solubilized in solvent A (0.1% formic acid and 3% acetonitrile), and analyzed using an Easy-nLC 1000 system coupled to a Q-Exactive Orbitrap Plus MS (Thermo Fisher Scientific, Bremen, Germany) according to Mostafa et al. (2016) with minor modifications: The mobile phase gradient was ramped from 2 to 30% of solvent B (0.1% formic acid and 99.9% acetonitrile) in 57 min, then to 98% of solvent B in 6 min and maintained for 12 min. Mass analysis was performed in positive ion mode with high collision dissociation energy. The scan range was 400–2,000 m/z with full MS resolution of 70,000 and 200–2,000 m/z with MS2 resolution of 17,500. The first mass was fixed at 115 m/z, and 445.12003 m/z (polysiloxane ion mass) was used for real-time mass calibration.
Protein Identification and Quantification
The MS data were searched using Proteome Discoverer 1.4 (Thermo Scientific, Bremen, Germany) against the Arabidopsis TAIR10 database with 35,386 entries. The searching parameters were set to 300 and 5,000 Da as minimum and maximum precursor mass filters, digestion with trypsin with two missed cleavages, Carbamidomethylation of cysteine was set as a static modification, and TMT6plex of N terminus, TMT6plex of lysine, phosphorylation of STY (serine, threonine, and tyrosine) and methionine oxidation were set as dynamic modifications. Precursor mass tolerance was 10 ppm, fragment mass tolerance was 0.01 Da, spectrum grouping maximum retention time difference was 1.1 and false discovery rate was 0.01 at the peptide level. Proteins quantification based on labeled unique peptides intensities and statistical analyses were performed as previously described (Chen et al., 2012; Mostafa et al., 2016; Sun et al., 2017). The proteomics data were deposited to ProteomeXchange repository (accession number: PXD005781).
String Bioinformatics Analysis and Gene Ontology Enrichment
The relationship between the significantly changed proteins and Gls metabolic pathways (Chen et al., 2011; Mostafa et al., 2016) was analyzed using STRING bioinformatics tool (Baldrianová et al., 2015; Ji et al., 2016; Lim et al., 2017). The resulted networks were visualized in the confidence view relying on gene neighborhood, fusion, co-occurrence, co-expression, literature, and available data. To determine the enriched pathways, we performed Singular Enrichment Analysis (SEA) for the changed proteins and the results were compared using a cross comparison of SEA (SEACOMPARE) in the agriGO database (Silva-Sanchez et al., 2013).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
To determine whether protein expression levels were correlated with transcript levels, we conducted qRT-PCR of 44 genes selected based on the proteomics data (32 for cyp79B2/B3 and 22 for myb28/28). This list of primers used in qRT-PCR is provided in Supplementary Table 1. Total RNA was extracted using a RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) and cDNA was synthesized with ProtoScript® II Reverse Transcriptase (New England BioLabs, Ipswich, MA, USA). qRT-PCR was performed with VeriQuest SyBr and a fluorescein kit (Affymetrix, Santa Clara, CA, USA) using CFX96 (Bio-Rad, Hercules, CA, USA) as described previously (Koh et al., 2012). For each reaction, three technical and three biological replicates were included. Relative expression of the target genes was calculated using the comparative Ct method (Applied Biosystems, Framingham, USA). The differences in Ct values (ΔCt) between the target gene and two internal controls (AT4G34270 and AT5G44200) were calculated to normalize differences in the starting materials. The expression ratios of cyp79B2/B3 and myb28/29 to WT were calculated and compared to the ratios from the protein data using Pearson's r.
Results
cyp79B2/B3 and myb28/29 Membrane Proteomes
Based on the MS/MS spectra of high confidence peptides derived from the WT, cyp79B2/B3 and myb28/29, we identified 4673 proteins in two independent experiments using Proteome Discover (Supplementary Table 2). Out of these proteins, 3,132 were identified in both experiments, while 1,076 and 465 were unique to experiments 1 and 2, respectively (Figure 1A). A total of 4,655 proteins were available for quantification based on unique TMT labeled peptides, highlighting the high efficiency of labeling. PD enrichment analysis (based on TAIR and Uniprot annotations) of the identified proteins showed 2,171 to be membrane proteins (Figure 1B and Supplementary Table 2). Comparative analysis of the protein expression changes between the mutants and WT at a fold change cutoff (>1.2 and <0.8), a p < 0.05 and transmembrane domain analysis revealed 93 proteins to be increased (Figure 1C) and 99 to be decreased (Figure 1D). Transmembrane domain analysis revealed that 175 out of the 192 differentially expressed proteins contained at least one transmembrane domains. The rest deemed to be membrane associated proteins (Supplementary Table 3). Correlating the changed proteins to those involved in Gls metabolism using STRING showed new nodes and edges (Figures 2, 3). The new nodes can be categorized according to their positions in the network as directly correlated or indirectly correlated to Gls metabolism. They can also be classified according to their biological roles as secondary (stress related) and tertiary (other biological process) connections (Detailed in next sections).
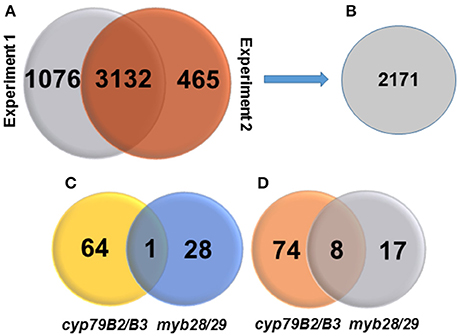
Figure 1. Venn diagrams showing numbers of identified proteins, membrane proteins, changed proteins, and their distributions. (A) Number of identified proteins in two independent TMT experiments at high peptide confidence. (B) Number of identified membrane proteins. (C) Number of significantly increased membrane proteins in cyp79B2/B3 and myb28/29 relative to WT at p < 0.05 and fold change >1.2. (D) Number of significantly decreased membrane proteins in cyp79B2/B3 and myb28/29 relative to WT at p < 0.05 and fold change < 0.8.
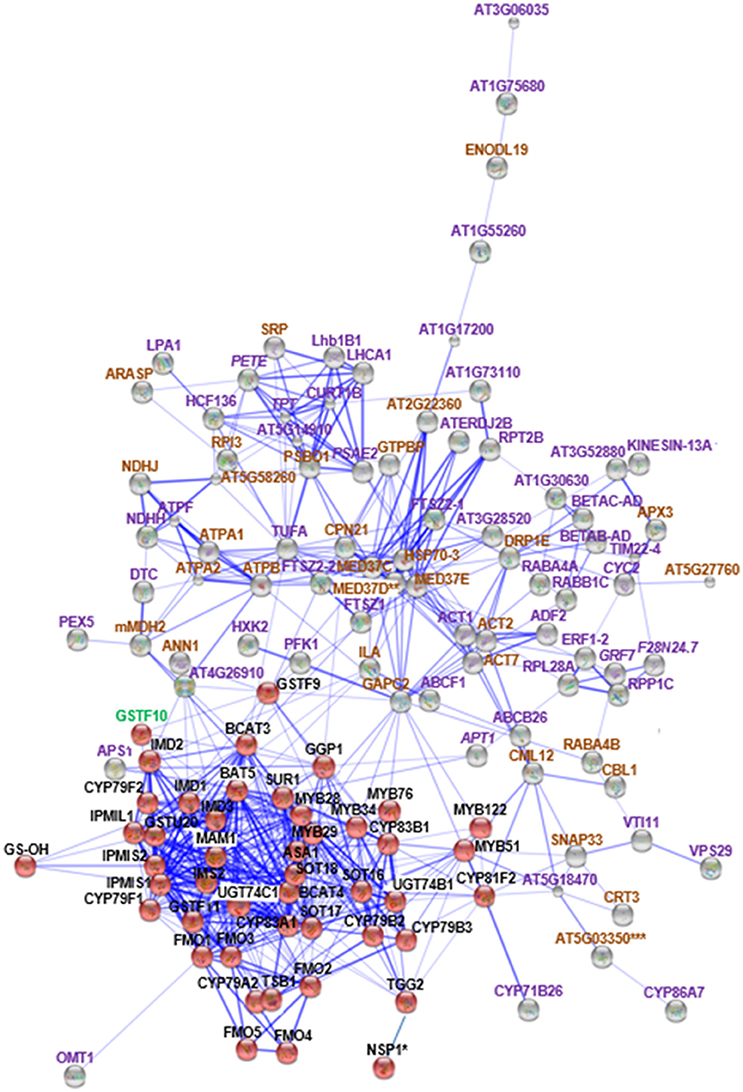
Figure 2. STRING analysis of cyp79B2/B3 changed proteins in relation to known proteins in Gls biosynthesis. Known Gls biosynthetic proteins are indicated by red balls, new proteins in the GMN are indicated by gray balls, proteins changed in both mutants are indicated by italic labeling, and uniquely changed proteins in cyp79B2/B3 are indicated by non-italic labeling. Proteins involved in Gls biosynthesis, stress and defense, and other processes are labeled with green, brown, and violet labels, respectively. Connections strength are proportional to edges thickness as derived from neighborhood, gene fusion, co-occurrence, co-expression, previous experiments, and text-mining information at medium confidence score. Asterisk (*) indicates manual connections based on literature. Double asterisk (**) indicates known nodes in both mutants (Mostafa et al., 2016), and triple asterisk (***) indicates known nodes in cyp79B2/B3 (Mostafa et al., 2016). Full names of the mapped proteins can be found in the abbreviation and protein name columns in Tables 1, 2.
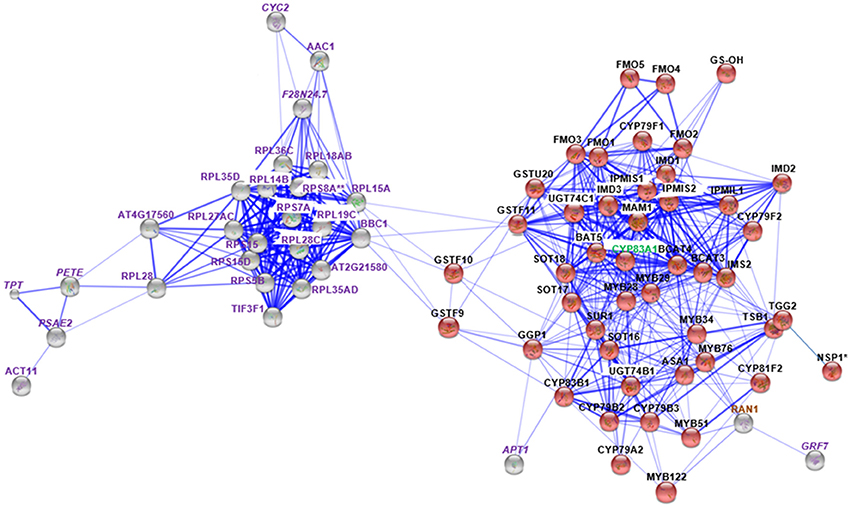
Figure 3. STRING analysis of myb28/29 changed proteins in relation to known proteins in Gls biosynthesis. Known Gls biosynthetic proteins are indicated by red balls, new proteins in GMN are indicated by gray balls, proteins changed in both mutants are indicated by italic labeling, and uniquely changed proteins in myb28/29 are indicated by non-italic labeling. Proteins involved in Gls biosynthesis, stress and defense, and other processes are labeled with green, brown, and violet labels, respectively. Connections strength are proportional to edges thickness as derived from neighborhood, gene fusion, co-occurrence, co-expression, previous experiments, and text-mining information at medium confidence score. Asterisk (*) indicates manual connections based on literature. Double asterisk (**) indicates known nodes in both mutants (Mostafa et al., 2016). Full names of the mapped proteins can be found in the abbreviation and protein name columns in Tables 1, 2.
