- Key Laboratory of Biology and Genetic Improvement of Oil Crops, Ministry of Agriculture, Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences, Wuhan, China
Drought and heat stress are major causes of lost plant crop yield. In the future, high levels of CO2, in combination of other abiotic stress factors, will become a novel source of stress. Little is known of the mechanisms involved in the acclimation responses of plants to this combination of abiotic stress factors, though it has been demonstrated that heat shock transcription factors (Hsfs) are involved in plant response to various abiotic stresses. In this study, we performed a genome-wide identification and a systematic analysis of genes in the Hsf gene family in Brassica napus. A total of 64 genes encoding Hsf proteins were identified and classified into 3 major classes: A, B and C. We found that, unlike in other eudicots, the A9 subclass is absent in rapeseed. Further gene structure analysis revealed a loss of the only intron in the DBD domain for BnaHsf63 and -64 within class C, which is evolutionarily conserved in all Hsf genes. Transcription profile results demonstrated that most BnaHsf family genes are upregulated by both drought and heat conditions, while some are responded to a high CO2 treatment. According to the combined RNA-seq and qRT-PCR analysis, the A1E/A4A/A7 subclasses were upregulated by both drought and heat treatments. Members in class C seemed to be predominantly induced only by drought. Among BnaHsf genes, the A2/A3/B2 subclasses were regulated by all three abiotic stresses. Members in A2/B2 subclasses were upregulated by drought and heat treatments, but were downregulated under high CO2 conditions. While the A3 subclass was upregulated by all the three abiotic stresses. Various stress-related cis-acting elements, enriched in promoter regions, were correlated with the transcriptional response of BnaHsfs to these abiotic stresses. Further study of these novel groups of multifunctional BnaHsf genes will improve our understanding of plant acclimation response to abiotic stresses, and may be useful for improving the abiotic stress resistance of crop varieties.
Introduction
Environmental stresses, such as drought and heat, cause substantial loss to plant growth and production (Hu and Xiong, 2014; Fang and Xiong, 2015; Pereira, 2016; Zhu, 2016). Increasing CO2 levels result in lower concentrations of zinc, iron and protein for most C3 crops, and threaten human food sources; these results suggest that high CO2 concentrations (hereafter abbreviated to high CO2) may be a novel stress in the future (Myers et al., 2014). However, the basic molecular mechanisms driving plant responses to high CO2 remain elusive (Becklin et al., 2017). To cope with abiotic stresses, plants have evolved diverse adaptive strategies and signaling mechanisms. Transcription factors play crucial regulatory roles in the signal transduction process under these stresses (Hu and Xiong, 2014; Yoshida et al., 2014; Song et al., 2016). Among the transcription factors, heat shock transcription factors (Hsfs) serve as the terminal components in the signal transduction chain mediating the activation of genes responsive to heat and other stresses by recognizing palindromic binding motifs conserved in promoters of heat stress inducible genes called heat stress elements (HSEs: 5′-AGAAnnTTCT-3′) (von Koskull-Döring et al., 2007; Scharf et al., 2012; Guo et al., 2016).
As many other transcription factors, Hsf proteins possess a conserved modular structure (von Koskull-Döring et al., 2007; Scharf et al., 2012). The highly-structured DNA-binding domain (DBD) is located close to the N-terminal of all Hsfs and is responsible for selective interaction with HSE. An oligomerization domain (OD or HR-A/B region) is connected to the DBD by a flexible linker of variable length (15–80 amino acid residues) with a bipartite heptad pattern of hydrophobic amino acid residues, leading to the formation of a coiled-coil domain for protein interaction of Hsfs. Based on the length of the flexible linker and the number of amino acid residues inserted into the HR-A/B region, plant Hsfs are grouped into three main classes (HsfA, -B, and -C) (von Koskull-Döring et al., 2007; Scharf et al., 2012; Guo et al., 2016). The nuclear localization signal (NLS) and the nuclear export signal (NES) control the intracellular distribution of Hsfs between nucleus and cytoplasm. AHA motifs are usually present in the HsfA subfamily and confer transcriptional activator function. However, HsfB members, except HsfB5, are characterized by a repressor domain (RD) consisting of tetrapeptide LFGV in the C-terminal region.
Heat shock transcription factor genes were first cloned in yeast in 1988 (Wiederrecht et al., 1988). Unlike the small Hsf family in yeast and animals, plants hold complex and large Hsf gene families. 21, 25, 38, 40, 25, 32 Hsf genes have been found in Arabidopsis, rice, soybean, cotton, pepper and poplar, respectively (Nover et al., 2001; Chauhan et al., 2011; Li et al., 2014; Wang et al., 2014; Guo et al., 2015; Zhang et al., 2016). Among monocots and eudicots, the largest number of Hsf genes were found in wheat (56 members) (Xue et al., 2014).
Plant Hsfs do not only play a role in heat stress response, but also function both in response to other stressers and during organ development. In addition, structural characteristics and diverse expression patterns of Hsf family genes have revealed functional diversification (von Koskull-Döring et al., 2007). Our understanding of plant Hsf gene function comes predominantly from studies of tomato and Arabidopsis thaliana. In tomato, constitutively expressed HsfA1a functions as a master regulator for induced thermotolerance, and cannot be replaced by any other Hsfs (Mishra et al., 2002). However, in Arabidopsis, due to functional redundancy no comparable master regulator role could be identified for any of its four HsfA1 genes (Liu et al., 2011; Nishizawa-Yokoi et al., 2011; Guo et al., 2016). HsfA2 is structurally and functionally similar to HsfA1, but is only strongly accumulated in stressed plants. Its interaction with HsfA1 and B1 forms a functional complex, responsible for regulating core aspects of heat stress response and recovery (Nishizawa et al., 2006; Charng et al., 2007; Scharf et al., 2012; Guo et al., 2016). Additionally, HsfA2 is also involved as a key regulator in other environmental stresses such as osmotic and oxidative stress (Ogawa et al., 2007; Wang et al., 2017). Previous investigations showed that heat- and drought-induced expression of HsfA3 depends on transcription factor DREB2A in Arabidopsis, this indicates that HsfA3 plays a role in drought stress signaling (Sakuma et al., 2006; Schramm et al., 2008). Ectopic expression of HsfA3 has also been shown to enhance thermotolerance in Arabidopsis (Li et al., 2013). Group A4 Hsfs are involved in controlling reactive oxygen species homeostasis of plants, and group A5 Hsfs act as specific repressors of HsfA4 (Yamanouchi et al., 2002; Baniwal et al., 2007). Orthologous HsfA4a was reported to confer cadmium tolerance in wheat and rice (Shim et al., 2009). HsfA9 participates in embryo development and seed maturation in Arabidopsis and sunflower (Almoguera et al., 2002; Kotak et al., 2007); in addition, overexpression of seed-specific HaHSFA9 in tobacco confers tolerance to severe dehydration at vegetative stage (Prieto-Dapena et al., 2008).
Unlike class A Hsfs, class B Hsfs lack an AHA motif and show no independent function as transcriptional activators (von Koskull-Döring et al., 2007). However, in tomato, heat-induced HsfB1 acts as a coactivator with HsfA1a (Bharti et al., 2004). Moreover, Arabidopsis HsfB1 is inactive as a coactivator, although AtHsfB1 can act as a repressor of heat stress-inducible Hsfs (Czarnecka-Verner et al., 2000, 2004). To date, limited information is available for class C Hsfs. The results from tomato and Arabidopsis revealed striking species-specific deviations in the functional diversification of Hsf family members (von Koskull-Döring et al., 2007; Scharf et al., 2012; Guo et al., 2016). Overall, comprehensive characterization is still needed to investigate the functions of Hsfs in plant abiotic stress responses, and in particular in response to high CO2 conditions.
Brassica napus (rapeseed) is one of the most important oil crops in the world. Given the recent publication of the B. napus genome (Chalhoub et al., 2014), rapeseed is becoming an important crop model system (Zhu et al., 2016). While rapeseed plants are sensitive to water deficit and high temperature during all stages of growth and development, the typical stress-related Hsf transcription factors have not yet been characterized in B. napus. To investigate the potential roles of rapeseed Hsfs in abiotic stress responses, the present study identifies 64 genes encoding BnaHsf proteins in the B. napus genome and analyzes their phylogenetic relationships, gene and domain structures, putative cis-acting elements, and expression patterns across different tissues and under heat, drought and high CO2 stresses. The results of this study will help to provide a foundation for further functional studies of BnaHsf genes, and improve our understanding of the functional diversification of plant Hsf genes under various environmental stresses.
