- 1Laboratory of Photosynthesis and Stress Signaling, Center for Excellence in Molecular Plant Sciences, Shanghai Center for Plant Stress Biology, Chinese Academy of Sciences, Shanghai, China
- 2Shanghai Center for Plant Stress Biology, Chinese Academy of Sciences, Shanghai, China
Photosystem II reaction center (PSII RC) and light-harvesting complex inevitably generate highly reactive singlet oxygen (1O2) that can impose photo-oxidative damage, especially when the rate of generation exceeds the rate of detoxification. Besides being toxic, 1O2 has also been ascribed to trigger retrograde signaling, which leads to nuclear gene expression changes. Two distinctive molecular components appear to regulate 1O2 signaling: a volatile signaling molecule β-cyclocitral (β-CC) generated upon oxidation of β-carotene by 1O2 in PSII RC assembled in grana core, and a thylakoid membrane-bound FtsH2 metalloprotease that promotes 1O2-triggered signaling through the proteolysis of EXECUTER1 (EX1) proteins associated with PSII in grana margin. The role of FtsH2 protease in 1O2 signaling was established recently in the conditional fluorescent (flu) mutant of Arabidopsis thaliana that generates 1O2 upon dark-to-light shift. The flu mutant lacking functional FtsH2 significantly impairs 1O2-triggered and EX1-mediated cell death. In the present study, the role of FtsH2 in the induction of 1O2 signaling was further clarified by analyzing the FtsH2-dependent nuclear gene expression changes in the flu mutant. Genome-wide transcriptome analysis showed that the inactivation of FtsH2 repressed the majority (85%) of the EX1-dependent 1O2-responsive genes (SORGs), providing direct connection between FtsH2-mediated EX1 degradation and 1O2-triggered gene expression changes. Furthermore, the overlap between β-CC-induced genes and EX1-FtsH2-dependent genes was very limited, further supporting the coexistence of two distinctive 1O2 signaling pathways.
Background
Under a multitude of environmental factors, altered levels of reactive oxygen species (ROS) in chloroplasts, which have long been implicated with damaging of macromolecules, appear to trigger certain signaling cascades leading to nuclear gene expression changes via a process known as retrograde signaling (Apel and Hirt, 2004). The transcriptional reprogramming seems to be essential for plants to sustain and adapt to the environmental changes. Among various ROS, chloroplasts generate 1O2 during oxygenic photosynthesis at the active PSII RC residing in the grana core (appressed grana region) of the thylakoids (Triantaphylidès et al., 2008). In order to minimize the photo-oxidative damage caused by 1O2, plants utilize molecular components residing in PSII RC, such as β-carotene and D1 protein, to quench 1O2 (Triantaphylidès et al., 2008). Oxidation of D1 protein leads to PSII damage, which subsequently undergoes disassembly and reassembly processes through the proteolysis of the damaged D1 proteins by ATP-dependent hexameric FtsH metalloprotease and the concurrent de novo synthesis of D1 proteins, respectively (Kato and Sakamoto, 2009; Kato et al., 2012).
1O2 has also been reported to trigger chloroplast-to-nucleus retrograde signaling, which is manifested by altered nuclear gene expression, acclimation response and programmed cell death (PCD) (op den Camp et al., 2003; Wagner et al., 2004; Lee et al., 2007; Kim et al., 2008, 2012; Ramel et al., 2012, 2013; Wang et al., 2016). Because 1O2 has an extremely short lifespan (∼200 ns) in a cellular environment (Li et al., 2012; Telfer, 2014), it is unlikely to serve as a signaling molecule. Therefore, chloroplasts may contain 1O2 sensor(s) to translate the levels of 1O2, either by self-oxidation or by monitoring the oxidation of other molecule(s), to subsequent changes in cellular processes. As 1O2 is generated in PSII, it is reasonable to assume that 1O2 sensor might be physically associated with PSII. Accordingly, the PSII components, such as D1 protein and β-carotene have long been supposed to act as primary scavengers of 1O2 (Triantaphylidès and Havaux, 2009). Even though the degradation product of D1 has been shown to control the synthesis of D1 in cyanobacteria (Stelljes and Koenig, 2007), there is no evidence to support that the degradation product of D1 acts as a signaling molecule in plants.