Common Changes of Membrane Proteins between the cyp79B2/B3 and myb28/29
Nine membrane proteins showed common changes between the two mutants relative to WT, with only one protein increased while the other eight decreased (Table 1). By STRING mapping of the significantly changed proteins (Figures 2, 3), we found seven of the nine proteins represented new connections with the glucosinolate metabolic network (GMN). The role of probable cytochrome c (CYC2) and plastocyanin minor isoform (PETE) in electron transport process (Pesaresi et al., 2009; Welchen et al., 2012) makes them biologically relevant tertiary connections in GMN in a way similar to cytochrome B5 isoform C and cytochrome c oxidase subunit 5b-2 (Mostafa et al., 2016). Photosystem I reaction center subunit IV B (PSAE2), 14-3-3-like protein GF14 nu (GRF7), adenine phosphoribosyltransferase 1 (APT1), alba DNA/RNA-binding protein (F28N24.7) and triose phosphate/phosphate translocator (APE2) form other tertiary nodes. Out of this group, APT1 was the only protein directly connected to the GMN (Figures 2, 3).
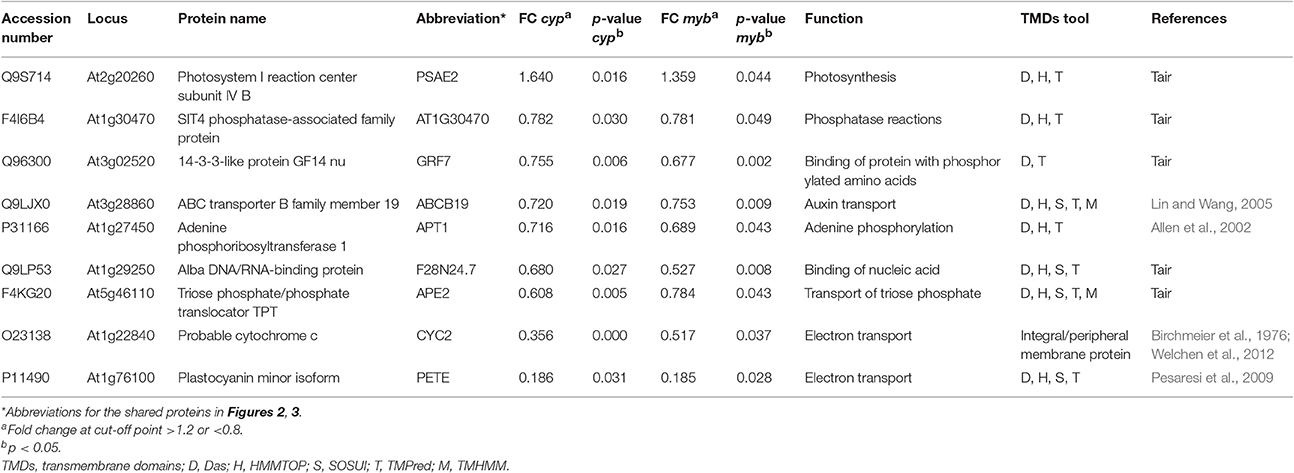
Table 1. List of common membrane proteins showing significant changes in cyp79B2/B3 and myb28/29 mutants relative to WT and their biological functions.
Specific Changes of cyp79B2/B3 Membrane Proteins
Sixty-four and 74 membrane proteins showed unique increases and decreases, respectively, in the cyp79B2/B3 mutant (Table 2). Seventy-seven new nodes were discovered by the STRING mapping of these cyp79B2/B3 proteins to the GMN (Figure 2). It was obvious that perturbation of the indolic Gls metabolism affected a group of stress-related membrane proteins forming new secondary nodes. Representative examples from this group are calmodulin-like protein 12 (CML12; Cazzonelli et al., 2014), mediator of RNA polymerase II transcription subunit 37c (MED37C; Lee et al., 2009), SNAP25 homologous protein (SNAP33; Eschen-Lippold et al., 2012), dynamin-related protein 1E (DRP1E; Minami et al., 2015), protein ILITYHIA (ILA; Monaghan and Li, 2010), glyceraldehyde-3-phosphate dehydrogenase (GAPC2; Guo et al., 2012), L-ascorbate peroxidase 3 (APX3; Narendra et al., 2006), Ras-related protein (RABA4B; Antignani et al., 2015), annexin D1 (ANN1; Gorecka et al., 2005; Jia et al., 2015), hypoxia-responsive family protein (At5g27760), and malate dehydrogenase 2 (mMDH2; Jones et al., 2006).
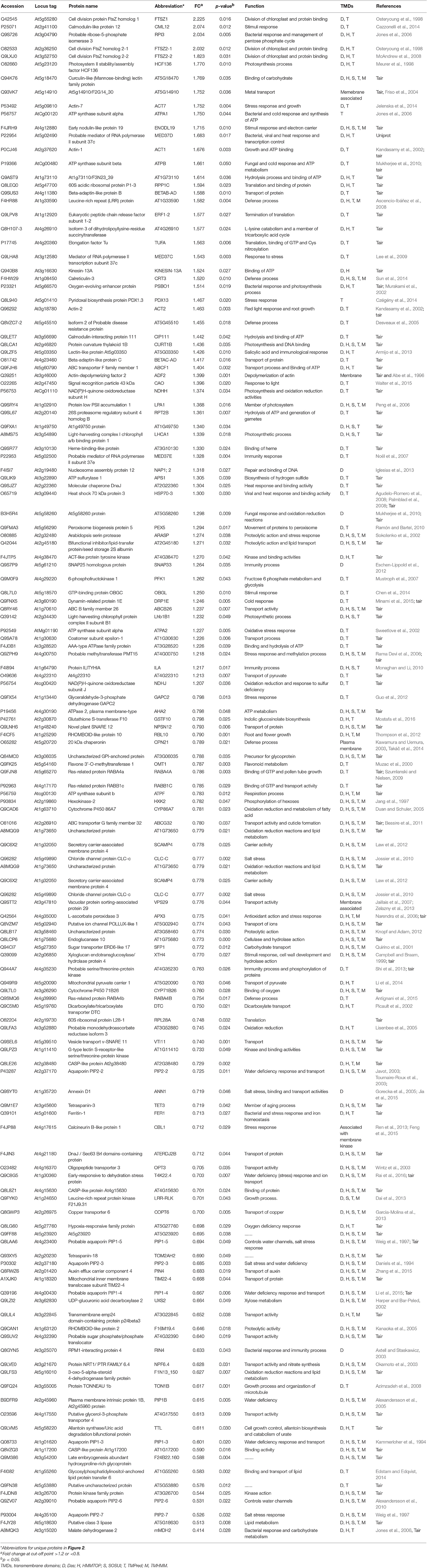
Table 2. List of membrane proteins in the cyp79B2/B3 mutant showing significant changes relative to WT and their biological functions.
The role of cytochromes P450s 86A7 (CYP86A7) and 71B26 (CYP71B26) in oxidation reduction reaction and oxygen binding (Duan and Schuler, 2005) makes them biologically relevant tertiary nodes in the GMN (Figures 2, 4). Other interesting new tertiary nodes related to Gls metabolism include division protein FtsZ homologs 1 (FTSZ1), 2-1 (FTSZ2-1), 2-2 (FTSZ2-2), curculin-like (mannose-binding) lectin family protein (At5g18470), isoform 3 of dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2 (At4g26910), elongation factor Tu (TUFA), ATP sulfurylase 1 (APS1), ABC transporter B family member 26 (ABCB26), flavone 3'-O-methyltransferase 1 (OMT1), Ras-related protein (RABA4A), Ras-related protein (RABB1C), endoglucanase 10 (AT1G75680), dicarboxylate/tricarboxylate transporter (DTC), probable monodehydroascorbate reductase isoform 3 (At3g52880), vesicle transport v-SNARE 11 (VTI11), DnaJ/Sec63 Brl domains-containing protein (ATERDJ2B), and glycosylphosphatidylinositol-anchored lipid protein transfer 6 (At1g55260). Myrosin cells (myrosinase storage sites) endocytosis is controlled by SYP22 from SNARE complex and VPS9A (Shirakawa et al., 2016). Here the decrease of VTI11 from this family is in agreement with the reduced myrosinase, nitrile specifier protein and Gls levels in the soluble proteome (Mostafa et al., 2016) and supports the cross talk between Gls and its hydrolyzing enzymes.
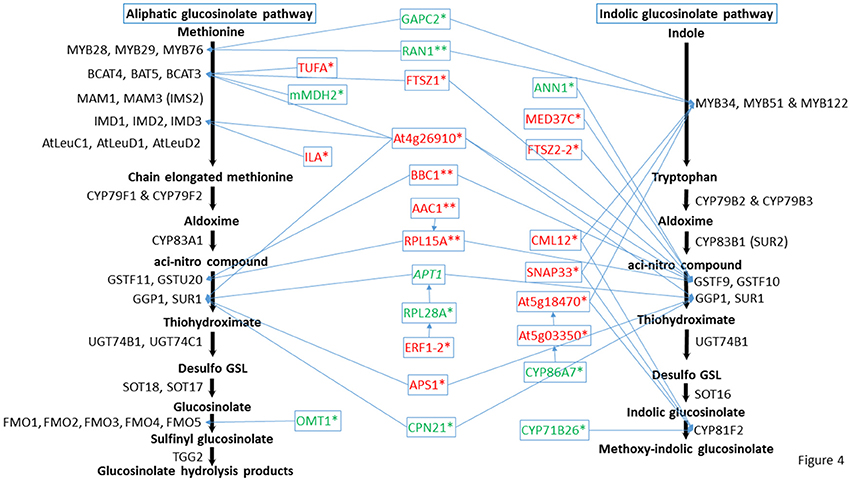
Figure 4. Predicted positions of the directly connected nodes and connected cytochrome nodes on the glucosinolate metabolic pathway. Italic indicates proteins changed in both mutants, * indicates proteins changed in cyp79B2/B3, and ** indicates proteins changed in myb28/29. Red color means increased proteins, green color means decreased proteins. Full names of the directly connected proteins can be found in the abbreviation and protein name columns in Tables 1–3.