Materials and Methods
Identification of Hsf Genes in Brassica napus
The gene sequence of B. napus were downloaded from the genome database1, to gather the probable candidate B. napus Hsf amino acid sequences, the Hsf-type DBD domain (Pfam: PF00447) was submitted as a query in a protein BLAST search of the B. napus genome database. Hsf gene sequences from Arabidopsis (Nover et al., 2001) were retrieved from the TAIR database (Lamesch et al., 2012) and used as queries to perform repetitive BLAST searches against the Phytozome database v9.1 (Goodstein et al., 2012). BLAST searches were also performed against the NCBI nucleic-acid sequence data repositories. All protein sequences derived from the BLAST searches were examined using domain-analysis programs. Molecular weight, iso-electric point, functional domains, and amino acid signal peptides of BnaHsfs were calculated using the ExPASy online servers2.
Multiple Sequence Alignments and Phylogenetic Analysis
Multiple sequence alignment of Hsf proteins from B. napus were performed using the program ClustalX 1.83 (Thompson et al., 1997). The phylogenetic tree was constructed using the neighbor-joining (NJ) method by MEGA 6 program (Tamura et al., 2013), the bootstrap value was set at 1000 replications to assess tree reliability.
Domain and Gene Structure Analysis
The MEME program3 (Bailey et al., 2009) was used for identification of conserved motifs, with the following parameters: number of repetitions: any; maximum number of motifs:15; and the optimum motif widths: 6–200 amino acid residues. Exton/intron organization of the Hsf genes in B. napus was illustrated using Gene Structure Display Server program (GSDS4) (Hu et al., 2015) by alignment of the cDNAs with their corresponding genomic DNA sequences.
Regulatory Cis-Element Analysis
Prediction of putative cis-elements was performed using Signal Scan Search against the plant cis-acting regulatory DNA elements (PLACE) database (Higo et al., 1999). A 2-kb sequence upstream of ATG initiation codon of BnaHsf genes was selected as the promoter region for this analysis.
Plant Materials and Growth Conditions
Rapeseed seeds were germinated on a filter paper, and then transplanted to soil pots growing in the greenhouse at Oil crops research institute (Wuhan, China) with conditions of a temperature of 22°C, LED sodium lamp and a humidity of about 50–70% for 6 weeks. The plants were then transferred to growth chamber programmed under specific environmental conditions for 2–3 days before stress treatment. The conditions in growth chamber were set as follows: temperature of 25°C and humidity of 50% in 16 h light; temperature of 22°C and humidity of 60% in 8 h dark.
Stress Treatments
The high CO2 stress was performed in a growth chamber (AR-41L2, United States) in which CO2 gas can be accurately and stably controlled in the range of 100–3000 ppm. The conditions of growth chamber were set as follows: CO2 concentration of 1000 ppm, light intensity of 100 umol/m2/s, temperature of 25°C and 60% relative humidity. Leaf samples were collected at 1, 3, and 6 h during the treatment.
The heat and drought stress were performed in a common growth chamber. For heat stress, the chamber was set with temperature of 40°C and humidity of 60%. Leave samples under heat were collected at 1, 3, and 6 h during treatment. For drought stress, the chamber was set as follows: temperature of 25°C and humidity of 40% in 16 h light; temperature of 22°C and humidity of 50% in 8 h dark; withholding water for 7 days, leaf samples were collected at 1, 2, and 3 days during drought treatment.
All the collected leaf samples were immediately frozen in liquid nitrogen, and stored at -70°C for further analysis.
RNA Isolation and Quantitative Real-Time PCR (qRT-PCR) Analysis
The RNA was isolated from leaf tissues using an RNA extraction kit (Takara, Dalian), according to the manufacturer’s instructions. The first-strand cDNA was synthesized by the Prime Script RT reagent Kit (Takara, Dalian). Real-time quantitative PCR was performed using 2 μl of cDNA in a 20 μl reaction volume with SYBR Premix Ex Taq (Takara) on a 7500-Fast real-time PCR System (Applied Biosystems). Gene-specific primers were designed (Supplementary Table 1). The rapeseed TMA7 gene (BnaC05g11560D) was used as an internal control to normalize the expression level of the target gene, which has highly stable expression level in different tissues and under various growth conditions. Each treatment was repeated three times independently. The thermal cycler was set as follows: an initial incubation at 50°C for 2 min and 95°C for 5 min, followed by 40 cycle at 95°C for 30 s, 55°C for 30 s and 72°C for 30 s. The relative quantification of BnaHsfs transcription levels was determined by the methods described previously (Livak and Schmittgen, 2001).
Results
Genome-Wide Identification of Hsf Genes in Brassica napus
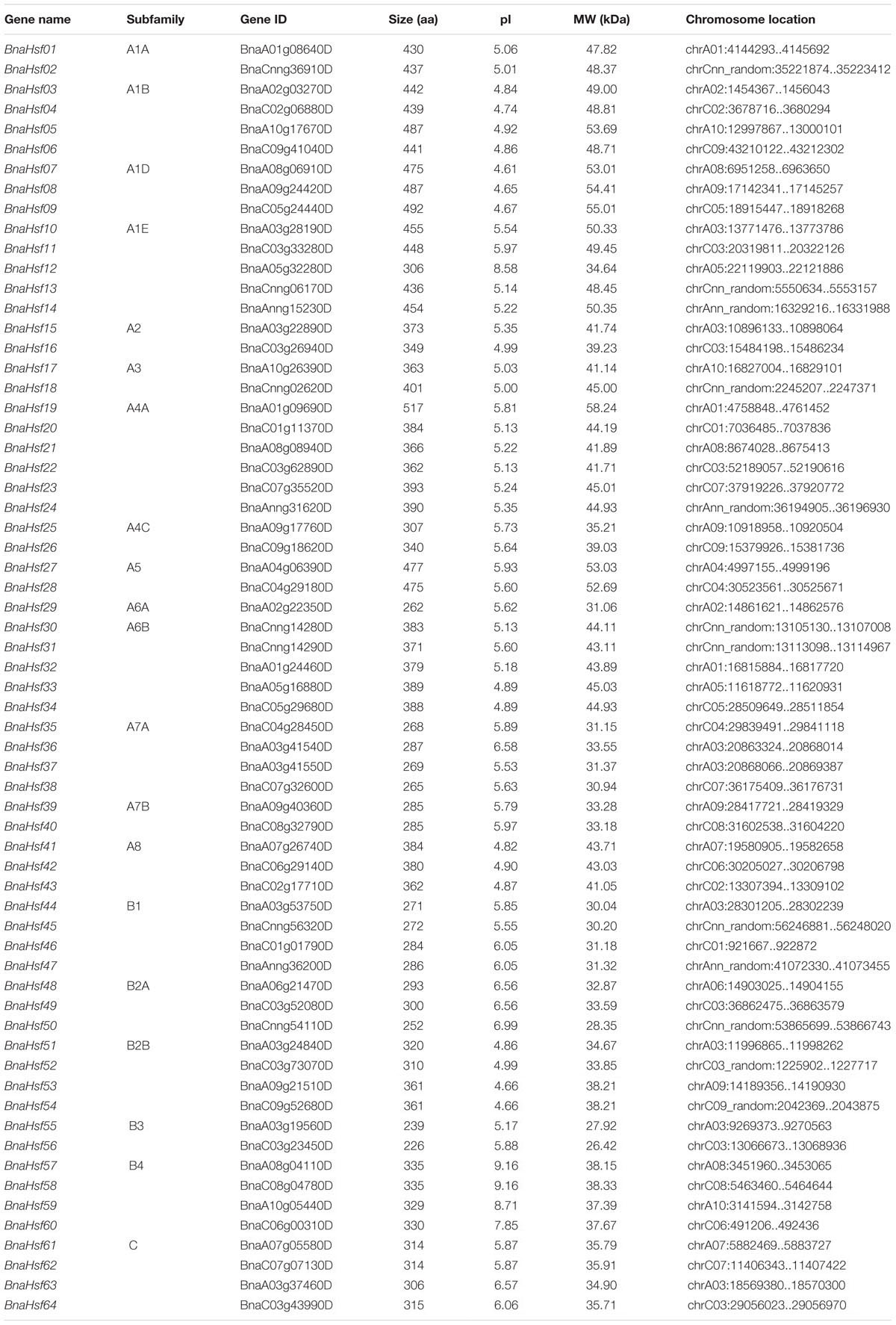
To identify Hsf genes in B. napus, candidate genes were selected by using the conserved Hsf domain (PF00447) from the Pfam database to query the B. napus genome. Meanwhile, the amino acid sequences of 21 AtHsfs from Arabidopsis were used to protein BLAST the B. napus genome. A total of 64 BnaHsfs (BnaHsf01-BnaHsf64) were identified as members of the Hsf gene family in B. napus, through simultaneous consideration of the conservation of the DBD domain, the coiled-coil structure from the SMART database, and the CD-search of the NCBI database (Table 1).
