- 1Plant Lab, Institute of Life Sciences, Scuola Superiore Sant'Anna, Pisa, Italy
- 2Biology Department, University of Pisa, Pisa, Italy
Plants are known to respond to variations in cellular oxygen availability and distribution by quickly adapting the transcription rate of a number of genes, generally associated to improved energy usage pathways, oxygen homeostasis and protection from harmful products of anaerobic metabolism. In terrestrial plants, such coordinated gene expression program is promoted by a conserved subfamily of ethylene responsive transcription factors called ERF-VII, which act as master activators of hypoxic gene transcription. Their abundance is directly regulated by oxygen through a mechanism of targeted proteolysis present under aerobic conditions, which is triggered by ERF-VII protein oxidation. Beside this, in Arabidopsis thaliana, the activity of the ERF-VII factor RAP2.12 has been shown to be restrained and made transient by the hypoxia-inducible transcription factor HRA1. This feedback mechanism has been proposed to modulate ERF-VII activity in the plant under fluctuating hypoxia, thereby enhancing the flexibility of the response. So far, functional balancing between RAP2.12 and HRA1 has been assessed in isolated leaf protoplasts, resulting in an inverse relationship between HRA1 amount and activation of RAP2.12 target promoters. In the present work, we showed that HRA1 is effective in balancing RAP2.12 activity in whole arabidopsis plants. Examination of a segregating population, generated from RAP2.12 and HRA1 over-expressing plants, led to the first quantitative proof that, over a range of either transgene expression levels, HRA1 counteracts the phenotypic and transcriptional effects of RAP2.12. This report supports the occurrence of fine-tuned regulation of the hypoxic response under physiological growth conditions.
Introduction
Conditions characterized by sub-optimal oxygen levels are considered common in plants (van Dongen and Licausi, 2015). Cellular oxygen concentrations can fall below the threshold set by the mitochondrial complexes for optimal aerobic respiration in several situations, ranging from soil waterlogging, with consequent root asphyxia, and flooding stress (Drew, 1997; Voesenek et al., 2006), to tight packaging of cells inside compact structures like bulky tissues and fruits (Ho et al., 2011; Licausi et al., 2011a), biogenesis of gas-impermeable layers in some seeds (Borisjuk and Rolletschek, 2009), or the existence of underground organs (De Willigen and van Noordwijk, 1989). Terrestrial plants have evolved a wide range of adaptations to either prevent the onset of severe hypoxia in their organs, or improve metabolism during a shortage of oxygen (Bailey-Serres and Voesenek, 2008). On the other hand, in some cases, hypoxia can help cells control the production of dangerous oxidative conditions, so that it becomes even required for the correct progression of specific developmental processes, such as and pollen differentiation inside maize anthers (Kelliher and Walbot, 2012, 2014).
Hypoxic responses have been associated to a wide reconfiguration of plant transcriptomes (Branco-Price et al., 2005; Loreti et al., 2005; Mustroph et al., 2010; Lee et al., 2011). Regulation of transcription upon oxygen deprivation relies on ERF-VII transcription factor family members (Nakano et al., 2006), which redundantly activate the expression of the complete set of hypoxia-responsive genes by direct promoter recognition (Bui et al., 2015; Papdi et al., 2015; Gasch et al., 2016). Over-expression of the RAP2.12 member of the ERF-VII subfamily, for instance, is sufficient to trigger the core transcriptional response to hypoxia in Arabidopsis thaliana, as previously defined (Mustroph et al., 2009), even in the absence of the corresponding external stimulus (Licausi et al., 2011b). The activity of RAP2.12 is tightly regulated by oxygen through a targeted proteolytic pathway, whereby the transcription factor is made oxygen-labile, coupled to a subcellular localization mechanism that guarantees the cell the presence of quickly available RAP2.12 as hypoxia arises (Kosmacz et al., 2015). Indeed, RAP2.12 is post-transcriptionally regulated by oxygen through a direct biosensing mechanism that deploys the Cys/Arg branch of the N-end rule pathway for proteasomal degradation (composed, in Arabidopsis, by the arginyl aminotransferase enzymes ATE1 and ATE2 and by the E3 ubiquitin ligase PRT6) and plant cysteine oxidase enzymes (Gibbs et al., 2011; Weits et al., 2014).
Beside the basic working principle of this oxygen biosensor, it has been shown that further mechanisms of regulation exist that empower plant cells to achieve improved control of RAP2.12 activity. In detail, a feedback loop has been described, in which the transcription factor HRA1 can act on RAP2.12 to restrain its transactivation power on target genes (Giuntoli et al., 2014). Intriguingly, the up-regulation of HRA1 homologs in response to low oxygen in several species allows the attribution of HRA1 to the set of plant core conserved hypoxia-responsive genes (Mustroph et al., 2010). It has been put forward that the induction of HRA1 might be used by plants to produce transient pulses of anaerobic gene expression promoted by RAP2.12, which would enable dynamic and fast regulation in response to conditions of fluctuating hypoxia (Giuntoli et al., 2014). However, an assessment of the influence of this mechanism in planta is still needed.
In order to understand whether the interaction of RAP2.12 and HRA1 transcription factors results in a functional balancing in the plant, we decided to study how plant morphology and gene expression are affected when both genes were over-expressed in a constitutive fashion. Our results report that HRA1 had measurable effects on the processes downstream of RAP2.12. Our findings give way to future experiments to gain more in-depth knowledge regarding the range of action of the HRA1 fine-tuning function in the plant.
