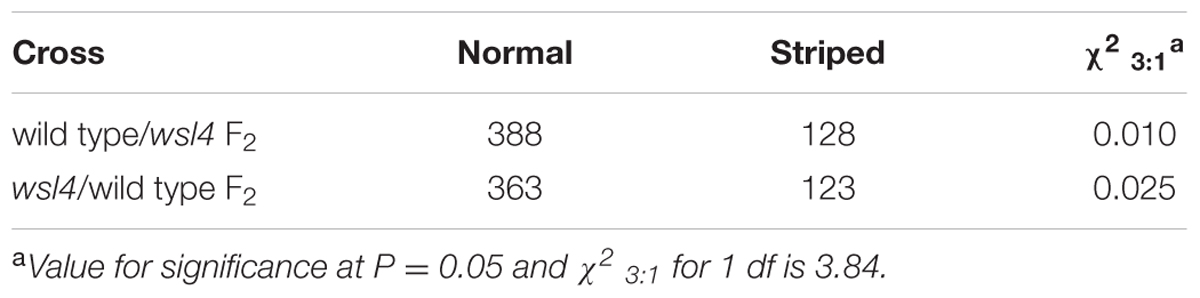- 1National Key Facility for Crop Gene Resources and Genetic Improvement, Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing, China
- 2National Key Laboratory for Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing, China
Pentatricopeptide repeat (PPR) proteins comprise a large family in higher plants and perform diverse functions in organellar RNA metabolism. Despite the rice genome encodes 477 PRR proteins, the regulatory effects of PRR proteins on chloroplast development remains unknown. In this study, we report the functional characterization of the rice white stripe leaf4 (wsl4) mutant. The wsl4 mutant develops white-striped leaves during early leaf development, characterized by decreased chlorophyll content and malformed chloroplasts. Positional cloning of the WSL4 gene, together with complementation and RNA-interference tests, reveal that it encodes a novel P-family PPR protein with 12 PPR motifs, and is localized to chloroplast nucleoids. Quantitative RT-PCR analyses demonstrate that WSL4 is a low temperature response gene abundantly expressed in young leaves. Further expression analyses show that many nuclear- and plastid-encoded genes in the wsl4 mutant are significantly affected at the RNA and protein levels. Notably, the wsl4 mutant causes defects in the splicing of atpF, ndhA, rpl2, and rps12. Our findings identify WSL4 as a novel P-family PPR protein essential for chloroplast RNA group II intron splicing during early leaf development in rice.
Introduction
Chloroplasts, thought to have originated from cyanobacteria through endosymbiosis, are the exclusive organelles for photosynthesis in plants and algae, and also have important roles in synthesis and storage of many key metabolites, such as lipids, terpenoids, and amino acids (Mullet, 1993; Moreira et al., 2000; Sugimoto et al., 2004). Extensive studies have uncovered the basic photosynthetic and metabolic processes of chloroplast development (Kangasjärvi et al., 2014; Kaur, 2014; de Souza et al., 2016).
Chloroplast development from proplastids is subdivided into three stages; that are coordinately regulated by both plastid- and nuclear-encoded genes (Kusumi et al., 2004; Munekage et al., 2004; Jarvis and Lopez-Juez, 2013; Kusumi and Iba, 2014; Pogson et al., 2015). The first step is activation of plastid DNA replication and plastid DNA synthesis. The second step known as the chloroplast “build-up” stage establishes the chloroplast genetic system, in which a nuclear-encoded plastid RNA polymerase (NEP) preferentially transcribes genes encoding plastid gene expression machinery that promotes the transcription and translation in chloroplasts (Hajdukiewicz et al., 1997). In the third stage, plastid genes, predominantly transcribed by a plastid-encoded plastid RNA polymerase (PEP), in combination with imported nuclear-encoded proteins, constitute the photosynthetic and metabolic machinery to control chloroplast development (Kanamaru et al., 1999). Taking rice (Oryza sativa) as an example, the plastid genome is about 135 kb composed of 34 RNA-coding genes and 120 protein-coding genes (Hiratsuka et al., 1989; Pfalz and Pfannschmidt, 2013). About 3,000 proteins function in chloroplasts, more than 95% of which are encoded by nuclear genes (Reumann et al., 2005), suggesting that chloroplast development is predominantly under control of nuclear genes. Therefore cloning and characterization of such nuclear genes should help elucidate the complex regulatory mechanisms of chloroplast development in plants.
Pentatricopeptide repeat (PPR) proteins, characterized by tandem arrays of degenerate PPR motifs, which are a type of commonly existing, evolutionarily widespread, plant protein segments composed of 35 canonical amino acids (Small and Peeters, 2000; Yin et al., 2013; Barkan and Small, 2014). According to the tandem motifs, PPR proteins have been classified into two subfamilies: P- and PLS-type. Based on the structure of C-terminal motifs, the latter was further classified into PLS, E, E+, and DYW subgroups (Lurin et al., 2004). Accumulating evidence shows that nuclear-encoded PPR proteins perform diverse functions in post-transcriptional modulation of gene transcripts, such as RNA processing (Kazama and Toriyama, 2003; Nakamura et al., 2003; Hattori et al., 2007), RNA editing (Hammani et al., 2009; Fujii and Small, 2011; Yap et al., 2015; Xie et al., 2016), RNA splicing (Tan et al., 2014; Hsieh et al., 2015; Yap et al., 2015), RNA stability (Beick et al., 2008; Tavares-Carreon et al., 2008; Hammani et al., 2016; Zoschke et al., 2016), RNA translation (Schmitz-Linneweber et al., 2005; Barkan et al., 2012; Zoschke et al., 2016), and RNA maturation (Wu et al., 2016; Zhou et al., 2017). Genetic evidence shows little or no redundancy of function between PPR proteins, although gene family has expanded in higher plants, with 450 members in Arabidopsis and 477 members in rice (Small and Peeters, 2000; Lurin et al., 2004; O’Toole et al., 2008). This evidence suggests that the functions of PPR proteins are highly diversified in higher plants.
Of the 477 PPR members in the rice genome, only a limited number has been cloned. Among them, RF5 and RF6 associate with GRP162 and hexokinase 6 to regulate mitochondrial RNA metabolism and fertility restoration, respectively (Hu et al., 2012; Huang et al., 2015). OGR1 and MPR25 encode a DYW motif-containing PPR protein and an E subgroup member of PPR, respectively, and both are involved in RNA editing in mitochondria (Kim et al., 2009; Toda et al., 2012). Their loss-of-function mutations caused retarded growth (Kim et al., 2009; Toda et al., 2012). In addition to the mitochondrially-localized PPR proteins listed above, previous studies also identified several chloroplast-targeted PPR members, including OsPPR1 (Gothandam et al., 2005), YSA (Su et al., 2012), OsV4 (Gong et al., 2014), WSL (Tan et al., 2014), ASL3 (Lin et al., 2015a), and OspTAC2 (Wang D. et al., 2016). A conspicuous feature of these rice mutants or their corresponding RNAi transgenics is the chlorophyll (Chl) deficient phenotype and abnormally developed chloroplasts at seedling stage, suggesting essential roles in chloroplast development at an early leaf growth stage. Due to lack of in-depth functional studies on these genes, only WSL has thus far been implicated in RNA splicing of chloroplast transcript rpl2 (Tan et al., 2014). Additionally, OspTAC2 may act as the core subunit of the PEP complex based on studies of the Arabidopsis homolog pTAC2 (Wang D. et al., 2016). Despite significant advances, the roles of the PPR proteins in regulating post-transcriptional modification of organelle genes, especially chloroplast transcripts, remain obscure. It is expected that cloning and more in-depth studies of PPR proteins will help to decipher the regulatory network of post-transcriptional modulation of gene expression in rice.
Here, we identified rice Chl deficient mutant wsl4 that develops striped leaves with decreased Chl contents and impaired chloroplast structures at the early leaf developmental stage. We show that WSL4 encodes a novel P-family PPR protein that targets to the chloroplast nucleoid. Expression analyses suggested that WSL4 coordinates expression of many nuclear- and plastid-encoded genes involved in chloroplast development. We found that WSL4 could be involved in chloroplast development by affecting RNA splicing at an early stage of leaf development.
Materials and Methods
Plant Materials and Growth Conditions
The wsl4 mutant was isolated from a 60Co-irradiated mutant pool of japonica cultivar RX69. The mutagenesis screen was performed as described previously (Wang L. et al., 2016; Zhou et al., 2016). A cross was made between the wsl4 mutant and RX69 for preliminary genetic analysis and a large F2 segregation population was generated from wsl4 × Yue13 (ssp. indica) for fine mapping. Plants were grown in a paddy field during the normal rice growing season in Beijing (39°54′N, summer season) or in a growth chamber with a 12 h photoperiod at 30/25°C (L30/D25). Seedlings for differential temperature studies were grown in a growth chamber with a 12 h photoperiod and constant 30°C (C30) and 20°C (C20).
Chlorophyll Contents and Transmission Electron Microscopy (TEM) Analyses
Chlorophyll contents were measured according to a method described by Wu et al. (2007). Briefly, fresh leaves of WT and wsl4 mutant plants were collected at the two-, three-, four-, and five-leaf stages, and then cut and marinated in 5 ml of 95% ethanol for 48 h in darkness. After centrifugation, the residual plant debris was removed. The supernatants were analyzed with a DU 800 UV/Vis Spectrophotometer (Beckman Coulter) at 665, 649, and 470 nm, respectively.
For TEM analyses, third leaves from WT and green and white sectors of wsl4 mutant were cut into slices of 0.5 cm and three slices of each tissue were fixed in a solution of 2.5% glutaraldehyde in phosphate buffer at 4°C for 4 h, and incubated overnight at 4°C in 1% OsO4. The tissues were subsequently dehydrated in an ethanol series, infiltrated in a gradient series of epoxy resin, and finally embedded in Spurr’s medium prior to thin sectioning (50–80 nm). Samples were stained again and examined with a Hitachi H-7650 transmission electron microscope.
Fine-Mapping of WSL4 Locus
Sequence polymorphisms between Nipponbare (japonica) and 93-11 (indica) were identified from a public database1 and used to develop Indel markers for fine mapping. Primer pairs were designed with Primer Premier 3.0. Newly developed PCR-based molecular markers used in this study are listed in Supplementary Table S1. The PCR procedure was as follows: 94°C for 5 min, followed by 34 cycles of 94°C for 30 s, annealing for 30 s, 72°C for 30 s, and a final elongation step at 72°C for 7 min.
Complementation Test and RNAi Suppression of WSL4
For complementation of the wsl4 mutation a 5.7 kb WT genomic fragment (primer pairs PPR-G, Supplementary Table S1) containing a 2.2 kb upstream sequence, the entire coding region of WSL4, and a 2.0 kb downstream sequence was amplified from RX69 with Prime STAR HS DNA Polymerase (TaKaRa) and cloned into the binary vector pCAMBIA1305 to generate the vector pCAMBIA1305-GWSL4. This vector was introduced into Agrobacterium tumefaciens EHA105, which was then used to infect wsl4 mutant calli (Hiei et al., 1994).
For RNAi test, the construct pCUbi1390-DFAD2 (ubiquitin promoter and a FAD2 intron inserted into pCAMBIA1390) was used as an RNAi vector (Stoutjesdijk et al., 2002; Wu et al., 2007). Both anti-sense and sense versions of a specific 332 bp fragment from the coding region of the WSL4 5′-end were amplified (primer pairs WSL4-RNAi-SacI-InF and WSL4-RNAi-BamHI-InR, Supplementary Table S1), and successively inserted into pCUbi1390-DFAD2, to form the RNAi construct pUbi-dsRNAiWSL4. The construct was then used to infect the calli produced from RX69. Transformation was conducted according to the method described above.
Sequence and Phylogenetic Analysis
Gene prediction was performed using the GRAMENE database2. Homologous sequences of WSL4 were identified using the Blastp search program of the National Center for Biotechnology Information (NCBI3). A chloroplast transit peptide at the N-terminus of WSL4 was predicted by ChloroP4 and TargetP5. A phylogenetic tree was constructed using MEGA v4.1 software6 by the bootstrap method with 1,000 replicates. Multiple sequence alignments were conducted with BioEdit software (Borland Company).
Subcellular Localization of WSL4 Protein
For subcellular localization of WSL4 and wsl4 proteins in rice the coding sequences of WSL4 and wsl4 were amplified using specific primer pairs (listed in Supplementary Table S1) and cloned into the transient expression vector pA7-GFP to generate the fusion genes WSL4-GFP and wsl4-GFP driven by the CaMV 35S promoter. All transient expression constructs were separately transformed into rice protoplasts and incubated in darkness at 28°C for 16 h before examination according to the protocols described previously (Chiu et al., 1996; Chen et al., 2006). Fluorescence of GFP in transformed protoplasts was visualized using a confocal laser scanning microscope (Leica TCS SP5).