Fifty-four BnaHsf genes were distributed unevenly among the 19 chromosomes of B. napus from A01 to A10 and C01 to C09; however, 10 members (including BnaHsf02) were located on unanchored scaffolds that could not be mapped to a specific chromosome. Most BnaHsf genes were localized to chromosome A03 and C03 (8 and 7 Hsf genes), while there was only one BnaHsf on chromosome A04 and A06 (Table 1). The deduced proteins of the BnaHsfs ranged from 226 amino acids (aa; BnaHsf56) to 517 aa (BnaHsf19) in length, with predicted isoelectric points (pI) varying from 4.61 (BnaHsf07) to 9.16 (BnaHsf57 and BnaHsf60) and molecular weight (MW) from 26.42 kDa (BnaHsf56) to 58.24 kDa (BnaHsf19) (Table 1).
Phylogenetic Analysis and Multiple Sequence Alignment of BnaHsf Proteins
Among the best-studied 21 Arabidopsis Hsfs, 15 members belong to group A and 5 members belong to Group B, and only one Hsf has been classified as part of group C (Nover et al., 2001). To explore the classification and the evolutionary characteristics of the BnaHsf genes, an unrooted phylogenetic tree was generated using protein sequences of BnaHsfs (Figure 1 and Supplementary Datasheet 1).
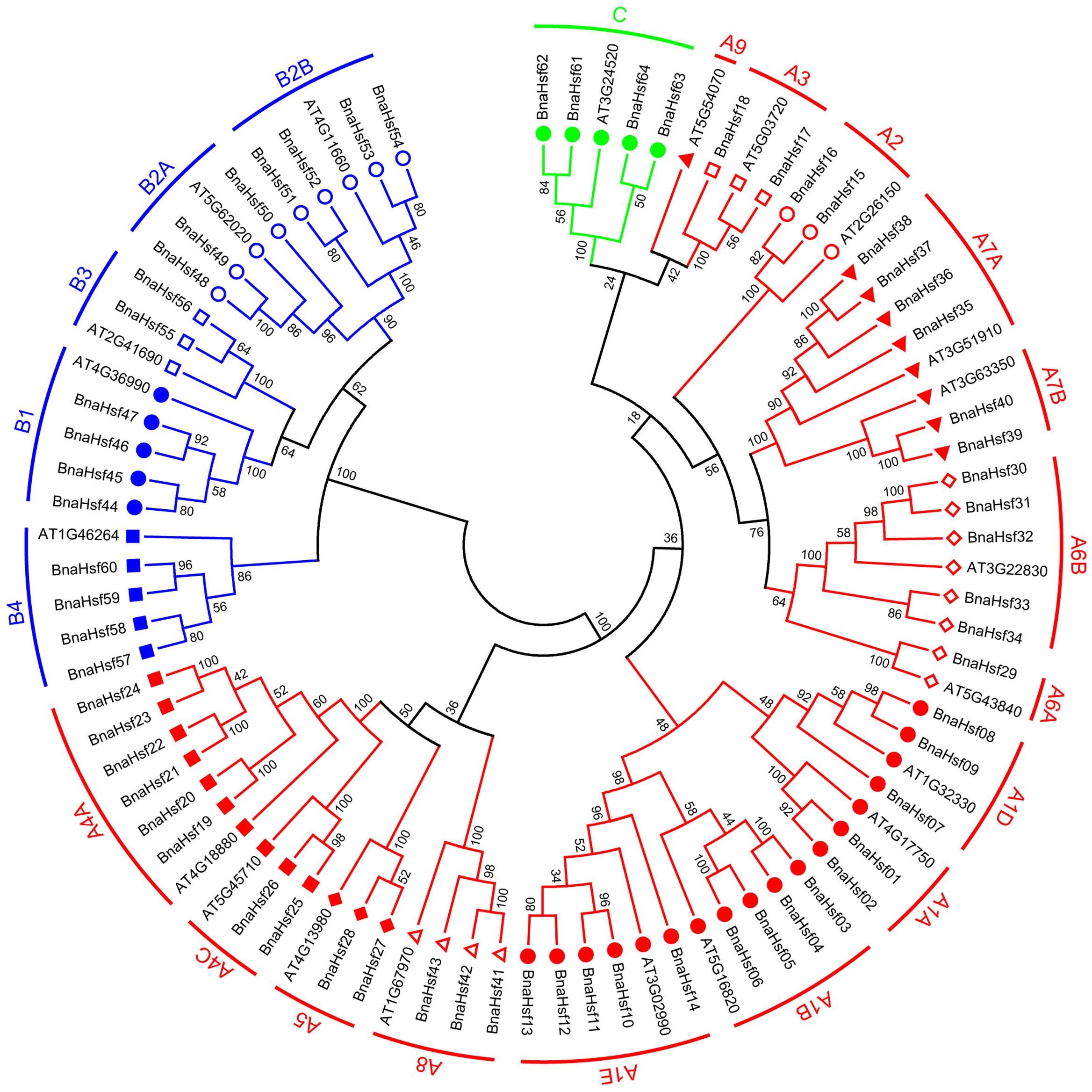
FIGURE 1. Unrooted phylogenetic tree of rapeseed and Arabidopsis Hsf family. Amino acid sequences of Hsf proteins were analyzed using the neighbor-joining method with genetic distance calculated by MEGA6.0. The numbers at the nodes represent percentage bootstrap values based on 1,000 replications. The length of the branches is proportional to the expected number of amino acid substitutions per site.
According to this phylogenetic analysis, the BnaHsfs genes were grouped into three classes, A (BnaHsf01-BnaHsf43), B (BnaHsf44-BnaHsf60), and C (BnaHsf61-BnaHsf64), as in Arabidopsis. Class A was the largest and consisted of 43 members from eight subclasses (A1–A8). In Arabidopsis, class A is further subdivided into 9 subclasses, A1-A9, with A9 (At5g54070) appearing as a single branch of the AtHsfs molecular phylogeny. However, no orthologous HsfA9 genes were found in B. napus, indicating that Hsf genes in this subgroup were lost. Class B consisted of 17 members from four subclasses (B1–B4), and class C was the smallest, containing only 4 members (Figure 1 and Table 1).
Multiple sequence alignment analysis of BnaHsfs proteins showed that a typical B. napus Hsf protein contains 5 conserved domains, including of DBD, OD, NLS, NES, and AHA domains in order from N-terminal to C-terminal. Among these, the highly structured DBD and OD domains are the most conserved sections in each BnaHsf. The OD domain (HR-A/B region) served as an important basis for classification of BnaHsfs. B. napus class B Hsf proteins, like other plant Hsf proteins, are compact and lack an insertion between the HR-A and HR-B regions (Figure 2), while an insertion of 21 aa in length was found in BnaHsfA and an insertion of 7 aa in length was found in BnaHsfC between the HR-A and HR-B regions. Thus, class A members are less conserved than class B and C members.
Structure and Motif Analysis of BnaHsf Proteins
To study the structural diversity of BnaHsf genes, the exon/intron organization of individual BnaHsf genes was analyzed by comparing cDNA sequences with the corresponding genomic DNA sequence. The detailed gene structures are shown in Figure 3A. The number of introns ranges from zero to five in BnaHsf genes. Five introns were found in BnaHsf31, while none were found in the BnaHsf63/64 gene pair. Most BnaHsfs contained one or two introns (Figure 3A).