Materials and Methods
Generation of Δ13RAP2.12xHRA1 Double Over-Expressors in Arabidopsis thaliana
Two stabilized transgenic lines over-expressing the individual genes, isolated in previous works, were crossed (Licausi et al., 2011b; Giuntoli et al., 2014). 35S:HRA1 plants express the coding sequence of HRA1 fused to a C-terminal FLAG tag sequence, under control of the CaMV 35S promoter. 35S:Δ13RAP2.12 plants, instead, encode an N-end rule insensitive version of RAP2.12 lacking the first 13 N-terminal residues. Both lines were generated in the Columbia-0 background. Homozygous parental plants were crossed manually and the hybrid progeny was propagated to the following F2 generation.
Plant Growth Conditions and Sampling
Seeds were sown in a moist mixture of soil perlite 3:1 and stratified at 4°C in the dark for 48 h. Plants were grown at 23°C day/18°C night under a neutral day cycle (12 h light/12 h darkness, ~100 μmol photons m−2s−1 light intensity). Upon attainment of the developed rosette stage (stage 3.50 Boyes et al., 2001), corresponding to 4–5 weeks of growth in our conditions, plants were evaluated phenotypically and subsequently sampled for gene expression analyses.
RT-qPCR
Transcript abundance was measured in whole rosettes of stage 3.50 (Boyes et al., 2001) arabidopsis plants, by means of real time quantitative PCR. RNA extraction, removal of genomic DNA, cDNA synthesis and RT–qPCR analyses were performed as described previously (Licausi et al., 2011c). The sequences of the primers used for cDNA amplification are listed in Table 1. Steady-state mRNA levels were normalized using UBQ10 as the reference gene, and relative expression values were calculated using the comparative Ct method (Schmittgen and Livak, 2008). Total RAP2.12 and HRA1 expression was assessed with primers annealing on the respective coding sequences. In non-transgenic plants, total gene expression coincided with the level of the endogenous transcripts encoded by the wild type genome. On the opposite, in transgenic plants, 3′-UTR sequences were exploited in order to discriminate between the expression of transgenes and endogenous genes. Specifically, expression of the RAP2.12 transgenic sequence (referred to as transRAP2.12) was measured directly, through an amplification product spanning over the 3′-UTR region encoded by the transgenic construct. On the other hand, HRA1 transgene expression (transHRA1) was calculated by subtracting the endogenous HRA1 expression level, measured with specific HRA1 3′-UTR genomic primers, from the total amount of HRA1 transcript, measured with primers annealing on the coding sequence. Transgenic mRNA abundance was subsequently expressed as relative to the level measured in one selected plant from the relative parental line, in which it was set to 100%.
Statistical Analysis
In the segregating F2 population (n = 32), resulting from the cross of 35S:HRA1 and 35S:Δ13RAP2.12 parental lines of arabidopsis, the interaction between the two factors under investigation was evaluated upon measurement of the expression level of eight anaerobic marker genes, known from the literature as targets of RAP2.12. Specifically, a linear model was fit to the scatterplot expression of every marker gene, used as output variable, in dependence of two chosen predictors, namely total RAP2.12 expression values and transHRA1 presence. The analyses of covariance and linear regressions were performed with the R statistical software (R Development Core Team, 2010).
Results
Over-Expression of HRA1 Restrains the Impact of Δ13RAP2.12 on Plant Phenotype
We chose to investigate the effects of HRA1 on the transcriptional activity of RAP2.12 by crossing homozygous 35S:Δ13RAP2.12 and 35S:HRA1 parental plants. The former parental genotype expressed an N-terminally mutated form of RAP2.12 that, by escaping the oxygen-dependent degradation, allowed us to study a constitutive hypoxic response in plants kept in aerobic conditions (Licausi et al., 2011b). Constitutive expression of an oxygen-insensitive RAP2.12 form leads to widespread morphological changes in arabidopsis (Weits et al., 2014). 35S:Δ13RAP2.12 plants developed abnormal lateral organs in the vegetative rosette, where leaves often displayed irregular margins, bent and twisted petioles associated with downwards curling leaves, and enhanced wax deposition that results in higher stiffness and glossy appearance. Bleaching and necrosis of leaves was also commonly observed in this parental line (Figure 1A).
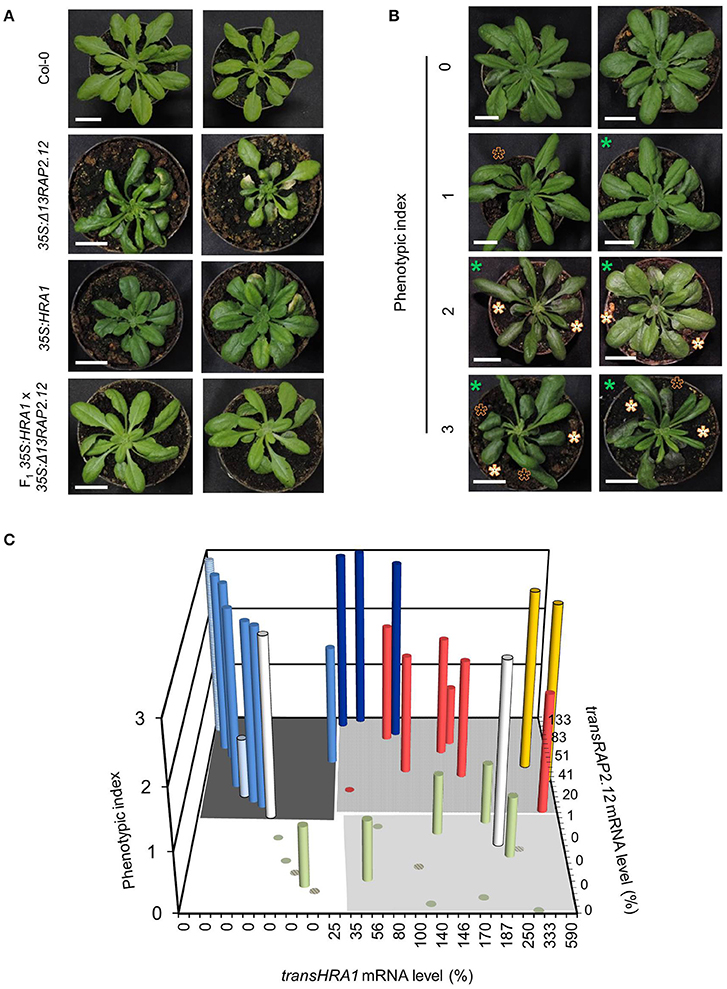
Figure 1. Modulation of the Δ13RAP2.12 phenotype by high levels of HRA1 expression. (A) Representative morphology of parental 35S:Δ13RAP2.12 and 35S:HRA1 plants, first generation hybrids and wild type Col-0 plants at the adult stage of rosette development. Scale bar = 2 cm. (B) Sample output of the visual ranking procedure applied for the phenotypization of the F2 population. Appearance of the glossy leaves feature is marked with white asterisks, curved leaves are indicated with black ones, while smaller plant diameter can be inferred from the scale bar (2 cm) and is marked by green asterisks. (C) Bar plot of phenotypic index as a function of transRAP2.12 mRNA abundance and presence of transgenic HRA1. Each column represents an F2 individual, or a plant of a reference genotype (hatched columns). transRAP2.12 and transHRA1 were expressed as percent relatively to one 35S:Δ13RAP2.12 or 35S:HRA1 parental plant, respectively. Support data for the diagram can be found in Table 2. The grouping of the bars in different colors is discussed in the main text.
