- 1State Key Laboratory of Hybrid Rice, Key Laboratory of Major Crop Diseases & Collaborative Innovation Center for Hybrid Rice in Yangtze River Basin, Sichuan Agricultural University at Wenjiang, Chengdu, China
- 2Rice Research Institute, Agricultural Academy of Sciences at Mianyang, Mianyang, China
The brassinosteroid-SIGNALING KINASE (BSK) belongs to the receptor-like cytoplasmic kinase XII subgroup. BSK1 regulates development and immunity in Arabidopsis. However, the function of rice (Oryza sativa) BSK1 is largely unknown. Here, we report that the expression level of OsBSK1-2 is induced after a chitin or fagellin22 (flg22) treatment. Silencing OsBSK1-2 in rice results in compromised responses to chitin- or flg22-triggered immunity and resistance to Magnaporthe oryzae, but does not alter the plant’s architecture nor reduce plant responses to brassinosteroid signaling. Our study reveals that OsBSK1-2 functions as a major regulator in rice plant immunity.
Introduction
Receptor-like cytoplasmic kinases (RLCKs) are a subgroup of the receptor-like kinase family, but unlike the receptor-like kinases, which have an extracellular receptor domain, a transmembrane domain, and an intracellular kinase domain (Shiu and Bleecker, 2001), RLCKs just have a typical catalytic kinase, and they localize to the cytoplasm. Several RLCKs are predicted as regulators of plant development and immunity (Vij et al., 2008).
Receptor-like cytoplasmic kinases VII is an important gene subfamily in plant. In Arabidopsis, BOTRYTIS INDUCED KINASE 1 (BIK1) was first isolated as an early-induced kinase in response to infection by Botrytis cinerea (Veronese et al., 2006). BIK1 was then revealed to play an essential role in regulating immune responses mediated by pattern recognition receptors (PRRs), including flagellin-sensitive-2 (FLS2), EF-Tu receptor (EFR), and PEP-RECEPTOR1 (PEPR1) (Zhang et al., 2010; Eckardt, 2011; Liu et al., 2013). The orthologs of BIK1, PBS1, PBL1, PBL2 and PBL27, were also identified as regulators for pattern-triggered immunity (PTI) (Zhang et al., 2010; Lin et al., 2013; Liu et al., 2013; Yamaguchi et al., 2013a,b; Shinya et al., 2014). Other members of the RLCK VII subfamily have also been characterized as regulators in rice immunity. For examples, OsRLCK185, which is phosphorylated by OsCERK1 upon recognition of chitin and then dissociates from the OsCERK1 complex to activate MAPK cascades, is involved in an immune response (Yamaguchi et al., 2013a,b). OsRLCK57, OsRLCK107, OsRLCK118 and OsRLCK176, four homologs of BIK1, positively regulate immune responses by contributing to the expression of the immune receptor XA21 (Zhou et al., 2016).
Recently, brassinosteroid (BR)-signaling kinase1 (BSK1), a member of RLCK-XII, which belongs to another RLCK subfamily, was reported to play vital roles in plant immunity. BSK1, first isolated as a phosphorylation substrate of the BR receptor kinase BRI1, plays critical role in BR signaling (Tang et al., 2008). In Arabidopsis, 12 BSK members were predicted (BSK1–12). Biochemical assays revealed that BRI1 targets many BSKs for phosphorylation, including BSK1, BSK3, BSK5, BSK6 and BSK11, suggesting that these BSKs might have redundant functions in response to BRs (Tang et al., 2008; Sreeramulu et al., 2013). Moreover, the bsk1 mutant showed compromised resistance to various pathogens and responses to PTI. BSK1 associates with the PRR FLAGELLIN SENSING2 (FLS2) to activate downstream signal transduction in plant immunity (Shi et al., 2013). In rice, a member of the RLCK-XII subgroup, OsBSK3, plays a positive role in BR signaling, which can be phosphorylated by OsBRI1 and then associates with BRI1 SUPPRESSOR1 to transduce the BR signal (Zhang et al., 2016). However, the biological roles of their functions in rice immunity are poorly known.
In this study, we report that the enrichment of the transcripts levels of OsBSK1-2 was induced by both flg22 and chitin. Silencing OsBSK1-2 reduced the transcriptional expression of the pathogenesis-related genes, Os04g10010 and OsPR10b, in response to the treatment of flg22 or chitin and compromised plant resistance to Magnaporthe oryzae. However, plants with reduced OsBSK1-2 expression levels did not alter plant architecture or BR signaling. Thus, our study reveals that OsBSK1-2 functions as a positive regulator in rice immunity.
Materials and Methods
Plant Growth
All of the transgenic plants were created in the ‘Kitaake’ background. All of the plants were grown in a controlled rice field in Chengdu, Sichuan, China.
Construction
All of the primers used here are listed in Supplementary Table S1. To generate RNA interference (RNAi) construction, the unique cDNA sequence of OsBSK1-2 was amplified and cloned into the pCRR8TM/GW/TOPO® (Invitrogen, Carlsbad, CA, United States) vector to create the pCR8-OsBSK1-2Ri constructs. Then, the RNAi fragments were inserted into the pANDA vector (Miki and Shimamoto, 2004) using LR recombination to create the RNAi construct pANDA-OsBSK1-2Ri (OsBSK1-2Ri).
Generation of Transgenic Rice
OsBSK1-2Ri was introduced into ‘Kitaake’ plants through Agrobacterium-mediated transformation, and hygromycin was used to select the regenerated plants (Chern et al., 2005).
RNA Extraction and Quantitative Real Time RT-PCR Analyses
Total RNA was prepared from samples collected using Trizol (Invitrogen) according to the manufacturer’s instructions. The RNA samples were then subjected to cDNA synthesis system with RT reagent Kit with gDNA Eraser (Takara, Dalian, China). A Quantitative RT-PCR Kit was used to establish the qRT-PCR reactions (Qiagen, Valencia, CA, United States). The gene expression levels were normalized to the transcript level of the reference gene ubiquitin5 (LOC_06g46770) gene. All of the primer pairs used for qRT-PCR, which are listed in Supplementary Table S2, have been tested for efficiency and specificity following the guidelines for successful real time1. qRT-PCR analysis were performed follow the MIQe guidelines (Bustin et al., 2009).
Defense Gene Expression Analysis
The 2–3 cm long leaf strips from fully developed leaves of 4-week-old seedlings were incubated for 12 h in ddH2O. They were then treated with 1 μM flg22 peptide (Pacific Immunology, San Diego, CA, United States) or 20 μg/ml chitin from shrimp shells (Sigma, St. Louis, MO, United States) according to a method described previously (Felix et al., 1999). The samples were collected after 12 h for further investigation.