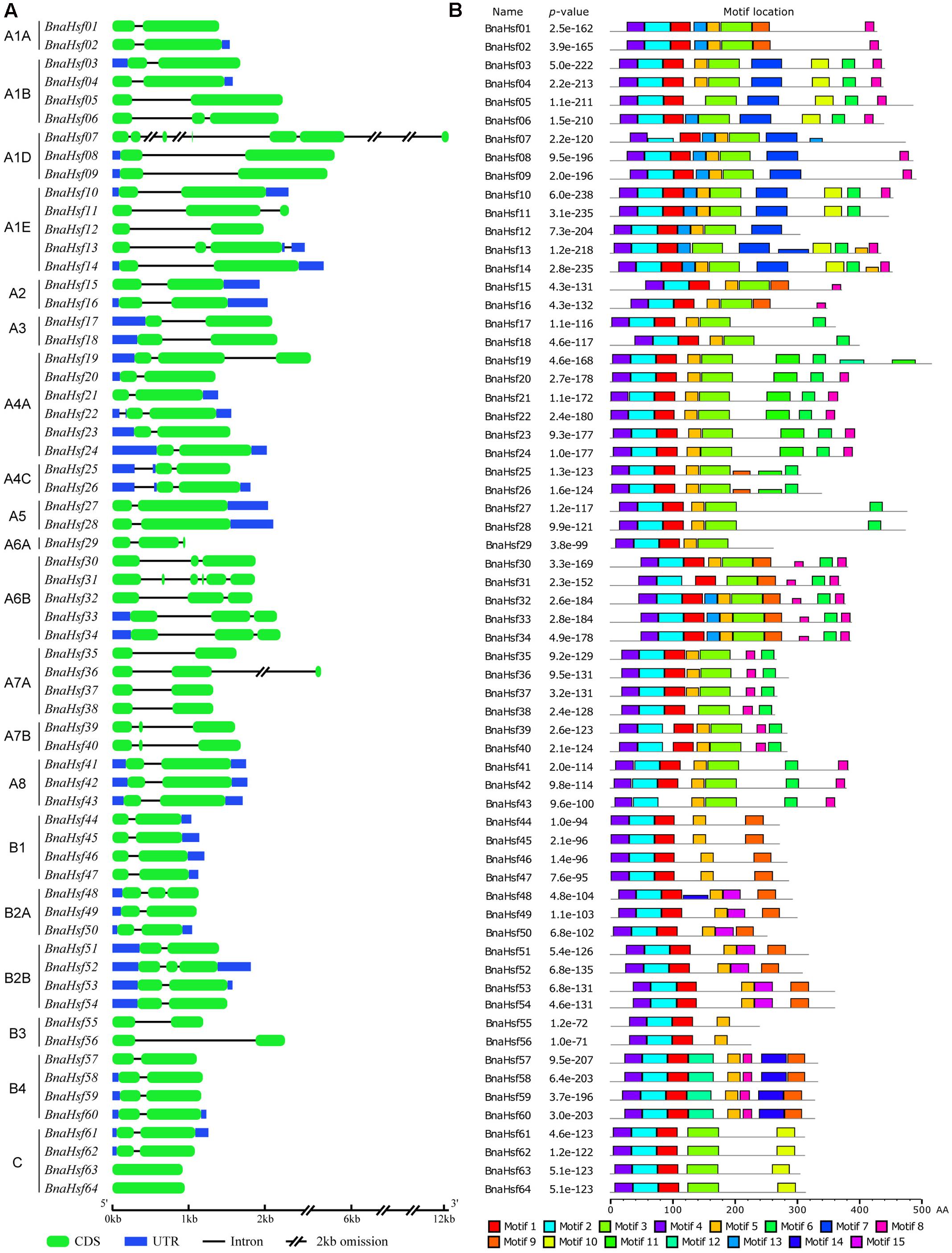
FIGURE 3. Gene organization of BnaHsfs (A) and motifs identified by MEME tools in rapeseed Hsf proteins (B). Fifteen motifs (1–15) were identified and are here indicated by different colors. The combined p-value is shown.
Conserved motif analysis was conducted by using MEME, and 15 motifs were detected in BnaHsf proteins (Figure 3B and Supplementary Figure 1). The DBD and OD domains, composed of motif 4, motif1 and motif 2, were the most conserved and were found in almost all 64 BnaHsf members, while motif 1 was absent in BnaHsf43. The NLS motif (motif 9) and NES motif (motif 15) were found in most class A and B members but not in class C BnaHsf proteins. Motif 3 is the insertion between the HR-A and HR-B regions that was found only in class A and class C members. The AHA motif (motif 6) was found in most class A members but not in classes B and C. In general, the structure of Hsf proteins was conserved throughout the BnaHsfs gene family.
Tempo-Spatial Expression Profiles of Hsf Genes in Brassica napus
To examine spatial and temporal expression profiles of BnaHsfs across different tissues and organs, an expression pattern map was drawn based on RNA-seq data (Supplementary Table 2) from twelve rapeseed tissues (leaf, root, stem, sepal, silique, pericarp, bud, stamen, ovule, new pistil, mature pistil and wilting pistil).
We found that BnaHsf genes were differently expressed among the subclasses in 14 tissues and at different developmental stages (Figure 4). BnaHsfA6 subclass (BnaHsf29-BnaHsf34) exhibited root specific expression but was hardly detected in other tissues. Subclass A7 (BnaHsf35-BnaHsf40) was also root specific but BnaHsf35 and BnaHsf37 were also detected in reproductive organs such as petal, stamen and pistil. The BnaHsf A2, A3, A4C and A5 subclasses were the most abundant BnaHsf genes and were constitutively expressed among all tissues, as were BnaHsf23 and -24 in subclass A4A, BnaHsf46 and -47 in subclass B1, and BnaHsf48 and -49 in subclass B2A. Subclass B3 (BnaHsf55-56) showed a higher transcription level in root, sepal and bud tissues. BnaHsf55 and -56 in subclass B4 were specifically expressed in ovule tissues. In contrast, class BnaHsfC was inactive in ovule tissues.
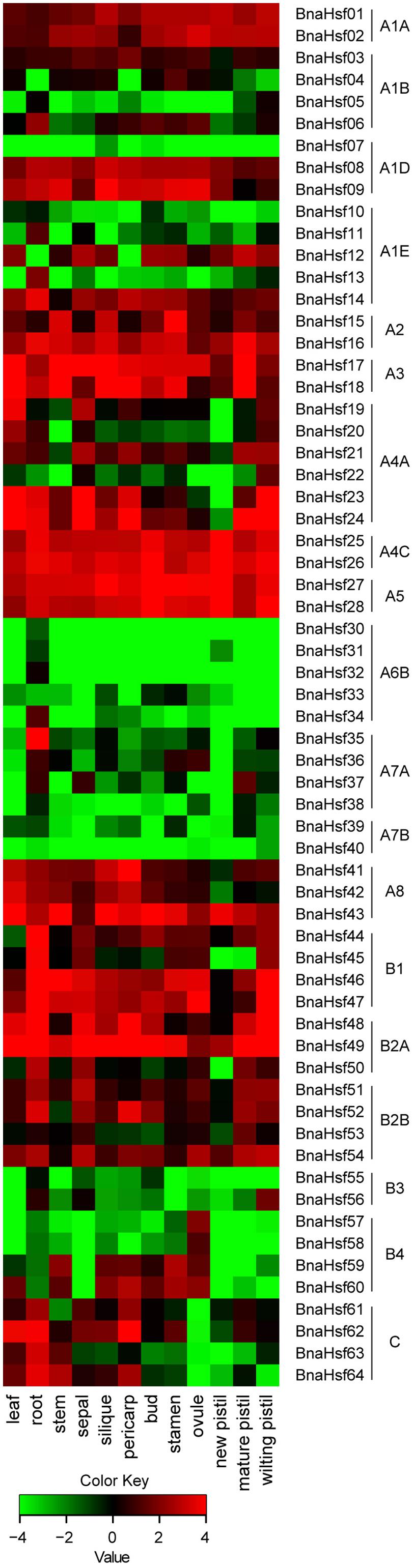
FIGURE 4. The tempo-spatial transcription profiles of BnaHsf family genes in various tissues at different developmental stages. The different colors correspond to log2 transformed value.