In contrast, strong evidence has been found to support that β-carotene, a primary quencher of 1O2 and a constituent of PSII RC, is a 1O2 sensor. β-cyclocitral (β-CC), a volatile oxidized derivatives of β-carotene, has been found to act as a signaling molecule in Arabidopsis wild-type plants exposed to excess light stress (Ramel et al., 2013). Moreover, plants treated with β-CC upregulate a set of 1O2-responsive genes (SORGs) (Ramel et al., 2012) identified previously using conditional Arabidopsis flu mutant generating 1O2 upon dark-to-light shift (op den Camp et al., 2003). In addition, β-CC has also been implicated with the acclimation responses in chlorina 1 (ch1) mutant lacking light-harvesting complex (LHC) of PSII, in which the level of 1O2 under light stress was escalated (Ramel et al., 2013). Although the volatile can diffuse freely into the nucleus, the molecular mechanism underlying the β-CC-mediated transcriptional reprogramming is largely unknown. Nonetheless, the earlier genetic screen using the unicellular green alga Chlamydomonas reinhardtii uncovered Methylene Blue Sensitivity 1 (MBS1), a small Zinc finger protein, as a mediator of 1O2 signaling (Shao et al., 2013). While the MBS1 protein is present in both cytosol and nucleus under normal conditions, it concentrates in specific stress granules and processing bodies under oxidative stress (Shao et al., 2013). A recent study revealed that MBS1 was upregulated in wild-type Arabidopsis plants under β-CC treatment even in normal light conditions, which led to gene expression changes for photo-acclimation (Shumbe et al., 2017). However, a further study is needed to show whether MBS1 can directly bind to DNA or requires other mediators to alter the nuclear gene expression.
Another distinct 1O2 signaling has been identified earlier in Arabidopsis flu mutant (op den Camp et al., 2003). In the dark, FLU protein acts as a negative feedback regulator of tetrapyrrole synthesis in Mg2+-branch. As the reduction of protochlorophyllide (Pchlide) to chlorophyllide is catalyzed by light-dependent enzyme protochlorophyllide oxidoreductase (POR), the flu mutant overaccumulates Pchlide in the dark. As a potent photosensitizer, the accumulated Pchlide upon illumination absorbs light energy and subsequently transfers the absorbed light energy to stable molecular oxygen, resulting in the formation of highly reactive 1O2 in chloroplasts (Meskauskiene et al., 2001). The flu mutant, therefore, offers a conditional system that allows generating 1O2 in a controlled and non-invasive manner (op den Camp et al., 2003). Various studies using the flu mutant revealed that the chloroplast-generated 1O2 can induce the nuclear gene expression changes and PCD (op den Camp et al., 2003; Lee et al., 2007). The previous genetic screen using flu mutant has revealed that nuclear-encoded and chloroplast-targeted protein EX1 mediates the 1O2-triggered stress responses (Wagner et al., 2004). Inactivation of EX1 significantly abolishes the 1O2-induced cell death and nuclear gene expression changes in flu mutant (Wagner et al., 2004; Lee et al., 2007). Besides EX1, its close homolog EXECUTER2 (EX2) has also been implicated with mediating 1O2 signaling. While inactivation of EX1 significantly attenuates the SORGs, inactivation of both EX1 and EX2 leads to almost complete repression of SORGs (Lee et al., 2007). In addition, EX1 and EX2 also appear to regulate the local and systemic gene expression changes in response to high light stress (Carmody et al., 2016).
Initially, it was contemplated that the two signaling cascades might be dependent on each other. However, in ch1 mutant the β-CC was found to relay the signal under photoinhibitory conditions in an EX1-independent manner (Ramel et al., 2012), implying that the two signaling cascades might be independent. Considering that both signaling events are instigated from PSII, it is obscure how these two signaling events are triggered independently. This seeming paradox was resolved in our recent study in which EX1 proteins were found mainly in ‘grana margin’ where the repair of PSII and the chlorophyll synthesis happen, implying that EX1-mediated 1O2 signaling is initiated in grana margin rather than the grana core (Wang et al., 2016). The study has also shown that light-adapted flu plants accumulated Pchlide in the dark almost evenly in different fractions across the thylakoid membrane, such as grana core, grana margin, and stroma lamellae (Wang et al., 2016). Given that ex1/flu abrogates the 1O2-triggered stress responses including growth inhibition and cell death, 1O2 generated in either grana core or stroma lamellae may not irreversibly damage the photosynthetic apparatus.