Brassinolide (BL) Treatment
To analyze the effects of BL on the lamina joint angles of the transgenic plants, 1 μl 2,4-epiBL (100 ng/μl) was applied to the leaf angles and the lamina joint angles were measured following a method described previously (Chen et al., 2013).
Phylogenetic and Molecular Evolutionary Analyses
To identify ortholog(s) of AtBSK1 in the rice genome, the amino acid sequence of AtBSK1 was used as the seed sequence to perform a BLAST algorithm-related search of the rice protein database2. The proteins with hit scores more than 1,000 and E-value less than 1.6e-100, were chosen for further study. Twelve AtBSK1 and five OsBSKs were first aligned using ClusterX software. Then, a phylogenetic tree was constructed using MEGA5.10 software. The evolutionary history was inferred using the Maximum Likelihood method based on the Poisson correction model. The tree with the highest log likelihood (-6702.6606) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured using the number of substitutions per site (Tamura et al., 2011).
Rice Blast Incubation
Magnaporthe oryzae isolates, ZHONG1 and ZB25, were used for the inoculations. Leaf strips from 8-week-old plants were lightly wounded with a mouse ear punch, and 5 μl of spore suspension (5 × 105 spores/ml) was added to the wound. The lesion size was measured after incubating for 8–14 days at 28°C. The DNA was then extracted to calculate the expression level of the Pot2 gene of M. oryzae relative to the genomic Ubiquitin in rice (Park et al., 2012).
Results
Characterization of OsBSK1-1 and OsBSK1-2
Many BR signaling partners have been characterized in rice, and they have functions that are similar to those of their orthologs in Arabidopsis (Tong and Chu, 2012). AtBSK1 was reported to regulate Arabidopsis immunity. We therefore presumed that the rice ortholog(s) of AtBSK1 should function in rice immunity. To test this hypothesis, we identified five OsBSKs, OsBSK1-1 (LOC_Os03g04050), OsBSK1-2 (LOC_Os10g39670), OsBSK2 (LOC_Os10g42110), OsBSK3 (LOC_Os04g58750), and OsBSK4 (LOC_Os03g61010) from rice genome by blast research using the amino acid sequence of AtBSK1 as the seed sequence. The result showed that two proteins, OsBSK1-1 (LOC_Os03g04050) and OsBSK1-2 (LOC_Os10g39670) which share more than 79% similarity with each other in amino acid sequence were sub-grouped with AtBSK1 in phylogenetic and molecular evolutionary analyses (Figure 1).
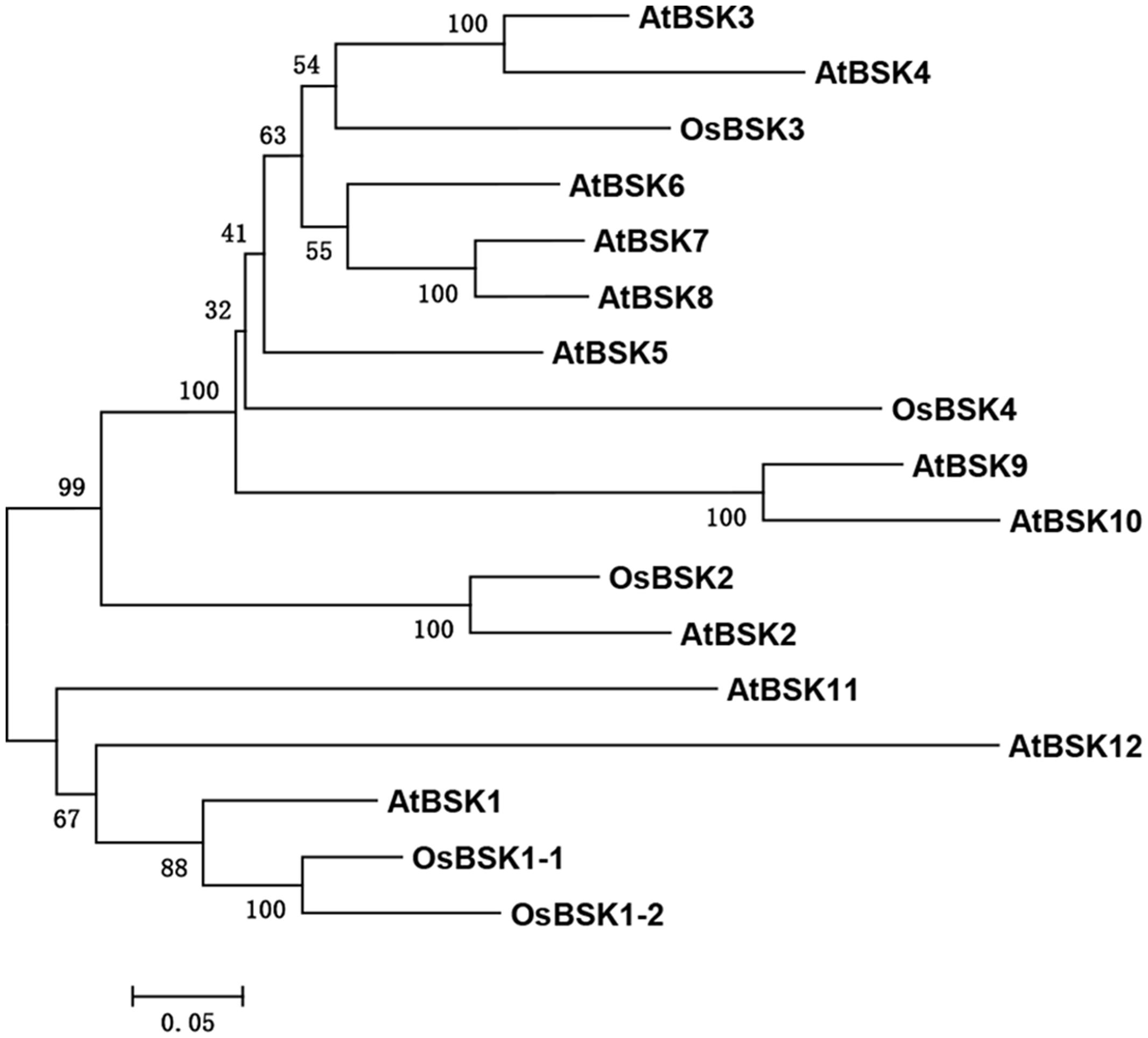
FIGURE 1. Phylogenetic analysis on five OsBSKs and 12 Arabidopsis BSKs. Five OsBSKs and 12 AtBSKs were subjected to the ClustalX analysis with default parameters. Phylogenetic was built using MEGA version5.10. Two orthologous of AtBSK1 were identified as OsBSK1-1 and OsBSK1-2.