RT-PCR and Real-Time RT-PCR Analyses
Total RNA was extracted from flag leaves, culms, young panicles, and leaf sheaths at the booting stage, as well as shoot bases, leaves, and seedling roots using an RNA Prep Pure Plant kit (Tiangen Co., Beijing). Each RNA sample (about 3 μg) was reverse transcribed using primer Script I (TaKaRa) and an oligo(dT)18 primer for nuclear-encoded genes or random primers for plastid-encoded genes since mRNA from most plastid-encoded genes carried no poly-A tail. The RT-PCR procedure was as follows: 94°C for 5 min, followed by 33 cycles of 94°C for 30 s, annealing for 30 s, 72°C for 1 kb/s, and a final elongation step at 72°C for 7 min. The primer pairs are listed in Supplementary Table S1. For analysis of RNA splicing we sequenced full-length RT-PCR products with special primers flanking the introns (Supplementary Table S1). For Real-Time RT-PCR analysis of RNA splicing efficiency, specific primers (Supplementary Table S1) were designed for intron-exon (unspliced forms) and exon–exon (spliced forms) links of each gene (de Longevialle et al., 2007; Koprivova et al., 2010; Hsieh et al., 2015; Yap et al., 2015).
Real-Time RT-PCR was performed using a SYBR Premix Ex TaqTM Kit (TaKaRa) on an ABI prism 7500 Real-Time PCR System. The program was as follows: initial polymerase activation for 30 s at 95°C followed by 40 cycles of 95°C for 5 s and 60°C for 34 s. The 2-ΔΔCT method was used to analyze relative transcript levels of genes (Livak and Schmittgen, 2001). The primer sequences for qRT-PCR are listed in Supplementary Table S1. The rice Ubiquitin gene (LOC_Os03g13170) was used as a reference in qRT–PCR (primer pairs Ubq).
Northern Blot Analyses
Total RNA from leaves (L3-3) at the seedling stage was isolated using TransZol Up (TransGen) following recommendations by the manufacturer. About 2 μg of RNA was fractionated in a 1.2% (w/v) denaturing formaldehyde agarose gel and transferred onto Hybond N+ nylon membranes (GE Healthcare Biosciences). Digoxigenin-labeled probes were obtained by PCR using a DIG Northern Starter Kit (Roche). The primers are listed in Supplementary Table S1. Prehybridization of the membrane was carried out for 1 h at 68°C in PerfectHyb Plus Hybridization Buffer (SIGMA). Hybridization was performed overnight in the same buffer at 68°C. Signal was detected using the DIG Wash and Block Buffer Set (Roche), and eventually read in an imaging system (Tanon 5200).
Protein Extraction and Western Blot Analyses
Fresh leaves (L3-3) were standardized by fresh weight and ground into fine powder. Total plant proteins were extracted with an appropriate volume (2 mL g-1) of NB1 buffer [50 mM Tris-MES, 1 mM MgCl2, 0.5 M sucrose, 10 mM EDTA, 5 mM DTT, and protease inhibitor cocktail (Roche, Cat. No. 11 836-153-001; the used concentration was 1 tablet for a volume of 50 ml solution), pH 8.0]. The protein samples were resolved in 10% SDS–polyacrylamide gel electrophoresis (PAGE) gels, and then transferred onto PVDF membranes (0.45 μm, Millipore), followed by incubation with antibodies. Signals were detected using a LuminataTM Forte Western HRP Substrate (Millipore) and visualized by an imaging system (ChemiDocTMX- RS; Bio-Rad). The polyclonal antibodies used in this study were obtained from BGI7. The intensities of chloroplast proteins were quantified by “ImageJ” software.
Statistical Analysis
All statistical analyses were performed using Student’s t-test. The number of biological replicates (n) in each experiment is indicated in the corresponding figure legends. Values were considered statistically significant at P < 0.05, and very significant at P < 0.01.
Results
Phenotypic Characterization of the wsl4 Mutant
To characterize the wsl4 mutant in detail, we conducted a time-course examination of the mutant phenotype from the two to five-leaf stages. The leaves of seedlings at the 3–4 leaf stage exhibited a bleached appearance with the most extreme symptoms observed in the third leaf and distal half of the fourth leaf (Figures 1A–C). All leaves after the 5-leaf stage were as green as WT plants (Figure 1D). Young leaves on newly emerging tillers of the wsl4 mutant also exhibited the green/white leaf color defect (Figure 1E). No difference was observed between WT and wsl4 mutant after the heading (Figure 1F). Consistent with these observations, levels of Chl a and b in striped leaves of wsl4 plants were much lower than those in WT (Figures 1G,H) Compared with WT plants, wsl4 mutants showed no statistically significant differences in plant height, number of tillers, number of branches per panicle, number of spikelets per panicle, and 1000-grain weight (Table 1).
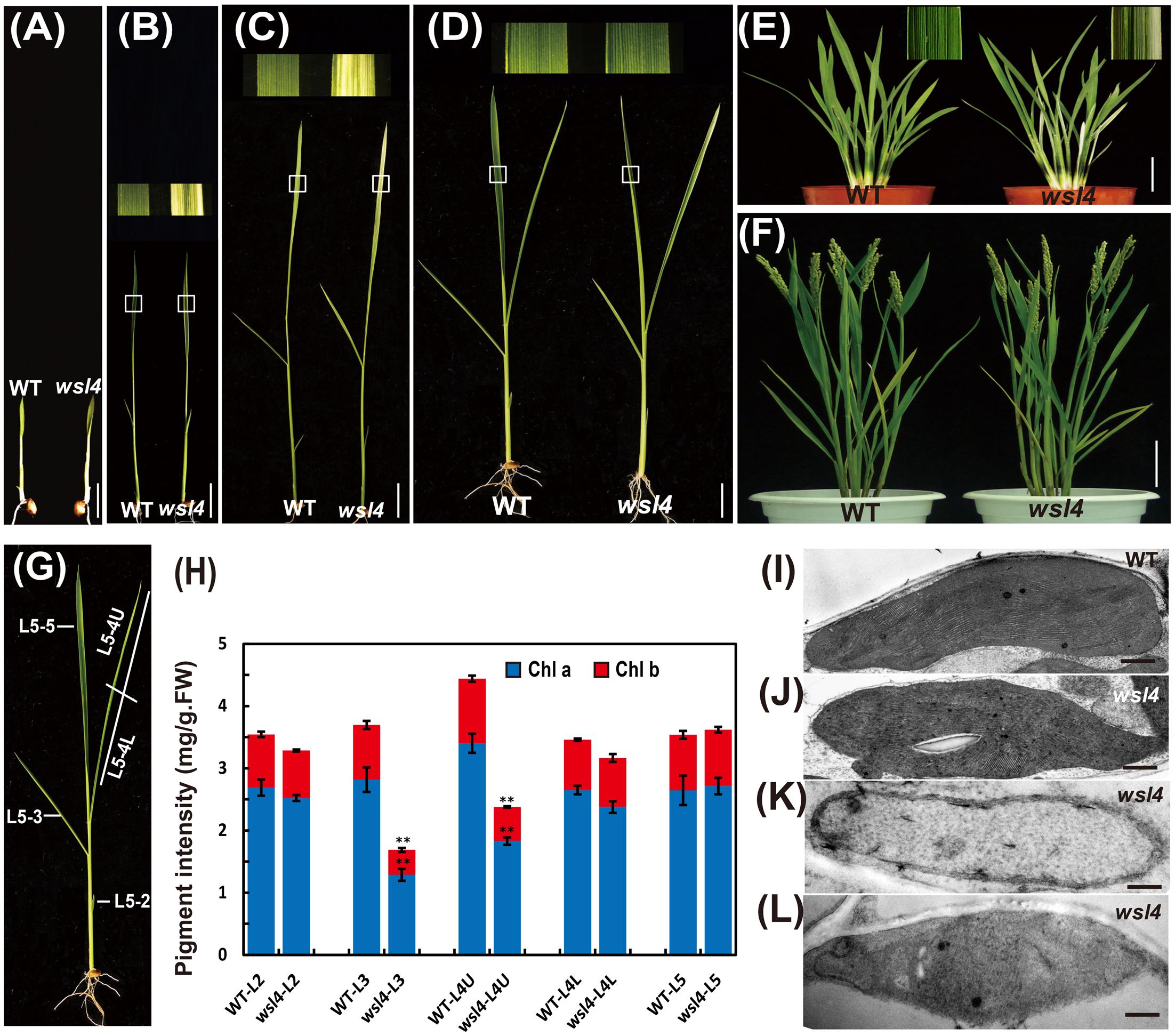
FIGURE 1. Phenotypic comparison of WT and wsl4 mutant plants. Comparisons of leaves of WT and wsl4 mutant plants grown in a growth chamber with a 12 h photoperiod at 30°C/25 °C (L30/D25) at the two- (A), three- (B), four- (C), and five- leaf (D) stages. The insets represent magnified views of the selected areas in (B–D). Bars, 0.5 cm. (E,F) Leaves of WT and wsl4 mutant plants grown in field at the tillering stage (E) and after heading stage (F). Note the white-striped leaves from the tillers of the wsl4 mutant at the tillering stage. Bars, 5 cm (E); 10 cm in (F). (G) A rice shoot with fully emerged fifth leaf at the five-leaf stage. L5-2, L5-3, L5-4, and L5-5 represent the second, third, fourth, and fifth leaf at the five-leaf stage, respectively. (H) Measurements of chlorophyll a and b contents of all leaves (L5-2, L5-3, L5-4U, L5-4L, and L5-5) from five-leaf stage WT and wsl4 mutant plants grown in a growth chamber at L30/D25. L5-4U represents the upper half of the fourth leaf, and L5-4L represents the basal half of the fourth leaf. Values are means ± SD from three independent repeats. Student’s t-test: ∗P < 0.05; ∗∗P < 0.01. Chl a, chlorophyll a, Chl b, chlorophyll b, FW, fresh weight. (I–L) Electron micrographs of chloroplasts from L3-3 of WT (I) and wsl4 mutant (J–L) plants grown in a growth chamber at L30/D25. Chloroplasts from the WT plants (I) and chloroplasts from the green portions of the wsl4 mutant (J) plants have abundant and well-ordered thylakoids and stacked membranes, whereas chloroplasts from white striped sectors of the wsl4 mutant plants had almost no normal stacked membrane structures (K,L). L3-3 indicates the third leaf at the three-leaf stage. Bars, 0.5 μm (I,J,L); 0.2 μm in (K).
In contrast to the moderate striped phenotype and Chl accumulations under optimum temperature (L30/D25), the wsl4 mutant exhibited less color deficiency and accumulated much higher Chl contents when grown at a constant 30°C (C30) (Figure 2). At a constant 20°C the wsl4 mutant showed more extreme symptoms and Chl content that were barely detectable (Figure 2). Our results show that the wsl4 mutant is sensitive to low temperatures.
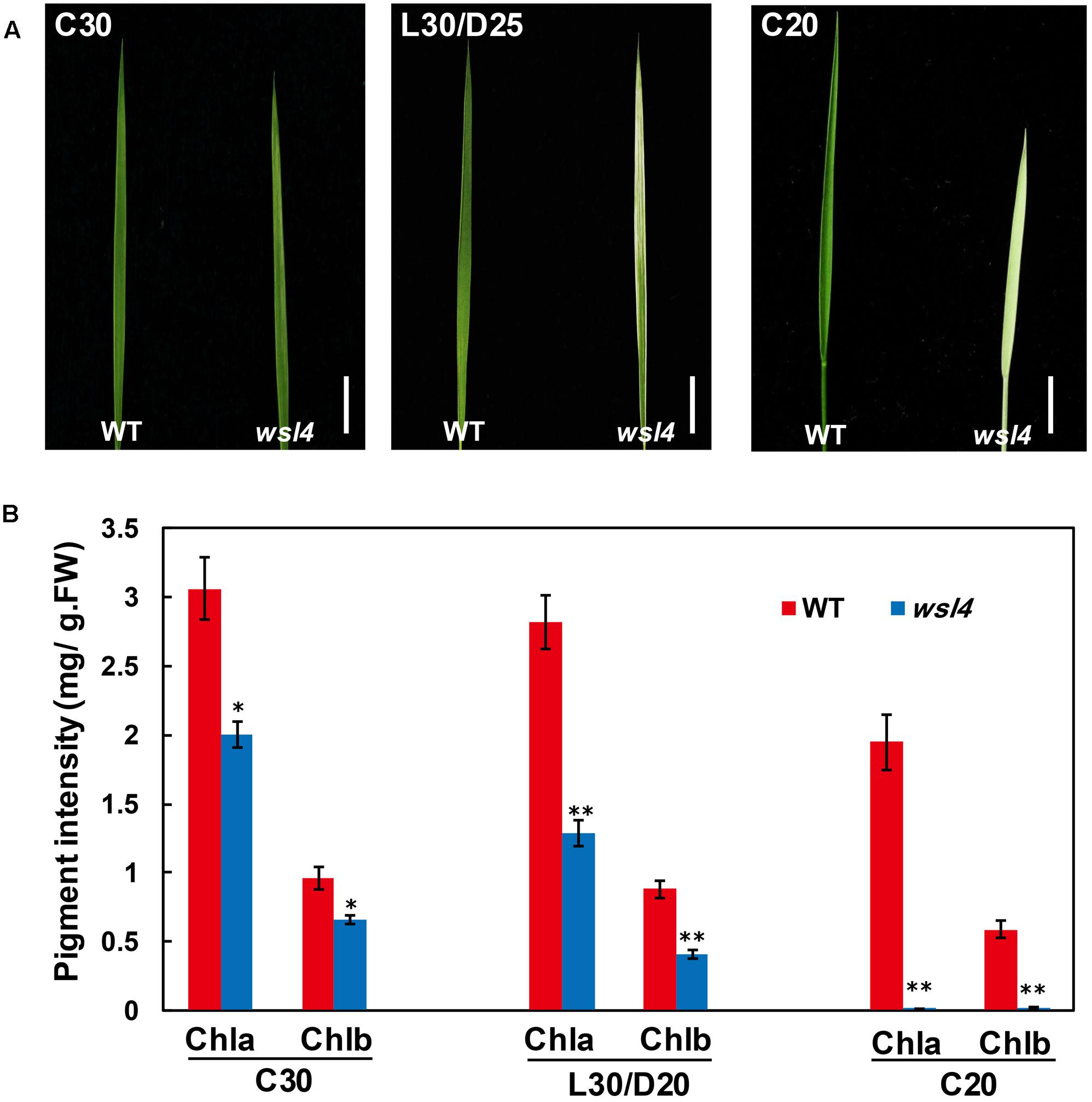
FIGURE 2. Response of the wsl4 mutant to temperature. (A) WT and wsl4 mutant seedlings grown in a growth chamber with 12 h photoperiod and 30°C (C30), 30/25°C (L30/D25), and 20°C (C20). Bars, 0.5 cm. (B) Chl a and Chl b contents in L3-3 from WT and wsl4 mutant seedlings (n = 6) grown in a growth chamber with the same conditions as (A). Student’s t-test: ∗P < 0.05; ∗∗P < 0.01.