In this genetic background, we assessed whether and to which extent the over-expression of HRA1 modulated the activity of RAP2.12. We analyzed the F2 progeny of the cross, which, as a segregating population, enabled us to observe the combinatorial effect of the two loci in a uniform genetic background. Among 32 F2 plants, the expression of the transgenes ranged from 0 to 133% for Δ13RAP2.12 (indicated as transRAP2.12) and from 0 to 590% for HRA1 (transHRA1), as compared to a reference parental plant whose expression was set at 100% (Table 2). Variable transgene expression levels could be explained by the segregation of the two T-DNAs, as well as by an intrinsic degree of individual variation derived from the parental lines.
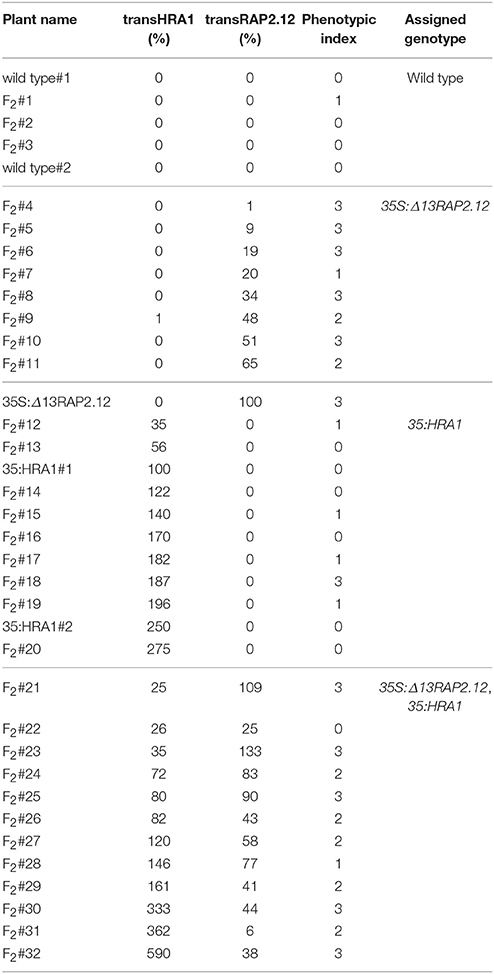
Table 2. Expression values of the two transgenes and phenotypic index value of the plants used in the analysis.
As a first remark, the strong phenotype displayed by the 35S:Δ13RAP2.12 parental was attenuated in the F2 population, which showed a variable extent of reversion to the wild type phenotype. This observation prompted us to look for a correlation between abundance of the two transgene products and phenotypic aspect of the plants. We ranked the phenotypes displayed by the hybrid progeny by means of three main qualitative hallmarks of the Δ13RAP2.12-related morphology: smaller rosette (parameter P1), petiole bending (P2), and increased waxiness of leaf adaxial surfaces (P3). The evaluation of each qualitative parameter describing the Δ13RAP2.12-related phenotype was carefully made. For P1, smaller rosette size had to be coupled with normal petiole and leaf blade length, to avoid confusion with the 35S:HRA1 phenotype (compact rosette with contracted petioles and rounder leaf shape (Figure 1A and Giuntoli et al., 2014). For P2, bending was scored when it coincided with altered leaf margin shape and curling of the leaf blade. In parameter P3, finally, the presence of leaf gloss and enhanced thickness were both required. The presence or absence of each parameter was scored upon visual inspection and expressed as a binary value (0 or 1), the three scores were summed and each plant's phenotype was expressed by a lumped index ranging from 0 (near-wild type morphology) to 3 (near-Δ13RAP2.12 morphology) (Figure 1B).
The first hybrid generation presented a uniform morphology with intermediate Δ13RAP2.12 traits, namely bent and curled leaves with rounder blades (Figure 1A). Such outcome might be due either to an incomplete dominance of the transRAP2.12 allele in the heterozygous configuration, or to a functional balancing between transRAP2.12 and transHRA1 alleles. In the subsequent F2 generation, the observed phenotypes segregated and their distribution was plotted against the expression level of the two transgenes (Figure 1C and Table 2).
