- 1Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, Beijing, China
- 2Institute of Botany, University of Chinese Academy of Sciences, Beijing, China
- 3Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI, USA
- 4Key Laboratory of Marine Food Quality and Hazard Controlling Technology of Zhejiang Province, College of Life Sciences, China Jiliang University, Hangzhou, China
- 5Beijing Engineering Research Center for Biofuels, Tsinghua University, Beijing, China
Laccase is a key enzyme in plant lignin biosynthesis as it catalyzes the final step of monolignols polymerization. Sweet sorghum [Sorghum bicolor (L.) Moench] is considered as an ideal feedstock for ethanol production, but lignin greatly limits the production efficiency. No comprehensive analysis on laccase has ever been conducted in S. bicolor, although it appears as the most promising target for engineering lignocellulosic feedstock. The aim of our work is to systematically characterize S. bicolor laccase gene family and to identify the lignin-specific candidates. A total of twenty-seven laccase candidates (SbLAC1-SbLAC27) were identified in S. bicolor. All SbLACs comprised the equivalent L1-L4 signature sequences and three typical Cu-oxidase domains, but exhibited diverse intron-exon patterns and relatively low sequence identity. They were divided into six groups by phylogenetic clustering, revealing potential distinct functions, while SbLAC5 was considered as the closest lignin-specific candidate. qRT-PCR analysis deciphered that SbLAC genes were expressed preferentially in roots and young internodes of sweet sorghum, and SbLAC5 showed high expression, adding the evidence that SbLAC5 was bona fide involved in lignin biosynthesis. Besides, high abundance of SbLAC6 transcripts was detected, correlating it a potential role in lignin biosynthesis. Diverse cis regulatory elements were recognized in SbLACs promoters, indicating putative interaction with transcription factors. Seven SbLACs were found to be potential targets of sbi-miRNAs. Moreover, putative phosphorylation sites in SbLAC sequences were identified. Our research adds to the knowledge for lignin profile modification in sweet sorghum.
Introduction
Sorghum [Sorghum bicolor (L.) Moench], as a widely adapted C4 cereal crop, is the fifth most cultivated crop around the world. It can be planted on marginal or non-arable lands owing to its favorable traits of low-input cost but high-yielding, drought-tolerance, high nutrient-, and water-use efficiency (Yuan et al., 2008). Sweet sorghum is a natural variant of common grain sorghum with greater height, higher biomass, and especially higher level of fermentable sugar in stems (Rooney et al., 2007; Calviño and Messing, 2012). It has been increasingly grown as a dedicated bioenergy feedstock offering grain, forage, sugar, and fiber simultaneously, therefore adds a new member to the family of bioenergy crops (Gill et al., 2014). In industrial production, both the starch in grain and the sugar in stem can be directly fermented for ethanol, while crop residuals are favorable lignocellulosic feedstock for ethanol production, i.e., the second generation biofuel.
The major component of lignocellulosic biomass is plant cell walls which mainly consist of cellulose, hemicellulose, and lignin. Lignin is a complex heteropolymer derived primarily from three monolignol units: p-coumaryl alcohol (H), coniferyl alcohol (G), and sinapyl alcohol (S) (Boerjan et al., 2003; Vanholme et al., 2010). It is usually covalently linked to cellulose and hemicellulose, conferring mechanical strength, and hydrophobicity to cell wall but increased recalcitrance to lignocellulose (Chang, 2007; Zeng et al., 2014). Several studies have demonstrated the link between reduced lignin levels and decreased recalcitrance with improved saccharification efficiency (Chen and Dixon, 2007; Jackson et al., 2008). Hence, if the lignocellulose composition of sweet sorghum can be manipulated toward lower lignin but higher cellulose, great economic, and environmental benefits will be achieved (Sticklen, 2006; Wang P. et al., 2015). And in the production practice, pretreatment of raw materials with lignolytic enzymes or biomass lignin modification has been frequently attempted in order to increase lignocellulose digestibility for higher biofuel yields. Genetic attempts always focused on down-regulating individual monolignol biosynthetic genes such as PAL, C4H, 4CL, HCT, COMT, CCR, and CAD, where altered lignin profile were achieved but accompanied by defected morphology (Boudet et al., 2003; Bonawitz and Chapple, 2010). By comparison, a little effort has been focused on laccase, which has newly certified function in catalyzing monolignols oxidation and polymerization in plant lignin synthesis (Berthet et al., 2011; Zhao et al., 2013).
Laccase (p-diphenol:dioxygen oxidoreductase, EC 1.10.3.2) is a member of the multicopper oxidases (MCOs) family. It has been reported to catalyze the one-electron oxidation of a wide range of substrates, coupled with the reduction of oxygen to water (Mot and Silaghi-Dumitrescu, 2012). Typical laccase contains three conserved Cu-oxidase domains, coupled with four copper ions: a mononuclear blue copper ion (Cu1) at the T1 site conferring the typical blue color, and a trinuclear copper cluster at the T2/T3 site consisting of one T2 copper ion (Cu2) and two T3 copper ions (Cu3) (Morozova et al., 2007; Giardina et al., 2010; Dwivedi et al., 2011). A total of 12 amino acid residues, including 10 histidines, and one cysteine as well as an axial methionine or leucine, have been thought to serve as copper ligands. They are housed within a set of four ungapped sequence regions L1-L4, which have been identified as signature sequences that distinguish laccase among the broader class of multicopper oxidases (Kumar et al., 2003). Laccase is widely distributed in plants, bacteria, fungi, and insects, while plant laccase is clustered in a separate clade in phylogenetic tree (Wang J. H. et al., 2015). To date, laccases have been characterized in many plants, such as Anacardiaceae, Arabidopsis thaliana (McCaig et al., 2005; Turlapati et al., 2011), Brachypodium distachyon (Wang Y. et al., 2015), Brassica napus (Zhang K. et al., 2013), cotton (Gossypium arboreum) (Wang et al., 2004), loblolly pine (Pinus taeda) (Bao et al., 1993), maize (Zea mays) (Caparrós-Ruiz et al., 2006; Liang et al., 2006b), poplar (Populus trichocarpa) (Ranocha et al., 1999), rice (Oryza Sativa) (Cho et al., 2014), ryegrass (Lolium perenne) (Gavnholt et al., 2002), sugarcane (Saccharum officenarum) (Cesarino et al., 2013), sycamore maple (Acer pseudoplatanus) (LaFayette et al., 1995), tobacco (Nicotiana tabacum) (Kiefer-Meyer et al., 1996), and yellow poplar (Liridendron tulipifera) (LaFayette et al., 1999). Diverse temporal and spatial expression patterns have been previously reported for plant laccases. In Arabidopsis, LAC4 was uniquely expressed in interfascicular fibers and seed coat columella while LAC7 in hydathodes and root hairs, LAC8 in pollen grains and phloem, LAC15 in seed coat cell walls, and LAC17 in interfascicular fibers (Berthet et al., 2011; Turlapati et al., 2011). In B. distachyon, BdLAC5 and BdLAC6 were mainly expressed in lignified tissues (Wang Y. et al., 2015). Such different expression profile indicates tissue specific physiological/biochemical roles for laccase genes. Besides, expression of multiple laccases within one tissue has been detected as well. For example in P. trichocarpa, 30 PtrLAC transcripts were expressed in stem differentiating xylem, of which 17 are abundant, suggesting a certain level of functional redundancy (Lu et al., 2013).