The wsl4 Mutant Has Disrupted Chloroplast Development
To determine whether the color deficiency resulting from the decreased photosynthetic pigment accumulation in the wsl4 mutant were associated with ultrastructural damages in chloroplasts, we compared the ultrastructural features of chloroplasts from WT and wsl4 mutant leaves using TEM. As shown in Figures 1I–L, the lamellar structures of WT chloroplasts were well developed and were equipped with normally stacked grana and thylakoid membranes (Figure 1I). While green sectors of wsl4 mutant seedlings were still able to develop normal chloroplasts (Figure 1J), the white striped regions had abnormal chloroplasts that lacked organized lamellar structures (Figures 1K,L). Thus the stripe leaf phenotype and the reduced Chl contents in the wsl4 mutant leaves were probably due to developmentally defective chloroplasts.
The WSL4 Gene Encodes a Novel P-Family PPR Protein
Genetic analyses of reciprocal crosses between the WT and wsl4 mutant showed that the wsl4 phenotype was inherited by a single recessive nuclear mutation (Table 2). Using the F2 population derived from the wsl4/Yue13 cross, the wsl4 locus was initially mapped to the long arm of rice chromosome 2, between the insertion/deletion markers L-3 and L-9 (Figure 3A). Based on 978 F2 recessive homozygous wsl4 background individuals, the WSL4 locus was ultimately narrowed down to an 86-kb genomic region flanked by the insertion/deletion markers L-37 and L-26 on the BAC clone P0020C11, in which nine putative open reading frames (ORFs) were predicted (Figure 3B). Comparison with WT cDNA sequences revealed a 2 bp deletion of positions 1,330 and 1,331 bp from the ATG start codon in the 7th ORF (LOC_Os02g35750) (Figure 3C), generating a premature stop codon. Sequence comparison between genomic and cDNA showed that the WSL4 gene contained a single exon, which was predicted to encode a 554 amino acid residue polypeptide with a calculated molecular mass of approximately 60 kDa. By contrast, the wsl4 gene, if translated, would encode a truncated protein, named wsl4, and missing the C-terminal 111 amino acid residues, but with five added unrelated amino acid residues resulting from translation of the frame-shift (Supplementary Figure S1).
FIGURE 3. Map-based cloning of the WSL4 gene. (A) The WSL4 locus was initially mapped on the long arm of chromosome 2 between the insertion/deletion markers L-3 and L-9. (B) The WSL4 locus was ultimately narrowed to an 86-kb region between the insertion/deletion markers L-37 and L-26 on BAC clone P0020C11 using 978 F2 homozygous mutant background plants. Nine open reading frames (ORFs) were predicted in the region. (C) Comparison with WT cDNA sequence revealed a 2 bp deletion (underlined) in ORF7, generating a premature stop codon. (D) Phenotypic comparison at the three-leaf stage of WT, wsl4 mutant, complemented T1 (Com), and RNAi T1 plants grown in a growth chamber at L30/D25. Bars, 0.5 cm. (E) Chlorophyll contents of L3-3 from plants corresponding to (D). L3-3 indicates the third leaf at the three-leaf stage. Values are means ± SD from three independent repeats. Student’s t-test: ∗P < 0.05; ∗∗P < 0.01. Chl a, chlorophyll a, Chl b, chlorophyll b, FW, fresh weight. (F) Real-time RT-PCR analyses of expression levels of the WSL4 gene in L3-3 of plants corresponding to (D). The ubiquitin gene was used as an internal control. Data are means ± SD of three repeats. Student’s t-test: ∗P < 0.05; ∗∗P < 0.01.
To test whether the 2 bp deletion conferred the wsl4 mutant phenotype, a 5.7 kb WT genomic fragment harboring the transcriptional regulation elements and putative full-length coding sequence of LOC_Os02g35750 was introduced into the wsl4 mutant. All six positive transgenic lines exhibited green leaves, and had comparable levels of Chl a and b to the WT (Figures 3D,E). To further confirm that disruption of LOC_Os02g35750 function was responsible for the wsl4 mutant phenotype, we generated RNAi transgenic lines in WT background and obtained four independent transgenic lines, that phenocopied the wsl4 mutant with respect to leaf color and Chl contents (Figures 3D,E). Thus LOC_Os02g35750 corresponds to the WSL4 gene.
An analysis of WSL4 protein in the Pfam database (Finn et al., 2010) showed that it contained a tandem repeat of 12 PPR motifs and belonged to a member of the P subfamily (Figure 4A). The 12 PPR motifs in WSL4 protein were sequentially arranged, with an average 31.5% sequence identity among them (Figure 4B). The 2 bp deletion in wsl4 mutant occurred at the middle of the 10th PPR motif site, resulting in loss of the last three PPR repeats (Figure 4A and Supplementary Figure S1). The fact that wsl4 mutant exhibited the same or a slightly weaker phenotype (Figures 3D–F) and splicing defects (see below) as the WSL4 knock-down transgenic lines suggests that the 2 bp deletion in wsl4 mutant causes the partial loss-of-function mutation.
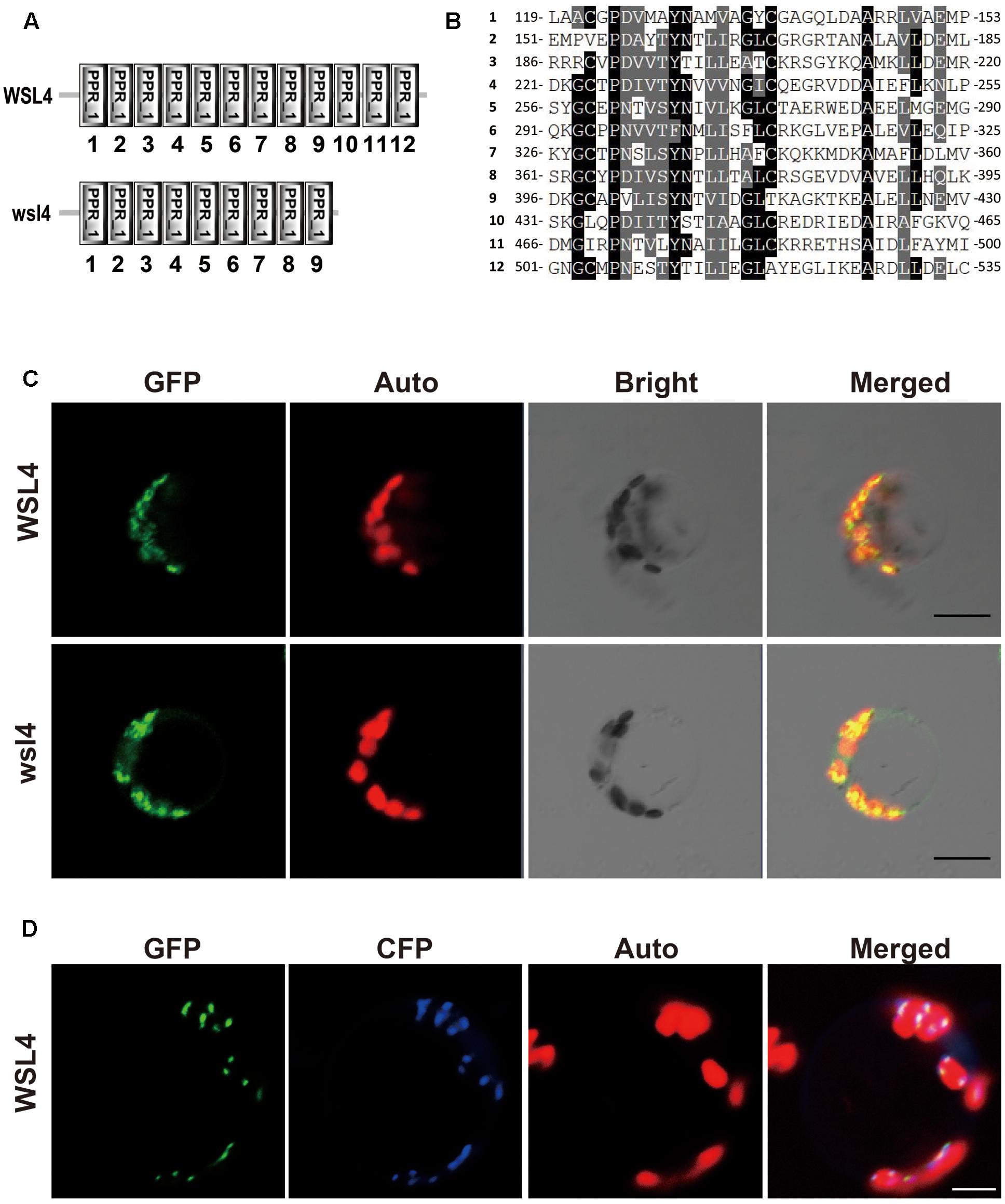
FIGURE 4. Sequence analysis and subcellular localization of the WSL4 protein. (A) WSL4 protein has 12 PPR motifs, whereas the wsl4 mutant protein contains only 9. (B) The 12 PPR motifs of WSL4. (C) Transient expression of WSL4–GFP and wsl4–GFP fusion proteins in rice protoplasts. (D) Colocalization of WSL4 (GFP) and PEND (CFP) within chloroplast nucleoids. GFP, GFP signals of WSL4 fusions; CFP, CFP signals of chloroplast nucleoid marker PEND; Auto, Chl autofluorescence; Merged, Merged images of GFP, CFP, and Auto. Bars, 5 μm.
A BLAST search of the NCBI database showed that WSL4 is a single-copy gene in the rice genome, and homologs can be found in many other plant genomes. We performed phylogenetic analyses of WSL4 and these homologs to determine their evolutionary relatedness. The WSL4 protein shared high sequence homology with homologs in Brachypodium distachyon, Hordeum vulgare, Setaria italic, Zea mays, Sorghum bicolor, and Aegilops tauschii (Supplementary Figures S2, 3); however, the functions of these genes including the Arabidopsis homolog were unknown. The WSL4 protein thus defined a new member of the P-superfamily of PPR proteins.
Subcellular Localization of WSL4 Protein
The prediction of a chloroplast transit peptide at the N-terminus of WSL4 by ChloroP8 and TargetP9, together with the Chl-deficient phenotype of wsl4 mutant, suggests that WSL4 is localized to chloroplasts. To determine its actual localization, we transiently expressed p35S:WSL4-GFP in rice protoplasts. As shown in Figure 4C confocal microscopy observations showed that WSL4-GFP was localized to punctate compartments associated with Chl autofluorescence. Moreover, we noted that wsl4-GFP was still targeted to the chloroplast, suggesting that the 111 missing amino acid residues at the C-terminus of WSL4 were not necessary for chloroplast localization (Figure 4C). To further determine the nature of these small dot-like structures, we coexpressed WSL4-GFP and CFP-tagged plastid binding protein PEND-CFP known to be a fluorescent marker protein for plastid nucleoids (Terasawa and Sato, 2005). As expected, the GFP signals colocalized with the CFP signals, indicating that WSL4 was localized to the chloroplast nucleoids (Figure 4D). These results show that WSL is indeed a chloroplast nucleoid-localized protein.
Expression Analyses of WSL4 and Genes Associated with Chloroplast Development
A survey of the rice expression profile database10,11 revealed that WSL4 was constitutively expressed in all tissues examined, with the strongest expression in the young leaves. For validation, expression analyses performed by quantitative RT-PCR (qRT-PCR) using RNA samples prepared from culms (C), flag leaves (FL), young leaves (YL), shoot bases (SB), young panicles (P), young roots (R), and leaf sheaths (LS) showed that the abundance of WSL4 transcript was significantly higher in young leaves compared to other tissues (Figure 5A). These expression data together with the observed wsl4 phenotype support the hypothesis that WSL4 has an important role at chloroplast development stage during the early leaf development in rice. Interestingly, accumulation of WSL4 transcript was enhanced under low temperature conditions (Figure 5B).
