Corrigendum: Phytoplasmas—The “Crouching Tiger” Threat of Australian Plant Pathology
- 1State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Agriculture and Forestry University, Fuzhou, China
- 2Institute of Applied Ecology, Fujian Agriculture & Forestry University, Fuzhou, China
- 3Graham Centre for Agricultural Innovation (Charles Sturt University & NSW Department of Primary Industries), Orange, NSW, Australia
- 4NSW Department of Primary Industries, Wagga Wagga Agricultural Institute, Wagga Wagga, NSW, Australia
Phytoplasmas are insect-vectored bacteria that cause disease in a wide range of plant species. The increasing availability of molecular DNA analyses, expertise and additional methods in recent years has led to a proliferation of discoveries of phytoplasma-plant host associations and in the numbers of taxonomic groupings for phytoplasmas. The widespread use of common names based on the diseases with which they are associated, as well as separate phenetic and taxonomic systems for classifying phytoplasmas based on variation at the 16S rRNA-encoding gene, complicates interpretation of the literature. We explore this issue and related trends through a focus on Australian pathosystems, providing the first comprehensive compilation of information for this continent, covering the phytoplasmas, host plants, vectors and diseases. Of the 33 16Sr groups reported internationally, only groups II, XI, XII, XXIII, XXV, and XXXIII have been recorded in Australia and this highlights the need for ongoing biosecurity measures to prevent the introduction of additional pathogen groups. Many of the phytoplasmas reported in Australia have not been sufficiently well studied to assign them to 16Sr groups so it is likely that unrecognized groups and sub-groups are present. Wide host plant ranges are apparent among well studied phytoplasmas, with multiple crop and non-crop species infected by some. Disease management is further complicated by the fact that putative vectors have been identified for few phytoplasmas, especially in Australia. Despite rapid progress in recent years using molecular approaches, phytoplasmas remain the least well studied group of plant pathogens, making them a “crouching tiger” disease threat.
Introduction
Phytoplasmas are insect-vectored bacteria that cause disease in a wide range of plant species (Lee et al., 2000; IRPCM, 2004; Bertaccini et al., 2014; Marcone, 2014). They contrast with other phloem-limited bacteria (Gram-negative proteobacteria such as liberibacters and phlomobacters Bove and Garnier, 2003) which are vectored by the same types of insects, in lacking a cell wall and in having a much reduced genome size (0.53–1.2 kb; Streten and Gibb, 2006). Spiroplasmas, another group of insect vectored plant pathogenic microbes, share the absence of a cell wall but differ from phytoplasmas in that some are culturable in vitro. In this mini review we seek to raise awareness of the importance of this group of plant pathogens, summarizing five key issues that mean the threat they pose to agricultural, ornamental and natural vegetation is not fully appreciated, making them worthy of the “crouching tiger” description. We focus principally on the phytoplasmas of Australia, as a sub-set of the large and rapidly-expanding global literature. We consider the phytoplasmas, associated plant diseases, plant hosts and the putative insect vectors (Table 1). Work from locations other than Australia is mentioned where appropriate to provide context and show where the issues we identify are generic rather than Australian-specific as well as to point out opportunities that are open for future study of Australian phytoplasma pathosystems. The only other reviews of phytoplasmas in Australia are at least a decade old and confined to tropical (Wilson et al., 2001) and sub-tropical regions (Streten and Gibb, 2006) whereas the scope of the present review includes the agriculturally important temperate zone. Reviews of varying levels of comprehensiveness are available for other geographical areas: New Zealand (Veerakone et al., 2015), Latin America (Pérez-López et al., 2016), the Pacific region (Davis and Ruabete, 2010) and Southern Italy (Marcone, 2011). Reflecting the large body of literature now available on phytoplasmas, the only reviews with an international scope are confined to particular crops such as coconut palm (Gurr et al., 2016), date palm (Gurr et al., 2015), fruit trees (Adams et al., 2001) and sugar cane (Smith et al., 2001).
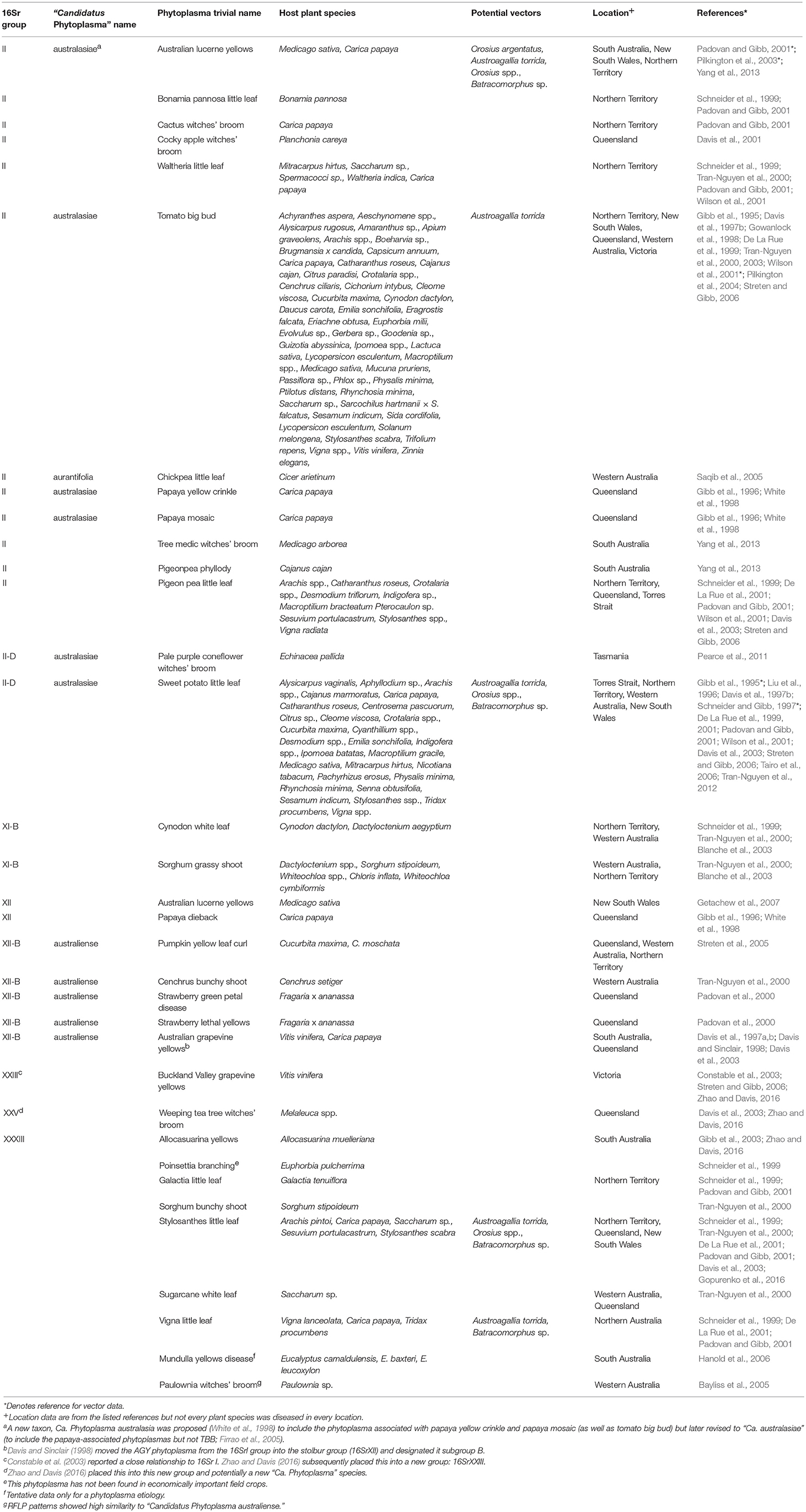
Table 1. Taxonomic and biological information on phytoplasmas in Australia (empty cells denote the absence of available information).
Issue 1: Phytoplasmology is a Young Science