Recently in 2011, plant laccase has been genetically demonstrated to participate in lignin biosynthesis, while peroxidase has always been deemed to play the major role of catalyzing monolignols oxidative polymerization (Shigeto and Tsutsumi, 2016). Experimental evidence was preliminarily derived from the Arabidopsis LAC4 and LAC17, since lignin content reduced by 20 and 40% in double mutant lac4-1lac17 and lac4-2lac17, respectively; On the other hand, complementation with LAC17 restored the lignin profile of lac17 to normal. This provided the first genetic evidence that both LAC4 and LAC17 contribute to the constitutive lignification of Arabidopsis stems (Berthet et al., 2011). Researches in S. officenarum discovered that SofLAC mRNA is preferentially accumulated in sclerenchymatic bundle sheaths of young internodes, and in the meanwhile, heterogenous expression of SofLAC in Arabidopsis was able to restore the lignin content of lac17 mutant, demonstrating the role of SofLAC in lignification of sugarcane (Cesarino et al., 2013). In B. distachyon, the BdLAC5-misregulated Bd4442 mutant line exhibited significant alterations in lignification of mature culms, with a 10% lower lignin level, a slight increase of S lignin unit frequency, and a substantial increase of measurable FA esters, indicating that BdLAC5 is required for B. distachyon lignifications (Wang Y. et al., 2015). All these findings suggest that genetic manipulation of lignin biosynthesis-specific laccases is a feasible strategy for fine-tuning lignin content and/or composition.
It should be a bold and promising attempt to achieve better degradable S. bicolor biomass through manipulation of S. bicolor laccase. However, no research has ever been conducted. The objective of our work is to characterize S. bicolor laccases, with the long-term goal to identify bona fide laccases involved in monolignols oxidative polymerization. In this work, gene structure and protein domains as well as putative promoter cis regulatory elements were analyzed. A phylogenetic tree was constructed using the neighbor-joining method. In addition, the expression patterns of S. bicolor laccase genes were analyzed by quantitative RT-PCR. To sum up, twenty-seven laccase candidates were identified in S. bicolor genome. All laccase members have conserved copper-binding domains but are different in gene structures, indicating similar genetic origin but divergent biological functions. The potential regulation of SbLAC genes by TFs, miRNAs and phosphorylation was discussed. More efforts are needed to find out the bona fide lignin-specific laccase gene, which will shed light on modification of lignin profile in S. bicolor.
Materials and Methods
Plant Materials and Growth Conditions
In previous researches of our laboratory, comparative transcriptome combined with morpho-physiological analyses were performed to reveal the key factors responsible for differential cadmium accumulation in two contrasting sweet sorghum genotypes, among which the high-Cd accumulation one UMM EL TEIMAN (accession: PI 152873) was designated as H18 (Feng et al., unpublished data). Seeds of the sweet sorghum genotype H18 were obtained from Plant Genetic Resources Conservation Unit, the United States Department of Agriculture, Griffin, USA. The plants were grown in greenhouse with a day/night temperature regime of 25/20°C, a photoperiod of 16/8 h (light/dark), and a relative humidity of 50 ± 10%.
Genome-Wide Characterization of Laccase Genes in S. bicolor
The amino acid sequences of AtLAC1 to AtLAC17, ZmLAC1 to ZmLAC5, GaLAC1, SofLAC, BnTT10-1, BdLAC5, PtLAC3, PtLAC90, and PtLAC110 (Database accession numbers were listed in Supplemental Table 1) were used as queries for local BLASTP search against the Phytozome S. bicolor v3.1 proteome database (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Sbicolor). The resulted peptide sequences were verified while re-blasted in NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and checked on SMART (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1). Those possessing typical Cu-oxidase domain were predicted to be laccase candidates after exclusion of monocopper oxidase-like proteins and L-ascorbate oxidase homologs.
Putative signal peptide cleavage sites and subcellular locations were predicted by SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/) and TargetP 1.1 server (http://www.cbs.dtu.dk/services/TargetP/), respectively. Potential glycosylation sites and phosphorylation sites were separately analyzed through online NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/), YinOYang 1.2 server (http://www.cbs.dtu.dk/services/YinOYang/), and NetPhos 2.0 Sever (http://www.cbs.dtu.dk/services/NetPhos/). Visualization of the intron-exon structure of SbLAC genes was conducted by GSDS 2.0 server (http://gsds.cbi.pku.edu.cn/). Multiple amino acid sequences were aligned by Genestudio software (http://www.genestudio.com/).
Phylogenetic Analysis
A neighbor-joining phylogenetic tree was constructed by MEGA (http://www.megasoftware.net/) to gain insights into the evolutionary relationships between SbLACs and other plant laccases, with bootstrap tests for 1,000 replicates. Those previously characterized laccases like AtLAC1 to 17, ZmLAC1 to 5, GaLAC1, SofLAC, BnTT10-1, BdLAC5, PtLAC3, PtLAC90, and PtLAC110 were included in the analysis.
RNA Extraction and Quantitative RT-PCR
Roots and internodes at three developmental stages (internodes 2, 6, and 12 from the bottom up, corresponding to the mature, developing and young phase, respectively) as well as leaves to corresponding internodes were collected from 50-days old H18 plants and were put into liquid nitrogen immediately. Total RNA was extracted with Trizol reageant (Transgen, China) followed by RNase-free DNase I (Fermentas, Lithuania) digestion. The first strand cDNA was afterwards synthesized by TransScript® reverse transcriptase (Transgen, China), all according to the manufacturer's instructions. Gene-specific primers (Information is available in Supplemental Table 2) were designed for analysis of laccase expression profile, with specificity being confirmed. The qRT-PCR experiment was performed with an Mx3000P™ real-time PCR system (Agilent, USA), using THUNDERBIRD SYBR® qPCR mix (Toyobo, Japan). The relative gene expression levels were calculated by the 2−ΔΔCt method (Livak and Schmittgen, 2001), while 18S rRNA was chosen as the internal control. Each sample has three independent replicates.
In silico Analysis of SbLAC Promoter Sequences
The promoter sequences of S. bicolor laccases were investigated for potential cis-acting regulatory elements with PlantCARE (Plant cis-Acting Regulatory Elements, http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The identified elements were sort out according to their reported functions.
Prediction of sbi-miRNA Target Laccase Genes
The transcript sequences of 27 SbLACs were uploaded to web-based psRNATarget server (http://plantgrn.noble.org/psRNATarget/?function=3) for identification of potential targets corresponding to the preloaded S. bicolor miRNAs (241 published miRNAs from miRBase Release 21, June 2014). Sequences with a cut-off score ≤3 were chosen as putative targets.