Protein immunoprecipitation (IP) coupled with mass spectrometry (MS) unveiled a group of proteins potentially forming the EXECUTER complex in grana margin, including enzymes involved in chlorophyll biosynthesis, PSII RC proteins and FtsH proteases (Wang et al., 2016). Because EX1 is associated with PSII and FtsH protease, upon 1O2 generation, EX1 possibly undergoes similar oxidative post-translational modification as PSII RC proteins including D1 and CP43 (Dreaden et al., 2012). If the oxidation occurs in EX1, the modified EX1 by 1O2 may become a target of FtsH protease, which is shown to be the case of D1 (Kato and Sakamoto, 2009). This proteolysis of EX1 may lead to the release of a signal that might be a part of EX1 complex or EX1 itself. Indeed, FtsH2-dependent proteolysis of EX1 in response to 1O2 and the concurrent induction of cell death are shown to be attenuated in the FtsH2-defecient flu mutant, namely var2/flu, implying that the proteolysis of EX1 is crucial for initiating the retrograde signaling (Wang et al., 2016). To investigate the underlying molecular mechanisms, we examined the effect of EX1 proteolysis on the induction of SORGs in flu mutant.
Experimental Results
Identification of a True Set of SORGs
In earlier studies, the SORGs identified in flu mutant (Danon et al., 2006; Laloi et al., 2007; Lee et al., 2007) were employed to address the probable activation of 1O2 signaling in various mutants and wild-type plants under photooxidative stress conditions (e.g., Ramel et al., 2013). Afterward, the detection of the rapid loss of chloroplast integrity following 1O2 burst, as evidenced by the appearance of stroma proteins into cytosol (Kim et al., 2012; Chen et al., 2015), prompted us to redefine SORGs as a group of genes that were upregulated prior to the loss of chloroplast integrity (Chen et al., 2015) (399 genes; Supplementary Table S1). In addition, in all of those previous transcriptome data, the confounding effect of dark on the gene expression changes was not excluded. Hence, in the present study, we carried out RNA-seq-based gene expression analysis in 5-day-old seedlings of flu and FtsH2-deficient var2/flu, initially grown under continuous light (LL), and then subjected to 4 hours dark (4hD) followed by re-illumination for 30 (L’30) and 60 min (L’60). Samples harvested at the end of dark were included as additional control to exclude genes upregulated in the dark. As a result, we found that a large proportion (215 genes, 53%) of previously identified SORGs (399) appeared to be upregulated in the dark followed by a gradual repression during re-illumination (Figure 1A). These genes were reclassified as dark-induced genes (DIGs) and were excluded from the SORGs, together with 26 more genes that were either not expressed or significantly changed in the expression. The remaining 168 SORGs were thus considered as early 1O2-responsive genes (hereafter ESORGs) (Figure 