Out of these new nodes, 15 formed direct edges with the GMN: FTSZ1, CML12, FTSZ2-2, At5g18470, At4g26910, TUFA, MED37C, APS1, SNAP33, ILA, GAPC2, OMT1, CYP71B26, ANN1, and mMDH2 in addition to the membrane associated protein (20 kDa chaperonin, CPN21). As we detected a side network correlated to indolic GMN (Mostafa et al., 2016), here we also found a side network strongly correlated to indolic Gls metabolism as it contains nine stress-related proteins out of eleven. These proteins are xyloglucan endotransglucosylase/hydrolase protein 4 (XTH4; Campbell and Braam, 1999), aquaporin PIP2-2 (Javot, 2003; Tournaire-Roux et al., 2003), probable aquaporin PIP1-5 (Weig et al., 1997), aquaporin PIP2-3 (Daniels et al., 1994), probable aquaporin PIP1-4 (Li et al., 2015), plasma membrane intrinsic protein 1B (PIP1B; Alexandersson et al., 2005), aquaporin PIP1-3 (Kammerloher et al., 1994), probable aquaporin PIP2-6 (Alexandersson et al., 2010), and aquaporin PIP2-7 (Weig et al., 1997). Other members in this side network are bifunctional inhibitor/lipid-transfer protein (At2g45180; which has a proteolytic action) and a tetraspanin-18 (TOM2AH2) with unknown functions.
Specific Changes of myb28/29 Membrane Proteins
Membrane proteomics of the myb28/29 mutant showed 28 and 17 proteins to be significantly increased and decreased, respectively (Table 3). STRING analysis of the increased and decreased myb28/29 specific membrane proteins revealed 21 new nodes in the GMN (Figure 3). Except for the directly connected and stress-related GTP-binding nuclear protein (RAN1; Jiang et al., 2007), other connections including 17 ribosomal proteins [e.g., 60S ribosomal protein L14-2 (RPL14B), 40S ribosomal protein S15-1 (RPS15) and 40S ribosomal protein S15-4 (RPS15D)], and actin-11 (ACT11), ADP/ATP carrier protein 1 (AAC1) and eukaryotic translation initiation factor 3 subunit F (TIF3F1) formed tertiary nodes. These tertiary nodes are connected to the GMN through two bridges (directly connected nodes) which are 60S ribosomal protein L15-1(RPL15A) and 60S ribosomal protein L13-1 (BBC1). The expression changes in ribosomal proteins reflect a correlation between aliphatic Gls perturbation and the translation process in A. thaliana.
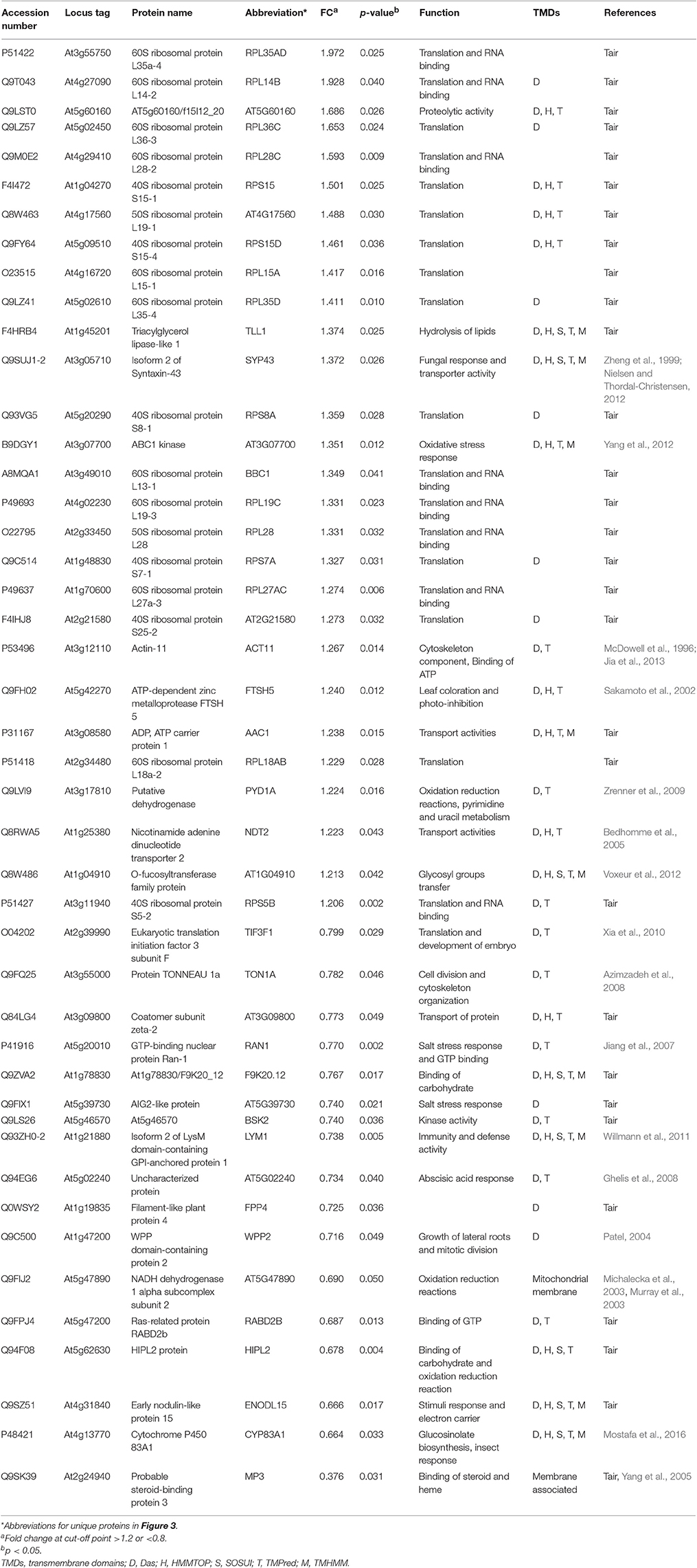
Table 3. List of membrane proteins the myb28/29 mutant showing significant level changes relative to WT and their biological functions.
Gene Ontology Analysis of the Significantly Changed Membrane Proteins
AgriGO enrichment analysis of the changed proteins was conducted at the biological processes (BP), cellular components (CC), and molecular functions (MF) levels. By annotating 147 changed membrane proteins in the cyp79B2/B3 using SEA, we got 302 enriched GO terms for BP (Supplementary Figure 1), 63 for CC (Supplementary Figure 2), and 47 for MF (Supplementary Figure 3). SEA of 54 changed membrane proteins in the myb28/29 showed 45 enriched GO terms for BP (Supplementary Figure 4), 56 for CC (Supplementary Figure 5) and 2 for MF (Supplementary Figure 6). SEACOMPARE of the mutant revealed 271 BP, 21 CC, and 46 MF GO terms to be enriched specifically in cyp79B2/B3, while 14 BP, 14 CC, and one MF were the specifically enriched GO terms in myb28/29 (Supplementary Table 4). From this BP analysis, it was obvious that responses to stimuli including abiotic, chemical and stress were highly enriched in cyp79B2/B3 in addition to transport, photosynthesis and metabolic processes. In myb28/29, the most enriched BP terms were those related to translation process. This observation supported our results concerning the stimuli and translation-related proteins in the cyp79B2/B3 and myb28/29, respectively (Supplementary Table 4). On the level of CC, the high enrichment of membrane GO terms supported the effectiveness of our membrane preparation procedure (Supplemental Figures 2, 5).
Comparison of Protein Expression Data with Transcription Data
To determine whether protein level changes correlated with gene transcription changes, we examined the transcript levels of 32 and 22 genes from cyp79B2/B3 and myb28/28, respectively (Supplementary Table 5). The two mutants exhibited different patterns of correlation. In comparison of cyp79B2/B3 to WT, the genes investigated showed a positive correlation between transcript and protein levels in both direction and degree of expression (r = 0.6579, p = 4.269e−05; Supplementary Figure 7). However, in comparison of myb28/29 to WT, the genes did not show correlation between the transcript and protein levels (r = 0.0887, p = 0.6945; Supplementary Figure 7), only three out of the 22 genes showed similar regulation at both transcript and protein levels. For example, At4g13770 encoding cytochrome P450 83A1, exhibited down-regulation in myb28/29 compared to WT (Supplementary Table 5). The difference in the degree of correlation in these two mutants implies that different regulatory mechanisms are involved in the transcriptional and posttranscriptional processes in different genotypes (Marmagne et al., 2010; Koh et al., 2012).
Discussion
As a result of Gls metabolism perturbation, many changes in the levels of soluble (Mostafa et al., 2016) and membrane proteins took place. It was interesting to discover new cytochromes to be involved in the GMN. In addition, several groups of stress and defense-related proteins as well as binding and transport activity proteins were related to the indolic and aliphatic GMNs, in addition to a group of ribosomal proteins in the myb28/29 mutant.
Three New Cytochromes in the Glucosinolate Molecular Network
Cytochromes play a key role in Gls biosynthesis. In aliphatic Gls biosynthesis, CYP79F1 and CYP79F2 catalyze the conversion of chain-elongated methionines to aldoximes, which are metabolized by another cytochrome (CYP83A1) to aci-nitro compounds, precursors of desulphoglucosinolates (Grubb and Abel, 2006). As to indolic Gls biosynthesis, CYP79B2 and CYP79B3 convert tryptophan to aldoximes, that are metabolized by CYP83B1 to form the aci-nitro compounds (Grubb and Abel, 2006). In addition, there is another CYP81F2 catalyzing the conversion of indolic-3-glucosinolate to 4-hydroxy-indolic-3-glucosinolate (Sønderby et al., 2010). Furthermore, CYP71A12 and CYP71A13 can metabolize indolic aldoximes to indole acetonitrile and subsequently indole acetic acid derivatives (Nafisi et al., 2007). In our previous study, we reported cytochrome B5 isoform C and cytochrome c oxidase subunit 5b-2 to be new nodes in the aliphatic and indolic GMNs, respectively (Mostafa et al., 2016). Here we discovered cytochrome P450 86A7 (CYP86A7) in redox reaction and metabolism of fatty acids (Duan and Schuler, 2005), and cytochrome P450 71B26 (CYP71B26) as new nodes in the indolic GMN. Based on STRING analysis, CYP71B26 is connected to CYP81F2 through a direct edge, while CYP86A7 is connected indirectly to CYP81F2 through lectin family proteins (At5g03350 and At5g18470; Figures 2, 4). Given that their connection to a specific and key enzyme in indolic Gls biosynthetic pathway (CYP81F2) and their expression levels were decreased in the cyp79B2/B3 mutant (Table 2), it is reasonable to hypothesize that CYP86A7 and CYP71B26 play specific roles in 4-hydroxy indolic-3-glucosinolate production (Figure 4). Especially their precursor (indolic-3-glucosinolate) and the product were decreased in cyp79B2/B3 mutant as revealed in our previous study (Mostafa et al., 2016). Also by similarity, we can predict a role for the enzymes in hydroxy indolic-1-glucosinolate production (Figure 5) as its synthesizing enzymes are not known (Sønderby et al., 2010). The third new cytochrome discovered in this study is a probable cytochrome c At1Gg22840 (CYC2), which plays a role in electron transport process (Welchen et al., 2012). CYC2 is in the shared decreased protein category, forming new connections with aliphatic GMN through ADP/ATP carrier protein 1 (AAC1) and 60S ribosomal protein L15-1 (RPL15A), which is connected to GSTF9, GSTF10 and GSTF11, and with indolic GMN through eukaryotic peptide chain release factor subunit 1–2 (ERF1-2), 60S ribosomal protein L28-1 (RPL28A) and adenine phosphoribosyltransferase 1 (APT1). APT1 is connected to GGP1 and SUR1. Although the CYC2 function awaits for further studies, it might play a role in the conversion of aci-nitro compounds to thiohydroximates.