Results
Twenty Seven Laccase Genes Were Identified in S. bicolor Genome
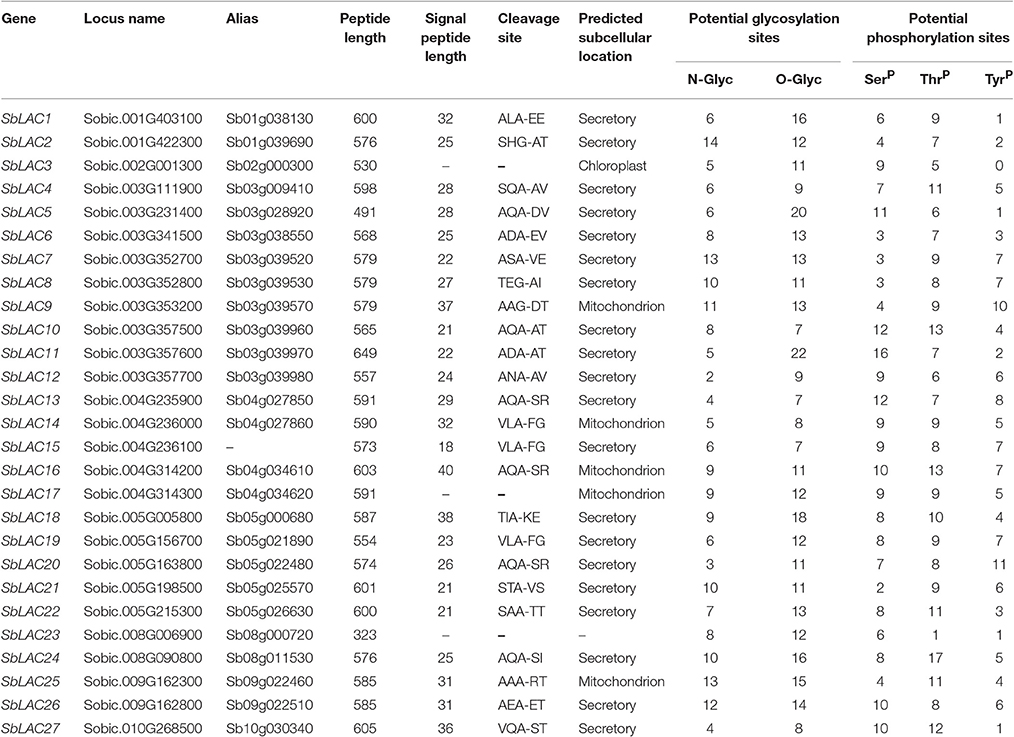
In order to identify S. bicolor laccases in silico, blastp search was performed using those well-characterized laccases from Arabidopsis, B. distachyon, B. napus, G. arboretum, Z. mays, P. trichocarpa, S. officenarum as queries against S. bicolor protein database, which output 57 S. bicolor hits. Then they were blasted in NCBI and checked on SMART, the results of which showed that 9 of them were monocopper oxidase-like proteins, 15 were deduced as L-ascorbate oxidase homologs (Details in Supplemental Table 3), while the other 33 with typical Cu-oxidase domains were considered as potential laccases. Taken alternative splicing in Sobic.003G111900, Sobic.003G357500, and Sobic.003G357700 into consideration, the number of laccase genes in S. bicolor genome was finally predicted to be 27, i.e., SbLAC1 to SbLAC27 numbered according to their distribution on chromosomes 1, 2, 3, 4, 5, 8, 9, 10. Generally, SbLACs consist of 500–600 amino acids and the majority were probably secreted proteins as indicated by a cleavable N-terminal signal peptide, with a few exceptions predicted to be located in chloroplast (SbLAC3) or mitochondria (SbLAC9, SbLAC14, SbLAC16, SbLAC17, and SbLAC25). Additionally, variable N- or O-glycosylation sites and phosphorylation sites were predicted to present in all SbLAC proteins, indicating potential post-translational modifications (Table 1).
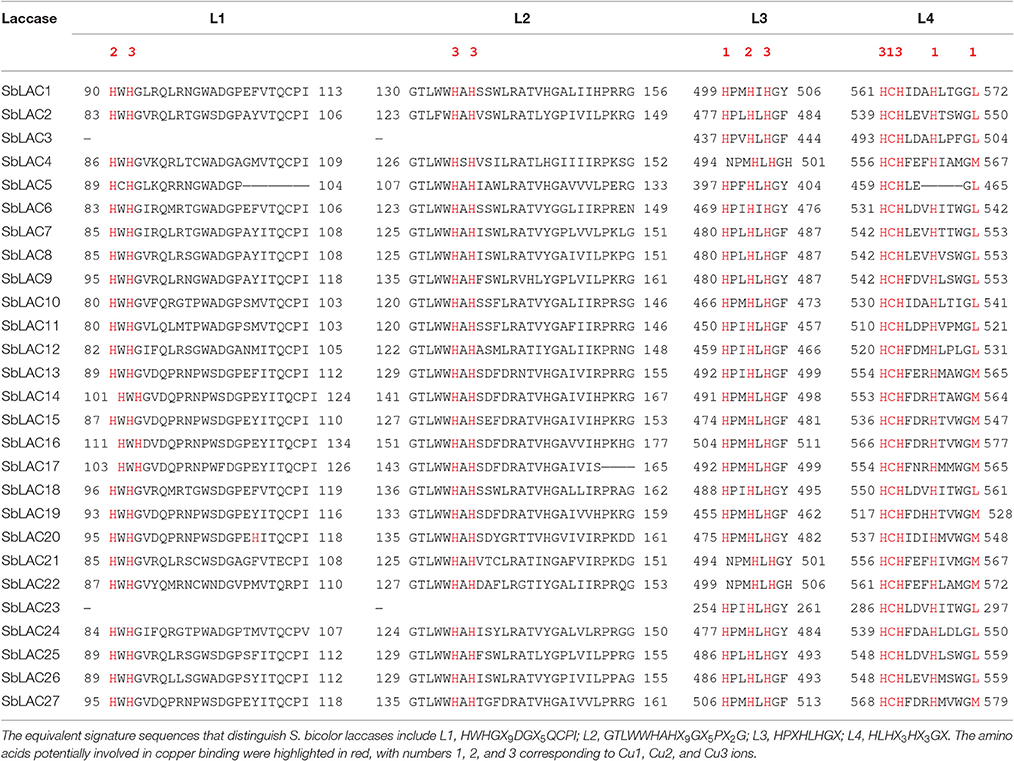
S. bicolor Laccases Had Conserved Copper-Binding Domains but Potentially Distinct Functions
The structure of S. bicolor laccase genes exhibited diverse intron-exon patterns, as the number of exons ranged from 2 to 7 (Figure 1). Likewise, amino acid sequence alignment showed relatively low sequence identity, as the identity percentage varied from 28 to 80% for most ones, with the highest homology between SbLAC14 and SbLAC15 (86.51%). Though, the equivalent L1-L4 signature sequences that distinguish laccase within the broader class of multicopper oxidases were present among individual S. bicolor laccase members (Table 2). The amino acids potentially involved in copper binding, including ten histidines and one cysteine as well as an axial ligand of methionine or leucine, were housed in the four conserved regions (Table 2). A total of 11 S. bicolor laccases (SbLAC4, SbLAC13, SbLAC14, SbLAC15, SbLAC16, SbLAC17 SbLAC19, SbLAC20, SbLAC21, SbLAC22, and SbLAC27) were provisionally grouped into the low-redox potential class (Met as the axial ligand), while the others belong to the high-redox potential class (Leu as the axial ligand).
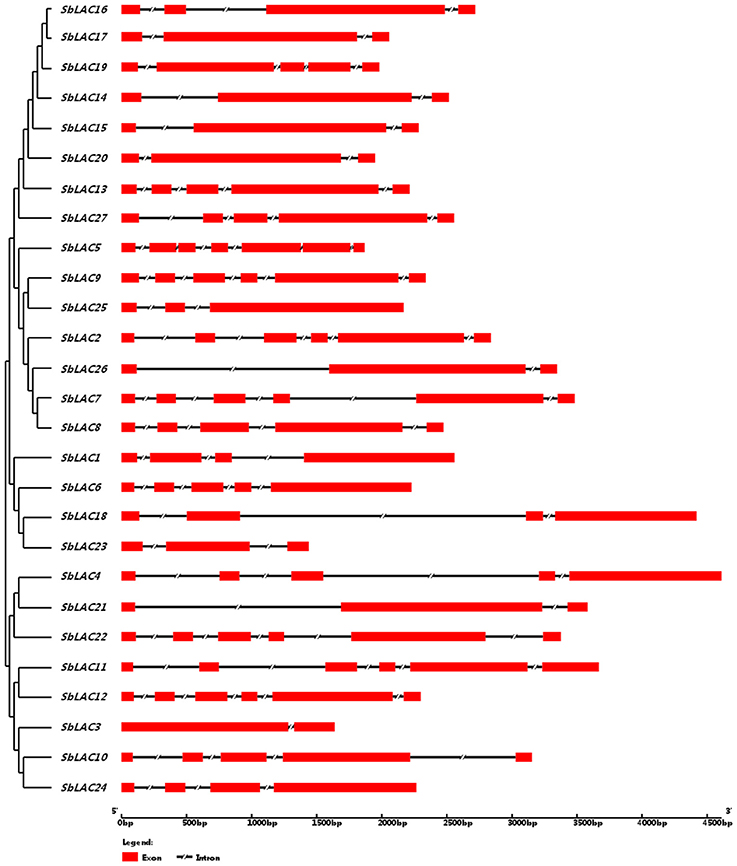
Figure 1. The intron-exon patterns of S. bicolor laccase genes. Exons and introns are represented by red rectangle and shrinked lines, respectively.
