- 1The Genetic Engineering International Cooperation Base of Chinese Ministry of Science and Technology, Key Laboratory of Molecular Biophysics of Chinese Ministry of Education, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan, China
- 2Hubei University of Education, Wuhan, China
Calcineurin B-like protein interacting protein kinases (CIPKs) are vital elements in plant abiotic stress signaling pathways. However, the functional mechanism of CIPKs has not been understood clearly, especially in Brachypodium distachyon, a new monocot model plant. In this study, BdCIPK31, a CIPK gene from B. distachyon was characterized. BdCIPK31 was downregulated by polyethylene glycol, NaCl, H2O2, and abscisic acid (ABA) treatments. Transgenic tobacco plants overexpressing BdCIPK31 presented improved drought and salt tolerance, and displayed hypersensitive response to exogenous ABA. Further investigations revealed that BdCIPK31 functioned positively in ABA-mediated stomatal closure, and transgenic tobacco exhibited reduced water loss under dehydration conditions compared with the controls. BdCIPK31 also affected Na+/K+ homeostasis and root K+ loss, which contributed to maintain intracellular ion homeostasis under salt conditions. Moreover, the reactive oxygen species scavenging system and osmolyte accumulation were enhanced by BdCIPK31 overexpression, which were conducive for alleviating oxidative and osmotic damages. Additionally, overexpression of BdCIPK31 could elevate several stress-associated gene expressions under stress conditions. In conclusion, BdCIPK31 functions positively to drought and salt stress through ABA signaling pathway. Overexpressing BdCIPK31 functions in stomatal closure, ion homeostasis, ROS scavenging, osmolyte biosynthesis, and transcriptional regulation of stress-related genes.
Introduction
Abiotic stress severely constrains the growth and development of plants, causes crop yield losses and even death (Bartels and Sunkar, 2005). Plants have developed a complex mechanism to prevent damages caused by environmental changes. ABA is a pivotal element in this mechanism. ABA functions in seed germination inhibition, growth regulation, fruit abscission, and stomatal closure (Raghavendra et al., 2010). Ca2+ serves as a second messenger in ABA signaling pathway (Luan, 2009). The fluctuation of cytoplasmic calcium concentration ([Ca2+]cyt) initiates a series of intracellular biochemical and physiological changes (Luan, 2009). Ca2+ sensors detect the change in [Ca2+]cyt and transduce the signal to sensor responder proteins (Yu et al., 2014). During the past 20 years, multiple families of plant Ca2+ sensors and responders are identified, including calmodulins, CDPKs, CBLs, and CIPKs (Asano et al., 2005; Luan, 2009). Among these proteins, CBL-CIPK networks and CDPKs are unique to higher plants (Liu and Zhu, 1998; Kudla et al., 1999).
The typical structure of the CIPK protein contains a kinase activation domain, a regulatory domain, and a junction domain connecting the two domains (Batistic and Kudla, 2004). In the CBL-CIPK network, Ca2+-bound CBL interacts with the CIPK NAF/FISL motif and activates CIPK. Afterward, the CIPK can phosphorylate its targets and transduce the Ca2+ signal downstream (Luan, 2009). Genome-wide analyses have suggested 26 CIPKs in Arabidopsis (Kolukisaoglu et al., 2004), 33 CIPKs in rice (Kolukisaoglu et al., 2004; Piao et al., 2010), 43 CIPKs in maize (Chen et al., 2011), and 23 CIPKs in canola (Zhang et al., 2014).