1A and Supplementary Table S2). The 168 ESORGs include SIGMA FACTOR BINDING PROTEIN 1 (SIB1), WRKY33 (WRKY DNA-BINDING PROTEIN 33), WRKY40 (WRKY DNA-BINDING PROTEIN 40), ETHYLENE RESPONSE FACTOR 104 (ERF104), MAP KINASE KINASE 9 (MKK9), SENESCENCE ASSOCIATED GENE 20 (SAG20), TOLL/INTERLEUKIN-1 RECEPTOR-LIKE (TIR) and many other stress-responsive genes (Figure 1A and Supplementary Table S2). This set of true ESORGs was used for further comparative analysis between flu and var2/flu.
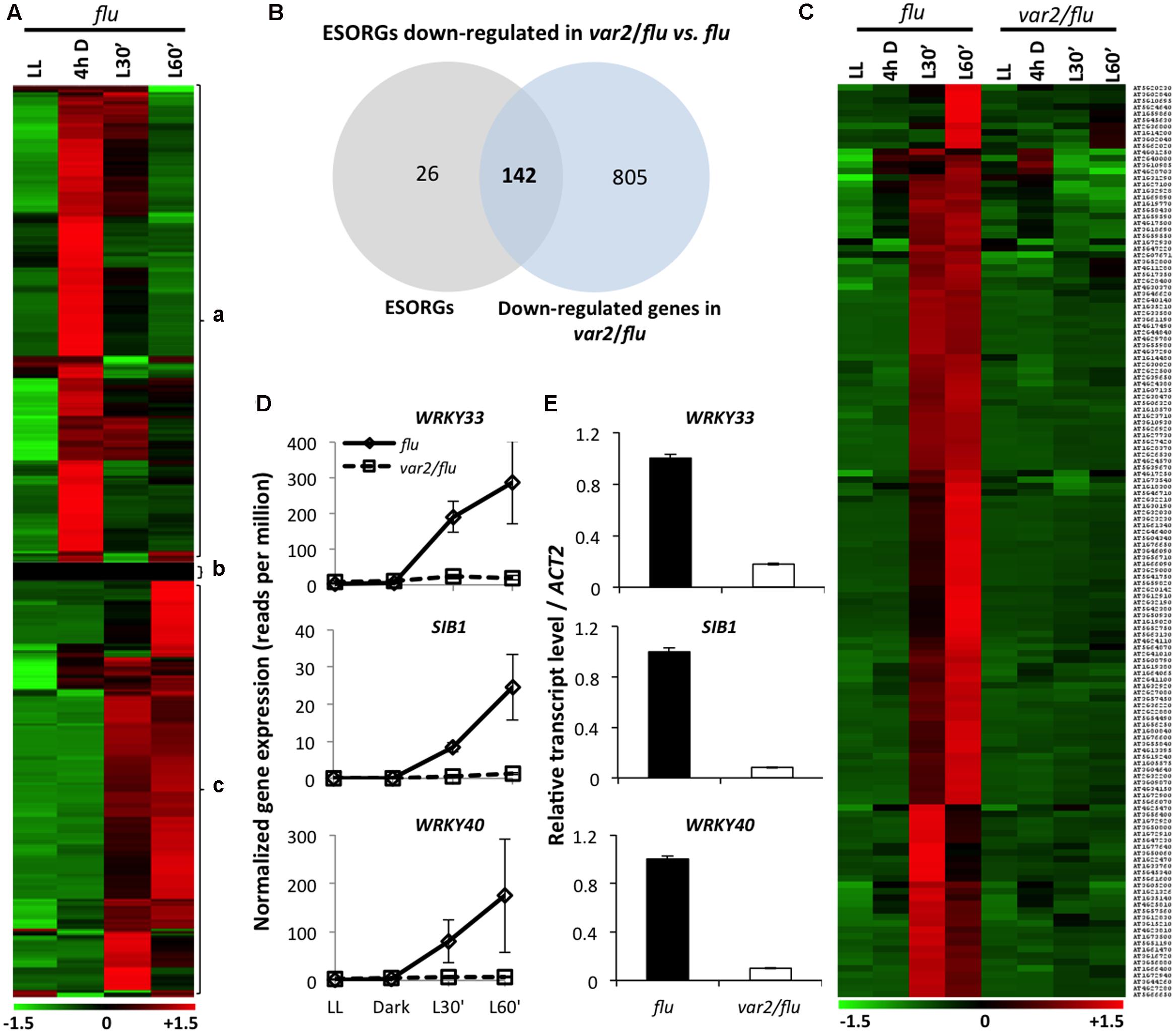
FIGURE 1. FtsH2 is an integral component of EX1-mediated 1O2 signaling. RNA- seq-based comparison of gene expression was performed in 5-day-old var2/flu and flu seedlings that were initially grown under continuous light (LL), transferred to dark for 4 hours (4 h D), and then re-exposed to light for 30 (L30′) and 60 min (L60′). (A) Heat map reclassified the expression of previously reported SORGs. After excluding 215 genes induced during dark incubation (a) and 26 genes that either exhibited no expression or were not expressed differentially (b), 168 true ESORGs (c) were identified. (B,C) Venn diagram and heat map revealed that the majority (142 genes; 85%) of ESORGs were downregulated in var2/flu in comparison to flu upon dark-to-light shift. (D) Expression of 1O2-specific and EX1/EX2-dependent genes SIB1, WRKY33 and WRKY40 in var2/flu and flu. The average values and standard error of the normalized gene expression (reads per million; rpm) of three independent biological samples obtained by RNA-seq analysis were shown. (E) Validation of RNA-seq using those selected genes (D) by qRT-PCR. Results represent means of three independent biological replicates. Actin2 was used as a control for normalization.
FtsH2 Is Essential for the Induction of ESORGs
The comparison of transcriptome profiles of flu and var2/flu revealed that approximately 85% of ESORGs (142 out of 168 genes) were repressed in var2/flu as compared to flu (Figures 1B,C and Supplementary Table S3). In an earlier study, only three SORGs, namely SIB1, WRKY33 and WRKY40, were shown to be upregulated not only in wild-type seedlings under photoinhibitory condition (Chen et al., 2015) but also in flu mutant upon dark-to-light shift in EX1/EX2-dependent manner. Hence, these genes were considered as 1O2-specific and EX1/EX2-dependent genes (Figure 1D). The RNA-seq result was further validated by quantitative real time PCR (qRT-PCR) using these genes (Figure 1E). In consistent with the phenotype of var2/flu seedlings, which appear to be more tolerant to 1O2 as compared to flu mutant seedlings (Wang et al., 2016), the global gene expression levels were less responsive to 1O2 in var2/flu seedlings than in flu seedlings. The repressed expression of ESORGs following re-illumination and the concurrent attenuation of stress responses suggest that FtsH2 is a positive regulator of EX1-mediated 1O2 signaling in flu mutant.