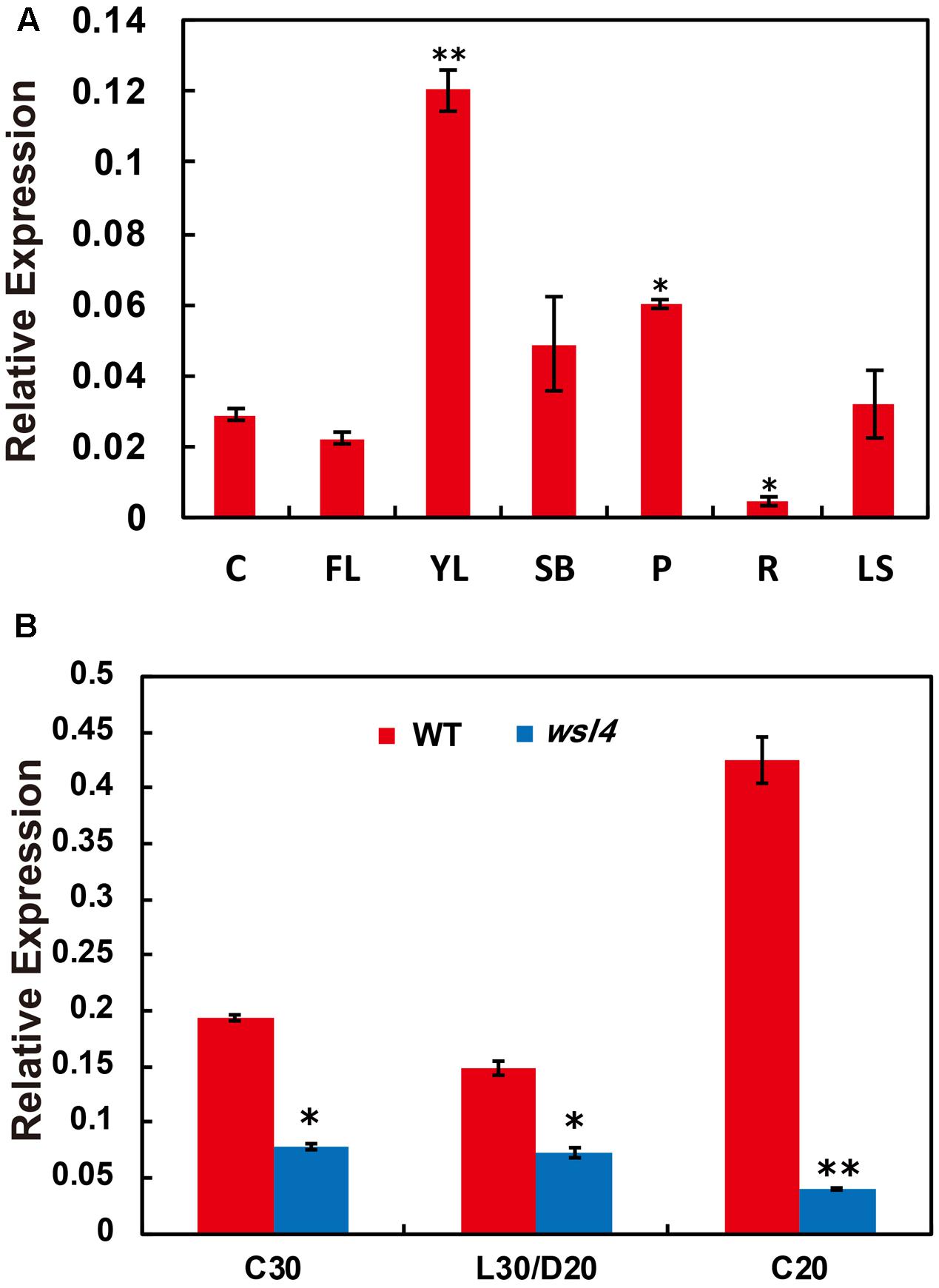
FIGURE 5. Expression analyses of WSL4, chloroplast-encoded, and nuclear-encoded genes as well as immunoblot analyses of plastidic proteins. (A) Relative expression of WSL4 in culms (C), flag leaf (FL), young panicles (P), and leaf sheaths (LS) from field-grown WT plants grown in the field at the booting stage, as well as young leaves (YL), shoot bases (SB), and young roots (R) from WT seedlings grown in a growth chamber at L30/D25. For t-test, values were compared with expression level of WSL4 in culms. Student’s t-test: ∗P < 0.05; ∗∗P < 0.01. (B) qRT-PCR analyses of WSL4 transcript in L3-3 from WT and wsl4 mutant seedlings grown in a growth chamber with a 12 h photoperiod at C30, L30/D25, and C20. Values are means ± SD of three replicates. Student’s t-test: ∗P < 0.05; ∗∗P < 0.01.
Multiple chloroplast-associated genes were investigated to further determine whether the defective chloroplast development was associated with the altered gene expression. As shown in Figure 6A, key photosynthesis-associated nuclear genes (cab1R, cab2R, and rbcS) were significantly down-regulated in wsl4 mutant, suggesting that photosynthesis in wsl4 mutant was compromised. Plastidic genes can be grouped into three types. Class I genes are mainly transcribed by PEP, whereas Class III genes are absolutely transcribed by NEP. In contrast, NEP functions cooperatively with PEP to regulate the transcription of Class II genes (Hedtke et al., 1997; Yu et al., 2014). We next examined expression levels of Classes I and plastid genes. Expression of PEP-dependent Class I genes (e.g., psaA, psbA, and rbcL) was significantly down-regulated in the wsl4 mutant, whereas expression of NEP-dependent Class III genes (e.g., rpoB, rpoC1, and rpoC2) was significantly up-regulated (Figure 6A). These results suggest that PEP complex activity was also compromised in the wsl4 mutant as well. Consistent with the reduced Chl content, expression of Chl synthesis genes CHLD, CHLI, CHLH, and PORA, but not YGL1, was significantly inhibited in wsl4 mutant (Supplementary Figure S4). We therefore propose that WSL4 might coordinate the expression of numerous nuclear- and plastid-encoded genes to regulate chloroplast development at the early leaf development stage.
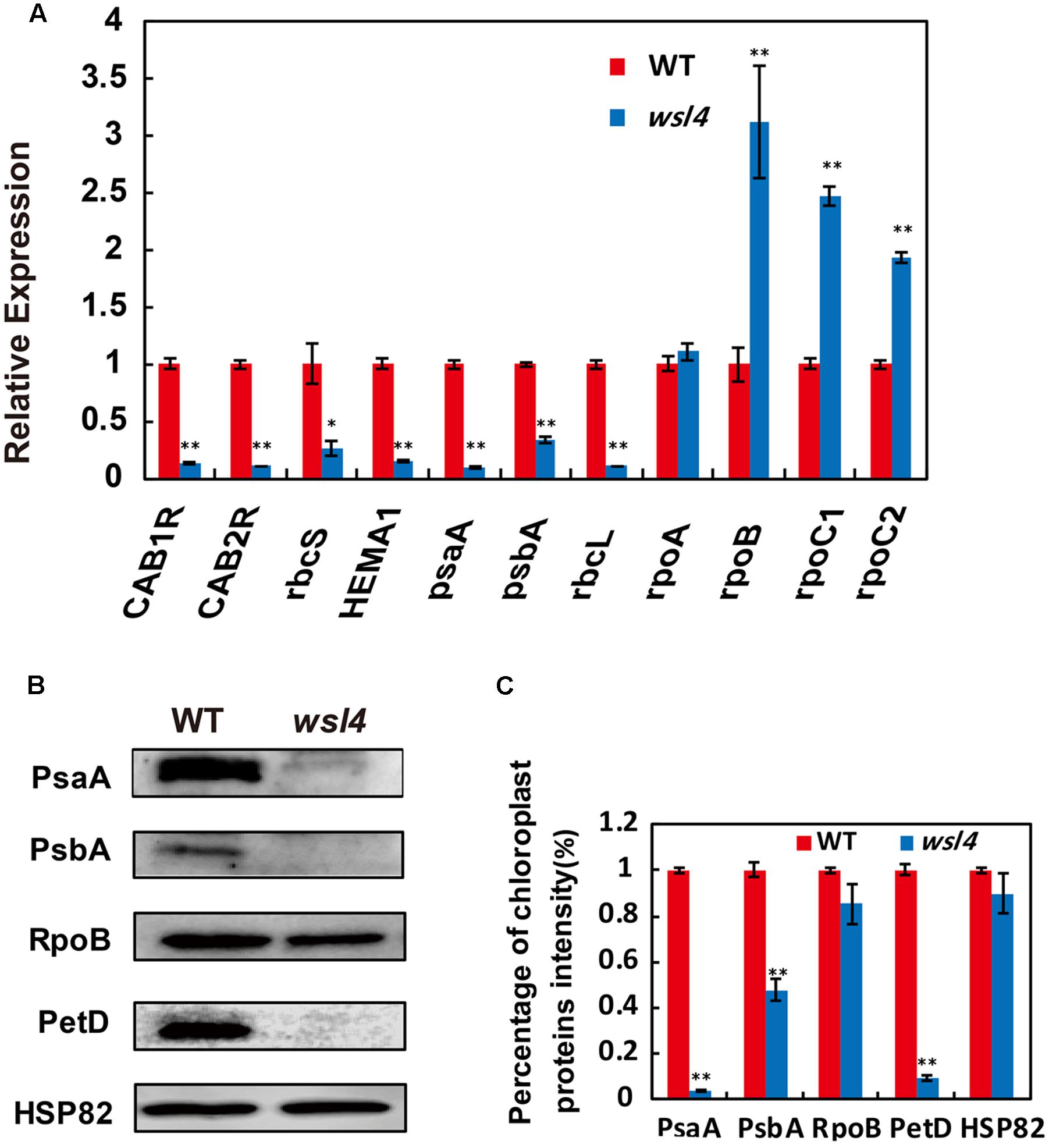
FIGURE 6. Expression analyses of chloroplast-encoded, and nuclear-encoded genes as well as immunoblot analyses of plastidic proteins. (A) qRT-PCR analyses of genes associated with chloroplast development in WT and wsl4. Total RNA was extracted from L3-3 of the corresponding plants grown in a growth chamber at L30/D25. (B) Immunoblot analyses of photosynthetic proteins in WT and the wsl4 mutant. Total proteins were extracted from L3-3 of WT and wsl4 mutant plants grown in a growth chamber at L30/D25. L3-3 indicates the third leaf at the three-leaf stage. HSP82 was used as an internal control. (C) Quantification of the band intensity of photosynthetic proteins in the wsl4 mutant compared- to WT corresponding to (B). Values are means ± SD of three replicates. Student’s t-test: ∗P < 0.05, ∗∗P < 0.01.
We finally assessed the accumulation of core subunits of photosynthetic enzyme complexes in WT and the wsl4 mutant, including photosystem I (PsaA and PsaB) and photosystem II (PsbA and PsbD) subunits, RNA polymerase subunits (RpoB), ATP synthase CF1 β subunit (AtpB), and cytochrome b6f (PetD). All assayed proteins in bleached tissues, except RpoB, were barely detectable in the wsl4 mutant compared to WT (Figures 6B,C).
WSL4 Affects the Editing Efficiency of rpoB Gene
Accumulating evidence showed that a large group of nuclear-encoded PPR proteins required for RNA editing, splicing, stability, and translation were critical for chloroplast development (Hattori et al., 2007; Beick et al., 2008; Schmitz-Linneweber and Small, 2008; Barkan et al., 2012; Tan et al., 2014; Hsieh et al., 2015; Hammani et al., 2016; Xie et al., 2016). Firstly, we determined whether loss of WSL4 function affected editing at 21 identified RNA editing sites in chloroplast RNA (Corneille et al., 2000). The results showed that the editing efficiency of rpoB at C545 and C560 exhibited a significant increase in wsl4 mutant compared with WT (Figure 7), while the rest 11 genes and the corresponding 19 editing sites were normally edited in wsl4 mutant (Supplementary Figure S5). Furthermore, we analyzed the editing efficiency of rpoB at C545 and C560 in complemented and RNAi transgenic plants. As expected, the editing efficiency of rpoB at C545 and C560 showed a markedly reduction in complemented plant, while the RNAi plant mimicked a similar increase in RNA editing as the wsl4 mutant (Figure 7). These data further suggest that the mutation in WSL4 affects the editing efficiency of rpoB gene.