With two exceptions (yellow columns), top phenotype index scores were assigned to plants that expressed transRAP2.12 alone (light blue columns, transRAP2.12 = 9–100%) or to such an extent that transHRA1 expression could be overcome by RAP2.12 (dark blue columns, transRAP2.12 = 90–133%). The absence of transgenic Δ13RAP2.12 expression, on the other hand, translated into a low phenotype index (olive green columns). Only one plant showed low phenotype in spite of detectable transRAP2.12 expression and absence of the HRA1 transgene (pale light blue column, transRAP2.12 = 25%), indeed all RAP2.12 targets analyzed in the subsequent gene expression analysis proved to be lowly expressed for this individual, hinting at a reduced activity of the stabilized transcription factor as the cause of the phenotype in this plant. On the opposite, two plants (white bars) presented a strong phenotype, in front of very low transRAP2.12) or undetectable transRAP2.12 expression; even assuming transRAP2.12 to be already active in the first case, we could not explain the observed phenotype in the second. In all the remaining plants (8/32, red columns), concurrent HRA1 over-expression was able to restrict the impact of Δ13RAP2.12 expression (phenotype index = 0–2, transRAP2.12 = 6–83%), defining the borders of a RAP2.12-HRA1 balancing zone in our diagram. An average phenotypic index, obtained as the sum of the index of all plants in a set divided by their number, passed from 2.6 in plants only expressing transRAP2.12 (dark gray-shaded quarter in the diagram in Figure 1C, transHRA1 = 0–1%) to 2.2 in the set of plants also expressing transHRA1 (light gray-shaded quarter, transHRA1 >25%), while it reached 0.2 in wild type plants (white quarter) and 0.6 when only transHRA1 was expressed (pale gray-shaded quarter). Overall, we consider this assessment in favor of the hypothesis that abundant HRA1 protein could contrast the activity of the oxygen-insensitive version of RAP2.12, assumed as correlated to the phenotype index in 35S:Δ13RAP2.12 plants.
Activation of RAP2.12 Target Genes is Affected by HRA1 in planta
After assessing the impact of HRA1 on the Δ13RAP2.12 phenotype, we moved forward and analyzed the impact in terms of molecular markers. RAP2.12 stabilization and over-expression is known to cause constitutive expression of core hypoxia-responsive genes in arabidopsis (Licausi et al., 2011b). Therefore, we considered appropriate to evaluate the correlation between RAP2.12 over-expression and expression of hypoxic targets in our F2 population, and verify to which extent it might be affected by HRA1 over-expression. The set of marker genes included in the analysis encompassed the transcription factor LBD41 (At5g02550), the acyl-CoA desaturase HUP7 (At1g43800), the cysteine oxidase PCO1 (At5g15120), the two fermentative genes PDC1 (At4g33070) and ADH (At1g77120), the non-symbiontic hemoglobin HB1 (At2g16060), and HRA1 (At3g10040) itself. The anoxia-responsive heat shock factor HSP18.2 (At5g59720) was, moreover, selected as a negative control gene, since a survey of public trascriptomic data suggested it not to be activated in the N-end rule mutant backgrounds ate1/2 and prt6 (Pucciariello et al., 2012).
Two populations of transcripts corresponding to RAP2.12 were quantified and correlated to target gene expression. The mRNA encoded by the Δ13RAP2.12 transgene was measured with transRAP2.12 specific primers, while total RAP2.12 expression represented the cumulative amount of the transcript encoded by the endogenous genomic locus and by the transgene, when present. Predictably, total RAP2.12 expression displayed poor correlation with the aerobic levels of the targets in plants lacking Δ13RAP2.12 transgene expression (Figure 2A), where the endogenous RAP2.12 mRNA was translated in the oxygen-labile, inactive form of the transcription factor. Instead, a closer relationship was found in Δ13RAP2.12 over-expressors, when target transcript levels were plotted either against total RAP2.12 (Figure 2B) or transRAP2.12 expression values (Figure 2C). Total RAP2.12 was, therefore, assumed as a suitable predictor of target expression and kept for the subsequent analysis. Incidentally, HSP18.2 proved to be activated by Δ13RAP2.12 in a similar fashion to the other well-known core hypoxia-responsive genes and was, therefore, assimilated to the other target genes.