S. bicolor laccases were clustered into six phylogenetic groups (Figure 2). To be concrete, SbLAC2, SbLAC7, SbLAC8, and SbLAC26 were clustered in Group I with AtLAC17, SofLAC and BdLAC5, while SbLAC5, SbLAC9, and SbLAC25 were gathered in Group II together with AtLAC4, AtLAC11, and PtLAC3, all the latter ones have been genetically proved to be monolignol laccases involved in lignin biosynthesis, implying similar role for SbLAC2, SbLAC7, SbLAC8, and SbLAC26 in Group I and SbLAC5 in Group II. Particularly, SbLAC2 was the closest homolog to SofLAC (95.59% identity) and BdLAC5 (84.95% identity), while SbLAC5 was also tightly clustered with AtLAC11 (a lower identity of 65.20%), thus added the probability of catalyzing lignin biosynthesis. Taken that monolignol laccases AtLAC4, AtLAC17, and BdLAC5 have been proved to be localized in secondary cell wall (Schuetz et al., 2014; Wang Y. et al., 2015), SbLAC9 and SbLAC25 were more likely to participate in mitochondria oxidation-reduction cycle or similar processes due to their predicted location in mitochondria. Interestingly, Group IV comprised eight sorghum laccases from the 11 putative low-redox potential members (except SbLAC4, SbLAC21, and SbLAC22), along with AtLAC15, BnTT10-1, GaLAC1, and ZmLAC3, all of which have similar low-redox potential and have been previously reported to take part in polymerization of phenolic compounds (Ranocha et al., 2002; Cai et al., 2006; Caparrós-Ruiz et al., 2006; Liang et al., 2006a; Zhang K. et al., 2013). The classification suggested that the eight members were likely to catalyze oxidation of phenolic compounds. Meanwhile, the remaining three with low-redox potential, as well as five other S. bicolor laccases, were distributed together in Group V with stress-induced AtLAC7, AtLAC8, AtLAC9, and ZmLAC1. Besides the above, Group III contained four SbLACs and three Arabidopsis laccase members with unknown functions. Group VI included only AtLAC1, with none of the 27 S. bicolor laccase members.
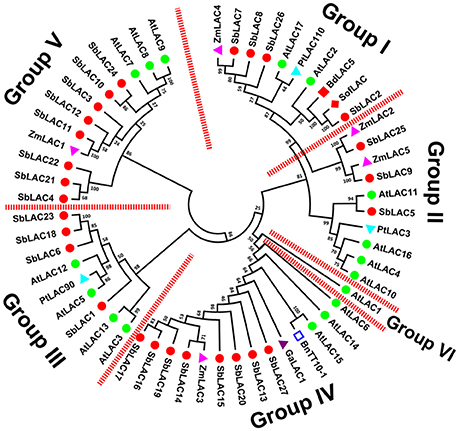
Figure 2. Phylogeny of S. bicolor laccases with the ones previously characterized from other plants. A total of fifty six plant laccases were used for MEGA analysis.  , SbLAC1 to 27;
, SbLAC1 to 27;  , AtLAC1 to 17;
, AtLAC1 to 17;  , ZmLAC1 to 5;
, ZmLAC1 to 5;  , GaLAC1;
, GaLAC1;  , PtLAC3, PtLAC90, and PtLAC110;
, PtLAC3, PtLAC90, and PtLAC110;  , BdLAC5;
, BdLAC5;  , SofLAC;
, SofLAC;  , BnTT10-1.
, BnTT10-1.
SbLAC Genes Were Preferentially Expressed in Roots and Young Internodes
To investigate the organ and development-specific expression patterns of SbLAC genes, qRT-PCR was used to detect the expression of SbLACs in roots, mature, developing and young internodes as well as the leaves to corresponding internodes. The results showed that SbLAC members have different expression levels and differential organ expression patterns. Among them, the expressions of SbLAC4/9/14/18/21/22 were hardly detected. All the remaining members showed highest expression levels in roots except for SbLAC6 and SbLAC25, which exhibited highest level in young internodes (Figure 3). Such pattern of highest expression of LAC genes in roots has been reported in Arabidopsis and Z. mays (McCaig et al., 2005; Caparrós-Ruiz et al., 2006; Abdel-Ghany and Pilon, 2008), which may be due to the main accumulation of copper ions in plant roots (Burkhead et al., 2009). Moreover, 13 SbLACs showed higher expression in young internodes but reduced level in pace with maturity (Figure 3), similarly to the previously reported sugarcane SofLAC, which was preferentially expressed in sclerenchymatic and parenchymatic cells of young internodes (Cesarino et al., 2013). This kind of expression pattern reconfirmed the hypothesis that laccases may function in early stages of lignification to polymerize monolignols into oligo-lignols (Sterjiades et al., 1993). Among all the SbLAC genes, SbLAC5 and SbLAC6 were highly expressed (the expression of SbLAC6 was higher than that of SbLAC5), while the other ones exhibited lower abundance.
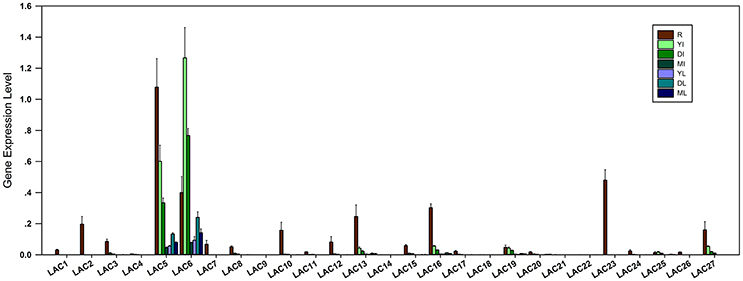
Figure 3. Expression profiles of SbLACs in H18 internodes. The expression levels are represented by means ± SD of three independent replicates. R, roots; YI, young internodes; DI, developing internodes; MI, mature internodes; YL, DL, and ML, leaves to corresponding young, developing and mature internodes, respectively.
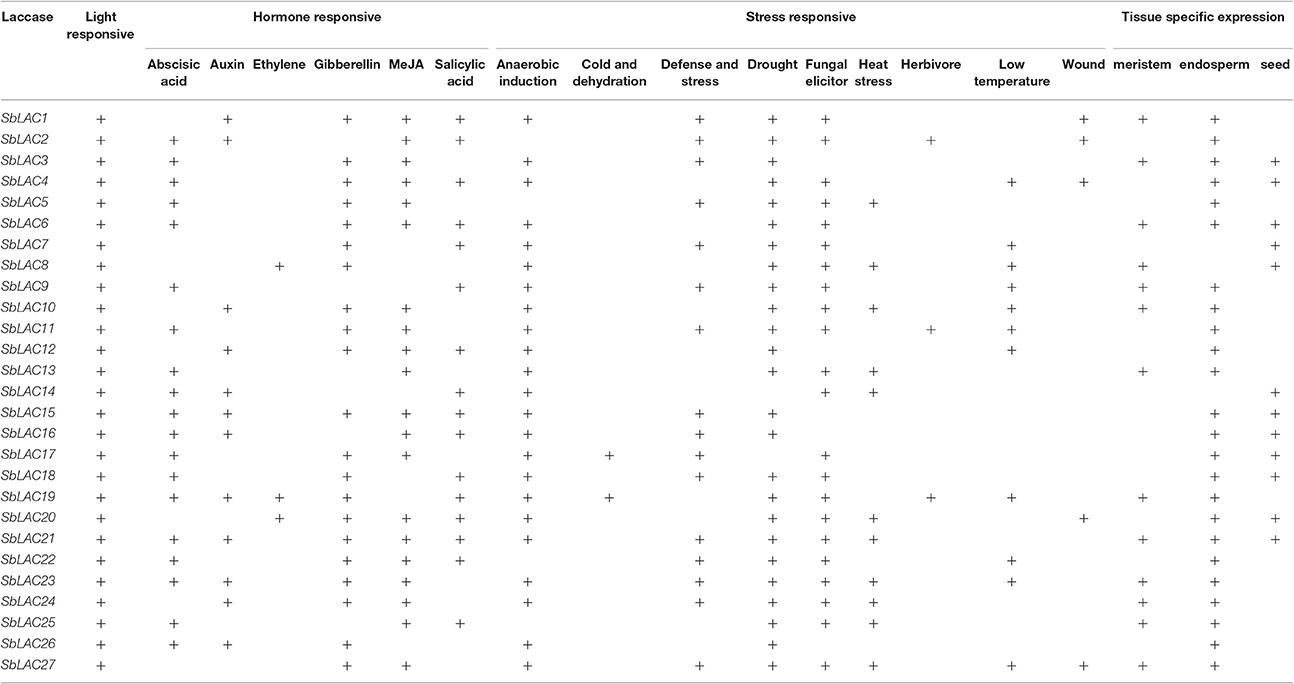
Diverse cis Regulatory Elements Were Recognized in SbLAC Promoters
Various numbers of putative cis-acting elements, including the core TATA box and CAAT box, were detected in the promoters of S. bicolor laccase genes by PlantCARE (Table 3). All 27 SbLAC promoter sequences had many light responsive elements, such as G-box (Arguello-Astorga and Herrera-Estrella, 1998), revealing an essential role of SbLACs in plant morphogenesis. Besides, there are three types of representative DNA regulatory elements: hormone responsive elements involved in response to various plant hormones, such as abscisic acid (ABA), auxin, ethylene, gibberellins (GA), methyl jasmonate (MeJA), salicylic acid; stress responsive elements responding to diverse biotic (fungal elicitor and herbivore) and abiotic (anaerobic induction, cold and dehydration, defense and stress, drought, heat stress, low temperature, and wound) stresses; tissue specific expressed elements related to endosperm-, meristem- or seed-specific activation, and regulation. Moreover, two classes of AC elements (AC-I and AC-II), which has been reported in AtLAC promoters (Turlapati et al., 2011), were also discovered in promoters of seven SbLAC genes.