Comparative Analysis of β-CC-Induced and EX1-FtsH2-Dependent SORGs
Previous studies have established the role of β-CC in 1O2-triggered acclimation responses (Ramel et al., 2012, 2013), whereas the EX1-mediated 1O2 signaling has been mainly attributed to PCD in young seedlings and growth inhibition in mature plants (Wagner et al., 2004; Kim et al., 2012). In a very recent study, however, the role of EX1 and EX2 in an acclimation response to high light-induced transcriptional reprogramming has also been demonstrated (Carmody et al., 2016). To get insight into these distinctive cascades, we compared the β-CC-induced SORGs and EX1-FtsH2-dependent ESORGs, with the aim to explain the common and distinct molecular characteristics and enlighten their coexistence. Plants treated with β-CC for 4 h under normal light conditions exhibited a rapid reprogramming of nuclear gene expression, resulting in the altered expression of around 380 genes with at least twofold up- or down-regulation (Ramel et al., 2012). Among these 380 genes, the upregulated genes (292) are considered as β-CC-induced SORGs (Supplementary Table S4). Comparative analysis revealed that β-CC-induced (292) and EX1-FtsH2-dependent (142) genes shared only a small number of genes (20) (Figure 2). The shared genes include above-mentioned 1O2-responsive genes, such as WRKY33, WRKY40, and SIB1. However, the expressions of these genes were comparatively higher in flu mutant (Supplementary Table S5). Taken together, these results further support the coexistence of the two independent 1O2-signalings in Arabidopsis. Considering that 1O2-specific and EX1/EX2-dependent genes including WRKY33, WRKY40 and SIB1, appear to be upregulated in response to β-CC, we cannot rule out the possibility that these two signaling cascades may share a certain downstream component to regulate those shared genes.
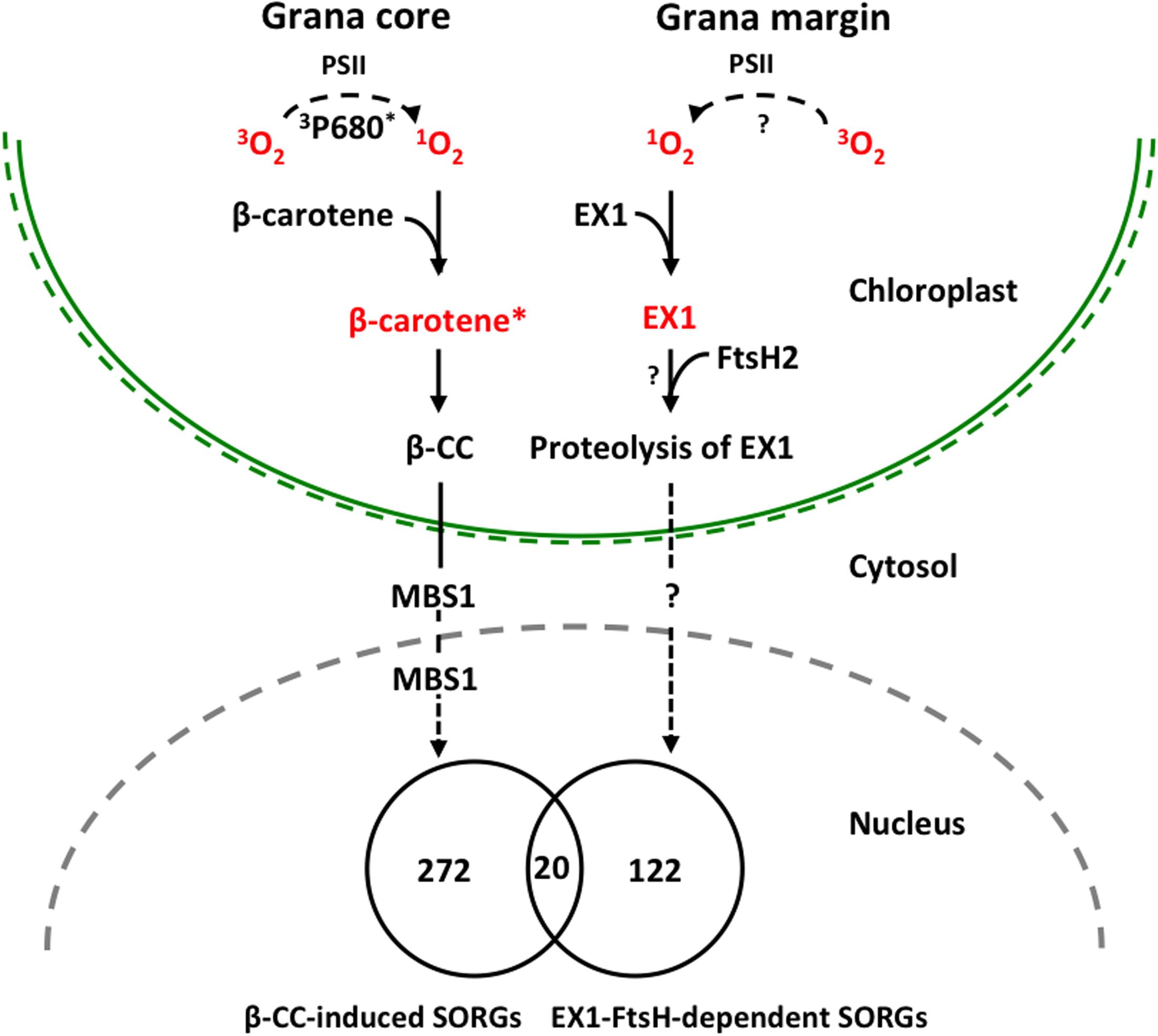
FIGURE 2. Proposed model illustrating 1O2-triggered EX1-independent and EX1-dependent chloroplast-to-nucleus retrograde signaling pathways. Under severe light stress, β-carotene from PSII RC enriched in grana core acts as a sensor of 1O2 generated by the excited triplet P680 chlorophyll (3P680∗) through the energy transfer from 3P680∗ to stable oxygen molecule (3O2). The subsequent oxidation of β-carotene (β-carotene∗) by 1O2 concomitantly release β-CC, a volatile oxidized derivative, which functions as a messenger involved in the 1O2 signaling pathway. MBS1 protein, a zinc finger protein located in cytoplasm and nucleus, acts downstream of β-CC. In grana margin, EX1 may sense 1O2 generated through an as yet unknown process and initiates 1O2 signaling in an FtsH2-dependent manner. FtsH2-dependent proteolysis of EX1 appears to be essential in the induction of nuclear gene expression changes. β-CC-induced and EX1-FtsH2-dependent SORGs are reasonably distinct as evidenced that these transcriptomes share relatively a very small portion of genes, signifying the coexistence of these two 1O2 signaling cascades in plants.