A few studies have indicated that the CIPKs function in plant development (Tripathi et al., 2009; Held et al., 2011), nutrient uptake (Li et al., 2006; Xu et al., 2006; Cheong et al., 2007; Chen et al., 2012), and pollen tube elongation (Zhou et al., 2015). However, the CIPKs perform major functions in stress responses. The first characterized CBL-CIPK network is the SOS network. Plasma membrane-localized SOS3 (AtCBL4) recruits SOS2 (AtCIPK24). Subsequently, the CBL-CIPK complex activates a Na+/H+ antiporter SOS1 (AtNHX7), enhances sodium export, and promotes salt tolerance (Liu and Zhu, 1997, 1998; Qiu et al., 2002). The atcipk3 mutant shows a hypersensitive phenotype when treated with salt and ABA (Kim et al., 2003). By contrast, the atcipk1 mutant is hypersensitive to osmotic treatment, but insensitive to ABA (D’Angelo et al., 2006). The atcipk23 mutant exhibits enhanced drought tolerance by regulating leaf transpiration (Cheong et al., 2007), whereas the atcipk21 mutant presents impaired salt and osmotic tolerance (Pandey et al., 2015). Similar CBL-CIPK networks are also found in other plant species including wheat. TaCIPK14 and TaCIPK29, which enhance salt and low temperature resistance in transgenic tobacco, respectively (Deng et al., 2013a,b), whereas TaCIPK25 negatively regulated salt tolerance in transgenic wheat (Jin et al., 2016). The overexpression of the constitutively activated form of BnCIPK6 enhances salt and low-K+ tolerances, as well as ABA sensitivity in Arabidopsis (Chen et al., 2012). The MdCIPK6L improves tolerance to salt, drought, and chilling stress in transgenic apples and Arabidopsis (Wang et al., 2012). Notably, ABA participates extensively in the responses of CIPK genes to abiotic stress. Among the 21 stress-responsive OsCIPK genes, 17 OsCIPKs are also responsive to ABA (Xiang et al., 2007).
Brachypodium distachyon, belonging to the Pooideae subfamily, has close evolutionary relationship with rice, wheat, and sorghum (Vogel et al., 2010). Therefore, the characterization of B. distachyon genes could provide reference for studies of other species in Pooideae subfamily. Our previous study identified 32 CIPK (SnRK3) genes in the Brachypodium genome (Wang et al., 2015). However, none of these BdCIPKs has been functionally characterized. In the current study, we characterized a BdCIPK gene, designated BdCIPK31. Transgenic tobacco overexpressing BdCIPK31 exhibited tolerance to drought and salt stress and hypersensitivity to ABA. These findings indicated that BdCIPK31 participates in plant’s response to abiotic stress and ABA signaling.
Materials and Methods
Plant Materials Preparation
Brachypodium distachyon inbred line Bd21 was employed as plant material. Sterilized seeds were germinated in Petri dishes with water at 25°C. For stress and signal-molecule treatments, 10-day-old seedlings were treated with 20% (w/v) PEG6000, 200 mM NaCl, 10 mM H2O2 or 100 μM ABA solutions. Normal growing seedlings were employed as untreated control. The leaves of these seedlings were harvested in time for total RNA extraction. For organ-specific expression analysis, leaves, stems, roots, and spikelets were collected from 6-week-old mature Brachypodium plants grown in soil. All the seedlings were cultured in 25°C, 14 h light/10 h dark cycle.
Cloning and Bioinformatics Analysis of the BdCIPK31 Gene
The cDNA of the BdCIPK31 gene (accession No. XM_003562376) was amplified by primer P1 (Supplementary Table S1) using mRNA extracted from 10-day-old Brachypodium seedlings. Homologs of BdCIPK31 were searched using NCBI BlastP1 and aligned by ClustalX. The phylogenetic analysis was performed by MEGA 6.0 using the neighbor-joining method.
Yeast-Two-Hybrid Assay
The CDS of BdCIPK31 gene was introduced into pGADT7 while the BdCBLs were introduced into pGBKT7 (The primers are shown in Supplementary Table S1). Subsequently, The AD-BdCIPK31 and the BD-BdCBLs constructs were co-transformed into yeast strain AH109 by using YeastmakerTM Yeast Transformation System 2 (Clontech, CA, United States). The transformants grown on DDO/-Leu/-Trp were diluted and dotted on TDO/-Leu/-Trp/-His for screening.
Subcellular Localization of BdCIPK31 Protein
The CDS of the BdCIPK31 gene was introduced into pCAMBIA1303 using BglII and SpeI restriction enzyme sites. The recombinant plasmid was transformed into tobacco leaf epidermis via Agrobacterium-mediated transformation as described (Sparkes et al., 2006). A fluorescence microscope (Olympus IX71, Japan) was used to detect the fluorescence.
qRT-PCR and Semi-Quantitative PCR Analysis
RNA samples were prepared with the RNAprep Pure Plant Kit (TIANGEN, Beijing, China), and cDNA was prepared with the FastQuant RT Kit (TIANGEN, Beijing, China). The semi-quantitative PCR was used to detect the expression of BdCIPK31 in different transgenic tobacco lines. The Nicotiana tabacum ubiquitin gene was employed as endogenous control. The qRT-PCR analysis was applied with the SYBR Green PreMix (TIANGEN, Beijing, China) and the CFX96TM Real-Time Detection System (Bio-Rad, United States). The Brachypodium β-actin gene and the N. tabacum ubiquitin gene were employed as endogenous controls. All primers applied in these assays are listed in Supplementary Table S2. The 2-ΔΔCt method was used to analyze the qRT-PCR results.
