- 1The State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China
- 2College of Forestry, Henan Agricultural University, Zhengzhou, China
- 3College of Life Sciences, Hunan Agricultural University, Changsha, China
- 4Department of Botany, Shah Abdul Latif University, Khairpur, Pakistan
- 5The Collaborative Center for Grain Crops in Henan, College of Agronomy, Henan Agricultural University, Zhengzhou, China
Wheat (Triticum aestivum L.) is a major crop worldwide. The utilization of heterosis is a promising approach to improve the yield and quality of wheat. Although there have been many studies on wheat cytoplasmic male sterility, its mechanism remains unclear. In this study, we identified two MADS-box genes from a wheat K-type cytoplasmic male sterile (CMS) line using homology-based cloning. These genes were localized on wheat chromosomes 3A and 3B and named TaAG-A and TaAG-B, respectively. Analysis of TaAG-A and TaAG-B expression patterns in leaves, spikes, roots, and stems of Chinese Spring wheat determined using quantitative RT-PCR revealed different expression levels in different tissues. TaAG-A had relatively high expression levels in leaves and spikes, but low levels in roots, while TaAG-B had relatively high expression levels in spikes and lower expression in roots, stems, and leaves. Both genes showed downregulation during the mononucleate to trinucleate stages of pollen development in the maintainer line. In contrast, upregulation of TaAG-B was observed in the CMS line. The transcript levels of the two genes were clearly higher in the CMS line compared to the maintainer line at the trinucleate stage. Overexpression of TaAG-A and TaAG-B in Arabidopsis resulted in phenotypes with earlier reproductive development, premature mortality, and abnormal buds, stamens, and stigmas. Overexpression of TaAG-A and TaAG-B gives rise to mutants with many deformities. Silencing TaAG-A and TaAG-B in a fertile wheat line using the virus-induced gene silencing (VIGS) method resulted in plants with green and yellow striped leaves, emaciated spikes, and decreased selfing seed set rates. These results demonstrate that TaAG-A and TaAG-B may play a role in male sterility in the wheat CMS line.
Introduction
Wheat (Triticum aestivum L.) is a widely cultivated and consumed food crop worldwide. High yield and quality are the ultimate objectives of grain production. The utilization of heterosis is a promising approach to improving the yield and quality of wheat. Cytoplasmic male sterility, characterized by the inability to produce viable pollen while vegetative and female development remain unaffected (Duvick, 1959), is an essential tool in the utilization of heterosis. K-type cytoplasmic male sterile (CMS) lines (male sterile lines with Aegilops kotschyi cytoplasm) are potential candidates for use in experiments because they suffer few side effects and have abundant restoring resources among common wheat cultivars (Tsunewaki et al., 1978). The mechanism of wheat male sterility remains unclear. The CMS phenotype is mainly reflected in flowers. The ABC(DE) model of flower development in higher plants proposes that class A, B, C, D, and E homeotic genes specify floral organ identity (Theißen, 2001). According to this model, class A and E genes in combination control the development of sepals; class A, B, and E genes control petal development; class B, C, and E genes control stamen development; and class C and E genes control carpel development. The wheat inflorescence (spike or ear) is composed of spikelets, whereas an individual wheat flower contains one pistil, three stamens, and two lodicules, which are modified petals. Though the floral organs of wheat differ from those of dicotyledons, the ABC(DE) model can also be applied to wheat flower development (Murai, 2013).
In the typical ABC model (Weigel and Meyerowitz, 1994) of Arabidopsis, APETALA1 (AP1, Mandel et al., 1992) and APETALA2 (AP2, Jofuku et al., 1994) are classified as A genes, APETALA3 (AP3, Jack et al., 1992) and PISTILLATA (PI, Goto and Meyerowitz, 1994) are B genes, and AGAMOUS (AG, Yanofsky et al., 1990) is the only class C gene. With the exception of AP2, the other four genes AP1, AP3, PI, and AG, are all MADS-box genes. MADS-box genes encode a eukaryotic transcription factor family that plays a fundamental role in very diverse and important biological functions, including vegetative and reproductive development, floral organ development, and signal transduction (Ng and Yanofsky, 2001; De Bodt et al., 2003; Duan et al., 2006). In the past few decades, various proteins have been identified with highly conserved DNA binding and dimerized MADS-box domains (Schwarz-Sommer et al., 1990; Coen and Meyerowitz, 1991) in yeasts, plants, insects, nematodes, lower vertebrates, and mammals. Moreover, several ABC(DE) MADS-box genes, such as WAG (Meguro et al., 2003; Hirabayashi and Murai, 2009; Wei et al., 2011; Wang et al., 2015), WAP1 (Murai et al., 2003), and TaMADS1 (Zhao et al., 2005), have been identified and characterized in wheat. The MADS-box gene family was further split into type I (SRF-like) and type II (MEF2-like) genes through phylogenetic analysis. Type II proteins, which are also called MIKC-type proteins, are mostly found in higher plants (Münster et al., 1997). Some of them are related to male sterility (Ai et al., 2016).
MIKC-type proteins play a vital role in floral organ formation (Ng and Yanofsky, 2001). Our previous microarray results showed higher expression levels of a MIKC-type MADS-box transcription factor, Ta.4147, in a wheat K-type CMS line, in contrast to the maintainer line. To understand the relationship between the MADS-box gene and male sterility in the CMS line, in this study we identified two MADS-box genes from the uninucleate pollen of the CMS line using homology-based cloning. The two genes were named TaAG-A and TaAG-B based on their localization on wheat chromosomes 3A and 3B, respectively. The objective of the present study was to investigate the functions of the TaAG-A and TaAG-B genes. We conducted expression analysis of the genes in different tissues at different stages. Overexpression in Arabidopsis and silencing in a wheat fertile line using BSMV-VIGS were also performed. The results suggest that TaAG-A and TaAG-B may play central roles in male sterility of the wheat CMS line.
Materials and Methods
Plant Materials
A wheat K-type CMS line (Yumai 3, KA), a maintainer line (Yumai 3, KB), a restorer (Yumai 2) line, and F1 lines were grown in the experimental field of the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences (Beijing, China) until the heading stage. Anthers at the mononucleate, dinucleate, and trinucleate stages were sampled, quickly frozen in liquid nitrogen, and stored at -80°C until RNA isolation.