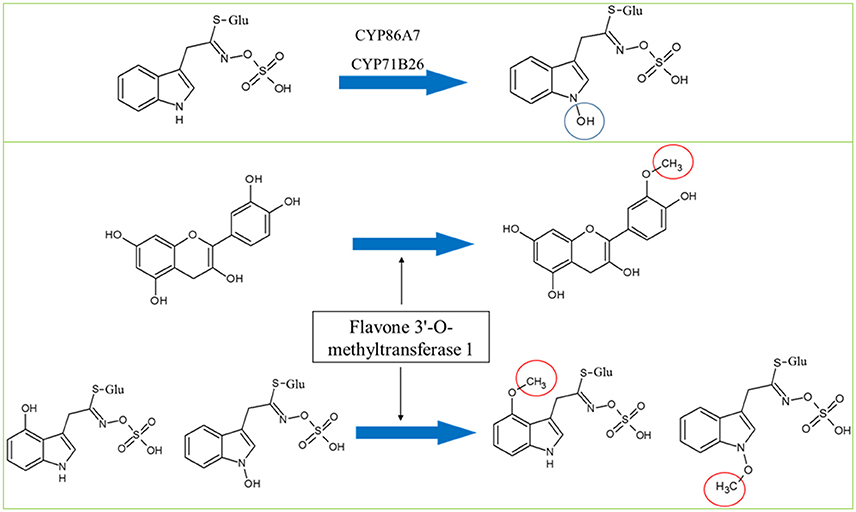
Figure 5. Hypothesized roles of CYP86A7 and CYP71B26 in the hydroxylation of indolic-1-glucosinolate (top panel) and the potential dual functions of flavone 3′-O-methyltransferase in flavonoid and Gls metabolism. Circles indicate chemical modifications to the substrates.
Stress Related Membrane Protein Changes as a Secondary Result of Glucosinolate Metabolism Perturbation
Plant Gls metabolism is responsive to stress conditions, e.g., temperature and light stress (Martínez-Ballesta et al., 2013), water stress (Khan et al., 2010), salt stress (Guo et al., 2013), and microbial stress (Clay et al., 2009). In our previous study, glucan endo-1,3-beta-glucosidase, glutathione S-transferase F2 and glutathione S-transferase F7 in addition to others as stress-related proteins were found to connect to the Gls pathway (Mostafa et al., 2016). Here we found the levels of 51 stress-related proteins changed significantly in the cyp79B2/B3 mutant and six with changes in the myb28/29 mutant. In the cyp79B2/B3 membrane proteome, a group of general stimuli response-related proteins exhibited significant changes compared to WT (Table 2). Among them, the following are examples to directly connect with Gls enzymes: calmodulin-like protein 12 (CML12; Cazzonelli et al., 2014; connected to the indolic GMN via MYB122 and CYP81F2), mediator of RNA polymerase II transcription subunit 37c (MED37C; Lee et al., 2009; connected via GSTF9 to GMN, with possible role in thiohydroximate formation), and glyceraldehyde-3-phosphate dehydrogenase (GAPC2; Guo et al., 2012; formed edges with GMN through MYB28, MYB29, MYB76, and MYB34, suggesting roles in methionine chain-elongation and tryptophan synthesis; Figures 2, 4). It is known that GAPC2 participates in the oxidation of glyceraldehydes-3-phophate to glycerate from which pyruvate is formed. The pyruvate can be converted to acetylCoA for methionine chain-elongation in aliphatic Gls biosynthesis or for synthesis of tryptophan in indolic Gls pathway (Mann, 1987). Both glucosinolate classes were decreased in the cyp79B2/B3 mutant in our previous study (Mostafa et al., 2016) together with GAPC2 in this study. Therefore, the connection between GAPC2 and MYBs in the STRING maps reflects functional relationship and does not necessarily indicate direct physical interaction. Another stress related group showing expression level changes was the salt stress and water deficiency group represented by chloride channel protein CLC-c (Jossier et al., 2010), aquaporin PIP2-2 (Javot, 2003; Tournaire-Roux et al., 2003), annexin D1 (ANN1; Gorecka et al., 2005; Jia et al., 2015; formed edge with GSTF9), early-responsive to dehydration stress protein (Rai et al., 2016), probable aquaporin PIP1-5 (Weig et al., 1997), aquaporin PIP2-3 (Daniels et al., 1994), probable aquaporin PIP1-4 (Li et al., 2015), plasma membrane intrinsic protein 1B (Alexandersson et al., 2005), aquaporin PIP1-3 (Kammerloher et al., 1994), probable aquaporin PIP2-6 (Alexandersson et al., 2010), and aquaporin PIP2-7 (Weig et al., 1997; Figures 2, 4). The decreased expression of this group of aquaporins (Table 2) confirms crosstalk between indolic Gls production and water deficiency enzymes (Khan et al., 2010). The mechanism underlying such crosstalk is intriguing. The reduction in aquaporins potentiates our observation of retarded growth of Gls mutants (Mostafa et al., 2016). The decreased Gls production resulted in stress status, which led to decreased water uptake and decreased expression of aquaporins, and thus growth retardation.
The immunity and defense process was also affected by Gls perturbation, and it is represented by changes in the directly connected nodes: SNAP25 homologous protein (SNAP33; Eschen-Lippold et al., 2012; connected by MYB51 in tryptophan synthesis and CYP81F2 to GMN), protein ILITYHIA (ILA; Monaghan and Li, 2010; playing a role in methionine chain elongation by forming edges with IMD1, IMD2, and IMD3) and a 20 kDa chaperonin (CPN21; Takáč et al., 2014; connected to GMN by the edge GGP1; Figures 2, 4). Another protein exhibiting expression changes and connected to GMN is malate dehydrogenase 2 (mMDH2), which participates in bacterial defense (Jones et al., 2006; Figures 2, 4). In myb28/29, a GTP-binding nuclear protein Ran-1 (Jiang et al., 2007) was found to connect MYB28, MYB29, MYB76, MYB34, MYB51, and MYB122, suggesting its role in methionine chain-elongation and tryptophan synthesis (Figures 3, 4 and Table 3).
Effects of Glucosinolate Metabolism Perturbation on Other Processes and Nodes
Gls biosynthetic pathway is organelle specific and involves transport starting from methionine chain-elongation, sulfate transport, and ending with Gls storage in the seeds (Sønderby et al., 2010; Gigolashvili and Kopriva, 2014; Jørgensen et al., 2015). Here we report a decrease in ABC transporter B family member 19 (Lin and Wang, 2005) in both mutants (Table 1). In addition to their role in sulfate transport, ABC transporters are involved in transporting Gls hydrolysis products (Kang et al., 2011). This result indicates the decrease in glucosinolate levels in the mutants feedback regulate the ABC transporter level. In cyp79B2/B3, a curculin-like (mannose-binding) lectin family protein (At5g18470) involved in carbohydrate binding forms connections with MYB51 and CYP81F2 (Figures 2, 4). How this lectin family protein function is not known. Another biological process affected by the Gls perturbation is photosynthesis as revealed by the increase of photosystem I reaction center subunit IV B in both mutants (Table 1), and increases in cyp79B2/B3 photosystem II stability/assembly factor HCF136 (Meurer et al., 1998), protein curvature thylakoid 1B, NAD(P)H-quinone oxidoreductase subunit H, light-harvesting complex I chlorophyll a/b binding protein 1 and light-harvesting chlorophyll protein complex II subunit B1 (Table 2). The increased activity in the photosynthetic process could be a strategy to compensate for the internal stress in the mutants as indicated by changes of many stress-related proteins (Tables 2, 3; Mostafa et al., 2016). It was obvious that aliphatic Gls metabolism perturbation activated the ribosomal protein expression as reflected by the increased levels of 18 ribosomal proteins in the myb28/29 (Table 3). The biological implication of this change is not known although we can correlate it to the regulation of aliphatic Gls biosynthetic pathway by MYB28 and MYB29 (Li et al., 2013).
In both mutants, adenine phosphoribosyltransferase 1 (APT1) acting on adenine phosphorylation (Allen et al., 2002) showed connections with GGP1 and SUR1, so it might have a role in thiohydroximate formation (Figures 2–4). Its decrease in levels may be a feedback of the decreased Gls production in the mutants. In cyp79B2/B3, FtsZ homolog 1 (FTSZ1) involved in chloroplast division and protein binding (Osteryoung et al., 1998) was found to connect with BCAT3 and GSTF9, suggesting it may affect methionine chain-elongation and thiohydroximate synthesis. Interestingly, another FtsZ homolog 2-2 (FTSZ2-2; McAndrew et al., 2008) was also connected with GSTF9 (Figures 2, 4). Isoform 3 of dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2 (At4g26910) is a member of tricarboxylic acid cycle and can affect methionine biosynthesis and its coupling to acetylCoA in the chain elongation process. Interestingly, it was found to form multiple connections with GMN via BAT5, BCAT3, IMD1, IMD2, IMD3, GSTF9, and SUR1 (Figures 2, 4). In addition, ATP sulfurylase 1 (APS1), a hydrogen sulfide biosynthesis enzyme, formed edges with GGP1 and SUR1, suggesting its potential role in thiohydroximate synthesis (Figures 2, 4). The increased levels of the aforementioned proteins may reflect a feedback mechanism to compensate for reduced Gls levels in the cyp79B2/B3. Flavone 3'-O-methyltransferase 1 (OMT1) in flavonoid metabolism (Muzac et al., 2000) was connected with FMO1, so it could participate in sulfinyl Gls formation (Figures 2, 4). This finding provides another line of evidence for the pathway interaction between phenylpropanoids and glucosinolates. Previously, methionine derived aldoximes were shown to directly or indirectly inhibit caffeic acid O-methyltransferase (COMT) and caffeoyl-CoA O-methyltransferase CCoAOMT), leading to low levels of phenylpropanoid metabolites (Hemm et al., 2003). Here the decreased levels of OMT1 in cyp79B2/B3 may contribute to the decreased production of sulfinyl Gls in the mutant. The data support our metabolomics finding concerning the decreased shikimate level (Mostafa et al., 2016). Another possibility of the OMT1 activity is methylation of hydroxy-indolyl Gls to form methylated indolic Gls (unknown before, Sønderby et al., 2010) in a way similar to methylation of quercetin into isorhamnetin (Figure 5). In myb28/29, 60S ribosomal proteins L13-1 (BBC1) and L15-1 (RPL15A) might be a component in thiohydroximate synthesis through the connections with GSTF9, GSTF10 and/or GSTF11. Both proteins were increased, presumably to compensate for the deficiency of aliphatic Gls in the mutant (Mostafa et al., 2016).