Summary and Future Directions
In summary, our previous study revealed the important role of ATP-dependent FtsH metalloprotease, assembled in thylakoid membrane and functioning in PSII repair cycle, in the induction of EX1-mediated stress response upon 1O2 generation in chloroplast. The FtsH protease is co-localized with EX1 in grana margin where the PSII repair process undergoes, and it coordinates the retrograde signaling by promoting the proteolytic degradation of EX1 proteins through a yet unknown mechanism (Figure 2). FtsH-dependent EX1 degradation was found to occur upon 1O2 burst in chloroplast, which possibly triggers the signaling. In agreement, inactivation of FtsH2, a subunit of the FtsH protease complex, appears to considerably compromise the proteolysis of EX1 and the concurrent 1O2-triggered PCD in Arabidopsis flu mutant. Furthermore, the loss of FtsH2 in flu resulted in the repression of the majority of SORGs. β-CC-induced and EX1-FtsH2-dependent SORGs are largely distinct, sharing relatively a very small portion of genes. These results provide compelling evidence that chloroplasts may operate two distinctive 1O2 signaling: one operated in grana core by β-carotene and one in grana margin by the coordination of EX1 and FtsH2 (Figure 2). Based on this foundation, further investigation on EX1-FtsH2-dependent signaling is needed in order answer questions such as (i) what is the source of 1O2 in the grana margin? (ii) how EX1 senses and mediates this signaling in coordination with FtsH2? (iii) what is the genuine signaling molecule that is generated upon EX1 proteolysis? (Figure 2). Furthermore, the reasons for the coexistence of two distinctive 1O2-triggered retrograde signaling pathways in plants are still unknown, which probably opens new perspectives of research in this direction.
Materials and Methods
Plant Materials, Growth Conditions, and RNA Isolation
Seedlings of flu and var2/flu mutant lines (Wang et al., 2016), all in the Columbia-0 (Col-0) background, were grown under continuous light (40 μmol photons m-2 s-1) at 20 ± 1°C on 0.6% agar plates containing ½ MS medium and 1xGamborg vitamins. Total RNA was isolated using RNA easy Plant mini kit (Qiagen, Germany). DNase I (Qiagen, Germany) treatment was performed to remove potential DNA contamination. RNA was examined by a Nano Photometer spectrophotometer (Implen, Westlake Village, CA, United States) for RNA purity. Qubit RNA Assay Kit in Qubit 2.0 Flurometer (Life Technologies, Foster City, CA, United States) was used to measure RNA concentration, and RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, United States) was used to evaluate RNA integrity. Only RNA samples that passed the quality control were used for RNA-Seq analyses. Three biological replicates were subject to RNA-Seq based gene expression analysis for each genotype/treatment.