Wheat variety Chinese Spring and 21 Chinese Spring nullisomic-tetrasomic lines were grown in a greenhouse at 22°C under a 16-h light/8-h dark regime. Young leaves were collected for DNA extraction. Roots, stems, flag leaves, and young spikes at the heading stage were sampled for RNA isolation.
Arabidopsis wild-type Columbia (Col-0) and transgenic pEarleyGate 101 plants (pEarleyGate 101-TaAG-A and pEarleyGate 101-TaAG-B) were grown in a greenhouse at 22°C under a 16-h light/8-h dark regime. Leaves were collected for RNA isolation.
Normal wheat and wheat inoculated with RNAγb:TaAG-A or RNAγb:TaAG-B were grown in a greenhouse at 25/15°C under a 16-h light/8-h dark regime. Spikes were collected for RNA isolation.
Genomic DNA and RNA Extraction
Genomic DNA was extracted using the cetyltrimethylammonium bromide (CTAB) method (Saghai-Maroof et al., 1984). Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) and treated with RNase-free DNaseI (Promega, Madison, WI, United States) as described by Huang et al. (2012). First strand cDNA was synthesized using the FastQuant RT Kit (with gDNase) (TIANGEN, Beijing) following the manufacturer’s instructions.
Cloning of Wheat MADS-box Genes
Ta.4147 was used to conduct a BLAST search of the Triticum urartu genome database1. Then two pairs of gene specific primers, TaMADS1.CF/CR and TaMADS2.CF/CR (Supplementary Table S1), were designed and used to clone MADS-box genes from the cDNA of anthers from the male sterile line at the mononucleate stage. The following PCR reaction conditions were used: 5 min at 94°C; followed by 35 cycles of 30 s at 94°C, 30 s at 58°C, and 1 min at 72°C; with a final extension at 72°C for 10 min using LA-Taq DNA polymerase (Takara, Biotechnology Co., Ltd., Dalian, China). The PCR products were individually cloned into pGEM-T easy vectors (Promega, Madison, WI, United States) and introduced into Escherichia coli. Ten positive independent clones were commercially sequenced. The GenBank/EMBL accession numbers are as follows: KX354939–KX354940. The sequences were analyzed using DNAMAN software and the Conserved Domain Search in NCBI2. The phylogenetic trees were drawn using Clustal W and MEGA software.
Chromosomal Localization Analysis
Nulli-tetrasomic lines of Chinese Spring were employed to determine the chromosomal locations of the TaMADS1 and TaMADS2 genes with the gene-specific primers TaMADS1.LEF/LER and TaMADS2.LEF/LER (Supplementary Table S1). The PCR products were resolved by electrophoresis on a 1% agarose gel containing ethidium bromide and visualized under a UV spectrometer.
Quantitative Real-time PCR Analysis
Quantitative real-time PCR (qRT-PCR) was performed to study relative gene expression levels using the LightCycler 480 system (Roche, Indianapolis, IN, United States) with SYBR Green I Master (Roche) according to the manufacturer’s protocol. Two pairs of gene-specific primers, TaMADS1.LEF/LER and TaMADS2.LEF/LER (Supplementary Table S1), were used for gene expression analysis with the Ta4045 gene, which encodes ubiquinol-cytochrome C reductase iron-sulfur subunit as described by Paolacci et al. (2009), as a control. The expression profiles of TaAG-A and TaAG-B in various wheat organs (roots, stems, flag leaves, and young spikes) and at different stages (mononucleate, dinucleate, and trinucleate) were investigated. The qRT-PCR experiment was performed independently with three biological replicates and three technical replicates according to the method of Li et al. (2013).
Transformation Assays
The entire coding sequences of the TaAG-A and TaAG-B genes were individually cloned into the pEarleyGate101 vector using the Gateway-compatible vector cloning system (Invitrogen, Carlsbad, CA, United States) to produce the plasmid constructs pEarleyGate 101- TaAG-A and pEarleyGate 101- TaAG-B. The constructs were used to transform Arabidopsis wild-type Col-0 by the Agrobacterium- mediated floral-dip method (Clough and Bent, 1998). Transgenic lines were selected on MS (Murashige and Skoog, 1962) plates with phosphinothricin (5 μg/L), and more than 20 transgenic lines were obtained. Transgenic plants were selected for phenotypic analysis and semi-quantitative RT-PCR with Actin as a control. The primers used for plasmid construction (TaAG-AB.101F/101R), transformant identification (101.F/R), and semi-quantitative RT-PCR (TaAG-AB.SF/SR, Actin.F/R) are listed in Supplementary Table S1.
Virus-Induced Gene Silencing with the Barley Stripe Mosaic Virus
The NheI restriction site was introduced into TaAG-A and TaAG-B using the primers TaAG-A-NheI-F/R and TaAG-B-NheI-F/R (Supplementary Table S1). The amplified fragments were used to replace the GFP coding sequence in RNAγb:GFP, resulting in RNAγb:TaAG-A and RNAγb:TaAG-B. The preparation of BSMV transcripts for wheat inoculation was consistent with the method of Zhou et al. (2007). The fourth leaf and spikes of inoculated plants were observed at 12, 16, and 20 days after inoculation. The relative transcript levels of the TaAG-A and TaAG-B genes in the spikes were assessed by semi-quantitative RT-PCR, which were performed independently with three biological replicates and three technical replicates.
Results
Cloning and Sequence Analysis of Two MADS-box Genes from a Wheat K-type CMS Line
Our previous microarray results showed that the expression level of the MIKC-type MADS-box transcription factor Ta.4147 in the CMS line was higher than in the maintainer line. Therefore, the Ta.4147 sequence was used to conduct a BLAST search of the T. urartu genome database and we obtained a very similar sequence, TRIUR3_34584 (GenBank accession number KD145636). We designed gene-specific primers (Supplementary Table S1) and performed RT-PCR analysis with the purified RNA extracted from uninucleate pollen of the wheat K-type CMS line. The PCR products were separated on a 1% agarose gel and purified with the Agarose Gel DNA Purification kit (TIAGEN, Beijing) according to the manufacturer’s instructions. The purified fragment was then cloned into the pGEM T easy vector (Promega) and transformed into E. coli TOP10. Positive clones were selected for sequencing. The results of sequence analysis (Supplementary Table S2) revealed two types of sequences derived from positive clones. One sequence had an open reading frame (ORF) of 819 bp encoding a peptide of 272 amino acid residues with a molecular weight of 30.8 kDa and a pI of 9.072. The other type had an ORF of 831 bp encoding a peptide of 276 amino acid residues with a molecular weight of 31.4 kDa and a pI of 8.794. The predicted proteins (Figure 1) have conserved MADS-MEF2-like and K-box domains, which are typical characteristics of MIKC-type MADS-box proteins, there are ten amino acids different between these two MADS-box proteins

FIGURE 1. Multiple alignment of the TaMADS1 and TaMADS2 amino acid sequences. Wheat TaWM29A (CAM59076) and TaAGL39 (ABF57939), barley HvWM29B (ACB45306), Brachypodium distachyon BdMADS (XP_003565181), and rice OsMADS3 (ACY26070). Underlines indicate the conserved MADS-MEF2-like region and the K-box motif.