To determine the expression patterns of OsBSK1-1 and OsBSK1-2, we measured their transcription levels in different developmental stages using quantitative reverse transcription-PCR (qRT-PCR). Both OsBSK1-1 and OsBSK1-2 were predominantly expressed in the four-leaf stage of development, and the transcripts of OsBSK1-2 were more abundant than those of OsBSK1-1 (Figure 2).
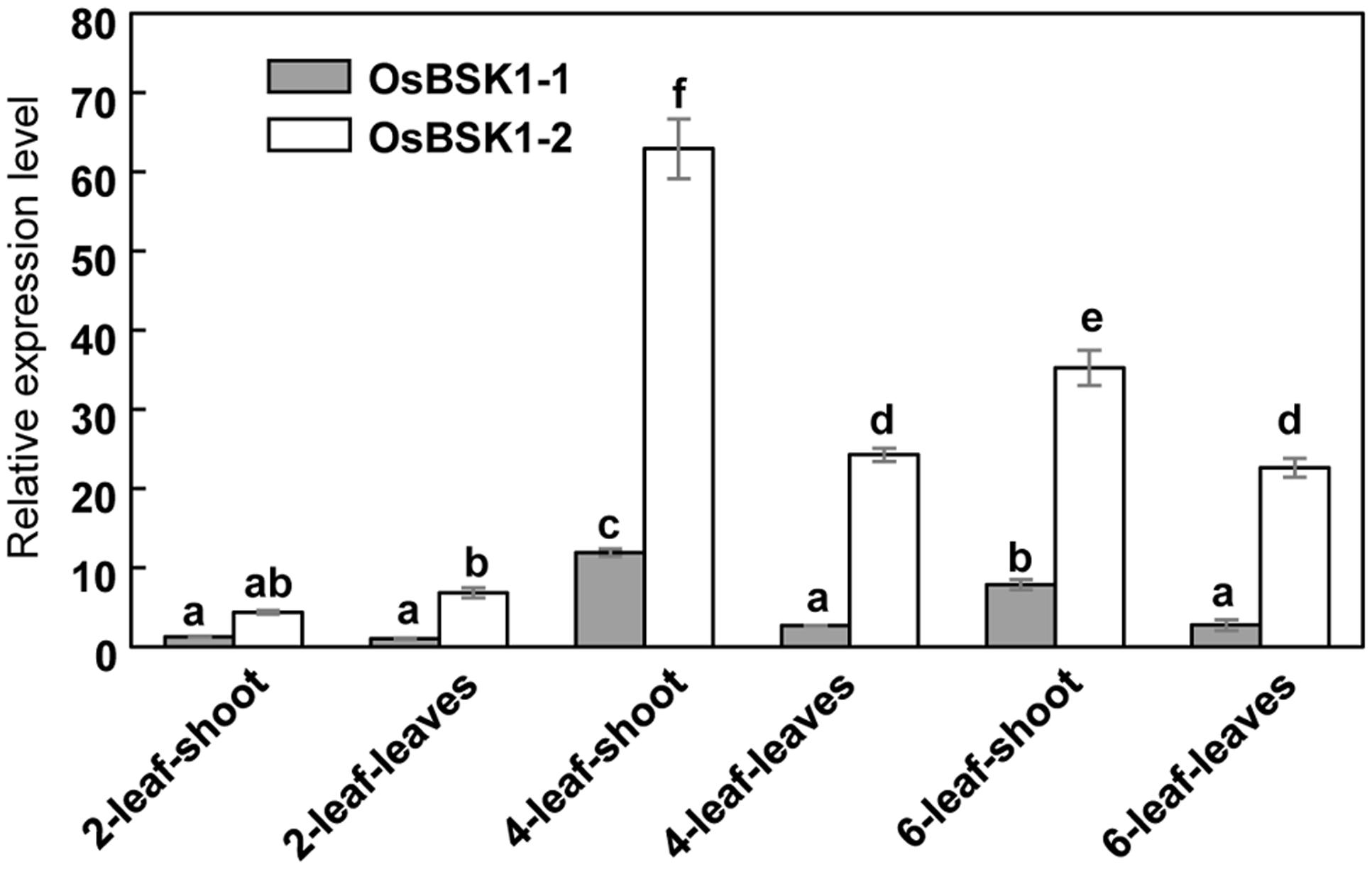
FIGURE 2. Determination of the expression patterns of OsBSK1-1 and OsBSK1-2. qRT-PCR analysis was performed on cDNA synthesized from RNA samples. The RNA samples were extracted from tissues of rice cultivar Kitaake at the development stages as indicated. The expressions of the OsBSK1-1 and OsBSK1-2 were normalized to level of the ubiquitin reference gene. Error bars indicate SD obtained from three technical replicates. The letters indicate significant differences as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis.
Silencing OsBSK1-2 Inhibits Flagellin- and Chitin-Triggered Immune Responses in Rice
BSK1 associates with FLS2 to regulate plant PTI in Arabidopsis (Shi et al., 2013). To test whether OsBSK1-1 and OsBSK1-2 regulate PTI in rice, we collected leaf strips that were 2–3 cm in length from fully developed leaves of 4-week-old Kitaake seedlings and treated them with flg22 peptide or chitin. Then, we detected the transcription levels of OsBSK1-1 and OsBSK1-2, and found that OsBSK1-2 was induced after the flg22 and chitin treatments, while OsBSK1-1 was not (Figure 3). We then chose OsBSK1-2 for further studies.