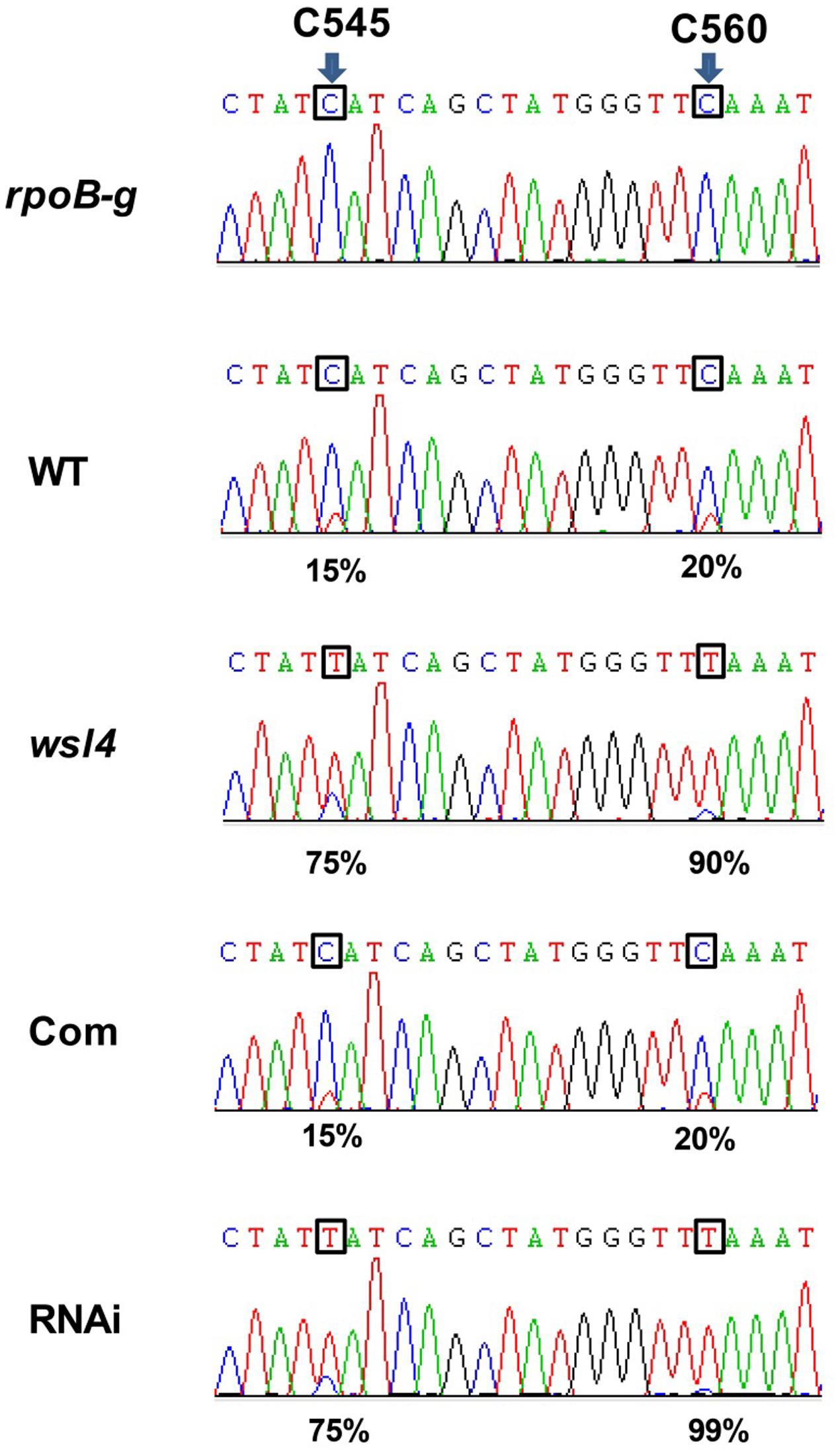
FIGURE 7. Editing efficiencies of rpoB genes in WT and the wsl4 mutant. RT-PCR products of rpoB transcripts from L3-3 of WT, wsl4 mutant (white striped sector), complemented (Com), and RNAi (white striped sector) plants grown in a growth chamber at L30/D25 were sequenced. In wsl4 mutant and RNAi plants, the editing efficiency of rpoB at C545 and C560 were significantly increased compared to WT and Com. The top panel corresponds to the genomic nucleotide sequence. Green, black, red, and blue peak represent A, G, T, and C, respectively. Arrowheads indicate the editing sites and their position in the corresponding chloroplast cDNA strand. L3-3 indicates the third leaf at the three-leaf stage.
The wsl4 Mutant Is Defective in the Splicing of Chloroplast Group II Introns
Accumulating evidence shows that a large group of nuclear-encoded PPR proteins required for RNA editing, splicing, stability, maturation, and translation are critical for chloroplast development (Hattori et al., 2007; Beick et al., 2008; Barkan et al., 2012; Tan et al., 2014; Hsieh et al., 2015; Hammani et al., 2016; Wu et al., 2016; Xie et al., 2016; Zoschke et al., 2016; Zhou et al., 2017). To determine whether loss of WSL4 function affected RNA splicing of chloroplast genes containing 17 group II introns and one group I intron (Hiratsuka et al., 1989), we carried out RT-PCR analyses using primers situated in exons flanking intron 1, and then compared the lengths of the amplified products between WT and wsl4 mutant plants. Four chloroplast transcripts containing group II introns were spliced with much reduced efficiency in wsl4 mutant compared to WT (Figures 8, 9A). The splicing defects were largely rescued in complemented transgenic plants and were mimicked in RNAi transgenic plants (Figure 9A). As a control, the splicing efficiencies of atpF, ndhA, rpl2, and rps12 intron 2 were not significantly affected in the wsl3 mutant (Figure 9A), a rice chloroplast translation-defective mutant lacking an essential peripheral subunit of the PEP complex (Wang L. et al., 2016). Splicing of other chloroplast transcripts was not significantly impeded in wsl4 mutant (Figure 8).
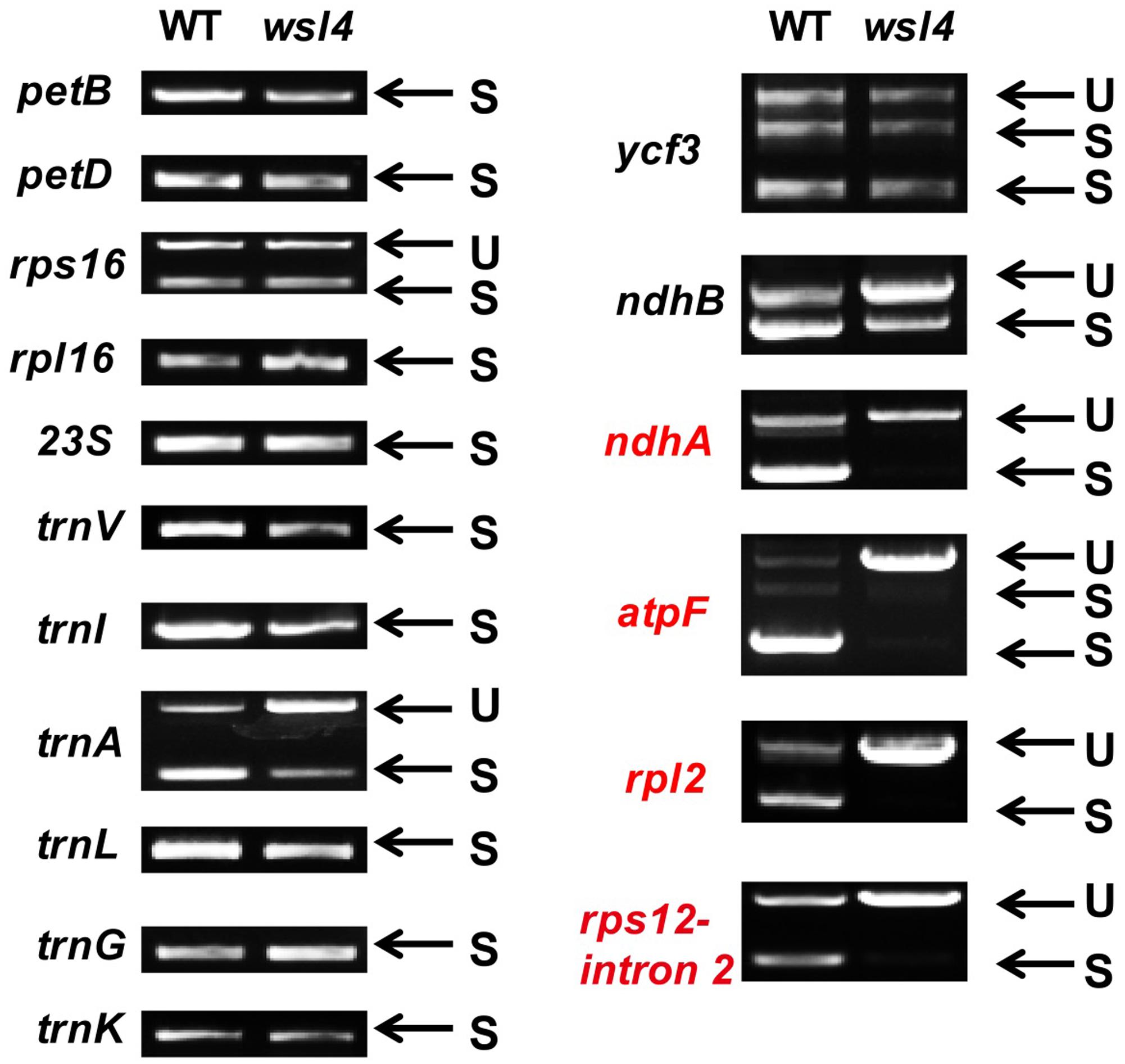
FIGURE 8. Splicing analyses of rice chloroplast transcripts in WT and wsl4 mutant (white striped sectors). Gene transcripts are labeled at the left. Spliced (S) and unspliced (U) transcripts are shown at the right. RNA was extracted from L3-3 of WT and wsl4 mutant plants grown in a growth chamber at L30/D25. L3-3 indicates the third leaf at the three-leaf stage.
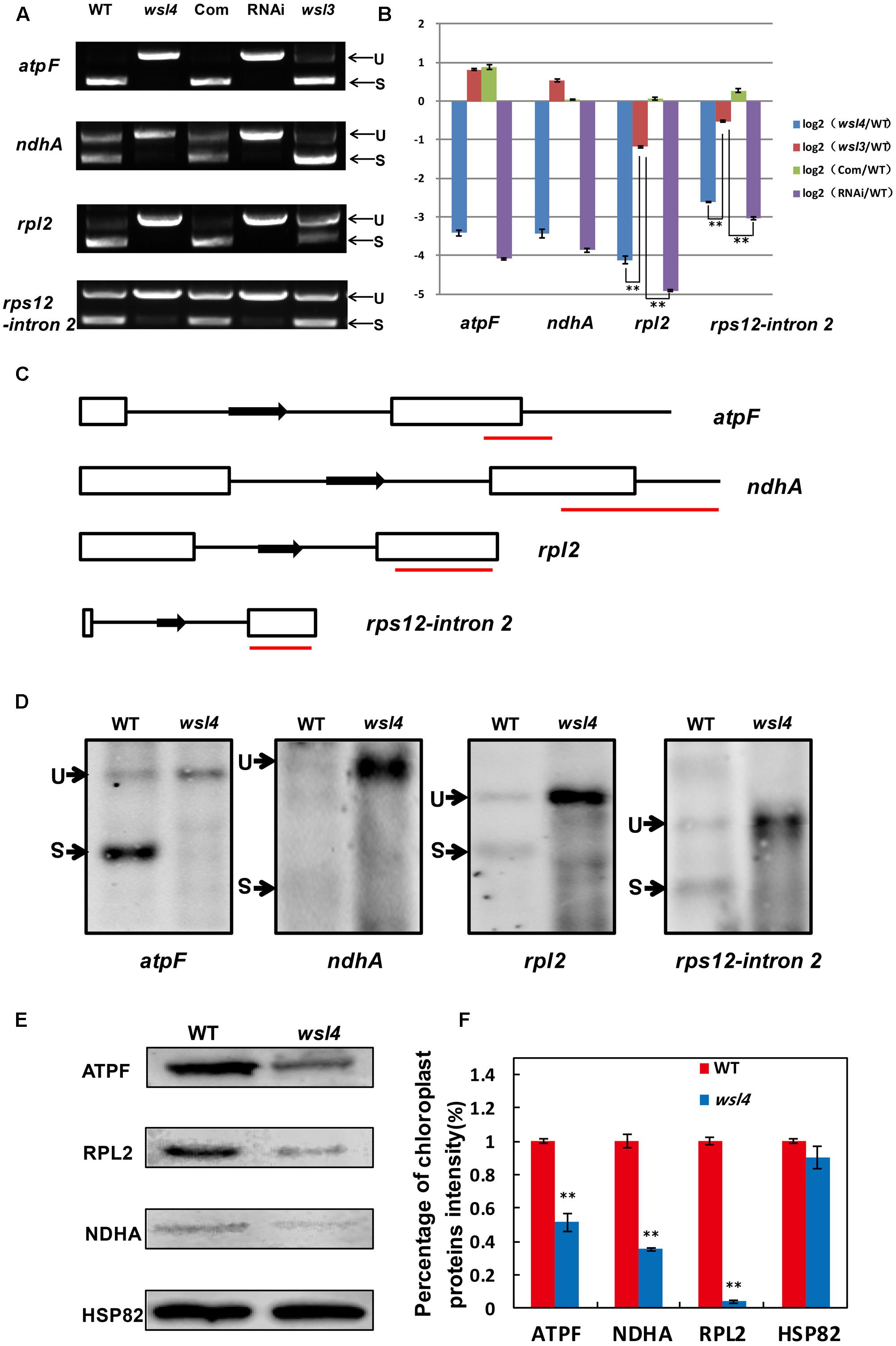
FIGURE 9. Splicing analyses of four chloroplast group II introns in WT and wsl4 mutant. (A) RT-PCR analyses of atpF, ndhA, rpl2, and rps12 transcripts in WT, wsl4 mutant (white striped sector), complemented (Com), RNAi (white striped sector), and a control wsl3 mutant with a transitory variegation phenotype. RT–PCR was performed with RNA extracted from L3-3 of the corresponding plants grown in a growth chamber at L30/D25. L3-3 indicates the third leaf at the three-leaf stage. (B) Quantitative RT-PCR analyses of atpF, ndhA, rpl2, and rps12 transcripts in WT, wsl4 mutant (white striped sector), Com, RNAi (white striped sector), and the wsl3 mutant plants grown in a growth chamber at L30/D25. Histograms show log2 ratios of spliced to unspliced RNA in wsl4, wsl3, Com, and RNAi plants as compared with WT. Values are means ± SD of three replicates. (C) Sketch map of the atpF, ndhA, rpl2, and rps12 transcripts. The red lines represent the probes used in (D). (D) RNA gel blot assays of atpF, ndhA, rpl2, and rps12 splicing in WT and wsl4 mutant. Positions of spliced (S) and unspliced (U) transcripts are shown at the left. RNA was extracted from L3-3 of WT and wsl4 mutant plants grown in a growth chamber at L30/D25. L3-3 indicates the third leaf at the three-leaf stage. (E) Immunoblot analyses of ATPF, NDHA, and RPL2 in WT and wsl4 mutant at the three-leaf stage. Total proteins were extracted from L3-3 of WT and wsl4 mutant plants grown in a growth chamber at L30/D25. L3-3 indicates the third leaf at the three-leaf stage. HSP82 was used as an internal control. (F) Quantification of the band intensity of ATPF, NDHA, and RPL2 in wsl4 mutant compared to WT corresponding to (E). Data are means ± SD of three repeats. Student’s t-test: ∗P < 0.05; ∗∗P < 0.01.