The Proteome and Transcriptome Correlation
In the cyp79B2/B3, the defense and stress-related genes calreticulin 3 (At1g08450; Sun et al., 2014), calmodulin (At2g41100; Cazzonelli et al., 2014), lectin (At5g03350; Armijo et al., 2013), and SNAP25 (At5g61210; Eschen-Lippold et al., 2012) showed significant upregulation in the transcriptome and increases in the proteome. Malate dehydrogenase 2 expression was decreased at both the transcript and protein levels, and it is known to be involved in bacterial defense (Jones et al., 2006). These data have provided additional evidence for the relationship between indolic glucosinolates and stress responses. The overall positive correlation between protein and gene expression levels in the cyp79B2/B3 indicates transcriptional regulation of indole glucosinolates. In myb28/29, although there was no overall correlation between transcript and protein levels, isoform 2 of LysM (At1g21880; Willmann et al., 2011) and AIG2 (avirulence induced gene, At5g39730) exhibited similar downregulation patterns as their corresponding proteins. Both genes are involved in cellular stress responses (Jiang et al., 2007; Willmann et al., 2011). Post-transcriptional and post-translational regulations may contribute to the non-correlation between the expression of some of the genes and their encoded proteins in myb28/29.
Conclusions
Glucosinolate biosynthetic process is controlled by several cytochrome proteins known to be localized to the membrane, but little is known about how Gls metabolism would affect the membrane proteome. In this study, we aim to address this important question utilizing the TMT labeling based quantitative proteomics of two genetic mutants, i.e., cyp79B2/B3 as the indolic Gls mutant and myb28/29 as the aliphatic Gls mutant. We identified 4,673 proteins, out of which 2,171 were membrane proteins. From these membrane proteins and after transmembrane domain analysis, 192 exhibited different levels relative to WT, with cytochrome P450 86A7, cytochrome P450 71B26 and probable cytochrome c representing new cytochromes potentially involved in GMN. Based on our analyses, the first two might play a role in hydroxyl-indolic Gls production. In addition, a flavone 3′-O-methyltransferase 1 is hypothesized to participate in the methylation process of the hydroxyl-indolic Gls to form methoxy-indolic Gls. GO functional enrichment revealed important processes related to stress response, transport activities and photosynthesis in the cyp79B2/B3 and those related to protein translation in the myb28/29. A transcription profiling of both mutants showed a strong correlation between transcript and protein levels in cyp79B2/B3, and no significant correlation in myb28/29. Overall, the new nodes and edges discovered in the GMNs are useful resources for future hypothesis-testing experiments and ultimately toward engineering and breeding of Gls profiles with positive impacts on human health and plant defense.
Author Contributions
IM performed the experiments, data analysis and paper drafting; MY performed qRT-PCR experiment and data analysis; NZ participated in protein extraction and peptides labeling; SG conducted the statistical analysis; CD contributed in LC/MS analysis of peptides; MA and ME provided supervision and advice, and SC designed the experiments, supervised the work and finalized the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer XH and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
We would like to thank Chen laboratory members for their support and co-operation. The US National Science Foundation (NSF CAREER 0845162), University of Florida, and the Egyptian Government represented by the Egyptian Cultural and Educational Bureau at Washington DC are acknowledged for funding this project.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00534/full#supplementary-material
Supplementary Figure 1. Biological process GO enrichment of membrane proteins differentially expressed in cyp79B2/B3 compared to WT.
Supplementary Figure 2. Cellular component GO enrichment of membrane proteins differentially expressed in cyp79B2/B3 compared to WT.
Supplementary Figure 3. Molecular function GO enrichment of membrane proteins differentially expressed in cyp79B2/B3 compared to WT.
Supplementary Figure 4. Biological process GO enrichment of membrane proteins differentially expressed in myb28/29 compared to WT.
Supplementary Figure 5. Cellular component GO enrichment of membrane proteins differentially expressed in myb28/29 compared to WT.
Supplementary Figure 6. Molecular function GO enrichment of membrane proteins differentially expressed in myb28/29 compared to WT.
Supplementary Figure 7. Correlation between transcript and protein levels inferred from 32 to 22 genes for cyp79B2/B3 and myb28/29, respectively. Pearson correlation r = 0.6579 (p = 4.269e−05) for cyp79B2/B3 and r = 0.0887 (p = 0.6945) for myb28/29.
Supplementary Table 1. Primer information used in qRT-PCR.
Supplementary Table 2. Proteomics data from two independent experiments (the data were generated using Proteome Discoverer 1.4 by searching the raw data against the Arabidopsis tair 10 database).
Supplementary Table 3. Transmembrane domains predection analyses.
Supplementary Table 4. GO enrichment of proteins differentially expressed in cyp79B2/B3 and myb28/29 mutants relative to WT using AgriGO SEACOMPARE.
Supplementary Table 5. Gene expression at transcript and protein levels in cyp79B2/B3 and myb28/29 relative to WT.
Abbreviations
Gls, glucosinolate; GMN, glucosinolate molecular network; GO, Gene Ontology; TMT, tandem mass tags; WT, Arabidopsis thaliana wild type.
References
Abe, H., Obinata, T., Minamide, L. S., and Bamburg, J. R. (1996). Xenopus laevis actin-depolymerizing factor/cofilin: a phosphorylation-regulated protein essential for development. J. Cell Biol. 132, 871–885. doi: 10.1083/jcb.132.5.871
Agudelo-Romero, P., Carbonell, P., De La Iglesia, F., Carrera, J., Rodrigo, G., Jaramillo, A., et al. (2008). Changes in the gene expression profile of Arabidopsis thaliana after infection with Tobacco etch virus. Virol. J. 5, 1. doi: 10.1186/1743-422X-5-92
Alexandersson, E., Danielson, J. A., Råde, J., Moparthi, V. K., Fontes, M., Kjellbom, P., et al. (2010). Transcriptional regulation of aquaporins in accessions of Arabidopsis in response to drought stress. Plant J. 61, 650–660. doi: 10.1111/j.1365-313X.2009.04087.x
Alexandersson, E., Fraysse, L., Sjövall-Larsen, S., Gustavsson, S., Fellert, M., Karlsson, M., et al. (2005). Whole gene family expression and drought stress regulation of aquaporins. Plant Mol. Biol. 59, 469–484. doi: 10.1007/s11103-005-0352-1
Allen, M., Qin, W., Moreau, F., and Moffatt, B. (2002). Adenine phosphoribosyltransferase isoforms of Arabidopsis and their potential contributions to adenine and cytokinin metabolism. Physiol. Plant. 115, 56–68. doi: 10.1034/j.1399-3054.2002.1150106.x
Antignani, V., Klocko, A. L., Bak, G., Chandrasekaran, S. D., Dunivin, T., and Nielsen, E. (2015). Recruitment of plant U-BOX13 and the PI4Kβ1/β2 phosphatidylinositol-4 kinases by the small GTPase RabA4B plays important roles during salicylic acid-mediated plant defense signaling in Arabidopsis. Plant Cell 27, 243–261. doi: 10.1105/tpc.114.134262
Armijo, G., Salinas, P., Monteoliva, M. I., Seguel, A., García, C., Villarroel-Candia, E., et al. (2013). A salicylic acid-induced lectin-like protein plays a positive role in the effector-triggered immunity response of Arabidopsis thaliana to Pseudomonas syringae Avr-Rpm1. Mol. Plant Microbe Interact. 26, 1395–1406. doi: 10.1094/MPMI-02-13-0044-R
Ascencio-Ibáñez, J. T., Sozzani, R., Lee, T. J., Chu, T. M., Wolfinger, R. D., Cella, R., et al. (2008). Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148, 436–454. doi: 10.1104/pp.108.121038
Axtell, M. J., and Staskawicz, B. J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112, 369–377. doi: 10.1016/S0092-8674(03)00036-9
Azimzadeh, J., Nacry, P., Christodoulidou, A., Drevensek, S., Camilleri, C., Amiour, N., et al. (2008). Arabidopsis TONNEAU1 proteins are essential for preprophase band formation and interact with centrin. Plant Cell 20, 2146–2159. doi: 10.1105/tpc.107.056812
Baldrianová, J., Černý, M., Novák, J., Jedelský, P. L., Divíšková, E., and Brzobohatý, B. (2015). Arabidopsis proteome responses to the smoke-derived growth regulator karrikin. J. Proteomics 120, 7–20. doi: 10.1016/j.jprot.2015.02.011