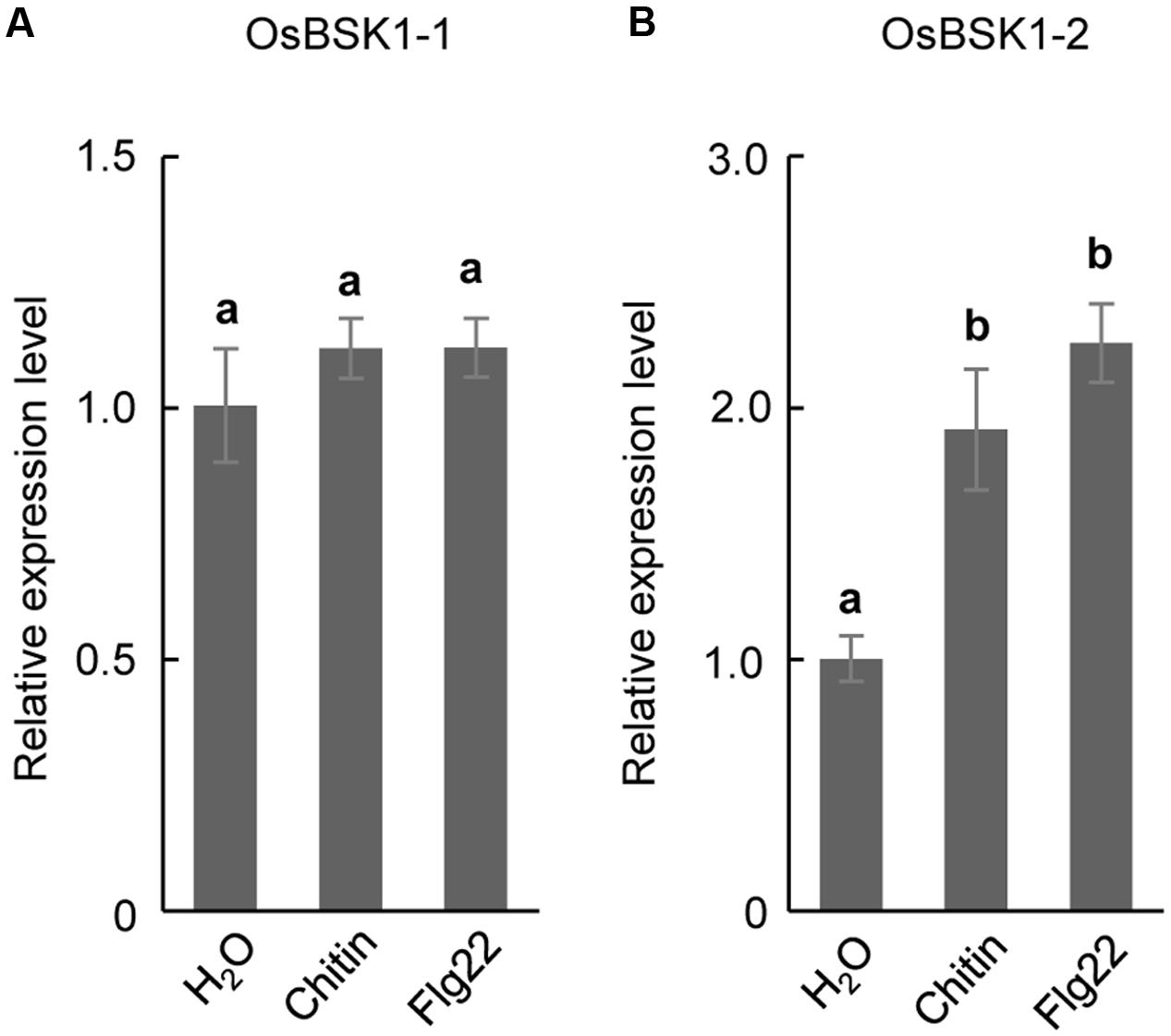
FIGURE 3. The expression of OsBSK1-2 is induced by PAMPs. The expression analyses on OsBSK1-2 (A) and OsBSK1-1 (B), respectively, in rice leaves underflg22- and chitin- treatments. The Results from two independent biological experiments were combined with statistical analysis. The letters indicate significant differences determined by a one-way ANOVA followed by post hoc Tukey HSD analysis.
To analyze whether OsBSK1-2 functions in rice immunity, we generated OsBSK1-2 RNAi transgenic rice lines. Four independent transgenic lines (OsBSK1-2Ri) with reduced expression levels of OsBSK1-2 were obtained (Supplementary Figure S1).
The genes Os04g10010 and OsPR10b function as markers involved in the downstream responses associated with PTI (Park et al., 2012; Chen et al., 2014). We thus examined the transcription levels of these two genes in OsBSK1-2Ri transgenic plants after the flg22 or chitin treatments. In Kitaake plants, the expressions of Os04g10010 and OsPR10b were induced by 17.0 ± 0.9- and 10.3 ± 0.6-fold, respectively, after the chitin treatment (Figure 4). However, the expression levels of Os04g10010 and OsPR10b were induced only about 12.7 ± 0.7- and 5.1 ± 0.8-fold in OsBSK1-2Ri-1 and 10.6 ± 0.7- and 4.2 ± 0.5-fold in OsBSK1-2Ri-2 plants, respectively. Similarly, the expression levels of Os04g10010 and OsPR10b were induced only about 25.6 ± 0.1- and 8.2 ± 0.1-fold in OsBSK1-2Ri-1 and 26.1 ± 2.2- and 8.6 ± 0.8-fold in OsBSK1-2Ri-2 plants, respectively, whereas induced about 41.0 ± 2.1- and 12.4 ± 0.2-fold in the wild type Kitaake after the flg22 treatment (Figure 4). Taken together, these results suggest that the silencing of OsBSK1-2 compromises the base immune response in rice. Thus, OsBSK1-2 might be involved in the regulation of rice PTI.