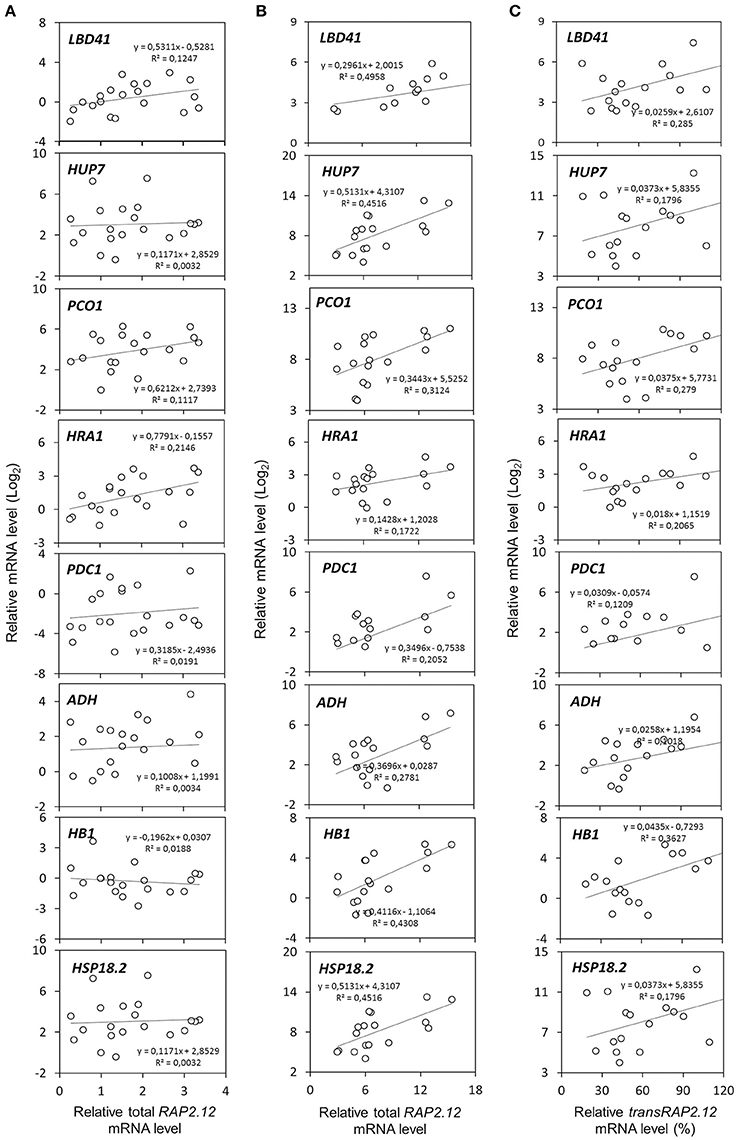
Figure 2. Correlation between RAP2.12 and target gene expression. Linear regressions of anaerobic mRNA levels in F2 (n = 32), parental (n = 3), and Col-0 plants (n = 2). The full set of plants was split into two subsets, based on (A) the absence (n = 16) or (B,C) presence of transRAP2.12 transgene expression (n = 21). For the latter subset of plants, expressing the Δ13RAP2.12 T-DNA, target gene expression was correlated either with total RAP2.12 levels (B) or transRAP2.12 transgene levels (C). For the former, total RAP2.12 expression corresponded to the endogenous RAP2.12 transcript (A). Every dot represents an individual plant.
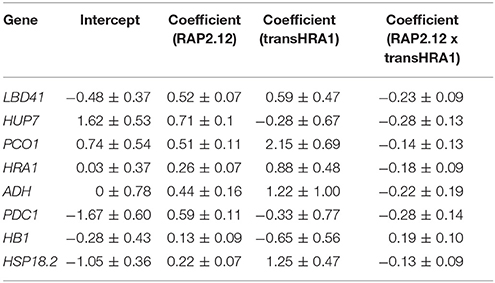
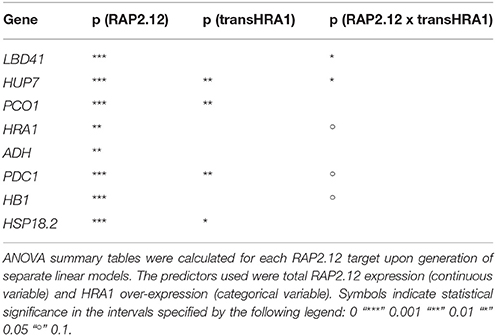
To assess the effect of HRA1 on the RAP2.12-mediated target activation (Figure 2) the F2 population was split into plants over-expressing HRA1 (“+transHRA1” plants) and those not (“-transHRA1”). With an ANCOVA, the tendency of HRA1 to limit RAP2.12 activation power on the anaerobic targets was measured. A linear model was fit to the scatterplot expression of every RAP2.12 target (Figure 3), using total RAP2.12 expression values and transHRA1 presence as predictors (Table 3). The analysis confirmed the existence of a significant effect exerted by total RAP2.12 expression over the steady state mRNA levels of all selected markers and furthermore highlighted a contribution by HRA1 transgene expression (Table 4). HRA1 importance was especially apparent from the significant interaction terms between the two predictor variables in the case of LBD41, HUP7, and marginally for HB1, HRA1, and PDC1. However, no HRA-RAP2.12 interaction occurred for ADH, HSP18.2, and PCO1 activation. In four of the five cases where the interaction took place, the expression of transHRA1 had an antagonistic effect in respect to RAP2.12, as indicated by the negative coefficients for the “RAP2.12 x transHRA1” term in the linear model (Table 3).
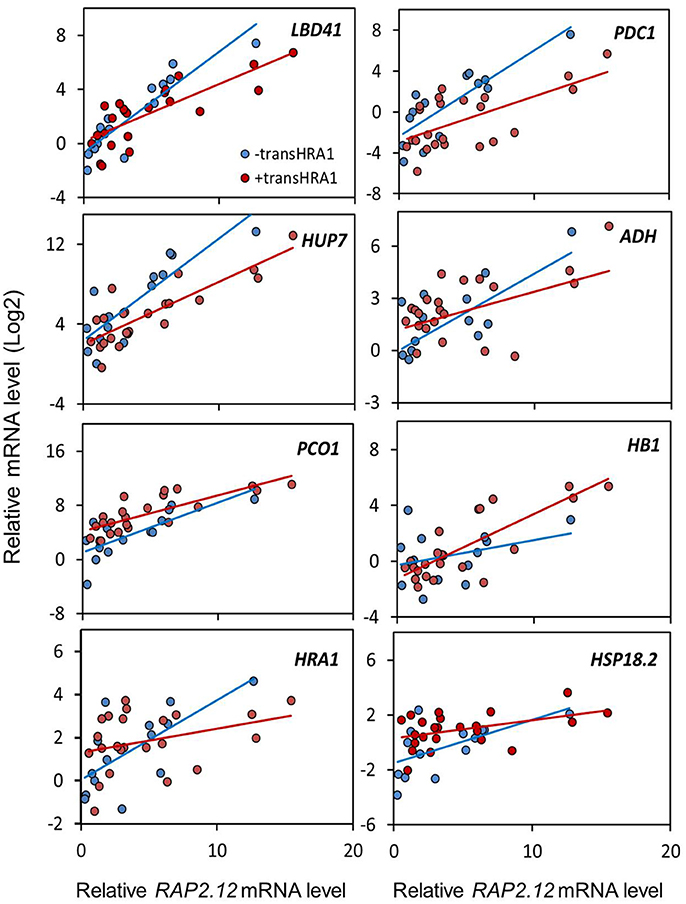
Figure 3. Effect of HRA1 over-expression on RAP2.12 targets. F2 (n = 32), parental (n = 3) and Col-0 plants (n = 2) were clustered into two groups, distinguishing HRA1 over-expressors (“+transHRA1,” transHRA1 >25%, n = 23) from plants with wild type HRA1 levels (“-transHRA1,” transHRA = 0–1%, n = 14), and the expression of the targets was plotted against total RAP2.12 expression.