Bedhomme, M., Hoffmann, M., McCarthy, E. A., Gambonnet, B., Moran, R. G., Rébeillé, F., et al. (2005). Folate metabolism in plants: an Arabidopsis homolog of the mammalian mitochondrial folate transporter mediates folate import into chloroplasts. J. Biol. Chem. 280, 34823–34831. doi: 10.1074/jbc.M506045200
Bessire, M., Borel, S., Fabre, G., Carraça, L., Efremova, N., Yephremov, A., et al. (2011). A member of the pleiotropic drug resistance family of ATP binding cassette transporters is required for the formation of a functional cuticle in Arabidopsis. Plant Cell 23, 1958–1970. doi: 10.1105/tpc.111.083121
Birchmeier, W., Kohler, C. E., and Schatz, G. (1976). Interaction of integral and peripheral membrane proteins: affinity labeling of yeast cytochrome oxidase by modified yeast cytochrome c. Proc. Natl. Acad. Sci. U.S.A. 73, 4334–4338. doi: 10.1073/pnas.73.12.4334
Campbell, P., and Braam, J. (1999). In vitro activities of four xyloglucan endotransglycosylases from Arabidopsis. Plant J. 18, 371–382. doi: 10.1046/j.1365-313X.1999.00459.x
Cazzonelli, C. I., Nisar, N., Roberts, A. C., Murray, K. D., Borevitz, J. O., and Pogson, B. J. (2014). A chromatin modifying enzyme, SDG8, is involved in morphological, gene expression, and epigenetic responses to mechanical stimulation. Front. Plant Sci. 5:533. doi: 10.3389/fpls.2014.00533
Chen, J., Bang, W. Y., Lee, Y., Kim, S., Lee, K. W., Kim, S. W., et al. (2014). AtObgC-AtRSH1 interaction may play a vital role in stress response signal transduction in Arabidopsis. Plant Physiol. Biochem. 74, 176–184. doi: 10.1016/j.plaphy.2013.10.022
Chen, Y., Yan, X., and Chen, S. (2011). Bioinformatic analysis of molecular network of glucosinolate biosynthesis. Comput. Biol. Chem. 35, 10–18. doi: 10.1016/j.compbiolchem.2010.12.002
Chen, Y. Z., Pang, Q. Y., He, Y., Zhu, N., Branstrom, I., Yan, X. F., et al. (2012). Proteomics and metabolomics of Arabidopsis responses to perturbation of glucosinolate biosynthesis. Mol. Plant 5, 1138–1150. doi: 10.1093/mp/sss034
Clay, N. K., Adio, A. M., Denoux, C., Jander, G., and Ausubel, F. M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323, 95–101. doi: 10.1126/science.1164627
Czégény, G., Wu, M., Dér, A., Eriksson, L. A., Strid, Å., and Hideg, É. (2014). Hydrogen peroxide contributes to the ultraviolet-B (280–315nm) induced oxidative stress of plant leaves through multiple pathways. FEBS Lett. 588, 2255–2261. doi: 10.1016/j.febslet.2014.05.005
Dai, N., Wang, W., Patterson, S. E., and Bleecker, A. B. (2013). The TMK subfamily of receptor-like kinases in Arabidopsis display an essential role in growth and a reduced sensitivity to auxin. PLoS ONE 8:e60990. doi: 10.1371/journal.pone.0060990
Daniels, M. J., Mirkov, T. E., and Chrispeels, M. J. (1994). The plasma membrane of Arabidopsis thaliana contains a mercury-insensitive aquaporin that is a homolog of the tonoplast water channel protein TIP. Plant Physiol. 106, 1325–1333. doi: 10.1104/pp.106.4.1325
Desveaux, D., Maréchal, A., and Brisson, N. (2005). Whirly transcription factors: defense gene regulation and beyond. Trends Plant Sci. 10, 95–102. doi: 10.1016/j.tplants.2004.12.008
Duan, H., and Schuler, M. A. (2005). Differential expression and evolution of the Arabidopsis CYP86A subfamily. Plant Physiol. 137, 1067–1081. doi: 10.1104/pp.104.055715
Edstam, M. M., and Edqvist, J. (2014). Involvement of GPI-anchored lipid transfer proteins in the development of seed coats and pollen in Arabidopsis thaliana. Physiol. Plant. 152, 32–42. doi: 10.1111/ppl.12156
Eschen-Lippold, L., Landgraf, R., Smolka, U., Schulze, S., Heilmann, M., Heilmann, I., et al. (2012). Activation of defense against Phytophthora infestans in potato by down-regulation of syntaxin gene expression. New Phytol. 193, 985–996. doi: 10.1111/j.1469-8137.2011.04024.x
Feng, J., Li, J., Gao, Z., Lu, Y., Yu, J., Zheng, Q., et al. (2015). SKIP confers osmotic tolerance during salt stress by controlling alternative gene splicing in Arabidopsis. Mol. Plant 8, 1038–1052. doi: 10.1016/j.molp.2015.01.011
Frerigmann, H., Böttcher, C., Baatout, D., and Gigolashvili, T. (2012). Glucosinolates are produced in trichomes of Arabidopsis thaliana. Front. Plant Sci. 3:242. doi: 10.3389/fpls.2012.00242
Friso, G., Giacomelli, L., Ytterberg, A. J., Peltier, J. B., Rudella, A., Sun, Q., et al. (2004). In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16, 478–499. doi: 10.1105/tpc.017814
Garcia-Molina, A., Andrés-Colás, N., Perea-García, A., Neumann, U., Dodani, S. C., Huijser, P., et al. (2013). The Arabidopsis COPT6 transport protein functions in copper distribution under copper-deficient conditions. Plant Cell Physiol. 54, 1378–1390. doi: 10.1093/pcp/pct088
Ghelis, T., Bolbach, G., Clodic, G., Habricot, Y., Miginiac, E., Sotta, B., et al. (2008). Protein tyrosine kinases and protein tyrosine phosphatases are involved in ABA-dependent processes in Arabidopsis thaliana seeds and suspension cells. Plant Physiol. 148, 1668–1680. doi: 10.1104/pp.108.124594
Gigolashvili, T., and Kopriva, S. (2014). Transporters in plant sulfur metabolism. Front. Plant Sci. 7:442. doi: 10.3389/fpls.2014.00442
Gorecka, K. M., Konopka-Postupolska, D., Hennig, J., Buchet, R., and Pikula, S. (2005). Peroxidase activity of annexin 1 from Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 336, 868–875. doi: 10.1016/j.bbrc.2005.08.181
Grubb, C. D., and Abel, S. (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11, 89–100. doi: 10.1016/j.tplants.2005.12.006
Guo, L., Devaiah, S. P., Narasimhan, R., Pan, X., Zhang, Y., Zhang, W., et al. (2012). Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Dδ to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell 24, 2200–2212. doi: 10.1105/tpc.111.094946
Guo, R. F., Yuan, G. F., and Wang, Q. M. (2013). Effect of NaCl treatments on glucosinolate metabolism in broccoli sprouts. J. Zhejiang Univ. Sci. B 14, 124–131. doi: 10.1631/jzus.B1200096
Halkier, B. A., and Gershenzon, J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57, 303–333. doi: 10.1146/annurev.arplant.57.032905.105228
Harper, A. D., and Bar-Peled, M. (2002). Biosynthesis of UDP-xylose. Cloning and characterization of a novel Arabidopsis gene family, UXS, encoding soluble and putative membrane-bound UDP-glucuronic acid decarboxylase isoforms. Plant Physiol. 130, 2188–2198. doi: 10.1104/pp.009654
Hemm, M. R., Ruegger, M. O., and Chapple, C. (2003). The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell 15, 179–194. doi: 10.1105/tpc.006544
Iglesias, J., Trigueros, M., Rojas-Triana, M., Fernández, M., Albar, J. P., Bustos, R., et al. (2013). Proteomics identifies ubiquitin–proteasome targets and new roles for chromatin-remodeling in the Arabidopsis response to phosphate starvation. J. Proteomics 94, 1–22. doi: 10.1016/j.jprot.2013.08.015
Jaillais, Y., Santambrogio, M., Rozier, F., Fobis-Loisy, I., Miège, C., and Gaude, T. (2007). The retromer protein VPS29 links cell polarity and organ initiation in plants. Cell 130, 1057–1070. doi: 10.1016/j.cell.2007.08.040
Jang, J. C., León, P., Zhou, L., and Sheen, J. (1997). Hexokinase as a sugar sensor in higher plants. Plant Cell 9, 5–19. doi: 10.1105/tpc.9.1.5
Javot, H. (2003). Role of a single aquaporin isoform in root water uptake. Plant Cell Online 15, 509–522. doi: 10.1105/tpc.008888
Jelenska, J., Kang, Y., and Greenberg, J. T. (2014). Plant pathogenic bacteria target the actin microfilament network involved in the trafficking of disease defense components. Bioarchitecture 4, 149–153. doi: 10.4161/19490992.2014.980662
Ji, W., Cong, R., Li, S., Li, R., Qin, Z., Li, Y., et al. (2016). Comparative proteomic analysis of soybean leaves and roots by iTRAQ provides insights into response mechanisms to short-term salt stress. Front. Plant Sci. 7:573. doi: 10.3389/fpls.2016.00573
Jia, F., Wang, C., Huang, J., Yang, G., Wu, C., and Zheng, C. (2015). SCF E3 ligase PP2-B11 plays a positive role in response to salt stress in Arabidopsis. J. Exp. Bot. 66, 4683–4697. doi: 10.1093/jxb/erv245
Jia, H., Li, J., Zhu, J., Fan, T., Qian, D., Zhou, Y., et al. (2013). Arabidopsis CROLIN1, a novel plant actin-binding protein, functions in cross-linking and stabilizing actin filaments. J. Biol. Chem. 288, 32277–32288. doi: 10.1074/jbc.M113.483594
Jiang, Y., Yang, B., Harris, N. S., and Deyholos, M. K. (2007). Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J. Exp. Bot. 58, 3591–3607. doi: 10.1093/jxb/erm207
Jones, A. M., Thomas, V., Bennett, M. H., Mansfield, J., and Grant, M. (2006). Modifications to the Arabidopsis defense proteome occur prior to significant transcriptional change in response to inoculation with Pseudomonas syringae. Plant Physiol. 142, 1603–1620. doi: 10.1104/pp.106.086231