Seven SbLACs Were Found to Be Potential Targets of sbi-miRNAs
Seven SbLACs were predicted to be potential sbi-miRNA targets (Details in Supplemental Table 4), among which SbLAC8 was possibly targeted by sbi-miRNA164 members (a, b, d, and e) in its N-terminal signal peptide. The mode of action was nearly the same for sbi-miR528 targeted SbLAC9 and SbLAC21. Additionally, SbLAC2, SbLAC7, SbLAC8, and SbLAC26 were putative targets of sbi-miRNA397-5p and shared similar interaction with sbi-miR397-5p at the position of Cu-oxidase domain. In addition, sbi-miRNA6235-5p had only one target SbLAC17.
Discussion
The availability of the whole genome sequences for grain sorghum facilitated characterization of laccase genes in sweet sorghum, since grain sorghum and sweet sorghum are clustered together in diversity studies despite certain genetic variation (Ritter et al., 2007; Zheng et al., 2011). Laccase is encoded by a multigene family in plants. For instance, the Arabidopsis genome encodes 17 laccases dispersed across chromosome 1, 2, 3, and 5, and the number of putative laccase genes in S. officinarum and B. distachyon is 12 and 29, respectively (McCaig et al., 2005; Cesarino et al., 2013; Wang Y. et al., 2015). Here, we characterized 27 laccase candidates in S. bicolor. They exhibited the typical characteristics of three conserved Cu-oxidase domains, four signature sequences and twelve housed copper ligands. Even so, relatively low sequence similarities and quite different intron-exon structures were presented among the 27 members, implicating potentially functional divergence.
S. bicolor Internodes Were Favorable for Identification of Cell-Wall Related Genes
Internodes of grass stalks have been considered as a useful model for identification of cell wall-related genes, since the successive internodes from the apex to the base represent a developmental profile of young to mature (Sattler et al., 2009; Bosch et al., 2011). Therefore, we chose internodes 2, 6, and 12 from the bottom up in sweet sorghum plants as representative samples belonging to different developmental stages. Case studies of histochemical staining of sugarcane and maize internodes indicated that lignification in young internodes was restricted to tracheary elements while in developing and mature internodes, lignin accumulated significantly in different cell types (Bosch et al., 2011; Cesarino et al., 2012), establishing the association of internodes anatomical changes with lignification during plant development. On the other hand, it has been reported that the expression of genes involved in lignin biosynthetic pathway, including PAL, 4CL, CCR, CCoAOMT, F3'H, LAC, etc., were higher in mature internodes with active secondary cell wall synthesis and lignifications (Sattler et al., 2009), which build the basis for functional analysis of lignin-related genes within mature internodes in plants with stalks that are attached to grass family.
SbLAC5 and SbLAC6 Were bona fide Correlated to Lignin Biosynthesis
It would be a formidable task to determine the exact role of each SbLAC member on account of expression and functional redundancy, but hints can be acquired from phylogenetic analysis. For example, eight SbLAC members were clustered with stress-induced AtLAC7, AtLAC8, AtLAC9, and ZmLAC1. AtLAC7 was reported to be up-regulated under iron deficiency while AtLAC8 and AtLAC9 were induced by 150 mM NaCl treatment and wounding in Arabidopsis (McCaig et al., 2005). And in maize primary roots treated with varied NaCl concentrations, increase of ZmLAC1 transcripts was observed (Liang et al., 2006b). The clustering revealed potential involvement of sorghum laccases in responses to environmental stresses. In another research of our laboratory (Feng et al., unpublished data), changed or different expression of SbLAC4, SbLAC21, and SbLAC22 genes has been detected in 10 μM Cd treated sweet sorghum H18 and L69, indicating that the three laccases may participates in Cd stress response.
The role of catalyzing lignin biosynthesis has always been expected to be acted by class III peroxidases, while the demonstration of laccases functioning in monolignols polymerization has raised the viewpoint that laccases might act redundantly with peroxidases. However, simultaneous disruption of the Arabidopsis LAC4, LAC11, and LAC17 brought almost abolished lignin deposition and severe growth defect in lac4lac11lac17 triple mutant, while casparian strip was still lignified through the activity of peroxidase, suggesting that laccase is necessary and non-redundant with peroxidase for lignin polymerization during vascular development in Arabidopsis (Zhao et al., 2013). It has been suggested by Sterjiades that laccase might function during early lignification stages whereas cell-wall peroxidases play the role in the follow-up proceedings of xylem development (Sterjiades et al., 1993), which can partially explain the reduced expression of SbLACs following maturity of internodes.
Based on phylogenetic analysis, SbLAC2 and SbLAC5 were considered to catalyze lignin biosynthesis. The results of qRT-PCR detected high abundance of SbLAC5 transcripts, making it the closest lignin-specific candidate (Figure 3). What's more, it's interesting to find that SbLAC6 also showed high expression, indicating a necessary but unclear role in lignin formation. In order for functional verification, more researches such as in situ hybridization or fluorescence microscopy should be performed for accurate protein localization. Examples of references are limited but come to a similar conclusion that monolignol laccases are potentially localized in apoplast of lignified tissues. It has been reported that AtLAC4 and AtLAC17 are located in secondary cell walls throughout protoxylem tracheary element differentiation in Arabidopsis (Schuetz et al., 2014). And in B. distachyon, BdLAC5 were detected in apoplasm in lignified interfascicular fibers (Wang Y. et al., 2015). Besides, genetic evidence is indispensable from either heterologous expression or self-transformation. The demonstration of bona fide SbLACs catalyzing lignin biosynthesis will point out the direction of generating feedstock with genetically alleviated recalcitrance but improved digestibility and bioethanol yields.
Regulation of SbLACs
While much progress has been made in characterization of plant laccase genes, less is clearly defined concerning the precise regulatory mechanisms. We proposed that the expression of SbLACs may be regulated by transcription factors at transcriptional level, by sbi-miRNAs at post-transcriptional level or by post-translational modifications.