Difficulties in studying phytoplasmas greatly limited early progress because phytoplasmas cannot be grown in axenic culture. Researchers were reliant initially on symptomology and transmission experiments, sometimes using grafting or the parasitic plant dodder (Cuscuta spp.) as in Australian work by Gibb et al. (1995), to study symptoms and host ranges but were unable to determine the nature of the pathogen or differentiate phytoplasmas from plant pathogenic viruses. Electron microscopy allowed phytoplasma bodies to be visualized in plant and insect vector tissue and differentiation of phytoplasmas from viruses. Bertaccini and Duduk (2010) provide a useful summary of the development of methods in phytoplasmology. Enzyme-linked immunosorbent assay (ELISA)-based methods began to emerge in the 1980s allowing more rapid detection and identification. The development of polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) methods for the detection and identification of phytoplasmas since the early 1990s allowed major advances, particularly in diagnostics and development of a genetic system for phenetic group classifications of phytoplasmas. By 1998, the international total of 34 representative phytoplasma strains were differentiated into 14 groups and 32 sub-groups based on similarity coefficients derived from RFLP analyses (Lee et al., 1998; Duduk and Bertaccini, 2011). More recent work has extended these counts to 33 groups and at least 100 sub-groups (Dickinson and Hodgetts, 2013; Davis et al., 2015; Zhao and Davis, 2016).
The increasingly widespread availability of molecular methods, equipment and expertise in recent decades has led to a proliferation of discoveries of phytoplasma-plant host associations and in taxonomic groupings for phytoplasmas. Many articles on phytoplasma pathosystems published this century are “first report” or “first record” articles for phytoplasmas in geographical regions or report known phytoplasmas from new host plant species. More fundamentally, new taxa of phytoplasma are being reported on a frequent basis. Progress has accelerated with the development of new approaches for detection and study of phytoplasmas. A prominent example is loop-mediated isothermal amplification (LAMP; Obura et al., 2011; Dickinson, 2015). This approach offers the advantages of low cost and high sensitivity, low risk of cross-contamination because reaction vessels do not need to be opened and, most especially, use in a kit form that requires very little training and can be used in the field (Hodgetts et al., 2011; Dickinson, 2015; Kogovšek et al., 2015). Though LAMP assays have yet to be used in studies of Australian phytoplasma pathosystems they have recently proven useful in studies of Bogia coconut syndrome in nearby Papua New Guinea (Lu et al., 2016).
In parallel to the use of simple LAMP diagnostics, next-generation sequencing (NGS) technologies have provided genomic characterization of phytoplasmas, and this has profoundly advanced the understanding of phytoplasma evolution and pathogenicity over recent years (Marcone, 2014). To date, complete genomes of six phytoplasma strains (Oshima et al., 2004; Bai et al., 2006; Kube et al., 2008; Tran-Nguyen et al., 2008; Andersen et al., 2013; Orlovskis et al., 2017) and an additional 16 draft or incomplete genomes have been reported (refer Genomes OnLine database [GOLD]; https://gold.jgi.doe.gov).
Comparative genomic analyses of phytoplasma and other mollicutes have provided direct evidence of reduced genomic complexity in phytoplasmas, characterized by encoding for fewer metabolic functions and pathways (Oshima et al., 2004). This highlights the obligate adaptation to, and reliance on, varied metabolic substrates available in hosts and vectors by phytoplasmas (Kube et al., 2013). Genomic studies have also detailed genes putatively encoding for phytoplasma virulence factors, including “effector” proteins which phytoplasmas produce and secrete to alter host cell activities, thereby modifying host development and reducing host defences against herbivorous arthropod vectors (Hogenhout and Loria, 2008; Hogenhout et al., 2008, 2009). Interestingly, comparative genomic analysis of five diverse phytoplasma groups failed to detect a consistent shared set of predicted secreted effector encoding genes (Wang et al., 2014). This suggests virulence encoding genes are likely to be diverse among phytoplasma strains, and may explain the wide range of pathogenicity in different 16Sr groups. Other work (Chung et al., 2013) indicates that horizontal gene flow of mobile genetic elements among some divergent phytoplasma strains may have facilitated horizontal transfer of effector genes linked to the mobile elements, adding a novel pathway for increasing the adaptive potential of phytoplasmas with regards to their hosts. NGS methods have been applied also for population metagenomics, allowing valuable insight into the ecology and dynamics of phytoplasmas and their relationships to hosts (Nicolaisen et al., 2011). Finally, phylogenetic analyses of phytoplasmas using NGS data are likely to improve understanding of systematic and taxonomic relationships in the genus, traditionally reliant on analyses of the 16S rRNA gene region (see Issue 3) and (in some cases) use of additional informative loci for comparison of very recently diverged strains (Al-Abadi et al., 2016).