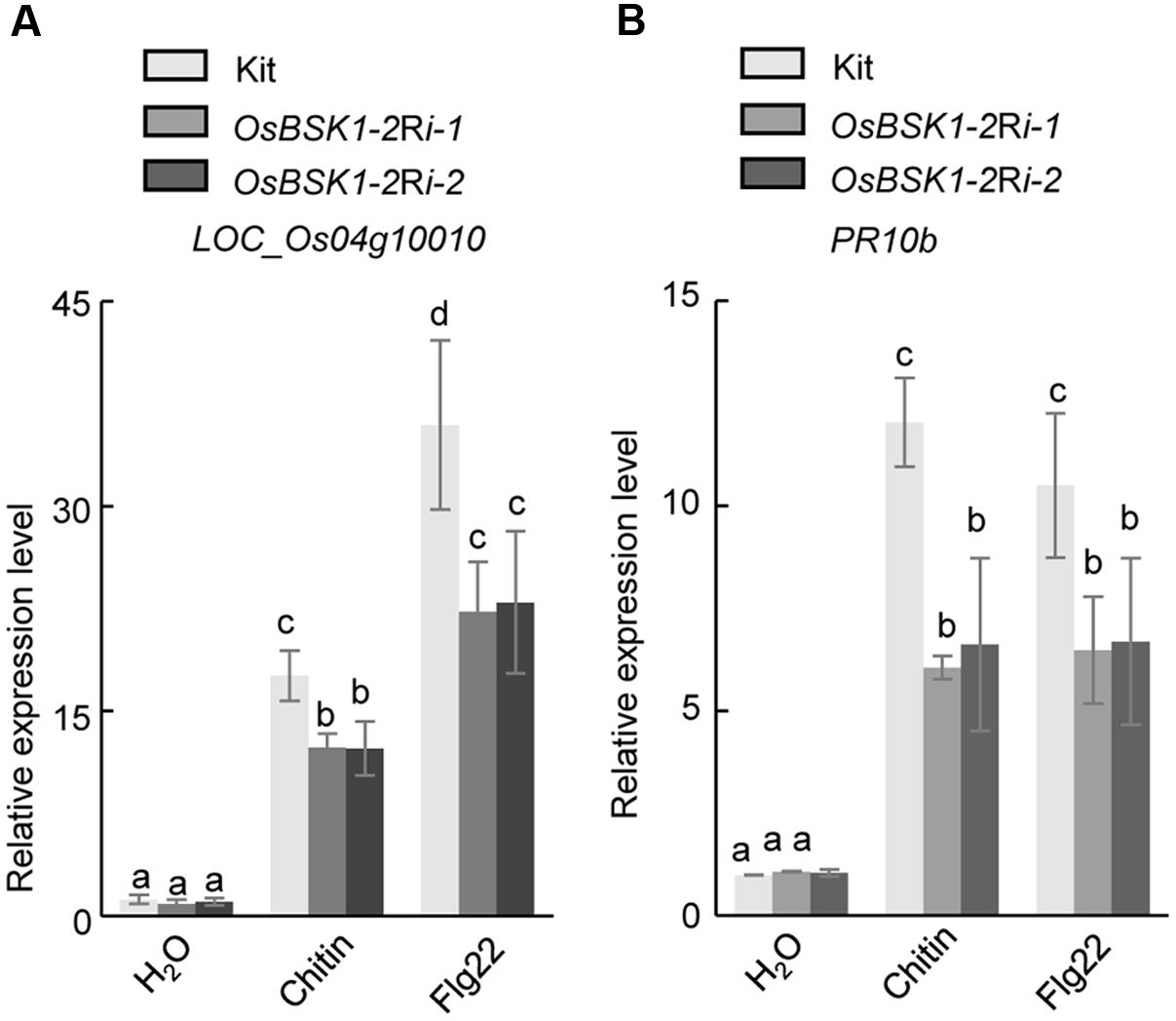
FIGURE 4. Effects of silencing OsBSK1-2 on flg22- and chitin-triggered immune responses. Leaf strips of 4-week-old plants were treated with 1 μM flg22 peptide (Pacific immunology) or 20 ug/ml chitin (Sigma) for 12 h. The mock treatment was ddH2O. The expression levels of two defense marker genes -Os04g10010 (A) and -OsPR10b (B) were measured by qRT-PCR. Gene expression levels for the target gene were normalized to the ubiquitin gene. Data shown were normalized to the Kitaake mock-treated (12 h) samples (100%). The Results from two independent biological experiments were combined with statistical analysis. The letters indicate significant differences (one-way ANOVA followed by post hoc Tukey HSD analysis).
Silencing of OsBSK1-2 Compromises Plant Resistance to M. oryzae
To further investigate the genetic role of OsBSK1-2 in rice immunity, we inoculated the OsBSK1-2Ri transgenic and wild type plants with the M. oryzae isolate ZB25. Ten days post-inoculation, the lesion size of OsBSK1-2Ri-1 (12.6 ± 1.7 cM for lesion length and 2.2 ± 0.1 cM for lesion width) and OsBSK1-2Ri-2 (12.2 ± 1.0 cM and 2.3 ± 0.2 cM) plants displayed larger lesions than the Kitaake plants (5.1 ± 0.9 cM and 1.5 ± 0.2 cM) (Figures 5A–C). To determine the fungal biomass in the inoculated leaves precisely, we isolated the total DNA from the infected leaves and quantified it using a DNA-based quantitative PCR method (Park et al., 2012). Relative to the reference rice Ubiquitin gene, the DNA accumulation level of fungal gene Pto2 in OsBSK1-2Ri-1/-2 plants was higher than in Kitaake plants (Figure 5D). This result revealed that the fungal biomass was more accumulated in the infected OsBSK1-2Ri-1/-2 than in the wild type Kittake leaves. We then inoculated OsBSK1-2Ri and wild type plants with another compatible M. oryzae isolate, ZHONG1, and obtained similar results (Supplementary Figure S2).
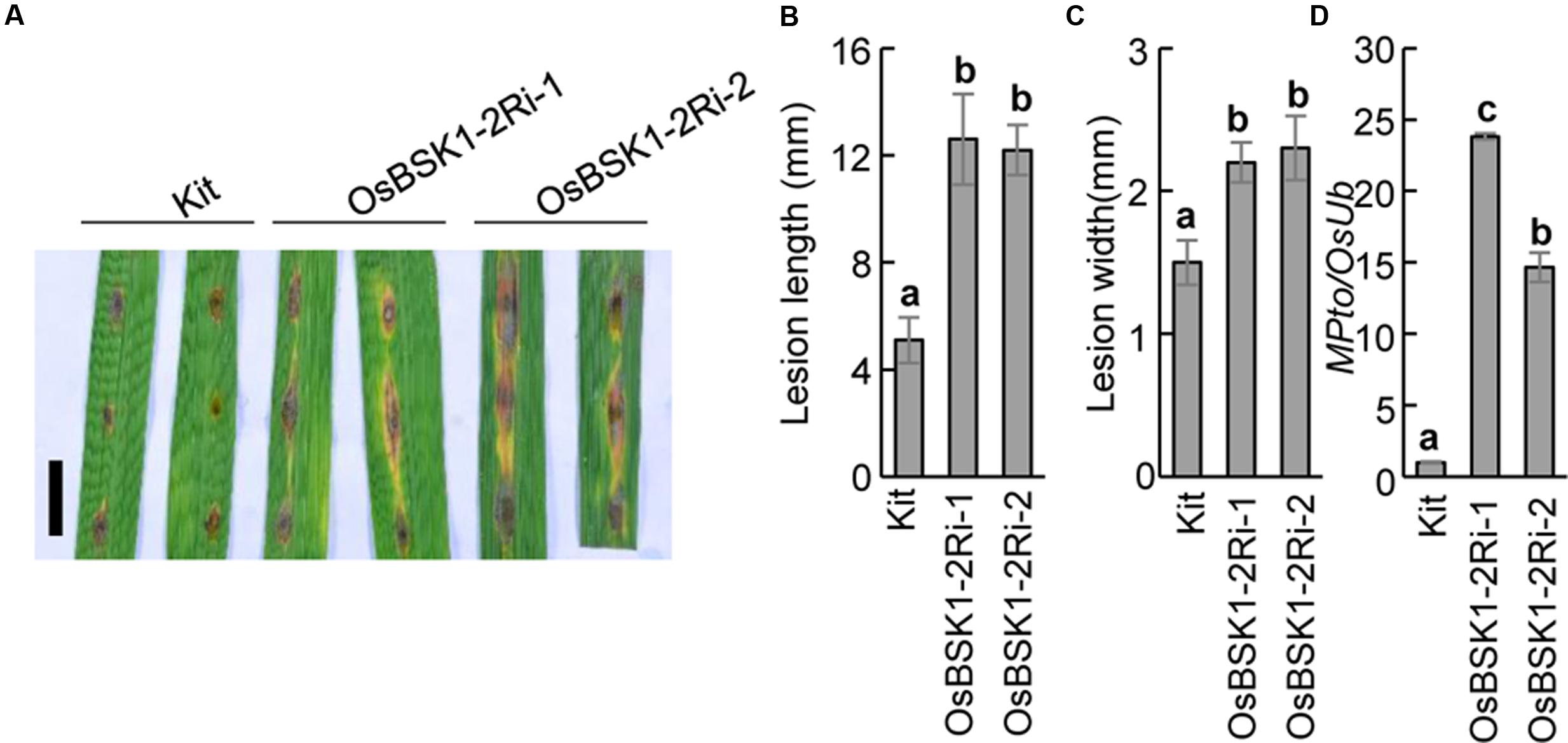
FIGURE 5. Silencing of OsBSK1-2 compromises plant resistance to Magnaporthe oryzae. The leaf strips from 8-week-old plants were inoculated the suspension of M. oryzae isolate ZB25 after 12 h post lightly wounding. (A) Photographs of rice leaves 10 days after M. oryzae inoculated. Bars = 10 mm. The length (B) and width (C) of lesion size were measured 10 days after M. oryzae inoculated (n = 10). (D) The relative fungal growth was determined on the inoculated leaves. Gene expression levels for the target gene were normalized to the ubiquitin gene (n = 3). The letters indicate significant differences as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis. Three independent biological repeats were performed and similar results were obtained.