Putative interaction of SbLACs with transcription factors (TFs) can be indicated by varied cis elements in SbLAC promoter sequences. For example, the G-box elements are usually present in promoters of light-responsive genes, serving as binding sites for bZIP, bHLH, and NAC TFs (Toledo-Ortiz et al., 2003; Guo and Gan, 2006; Shen et al., 2008). They have been reported to confer salt tolerance in plants, and play roles in Arabidopsis jasmonate (JA) response and early senescence of rice flag leaf and so on. Besides, it has been reported in Arabidopsis that MYB58 was able to bind to AC elements and directly activate the expression of LAC4 gene (Zhou et al., 2009). The existence of diverse cis elements in promoters of SbLAC genes provides valuable tips in future understanding of mechanisms regulating SbLAC expression.
It has been reported as a common mechanism in flowering plants that miRNA can negatively regulate laccase expression by degrading target mRNA. In Arabidopsis, seven laccases were validated to be targets for miR408, miR397, and miR857 under Cu deficient conditions (Abdel-Ghany and Pilon, 2008). Similarly in P. trichocarpa and O. Sativa, Ptr-miR397a and Os-miR397 were verified as negative regulators of PtrLACs and OsLAC, respectively (Lu et al., 2013; Zhang Y. C. et al., 2013). These works together suggest the strategy of indirect modification of laccase expression through modulating miRNAs expression. Here in our work, seven SbLACs were predicted to be sbi-miRNA targets, among which SbLAC2, SbLAC7, SbLAC8, and SbLAC26 were clusterd within Group I in phylogenetic tree, together with lignin-related AtLAC17, BdLAC5, and SofLAC, suggesting the role of sbi-miRNA397-5p in modulating cell wall lignin biosynthesis. Therefore, it can be applied to engineer sorghum lignin profile via genetic manipulation of sbi-miRNA397-5p by reference to Lu et al. and Wang et al., where overexpression of Ptr-miR397a and miR397b led to reduced lignin content in transgenic P. trichocarpa and Arabidopsis, respectively (Lu et al., 2013; Wang et al., 2014). What is important to note is that the putative sbi-miRNA/laccase pairs should be experimentally confirmed by modified 5′-rapid amplification of cDNA ends (RACE). Genetic evidences are essentially required as well.
Plant laccases are glycoproteins with relatively high carbohydrate content (20–45%) (Wang J. H. et al., 2015). It's not a surprise to find variable N- or O-glycosylation sites in S. bicolor laccase proteins, which might be effective in copper retention, enzyme stability (Ceriotti et al., 1998) and activity (Graziani et al., 1990). In addition, there exist a number of predicted serine, threonine, and tyrosine phosphorylation sites, symbols of underlying regulation by phosphorylation (Johnson and Lewis, 2001). Protein phosphorylation is one of the most widespread post-translational modifications that regulate protein activity, location, stability, or interactions (Guérinier et al., 2013; Meng et al., 2013; Umezawa et al., 2013). Numerous phosphorylation events have been identified in cellulose biosynthetic CESA proteins and CSC-associated subunits (Taylor, 2007; Chen et al., 2010; Bischoff et al., 2011; Jones et al., 2016), whereas the involvement of phosphorylation in monolignol biosynthesis has been rarely demonstrated. It was discovered only in P. trichocarpa that phosphorylation performed as an on/off switch for 5-hydroxyconiferaldehyde O-methyltransferase (PtrAldOMT2) activity in poplar monolignol biosynthesis (Wang J. P. et al., 2015). Elucidation of phosphorylation patterns in laccases will add new clues to the regulatory network for plant lignin biosynthetic pathway.
To sum up, plant laccase has drawn more and more attention in recent years with focus on its involvement in lignin biosynthesis, making it an ideal target for lignocellulose modification, especially in those bioenergy plants. It should be valuable to engineer sweet sorghum biomass through manipulation of laccase or sbi-miRNA for fine-tuning lignin profile. Our work adds to the knowledge for genetic engineering of sweet sorghum, and demonstrates a promising future in sweet sorghum cultivation for biofuel production on marginal lands such as saline or heavy metal polluted soils.
Author Contributions
YL and SL initiated the research. JF, JW, and YL designed the experiments. JW performed the experiments, analyzed the data, and drafted the manuscript. JF and YL helped draft and revise the manuscript. WJ contributed to plant material cultivation. PF and HB participated in revising the manuscript. All authors read and approved the final manuscript.
Funding
This work was financially supported by the Key Project of Intergovernmental International Cooperation on S&T Innovation, National Key R&D Program (Project number: S2016G9072).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00714/full#supplementary-material
Abbreviations
4CL, 4-coumarate: CoA, ligase; miRNA, micro-RNA; C4H, cinnamate 4-hydroxylase; CAD, cinnamyl alcohol dehydrogenase; CCR, (hydroxy)cinnamoyl CoA reductase; COMT, caffeic acid (5-hydroxyconiferaldehyde) O-methyltransferase; FA, ferulic acid; G, guaiacyl; H, p-hydroxyphenyl; H18, high-Cd accumulation genetype UMM EL TEIMAN; HCT, hydroxycinnamoyl CoA shikimate hydroxycinnamoyl transferase; Leu, leucine; Met, methionine; MCOs, multicopper oxidases; NCBI, National Center for Biotechnology Information; PAL, phenylalanine ammonia lyase; PlantCARE, Plant cis-Acting Regulatory Elements; RACE, rapid amplification of cDNA ends; S, syringyl; SMART, Simple Modular Architecture Research Tool; TF, transcription factors.
References
Abdel-Ghany, S. E., and Pilon, M. (2008). MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 283, 15932–15945. doi: 10.1074/jbc.M801406200
Arguello-Astorga, G., and Herrera-Estrella, L. (1998). Evolution of light-regulated plant promoters. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 525–555. doi: 10.1146/annurev.arplant.49.1.525
Bao, W., Omalley, D. M., Whetten, R., and Sederoff, R. R. (1993). A laccase associated with lignification in loblolly pine xylem. Science 260, 672–674. doi: 10.1126/science.260.5108.672
Berthet, S., Demont-Caulet, N., Pollet, B., Bidzinski, P., Cézard, L., Le, B. P., et al. (2011). Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23, 1124–1137. doi: 10.1105/tpc.110.082792
Bischoff, V., Desprez, T., Mouille, G., Vernhettes, S., Gonneau, M., and Hofte, H. (2011). Phytochrome regulation of cellulose synthesis in Arabidopsis. Curr. Biol. 21, 1822–1827. doi: 10.1016/j.cub.2011.09.026
Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. doi: 10.1146/annurev.arplant.54.031902.134938
Bonawitz, N. D., and Chapple, C. (2010). The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu. Rev. Genet. 44, 337–363. doi: 10.1146/annurev-genet-102209-163508