RNA-seq Library Construction, Sequencing, and Analysis
RNA-seq libraries were constructed using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, United States) following manufacturer’s instructions. RNA-seq libraries were sequenced on an Illumina HiSeq 2500 platform to generate 100 bp paired-end reads as described previously (Zhao et al., 2016). Lowly expressed genes were removed and genes with an expression level of at least 1 transcripts per million (TPM) in at least three samples were selected for differential expression analysis. The R package edgeR, which uses counts per gene in different samples from HTSeq-count as input and performs data normalization using the trimmed mean of M-values (TMM) method, was used to identify differentially expressed genes (DEGs). The genes with at least twofold change in expression and the false discovery rate (FDR) of less than 0.05 were considered to be differentially expressed. Gene expression levels were normalized to TPM according to the total number of mapped clean reads in each of library. The log2 values of normalized expression levels were used to build expression matrix and subsequent clustering and visualization was conducted using Multi-Experiment Viewer (MeV 4.9.0).
qRT-PCR Analysis
Quantitative real time PCR assays were performed with samples that were collected independently of samples used for RNA-Seq analysis. Three biological replicates were used for each genotype/treatment. RNAs were treated with RQ1 RNase-Free DNase (Promega) and reverse-transcribed using Improm II reverse transcriptase (Promega) and oligo(dT)15 primer (Promega) according to the manufacturer’s recommendations. The qRT-PCR was performed by using the QuantStudioTM 6 Flex Real-Time PCR System (Applied Biosystems) and iTaq Universal SYBR Green PCR master mix (Bio-Rad). Relative transcript levels were calculated by the comparative delta-Ct method and normalized to the ACT2 (At3g18780) gene transcript level. The primers used in this study were designed by Primer Express Software for Real-time PCR, Version 3.0 (Applied Biosystems) and the primer sequences are included in Supplementary Table S6.
Author Contributions
VD, KL, JD, and CK planned and designed the research; VD, JD, and KL performed research; SL and VD analyzed RNA-seq data; and VD, RL, and CK wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the Core Facility of Genomics, Shanghai Center for Plant Stress Biology (PSC) for carrying out RNA sequencing. This research was supported by the 100-Talents Program from the Chinese Academy of Sciences (CAS) to CK and RL and by National Natural Science Foundation of China (NSFC) Grant 31570264 to CK.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01145/full#supplementary-material
References
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399.
Carmody, M., Crisp, P. A., D’Alessandro, S., Ganguly, D., Gordon, M., Havaux, M., et al. (2016). Uncoupling high light responses from singlet oxygen retrograde signaling and spatial-temporal systemic acquired acclimation in Arabidopsis. Plant Physiol. 171, 1734–1749.
Chen, S., Kim, C., Lee, J. M., Lee, H. A., Fei, Z., Wang, L., et al. (2015). Blocking the QB-binding site of photosystem II by tenuazonic acid, a non-host-specific toxin of Alternaria alternata, activates singlet oxygen-mediated and EXECUTER-dependent signalling in Arabidopsis. Plant Cell Environ. 38, 1069–1080. doi: 10.1111/pce.12462
Danon, A., Coll, N. S., and Apel, K. (2006). Cryptochrome-1-dependent execution of programmed cell death induced by singlet oxygen in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 103, 17036–17041.
Dreaden, K. T. M., Rexroth, S., and Barry, B. A. (2012). Light-Induced oxidative stress, N-formylkynurenine, and oxygenic photosynthesis. PLoS ONE 7:e42220. doi: 10.1371/journal.pone.0042220
Kato, Y., and Sakamoto, W. (2009). Protein quality control in chloroplasts: a current model of D1 protein degradation in the photosystem II repair cycle. J. Biochem. 146, 463–469. doi: 10.1093/jb/mvp073
Kato, Y., Sun, X., Zhang, L., and Sakamoto, W. (2012). Cooperative D1 degradation in the photosystem II repair mediated by chloroplastic proteases in Arabidopsis. Plant Physiol. 159, 1428–1439. doi: 10.1104/pp.112.199042
Kim, C., Meskauskiene, R., Apel, K., and Laloi, C. (2008). No single way to understand singlet oxygen signalling in plants. EMBO Rep. 9, 435–439. doi: 10.1038/embor.2008.57
Kim, C., Meskauskiene, R., Zhang, S., Lee, K. P., Ashok, M. L., Blajecka, K., et al. (2012). Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 24, 3026–3039. doi: 10.1105/tpc.112.100479
Laloi, C., Stachowiak, M., Pers-Kamczyc, E., Warzych, E., Murgia, I., and Apel, K. (2007). Cross-talk between singlet oxygen- and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 672–677.