Chromosomal Localization of TaMADS1 and TaMADS2
To assign the two MADS-box genes to the wheat genome, we used Chinese Spring nulli-tetrasomic lines for chromosomal localization. Specific primers for TaMADS1 and TaMADS2 were designed (Supplementary Table S1) and PCR was performed using DNA from the nulli-tetrasomic lines as templates. The results (Supplementary Figure S1) showed that N3A/T3B had no TaMADS1 band and the N3B/T3D lane had no TaMADS2 band, indicating that TaMADS1 and TaMADS2 were localized on wheat chromosomes 3A and 3B, respectively, thus these were designated as TaAG-A and TaAG-B.
Phylogenetic Analysis of TaAG-A and TaAG-B
We performed phylogenetic analysis using amino acid sequences derived from rice, Arabidopsis, and wheat MADS-box genes. The results (Figure 2A) showed that TaAG-A and TaAG-B belong to the AG subfamily C group of MADS-box family proteins and have very high identity to MIKC-type MADS-box transcription factors WM29B (CAM59077) and TaAGL39 (ABF57939) from wheat and 93% identity with OsMADS3 (ACY26070) from rice.
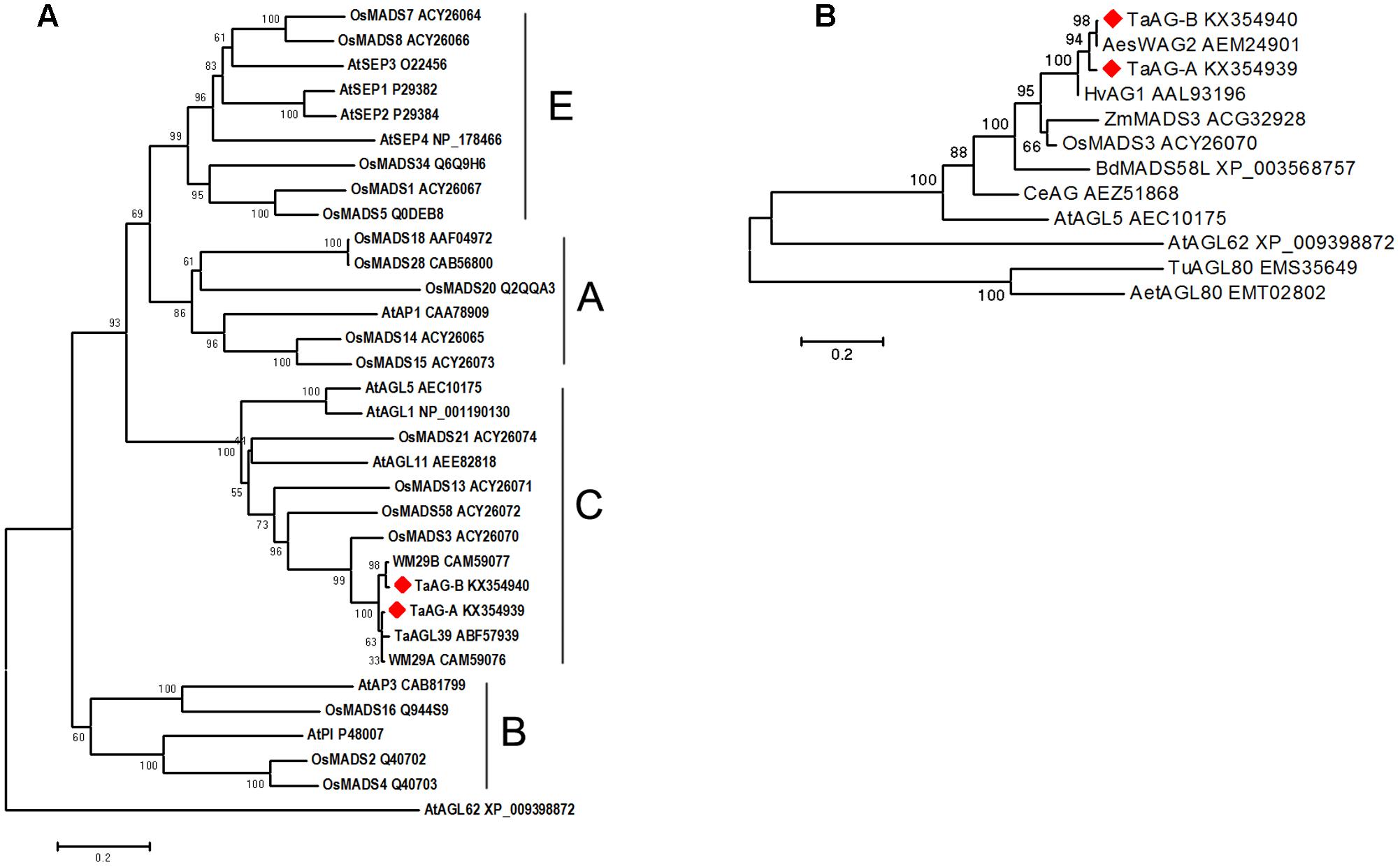
FIGURE 2. (A) Evolutionary relationships of MADS-box amino acid sequences in rice, Arabidopsis, and wheat. (B) Phylogenetic analysis of MADS-box proteins in different plants. Evolutionary analyses were conducted using MEGA 6 software with the neighbor-joining method [bootstrap test (1000 replicates) and Poisson correction method were used]. The phylogenetic tree was constructed using the MADS-box amino acid sequences from barley (Hv), maize (Zm), rice (Os), B. distachyon (Bd), Arabidopsis thaliana (At), Triticum urartu (Tu), Aegilops speltoides (Aes), Aegilops tauschii (Aet), and Cymbidium eburneum (Ce).