Quantitative RT-PCR was performed to quantify spliced (using primers situated in exons 1 and 2) and unspliced transcripts (using primers situated in exon 1 and intron 1) to further determine the extent of the splicing defects, Compared with wsl3, wsl4 and RNAi transgenic plants showed strong splicing defects in atpF, ndhA, rpl2, and rps12 transcripts (Figure 9B). Consistent with phenotypic expression, the splicing defects of atpF, ndhA, rpl2, and rps12 in wsl4 mutant appeared to be worse in plants frown at 20°C (Supplementary Figure S6) suggesting that loss of WSL4 function caused the temperature-sensitive leaf bleaching and splicing defects at the early leaf developmental stage. The defective splicing of these four chloroplast transcripts was confirmed by RNA gel blot hybridizations using specific probes (Figures 9C,D). As a means of further investigating the effects of impaired splicing of atpF, ndhA, and rpl2 transcripts on post-processing, western blots were performed to examine protein accumulations of atpF, ndhA, and rpl2 in wsl4 mutant. As shown in Figures 9E,F, atpF, ndhA, and rpl2 were present at lower levels in wsl4 mutant compared with WT. We propose that WSL4 functions in the splicing of chloroplast group II introns.
Discussion
Previous studies isolated and characterized numerous rice leaf color mutants. Based on phenotype such mutants have been subdivided into various classes, including albino, chlorina, stripe, virescent, and zebra12 (Jung et al., 2003). In this study, we isolated a rice stripe leaf mutant named wsl4. In contrast to mutants exhibiting Chl deficient phenotypes throughout plant development, such as ygl1 and vyl (Wu et al., 2007; Dong et al., 2013), wsl4 is characterized by a transient variegated phenotype during early seedling leaf and tiller development (Figure 1). Why wsl4 and some other mutants exhibit a stage-specific bleaching phenotype is an intriguing question. It is possible that WSL4 is not required at later leaf developmental stages when other genes compensate for its function in chloroplast development. Indeed, WSL4 is actually expressed at lower levels in more mature leaves such as flag leaves than in young leaves (Figure 5A). The ysa mutant likewise exhibits a seedling-specific albino phenotype, and the expression level of YSA, encoding a PPR protein, decreases with leaf development (Su et al., 2012).
The bleached tissue sectors in the wsl4 mutant stretch longitudinally (striping), but the stripes do not extend along the entire leaf (Figures 1, 2), suggesting that other factors might affect the expression of the wsl4 phenotype. The white tissue sections in the wsl4 mutant are devoid of normal chloroplasts and show significantly reduced expression of genes associated with Chl synthesis and chloroplast development. Low temperature is a key factor affecting chloroplast gene expression and particularly in exacerbating chloroplast translation defects caused by various mutations, such as ribosomal proteins (Gong et al., 2013; Song et al., 2014; Lin et al., 2015b) and RNA binding proteins (Gong et al., 2014; Tan et al., 2014). Our results show that wsl4 is a low temperature-sensitive mutant (Figure 2). Furthermore expression of WSL4 was increased at lower temperatures (Figure 5B). These results may be a reason for low temperature dependency of wsl4 phenotype. Similar low temperature dependency phenotypes have also been observed in several previously reported mutants, such as v1, v2, v3, and st1 (Sugimoto et al., 2007; Yoo et al., 2009; Kusumi et al., 2011).
PEP-dependent genes (e.g., psaA, psbA, and rbcL) and nuclear genes associated with photosynthesis (e.g., cab1R, cab2R, and rbcS) were significantly down-regulated in the wsl4 mutant, whereas expression levels of NEP-dependent genes were increased (Figure 6). These results together with decreased accumulation of the main photosynthetic protein (Figure 6) and the Chl deficient phenotype (Figure 1) suggest that PEP function may be impaired in wsl4 mutant. Similar phenotypes were observed in several previously reported PEP-deficient mutants including ptac2, clb19, ys1, and otp70 (Pfalz et al., 2006; Chateigner-Boutin et al., 2008; Zhou et al., 2009; Chateigner-Boutin et al., 2011). We also found there was a lack of correlation between gene expression and protein accumulation for some class I (such as psbA) and class III (rpoB) products. This discrepancy between transcript and protein levels may be due to feedback regulation or post-transcriptional modification.
RNA splicing is the process of removing introns between neighboring exons during translation (Lorkoviæ et al., 2000). Many RNA binding proteins have essential roles in RNA splicing of introns in plant plastids, including maturase proteins, CRM (Chloroplast RNA splicing and ribosome maturation) proteins, PPR proteins, and other nuclear-encoded factors (de Longevialle et al., 2010). To date, five PPR proteins have been reported to be involved in RNA splicing of group II introns in chloroplasts. Among them, the maize PPR4 protein acts as an rps12 trans-splicing factor (Schmitz-Linneweber et al., 2006), and PPR5, a maize P-class PPR protein, stabilizes trnG-UCC tRNA precursors in chloroplasts by binding and protecting the endonuclease-sensitive site; its mutation indirectly influences splicing of trnG-UCC RNAs (Beick et al., 2008). The Arabidopsis PPR protein OTP51 functions as a plastid ycf3-2 intron cis-splicing factor (de Longevialle et al., 2008). The Arabidopsis E-class PPR protein OTP70 has been implicated in splicing of the plastid transcript rpoC1 (Chateigner-Boutin et al., 2011). The maize short PPR protein THA8 is associated with splicing of specific ycf3-2 and trnA group II introns in chloroplasts (Khrouchtchova et al., 2012). In this study, the wsl4 mutant caused defects in the splicing of atpF, ndhA, rpl2, and rps12 (Figure 9), implying that WSL4 probably involves chloroplast RNA intron splicing during early leaf development in rice. To confirm this, further studies will be needed in future. Subcellular localization showed that WSL4 protein is a chloroplast nucleoid-localized protein (Figure 4), suggesting that it may participate in plastid RNA metabolism.
Plastidial maturase MatK is involved in RNA splicing by interacting with seven group II introns including trnV, trnI, trnA, trnK, atpF, rpl2, and rps12 intron 2 (Zoschke et al., 2010). It is possible that the splicing defects of atpF, rpl2, and rps12 transcripts (Figure 9) are due to lack of plastidial maturase MatK in the wsl4 mutant. A similar phenomenon was observed in the Arabidopsis emb2654 mutant (Aryamanesh et al., 2017). Given that ndhA is not a target of MatK, it is likely that WSL4 functions in the splicing of ndhA directly. Similar RNA splicing events of ndhA were observed in many other Chl deficient mutants, such as emb2654, ppr53, otp70, clb19, and sot1 (Chateigner-Boutin et al., 2008; Chateigner-Boutin et al., 2011; Wu et al., 2016; Zoschke et al., 2016; Aryamanesh et al., 2017). Therefore, it is also possible that the splicing defect of ndhA is a secondary effect of disrupted chloroplast development or reduced PEP activity in the wsl4 mutant.
RNA editing is a post-transcriptional process that alters RNA sequences by converting specific target cytidines to uridine in both plastid and mitochondrial transcripts (Takenaka et al., 2013). Our results showed that the editing efficiency of rpoB at C545 and C560 in WT is relatively low (15 and 20%, respectively), but almost complete in the wsl4 mutant (75 and 90%, respectively) (Figure 7 and Supplementary Figure S6). This inverse relationship between rpoB editing and the presence of WSL4 makes it extremely unlikely that WSL4 has a direct role in editing of rpoB. This phenotypic defect is most reminiscent of the previously reported iojap mutant in maize (that lacks chloroplast ribosomes) in which the editing sites in rpoB are highly edited (Halter et al., 2004). A possible explanation is that the lack of plastid translation may be indirectly responsible for efficient rpoB editing. In the Arabidopsis PEP-deficient mutant otp70 editing efficiency of unspliced rpoC1 transcripts is almost complete, but is relatively low in WT (Chateigner-Boutin et al., 2011). A reasonable explanation is that the editing of rpoC1 in otp70 mutant is influenced by the changes in splicing. Therefore, altered editing is most likely a secondary effect of defective splicing of chloroplast group II introns in the wsl4 mutant. Further studies on searching for interacting partners of WSL4- will help to uncover the regulatory mechanism of chloroplast development during the early leaf development.
Author Contributions
JW and YiW conceived the research. YiW, KZ, YR, YX, HZ, LZ, ZF, LW, and WM performed the experiments. JW provided the mutant material. YR, JW, XG, XZ, CL, YuW, and ZC provided the technical assistance. YiW, YR, and LL analyzed the data and wrote the manuscript. All authors declared no conflicting interest on the contents of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Oren Ostersetzer-Biran and three experts for their constructive comments on this manuscript. This work was supported by the Fundamental Research Funds for Excellent Young Scientists of ICS-CAAS (Grants to YR, 2014JB04-009; 1610092015003-08), Grants from the National Natural Science Foundation of China Grants 31671657, 31571629 and 31401406), and the National Transgenic Science and Technology Program (Grants 2015ZX08010-004 and 2016ZX08009003-003).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01116/full#supplementary-material
FIGURE S1 | Amino acid sequence alignment of the WSL4 and wsl4 proteins. Premature translation in the wsl4 mutant generated a truncated protein lacking 111 amino acids residues at the C-terminal, compared to WT WSL4 protein.
FIGURE S2 | Phylogenetic tree of WSL4 and its homologs in plants. Numbers near the major branches are bootstrap values. GenBank accession numbers for these proteins are: XP_010235566.1 (Brachypodium distachyon), BAK03057.1 (Hordeum vulgare), XP_004952727.1 (Setaria italica), NP_001141010.1 (Zea mays), KXG30467.1 (Sorghum bicolor), KQL30111.1 (Setaria italica), EMT15761.1 (Aegilops tauschii), XP_010917143.1 (Elaeis guineensis), XP_009402131.1 (Musa acuminata subsp. malaccensis), XP_015886049.1 (Ziziphus jujuba), XP_009800134.1 (Nicotiana sylvestris), XP_017981749.1 (Theobroma cacao), XP_008444958.1 (Cucumis melo), XP_010104578.1 (Morus notabilis), XP_002452305.1 (Sorghum bicolor), XP_011071341.1 (Sesamum indicum), XP_012840102.1 (Erythranthe guttata), XP_010671063.1 (Beta vulgaris subsp. vulgaris), XP_015935032.1 (Arachis duranensis), XP_017611896.1 (Gossypium arboreum), XP_016163747.1 (Arachis ipaensis), XP_009366374.1 (Pyrus x bretschneideri), XP_011649703.1 (Cucumis sativus), XP_010254618.1 (Nelumbo nucifera), XP_016576050.1 (Capsicum annuum), XP_012450936.1 (Gossypium raimondii), XP_016576051.1 (Capsicum annuum), XP_006433766.1 (Citrus clementina), XP_010321813.1 (Solanum lycopersicum), XP_016163746.1 (Arachis ipaensis), XP_006472405.1 (Citrus sinensis), XP_017189272.1 (Malus domestica), XP_015078222.1 (Solanum pennellii), XP_006364562.1 (Solanum tuberosum), XP_017178849.1 (Malus domestica), XP_002285611.1 (Vitis vinifera), XP_016464120.1 (Nicotiana tabacum), XP_008219211.2 (Prunus mume), XP_010544054.1 (Tarenaya hassleriana), XP_011016979.1 (Populus euphratica), XP_009595574.1 (Nicotiana tomentosiformis), XP_015579793.1 (Ricinus communis), XP_013696710.1 (Brassica napus), XP_014502416.1 (Vigna radiata var. radiata), XP_002889775.1 (Arabidopsis lyrata), XP_010061566.1 (Eucalyptus grandis), NP_172461.1 (Arabidopsis thaliana), XP_017422523.1 (Vigna angularis), XP_013640951.1 (Brassica napus), XP_009148305.1 (Brassica rapa), XP_010490447.1 (Camelina sativa), XP_011627875.1 (Amborella trichopoda), XP_004288876.1 (Fragaria vesca), XP_017249274.1 (Daucus carota), XP_013585784.1 (Brassica oleracea), XP_003523769.1 (Glycine max), XP_017244771.1 (Daucus carota), XP_003527866.1 (Glycine max), XP_012068070.1 (Jatropha curcas), XP_004501057.1 (Cicer arietinum), XP_006306156.1 (Capsella rubella), KVH98829.1 (Cynara cardunculus var), KYP46243.1 (Cajanus cajan), KHN08231.1 (Glycine soja), KMZ57347.1 (Zostera marina), ERN18052.1 (Amborella trichopoda).