Promoter Analysis and GUS Staining
A 2-kb fragment of the BdCIPK31 flanking 5′-upstream region before the translation initiation site (ATG) was identified in the Ensembl Plants database2 and amplified with primers containing the EcoRI and BamHI restriction sites (Primer P3, Supplementary Table S1). The cis elements were analyzed by the Plant-CARE3 database. The promoter fragment was introduced into the pBI121-GUS vector, thereby replacing the CaMV 35S promoter to recombine with the ProBdCIPK31::GUS construct. The construct and control pBI121-GUS were transformed into the Agrobacterium tumefaciens strain EHA105. Transgenic tobacco lines were generated by Agrobacterium-mediated transformation (Horsch et al., 1985). The transgenic seedlings were treated on 1/2 MS media supplied with 300 mM mannitol, 200 mM NaCl, or 5 μM ABA, respectively, for 24 h. For histochemical staining assay, the seedlings were infiltrated in GUS staining solutions (0.5 mg/mL 5-bromo-4-chloro-3-indolyl glucuronide, 0.1 M Na2HPO4, 10 mM Na2EDTA, 0.5 mM K4Fe(CN)6, and 0.06% Triton X-100, pH 7.0), vacuumized for 10 s, and placed at 37°C for 24 h. The stained seedlings were decolorized using ethanol. The quantitative GUS activity assay was performed by using 4-MUG method according to Jefferson et al. (1987).
Generation of BdCIPK31-Overexpression Transgenic Tobacco Plants and Stress Tolerance Assays
The CDS of BdCIPK31 gene was amplified and subcloned into pCAMBIA1303 vector by using primer P1 (Supplementary Table S1). The pCAMBIA1303-BdCIPK31 and vacant vector pCAMBIA1303 were transformed into the tobacco, respectively, as described above. The transformants were selected by 20 mg/L hygromycin and confirmed by reverse transcript PCR with primer P1. Three independent T2 homozygote lines were used in this experiment.
For the root length and fresh weight (FW) assay, the seeds were sown on 1/2 MS medium, then transferred into media that contained 150/300 mM mannitol, 150/200 mM NaCl, 2/5 μM ABA, or 0.1 mM Tu, respectively. The root length or FW of all tested seedlings were measured before and after treatment.
For stress tolerance assay, seeds were grown on 1/2 MS for 10 days in 25°C with 16 h light/8 h dark cycle, and then were transplanted into pots containing well-watered soil, each pot contains six tobacco seedlings. For normal growth control, all the tobacco plants were well-watered at 28°C with 16 h light/8 h dark cycle. For drought treatment, 4-week-old plants were grown without watering for 35 days. The survival rate was calculated after recovery for 7 days. To examine the water loss, leaves were detached and weighed at the indicated time points. Calculation of water loss rate was according to initial weight as described previously (Hu W. et al., 2013). In salt treatment, to ensure the consistency of NaCl concentration in different pots, all pots were placed in a basin containing 2 L 800 mM NaCl. The survival rate was calculated at 7 days after treatment.
Measurement of Stomatal Aperture
Leaves were detached from normal mature seedlings and floated in a buffer solution (30 mM KCl and 10 mM MES-KOH, pH 6.15), and incubated at 25°C under light for 5 h to fully open the stomata. For drought-induced stomatal closure, the leaves were exposed to air under light for 2 h. For ABA-induced stomatal closure, the appropriate concentrations of ABA was added. For Tu treatment, 0.3 mM Tu was applied in the buffer solution, and the treated leaves were exposed to air for 2 h after 5-h-incubation. The epidermal peels were stripped from the leaves and examined under a microscope (IX71, Olympus, Japan). The width and length of stomata were measured by Olympus cellSens software (Olympus, Japan).
Measurement of Ion Content and K+ Flux
Ten-day-old seedlings were treated on 1/2 MS containing 200 mM NaCl for 7 days. The seedlings was separated into the shoot and root samples. The sample pretreatment was performed according to Deng et al. (2013a). The ion content was measured by atomic absorption spectroscopy (180-50, GFAAS; HITACHI, Japan). The net K+ efflux was measured by the Xuyue-Science & Technology Co. (Shanghai, China) via a non-injuring technique (NMT, Younger USA Science and Technology Corp., Amherst, MA, United States) as previously described (Hu Y. et al., 2013).