Further phylogenetic analysis of AG subfamily proteins (Figure 2B) from wheat, barley, rice, maize, Brachypodium distachyon, T. urartu, and Aegilops tauschii showed that TaAG-A and TaAG-B are closely related to AesWAG-2 (AEM24901) from Aegilops tauschii and HvAG1 (AAL93196) from barley.
Expression Analysis of TaAG-A and TaAG-B
To assess the expression patterns of TaAG-A and TaAG-B in wheat, we performed qRT-PCR analysis using RNA extracted from roots, stems, leaves, and young spikes of Chinese Spring wheat. Anthers were collected from the wheat K-type CMS line at the uni-, di-, and tri-nucleate stages and the maintainer line. The results (Figure 3A) demonstrate that TaAG-A and TaAG-B had similar expression patterns in different wheat tissues, but their expression levels differed in different tissues. TaAG-A had very high expression levels in young spikes and leaves and very low expression levels in roots, while TaAG-B was most abundant in young spikes with very low expression in roots, stems, and leaves.
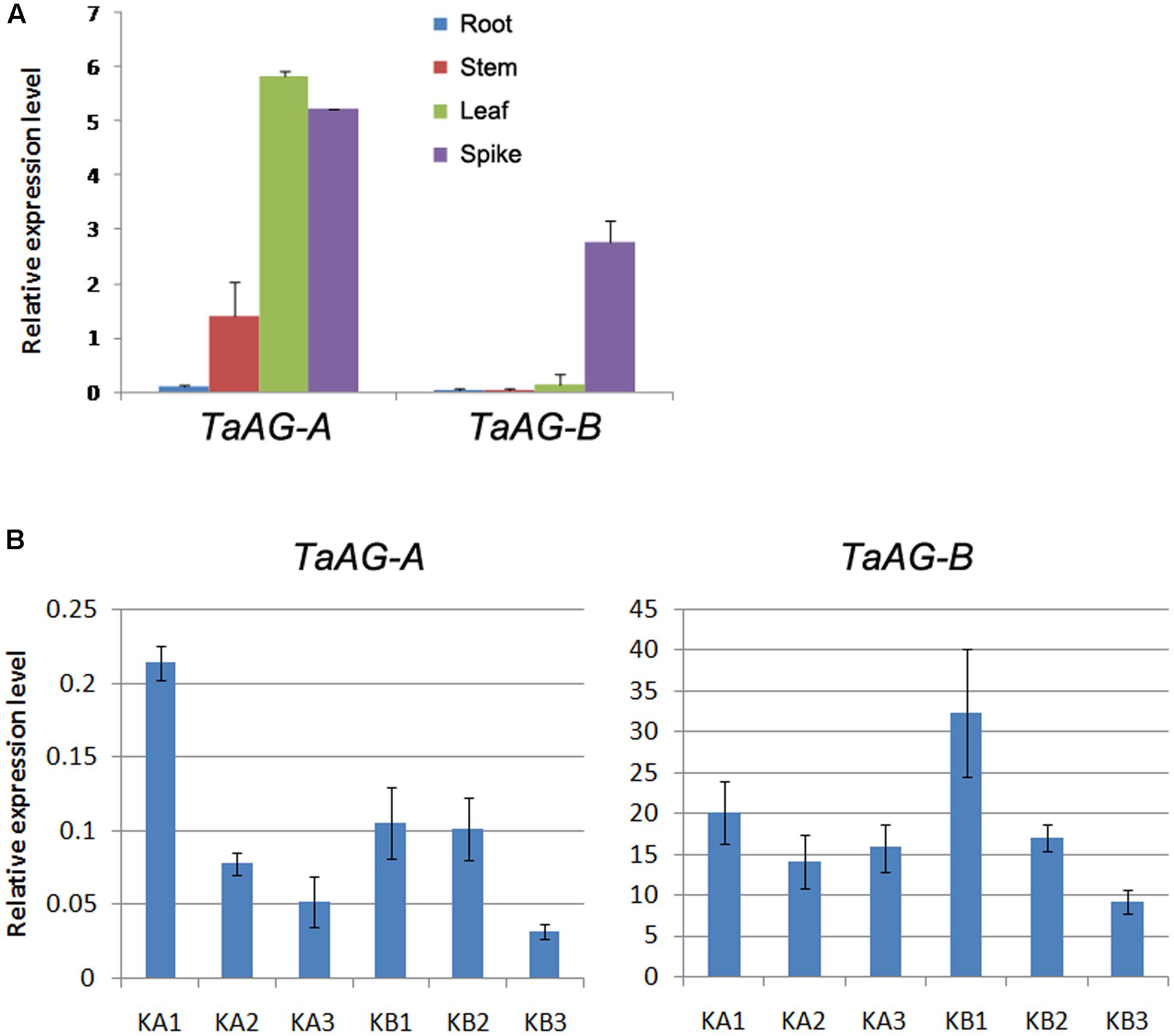
FIGURE 3. (A) Relative expression levels of TaAG-A and TaAG-B in tissues of Chinese Spring wheat. (B) Relative expression levels of TaAG-A and TaAG-B in wheat anthers at various developmental stages. KA1–3, mononucleate, dinucleate, and trinucleate stages of the wheat K-type CMS line, respectively. KB1–3, mononucleate, dinucleate, and trinucleate stages of the maintainer line, respectively.
The expression levels of TaAG-A and TaAG-B also showed similar trends at different developmental stages in anthers of the same wheat variety (Figure 3B). Both genes could be detected at all developmental stages in anthers, but TaAG-B transcripts were more abundant than those of TaAG-A. From the di-nucleate to tri-nucleate stages, the expression levels of TaAG-B were upregulated in the CMS line, but downregulated in the maintainer line. Additionally, the expression levels of TaAG-A and TaAG-B in the CMS line at the trinucleate stage were higher than in the maintainer line. These results demonstrate that TaAG-A and TaAG-B have different expression patterns between the CMS line and maintainer line at different anther developmental stages, especially at the trinucleate stage. Higher transcription of TaAG-A and TaAG-B may be closely related to male sterility in the CMS line.