To further confirm the results, we inoculated segregants derived from the OsBSK1-2Ri transgenic plants with the M. oryzae isolate ZB25 and determined the resistance. The segregants carrying the silencing constructs showed larger lesions than wild type Kittake, while segregants lacking the silencing constructs had a similar lesion size as the wild type (Supplementary Figure S3). Therefore, we concluded that silencing OsBSK1-2 resulted in compromised rice resistance to M. oryzae.
Silencing of OsBSK1-2 Does Not Alter Plant Architecture or Sensitivity to BR
To explore the biological role of OsBSK1-2 in plant development, we analyzed the plant architecture of OsBSK1-2Ri plants, including plant height or lamina joint. There were no obvious differences in plant height or lamina joint between OsBSK1-2Ri and Kittake plants (Figure 6 and Supplementary Figures S4A,B).

FIGURE 6. Plant architecture of Kitaake plants with reduced expressions of OsBSK1-2. (A) The gross morphological phenotypes of Kitaake and transgenic plants silenced for OsBSK1-2. Bars = 10 cm. (B) Photograph of a representative leaf of Kitaake and OsBSK1-2Ri plants. Bars = 5 cm. (C) Statistical analyses on the lamina joint angles of Kitaake and the plants silenced for OsBSK1-2. The averages and SDs were calculated from 10 plants of the representative transgenic OsBSKRi lines as indicated (n = 10). The letters indicate significant differences using a one-way ANOVA followed by post hoc Tukey HSD analysis.
To determine whether OsBSK1-2 regulated plant responses to the hormone BR, we treated the lamina joint of the wild type and OsBSK1-2Ri plants with BR. The lamina joint of OsBSK1-2Ri plants was seriously enlarged after the BL treatment, which was similar to the wild type (Figure 7 and Supplementary Figure S4C). These results suggest that silencing OsBSK1-2 does not alter plant architecture or responses to BRs.
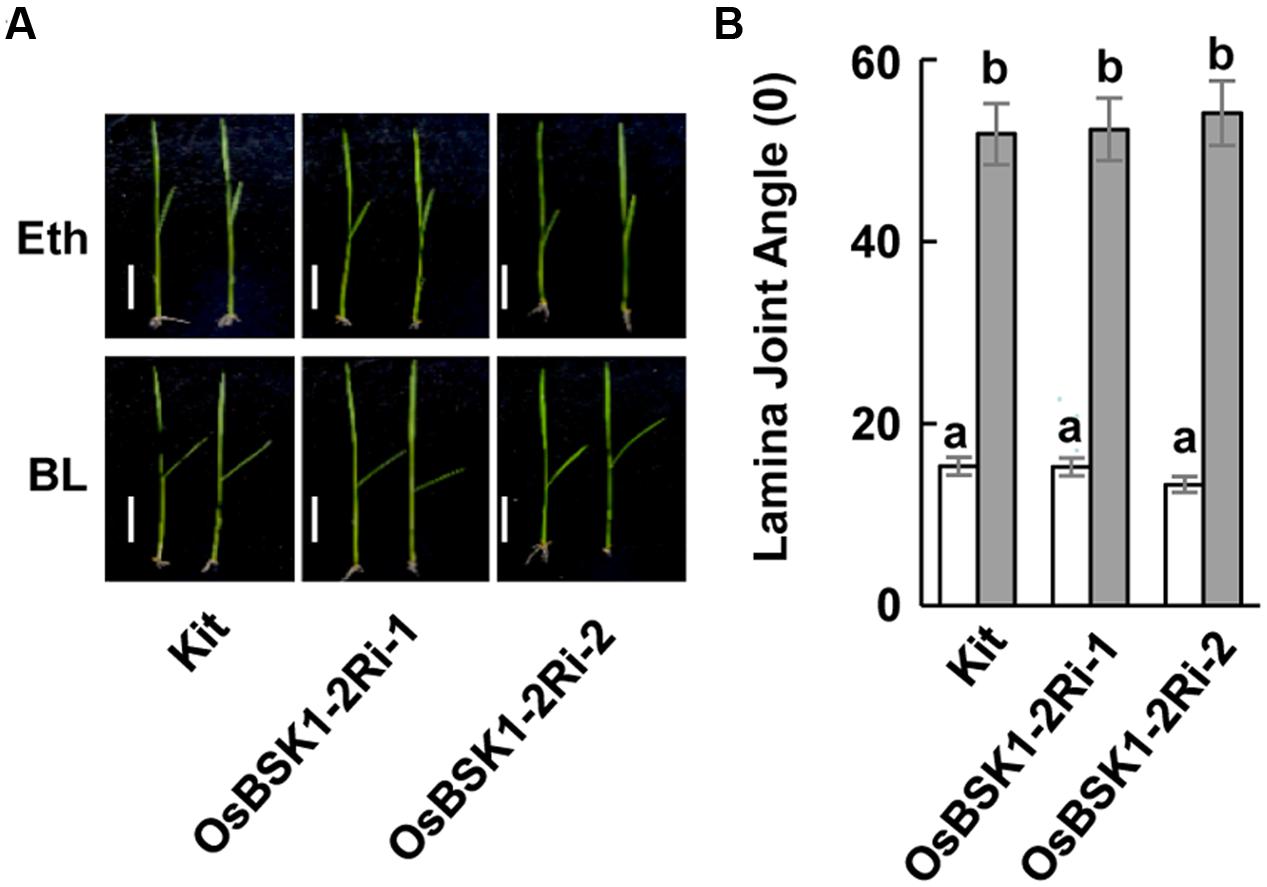
FIGURE 7. Effects of silencing the OsBSK1-2 on plants in response to BL hormone. (A) Photographs of representative seedlings showing the leaf angles for the Kitaake (Kit) plants and OsBSK1-2Ri after the mock-treatments (Upper panel) or BL treatment (Lower panel). The BL treatment contained 2,4-epiBL (100 ng), whereas Eth alone was used for the mock treatment. (B) Statistical analysis of the leaf angles of Kitaake and OsBSK1-2Ri after BL and mock-treatments, respectively. Each bar represents the average and SD of 10 leaves. The letters indicate significant differences as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis. Two independent repeats were performed with similar results produced.