Anticipated widespread use of affordable NGS services will result in a proliferation of the availability of published phytoplasma genomes and meta-genomic analyses, and this will ultimately lead to more advanced understanding of evolutionary relationships of species in this genus, and their interactive pathways with hosts and vectors including for Australian pathosystems.
Issue 2: Complex Taxonomic Nomenclature
A major impediment to comprehension of the phytoplasma literature and comparisons between studies, particularly for the non-specialist, is the taxonomic nomenclature with three systems currently in use. First, reflecting the history of phytoplasmology described above, the early literature uses disease common names based on symptoms (e.g., little leaf, yellows, witches' broom) often coupled with the host plant's name. These disease common names have been applied also to the phytoplasmas and continue to be used frequently in recent literature. Examples from the Australian literature include Buckland Valley grapevine yellows phytoplasma, tomato big bud phytoplasma and Cockey apple witches' broom phytoplasma (Table 1). This allows great scope for confusion because a given phytoplasma can be found in multiple plant species and can cause different disease symptoms in different hosts.
Second, as molecular methods became available, workers were able to group and phenetically classify phytoplasmas using restricted fragment length polymorphism (RFLP) analysis of a PCR amplified portion of the 16S rRNA gene with a defined set of restriction enzymes (Lee et al., 1998). The RFLP profiles generated for different phytoplasmas are generally consistent with sequence-based phylogenetic analyses of the 16S rRNA gene, particularly in the co-identification and grouping of related strains. The 33 16Sr groups currently defined each have a similarity of <85% compared with any representative phytoplasma from within an established 16Sr group (Zhao and Davis, 2016). Table 1 summarizes available information on the 16Sr groups reported in Australian studies. Of the 33 16Sr groups reported internationally, only groups II, XI, XII, XXIII, XXV, and XXXIII have been recorded in Australia and this highlights the need for ongoing biosecurity measures to prevent the introduction of additional pathogen groups.
Third, phytoplasmas are classified in the provisional genus “Candidatus Phytoplasma” (IRPCM, 2004). To date, there are 42 formally described species and ten potentially novel phytoplasma species (Davis et al., 2015). This number exceeds the current number of 16s rRNA groups because some of these groups contain several “Candidatus Phytoplasma” species. At least 100 subgroups are known (Dickinson and Hodgetts, 2013). According to Phytoplasma/Spiroplasma Working Team-Phytoplasma Taxonomy Group, a novel “Ca. Phytoplasma” species description should refer to a single, unique 16S rRNA gene sequence (>1,200 bp), and a strain can be recognized as a novel “Ca. Phytoplasma” species if its 16S rRNA gene sequence has <97.5% similarity to that of any previously described “Ca. Phytoplasma” species (Duduk and Bertaccini, 2011). Additional biological characters such as antibody specificity, host range and vector transmission specificity as well as genetic markers can also be used in an integrative taxonomy approach for species differentiation. Of the 42 recognized “Ca. Phytoplasma” species, only Ca. Phytoplasma aurantifolia, Ca. Phytoplasma australasiae and Ca. Phytoplasma australiense are reported in Australia (Table 1) but uncertainty exists because many papers appear without Ca. Phytoplasma names which are used consistently only in the case of the GenBank database.
The general literature uses a mix of Ca. Phytoplasma names, 16Sr group and sub-group names, and common names that often reflect the host plant or symptom (Table 1). Though formally named Ca. Phytoplasma species each align with a group or sub group in the 16Sr system, many groups and sub-groups do not currently have a “Candidatus Phytoplasma” species name.
Issue 3: Large and Poorly Understood Biodiversity among Pathogens and Wide Host Ranges
Whilst it is important to note that a given type of phytoplasma can attack more than one plant species, and that a given plant species can be attacked by multiple types of phytoplasma, the list of phytoplasma disease common names from Australia (Table 1) is instructive in indicating the wide range of plant species that are affected, even for specific phytoplasmas. Examples extend over forage, broadacre and horticultural crops of a perennial and annual nature, as well as ornamental and uncultivated (i.e., natural vegetation) species.
There is an especially poor knowledge of phytoplasma pathosystems in temperate Australia, largely because of the tropical and sub-tropical zone focus of most of the key Australian workers in the last 20 years. Many of the phytoplasmas reported in Australia have not been sufficiently well studied to assign them to 16Sr groups so it is likely that unrecognized groups—as well as sub-groups—are present, particularly in native vegetation.
Great efforts are currently being made to define the extent and diversity of phytoplasmas in Australia (most recently the discovery of Stylosanthes little leaf 16Sr XII phytoplasma from lucerne in New South Wales Gopurenko et al., 2016) and elsewhere but this remains a challenge because of the apparent high levels of biological diversity. As we approach a definitive list of 16Sr groups and sub groups, “Ca. phytoplasma” species names need to be assigned that avoid scope for confusion over the host plant, perhaps by wider use of names that reflect the country in which the phytoplasma was first discovered (as is the case for Ca. Phytoplasma australiense, though noting the potential confusion with Ca. Phytoplasma australasiae and the redundant Ca. Phytoplasma australasia Table 1) or a more neutral name based, perhaps, on the discoverer rather than the use of host plant taxon names such as pini, palmicola, pruni and oryzae, all of which are reported internationally though not from Australia.