FIGURE S3 | Amino acid sequence alignment of WSL4 and its close homologs. Sequences are from Oryza sativa WSL4, Brachypodium distachyon XP_010235566.1, Hordeum vulgare BAK03057.1, Sorghum bicolor XP_002452305.1, Sorghum bicolor KXG30467.1, Zea mays NP_001141010.1, Setaria italica XP_004952727.1, Setaria italica KQL30111.1, Aegilops tauschii EMT15761.1, and Arabidopsis thaliana NP_172461.1.
FIGURE S4 | Quantitative RT-PCR analyses of genes related to chlorophyll biosynthesis in WT and the wsl4 mutant. RNA was extracted from L3-3 of WT and wsl4 mutant plants grown in a growth chamber under L30/D25. L3-3 indicates the third leaf at the three-leaf stage. Values are means ± SD of three replicates. (Student’s t-test: ∗P < 0.05, ∗∗P < 0.01).
FIGURE S5 | RNA editing analyses of chloroplast genes in WT and wsl4 mutant. Arrowheads indicate editing sites and their positions in the corresponding chloroplast cDNA strand. RNA was extracted from L3-3 of WT and wsl4 mutant (white striped sector) plants grown in a growth chamber at L30/D25. L3-3 indicates the third leaf at the three-leaf stage.
FIGURE S6 | Low temperature exacerbated splicing defects of atpF, ndhA, rpl2, and rps12 transcripts in wsl4 mutant. qRT-PCR analyses of atpF, ndhA, rpl2, and rps12 transcripts in L3-3 from WT and wsl4 mutant (white striped sectors) seedlings grown in a growth chamber with 12 h photoperiod at C30, L30/D25, and C20. L3-3 indicates the third leaf at the three-leaf stage. Histograms show log2 ratios of spliced to unspliced RNA in wsl4 compared to WT. Values are means ± SD of three replicates. (Student’s t-test: ∗P < 0.05, ∗∗P < 0.01).
Abbreviations
Chl, chlorophyll; GFP, green fluorescent protein; PPR, pentatricopeptide repeat; RNAi, RNA interference; RT–PCR, reverse transcription–PCR; TEM, transmission electron microscopy; WT, wild type.
Footnotes
- ^http://www.gramene.org/
- ^www.gramene.org/
- ^http://www.ncbi.nlm.nih.gov/
- ^http://www.cbs.dtu.dk/services/ChloroP/
- ^http://www.cbs.dtu.dk/services/TargetP/
- ^http://www.megasoftware.net/
- ^http://www.genomics.cn/index
- ^http://www.cbs.dtu.dk/services/ChloroP/
- ^http://www.cbs.dtu.dk/services/TargetP/
- ^http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/
- ^http://ricexpro.dna.affrc.go.jp/GGEP/
- ^http://www.shigen.nig.ac.jp/rice/oryzabase/top/top.jsp
References
Aryamanesh, N., Ruwe, H., Sanglard, L. V., Eshraghi, L., Bussell, J. D., Howell, K. A., et al. (2017). The pentatricopeptide repeat protein EMB2654 is essential for trans-splicing of a chloroplast small ribosomal subunit transcript. Plant Physiol. 173, 1164–1176. doi: 10.1104/pp.16.01840
Barkan, A., Rojas, M., Fujii, S., Yap, A., Chong, Y. S., Bond, C. S., et al. (2012). A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8:e1002910. doi: 10.1371/journal.pgen.1002910
Barkan, A., and Small, I. (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65, 415–442. doi: 10.1146/annurev-arplant-050213-040159
Beick, S., Schmitz-Linneweber, C., Williams-Carrier, R., Jensen, B., and Barkan, A. (2008). The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cell. Biol. 28, 5337–5347. doi: 10.1128/MCB.00563-08
Chateigner-Boutin, A. L., des Francs-Small, C. C., Delannoy, E., Kahlau, S., Tanz, S. K., de Longevialle, A. F., et al. (2011). OTP70 is a pentatricopeptide repeat protein of the E subgroup involved in splicing of the plastid transcript rpoC1. Plant J. 65, 532–542. doi: 10.1111/j.1365-313X.2010.04441.x
Chateigner-Boutin, A. L., Ramos-Vega, M., Guevara-Garcia, A., Andres, C., de la Luz Gutierrez-Nava, M., Cantero, A., et al. (2008). CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 56, 590–602. doi: 10.1111/j.1365-313X.2008.03634.x
Chen, S., Tao, L., Zeng, L., Vega-Sanchez, M. E., Umemura, K., and Wang, G. L. (2006). A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol. Plant Pathol. 7, 417–427. doi: 10.1111/j.1364-3703.2006.00346.x
Chiu, W. L., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. doi: 10.1016/S0960-9822(02)00483-9
Corneille, S., Lutz, K., and Maliga, P. (2000). Conservation of RNA editing between rice and maize plastids: are most editing events dispensable? Mol. Gen. Genet. 264, 419–424. doi: 10.1007/s004380000295
de Longevialle, A. F., Hendrickson, L., Taylor, N. L., Delannoy, E., Lurin, C., Badger, M., et al. (2008). The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J. 56, 157–168. doi: 10.1111/j.1365-313X.2008.03581.x
de Longevialle, A. F., Meyer, E. H., Andres, C., Taylor, N. L., Lurin, C., Millar, A. H., et al. (2007). The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 Intron 1 in Arabidopsis thaliana. Plant Cell 19, 3256–3265. doi: 10.1105/tpc.107.054841
de Longevialle, A. F., Small, I. D., and Lurin, C. (2010). Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol. Plant 3, 691–705. doi: 10.1093/mp/ssq025
de Souza, A., Wang, J. Z., and Dehesh, K. (2016). Retrograde signals: integrators of interorganellar communication and orchestrators of plant development. Annu. Rev. Plant Biol. 68, 58–108. doi: 10.1146/annurev-arplant-042916-041007
Dong, H., Fei, G. L., Wu, C. Y., Wu, F. Q., Sun, Y. Y., Chen, M. J., et al. (2013). A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants. Plant Physiol. 162, 1867–1880. doi: 10.1104/pp.113.217604
Finn, R. D., Mistry, J., Tate, J., Coggill, P., Heger, A., Pollington, J. E., et al. (2010). The Pfam protein families database. Nucleic Acids Res. 38, D211–D222. doi: 10.1093/nar/gkp985
Fujii, S., and Small, I. (2011). The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 191, 37–47. doi: 10.1111/j.1469-8137.2011.03746.x
Gong, X., Jiang, Q., Xu, J., Zhang, J., Teng, S., Lin, D., et al. (2013). Disruption of the rice plastid ribosomal protein S20 leads to chloroplast developmental defects and seedling lethality. G3 3, 1769–1777. doi: 10.1534/g3.113.007856/-/DC1
Gong, X., Su, Q., Lin, D., Jiang, Q., Xu, J., Zhang, J., et al. (2014). The rice OsV4 encoding a novel pentatricopeptide repeat protein is required for chloroplast development during the early leaf stage under cold stress. J. Integr. Plant Biol. 56, 400–410. doi: 10.1111/jipb.12138
Gothandam, K. M., Kim, E. S., Cho, H., and Chung, Y. Y. (2005). OsPPR1, a pentatricopeptide repeat protein of rice is essential for the chloroplast biogenesis. Plant Mol. Biol. 58, 421–433. doi: 10.1007/s11103-005-5702-5
Hajdukiewicz, P. T., Allison, L. A., and Maliga, P. (1997). The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 16, 4041–4048. doi: 10.1093/emboj/16.13.4041
Halter, C. P., Peeters, N. M., and Hanson, M. R. (2004). RNA editing in ribosome-less plastids of iojap maize. Curr. Genet. 45, 331–337. doi: 10.1007/s00294-003-0482-4
Hammani, K., Okuda, K., Tanz, S. K., Chateigner-Boutin, A. L., Shikanai, T., and Small, I. (2009). A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell 21, 3686–3699. doi: 10.1105/tpc.109.071472
Hammani, K., Takenaka, M., Miranda, R., and Barkan, A. (2016). A PPR protein in the PLS subfamily stabilizes the 5’-end of processed rpl16 mRNAs in maize chloroplasts. Nucleic Acids Res. 44, 4278–4288. doi: 10.1093/nar/gkw270
Hattori, M., Miyake, H., and Sugita, M. (2007). A Pentatricopeptide repeat protein is required for RNA processing of clpP Pre-mRNA in moss chloroplasts. J. Biol. Chem. 282, 10773–10782. doi: 10.1074/jbc.M608034200
Hedtke, B., Börner, T., and Weihe, A. (1997). Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science 277, 809–811. doi: 10.1126/science.277.5327.809
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313X.1994.6020271.x
Hiratsuka, J., Shimada, H., Whittier, R., Ishibashi, T., Sakamoto, M., Mori, M., et al. (1989). The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol. Gen. Genet. 217, 185–194. doi: 10.1007/BF02464880
Hsieh, W. Y., Liao, J. C., Chang, C. Y., Harrison, T., Boucher, C., and Hsieh, M. H. (2015). The SLOW GROWTH3 pentatricopeptide repeat protein is required for the splicing of mitochondrial NADH dehydrogenase subunit7 intron 2 in Arabidopsis. Plant Physiol. 168, 490–501. doi: 10.1104/pp.15.00354
Hu, J., Wang, K., Huang, W., Liu, G., Gao, Y., Wang, J., et al. (2012). The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell 24, 109–122. doi: 10.1105/tpc.111.093211
Huang, W., Yu, C., Hu, J., Wang, L., Dan, Z., Zhou, W., et al. (2015). Pentatricopeptide-repeat family protein RF6 functions with hexokinase 6 to rescue rice cytoplasmic male sterility. Proc. Natl. Acad. Sci. U.S.A. 112, k14984–14989. doi: 10.1073/pnas.1511748112
Jarvis, P., and Lopez-Juez, E. (2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802. doi: 10.1038/nrm3702
Jung, K. H., Hur, J., Ryu, C. H., Choi, Y., Chung, Y. Y., Miyao, A., et al. (2003). Characterization of a rice chlorophyll-deficient mutant using the T-DNA gene-trap system. Plant Cell Physiol. 44, 463–472. doi: 10.1093/pcp/pcg064
Kanamaru, K., Fujiwara, M., Seki, M., Katagiri, T., Nakamura, M., Mochizuki, N., et al. (1999). Plastidic RNA polymerase σ factors in Arabidopsis. Plant Cell Physiol. 40, 832–842. doi: 10.1093/oxfordjournals.pcp.a029612
Kangasjärvi, S., Tikkanen, M., Durian, G., and Aro, E. M. (2014). Photosynthetic light reactions–an adjustable hub in basic production and plant immunity signaling. Plant Physiol. Biochem. 81, 128–134. doi: 10.1016/j.plaphy.2013.12.004
Kaur, J. (2014). A comprehensive review on metabolic syndrome. Cardiol. Res. Pract. 2014, 943162. doi: 10.1155/2014/943162
Kazama, T., and Toriyama, K. (2003). A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice. FEBS Lett. 544, 99–102. doi: 10.1016/s0014-5793(03)00480-0
Khrouchtchova, A., Monde, R. A., and Barkan, A. (2012). A short PPR protein required for the splicing of specific group II introns in angiosperm chloroplasts. RNA 18, 1197–1209. doi: 10.1261/rna.032623.112
Kim, S. R., Yang, J. I., Moon, S., Ryu, C. H., An, K., Kim, K. M., et al. (2009). Rice OGR1 encodes a pentatricopeptide repeat-DYW protein and is essential for RNA editing in mitochondria. Plant J. 59, 738–749. doi: 10.1111/j.1365-313X.2009.03909.x
Koprivova, A., des Francs-Small, C. C., Calder, G., Mugford, S. T., Tanz, S., Lee, B. R., et al. (2010). Identification of a pentatricopeptide repeat protein implicated in splicing of intron 1 of mitochondrial nad7 transcripts. J. Biol. Chem. 285, 32192–32199. doi: 10.1074/jbc.M110.147603
Kusumi, K., and Iba, K. (2014). Establishment of the chloroplast genetic system in rice during early leaf development and at low temperatures. Front. Plant Sci. 5:386. doi: 10.3389/fpls.2014.00386