Bosch, M., Mayer, C. D., Cookson, A., and Donnison, I. S. (2011). Identification of genes involved in cell wall biogenesis in grasses by differential gene expression profiling of elongating and non-elongating maize internodes. J. Exp. Bot. 62, 3545–3561. doi: 10.1093/jxb/err045
Boudet, A. M., Kajita, S., Grima-Pettenati, J., and Goffner, D. (2003). Lignins and lignocellulosics: a better control of synthesis for new and improved uses. Trends Plant Sci. 8, 576–581. doi: 10.1016/j.tplants.2003.10.001
Burkhead, J. L., Reynolds, K. A., Abdel-Ghany, S. E., Cohu, C. M., and Pilon, M. (2009). Copper homeostasis. New Phytol. 182, 799–816. doi: 10.1111/j.1469-8137.2009.02846.x
Cai, X. N., Davis, E. J., Ballif, J., Liang, M. X., Bushman, E., Haroldsen, V., et al. (2006). Mutant identification and characterization of the laccase gene family in Arabidopsis. J. Exp. Bot. 57, 2563–2569. doi: 10.1093/jxb/erl022
Calviño, M., and Messing, J. (2012). Sweet sorghum as a model system for bioenergy crops. Curr. Opin. Biotechnol. 23, 323–329. doi: 10.1016/j.copbio.2011.12.002
Caparrós-Ruiz, D., Fornalé, S., Civardi, L., Puigdomènech, P., and Rigau, J. (2006). Isolation and characterisation of a family of laccases in maize. Plant Sci. 171, 217–225. doi: 10.1016/j.plantsci.2006.03.007
Ceriotti, A., Duranti, M., and Bollini, R. (1998). Effects of N-glycosylation on the folding and structure of plant proteins. J. Exp. Bot. 49, 1091–1103. doi: 10.1093/jxb/49.324.1091
Cesarino, I., Araújo, P., Mayer, J. L. S., Leme, A. F. P., and Mazzafera, P. (2012). Enzymatic activity and proteomic profile of class III peroxidases during sugarcane stem development. Plant Physiol. Biochem. 55, 66–76. doi: 10.1016/j.plaphy.2012.03.014
Cesarino, I., Araújo, P., Mayer, J. L. S., Vicentini, R., Berthet, S., Demedts, B., et al. (2013). Expression of SofLAC, a new laccase in sugarcane, restores lignin content but not S:G ratio of Arabidopsis lac17 mutant. J. Exp. Bot. 64, 1769–1781. doi: 10.1093/jxb/ert045
Chang, M. C. (2007). Harnessing energy from plant biomass. Curr. Opin. Chem. Biol. 11, 677–684. doi: 10.1016/j.cbpa.2007.08.039
Chen, F., and Dixon, R. A. (2007). Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25, 759–761. doi: 10.1038/nbt1316
Chen, S. L., Ehrhardt, D. W., and Somerville, C. R. (2010). Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc. Natl. Acad. Sci. U.S.A. 107, 17188–17193. doi: 10.1073/pnas.1012348107
Cho, H. Y., Lee, C., Hwang, S. G., Park, Y. C., Lim, H. L., and Jang, C. S. (2014). Overexpression of the OsChI1 gene, encoding a putative laccase precursor, increases tolerance to drought and salinity stress in transgenic Arabidopsis. Gene 552, 98–105. doi: 10.1016/j.gene.2014.09.018
Dwivedi, U. N., Singh, P., Pandey, V. P., and Kumar, A. (2011). Structure-function relationship among bacterial, fungal and plant laccases. J. Mol. Catal. B Enzym. 68, 117–128. doi: 10.1016/j.molcatb.2010.11.002
Gavnholt, B., Larsen, K., and Rasmussen, S. K. (2002). Isolation and characterisation of laccase cDNAs from meristematic and stem tissues of ryegrass (Lolium perenne). Plant Sci. 162, 873–885. doi: 10.1016/S0168-9452(02)00035-3
Giardina, P., Faraco, V., Pezzella, C., Piscitelli, A., Vanhulle, S., and Sannia, G. (2010). Laccases: a never-ending story. Cell. Mol. Life Sci. 67, 369–385. doi: 10.1007/s00018-009-0169-1
Gill, J. R., Burks, P. S., Staggenborg, S. A., Odvody, G. N., Heiniger, R. W., Macoon, B., et al. (2014). Yield results and sability analysis from the sorghum regional biomass feedstock trial. Bioenergy Res. 7, 1026–1034. doi: 10.1007/s12155-014-9445-5
Graziani, M. T., Antonilli, L., Sganga, P., Citro, G., Mondovi, B., and Rosei, M. A. (1990). Biochemical and immunological studies of deglycosylated Rhus Vernicifera laccase. Biochem. Int. 21, 1113–1124.
Guérinier, T., Millan, L., Crozet, P., Oury, C., Rey, F., Valot, B., et al. (2013). Phosphorylation of p27(KIP1) homologs KRP6 and 7 by SNF1-related protein kinase-1 links plant energy homeostasis and cell proliferation. Plant J. 75, 515–525. doi: 10.1111/tpj.12218
Guo, Y. F., and Gan, S. S. (2006). AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46, 601–612. doi: 10.1111/j.1365-313X.2006.02723.x
Jackson, L. A., Shadle, G. L., Zhou, R., Nakashima, J., Chen, F., and Dixon, R. A. (2008). Improving saccharification efficiency of alfalfa stems through modification of the terminal stages of monolignol biosynthesis. Bioenergy Res. 1, 180–192. doi: 10.1007/s12155-008-9020-z
Johnson, L. N., and Lewis, R. J. (2001). Structural basis for control by phosphorylation. Chem. Rev. 101, 2209–2242. doi: 10.1021/cr000225s
Jones, D. M., Murray, C. M., Ketelaar, K. J., Thomas, J. J., Villalobos, J. A., and Wallace, I. S. (2016). The emerging role of protein phosphorylation as a critical regulatory mechanism controlling cellulose biosynthesis. Front. Plant Sci. 7:684. doi: 10.3389/fpls.2016.00684
Kiefer-Meyer, M. C., Gomord, V., O'Connell, A., Halpin, C., and Faye, L. (1996). Cloning and sequence analysis of laccase-encoding cDNA clones from tobacco. Gene 178, 205–207. doi: 10.1016/0378-1119(96)00381-2
Kumar, S. V., Phale, P. S., Durani, S., and Wangikar, P. P. (2003). Combined sequence and structure analysis of the fungal laccase family. Biotechnol. Bioeng. 83, 386–394. doi: 10.1002/bit.10681
LaFayette, P. R., Eriksson, K. E., and Dean, J. F. (1995). Nucleotide sequence of a cDNA clone encoding an acidic laccase from sycamore maple (Acer Pseudoplatanus L). Plant Physiol. 107, 667–668. doi: 10.1104/pp.107.2.667
LaFayette, P. R., Eriksson, K. E., and Dean, J. F. (1999). Characterization and heterologous expression of laccase cDNAs from xylem tissues of yellow-poplar (Liriodendron tulipifera). Plant Mol. Biol. 40, 23–35. doi: 10.1023/A:1026437406859
Liang, M. X., Davis, E., Gardner, D., Cai, X. N., and Wu, Y. J. (2006a). Involvement of AtLAC15 in lignin synthesis in seeds and in root elongation of Arabidopsis. Planta 224, 1185–1196. doi: 10.1007/s00425-006-0300-6
Liang, M. X., Haroldsen, V., Cai, X. N., and Wu, Y. J. (2006b). Expression of a putative laccase gene, ZmLAC1, in maize primary roots under stress. Plant Cell Environ. 29, 746–753. doi: 10.1111/j.1365-3040.2005.01435.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, S. F., Li, Q. Z., Wei, H. R., Chang, M. J., Tunlaya-Anukit, S., Kim, H., et al. (2013). Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl. Acad. Sci. U.S.A. 110, 10848–10853. doi: 10.1073/pnas.1308936110
McCaig, B. C., Meagher, R. B., and Dean, J. F. D. (2005). Gene structure and molecular analysis of the laccase-like multicopper oxidase (LMCO) gene family in Arabidopsis thaliana. Planta 221, 619–636. doi: 10.1007/s00425-004-1472-6
Meng, X. Z., Xu, J., He, Y. X., Yang, K. Y., Mordorski, B., Liu, Y. D., et al. (2013). Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25, 1126–1142. doi: 10.1105/tpc.112.109074
Morozova, O. V., Shumakovich, G. P., Gorbacheva, M. A., Shleev, S. V., and Yaropolov, A. I. (2007). “Blue” laccases. Biochemistry 72, 1136–1150. doi: 10.1134/S0006297907100112