Jørgensen, M. E., Olsen, C. E., Geiger, D., Mirza, O., Halkier, B. A., and Nour-Eldin, H. H. (2015). A functional EXXEK motif is essential for proton coupling and active glucosinolate transport by NPF2.11. Plant Cell Physiol. 56, 2340–2350. doi: 10.1093/pcp/pcv145
Jossier, M., Kroniewicz, L., Dalmas, F., Le Thiec, D., Ephritikhine, G., Thomine, S., et al. (2010). The Arabidopsis vacuolar anion transporter, AtCLCc, is involved in the regulation of stomatal movements and contributes to salt tolerance. Plant J. 64, 563–576. doi: 10.1111/j.1365-313X.2010.04352.x
Kammerloher, W., Fischer, U., Piechottka, G. P., and Schäffner, A. R. (1994). Water channels in the plant plasma membrane cloned by immunoselection from a mammalian expression system. Plant J. 6, 187–199. doi: 10.1046/j.1365-313X.1994.6020187.x
Kanaoka, M. M., Urban, S., Freeman, M., and Okada, K. (2005). An Arabidopsis rhomboid homolog is an intramembrane protease in plants. FEBS Lett. 579, 5723–5728. doi: 10.1016/j.febslet.2005.09.049
Kandasamy, M. K., McKinney, E. C., and Meagher, R. B. (2002). Functional nonequivalency of actin isovariants in Arabidopsis. Mol. Biol. Cell 13, 251–261. doi: 10.1091/mbc.01-07-0342
Kang, J., Park, J., Choi, H., Burla, B., Kretzschmar, T., Lee, Y., et al. (2011). Plant ABC transporters. Arabidopsis Book 9:e0153. doi: 10.1199/tab.0153
Kawamura, Y., and Uemura, M. (2003). Mass spectrometric approach for identifying putative plasma membrane proteins of Arabidopsis leaves associated with cold acclimation. Plant J. 36, 141–154. doi: 10.1046/j.1365-313X.2003.01864.x
Khan, M. A. M., Ulrichs, C., and Mewis, I. (2010). Influence of water stress on the glucosinolate profile of Brassica oleracea var. italica and the performance of Brevicoryne brassicae and Myzus persicae. Entomol. Exp. Appl. 137, 229–236. doi: 10.1111/j.1570-7458.2010.01059.x
Kissen, R., Rossiter, J. T., and Bones, A. M. (2009). The ‘mustard oil bomb’: not so easy to assemble?! Localization, expression and distribution of the components of the myrosinase enzyme system. Phytochem. Rev. 8, 69–86. doi: 10.1007/s11101-008-9109-1
Knopf, R. R., and Adam, Z. (2012). Rhomboid proteases in plants–still in square one? Physiol. Plant. 145, 41–51. doi: 10.1111/j.1399-3054.2011.01532.x
Koh, J., Chen, S., Zhu, N., Yu, F., Soltis, P. S., and Soltis, D. E. (2012). Comparative proteomics of the recently and recurrently formed natural allopolyploid Tragopogon mirus (Asteraceae) and its parents. New Phytol. 196, 292−305. doi: 10.1111/j.1469-8137.2012.04251.x
Law, A. H., Chow, C. M., and Jiang, L. (2012). Secretory carrier membrane proteins. Protoplasma 249, 269–283. doi: 10.1007/s00709-011-0295-0
Lee, S., Lee, D. W., Lee, Y., Mayer, U., Stierhof, Y. D., Lee, S., et al. (2009). Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in Arabidopsis. Plant Cell 21, 3984–4001. doi: 10.1105/tpc.109.071548
Li, C. L., Wang, M., Ma, X. Y., and Zhang, W. (2014). NRGA1, a putative mitochondrial pyruvate carrier, mediates ABA regulation of guard cell ion channels and drought stress responses in Arabidopsis. Mol. Plant 7, 1508–1521. doi: 10.1093/mp/ssu061
Li, L., Wang, H., Gago, J., Cui, H., Qian, Z., Kodama, N., et al. (2015). Harpin Hpa1 interacts with aquaporin PIP1; 4 to promote the substrate transport and photosynthesis in Arabidopsis. Sci. Rep. 5:17207. doi: 10.1038/srep17207
Li, Y., Sawada, Y., Hirai, A., Sato, M., Kuwahara, A., Yan, X., et al. (2013). Novel insights into the function of Arabidopsis R2R3-MYB transcription factors regulating aliphatic glucosinolate biosynthesis. Plant Cell Physiol. 54, 1335–1344. doi: 10.1093/pcp/pct085
Lim, T. K., Le, K. P. U., Lin, Q., and Nguyen, T. T. H. (2017). iTRAQ-based proteome analysis of fluoroquinolone-resistant Staphylococcus aureus. J. Glob. Antimicrob. Resist. 8, 82–89. doi: 10.1016/j.jgar.2016.11.003
Lin, R., and Wang, H. (2005). Two homologous ATP-binding cassette transporter proteins, AtMDR1 and AtPGP1, regulate Arabidopsis photomorphogenesis and root development by mediating polar auxin transport. Plant Physiol. 138, 949–964. doi: 10.1104/pp.105.061572
Lisenbee, C. S., Lingard, M. J., and Trelease, R. N. (2005). Arabidopsis peroxisomes possess functionally redundant membrane and matrix isoforms of monodehydroascorbate reductase. Plant J. 43, 900–914. doi: 10.1111/j.1365-313X.2005.02503.x
Marmagne, A., Brabant, P., Thiellement, H., and Alix, K. (2010). Analysis of gene expression in resynthesized Brassica napus allotetraploids: transcriptional changes do not explain differential protein regulation. New Phytol. 186, 216–227. doi: 10.1111/j.1469-8137.2009.03139.x
Martínez-Ballesta, M. D.-C., Moreno, D. A., and Carvajal, M. (2013). The physiological importance of glucosinolates on plant response to abiotic stress in Brassica. Int. J. Mol. Sci. 14, 11607–11625. doi: 10.3390/ijms140611607
McAndrew, R. S., Olson, B. J., Kadirjan-Kalbach, D. K., Chi-Ham, C. L., Vitha, S., Froehlich, J. E., et al. (2008). In vivo quantitative relationship between plastid division proteins FtsZ1 and FtsZ2 and identification of ARC6 and ARC3 in a native FtsZ complex. Biochem. J. 412, 367–378. doi: 10.1042/BJ20071354
McDowell, J. M., An, Y. Q., Huang, S., McKinney, E. C., and Meagher, R. B. (1996). The Arabidopsis ACT7 actin gene is expressed in rapidly developing tissues and responds to several external stimuli. Plant Physiol. 111, 699–711. doi: 10.1104/pp.111.3.699
Meurer, J., Plücken, H., Kowallik, K. V., and Westhoff, P. (1998). A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17, 5286–5297. doi: 10.1093/emboj/17.18.5286
Michalecka, A. M., Svensson, A. S., Johansson, F. I., Agius, S. C., Johanson, U., Brennicke, A., et al. (2003). Arabidopsis genes encoding mitochondrial type II NAD(P)H dehydrogenases have different evolutionary origin and show distinct responses to light. Plant Physiol. 133, 642–652. doi: 10.1104/pp.103.024208
Minami, A., Tominaga, Y., Furuto, A., Kondo, M., Kawamura, Y., and Uemura, M. (2015). Arabidopsis dynamin-related protein 1E in sphingolipid-enriched plasma membrane domains is associated with the development of freezing tolerance. Plant J. 83, 501–514. doi: 10.1111/tpj.12907
Monaghan, J., and Li, X. (2010). The heat repeat protein ILITYHIA is required for plant immunity. Plant Cell Physiol. 51, 742–753. doi: 10.1093/pcp/pcq038
Mostafa, I., Zhu, N., Yoo, M. J., Balmant, K. M., Misra, B. B., Dufresne, C., et al. (2016). New nodes and edges in the glucosinolate molecular network revealed by proteomics and metabolomics of Arabidopsis myb28/29 and cyp79B2/B3 glucosinolate mutants. J. Proteomics 138, 1–19. doi: 10.1016/j.jprot.2016.02.012
Mukherjee, A. K., Carp, M. J., Zuchman, R., Ziv, T., Horwitz, B. A., and Gepstein, S. (2010). Proteomics of the response of Arabidopsis thaliana to infection with Alternaria brassicicola. J. Proteomics 73, 709–720. doi: 10.1016/j.jprot.2009.10.005
Murakami, R., Ifuku, K., Takabayashi, A., Shikanai, T., Endo, T., and Sato, F. (2002). Characterization of an Arabidopsis thaliana mutant with impaired psbO, one of two genes encoding extrinsic 33-kDa proteins in photosystem II. FEBS Lett. 523, 138–142. doi: 10.1016/S0014-5793(02)02963-0
Murray, J., Zhang, B., Taylor, S. W., Oglesbee, D., Fahy, E., Marusich, M. F., et al. (2003). The subunit composition of the human NADH dehydrogenase obtained by rapid one-step immunopurification. J. Biol. Chem. 278, 13619–13622. doi: 10.1074/jbc.C300064200
Mustroph, A., Sonnewald, U., and Biemelt, S. (2007). Characterisation of the ATP-dependent phosphofructokinase gene family from Arabidopsis thaliana. FEBS Lett. 581, 2401–2410. doi: 10.1016/j.febslet.2007.04.060
Muzac, I., Wang, J., Anzellotti, D., Zhang, H., and Ibrahim, R. K. (2000). Functional expression of an Arabidopsis cDNA clone encoding a flavonol 3'-O-methyltransferase and characterization of the gene product. Arch. Biochem. Biophys. 375, 385–388. doi: 10.1006/abbi.1999.1681
Nafisi, M., Goregaoker, S., Botanga, C. J., Glawischnig, E., Olsen, C. E., Halkier, B. A., et al. (2007). Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19, 2039–2052. doi: 10.1105/tpc.107.051383
Narendra, S., Venkataramani, S., Shen, G., Wang, J., Pasapula, V., Lin, Y., et al. (2006). The Arabidopsis ascorbate peroxidase 3 is a peroxisomal membrane-bound antioxidant enzyme and is dispensable for Arabidopsis growth and development. J. Exp. Bot. 57, 3033–3042. doi: 10.1093/jxb/erl060
Neve, E. P., and Ingelman-Sundberg, M. (2010). Cytochrome P450 proteins: retention and distribution from the endoplasmic reticulum. Curr. Opin. Drug Discov. Dev. 13, 78–85.