We conclude that the assessment of RAP2.12 transcriptional activity, estimated from the expression of established marker genes, was in substantial agreement with the prior evaluation of its ability to affect plant morphology, when made stable in air and over-expressed. Broadly speaking, both pieces of evidence we collected, indeed, pointed at the ability of HRA1 to restrict RAP2.12 functionality, although the extent of HRA1 impact seems confined to a precise range of RAP2.12 protein abundance.
Discussion
Tight regulation of gene expression to withstand fluctuations in the intracellular oxygenation status is likely to be vital for organisms, like the terrestrial plants, that have not evolved specific systems for capillary oxygen delivery. In A. thaliana, transcription in response to low oxygen signals is redundantly triggered by the homologous ERF-VII transcription factors RAP2.2 and RAP2.12 (Hinz et al., 2010; Gibbs et al., 2011; Licausi et al., 2011c; Bui et al., 2015; Gasch et al., 2016). In our previous work, we have found evidence that the HRA1 transcription factor, whose constitutive expression in arabidopsis leads to marked reduction of hypoxic responses in oxygen-deprived plants, interacts with RAP2.12 and, in doing so, carries out a counterbalancing effect on the activation of RAP2.12 hypoxic target genes (Giuntoli et al., 2014). The presented research aimed at showing to which extent the hypoxic response attenuator HRA1 is effective in planta in modulating the transcription of RAP2.12 target genes and the production of phenotypes associated with RAP2.12 over-expression in arabidopsis. Previous demonstrations of the impact of this TF interaction are related to the response of isolated arabidopsis protoplast cells. Here, we combined the ectopic expression of HRA1 with that of an oxygen-insensitive form of RAP2.12, which enabled us to investigate the regulation of the anaerobic response without imposing external stress conditions on plants. Deployment of a segregating population made the correlation possible, in each individual plant, between the actual expression level of both transgenes, which spanned a range of combinations, and two marker traits describing RAP2.12 activity.
With this approach, we were able to spot the balancing action exerted by HRA1 on RAP2.12 by examining the phenotype of plants growing in normal conditions. Initial clues of the efficacy of such a mechanism in fully developed, unstressed plants had appeared previously, with the observation that stable transformation of 35S:HRA1 plants with a 35S:Δ13RAP2.12 T-DNA generated a progeny in which the phenotypic traits associated to HRA1 over-expression reverted to the wild type (Giuntoli et al., 2014). The achievement of comparable outcomes following two independent events of T-DNA insertion in the genome supports the conclusion that a causal link subsists between concurrent over-expression of HRA1 and reversion of the molecular and phenotypic effects of Δ13RAP2.12.
Furthermore, the present study provides the first quantitative description of RAP2.12-HRA1 balancing in whole developed plants, evaluated by means of anaerobic molecular markers. As highlighted before, this result was achieved by over-expression of the two transcription factors under control of the constitutive CaMV 35S promoter. Despite this simple strategy, whereby massive accumulation of either protein is allowed unrestrictedly during the entire plant lifespan, functional balancing proved to be still in place and amenable to quantitative modeling.
While analyzing plant responses, we decided to reconstruct the behavior of HRA1 as a function of the presence or absence of its expressed transgene (Figure 3), as quantified through specific qPCR amplification. This is because, in first place, total HRA1 transcript levels were so superior in the over-expressing plants (Log2 HRA1 = 10.2–13.8; values refer to the expression measured in one of the wild type reference plants, taken as reference and set to Log2 HRA1 = 0), as compared to those detected in plants with wild type HRA1 and RAP2.12 configuration (Log2 HRA1 = −1.2–2.6), or in Δ13RAP2.12 over-expressing plants (Log2 HRA1 = 3–5.8), that approximation to a categorical condition was allowed. In second place, we assumed that HRA1 mRNA steady state level could be bona fide considered as proportional to protein abundance, in the absence of any known mechanism of targeted post-transcriptional regulation specific for this gene. Therefore, a model where the HRA1 transcription factor was approximated as highly abundant, as in 35S:HRA1 individuals, or lowly abundant, as in all other genotypes, was considered acceptable to account for the balancing effect. The same did not apply to RAP2.12, which required treatment as a continuous quantity. A linear increase of marker gene expression was recorded with increasing total RAP2.12 (or transRAP2.12) abundance along the range of expression available in our measurements for this predictor (Figures 2B,C), suggesting that the over-expressed Δ13RAP2.12 protein was not abundant enough to saturate the target promoters.