Discussion
In Arabidopsis, a total of 12 BSKs have been isolated, and play various function in plant development and immunity (Tang et al., 2008; Sreeramulu et al., 2013). Although five OsBSKs identified in rice share a conserved kinase domain and a C-terminal tetratricopeptide repeat domain, their genetic functions in plant immunity and development might be different.
OsBSK1-1 and OsBSK1-2 are two homologs that also share similar expression patterns in rice developmental stages. However, the transcripts of OsBSK1-2 are more abundant than those of OsBSK1-1 in rice (Figure 2), suggesting that OsBSK1-2 might play more important roles in rice. In agreement, the expression level of OsBSK1-2, but not OsBSK1-1, is obviously induced after flg22 and chitin treatments (Figure 4). Therefore, OsBSK1-1 and OsBSK1-2 might play different roles in plant immunity. OsBSK1-2 is an ortholog of AtBSK1, which also regulate FLS2-mediated immunity as AtBSK1 (Shi et al., 2013). Although, no direct interaction between OsBSK1-2 and OsFLS2 was determined, OsBSK1-2 might still play a similar molecular mechanism as does AtBSK1 in plant immunity because silencing of OsBSK1-2 also compromised the base immune response in rice (Figure 4).
Brassinosteroid-SIGNALING KINASE are important regulators of BR signaling (Tang et al., 2008; Sreeramulu et al., 2013). However, we did not observe the BR-related phenotypes in rice plants in silence of OsBSK1-2. BSK1 is phosphorylated by BRI1 upon BR perception and disassociates from BRI1 to activate BRI1 SUPPRESSOR1 in Arabidopsis (Tang et al., 2008; Kim et al., 2009). BRI1 interacts with and phosphorylates the BSKs, BSK1, BSK3, BSK5, BSK6, BSK8 and BSK11, to activate downstream signaling (Sreeramulu et al., 2013). Nevertheless, knocking out any individual AtBSK in Arabidopsis does not affect plant architecture because of their redundant functions (Sreeramulu et al., 2013). LOC_Os04g58750 (OsBSK3) interacts with OsBRI1 in vivo, and the over-expression of OsBSK3 increases the hypersensitivity of rice plants to the hormone BR (Zhang et al., 2016). Although the reduced expression of OsBSK1-2 does not alter the BR-triggered response in rice plants (Figure 4), it does not rule out the possibility of its involvement in the regulation of BR-mediated signaling because of the possible redundancies of the other four OsBSKs. Thus, it needs to generate double or multiple knock-out mutants for the OsBSKs in rice to determine whether OsBSK1-2 regulates BR-mediated signaling and plant development for the future studies.
Author Contributions
JW and XC conceived this study. HS, LZ, CP, Dingyou Liu, XZ, WW, JY, HQ, WM, JCW performed the experiments. JW, MH, WL, Sigui Li, and XC analyzed data. JW and XC wrote the manuscript. All authors approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
XC was funded by NSFC (Grant Numbers: 31371705 and 31171622), Transgenic Projects from the Chinese Ministry of Agriculture (2014ZX0800903B), the Program for New Century Excellent Talents in University from the Ministry of Education in China, the “Hundred Talents Plan” Foundation, the Youth foundation (Grant Number: 13QNJJ0076), and the Youth Innovation Team Foundation (Grant Number: 14CXTD0039 from Sichuan, China); JW was funded by NSFC (Grant Number: 31401351); MH was funded by the Specialized Research Fund for doctoral program of higher education (Grant Number: 20135103120004 to JW); NSFC (Grant Number: 31301626) and International Cooperation and Exchange Program (Grant Number: 2014HH0066).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00908/full#supplementary-material
FIGURE S1 | Characterization of the transgenic plants silenced for OsBSK1-2. The relative expression levels of OsBSK1-2 (A) and OsBSK1-1 (B), were determined in the transgenic lines and the wild type Kitaake plants by qRT-PCR. All data were normalized to the reference ubiqutin gene. The average and SD for the relative expression of each line is shown. Data were obtained from three technical replicates. The letters indicate significant differences as determined by a one-way ANOVA followed by post hoc Tukey HSD analysis. Three independent biological experiments were performed and similar results were obtained.
FIGURE S2 | Silencing of OsBSK1-2 compromises plant resistance to Magnaporthe oryzae. The leaf strips from 8-week-old plants were inoculated with the suspension of M. oryzae isolate ZHONG1. (A) Photographs of rice leaves 10 days after M. oryzae inoculation. Bars = 10 mm. The length (B) and width (C) of lesion size are measured 10 days after M. oryzae inoculated (n = 10). The letters indicate significant differences, one-way ANOVA followed by post hoc Tukey HSD analysis.
FIGURE S3 | Co-segregation analysis on reduced resistance with the OsBSK1-2Ri transgene in plants. Segregants from the T1 progeny of transgenic OsBSK1-2Ri-1 line was analyzed for disease resistance post the inoculation with M. oryzae isolates, ZB25 (A) and ZHONG1 (B), respectively.