Nielsen, M. E., and Thordal-Christensen, H. (2012). Recycling of Arabidopsis plasma membrane PEN1 syntaxin. Plant Signal. Behav. 7, 1541–1543. doi: 10.4161/psb.22304
Noël, L. D., Cagna, G., Stuttmann, J., Wirthmüller, L., Betsuyaku, S., Witte, C.-P., et al. (2007). Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell 19, 4061–4076. doi: 10.1105/tpc.107.051896
Okamoto, M., Vidmar, J. J., and Glass, A. D. (2003). Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: responses to nitrate provision. Plant Cell Physiol. 44, 304–317. doi: 10.1093/pcp/pcg036
Osteryoung, K. W., Stokes, K. D., Rutherford, S. M., Percival, A. L., and Lee, W. Y. (1998). Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10, 1991–2004. doi: 10.2307/3870779
Palmblad, M., Mills, D. J., and Bindschedler, L. V. (2008). Heat-shock response in Arabidopsis thaliana explored by multiplexed quantitative proteomics using differential metabolic labeling. J. Proteome Res. 7, 780–785. doi: 10.1021/pr0705340
Pang, Q., Chen, S., Dai, S., Chen, Y., Wang, Y., and Yan, X. (2010). Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila. J. Proteome Res. 9, 2584–2599. doi: 10.1021/pr100034f
Patel, S. (2004). Arabidopsis WPP-domain proteins are developmentally associated with the nuclear envelope and promote cell division. Plant Cell Online 16, 3260–3273. doi: 10.1105/tpc.104.026740
Peng, L., Ma, J., Chi, W., Guo, J., Zhu, S., Lu, Q., et al. (2006). Low PSII accumulation1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18, 955–969. doi: 10.1105/tpc.105.037689
Pesaresi, P., Scharfenberg, M., Weigel, M., Granlund, I., Schröder, W. P., Finazzi, G., et al. (2009). Mutants, overexpressors, and interactors of Arabidopsis plastocyanin isoforms: revised roles of plastocyanin in photosynthetic electron flow and thylakoid redox state. Mol. Plant 2, 236–248. doi: 10.1093/mp/ssn041
Picault, N., Palmieri, L., Pisano, I., Hodges, M., and Palmieri, F. (2002). Identification of a novel transporter for dicarboxylates and tricarboxylates in plant mitochondria bacterial expression, reconstitution, functional characterization, and tissue distribution. J. Biol. Chem. 277, 24204–24211. doi: 10.1074/jbc.M202702200
Quirino, B. F., Reiter, W. D., and Amasino, R. D. (2001). One of two tandem Arabidopsis genes homologous to monosaccharide transporters is senescence-associated. Plant Mol. Biol. 46, 447–457. doi: 10.1023/A:1010639015959
Rai, A. N., Tamirisa, S., Rao, K. V., Kumar, V., and Suprasanna, P. (2016). Brassica RNA binding protein ERD4 is involved in conferring salt, drought tolerance and enhancing plant growth in Arabidopsis. Plant Mol. Biol. 90, 375–387. doi: 10.1007/s11103-015-0423-x
Rama Devi, S., Chen, X., Oliver, D. J., and Xiang, C. (2006). A novel high-throughput genetic screen for stress-responsive mutants of Arabidopsis thaliana reveals new loci involving stress responses. Plant J. 47, 652–663. doi: 10.1111/j.1365-313X.2006.02814.x
Ramón, N. M., and Bartel, B. (2010). Interdependence of the peroxisome-targeting receptors in Arabidopsis thaliana: PEX7 facilitates PEX5 accumulation and import of PTS1 cargo into peroxisomes. Mol. Biol. Cell 21, 1263–1271. doi: 10.1091/mbc.E09-08-0672
Ren, X. L., Qi, G. N., Feng, H. Q., Zhao, S., Zhao, S. S., Wang, Y., et al. (2013). Calcineurin B-like protein CBL10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis. Plant J. 74, 258–266. doi: 10.1111/tpj.12123
Sakamoto, W., Tamura, T., Hanba-Tomita, Y., and Murata, M. (2002). The VAR1 locus of Arabidopsis encodes a chloroplastic FtsH and is responsible for leaf variegation in the mutant alleles. Genes Cells 7, 769–780. doi: 10.1046/j.1365-2443.2002.00558.x
Shi, H., Shen, Q., Qi, Y., Yan, H., Nie, H., Chen, Y., et al. (2013). BR-signaling kinase1 physically associates with flagellin sensing2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25, 1143–1157. doi: 10.1105/tpc.112.107904
Shirakawa, M., Ueda, H., Shimada, T., and Hara-Nishimura, I. (2016). FAMA: a molecular link between stomata and myrosin cells. Trends Plant Sci. 21, 861–871. doi: 10.1016/j.tplants.2016.07.003
Silva-Sanchez, C., Chen, S., Zhu, N., Li, Q. B., and Chourey, P. S. (2013). Proteomic comparison of basal endosperm in maize miniature1 mutant and its wild-type Mn1. Front. Plant Sci. 4:211. doi: 10.3389/fpls.2013.00211
Sokolenko, A., Pojidaeva, E., Zinchenko, V., Panichkin, V., Glaser, V. M., Herrmann, R. G., et al. (2002). The gene complement for proteolysis in the cyanobacterium Synechocystis sp. PCC 6803 and Arabidopsis thaliana chloroplasts. Curr. Genet. 41, 291–310. doi: 10.1007/s00294-002-0309-8
Sønderby, I. E., Geu-Flores, F., and Halkier, B. A. (2010). Biosynthesis of glucosinolates - gene discovery and beyond. Trends Plant Sci. 15, 283–290. doi: 10.1016/j.tplants.2010.02.005
Sun, L., Xu, D., Xu, Q., Sun, J., Xing, L., Zhang, L., et al. (2017). iTRAQ reveals proteomic changes during intestine regeneration in the sea cucumber Apostichopus japonicus. Comp. Biochem. Physiol. Part D. 22, 39–49. doi: 10.1016/j.cbd.2017.02.004
Sun, T., Zhang, Q., Gao, M., and Zhang, Y. (2014). Regulation of SOBIR1 accumulation and activation of defense responses in bir1–1 by specific components of ER quality control. Plant J. 77, 748–756. doi: 10.1111/tpj.12425
Sweetlove, L. J., Heazlewood, J. L., Herald, V., Holtzapffel, R., Day, D. A., Leaver, C. J., et al. (2002). The impact of oxidative stress on Arabidopsis mitochondria. Plant J. 32, 891–904. doi: 10.1046/j.1365-313X.2002.01474.x
Szumlanski, A. L., and Nielsen, E. (2009). The Rab GTPase RabA4d regulates pollen tube tip growth in Arabidopsis thaliana. Plant Cell 21, 526–544. doi: 10.1105/tpc.108.060277
Takáč, T., Šamajová, O., Vadovič, P., Pechan, T., Košútová, P., Ovečka, M., et al. (2014). Proteomic and biochemical analyses show a functional network of proteins involved in antioxidant defense of the Arabidopsis anp2anp3 double mutant. J. Proteome Res. 13, 5347–5361. doi: 10.1021/pr500588c
Thompson, E. P., Smith, S. G., and Glover, B. J. (2012). An Arabidopsis rhomboid protease has roles in the chloroplast and in flower development. J. Exp. Bot. 63, 3559–3570. doi: 10.1093/jxb/ers012
Tournaire-Roux, C., Sutka, M., Javot, H., Gout, E., Gerbeau, P., Luu, D. T., et al. (2003). Cytosolic pH regulates root water transport during anoxic stress through gating of aquaporins. Nature 425, 393–397. doi: 10.1038/nature01853
Voxeur, A., André, A., Breton, C., and Lerouge, P. (2012). Identification of putative rhamnogalacturonan-II specific glycosyltransferases in Arabidopsis using a combination of bioinformatics approaches. PLoS ONE 7:e51129. doi: 10.1371/journal.pone.0051129
Walter, B., Pieta, T., and Schünemann, D. (2015). Arabidopsis thaliana mutants lacking cpFtsY or cpSRP54 exhibit different defects in photosystem II repair. Front. Plant Sci. 6:250. doi: 10.3389/fpls.2015.00250
Weig, A., Deswarte, C., and Chrispeels, M. J. (1997). The major intrinsic protein family of Arabidopsis has 23 members that form three distinct groups with functional aquaporins in each group. Plant Physiol. 114, 1347–1357. doi: 10.1104/pp.114.4.1347
Welchen, E., Hildebrandt, T. M., Lewejohann, D., Gonzalez, D. H., and Braun, H. P. (2012). Lack of cytochrome c in Arabidopsis decreases stability of Complex IV and modifies redox metabolism without affecting Complexes I and III. Biochim. Biophys. Acta 1817, 990–1001. doi: 10.1016/j.bbabio.2012.04.008
Willmann, R., Lajunen, H. M., Erbs, G., Newman, M., Kolb, D., and Tsuda, K. (2011). Mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 108, 19824–19829. doi: 10.1073/pnas.1112862108
Wintz, H., Fox, T., Wu, Y. Y., Feng, V., Chen, W., Chang, H. S., et al. (2003). Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J. Biol. Chem. 278, 47644–47653. doi: 10.1074/jbc.M309338200
Xia, C., Wang, Y. J., Li, W. Q., Chen, Y. R., Deng, Y., Zhang, X. Q., et al. (2010). The Arabidopsis eukaryotic translation initiation factor 3, subunit F (AteIF3f), is required for pollen germination and embryogenesis. Plant J. 63, 189–202. doi: 10.1111/j.1365-313X.2010.04237.x
Yan, X., and Chen, S. (2007). Regulation of plant glucosinolate metabolism. Planta 226, 1343–1352. doi: 10.1007/s00425-007-0627-7
Yang, S., Zeng, X., Li, T., Liu, M., Zhang, S., Gao, S., et al. (2012). AtACDO1, an ABC1-like kinase gene, is involved in chlorophyll degradation and the response to photooxidative stress in Arabidopsis. J. Exp. Bot. 63, 3959–3973. doi: 10.1093/jxb/ers072
Yang, X. H., Xu, Z. H., and Xue, H. W. (2005). Arabidopsis membrane steroid binding protein 1 is involved in inhibition of cell elongation. Plant Cell 17, 116–131. doi: 10.1105/tpc.104.028381
Zelazny, E., Santambrogio, M., Pourcher, M., Chambrier, P., Berne-Dedieu, A., Fobis-Loisy, I., et al. (2013). Mechanisms governing the endosomal membrane recruitment of the core retromer in Arabidopsis. J. Biol. Chem 288, 8815–8825. doi: 10.1074/jbc.M112.440503
Zhang, M., Wang, C., Lin, Q., Liu, A., Wang, T., Feng, X., et al. (2015). A tetratricopeptide repeat domain-containing protein SSR1 located in mitochondria is involved in root development and auxin polar transport in Arabidopsis. Plant J. 83, 582–599. doi: 10.1111/tpj.12911
Zhao, Y., Hull, A. K., Gupta, N. R., Goss, K. A., Alonso, J., Ecker, J. R., et al. (2002). Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16, 3100–3112. doi: 10.1101/gad.1035402
Zheng, H., Bassham, D. C., da Silva Conceição, A., and Raikhel, N. V. (1999). The syntaxin family of proteins in Arabidopsis : a new syntaxin homologue shows polymorphism between two ecotypes. J. Exp. Bot. 50, 915–924. doi: 10.1093/jxb/50.Special_Issue.915
Keywords: Arabidopsis, membrane proteome, glucosinolate, stress and defense, molecular networks
Citation: Mostafa I, Yoo M-J, Zhu N, Geng S, Dufresne C, Abou-Hashem M, El-Domiaty M and Chen S (2017) Membrane Proteomics of Arabidopsis Glucosinolate Mutants cyp79B2/B3 and myb28/29. Front. Plant Sci. 8:534. doi: 10.3389/fpls.2017.00534
Received: 02 February 2017; Accepted: 24 March 2017;
Published: 11 April 2017.
Edited by:
Wei Wang, Henan Agricultural University, ChinaReviewed by:
Xiuli Hu, Henan Agricultural University, ChinaOmar Pantoja, National Autonomous University of Mexico, Mexico
Copyright © 2017 Mostafa, Yoo, Zhu, Geng, Dufresne, Abou-Hashem, El-Domiaty and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sixue Chen, schen@ufl.edu
 Islam Mostafa
Islam Mostafa Mi-Jeong Yoo
Mi-Jeong Yoo Ning Zhu1,2
Ning Zhu1,2 Sisi Geng
Sisi Geng Craig Dufresne
Craig Dufresne Sixue Chen
Sixue Chen