Reflecting and compounding the three, related issues outlined above, there is no universally recognized taxonomic resource for phytoplasmas. For other higher taxa, there is a name-bearing type specimen lodged in an accessible scientific collection that can be checked by subsequent researchers, allowing comparison with unknown or new taxa. With phytoplasmas, however, the extreme difficulties associated with axenic culture mean that the “type” is a DNA sequence in GenBank®, the National Institutes of Health's annotated collection of all publicly available DNA sequences (Benson et al., 2013), and not a biological specimen. Further, there is no common platform for registering phytoplasma groups and sub-groups (Zhao and Davis, 2016). The International Journal of Systematics and Evolution (formerly the International Journal of Systematic Bacteriology) is the official journal of record for novel prokaryotic taxa since it is the official publication of the International Committee on Systematics of Prokaryotes and the Bacteriology and Applied Microbiology Division of the International Union of Microbiological Societies. Accordingly, new “Ca. Phytoplasma” species are published in this journal. Importantly, however, new 16Sr groups can be (and are) published in other journals provided that the group contains a previously described “Ca. Phytoplasma” species. This lack of a common platform for group names means that a given 16Sr group number can inadvertently be applied by different authors to different “Ca. Phytoplasma” taxa. Zhao and Davis (2016) provide the example that “Ca. Phytoplasma” allocasuarinae was assigned as 16SrXXXIII (Bertaccini et al., 2014) but the same 16Sr group name was also subsequently used for another new (Chilean grapevine phytoplasma) group (Pérez-López et al., 2016). This led to the recent suggestion (Zhao and Davis, 2016) that 16Sr groups and sub-groups should be registered via a web-based portal linked to iPhyClassifier, itself a web-based resource that allows identification and classification of phytoplasmas by simulating restriction enzyme digestions and electrophoresis to produce “virtual” RFLP patterns for submitted query sequences.
Issue 4: Challenges Associated with Vector Studies
The insect vectors of phytoplasmas are yet to be investigated thoroughly in Australia and more generally and this is a major constraint on the development of disease management approaches that focus on vector control. The Australian literature provides putative vector names only for Australian lucerne yellows, sweetpotato little leaf, tomato big bud, Stylosanthes little leaf and Vigna little leaf phytoplasmas (Table 1).
Since phytoplasmas are phloem-limited pathogens, they are transmitted by phloem-feeding hemipteran insects, especially leafhoppers and planthoppers (Weintraub and Beanland, 2006) though psyllids are responsible in some non-Australian pathosystems (Carraro et al., 1998). The identification of potential vector species involves field surveys to determine Hemiptera species that are spatially and temporally associated with plant symptoms followed by the use of PCR to detect phytoplasma DNA in the insects. However, the detection of phytoplasma DNA in an insect does not establish vector status because DNA may be confined to the gut as a result of feeding on an infected host plant rather than the pathogen having colonized the salivary glands making it a competent vector (Vega et al., 1993; Danielli et al., 1996). Definitive proof of vector status can be obtained from transmission tests confining putative vectors on phytoplasma-free host plants in insect-proof cages. Whilst this has been done in a preliminary manner for Australian lucerne yellows (e.g., Pilkington et al., 2004), vector transmission testing is logistically demanding, especially if host plants are large perennial species, if the pathogen is vectored inefficiently or if symptoms develop slowly (Gurr et al., 2016). A relatively new method that is intermediate in ease of use and the level of proof involves holding individual insects in vessels from which they can feed on a sucrose solution through a parafilm barrier with subsequent PCR-based detection of phytoplasma DNA in the medium (Tanne et al., 2001). Though this approach involves many samples (i.e., one per insect rather than one per plant as in the case of a cage transmission test) a recent study has employed LAMP to readily handle the large numbers of samples associated with screening multiple putative vector species of Bogia coconut syndrome (Lu et al., 2016).
Issue 5: Possibility of Seed Transmission
Among the crop species of great economic importance in Australia, tomato, canola and maize have all been experimentally shown to exhibit seed transmission of phytoplasmas, a phenomenon previously considered unlikely and this adds to the importance of ongoing biosecurity and research efforts. In that study, PCR was used to detect phytoplasma DNA in laboratory grown seedlings arising from seed of phytoplasma infected plants of oilseed rape, tomato and corn (Calari et al., 2011). Caution is required in extrapolating from those tests with herbaceous annuals, however, because earlier reports of phytoplasma DNA in embryos of various woody perennial plant species including coconut have not been followed-up with conclusive evidence of seed transmissibility (Nipah et al., 2007) but see Oropeza et al. (2017).