FIGURE S4 | Silencing of OsBSK1-2 does not effect plant response to BRs obviously. (A) The gross morphological phenotypes of Kitaake and transgenic plants silenced for OsBSK1-2. Bars = 10 cm. (B) Photograph of a representative leaf of Kitaake (Kit) and OsBSK1-2Ri plants. Bars = 5 cm. (C) Photographs of representative seedlings showing the leaf angles for the Kitaakeand OsBSK1-2Ri plants after the mock-treatments (Upper panel) or BL treatment (Lower panel). Eth solution containing 2,4-epiBL (100 ng) was used for the BL treatment, whereas Eth alone was used for the mock treatment.
Footnotes
References
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Chen, L., Xiong, G., Cui, X., Yan, M., Xu, T., Qian, Q., et al. (2013). OsGRAS19 may be a novel component involved in the brassinosteroid signaling pathway in rice. Mol. Plant 6, 988–991. doi: 10.1093/mp/sst027
Chen, X., Zuo, S., Schwessinger, B., Chern, M., Canlas, P. E., Ruan, D., et al. (2014). An XA21-associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors. Mol. Plant 7, 874–892. doi: 10.1093/mp/ssu003
Chern, M. S., Canlas, P. E., Fitzgerald, H., and Ronald, P. C. (2005). Rice NRR, a negative regulator of disease resistance in rice that interacts with Arabidopsis NPR1 and Rice NH1. Plant J. 43, 623–635. doi: 10.1111/j.1365-313X.2005.02485.x
Eckardt, N. A. (2011). BIK1 function in plant growth and defense signaling. Plant Cell 23:2806. doi: 10.1105/tpc.111.230811
Felix, G., Duran, J. D., Volko, S., and Boller, T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276. doi: 10.1046/j.1365-313X.1999.00265.x
Kim, T. W., Guan, S., Sun, Y., Deng, Z., Tang, W., Shang, J. X., et al. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11, 1254–1260. doi: 10.1038/ncb1970
Lin, W., Lu, D., Gao, X., Jiang, S., Ma, X., Wang, Z., et al. (2013). Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc. Natl. Acad. Sci. U.S.A. 110, 12114–12119. doi: 10.1073/pnas.1302154110
Liu, Z., Wu, Y., Yang, F., Zhang, Y., Chen, S., Xie, Q., et al. (2013). BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc. Natl. Acad. Sci. U.S.A. 110, 6205–6210. doi: 10.1073/pnas.1215543110
Miki, D., and Shimamoto, K. (2004). Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45, 490–495. doi: 10.1093/pcp/pch048
Park, C. H., Chen, S., Shirsekar, G., Zhou, B., Khang, C. H., Songkumarn, P., et al. (2012). The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24, 4748–4762. doi: 10.1105/tpc.112.105429
Shi, H., Shen, Q., Qi, Y., Yan, H., Nie, H., Chen, Y., et al. (2013). BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25, 1143–1157. doi: 10.1105/tpc.112.107904
Shinya, T., Yamaguchi, K., Desaki, Y., Yamada, K., Narisawa, T., Kobayashi, Y., et al. (2014). Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J. 79, 56–66. doi: 10.1111/tpj.12535
Shiu, S. H., and Bleecker, A. B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. U.S.A. 98, 10763–10768. doi: 10.1073/pnas.181141598
Sreeramulu, S., Mostizky, Y., Sunitha, S., Shani, E., Nahum, H., Salomon, D., et al. (2013). BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J. 74, 905–919. doi: 10.1111/tpj.12175
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tang, W., Kim, T. W., Oses-Prieto, J. A., Sun, Y., Deng, Z., Zhu, S., et al. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321, 557–560. doi: 10.1126/science.1156973
Tong, H., and Chu, C. (2012). Brassinosteroid signaling and application in rice. J. Genet. Genomics 39, 3–9. doi: 10.1016/j.jgg.2011.12.001
Veronese, P., Nakagami, H., Bluhm, B., Abuqamar, S., Chen, X., Salmeron, J., et al. (2006). The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18, 257–273. doi: 10.1105/tpc.105.035576
Vij, S., Giri, J., Dansana, P. K., Kapoor, S., and Tyagi, A. K. (2008). The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: organization, phylogenetic relationship, and expression during development and stress. Mol. Plant 1, 732–750. doi: 10.1093/mp/ssn047
Yamaguchi, K., Yamada, K., Ishikawa, K., Yoshimura, S., Hayashi, N., Uchihashi, K., et al. (2013a). A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe 13, 347–357. doi: 10.1016/j.chom.2013.02.007
Yamaguchi, K., Yamada, K., and Kawasaki, T. (2013b). Receptor-like cytoplasmic kinases are pivotal components in pattern recognition receptor-mediated signaling in plant immunity. Plant Signal. Behav. 8:e25662. doi: 10.4161/psb.25662
Zhang, B., Wang, X., Zhao, Z., Wang, R., Huang, X., Zhu, Y., et al. (2016). OsBRI1 activates BR signaling by preventing binding between the TPR and kinase domains of OsBSK3 via phosphorylation. Plant Physiol. 170, 1149–1161. doi: 10.1104/pp.15.01668
Zhang, J., Li, W., Xiang, T., Liu, Z., Laluk, K., Ding, X., et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7, 290–301. doi: 10.1016/j.chom.2010.03.007
Keywords: OsBSK, PTI, rice blast, brassinosteroids, rice
Citation: Wang J, Shi H, Zhou L, Peng C, Liu D, Zhou X, Wu W, Yin J, Qin H, Ma W, He M, Li W, Wang J, Li S and Chen X (2017) OsBSK1-2, an Orthologous of AtBSK1, Is Involved in Rice Immunity. Front. Plant Sci. 8:908. doi: 10.3389/fpls.2017.00908
Received: 13 January 2017; Accepted: 15 May 2017;
Published: 21 June 2017.
Edited by:
Jeremy Astier, Helmholtz Zentrum München, GermanyReviewed by:
Benjamin Schwessinger, Australian National University, AustraliaEls J. M. Van Damme, Ghent University, Belgium
Copyright © 2017 Wang, Shi, Zhou, Peng, Liu, Zhou, Wu, Yin, Qin, Ma, He, Li, Wang, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuewei Chen, xwchen88@163.com
†These authors have contributed equally to this work.
 Jing Wang
Jing Wang Hui Shi
Hui Shi Lian Zhou1†
Lian Zhou1† Xiaogang Zhou
Xiaogang Zhou