Expression Patterns of BnaHsf Genes under Abiotic Stresses
To determine the potential role BnaHsfs play in plant responses to different environmental stresses, the expression levels of BnaHsf genes under high temperature, drought and high CO2 stresses were analyzed using RNA-seq data (Supplementary Table 3). The mRNA for these transcriptomic analyses were extracted from the leaves of rapeseed plants both in normal growth conditions and after 3 h of heat treatment, 3 days of drought treatment and 3 h of high CO2 treatment. The results showed that BnaHsf genes were very sensitive to heat and drought stress (Figure 5). Members of subclasses A2, A4C, A5, and class B (except for the non-expressed subclasses B3 and B4) showed relatively higher basic transcription levels in leaf tissue under normal growth conditions. Among the subclasses with high basic expression levels, BnaHsf15 and -16, in subclass A2, were dramatically upregulated (>25-fold), becoming the predominant transcripts after 3 days of exposure to drought stress, but were suppressed after 3 h of heat treatment. B1 members, except for BnaHsf45, were strongly induced by drought stress, and BnaHsf46 and -47 were strongly induced by heat. All B2 members were significantly upregulated under drought conditions, but were only slightly induced by 3 h of heat treatment. As observed in the subclasses with low basic expression, a moderate induction was seen in the A1E subclass after exposure to drought and heat. Members of subclasses A3 and A4A were strongly induced by both drought and heat stress. Strikingly, the highest induction (>350-fold for BnaHsf36∼38) by drought was observed in A7A subclass, although expression of members of this subclass was hardly detectable under normal conditions. Heat treatment also resulted in a marked induction in A7A members. Members in class C were only upregulated by drought treatment. In the case of individual member genes, BnaHsf07 (in A1D) and BnaHsf40 (in A7B) were induced by drought stress, and BnaHsf43 in subclass A8 was strongly induced by heat stress.

FIGURE 5. Expression profiles of BnaHsf genes under drought, heat and high CO2 conditions. HT/CK, expression levels under heat vs. control; D/CK, expression levels under drought vs. control; HC/CK, expression levels under High CO2 vs. control. The different colors correspond to log2 transformed value.
Unlike drought and heat stress, high CO2 treatment did not cause a significant effect on the transcription level of most of the BnaHsf family genes (Figure 5). However, three member genes, BnaHsf18 (in A3), BnaHsf21 (in A4A) and BnaHsf43 (in A8), were clearly induced (3 to 9-fold) by exposure to high CO2 conditions. However, the expression of members of subclass A2 members (BnaHsf15/16) and of subclass B2 was largely suppressed by high CO2 treatment.
qRT-PCR Expression Analysis of Selected BnaHsf Genes under Abiotic Stresses
Twelve BnaHsf genes from three main classes were selected for examination of their function under three stress conditions using quantitative Real-Time PCR (qRT-PCR). These genes included BnaHsf04 from subclass A1, BnaHsf15 and -16 from subclass A2, BnaHsf17 and -18 from subclass A3, BnaHsf20 and BnaHsf23 from subclass A4, BnaHsf42 from subclass A8, BnaHsf45 from subclass B1, BnaHsf46 from subclass B2 and BnaHsf61 from class C. qRT-PCR was carried out using rapeseed plants exposed to heat (0, 1, 3, and 6 h), drought (0, 1, 2, and 3 days), and high CO2 (0, 1, 3, and 6 h).
The results of the qRT-PCR analyses were consistent with the expression patterns of selected BnaHsfs from RNA-seq data, and provided more details under progressive stresses (Figure 6 and Supplementary Table 4). The RNA-seq data showed that transcription levels of BnaHsf15-16 were decreased after exposure to 3 h heat (Figure 5), while in qRT-PCR results they were upregulated after 1 h of heat, and their expression then dropped to lower than basic levels (Figure 6). However, the strong induction of BnaHsf15-16 began with a marked reduction in transcription after 1 day of treatment under drought stress. In contrast to BnaHsf15-16, the expression levels of BnaHsf17-18 and BnaHsf43 were progressively induced by all prolonged drought, heat, and high CO2 stress conditions. A similar pattern was found in BnaHsf43 under drought and heat conditions. For BnaHsf21-22, the induction in their transcription was weakened as heat stress processed, while under progressive drought, the induction by stress showed an opposite enhancement (Figure 6).
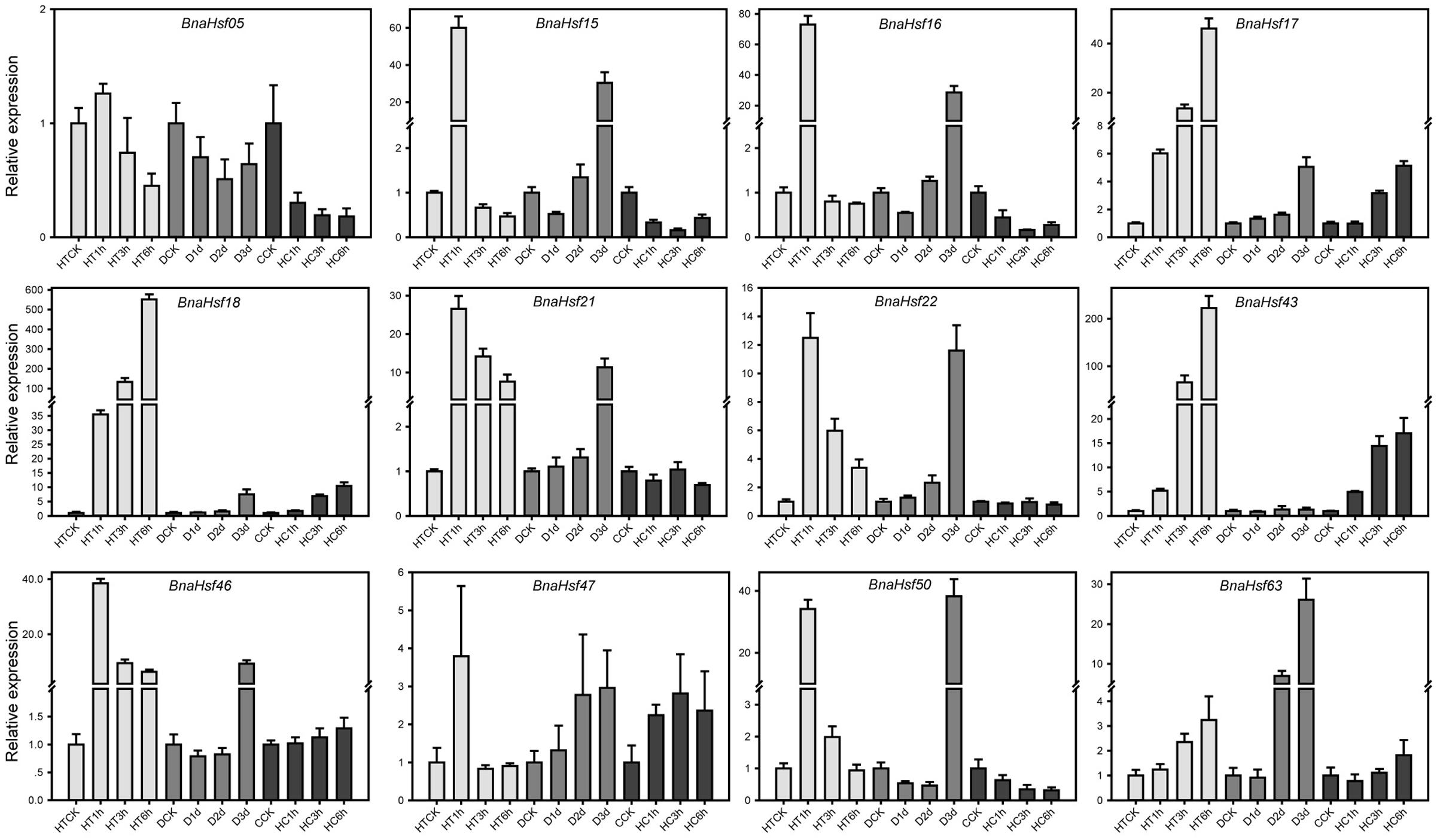
FIGURE 6. Real-time PCR analysis of the selected 12 representative BnaHsf genes responded to drought, heat and high CO2 treatments. HT, heat; D, drought; HC, High CO2.