Functional Analysis of TaAG-A and TaAG-B
To further investigate the functions of the TaAG-A and TaAG-B genes, we constructed overexpression plasmids (pEarleyGate 101-TaAG-A and pEarleyGate 101-TaAG-B) using the Gateway-compatible vector cloning system. Plasmid constructs were individually transformed into Arabidopsis wild-type Col-0. More than 20 transformants were obtained and T3 homozygous plants were used for phenotypic analysis (Figure 4A). We compared the vegetative growth of Col-0 and the pEarleyGate 101 transformants. Both the pEarleyGate 101-TaAG-A and pEarleyGate 101-TaAG-B transgenic plants were emaciated, dwarfed, and abnormal with inwardly curled young leaves, tiny yellow leaves, and advanced reproductive growth. At the ripening stage, pEarleyGate 101-TaAG-A and pEarleyGate 101-TaAG-B plants displayed abnormal growth and development, and exhibited delayed flowering and podding.

FIGURE 4. Phenotypes and expression analysis of TaAG-A and TaAG-B transgenic Arabidopsis. (A) Phenotypes of transgenic plants 16 days after transplantation. (B) Phenotypes of buds. (C) Expression analysis. CK, Arabidopsis wild-type Col-0 plants; 101, pEarleyGate 101 transgenic Arabidopsis plants; lanes 1–5, pEarleyGate 101-TaAG-B transgenic Arabidopsis plants (phenotype changed); lane 6, pEarleyGate 101-TaAG-B transgenic Arabidopsis plants (phenotype unchanged); lanes 7–8, pEarleyGate 101-TaAG-A transgenic Arabidopsis plants (phenotype changed and phenotype unchanged, respectively).
We observed the buds of transgenic Arabidopsis under a stereomicroscope (Figure 4B) in comparison with Col-0 buds. The pEarleyGate 101-TaAG-B plants had short and abnormal sepals, petals, and stamens; expanded pistils; and no selfing fructification.
We performed semi-quantitative RT-PCR analysis of flowers from transgenic plants compared with normal plants using Actin expression as a control (Figure 4C). TaAG-A and TaAG-B transcript levels were higher in plants with abnormal phenotypes, indicating that TaAG-A and TaAG-B expression had a great influence on pistil and stamen development of transgenic Arabidopsis. In addition, TaAG-B induced abnormal development more readily than TaAG-A in transgenic Arabidopsis.
Evidence for the Involvement of TaAG-A and TaAG-B in Wheat Male Sterility
To obtain more direct evidence of the functions of TaAG-A and TaAG-B, we performed virus-induced gene silencing (VIGS) developed with the barley stripe mosaic virus (BSMV) to decrease their transcript levels. Uninfected wheat plants, wheat plants infected by RNAγb, RNAγb:TaPDS, and RNAγb:GFP served as controls in the inoculation assays. The fourth leaf (Figure 5A) and spikes (Figure 5B) of inoculated plants were observed at 12, 16, and 20 days after inoculation. The leaves of the wheat plants infected by RNAγb had striped spots, while wheat plants inoculated with BSMV:TaPDS had white leaves, and the leaves of BSMV:GFP-inoculated plants produced yellow, diseased spots, indicating that the BSMV experiments were successful. In plants inoculated with BSMV:TaAG-A and BSMV:TaAG-B, leaf surfaces had striped diseased spots, which were more obvious at 16 and 20 days after inoculation.
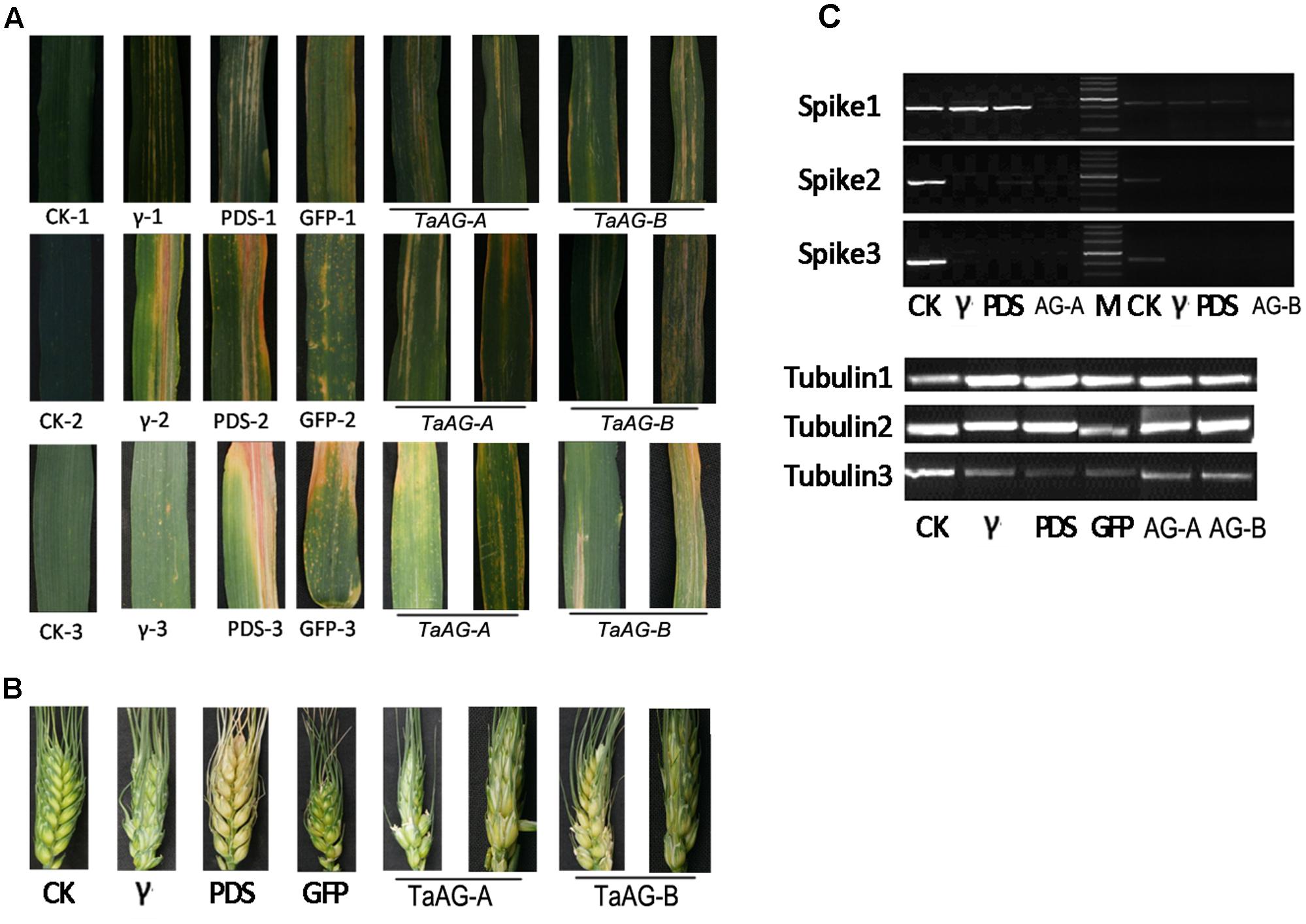
FIGURE 5. Phenotypes of virus-infected wheat leaves and spikes. (A) Leaf samples. (B) Spike samples. (C) Expression analysis of TaAG-A and TaAG-B genes in fertile wheat lines. CK, uninfected wheat plants; γ, wheat plants infected by RNAγb; PDS, wheat plants infected by RNAγb:TaPDS; GFP, wheat plants infected by RNAγb:GFP; 1, 2, and 3 represent different sampling stages.