Our investigation took advantage of striking phenotypic features that associate to the ectopic expression of an oxygen-insensitive variant of RAP2.12. Unraveling the downstream events that realize this specific ontogenetic program was beyond the aim of this work and might be worth focused investigation. Nonetheless, we might conclude that the phenotype originates from the accumulation of Δ13RAP2.12, rather than from spurious phenomena due to the untargeted process of T-DNA integration, because it can be at least partially rescued by the expression of a RAP2.12-specific repressor, HRA1. In the same way, the phenotypic consequences of 35S:HRA1 expression could be considerably reverted by enhancing RAP2.12 activity (this study and Giuntoli et al., 2014). Therefore, it is reasonable to think that the alterations visible in the Δ13RAP2.12 phenotype are caused by genes differentially regulated by 35S:Δ13RAP2.12 and subjected to HRA1-dependent negative regulation.
The interplay between HRA1 and RAP2.12 was revealed by the expression of transcriptional markers, namely known plant hypoxic targets identified from the broad specialized literature. Marginally, it could be noticed that our analysis provided further confirmation to the fact that aerobic transcription of RAP2.12 results in inactive protein accumulation. It has been noticed before that constitutive expression of the full version of RAP2.12 leads to minimal up-regulation of anaerobic gene expression in air and does not cause any detectable plant phenotype (Licausi et al., 2011b). Coherently, we found limited correlation between full-length RAP2.12 mRNA levels and RAP2.12 transcriptional targets (Figure 2A). Beside this, the ANCOVA highlighted a different degree of specificity in the targets we considered. More specifically, while some anaerobic genes are exclusively regulated by RAP2.12, cross-talk from different cell pathways is known to converge on other core hypoxia-responsive genes. Indeed, RAP2.12 function is known to be superimposed on unrelated regulatory pathways, such as the one brought about by heat shock factors on HSP18.2 (Nishizawa et al., 2006; Guo et al., 2008) and multiple ABA-mediated signaling events influencing ADH expression (de Bruxelles et al., 1996; Xiong et al., 2001; Papdi et al., 2015). We speculate that this might explain why the HRA1-mediated repression of RAP2.12 was not detectable on ADH and HSP18.2 (Table 4), being any additional regulation beyond the predictive power of our bifactorial model. Detailed promoter survey of representative genes for the two regulatory classes, aided by the recent identification of the cis-element recognized by RAP2.12 in its target promoters (Gasch et al., 2016), might unveil the near-exclusive presence of the RAP2.12-specific binding site in the first class of items and support our hypothesis.
In this first report of the effective balancing between HRA1 and RAP2.12 in the aerial tissues of arabidopsis, the equilibrium of the two transcription factors was moved to a non-physiological range, by deployment of over-expression constructs. Future steps of this research might take advantage of native gene promoters to understand whether, under physiological expression conditions, the transcriptional complex is actually able to modulate target gene expression by originating transient transcriptional responses.
Author Contributions
BG, FL, and PP designed the experiments that were carried out by BG and FL. HvV, and BG performed the statistical analysis. BG, wrote the manuscript. FL, HvV, and PP critically revised it.
Funding
Results have been achieved within the framework of the 1st call ERA-NET for Coordinating Plant Sciences, with funding from Scuola Superiore Sant'Anna.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Bailey-Serres, J., and Voesenek, L. A. (2008). Flooding stress: acclimations and genetic diversity. Annu. Rev. Plant Biol. 59, 313–339. doi: 10.1146/annurev.arplant.59.032607.092752
Borisjuk, L., and Rolletschek, H. (2009). The oxygen status of the developing seed. New Phytol. 182, 17–30. doi: 10.1111/j.1469-8137.2008.02752.x
Boyes, D. C., Zayed, A. M., Ascenzi, R., McCaskill, A. J., Hoffman, N. E., Davis, K. R., et al. (2001). Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13, 1499–1510. doi: 10.1105/tpc.13.7.1499
Branco-Price, C., Kawaguchi, R., Ferreira, R. B., and Bailey-Serres, J. (2005). Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann. Bot. 96, 647–660. doi: 10.1093/aob/mci217
Bui, L. T., Giuntoli, B., Kosmacz, M., Parlanti, S., and Licausi, F. (2015). Constitutively expressed ERF-VII transcription factors redundantly activate the core anaerobic response in Arabidopsis thaliana. Plant Sci. 236, 37–43. doi: 10.1016/j.plantsci.2015.03.008
de Bruxelles, G. L., Peacock, W. J., Dennis, E. S., and Dolferus, R. (1996). Abscisic acid induces the alcohol dehydrogenase gene in Arabidopsis. Plant Physiol. 111, 381–391. doi: 10.1104/pp.111.2.381
De Willigen, P., and van Noordwijk, M. (1989). Model calculations on the relative importance of internal longitudinal diffusion for aeration of roots of non-wetland plants. Plant Soil 113:9. doi: 10.1007/BF02181928
Drew, M. C. (1997). Oxygen deficiency and root metabolism: injury and acclimation under Hypoxia and Anoxia. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 223–250. doi: 10.1146/annurev.arplant.48.1.223
Gasch, P., Fundinger, M., Müller, J. T., Lee, T., Bailey-Serres, J., and Mustroph, A. (2016). Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell 28, 160–180. doi: 10.1105/tpc.15.00866