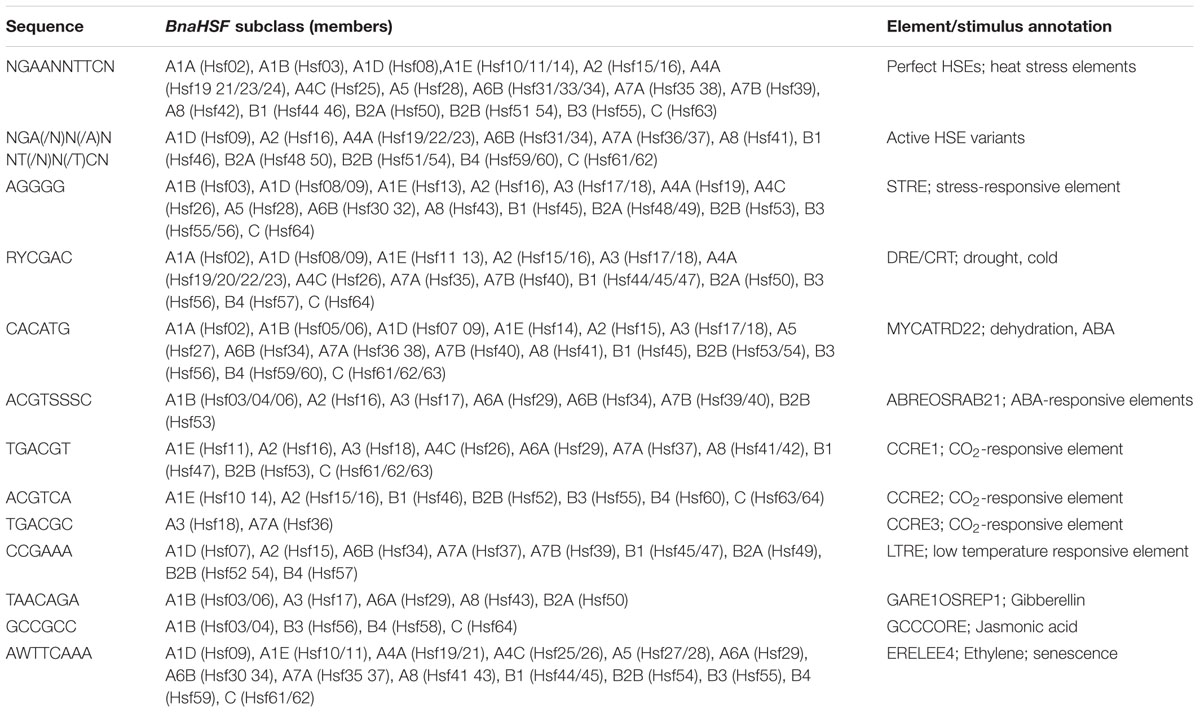
Regulatory Cis-Element Analysis of BnaHsf Genes
To identify the presence of putative regulatory cis-acting elements enriched in BnHsf genes, the promoter sequences upstream of their CDS were extracted and searched against the PLACE database (Higo et al., 1999). Analysis showed that HSEs were the most abundant cis-elements in promoter regions of BnHsf genes, including perfect type and active HSE variants (Table 2). Many of the subclasses of the BnHsf gene family possess the two types of HSE, although these are not present in subclasses A3 and A6A. Subclass A6A contains only one member, BnHsf29, which may be a pseudogene since its transcription cannot be detected in any tissues of rapeseed plants. The two members (BnHsf17 and -18) of subclass A3 were found to have three other major types of stress-related cis-elements present in the BnHsf family, STRE, DRE/CRT, and MYCATRD22. The STRE element was first found to be stress responsive in yeast and can serve as a specific binding site for HsfA1a in Arabidopsis (Martinez-Pastor et al., 1996; Haralampidis et al., 2002; Guo et al., 2008). STRE was present in most subclasses of BnaHsf except A1A, A6A, and A7. DRE/CRT and MYCATRD22, two types of cis-elements responsive to drought stress, also appeared in most BnHsf subclasses. Two other stress related cis-elements, ABREOSRAB21 and LTRE, were found in some of the BnaHsf family members. In addition, three CO2-responsive elements (CCRE1/2/3) (Ohno et al., 2012; Tanaka et al., 2016) were observed in all BnHsf subclasses except A1A/B/D, A4A, A5, A6B, A7B, and B2A. The presence of these stress related cis-elements is likely responsible for the regulative expression patterns of BnHsf genes under drought, heat, and high CO2 conditions. Moreover, some phytohormone responsive related cis-elements were enriched in promoter regions of BnHsf member genes, which may be involved in the stress acclimation response and development.
Discussion
High Number of Hsf Family Genes in Rapeseed Genome
Brassica napus (rapeseed, genome AACC) is an amphidiploid species formed by recent interspecific hybridization between ancestors of B. oleracea (genome CC) and B. rapa (genome AA) (Chalhoub et al., 2014). In this study, we identified 64 Hsf genes in the genome of B. napus. Unlike yeast and animals, plants usually have many Hsf coding genes. There are 21 Hsf member genes in the model plant Arabidopsis thaliana (Nover et al., 2001), 25 members in rice (Chauhan et al., 2011), 56 Hsf genes in wheat (Xue et al., 2014). To date, BnaHsfs represent the largest Hsf gene family in plant species of which Hsf member genes were analyzed. The diversification of plant Hsfs is presumably the result of gene- and whole-genome duplications (WGD) at different points in evolution, followed by gene loss (Chalhoub et al., 2014). In the case of rapeseed, the allopolyploid process, that followed from the fusion of genomes A and C, might also play a crucial role in the expansion of the BnaHsf gene family. In addition, the large size of the BnaHsf family may have been needed for adaptation of rapeseed to diverse climatic zones.
Structural Analysis of BnaHsf Genes
Similar to other plant Hsf families, the modular structure of rapeseed Hsf proteins is well conserved. While in comparison with that in Arabidopsis, there is no Hsf member in subclass A9 subclass in rapeseed. This differs from other eudicot plants, most of whom have sub classA9 Hsfs. This subclass is also lost in the Hsf family of B. rapa (Huang et al., 2015). The DBD is characterized as a central domain for the Hsf protein: it specifically binds to HSEs in the target promoter region, and subsequently activates the transcription of associated heat-inducible genes. The DBD domain of plant Hsfs is encoded by two regions separated by an evolutionarily conserved intron, which was inserted immediately adjacent to the HTH DNA binding motif (Nover et al., 2001; Scharf et al., 2012). Most BnaHsf genes have this intron in their DBD domains; however, no intron was found in BnaHsf63 and -64 genes from class C, as shown by their gene structure (Figure 3A). As far as we know, the fact that is this highly conserved intron in not present in the DBD domain of a plant Hsf is unique. Furthermore, this fact may indicate that BnaHsf63 and -64 have a novel regulation pattern relative to other Hsf genes.
Diverse Transcriptional Patterns of BnaHsf Family Genes during Development and Abiotic Stresses
The functional diversification of BnaHsf family genes was also found in the tempo-spatial expression profile of these genes during development and abiotic stress treatments. Among the tissues at different developmental stages, subclasses A1A, A2/3, A4C, A5, A8, and B2 were found to be constitutively expressed at relatively high levels in all the tissues examined. While almost all member genes from subclasses A6/7 and B3/4 were hardly detected in any tissue, BnaHsf35 from subclass A7A showed a high level of expression in root tissue. Subclasses B1 and B2A also showed high levels of expression in root tissue. Members of class C also showed increased expression in root tissue, but were not expressed in ovule tissue. These results suggest that these BnaHsf genes may be involved in root development. Under abiotic stress, many BnaHsf genes were upregulated in response to drought treatment. The number of drought induced BnaHsf genes was comparable to those induced by heat. This suggests that Hsf genes may also play an important role in the response and the acclimation to drought stress in rapeseed. Furthermore, the most inducible BnaHsf genes were upregulated by both drought and heat treatment, as shown by the combination of RNA-seq and qRT-PCR data. While BnaHsf43 of subclass A8 was only induced by heat, BnaHsf07 of subclass A1D and members of class C were predominantly upregulated by drought.