The VIGS assays also provided a visual indication of the occurrence of TaAG-A and TaAG-B gene silencing in spikes after virus inoculation. The spikes of plants inoculated with BSMV:TaAG-A and BSMV:TaAG-B were wizened and the ripening rate was notably reduced.
To further verify the involvement of TaAG-A and TaAG-B in wheat male sterility, we assessed the relative transcript levels of the TaAG-A and TaAG-B genes in spikes by semi-quantitative RT-PCR analysis with tubulin as a control (Figure 5C). TaAG-A and TaAG-B were silenced in spikes after viral inoculation. At 16 and 20 days after inoculation, the relative transcript levels in spikes inoculated with RNAγb and RNAγb:GFP were decreased compared to the control, indicating that BSMV infection affected the growth and development of plants.
Discussion
Cytoplasmic male sterility is a maternally inherited trait resulting in failure to produce functional pollen. It is generally thought that cytoplasmic male sterility is associated with the rearrangement of mitochondrial genomes (Schnable and Wise, 1998; Hanson and Bentolila, 2004). Recently, some MADS-box genes related to floral organ development were reported in wheat to be targets for floral organ homeotic transformation regulated by mitochondrial retrograde regulation, which is the communication pathway from mitochondria to the nucleus (Meguro et al., 2003; Hama et al., 2004). We found from our previous microarray results that the expression level of an MIKC-type MADS-box transcription factor, Ta.4147, in the CMS line was higher than in the maintainer line. To elucidate the relationship between the MADS-box gene and male sterility of the wheat K-type CMS line in this study, we isolated two MADS-box genes named TaAG-A and TaAG-B from uninucleate pollen and assigned them to chromosomes 3A and 3B, respectively. Their predicted amino acid sequences had typical conserved domains of MADS-MEF2-like and K-box of MADS-box proteins. Phylogenetic analysis showed that TaAG-A and TaAG-B belong to the AG subfamily C group of the MADS-box family. The hexaploid wheat (AABBDD) genome contains triplicate homoeologs of each gene derived from the ancestral diploid T. urartu (AA), Aegilops speltoides (BB), and Aegilops tauschii (DD). However, we only isolated two homoeologs, TaAG-A and TaAG-B, using RT-PCR in this study, indicating that transcription of the D genome homoeolog did not occur. Tanaka et al. (2015) also reported that the homoeologs of WAG1 (wheat AGAMOUS1) on the A and B genomes were expressed, while expression of its D genome homoeolog was suppressed.
According to the ABC(DE) and ‘floral quartets’ models, the combination BCE specifies the formation of stamens and the combination CE determines the development of carpels (Theißen, 2001). In Arabidopsis, AG is required to specify the identity of stamens and pistils (Yanofsky et al., 1990). Mutation in a class-C gene leads to homeotic transformations of the organs of region C (i.e., stamens and carpels) into perianth organs (i.e., petals and sepals, respectively). The wheat inflorescence, which is composed of spikelets, is quite different from the inflorescences of dicots. Stamens are transformed into pistil-like structures, which in turn contain ovule-like structures in the alloplasmic wheat line (cr)-CSdt7BS (Murai et al., 2002). Meguro et al. (2003) subsequently reported that an AG homolog, WAG, is associated with pistillody caused by nuclear-cytoplasm interaction in the alloplasmic lines of common wheat with the Aegilops crassa cytoplasm. The WAG gene reported by Meguro et al. (2003) is known as WAG1/TaAG-1 and WAG2 is known as TaAG-2/TaAGL39 (Zhao et al., 2006; Paolacci et al., 2007). Phylogenetic analysis showed that WAG1 and WAG2 are orthologs of rice OsMADS58 and OsMADS3, respectively. OsMADS3, ZMM2, WM29A, TaAGL39, WM29B, and HvAG1 were clustered in the WAG2 clade (Murai, 2013). In this study, TaAG-A and TaAG-B were closely related to WM29A, WM29B, TaAGL39, and OsMADS3, indicating that TaAG-A and TaAG-B may play similar roles to WAG2, OsMADS3, and ZMM2. OsMADS3 plays a predominant role in inhibiting lodicule development and in specifying stamen identity (Kang et al., 1995). ZMM2 participates in regulating the formation of stamens and carpels (Mena et al., 1996). Mizumoto et al. (2009) found that WAG-2 is preferentially expressed in the central region of pistils rather than developing stamens during floral organ development. Thus, the function of WAG-2 is also associated with pistillody. Based on these studies, we deduced that TaAG-A and TaAG-B may be involved in the development of pistil-like stamens in the K-type CMS line.
To verify our hypothesis, we performed qRT-PCR analysis to assess the expression patterns of TaAG-A and TaAG-B in wheat. In Chinese Spring wheat at the heading stage, TaAG-A had relatively high expression levels in leaves and spikes, and low levels in roots. Similarly, TaAG-B had relatively high expression levels in spikes and low levels in roots, stems, and leaves. Our results suggest that the product of the TaAG-A transcript is involved in leaf and spike development, while the product of TaAG-B functions mainly in spike development. The expression patterns of WAG1 homoeologs, with high expression of WAG1-A, very low expression of WAG1-B, and silencing of WAG1-D in young spikes, are consistent and reproducible features of hexaploid lines (Tanaka et al., 2015). However, AG orthologous genes, such as AG (Arabidopsis, Yanofsky et al., 1990), ZAG1 (maize, Schmidt et al., 1993), and OsMADS3 (rice, Kang et al., 1995), were not expressed in any vegetative tissues. The expression of TaAGL39 was detectable in 16-h, 20-h, and 28-h embryos of imbibed seeds, stems at the jointing stage, and developing seeds at 6 and 12 days after pollination (Zhao et al., 2006). In anthers at different stages (mononucleate, dinucleate, and trinucleate) collected from the K-type CMS line and the maintainer line, TaAG-B expression increased from the dinucleate to the trinucleate pollen stages in the CMS line, both TaAG-A and TaAG-B downregulated in the maintainer line. Meguro et al. (2003) also found that WAG was preferentially expressed during late stages in developing spikes.