Kusumi, K., Sakata, C., Nakamura, T., Kawasaki, S., Yoshimura, A., and Iba, K. (2011). A plastid protein NUS1 is essential for build-up of the genetic system for early chloroplast development under cold stress conditions. Plant J. 68, 1039–1050. doi: 10.1111/j.1365-313X.2011.04755.x
Kusumi, K., Yara, A., Mitsui, N., Tozawa, Y., and Iba, K. (2004). Characterization of a rice nuclear-encoded plastid RNA polymerase gene OsRpoTp. Plant Cell Physiol. 45, 1194–1201. doi: 10.1093/pcp/pch133
Lin, D., Gong, X., Jiang, Q., Zheng, K., Zhou, H., Xu, J., et al. (2015a). The rice ALS3 encoding a novel pentatricopeptide repeat protein is required for chloroplast development and seedling growth. Rice (N Y) 8, 17. doi: 10.1186/s12284-015-0050-9
Lin, D., Jiang, Q., Zheng, K., Chen, S., Zhou, H., Gong, X., et al. (2015b). Mutation of the rice ASL2 gene encoding plastid ribosomal protein L21 causes chloroplast developmental defects and seedling death. Plant Biol. (Stuttg) 17, 599–607. doi: 10.1111/plb.12271
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lorkoviæ, Z. J., Kirk, D. A. W., Lambermon, M. H., and Filipowicz, W. (2000). Pre-mRNA splicing in higher plants. Trends Plant Sci. 5, 160–167. doi: 10.1016/S1360-1385(00)01595-8
Lurin, C., Andres, C., Aubourg, S., Bellaoui, M., Bitton, F., Bruyere, C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103. doi: 10.1105/tpc.104.022236
Moreira, D., Le Guyader, H., and Philippe, H. (2000). The origin of red algae and the evolution of chloroplasts. Nature 405, 69–72. doi: 10.1038/35011054
Mullet, J. E. (1993). Dynamic regulation of chloroplast transcription. Plant Physiol. 103, 309–313. doi: 10.1104/pp.103.2.309
Munekage, Y., Hashimoto, M., Miyake, C., Tomizawa, K. I., Endo, T., Tasaka, M., et al. (2004). Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429, 579–582. doi: 10.1038/nature02598
Nakamura, T., Meierhoff, K., Westhoff, P., and Schuster, G. (2003). RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur. J. Biochem. 270, 4070–4081. doi: 10.1046/j.1432-1033.2003.03796.x
O’Toole, N., Hattori, M., Andres, C., Iida, K., Lurin, C., Schmitz-Linneweber, C., et al. (2008). On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 25, 1120–1128. doi: 10.1093/molbev/msn057
Pfalz, J., Liere, K., Kandlbinder, A., Dietz, K. J., and Oelmuller, R. (2006). pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18, 176–197. doi: 10.1105/tpc.105.036392
Pfalz, J., and Pfannschmidt, T. (2013). Essential nucleoid proteins in early chloroplast development. Trends Plant Sci. 18, 186–194. doi: 10.1016/j.tplants.2012.11.003
Pogson, B. J., Ganguly, D., and Albrecht-Borth, V. (2015). Insights into chloroplast biogenesis and development. Biochim. Biophys. Acta 1847, 1017–1024. doi: 10.1016/j.bbabio.2015.02.003
Reumann, S., Inoue, K., and Keegstra, K. (2005). Evolution of the general protein import pathway of plastids (review). Mol. Membr. Biol. 22, 73–86. doi: 10.1080/09687860500041916
Schmitz-Linneweber, C., and Small, I. (2008). Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 13, 663–670. doi: 10.1016/j.tplants.2008.10.001
Schmitz-Linneweber, C., Williams-Carrier, R., and Barkan, A. (2005). RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5’ region of mRNAs whose translation it activates. Plant Cell 17, 2791–2804. doi: 10.1105/tpc.105.034454
Schmitz-Linneweber, C., Williams-Carrier, R. E., Williams-Voelker, P. M., Kroeger, T. S., Vichas, A., and Barkan, A. (2006). A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 18, 2650–2663. doi: 10.1105/tpc.106.046110
Small, I. D., and Peeters, N. (2000). The PPR motif–a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 45–47. doi: 10.1016/S0968-0004(99)01520-0
Song, J., Wei, X., Shao, G., Sheng, Z., Chen, D., Liu, C., et al. (2014). The rice nuclear gene WLP1 encoding a chloroplast ribosome L13 protein is needed for chloroplast development in rice grown under low temperature conditions. Plant Mol. Biol. 84, 301–314. doi: 10.1007/s11103-013-0134-0
Stoutjesdijk, P. A., Singh, S. P., Liu, Q., Hurlstone, C. J., Waterhouse, P. A., and Green, A. G. (2002). hpRNA-mediated targeting of the Arabidopsis FAD2 gene gives highly efficient and stable silencing. Plant Physiol. 129, 1723–1731. doi: 10.1104/pp.006353
Su, N., Hu, M. L., Wu, D. X., Wu, F. Q., Fei, G. L., Lan, Y., et al. (2012). Disruption of a rice pentatricopeptide repeat protein causes a seedling-specific albino phenotype and its utilization to enhance seed purity in hybrid rice Production. Plant Physiol. 159, 227–238. doi: 10.1104/pp.112.195081
Sugimoto, H., Kusumi, K., Noguchi, K., Yano, M., Yoshimura, A., and Iba, K. (2007). The rice nuclear gene, VIRESCENT 2, is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. Plant J. 52, 512–527. doi: 10.1111/j.1365-313X.2007.03251.x
Sugimoto, H., Kusumi, K., Tozawa, Y., Yazaki, J., Kishimoto, N., Kikuchi, S., et al. (2004). The virescent-2 mutation inhibits translation of plastid transcripts for the plastid genetic system at an early stage of chloroplast differentiation. Plant Cell Physiol. 45, 985–996. doi: 10.1093/pcp/pch111
Takenaka, M., Zehrmann, A., Verbitskiy, D., Hartel, B., and Brennicke, A. (2013). RNA editing in plants and its evolution. Annu. Rev. Genet. 47, 335–352. doi: 10.1146/annurev-genet-111212-133519
Tan, J., Tan, Z., Wu, F., Sheng, P., Heng, Y., Wang, X., et al. (2014). A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Mol. Plant 7, 1329–1349. doi: 10.1093/mp/ssu054
Tavares-Carreon, F., Camacho-Villasana, Y., Zamudio-Ochoa, A., Shingu-Vazquez, M., Torres-Larios, A., and Perez-Martinez, X. (2008). The pentatricopeptide repeats present in Pet309 are necessary for translation but not for stability of the mitochondrial COX1 mRNA in yeast. J. Biol. Chem. 283, 1472–1479. doi: 10.1074/jbc.M708437200
Terasawa, K., and Sato, N. (2005). Visualization of plastid nucleoids in situ using the PEND-GFP fusion protein. Plant Cell Physiol. 46, 649–660. doi: 10.1093/pcp/pci070
Toda, T., Fujii, S., Noguchi, K., Kazama, T., and Toriyama, K. (2012). Rice MPR25 encodes a pentatricopeptide repeat protein and is essential for RNA editing of nad5 transcripts in mitochondria. Plant J. 72, 450–460. doi: 10.1111/j.1365-313X.2012.05091.x
Wang, D., Liu, H., Zhai, G., Wang, L., Shao, J., and Tao, Y. (2016). OspTAC2 encodes a pentatricopeptide repeat protein and regulates rice chloroplast development. J. Genet. Genomics 43, 601–608. doi: 10.1016/j.jgg.2016.09.002
Wang, L., Wang, C., Wang, Y., Niu, M., Ren, Y., Zhou, K., et al. (2016). WSL3, a component of the plastid-encoded plastid RNA polymerase, is essential for early chloroplast development in rice. Plant Mol. Biol. 92, 581–595. doi: 10.1007/s11103-016-0533-0
Wu, W., Liu, S., Ruwe, H., Zhang, D., Melonek, J., Zhu, Y., et al. (2016). SOT1, a pentatricopeptide repeat protein with a small MutS-related domain, is required for correct processing of plastid 23S-4.5S rRNA precursors in Arabidopsis thaliana. Plant J. 85, 607–621. doi: 10.1111/tpj.13126
Wu, Z., Zhang, X., He, B., Diao, L., Sheng, S., Wang, J., et al. (2007). A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol. 145, 29–40. doi: 10.1104/pp.107.100321
Xie, T., Chen, D., Wu, J., Huang, X., Wang, Y., Tang, K., et al. (2016). Growing Slowly 1 locus encodes a PLS-type PPR protein required for RNA editing and plant development in Arabidopsis. J. Exp. Bot. 67, 5687–5698. doi: 10.1093/jxb/erw331
Yap, A., Kindgren, P., Colas des Francs-Small, C., Kazama, T., Tanz, S. K., Toriyama, K., et al. (2015). AEF1/MPR25 is implicated in RNA editing of plastid atpF and mitochondrial nad5, and also promotes atpF splicing in Arabidopsis and rice. Plant J. 81, 661–669. doi: 10.1111/tpj.12756
Yin, P., Li, Q., Yan, C., Liu, Y., Liu, J., Yu, F., et al. (2013). Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504, 168–171. doi: 10.1038/nature12651
Yoo, S. C., Cho, S. H., Sugimoto, H., Li, J., Kusumi, K., Koh, H. J., et al. (2009). Rice virescent3 and stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol. 150, 388–401. doi: 10.1104/pp.109.136648
Yu, Q. B., Huang, C., and Yang, Z. N. (2014). Nuclear-encoded factors associated with the chloroplast transcription machinery of higher plants. Front. Plant Sci. 5:316. doi: 10.3389/fpls.2014.00316
Zhou, K., Ren, Y., Zhou, F., Wang, Y., Zhang, L., Lyu, J., et al. (2016). Young Seedling Stripe1 encodes a chloroplast nucleoid-associated protein required for chloroplast development in rice seedlings. Planta 245, 45–60. doi: 10.1007/s00425-016-2590-7
Zhou, W., Cheng, Y., Yap, A., Chateigner-Boutin, A. L., Delannoy, E., Hammani, K., et al. (2009). The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J. 58, 82–96. doi: 10.1111/j.1365-313X.2008.03766.x
Zhou, W., Lu, Q., Li, Q., Wang, L., Ding, S., Zhang, A., et al. (2017). PPR-SMR protein SOT1 has RNA endonuclease activity. Proc. Natl. Acad. Sci. U.S.A. 114, E1554–E1563. doi: 10.1073/pnas.1612460114
Zoschke, R., Nakamura, M., Liere, K., Sugiura, M., Borner, T., and Schmitz-Linneweber, C. (2010). An organellar maturase associates with multiple group II introns. Proc. Natl. Acad. Sci. U.S.A. 107, 3245–3250. doi: 10.1073/pnas.0909400107
Keywords: chloroplast, Oryza sativa L., pentatricopeptide repeat, RNA splicing, WSL4
Citation: Wang Y, Ren Y, Zhou K, Liu L, Wang J, Xu Y, Zhang H, Zhang L, Feng Z, Wang L, Ma W, Wang Y, Guo X, Zhang X, Lei C, Cheng Z and Wan J (2017) WHITE STRIPE LEAF4 Encodes a Novel P-Type PPR Protein Required for Chloroplast Biogenesis during Early Leaf Development. Front. Plant Sci. 8:1116. doi: 10.3389/fpls.2017.01116
Received: 03 March 2017; Accepted: 09 June 2017;
Published: 26 June 2017.
Edited by:
Gerrit T. S. Beemster, University of Antwerp, BelgiumReviewed by:
Yukihiro Ito, Tohoku University, JapanJosé Manuel Pérez-Pérez, Universidad Miguel Hernández de Elche, Spain
Gorou Horiguchi, Rikkyo University, Japan
Copyright © 2017 Wang, Ren, Zhou, Liu, Wang, Xu, Zhang, Zhang, Feng, Wang, Ma, Wang, Guo, Zhang, Lei, Cheng and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianmin Wan, wanjianmin@caas.cn;, wanjm@njau.edu.cn
†These authors have contributed equally to this work.
 Ying Wang
Ying Wang Yulong Ren
Yulong Ren Kunneng Zhou
Kunneng Zhou Linglong Liu
Linglong Liu Jiulin Wang
Jiulin Wang Yang Xu
Yang Xu Huan Zhang
Huan Zhang Long Zhang
Long Zhang Zhiming Feng
Zhiming Feng Liwei Wang
Liwei Wang Weiwei Ma
Weiwei Ma Yunlong Wang
Yunlong Wang Xiuping Guo
Xiuping Guo Xin Zhang
Xin Zhang Cailin Lei
Cailin Lei Zhijun Cheng
Zhijun Cheng Jianmin Wan
Jianmin Wan