According to the combined transcriptional analysis, heat inducible BnaHsf genes could be divided into three groups. The first group consisted of BnaHsf15 and -16 (subclass A2), BnaHsf47 (subclass B1) and BnaHsf50 (subclass B2A), in which the expression of member genes exhibited an immediate and strong induction after 1 h of heat to a high level of 40∼70-fold of that in non-stressed control, followed by a dramatic drop to the basic expression level after 3 and 6 h of heat treatment, even was slightly suppressed in subclass A2 after 3 and 6 h of heat. It may be that this group of genes governs early heat stress response. The second group contains BnaHsf21 and -22 of A4A, and BnaHsf46 of subclass B1. The transcription of the second group members showed also a fast and strong upregulation after 1 h of heat exposure. This upregulation of gene expression gradually declined after 3 and 6 h of heat, but still maintained at a high level relative to that in control. Genes from the second group might be involved in both early and late heat response. The last group greatly differed from the other two, comprised of BnaHsf17 and -18 of subclass A3 and BnaHsf43 of subclass A8. The genes from this third group were upregulated after 1 h of heat, and this induction was continuously enhanced as the stress treatment proceeded, finally peaking after 6 h of heat stress (46∼550-fold vs. control). The members of this group likely have some function to facilitate acclimation to prolonged heat stress.
Drought induced BnaHsf genes seemed to have a single expression pattern. The genes continuously increased transcription during exposure to drought, and reached peak expression after the 3 days drought treatment. BnaHsf genes also played an important role in response to high CO2 treatment, as BnaHsf18 (A3), BnaHsf21 (A4A) and BnaHsf43 (A8) were strongly upregulated, while members of A2 and B2 subclasses were downregulated.
Various Regulatory Cis-Elements Enriched in the Promoters of BnaHsf Genes
Regulatory element analysis revealed that there were many stress-related cis-acting elements enriched in the promoter regions of BnaHsf family genes. HSEs were found to be the dominant cis-elements (Table 2). Complex interactions may exist among BnaHsf genes, and these may come about via trans-acting regulation, as HSEs are marker binding sites for plant Hsf proteins. Previous work has supported this idea that HsfA1a/b target class B Hsf genes and are responsible for their induction during heat response in Arabidopsis (Lohmann et al., 2004; von Koskull-Döring et al., 2007), and that HsfA5 inhibits the activity of HsfA4 (Baniwal et al., 2007). Other abiotic stress-related cis-elements, including STRE, DRE/CRT, MYCATRD22, ABRE, CCRE, and LTRE were also major regulatory elements found in BnaHsf genes. The presence of these stress-related elements seemed to be correlated to the expression response of BnaHsfs to heat, drought, and high CO2 treatments. For example, many drought related DRE/CRT and MYCATRD22 elements found upstream were associated with a marked induction of BnaHsf genes by drought stress. The two CCRE elements situated in promoter region also agreed with our observation of high induction levels of BnaHsf18 under high CO2 treatment. Unlike BnHsf15 and -16 of subclass A2, the heat highly inducible genes BnHsf17 and -18 of subclass A3 do not have functional HSE elements, but rather an STRE element was found upstream of the target genes. The STRE element was identified to be stress responsive, and serves as a direct binding site for HsfA1a besides HSE in Arabidopsis (Guo et al., 2008). Furthermore, the deletion of STRE from the promoter of the Arabidopsis Hsp90-1 gene decreased its promoter activity under heat stress conditions (Haralampidis et al., 2002). These findings suggest that STRE also plays a crucial role in transcriptional regulation under heat conditions, as do HSEs. Moreover, the different heat induced expression patterns of subclass A3 and subclass A2 BnaHsf genes provides evidence for differential transcription regulation abilities of STRE and HSE element. Unexpectedly, rapeseed subclass A1A was not heat inducible, although HSE elements are found in the promoter. While HsfA1a serves as master regulator of thermotolerance in tomato (Mishra et al., 2002), it also functions actively in Arabidopsis under heat stress. These results may indicate differential gene regulation of rapeseed Hsf genes from those found in other plants, even those in the Brassicaceae.
Conclusion
Our genome-wide investigation of Hsf genes in B. napus reveals the largest plant Hsf gene family to date. With expression profile analysis, novel members of BnaHsf family were found to respond to high temperatures, as well as drought and high CO2 stresses. Further characterization of these novel multifunctional BnaHsf genes will improve our understanding of the acclimation response of plants to multifactorial and combinational abiotic stresses, and may also provide useful genetic resources for further research on abiotic stress resistance in crops.
Author Contributions
XZ and WH conceived and designed the research. XZ, CH, LZ, HL, JY, and ZH performed the experiments and bioinformatics. XZ and CH analyzed the data. XZ and CH wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Program on High Technology Development (2013AA102602), the National Program for Basic Research of China (2015CB150200) and the Natural Science Foundation of Hubei province (2015CFB348).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01174/full#supplementary-material
Footnotes
- ^ http://www.genoscope.cns.fr/brassicanapus/
- ^ http://cn.expasy.org/tools
- ^ http://meme-suite.org/tools/meme
- ^ http://gsds.cbi.pku.edu.cn
References
Almoguera, C., Rojas, A., Diaz-Martin, J., Prieto-Dapena, P., Carranco, R., and Jordano, J. (2002). A seed-specific heat-shock transcription factor involved in developmental regulation during embryogenesis in sunflower. J. Biol. Chem. 277, 43866–43872. doi: 10.1074/jbc.M207330200
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Baniwal, S. K., Chan, K. Y., Scharf, K. D., and Nover, L. (2007). Role of heat stress transcription factor HsfA5 as specific repressor of HsfA4. J. Biol. Chem. 282, 3605–3613. doi: 10.1074/jbc.M609545200
Becklin, K. M., Walker, S. M., Way, D. A., and Ward, J. K. (2017). CO2 studies remain key to understanding a future world. New Phytol. 214, 34–40. doi: 10.1111/nph.14336
Bharti, K., Von Koskull-Döring, P., Bharti, S., Kumar, P., Tintschl-Korbitzer, A., Treuter, E., et al. (2004). Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 16, 1521–1535. doi: 10.1105/tpc.019927
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Charng, Y. Y., Liu, H. C., Liu, N. Y., Chi, W. T., Wang, C. N., Chang, S. H., et al. (2007). A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 143, 251–262. doi: 10.1104/pp.106.091322
Chauhan, H., Khurana, N., Agarwal, P., and Khurana, P. (2011). Heat shock factors in rice (Oryza sativa L.): genome-wide expression analysis during reproductive development and abiotic stress. Mol. Genet. Genomics 286, 171–187. doi: 10.1007/s00438-011-0638-8
Czarnecka-Verner, E., Pan, S., Salem, T., and Gurley, W. B. (2004). Plant class B HSFs inhibit transcription and exhibit affinity for TFIIB and TBP. Plant Mol. Biol. 56, 57–75. doi: 10.1007/s11103-004-2307-3
Czarnecka-Verner, E., Yuan, C. X., Scharf, K. D., Englich, G., and Gurley, W. B. (2000). Plants contain a novel multi-member class of heat shock factors without transcriptional activator potential. Plant Mol. Biol. 43, 459–471. doi: 10.1023/A:1006448607740
Fang, Y., and Xiong, L. (2015). General mechanisms of drought response and their application in drought resistance improvement in plants. Cell. Mol. Life Sci. 72, 673–689. doi: 10.1007/s00018-014-1767-0
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Guo, L., Chen, S., Liu, K., Liu, Y., Ni, L., and Zhang, K. (2008). Isolation of heat shock factor HsfA1a-binding sites in vivo revealed variations of heat shock elements in Arabidopsis thaliana. Plant Cell Physiol. 49, 1306–1315. doi: 10.1093/pcp/pcn105
Guo, M., Liu, J.-H., Ma, X., Luo, D.-X., Gong, Z.-H., and Lu, M.-H. (2016). The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 7:114. doi: 10.3389/fpls.2016.00114
Guo, M., Lu, J. P., Zhai, Y. F., Chai, W. G., Gong, Z. H., and Lu, M. H. (2015). Genome-wide analysis, expression profile of heat shock factor gene family (CaHsfs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.). BMC Plant Biol. 15:151. doi: 10.1186/s12870-015-0512-7
Haralampidis, K., Milioni, D., Rigas, S., and Hatzopoulos, P. (2002). Combinatorial interaction of cis elements specifies the expression of the Arabidopsis AtHsp90-1 gene. Plant Physiol. 129, 1138–1149. doi: 10.1104/pp.004044
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hu, H., and Xiong, L. (2014). Genetic engineering and breeding of drought-resistant crops. Annu. Rev. Plant Biol. 65, 715–741. doi: 10.1146/annurev-arplant-050213-040000
Huang, X. Y., Tao, P., Li, B. Y., Wang, W. H., Yue, Z. C., Lei, J. L., et al. (2015). Genome-wide identification, classification, and analysis of heat shock transcription factor family in Chinese cabbage (Brassica rapa pekinensis). Genet. Mol. Res. 14, 2189–2204. doi: 10.4238/2015.March.27.5
Kotak, S., Vierling, E., Baumlein, H., and von Koskull-Döring, P. (2007). A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 19, 182–195. doi: 10.1105/tpc.106.048165
Lamesch, P., Berardini, T. Z., Li, D., Swarbreck, D., Wilks, C., Sasidharan, R., et al. (2012). The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210. doi: 10.1093/nar/gkr1090
Li, P. S., Yu, T. F., He, G. H., Chen, M., Zhou, Y. B., Chai, S. C., et al. (2014). Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genomics 15:1009. doi: 10.1186/1471-2164-15-1009
Li, Z., Zhang, L., Wang, A., Xu, X., and Li, J. (2013). Ectopic overexpression of SlHsfA3, a heat stress transcription factor from tomato, confers increased thermotolerance and salt hypersensitivity in germination in transgenic Arabidopsis. PLoS ONE 8:e54880. doi: 10.1371/journal.pone.0054880
Liu, H. C., Liao, H. T., and Charng, Y. Y. (2011). The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 34, 738–751. doi: 10.1111/j.1365-3040.2011.02278.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lohmann, C., Eggers-Schumacher, G., Wunderlich, M., and Schoffl, F. (2004). Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol. Genet. Genomics 271, 11–21. doi: 10.1007/s00438-003-0954-8
Martinez-Pastor, M. T., Marchler, G., Schüller, C., Marchler-Bauer, A., Ruis, H., and Estruch, F. (1996). The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15, 2227–2235.