To further investigate the function of TaAG-A and TaAG-B genes, we overexpressed TaAG-A and TaAG-B in Arabidopsis and silenced TaAG-A and TaAG-B in fertile wheat lines using BSMV-VIGS. The transgenic Arabidopsis showed earlier reproductive development, premature mortality, and abnormal buds, stamens, and stigmas. In addition, the higher the TaAG-A and TaAG-B expression levels were, the greater the deformity. Expression analysis of transgenic Arabidopsis indicated that TaAG-A and TaAG-B are involved in pistil and stamen development. BSMV-VIGS is an effective reverse genetics tool to investigate the genetic function of important genes in wheat (Scofield et al., 2005; Zhou et al., 2007). The TaAG-A and TaAG-B silenced plants had green and yellow striped leaves, emaciated spikes, and decreased selfing seed set rates. These results suggest that TaAG-A and TaAG-B may play roles in male sterility in the wheat CMS line.
Author Contributions
WY and AZ designed the study and provide guidance on the whole study. XL, JL, and MP carried out the plant materials culture, qRT-PCR analysis, data analysis, and drafted the manuscript. DL, JS, KZ, and LH prepared plant materials. DL and AM provided the value comments and revised the grammar of the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the National Key Research and Development Program of China (Grant No. 2016YFD0100804) and the National Natural Science Foundation of China (Grant No. 30971668).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01081/full#supplementary-material
Footnotes
- ^ http://plants.ensembl.org/Triticum_urartu/Tools/Blast?db=core
- ^ http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
References
Ai, Y., Zhang, Q., Wang, W., Zhang, C., Cao, Z., Bao, M., et al. (2016). Transcriptomic analysis of differentially expressed genes during flower organ development in genetic male sterile and male fertile Tagetes erecta by digital gene-expression profiling. PLoS ONE 11:e0150892. doi: 10.1371/journal.pone.0150892
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium- mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Coen, E. S., and Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
De Bodt, S., Raes, J., Van de Peer, Y., and Theißen, G. (2003). And then there were many: MADS goes genomic. Trends Plant Sci. 8, 475–483. doi: 10.1016/j.tplants.2003.09.006
Duan, K., Li, L., Hu, P., Xu, S. P., Xu, Z. H., and Xue, H. W. (2006). A brassinolide-suppressed rice MADS-box transcription factor, OsMDP1, has a negative regulatory role in BR signaling. Plant J. 47, 519–531. doi: 10.1111/j.1365-313X.2006.02804.x
Duvick, D. N. (1959). The use of cytoplasmic male-sterility in hybrid seed production. Econ. Bot. 13, 167–195. doi: 10.1007/BF02860581
Goto, K., and Meyerowitz, E. M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8, 1548–1560. doi: 10.1101/gad.8.13.1548
Hama, E., Takumi, S., Ogihara, Y., and Murai, K. (2004). Pistillody is caused by alterations to the class-B MADS-box gene expression pattern in alloplasmic wheats. Planta 218, 712–720. doi: 10.1007/s00425-003-1157-6
Hanson, M. R., and Bentolila, S. (2004). Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 16(Suppl.), S154–S169. doi: 10.1105/tpc.015966
Hirabayashi, C., and Murai, K. (2009). Class C MADS-box gene AGAMOUS was duplicated in the wheat genome. Wheat Inf. Serv. 107, 13–16.
Huang, Y., Yang, W., Pei, Z., Guo, X., Liu, D., Sun, J., et al. (2012). The genes for gibberellin biosynthesis in wheat. Funct. Integr. Genomics 12, 199–206. doi: 10.1007/s10142-011-0243-2
Jack, T., Brockman, L. L., and Meyerowitz, E. M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. doi: 10.1016/0092-8674(92)90144-2
Jofuku, K. D., den Boer, B. G., Van Montagu, M., and Okamuro, J. K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6, 1211–1225. doi: 10.1105/tpc.6.9.1211
Kang, H. G., Noh, Y. S., Chung, Y. Y., Costa, M. A., An, K., and An, G. (1995). Phenotypic alterations of petal and sepal by ectopic expression of a rice MADS box gene in tobacco. Plant Mol. Biol. 29, 1–10. doi: 10.1007/BF00019114
Li, A., Yang, W., Li, S., Liu, D., Guo, X., Sun, J., et al. (2013). Molecular characterization of three GIBBERELLIN-INSENSITIVE DWARF1 homologous genes in hexaploid wheat. J. Plant Physiol. 170, 432–443. doi: 10.1016/j.jplph.2012.11.010
Mandel, M. A., Gustafson-Brown, C., Savidge, B., and Yanofsky, M. F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273–277. doi: 10.1038/360273a0
Meguro, A., Takumi, S., Ogihara, Y., and Murai, K. (2003). WAG, a wheat AGAMOUS homolog, is associated with development of pistil-like stamens in alloplasmic wheats. Sex. Plant Reprod. 15, 221–230.