Mot, A. C., and Silaghi-Dumitrescu, R. (2012). Laccases: complex architectures for one-electron oxidations. Biochemistry 77, 1395–1407. doi: 10.1134/S0006297912120085
Ranocha, P., Chabannes, M., Chamayou, S., Danoun, S., Jauneau, A., Boudet, A. M., et al. (2002). Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in poplar. Plant Physiol. 129, 145–155. doi: 10.1104/pp.010988
Ranocha, P., McDougall, G., Hawkins, S., Sterjiades, R., Borderies, G., Stewart, D., et al. (1999). Biochemical characterization, molecular cloning and expression of laccases - a divergent gene family - in poplar. Eur. J. Biochem. 259, 485–495. doi: 10.1046/j.1432-1327.1999.00061.x
Ritter, K. B., McIntyre, C. L., Godwin, I. D., Jordan, D. R., and Chapman, S. C. (2007). An assessment of the genetic relationship between sweet and grain sorghums, within Sorghum bicolor ssp. bicolor (L.) Moench, using AFLP markers. Euphytica 157, 161–176. doi: 10.1007/s10681-007-9408-4
Rooney, W. L., Blumenthal, J., Bean, B., and Mullet, J. E. (2007). Designing sorghum as a dedicated bioenergy feedstock. Biofuels Bioprod. Biorefin. Biofpr 1, 147–157. doi: 10.1002/bbb.15
Sattler, S. E., Saathoff, A. J., Haas, E. J., Palmer, N. A., Funnell-Harris, D. L., Sarath, G., et al. (2009). A nonsense mutation in a cinnamyl alcohol dehydrogenase gene is responsible for the Sorghum brown midrib6 Phenotype. Plant Physiol. 150, 584–595. doi: 10.1104/pp.109.136408
Schuetz, M., Benske, A., Smith, R. A., Watanabe, Y., Tobimatsu, Y., Ralph, J., et al. (2014). Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol. 166, 798–807. doi: 10.1104/pp.114.245597
Shen, H. S., Cao, K. M., and Wang, X. P. (2008). AtbZIP16 and AtbZIP68, two new members of GBFs, can interact with other G group bZIPs in Arabidopsis thaliana. BMB Rep. 41, 132–138. doi: 10.5483/BMBRep.2008.41.2.132
Shigeto, J., and Tsutsumi, Y. (2016). Diverse functions and reactions of class III peroxidases. New Phytol. 209, 1395–1402. doi: 10.1111/nph.13738
Sterjiades, R., Dean, J. F. D., Gamble, G., Himmelsbach, D. S., and Eriksson, K. E. L. (1993). Extracellular laccases and peroxidases from sycamore maple (Acer pseudoplatanus) cell-suspension cultures - reactions with monolignols and lignin model compounds. Planta 190, 75–87. doi: 10.1007/BF00195678
Sticklen, M. (2006). Plant genetic engineering to improve biomass characteristics for biofuels. Curr. Opin. Biotechnol. 17, 315–319. doi: 10.1016/j.copbio.2006.05.003
Taylor, N. G. (2007). Identification of cellulose synthase AtCesA7 (IRX3) in vivo phosphorylation sites - a potential role in regulating protein degradation. Plant Mol. Biol. 64, 161–171. doi: 10.1007/s11103-007-9142-2
Toledo-Ortiz, G., Huq, E., and Quail, P. H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15, 1749–1770. doi: 10.1105/tpc.013839
Turlapati, P. V., Kim, K. W., Davin, L. B., and Lewis, N. G. (2011). The laccase multigene family in Arabidopsis thaliana: towards addressing the mystery of their gene function(s). Planta 233, 439–470. doi: 10.1007/s00425-010-1298-3
Umezawa, T., Sugiyama, N., Takahashi, F., Anderson, J. C., Ishihama, Y., Peck, S. C., et al. (2013). Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 6:rs8. doi: 10.1126/scisignal.2003509
Vanholme, R., Demedts, B., Morreel, K., Ralph, J., and Boerjan, W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153, 895–905. doi: 10.1104/pp.110.155119
Wang, C. Y., Zhang, S. C., Yu, Y., Luo, Y. C., Liu, Q., Ju, C. L., et al. (2014). MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis. Plant Biotechnol. J. 12, 1132–1142. doi: 10.1111/pbi.12222
Wang, G. D., Li, Q. J., Luo, B., and Chen, X. Y. (2004). Ex planta phytoremediation of trichlorophenol and phenolic allelochemicals via an engineered secretory laccase. Nat. Biotechnol. 22, 893–897. doi: 10.1038/nbt982
Wang, J. H., Feng, J. J., Jia, W. T., Chang, S., Li, S. Z., and Li, Y. X. (2015). Lignin engineering through laccase modification: a promising field for energy plant improvement. Biotechnol. Biofuels 8:145. doi: 10.1186/s13068-015-0331-y
Wang, J. P., Chuang, L., Loziuk, P. L., Chen, H., Lin, Y. C., Shi, R., et al. (2015). Phosphorylation is an on/off switch for 5-hydroxyconiferaldehyde O-methyltransferase activity in poplar monolignol biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 112, 8481–8486. doi: 10.1073/pnas.1510473112
Wang, P., Dudareva, N., Morgan, J. A., and Chapple, C. (2015). Genetic manipulation of lignocellulosic biomass for bioenergy. Curr. Opin. Chem. Biol. 29, 32–39. doi: 10.1016/j.cbpa.2015.08.006
Wang, Y., Bouchabké-Coussa, O., Lebris, P., Antelme, S., Soulhat, C., Gineau, E., et al. (2015). LACCASE 5 is required for lignification of the Brachypodium distachyon culm. Plant Physiol. 168, 192–204. doi: 10.1104/pp.114.255489
Yuan, J. S., Tiller, K. H., Al-Ahmad, H., Stewart, N. R., and Stewart, C. N. (2008). Plants to power: bioenergy to fuel the future. Trends Plant Sci. 13, 421–429. doi: 10.1016/j.tplants.2008.06.001
Zeng, Y. N., Zhao, S., Yang, S. H., and Ding, S. Y. (2014). Lignin plays a negative role in the biochemical process for producing lignocellulosic biofuels. Curr. Opin. Biotechnol. 27, 38–45. doi: 10.1016/j.copbio.2013.09.008
Zhang, K., Lu, K., Qu, C. M., Liang, Y., Wang, R., Chai, Y. R., et al. (2013). Gene silencing of BnTT10 family genes causes retarded pigmentation and lignin reduction in the seed coat of Brassica napus. PLoS ONE 8:e61247. doi: 10.1371/journal.pone.0061247
Zhang, Y. C., Yu, Y., Wang, C. Y., Li, Z. Y., Liu, Q., Xu, J., et al. (2013). Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 31, 848–852. doi: 10.1038/nbt.2646
Zhao, Q., Nakashima, J., Chen, F., Yin, Y. B., Fu, C. X., Yun, J. F., et al. (2013). LACCASE is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25, 3976–3987. doi: 10.1105/tpc.113.117770
Zheng, L. Y., Guo, X. S., He, B., Sun, L. J., Peng, Y., Dong, S. S., et al. (2011). Genome-wide patterns of genetic variation in sweet and grain sorghum (Sorghum bicolor). Genome Biol. 12:R114. doi: 10.1186/gb-2011-12-11-r114
Keywords: Sorghum bicolor, lignin, laccase, genetic engineering, lignin modification
Citation: Wang J, Feng J, Jia W, Fan P, Bao H, Li S and Li Y (2017) Genome-Wide Identification of Sorghum bicolor Laccases Reveals Potential Targets for Lignin Modification. Front. Plant Sci. 8:714. doi: 10.3389/fpls.2017.00714
Received: 15 February 2017; Accepted: 18 April 2017;
Published: 05 May 2017.
Edited by:
Junhua Peng, Center for Life Sci&Tech of China National Seed Group Co. Ltd., ChinaReviewed by:
Xiaoli Jin, Zhejiang University, ChinaAkiyoshi Kawaoka, Nippon Paper Industries Co. Ltd., Japan
Copyright © 2017 Wang, Feng, Jia, Fan, Bao, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juanjuan Feng, jjfeng@ibcas.ac.cn
Shizhong Li, szli@mail.tsinghua.edu.cn
Yinxin Li, yxli@ibcas.ac.cn
 Jinhui Wang1,2
Jinhui Wang1,2 Yinxin Li
Yinxin Li