Conclusion and Perspective
The ubiquity of PCR capacity and the advent of LAMP kits for simple diagnostics and NGS for advanced genomic and meta-genomic analyses will spur the discovery of many new phytoplasma diseases and allow rapid advances in understanding of phytoplasma biodiversity and biology in Australia and internationally. This has great practical relevance to address issues such as vector identity and the field-relevance of seed transmission. Because Australia is an island nation with well-established biosecurity measures, improvements in knowledge of phytoplasma biodiversity, host range and detection will be particularly valuable in the prevention of and appropriate responses to phytoplasma incursions. Further sustained research effort by specialists is important but this needs to be complemented by efforts to make this group of pathogens better known and easier to understand, especially taxonomically.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by a Graham Centre New Initiative Grant to the authors. GG is supported by a Chinese Government Thousand Talents Program fellowship (KX1452102).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adams, A. N., Davies, D. L., and Kirby, M. J. (2001). Virus and phytoplasma detection in fruit trees. Outlook Agric. 30, 45–54. doi: 10.5367/000000001101293454
Al-Abadi, S. Y., Al-Sadi, A. M., Dickinson, M., Al-Hammadi, M. S., Al-Shariqi, R., Al-Yahyai, A., et al. (2016). Population genetic analysis reveals a low level of genetic diversity of “Candidatus Phytoplasma aurantifolia” causing witches' broom disease in lime. BMC Microbiol. 5:1701. doi: 10.1186/s40064-016-3401-0
Andersen, M. T., Liefting, L. W., Havukkala, I., and Beever, R. E. (2013). Comparison of the complete genome sequence of two closely related isolates of “Candidatus Phytoplasma australiense” reveals genome plasticity. BMC Genomics 14:529. doi: 10.1186/1471-2164-14-529
Bai, X., Zhang, J., Ewing, E., Miller, S. A., Radek, A., Schevchenko, D., et al. (2006). Living with genome instability: the adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J. Bacteriol. 188, 3682–3696. doi: 10.1128/JB.188.10.3682-3696.2006
Bayliss, K. L., Saqib, M., Dell, B., Jones, M. G. K., and Hardy, G. E. S. J. (2005). First record of “Candidatus Phytoplasma australiense” in Paulownia trees. Australas. Plant Pathol. 34, 123–124. doi: 10.1071/AP04089
Benson, D. A., Cavanaugh, M., Clark, K., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., et al. (2013). GenBank. Nucleic Acids Res. 41, D36–D42. doi: 10.1093/nar/gks1195
Bertaccini, A., and Duduk, B. (2010). Phytoplasma and phytoplasma diseases: a review of recent research. Phytopathol. Mediterr. 48, 355–378. doi: 10.14601/Phytopathol_Mediterr-3300
Bertaccini, A., Duduk, B., Paltrinieri, S., and Contaldo, N. (2014). Phytoplasmas and phytoplasma diseases: a severe threat to agriculture. Am. J. Plant Sci. 5, 1763–1788. doi: 10.4236/ajps.2014.512191
Blanche, K. R., Tran-Nguyen, L. T. T., and Gibb, K. S. (2003). Detection, identification and significance of phytoplasmas in grasses in northern Australia. Plant Pathol. 52, 505–512. doi: 10.1046/j.1365-3059.2003.00871.x
Bove, J., and Garnier, M. (2003). Phloem-and xylem-restricted plant pathogenic bacteria. Plant Sci. 164, 423–438. doi: 10.1016/S0168-9452(03)00033-5
Calari, A., Paltrinieri, S., Contaldo, N., Sakalieva, D., Mori, N., Duduk, B., et al. (2011). Molecular evidence of phytoplasmas in winter oilseed rape, tomato and corn seedlings. Bull. Insectol. 64, S157–S158.
Carraro, L., Osler, R., Loi, N., Ermacora, P., and Refatti, E. (1998). Transmission of European stone fruit yellows phytoplasma by Cacopsylla pruni. J. Plant Pathol. 80, 233–239.
Chung, W.-C., Chen, L.-L., Lo, W.-S., Lin, C.-P., and Kuo, C.-H. (2013). Comparative analysis of the peanut witches'-broom phytoplasma genome reveals horizontal transfer of potential mobile units and effectors. Robinson-Rechavi, M., ed. PLoS ONE 8:e62770. doi: 10.1371/journal.pone.0062770
Constable, F. E., Whiting, J. R., Jones, J., Gibb, K. S., and Symons, R. H. (2003). The distribution of grapevine yellows disease associated with the buckland valley grapevine yellows phytoplasma. J. Phytopathol. 151, 65–73. doi: 10.1046/j.1439-0434.2003.00681.x
Danielli, A., Bertaccini, A., Vibio, M., Mori, N., Murari, E., Posenato, G., et al. (1996). Detection and molecular characterization of phytoplasmas in the planthopper Metcalfa pruinosa (Say) (Homoptera: Flatidae). Phytopathol. Mediterr. 35, 62–65.
Davis, R., Dally, E. L., Gundersen, D. E., Lee, I. M., and Habili, N. (1997a). “Candidatus Phytoplasma australiense,” a New Phytoplasma Taxon Associated with Australian Grapevine Yellows. Int. J. Syst. Evol. Microbiol. 47, 262–269. doi: 10.1099/00207713-47-2-262
Davis, R. E., and Sinclair, W. A. (1998). Phytoplasma identity and disease etiology. Phytopathology 88, 1372–1376. doi: 10.1094/PHYTO.1998.88.12.1372
Davis, R. I., Henderson, J., Jones, L. M., McTaggart, A. R., O'Dwyer, C., Tsatsia, F., et al. (2015). First record of a wilt disease of banana plants associated with phytoplasmas in Solomon Islands. Australas. Plant Dis. Notes 10:14. doi: 10.1007/s13314-015-0163-4
Davis, R. I., Jacobson, S. C., Rue, S. J., Tran-Nguyen, L., Gunua, T. G., and Rahamma, S. (2003). Phytoplasma disease surveys in the extreme north of Queensland, Australia, and the island of New Guinea. Australas. Plant Pathol. 32, 269–277. doi: 10.1071/AP03020
Davis, R. I., Jacobson, S. C., Waldeck, G. J., De La Rue, S. J., and Gibb, K. S. (2001). A witches' broom of cocky apple (Planchonia careya) in north Queensland. Australas. Plant Pathol. 30, 179–179. doi: 10.1071/AP01016
Davis, R. I., and Ruabete, T. K. (2010). Records of plant pathogenic viruses and virus-like agents from 22 Pacific island countries and territories: a review and an update. Australas. Plant Pathol. 39, 265–291. doi: 10.1071/AP10047
Davis, R., Schneider, B., and Gibb, K. (1997b). Detection and differentiation of phytoplasmas in Australia. Aust. J. Agric. Res. 48, 535–544. doi: 10.1071/A96114
De La Rue, S. J., Schneider, B., and Gibb, K. S. (1999). Genetic variability in phytoplasmas associated with papaya yellow crinkle and papaya mosaic diseases in Queensland and the Northern Territory. Australas. Plant Pathol. 28, 108–114. doi: 10.1071/AP99019
De La Rue, S., Padovan, A., and Gibb, K. (2001). Stylosanthes is a Host for Several Phytoplasmas, One of which Shows Unique 16S-23S Intergenic Spacer Region Heterogeneity. J. Phytopathol. 149, 613–619. doi: 10.1046/j.1439-0434.2001.00683.x
Dickinson, M. (2015). “Loop-mediated isothermal amplification (LAMP) for detection of phytoplasmas in the field,” in Plant Pathology, ed C. Lacomme (New York, NY: Springer), 99–111
Dickinson, M., and Hodgetts, J. (eds) (2013). Phytoplasma: Methods and Protocols, Dordrecht. Netherlands: Science+Business Media.