Lee, K. P., Kim, C., Landgraf, F., and Apel, K. (2007). EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 10270–10275.
Li, H., Melo, T. B., Arellano, J. B., and Naqvi, K. R. (2012). Temporal profile of the singlet oxygen emission endogenously produced by photosystem II reaction centre in an aqueous buffer. Photosynth. Res. 112, 75–79. doi: 10.1007/s11120-012-9739-4
Meskauskiene, R., Nater, M., Goslings, D., Kessler, F., op den Camp, R., and Apel, K. (2001). FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 98, 12826–12831.
op den Camp, R., Przybyla, D., Ochsenbein, C., Laloi, C., Kim, C., Danon, A., et al. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15, 2320–2332.
Ramel, F., Birtic, S., Ginies, C., Soubgou-Taconnat, L., Triantaphylidès, C., and Havaux, M. (2012). Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. U.S.A. 109, 5535–5540. doi: 10.1073/pnas.1115982109
Ramel, F., Ksas, B., Akkari, E., Mialoundama, A. S., Monnet, F., Krieger-Liszkay, A., et al. (2013). Light-induced acclimation of the Arabidopsis chlorina1 mutant to singlet oxygen. Plant Cell 25, 1445–1462. doi: 10.1105/tpc.113.109827
Shao, N., Duan, G. Y., and Bock, R. (2013). A mediator of singlet oxygen responses in Chlamydomonas reinhardtii and Arabidopsis identified by a luciferase-based genetic screen in algal cells. Plant Cell 25, 4209–4226. doi: 10.1105/tpc.113.117390
Shumbe, L., D’Alessandro, S., Shao, N., Chevalier, A., Ksas, B., Bock, R., et al. (2017). METHYLENE BLUE SENSITIVITY 1 (MBS1) is required for acclimation of Arabidopsis to singlet oxygen and acts downstream of β-cyclocitral. Plant Cell Environ. 42, 216–226. doi: 10.1111/pce.12856
Stelljes, C., and Koenig, F. (2007). Specific binding of D1 protein degradation products to the psbAI promoter in Synechococcus sp. strain PCC 7942. J. Bacteriol. 189, 1722–1726.
Telfer, A. (2014). Singlet oxygen production by PSII under light stress: mechanism, detection and the protective role of β-carotene. Plant Cell Physiol. 55, 1216–1223. doi: 10.1093/pcp/pcu040
Triantaphylidès, C., and Havaux, M. (2009). Singlet oxygen in plants: production, detoxification and signaling. Trends Plant. Sci. 14, 219–228. doi: 10.1016/j.tplants.2009.01.008
Triantaphylidès, C., Krischke, M., Hoeberichts, F. A., Ksas, B., Gresser, G., Havaux, M., et al. (2008). Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol. 148, 960–968. doi: 10.1104/pp.108.125690
Wagner, D., Przybyla, D., op den Camp, R., Kim, C., Landgraf, F., Lee, K. P., et al. (2004). The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306, 1183–1185.
Wang, L., Kim, C., Xu, X., Piskeuwicz, U., Dogra, V., Singh, S., et al. (2016). 1O2- and EXECUTER1-mediated signaling is initiated in grana margins and depends on FtsH2 protease. Proc. Natl. Acad. Sci. U.S.A. 113, E3792–E3800. doi: 10.1073/pnas.1603562113
Keywords: 1O2, retrograde signaling, EXECUTER 1, FtsH2 protease, β-carotene, β-cyclocitral, chloroplast
Citation: Dogra V, Duan J, Lee KP, Lv S, Liu R and Kim C (2017) FtsH2-Dependent Proteolysis of EXECUTER1 Is Essential in Mediating Singlet Oxygen-Triggered Retrograde Signaling in Arabidopsis thaliana. Front. Plant Sci. 8:1145. doi: 10.3389/fpls.2017.01145
Received: 13 February 2017; Accepted: 15 June 2017;
Published: 29 June 2017.
Edited by:
Fei Yu, Northwest A&F University, ChinaReviewed by:
John Froehlich, Michigan State University, United StatesAigen Fu, Northwest University, United States
Copyright © 2017 Dogra, Duan, Lee, Lv, Liu and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chanhong Kim, chanhongkim@sibs.ac.cn
 Vivek Dogra
Vivek Dogra Jianli Duan
Jianli Duan Keun Pyo Lee
Keun Pyo Lee Shanshan Lv
Shanshan Lv Renyi Liu
Renyi Liu Chanhong Kim
Chanhong Kim