Mishra, S. K., Tripp, J., Winkelhaus, S., Tschiersch, B., Theres, K., Nover, L., et al. (2002). In the complex family of heat stress transcription factors, HSfA1 has a unique role as master regulator of thermotolerance in tomato. Gene. Dev. 16, 1555–1567. doi: 10.1101/gad.228802
Myers, S. S., Zanobetti, A., Kloog, I., Huybers, P., Leakey, A. D. B., Bloom, A. J., et al. (2014). Increasing CO2 threatens human nutrition. Nature 510, 139–142. doi: 10.1038/nature13179
Nishizawa, A., Yabuta, Y., Yoshida, E., Maruta, T., Yoshimura, K., and Shigeoka, S. (2006). Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 48, 535–547. doi: 10.1111/j.1365-313X.2006.02889.x
Nishizawa-Yokoi, A., Nosaka, R., Hayashi, H., Tainaka, H., Maruta, T., Tamoi, M., et al. (2011). HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the Hsf signaling network in response to environmental stress. Plant Cell Physiol. 52, 933–945. doi: 10.1093/pcp/pcr045
Nover, L., Bharti, K., Döring, P., Mishra, S. K., Ganguli, A., and Scharf, K. D. (2001). Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperon. 6, 177–189. doi: 10.1379/1466-12682001006<0177:AATHST<2.0.CO;2
Ogawa, D., Yamaguchi, K., and Nishiuchi, T. (2007). High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J. Exp. Bot. 58, 3373–3383. doi: 10.1093/jxb/erm184
Ohno, N., Inoue, T., Yamashiki, R., Nakajima, K., Kitahara, Y., and Ishibashi, M. (2012). CO2-cAMP-responsive cis-elements targeted by a transcription factor with CREB/ATF-like basic zipper domain in the marine diatom Phaeodactylum tricornutum. Plant Physiol. 158, 499–513. doi: 10.1104/pp.111.190249
Pereira, A. (2016). Plant abiotic stress challenges from the changing environment. Front. Plant Sci. 7:1123. doi: 10.3389/fpls.2016.01123
Prieto-Dapena, P., Castano, R., Almoguera, C., and Jordano, J. (2008). The ectopic overexpression of a seed-specific transcription factor, HaHSFA9, confers tolerance to severe dehydration in vegetative organs. Plant J. 54, 1004–1014. doi: 10.1111/j.1365-313X.2008.03465.x
Sakuma, Y., Maruyama, K., Qin, F., Osakabe, Y., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2006). Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. U.S.A. 103, 18822–18827. doi: 10.1073/pnas.0605639103
Scharf, K.-D., Berberich, T., Ebersberger, I., and Nover, L. (2012). The plant heat stress transcription factor (Hsf) family: structure, function and evolution. BBA-Gene Regul. Mech. 1819, 104–119. doi: 10.1016/j.bbagrm.2011.10.002
Schramm, F., Larkindale, J., Kiehlmann, E., Ganguli, A., Englich, G., Vierling, E., et al. (2008). A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 53, 264–274. doi: 10.1111/j.1365-313X.2007.03334.x
Shim, D., Hwang, J. U., Lee, J., Lee, S., Choi, Y., An, G., et al. (2009). Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell 21, 4031–4043. doi: 10.1105/tpc.109.066902
Song, L., Huang, S.s.C., Wise, A., Castanon, R., Nery, J.R., Chen, H., et al. (2016). A transcription factor hierarchy defines an environmental stress response network. Science 354:aag1550. doi: 10.1126/science.aag1550
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tanaka, A., Ohno, N., Nakajima, K., and Matsuda, Y. (2016). Light and CO2/cAMP signal cross talk on the promoter elements of chloroplastic beta-carbonic anhydrase genes in the marine diatom Phaeodactylum tricornutum. Plant Physiol. 170, 1105–1116. doi: 10.1104/pp.15.01738
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
von Koskull-Döring, P., Scharf, K.-D., and Nover, L. (2007). The diversity of plant heat stress transcription factors. Trends Plant Sci. 12, 452–457. doi: 10.1016/j.tplants.2007.08.014
Wang, J., Sun, N., Deng, T., Zhang, L., and Zuo, K. (2014). Genome-wide cloning, identification, classification and functional analysis of cotton heat shock transcription factors in cotton (Gossypium hirsutum). BMC Genomics 15:961. doi: 10.1186/1471-2164-15-961
Wang, X., Huang, W., Liu, J., Yang, Z., and Huang, B. (2017). Molecular regulation and physiological functions of a novel FaHsfA2c cloned from tall fescue conferring plant tolerance to heat stress. Plant Biotechnol. J. 15, 237–248. doi: 10.1111/pbi.12609
Wiederrecht, G., Seto, D., and Parker, C. S. (1988). Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell 54, 841–853. doi: 10.1016/S0092-8674(88)91197-X
Xue, G. P., Sadat, S., Drenth, J., and McIntyre, C. L. (2014). The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 65, 539–557. doi: 10.1093/jxb/ert399
Yamanouchi, U., Yano, M., Lin, H., Ashikari, M., and Yamada, K. (2002). A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc. Natl. Acad. Sci. U.S.A. 99, 7530–7535. doi: 10.1073/pnas.112209199
Yoshida, T., Mogami, J., and Yamaguchi-Shinozaki, K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. doi: 10.1016/j.pbi.2014.07.009
Zhang, J., Jia, H., Li, J., Li, Y., Lu, M., and Hu, J. (2016). Molecular evolution and expression divergence of the Populus euphratica Hsf genes provide insight into the stress acclimation of desert poplar. Sci. Rep. 6:30050. doi: 10.1038/srep30050
Zhu, J.-K. (2016). Abiotic stress signaling and responses in plants. Cell 167, 313–324. doi: 10.1016/j.cell.2016.08.029
Keywords: Hsf gene family, abiotic stress, high CO2, gene expression, Brassica napus
Citation: Zhu X, Huang C, Zhang L, Liu H, Yu J, Hu Z and Hua W (2017) Systematic Analysis of Hsf Family Genes in the Brassica napus Genome Reveals Novel Responses to Heat, Drought and High CO2 Stresses. Front. Plant Sci. 8:1174. doi: 10.3389/fpls.2017.01174
Received: 10 April 2017; Accepted: 19 June 2017;
Published: 06 July 2017.
Edited by:
Ruth Grene, Virginia Tech, United StatesReviewed by:
Zhaoqing Chu, Shanghai Chenshan Plant Science Research Center (CAS), ChinaKazuo Nakashima, Japan International Research Center for Agricultural Sciences, Japan
Copyright © 2017 Zhu, Huang, Zhang, Liu, Yu, Hu and Hua. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Hua, huawei@oilcrops.cn
†These authors have contributed equally to this work.
 Xiaoyi Zhu
Xiaoyi Zhu Chunqian Huang†
Chunqian Huang†