Duduk, B., and Bertaccini, A. (2011). Phytoplasma classification: taxonomy based on 16S ribosomal gene, is it enough? Phytopathogenic Mollicutes 1, 3–13. doi: 10.5958/j.2249-4669.1.1.001
Firrao, G., Gibb, K., and Streten, C. (2005). Short taxonomic guide to the genus “Candidatus Phytoplasma.” J. Plant Pathol. 87, 249–263. doi: 10.4454/jpp.v87i4.926
Getachew, M. A., Mitchell, A., Gurr, G. M., Fletcher, M. J., Pilkington, L. J., and Nikandrow, A. (2007). First report of a “Candidatus phytoplasma australiense”-related strain in lucerne (Medicago sativa) in Australia. Plant Dis. 91, 111–111. doi: 10.1094/PD-91-0111A
Gibb, K., Padovan, A., and Mogen, B. (1995). Studies on sweet potato little-leaf phytoplasma detected in sweet potato and other plant species growing in Northern Australia. Phytopathology 85, 169–174. doi: 10.1094/Phyto-85-169
Gibb, K., Persley, D., Schneider, B., and Thomas, J. (1996). Phytoplasmas associated with papaya diseases in Australia. Plant Dis. 80, 174–178. doi: 10.1094/PD-80-0174
Gibb, K. S., Tran-Nguyen, L. T. T., and Randles, J. W. (2003). A new phytoplasma detected in the South Australian native perennial shrub, Allocasuarina muelleriana. Ann. Appl. Biol. 142, 357–364. doi: 10.1111/j.1744-7348.2003.tb00261.x
Gopurenko, D., Fletcher, M. J., Liu, J., and Gurr, G. M. (2016). Expanding and exploring the diversity of phytoplasmas from lucerne (Medicago sativa). Sci. Rep. 6:37746. doi: 10.1038/srep37746
Gowanlock, D. H., Ogle, H. J., and Gibb, K. S. (1998). Phytoplasma associated with virescence in an epiphytic orchid in Australia. Australas. Plant Pathol. 27, 265–268. doi: 10.1071/AP98031
Gurr, G. M., Bertaccini, A., Gopurenko, D., Krueger, R. R., Alhudaib, K. A., Liu, J., et al. (2015). “Phytoplasmas and their insect vectors: implications for date palm,” in Sustainable Pest Management in Date Palm: Current Status and Emerging Challenges, eds W. Wakil, J. Romeno Faleiro and A. T. Miller (Cham: Springer International Publishing), 287–314.
Gurr, G. M., Johnson, A., Ash, G., Wilson, B., Ero, M., Pilotti, C., et al. (2016). Coconut lethal yellowing diseases: a phytoplasma threat to palms of global economic and social significance. Front. Plant Sci. 7:5121. doi: 10.3389/fpls.2016.01521
Hanold, D., Gowanlock, D., Stukely, M. J. C., Habili, N., and Randles, J. W. (2006). Mundulla Yellows disease of eucalypts: descriptors and preliminary studies on distribution and etiology. Australas. Plant Pathol. 35, 199–215. doi: 10.1071/AP06013
Hodgetts, J., Tomlinson, J., Boonham, N., González-Martín, I., Nikolić, P., Swarbrick, P., et al. (2011). Development of rapid in-field loop-mediated isothermal amplification (LAMP) assays for phytoplasmas. Bull. Insectol. 64, S41–S42.
Hogenhout, S. A., and Loria, R. (2008). Virulence mechanisms of Gram-positive plant pathogenic bacteria. Curr. Opin. Plant Biol. 11, 449–456. doi: 10.1111/j.1364-3703.2008.00472.x
Hogenhout, S. A., Oshima, K., Ammar, E.-D., Kakizawa, S., Kingdom, H. N., and Namba, S. (2008). Phytoplasmas: Bacteria that manipulate plants and insects. Mol. Plant Pathol. 9, 403–423. doi: 10.1111/j.1364-3703.2008.00472.x
Hogenhout, S. A., Van der Hoorn, R. A. L., Terauchi, R., and Kamoun, S. (2009). Emerging concepts in effector biology of plant-associated organisms. Mol. Plant Microbe Interact. 22, 115–22. doi: 10.1111/j.1364-3703.2008.00472.x
IRPCM (2004). “Candidatus Phytoplasma,” a taxon for the wall-less, non-helical prokaryotes that colonize plant phloem and insects. Int. J. Syst. Evol. Microbiol. 54, 1243–1255. doi: 10.1099/ijs.0.02854-0
Kogovšek, P., Hodgetts, J., Hall, J., Prezelj, N., Nikolić, P., Mehle, N., et al. (2015). LAMP assay and rapid sample preparation method for on-site detection of flavescence dorée phytoplasma in grapevine. Plant Pathol. 64, 286–296. doi: 10.1111/ppa.12266
Kube, M., Schneider, B., Kuhl, H., Dandekar, T., Heitmann, K., Migdoll, A. M., et al. (2008). The linear chromosome of the plant-pathogenic mycoplasma ‘Candidatus Phytoplasma mali’. BMC Genomics 9:306. doi: 10.1186/1471-2164-9-306
Kube, M., Siewert, C., Migdoll, A. M., Duduk, B., Holz, S., Rabus, R., et al. (2013). Analysis of the complete genomes of Acholeplasma brassicae, A. palmae and A. laidlawii and their comparison to the obligate parasites from “Candidatus Phytoplasma.” J. Mol. Microbiol. Biotechnol. 24, 19–36. doi: 10.1159/000354322
Lee, I.-M., Davis, R. E., and Gundersen-Rindal, D. E. (2000). Phytoplasma: phytopathogenic mollicutes. Annu. Rev. Microbiol. 54, 221–255. doi: 10.1146/annurev.micro.54.1.221
Lee, I.-M., Gundersen-Rindal, D. E., Davis, R. E., and Bartoszyk, I. M. (1998). Revised classification scheme of phytoplasmas based on RFLP Analyses of 16S rRNA and ribosomal protein gene sequences. Int. J. Syst. Bacteriol. 48, 1153–1169. doi: 10.1099/00207713-48-4-1153
Liu, B., White, D., Walsh, K., and Scott, P. (1996). Detection of phytoplasmas in dieback, yellow crinkle, and mosaic diseases of papaya using polymerase chain reaction techniques. Aust. J. Agric. Res. 47, 387–394. doi: 10.1071/AR9960387
Lu, H., Wilson, B. A. L., Ash, G. J., Woruba, S. B., Fletcher, M. J., You, M., et al. (2016). Determining putative vectors of the Bogia Coconut Syndrome phytoplasma using loop-mediated isothermal amplification of single-insect feeding media. Sci. Rep. 6:35801. doi: 10.1038/srep35801
Marcone, C. (2011). Current status of phytoplasma diseases of medicinal and nutraceutical plants in Southern Italy. Bull. Insectol. 64, S233–S234.