Gibbs, D. J., Lee, S. C., Isa, N. M., Gramuglia, S., Fukao, T., Bassel, G. W., et al. (2011). Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479, 415–418. doi: 10.1038/nature10534
Giuntoli, B., Lee, S. C., Licausi, F., Kosmacz, M., Oosumi, T., van Dongen, J. T., et al. (2014). A trihelix DNA binding protein counterbalances hypoxia-responsive transcriptional activation in Arabidopsis. PLoS Biol. 12:e1001950. doi: 10.1371/journal.pbio.1001950
Guo, L., Chen, S., Liu, K., Liu, Y., Ni, L., Zhang, K., et al. (2008). Isolation of heat shock factor HsfA1a-binding sites in vivo revealed variations of heat shock elements in Arabidopsis thaliana. Plant Cell Physiol. 49, 1306–1315. doi: 10.1093/pcp/pcn105
Hinz, M., Wilson, I. W., Yang, J., Buerstenbinder, K., Llewellyn, D., Dennis, E. S., et al. (2010). Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 153, 757–772. doi: 10.1104/pp.110.155077
Ho, Q. T., Verboven, P., Verlinden, B. E., Herremans, E., Wevers, M., Carmeliet, J., et al. (2011). A three-dimensional multiscale model for gas exchange in fruit. Plant Physiol. 155, 1158–1168. doi: 10.1104/pp.110.169391
Kelliher, T., and Walbot, V. (2012). Hypoxia triggers meiotic fate acquisition in maize. Science 337, 345–348. doi: 10.1126/science.1220080
Kelliher, T., and Walbot, V. (2014). Maize germinal cell initials accommodate hypoxia and precociously express meiotic genes. Plant J. 77, 639–652. doi: 10.1111/tpj.12414
Kosmacz, M., Parlanti, S., Schwarzländer, M., Kragler, F., Licausi, F., and Van Dongen, J. T. (2015). The stability and nuclear localization of the transcription factor RAP2.12 are dynamically regulated by oxygen concentration. Plant Cell Environ. 38, 1094–1103. doi: 10.1111/pce.12493
Lee, S. C., Mustroph, A., Sasidharan, R., Vashisht, D., Pedersen, O., Oosumi, T., Voesenek, L. A., et al. (2011). Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol. 190, 457–471. doi: 10.1111/j.1469-8137.2010.03590.x
Licausi, F., Giorgi, F. M., Schmälzlin, E., Usadel, B., Perata, P., van Dongen, J. T., et al. (2011a). HRE-type genes are regulated by growth-related changes in internal oxygen concentrations during the normal development of potato (Solanum tuberosum) tubers. Plant Cell Physiol. 52, 1957–1972. doi: 10.1093/pcp/pcr128
Licausi, F., Kosmacz, M., Weits, D. A., Giuntoli, B., Giorgi, F. M., Voesenek, L. A., et al. (2011b). Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479, 419–422. doi: 10.1038/nature10536
Licausi, F., Weits, D. A., Pant, B. D., Scheible, W. R., Geigenberger, P., and van Dongen, J. T. (2011c). Hypoxia responsive gene expression is mediated by various subsets of transcription factors and miRNAs that are determined by the actual oxygen availability. New Phytol. 190, 442–456. doi: 10.1111/j.1469-8137.2010.03451.x
Loreti, E., Poggi, A., Novi, G., Alpi, A., and Perata, P. (2005). A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol. 137, 1130–1138. doi: 10.1104/pp.104.057299
Mustroph, A., Lee, S. C., Oosumi, T., Zanetti, M. E., Yang, H., Ma, K., et al. (2010). Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 152, 1484–1500. doi: 10.1104/pp.109.151845
Mustroph, A., Zanetti, M. E., Jang, C. J., Holtan, H. E., Repetti, P. P., Galbraith, D. W., et al. (2009). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 18843–18848. doi: 10.1073/pnas.0906131106
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Nishizawa, A., Yabuta, Y., Yoshida, E., Maruta, T., Yoshimura, K., and Shigeoka, S. (2006). Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 48, 535–547. doi: 10.1111/j.1365-313X.2006.02889.x
Papdi, C., Pérez-Salamó, I., Joseph, M. P., Giuntoli, B., Bögre, L., Koncz, C., et al. (2015). The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor VII genes RAP2.12, RAP2.2 and RAP2.3. Plant J. 82, 772–784. doi: 10.1111/tpj.12848
Pucciariello, C., Parlanti, S., Banti, V., Novi, G., and Perata, P. (2012). Reactive oxygen species-driven transcription in Arabidopsis under oxygen deprivation. Plant Physiol. 159, 184–196. doi: 10.1104/pp.111.191122
R Development Core Team (2010). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
van Dongen, J. T., and Licausi, F. (2015). Oxygen sensing and signaling. Annu. Rev. Plant Biol. 66, 345–367. doi: 10.1146/annurev-arplant-043014-114813
Voesenek, L. A., Colmer, T. D., Pierik, R., Millenaar, F. F., and Peeters, A. J. (2006). How plants cope with complete submergence. New Phytol. 170, 213–226. doi: 10.1111/j.1469-8137.2006.01692.x
Weits, D. A., Giuntoli, B., Kosmacz, M., Parlanti, S., Hubberten, H. M., Riegler, H., et al. (2014). Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat. Commun. 5:3425. doi: 10.1038/ncomms4425
Keywords: low oxygen, regulation of anaerobic gene expression, ERF-VII transcription factors, trihelix transcription factor family, transcription factor balancing
Citation: Giuntoli B, Licausi F, van Veen H and Perata P (2017) Functional Balancing of the Hypoxia Regulators RAP2.12 and HRA1 Takes Place in vivo in Arabidopsis thaliana Plants. Front. Plant Sci. 8:591. doi: 10.3389/fpls.2017.00591
Received: 01 February 2017; Accepted: 31 March 2017;
Published: 25 April 2017.
Edited by:
Ruth Grene, Virginia Tech, USACopyright © 2017 Giuntoli, Licausi, van Veen and Perata. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pierdomenico Perata, p.perata@sssup.it
 Beatrice Giuntoli
Beatrice Giuntoli Francesco Licausi
Francesco Licausi Hans van Veen
Hans van Veen Pierdomenico Perata
Pierdomenico Perata