Mena, M., Ambrose, B. A., Meeley, R. B., Briggs, S. P., Yanofsky, M. F., and Schmidt, R. J. (1996). Diversification of C-function activity in maize flower development. Science 274, 1537–1540. doi: 10.1126/science.274.5292.1537
Mizumoto, K., Hatano, H., Hirabayashi, C., Murai, K., and Takumi, S. (2009). Altered expression of wheat AINTEGUMENTA homolog, WANT-1, in pistil and pistil-like transformed stamen of an alloplasmic line with Aegilops crassa cytoplasm. Dev. Genes Evol. 219, 175–187. doi: 10.1007/s00427-009-0275-y
Münster, T., Pahnke, J., Di Rosa, A., Kim, J. T., Martin, W., Saedler, H., et al. (1997). Floral homeotic genes were recruited from homologous MADS-box genes preexisting in the common ancestor of ferns and seed plants. Proc. Natl. Acad. Sci. U.S.A. 94, 2415–2420. doi: 10.1073/pnas.94.6.2415
Murai, K. (2013). Homeotic genes and the ABCDE model for floral organ formation in wheat. Plants 2, 379–395. doi: 10.3390/plants2030379
Murai, K., Miyama, M. E., Kato, H., Takumi, S., and Ogihara, Y. (2003). WAP1, a wheat APETALA1 homolog, plays a central role in the phase transition from vegetative to reproductive growth. Plant Cell Physiol. 44, 1255–1265. doi: 10.1093/pcp/pcg171
Murai, K., Takumi, S., Koga, H., and Ogihara, Y. (2002). Pistillody, homeotic transformation of stamens into pistil-like structures, caused by nuclear-cytoplasm interaction in wheat. Plant J. 29, 169–181. doi: 10.1046/j.0960-7412.2001.01203.x
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Ng, M., and Yanofsky, M. F. (2001). Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2, 186–195. doi: 10.1038/35056041
Paolacci, A. R., Tanzarella, O. A., Porceddu, E., and Ciaffi, M. (2009). Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 10:11. doi: 10.1186/1471-2199-10-11
Paolacci, A. R., Tanzarella, O. A., Porceddu, E., Varotto, S., and Ciaffi, M. (2007). Molecular and phylogenetic analysis of MADS-box genes of MIKC type and chromosome location of SEP-like gene in wheat (Triticum aestivum L.). Mol. Genet. Genomics 278, 689–708. doi: 10.1007/s00438-007-0285-2
Saghai-Maroof, M. A., Soliman, K. M., Jorgensen, R. A., and Allard, R. W. (1984). Ribosomal DNA spacer-length polymorphism in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. U.S.A. 81, 8014–8018. doi: 10.1073/pnas.81.24.8014
Schmidt, R. J., Veit, B., Mandel, M. A., Mena, M., Hake, S., and Yanofsky, M. F. (1993). Identification and molecular characterization of ZAG1, the maize homolog of the Arabidopsis floral homeotic gene AGAMOUS. Plant Cell 5, 729–737. doi: 10.1105/tpc.5.7.729
Schnable, P. S., and Wise, R. P. (1998). The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3, 175–180. doi: 10.1016/S1360-1385(98)01235-7
Schwarz-Sommer, Z., Huijser, P., Nacken, W., Saedler, H., and Sommer, H. (1990). Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 250, 931–936. doi: 10.1126/science.250.4983.931
Scofield, S. R., Huang, L., Brandt, A. S., and Gill, B. S. (2005). Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol. 138, 2165–2173. doi: 10.1104/pp.105.061861
Tanaka, M., Tanaka, H., Shitsukawa, N., Kitagawa, S., Takumi, S., and Murai, K. (2015). Homoeologous copy-specific expression patterns of MADS-box genes for floral formation in allopolyploid wheat. Genes Genet. Syst. 90, 217–229. doi: 10.1266/ggs.15-00029
Theißen, G. (2001). Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 4, 75–85. doi: 10.1016/S1369-5266(00)00139-4
Tsunewaki, K., Mukai, Y., and Endo, E. R. (1978). “On the decent of the cytoplasm of polyploid species in Triticum and Aegilops,” in Proceedings of the 5th International Wheat Genetics Symposium, New Delhi, 261–272.
Wang, Q. H., Yang, Z. J., Wei, S. H., Jiang, Z. Y., Yang, Y. F., Hu, Z. S., et al. (2015). Molecular cloning, characterization and expression analysis of WAG-1 in the pistillody line of common wheat. Genet. Mol. Res. 14, 12455–12465. doi: 10.4238/2015.October.16.12
Wei, S., Peng, Z., Zhou, Y., Yang, Z., Wu, K., and Ouyang, Z. (2011). Nucleotide diversity and molecular evolution of the WAG-2 gene in common wheat (Triticum aestivum L) and its relatives. Genet. Mol. Biol. 34, 606–615. doi: 10.1590/S1415-47572011000400013
Weigel, D., and Meyerowitz, E. M. (1994). The ABCs of floral homeotic genes. Cell 78, 203–209. doi: 10.1016/0092-8674(94)90291-7
Yanofsky, M. F., Ma, H., Bowman, J. L., Drews, G. N., Feldman, K. A., and Meyerowitz, E. M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. doi: 10.1038/346035a0
Zhao, T., Ni, Z., Dai, Y., Yao, Y., Nie, X., and Sun, Q. (2006). Characterization and expression of 42 MADS-box genes in wheat (Triticum aestivum L.). Mol. Genet. Genomics 276, 334–350. doi: 10.1007/s00438-006-0147-3
Zhao, Y. X., Cheng, Z. J., and Zhang, X. S. (2005). Overexpression of TaMADS1, a SEPALLATA-like gene in wheat, causes early flowering and the abnormal development of floral organs in Arabidopsis. Planta 223, 698–707. doi: 10.1007/s00425-005-0123-x
Zhou, H., Li, S., Deng, Z., Wang, X., Chen, T., Zhang, J., et al. (2007). Molecular analysis of three new receptor-like kinase genes from hexaploid wheat and evidence for their participation in the wheat hypersensitive response to stripe rust fungus infection. Plant J. 52, 420–434. doi: 10.1111/j.1365-313X.2007.03246.x
Keywords: wheat, male sterile, MADS-box gene, cloning, functional analysis
Citation: Yang W, Lou X, Li J, Pu M, Mirbahar AA, Liu D, Sun J, Zhan K, He L and Zhang A (2017) Cloning and Functional Analysis of MADS-box Genes, TaAG-A and TaAG-B, from a Wheat K-type Cytoplasmic Male Sterile Line. Front. Plant Sci. 8:1081. doi: 10.3389/fpls.2017.01081
Received: 13 February 2017; Accepted: 06 June 2017;
Published: 20 June 2017.
Edited by:
Keqiang Wu, National Taiwan University, TaiwanReviewed by:
Turgay Unver, Izmir International Biomedicine and Genome Institute (iBG-izmir), TurkeyShabir Hussain Wani, Michigan State University, United States
Genying Li, Shandong Academy of Agricultural Sciences, China
Copyright © 2017 Yang, Lou, Li, Pu, Mirbahar, Liu, Sun, Zhan, He and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aimin Zhang, amzhang@genetics.ac.cn Wenlong Yang, wlyang@genetics.ac.cn
†These authors have contributed equally to this work.
 Wenlong Yang
Wenlong Yang Xueyuan Lou1,2†
Xueyuan Lou1,2† Dongcheng Liu
Dongcheng Liu Aimin Zhang
Aimin Zhang