Marcone, C. (2014). Molecular biology and pathogenicity of phytoplasmas. Annals of Applied Biology, 165, 199–221. doi: 10.1111/aab.12151
Nicolaisen, M., Contaldo, N., Makarova, O., Paltrinieri, S., and Bertaccini, A. (2011). Deep amplicon sequencing reveals mixed phytoplasma infection within single grapevine plants. Bull. Insectol. 64, S35–S36.
Nipah, J. O., Jones, P., Hodgetts, J., and Dickinson, M. (2007). Detection of phytoplasma DNA in embryos from coconut palms in Ghana, and kernels from maize in Peru. Bull. Insectol. 60, 385–386.
Obura, E., Masiga, D., Wachira, F., Gurja, B., and Khan, Z. R. (2011). Detection of phytoplasma by loop-mediated isothermal amplification of DNA (LAMP). J. Microbiol. Methods 84, 312–316. doi: 10.1016/j.mimet.2010.12.011
Orlovskis, Z., Canale, M. C., Haryono, M., Lopes, J. R. S., Kuo, C.-H., and Hogenhout, S. A. (2017). A few sequence polymorphisms among isolates of Maize bushy stunt phytoplasma associate with organ proliferation symptoms of infected maize plants. Ann. Bot. 119, 869–884. doi: 10.1093/aob/mcw213
Oropeza, C., Cordova, I., Puch-Hau, C., Castillo, R., Chan, J. L., and Sáenz, L. (2017). Detection of lethal yellowing phytoplasma in coconut plantlets obtained through in vitro germination of zygotic embryos from the seeds of infected palms. Ann. Appl. Biol. doi: 10.1111/aab.12351. [Epub ahead of print].
Oshima, K., Kakizawa, S., Nishigawa, H., Jung, H. Y., Wei, W., Suzuki, S., et al. (2004). Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nature Genet. 36, 27–29. doi: 10.1038/ng1277
Padovan, A. C., and Gibb, K. S. (2001). Epidemiology of phytoplasma diseases in papaya in Northern Australia. J. Phytopathol. 149, 649–658. doi: 10.1046/j.1439-0434.2001.00688.x
Padovan, A., Gibb, K., and Persley, D. (2000). Association of “Candidatus Phytoplasma australiense” with green petal and lethal yellows diseases in strawberry. Plant Pathol. 49, 362–369. doi: 10.1046/j.1365-3059.2000.00461.x
Pearce, T. L., Scott, J. B., and Pethybridge, S. J. (2011). First report of a 16SrII-D subgroup phytoplasma associated with pale purple coneflower Witches'-Broom disease in Australia. Plant Dis. 95, 773–773. doi: 10.1094/PDIS-03-11-0155
Pérez-López, E., Luna-Rodríguez, M., Olivier, C. Y., and Dumonceaux, T. J. (2016). The underestimated diversity of phytoplasmas in Latin America. Int. J. Syst. Evol. Microbiol. 66, 492–513. doi: 10.1099/ijsem.0.000726
Pilkington, L. J., Gibb, K. S., Gurr, G. M., Fletcher, M. J., Nikandrow, A., Elliott, E., et al. (2003). Detection and identification of a phytoplasma from lucerne with Australian lucerne yellows disease. Plant Pathol. 52, 754–762. doi: 10.1111/j.1365-3059.2003.00934.x
Pilkington, L. J., Gurr, G. M., Fletcher, M. J., Nikandrow, A., and Elliott, E. (2004). Vector status of three leafhopper species for Australian lucerne yellows phytoplasma. Aust. J. Entomol. 43, 366–373. doi: 10.1111/j.1440-6055.2004.00419.x
Saqib, M., Bayliss, K. L., Dell, B., Hardf, G. E. S., and Jones, M. G. K. (2005). First record of a phytoplasma-associated disease of chickpea (Cicer arietinum) in Australia. Australas. Plant Pathol. 34, 425–426. doi: 10.1071/AP05047
Schneider, B., and Gibb, K. S. (1997). Detection of phytoplasmas in declining pears in Southern Australia. Plant Dis. 81, 254–258. doi: 10.1094/PDIS.1997.81.3.254
Schneider, B., Padovan, A., Rue, S., Eichner, R., Davis, R., Bernuetz, A., et al. (1999). Detection and differentiation of phytoplasmas in Australia: an update. Aust. J. Agric. Res. 50, 333–342. doi: 10.1071/A98106
Smith, G. R., Braithwaite, K. S., and Cronjé, C. P. R. (2001). “The viral and phytoplasma forms of yellow leaf syndrome of sugarcane,” in Proceedings International Society Sugar Cane Technologists, ed D. M. Hogarth (Brisbane, QLD: International Society Of Sugar Cane Technologists), 614–617.
Streten, C., Conde, B., Herrington, M., Moulder, J., and Gibb, K. (2005). Candidatus Phytoplasma australiense is associated with pumpkin yellow leaf curl disease in Queensland, Western Australia and the Northern Territory. Australasian Plant Pathol. 34, 103–105. doi: 10.1071/AP04077
Streten, C., and Gibb, K. S. (2006). Phytoplasma diseases in sub-tropical and tropical Australia. Australas. Plant Pathol. 35, 129–146. doi: 10.1071/AP06004
Tairo, F., Jones, R. A. C., and Valkonen, J. P. T. (2006). Phytoplasma from little leaf disease affected sweetpotato in Western Australia: detection and phylogeny. Ann. Appl. Biol. 149, 9–14. doi: 10.1111/j.1744-7348.2006.00065.x
Tanne, E., Boudon-Padieu, E., Clair, D., Davidovich, M., Melamed, S., and Klein, M. (2001). Detection of phytoplasma by polymerase chain reaction of insect feeding medium and its use in determining vectoring ability. Phytopathology 91, 741–746. doi: 10.1094/PHYTO.2001.91.8.741
Tran-Nguyen, L., Blanche, K. R., Egan, B., and Gibb, K. S. (2000). Diversity of phytoplasmas in northern Australian sugarcane and other grasses. Plant Pathol. 49, 666–679. doi: 10.1046/j.1365-3059.2000.00498.x
Tran-Nguyen, L. T. T., Kube, M., Schneider, B., Reinhardt, R., and Gibb, K. S. (2008). Comparative genome analysis of “Candidatus Phytoplasma australiense” (subgroup tuf-Australia I; rp-A) and “Ca. Phytoplasma asteris” strains OY-M and AY-WB. J. Bacteriol. 190, 3979–3991. doi: 10.1128/JB.01301-07
Tran-Nguyen, L. T. T., Persley, D. M., and Gibb, K. S. (2003). First report of phytoplasma disease in capsicum, celery and chicory in Queensland, Australia. Australas. Plant Pathol. 32, 559–560. doi: 10.1071/AP03055
Tran-Nguyen, L. T. T., Smith, S. H., and Liberato, J. R. (2012). Sweet potato little leaf strain V4 phytoplasma associated with snake bean in the Northern Territory, Australia. Australas. Plant Dis. Notes 7, 147–150. doi: 10.1007/s13314-012-0071-9
Veerakone, S., Tang, J. Z., Ward, L. I., Liefting, L. W., Perez-Egusquiza, Z., Lebas, B. S. M., et al. (2015). A review of the plant virus, viroid, liberibacter and phytoplasma records for New Zealand. Australas. Plant Pathol. 44, 463–514. doi: 10.1007/s13313-015-0366-3
Vega, F. E., Davis, R. E., Barbosa, P., Dally, E. L., Purcell, A. H., and Lee, I. (1993). Detection of a plant pathogen in a nonvector insect species by the polymerase chain reaction. Phytopathology 83, 621–624. doi: 10.1094/Phyto-83-621
Wang, Q., Guo, Y., Wang, N., Li, Y., Chen, W., Chen, W., et al. (2014). Identification of a conserved core genome with group-specific genes from comparative genomics of ten different Candidatus Phytoplasma strains. J. Phytopathol. 162, 650–659. doi: 10.1111/jph.12239
Weintraub, P. G., and Beanland, L. (2006). Insect vectors of phytoplasmas. Annu. Rev. Entomol. 51, 91–111. doi: 10.1146/annurev.ento.51.110104.151039
White, D. T., Blackall, L. L., Scott, P. T., and Walsh, K. B. (1998). Phylogenetic positions of phytoplasmas associated with dieback, yellow crinkle and mosaic diseases of papaya, and their proposed inclusion in “Candidatus Phytoplasma australiense” and a new taxon, “Candidatus Phytoplasma australasia.” Int. J. Syst. Evol. Microbiol. 48, 941–951. doi: 10.1099/00207713-48-3-941
Wilson, D., Blanche, K. R., and Gibb, K. S. (2001). Phytoplasmas and disease symptoms of crops and weeds in the semi-arid tropics of the Northern Territory, Australia. Australas. Plant Pathol. 30, 159–163. doi: 10.1071/AP01015
Yang, S., Habili, N., Aoda, A., Dundas, I., Paull, J., and Randles, J. (2013). Three group 16SrII phytoplasma variants detected in co-located pigeonpea, lucerne and tree medic in South Australia. Australas. Plant Dis. Notes 8, 125–129. doi: 10.1007/s13314-013-0113-y
Keywords: “Candidatus Phytoplasma”, 16S rRNA, biosecurity, taxonomy, biodiversity, vector, seed transmission, host range
Citation: Liu J, Gopurenko D, Fletcher MJ, Johnson AC and Gurr GM (2017) Phytoplasmas–The “Crouching Tiger” Threat of Australian Plant Pathology. Front. Plant Sci. 8:599. doi: 10.3389/fpls.2017.00599
Received: 25 November 2016; Accepted: 03 April 2017;
Published: 26 April 2017.
Edited by:
Brigitte Mauch-Mani, University of Neuchâtel, SwitzerlandReviewed by:
Lefort François, University of Applied Sciences of Western Switzerland, SwitzerlandSaskia A. Hogenhout, John Innes Centre (BBSRC), UK
Copyright © 2017 Liu, Gopurenko, Fletcher, Johnson and Gurr. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Geoff M. Gurr, ggurr@csu.edu.au
 Jian Liu
Jian Liu David Gopurenko
David Gopurenko Murray J. Fletcher3
Murray J. Fletcher3 Anne C. Johnson
Anne C. Johnson Geoff M. Gurr
Geoff M. Gurr