Measurement of ABA Content
Four-week-old tobacco plants in soil were withheld watering for 7 days or treated by NaCl for 3 days. Fresh leaves were detached from treated plants and grinded in liquid nitrogen. Sample pretreatment was performed according to Yan et al. (2014). The ABA level was measured with an ELISA kit (Jiancheng, China).
Measurement of RWC, Water Loss Rate, IL, and the MDA, Chlorophyll, Proline, Ascorbic Acid, Soluble Sugar, Ascorbic Acid, and Anthocyanin Contents in Leaves
Four-week-old tobacco plants in soil were withheld watering for 7 days or treated by NaCl solution for 3 days. For RWC measurement, the FW of leaves was determineded. After incubation in water overnight, the turgid weight (TW) was measured. Subsequently, the samples were dried at 70°C for 2 days and the dried weight (DW) was tested. RWC (%) = [(FW - DW)/(TW-DW)] × 100%. For water loss rate calculation, the leaves from tobacco plants under normal condition were detached and exposed to air. The initial FW (W0) of the leaves was recorded, and the leave weight was also measured at indicated time points after detaching as Wt. The water loss rate (%) = (W0 - Wt)/W0 × 100%. For IL measurement, the leaves from plants were cut into small pieces and immersed in 8 mL deionized water. After 12 h incubation, the conductivity (C1) was detected with a conductivity meter (DDBJ-350, Shanghai, China). Subsequently, the samples were boiled for 15 min. After cooling, the conductivity (C2) was recorded again, such that IL (%) = (C1/C2) × 100%. The MDA level was measured using the TBA reaction (Heath and Packer, 1968). The UV spectrophotometry technique was used to measure the chlorophyll content (Yang et al., 2009). The proline and ascorbic acid contents in leaves were measured with the corresponding detection kits (Jiancheng, China). The level of soluble sugar in leaves was examined as described (Kong et al., 2011). The analysis of the relative anthocyanin content was done according to previous described method (Cui et al., 2014).
Measurement of Enzyme Activity
The activities of CAT, POD, SOD, and GST in leaves from tested plants were examined with the corresponding activity detection kit (Jiancheng, China) using leaves detached from the treated plants.
Expression Analysis of the Ion-Transporter Genes, ROS-Scavenging System Genes, Osmolyte Synthesis Genes, and Stress-Related Genes
Four-week-old tobacco plants in soil were withheld watering for 7 days or treated by NaCl solution for 3 days. The leaves from drought treated plants were detached and subjected to RNA extraction, and these RNA samples were used to analyze the TORK1 transcripts, as well as ROS-scavenging system genes, osmolyte synthesis genes, and stress-responsive genes. Two-week-old young seedlings were grown on 1/2 MS containing 200 mM NaCl for 2 days, and the samples were used for expression analyses of NKT1, NKT2, NtKC1, NtNHX2, and NtNHX4, respectively.
Statistical Analysis
The SPSS software (Chicago, IL, United States) was used to analyze the data. The data variance analysis was performed by the ANOVA Duncan’s test.
Results
Characterization of BdCIPK31
The cDNA of a CIPK gene was amplified by RT-PCR using mRNA isolated from B. distachyon seedlings. The coding sequence is 1,347 bp in length, which encodes a 448-amino-acid protein. Phylogenic analysis of the predicted BdCIPK and 33 OsCIPKs showed that the BdCIPK was clustered to the exon-rich group (group A), and exhibited closest relationship with OsCIPK31 and OsCIPK3 (Supplementary Figure S1). Since this BdCIPK shared highest identity with OsCIPK31 (82.2%), the BdCIPK was designated as BdCIPK31 (XM_003562376).
The BdCIPK31 presented high identities with AtCIPK3 (70%), OsCIPK31 (82%), and TaCIPK3 (89%). Multiple sequence alignment with these homologs indicated that BdCIPK31 contained all the typical features of CIPK domains, namely, an activation loop, NAF/FISL motif, and a protein–phosphatase interaction domain (Supplementary Figure S2).
The yeast-two-hybrid assay was performed to identify the BdCIPK31 interacting BdCBLs, the results showed that the BdCIPK31 interacted with BdCBL1 (XP_003559049), BdCBL2 (XP_003574350), and BdCBL5 (XP_003578877) (Figure 1A).
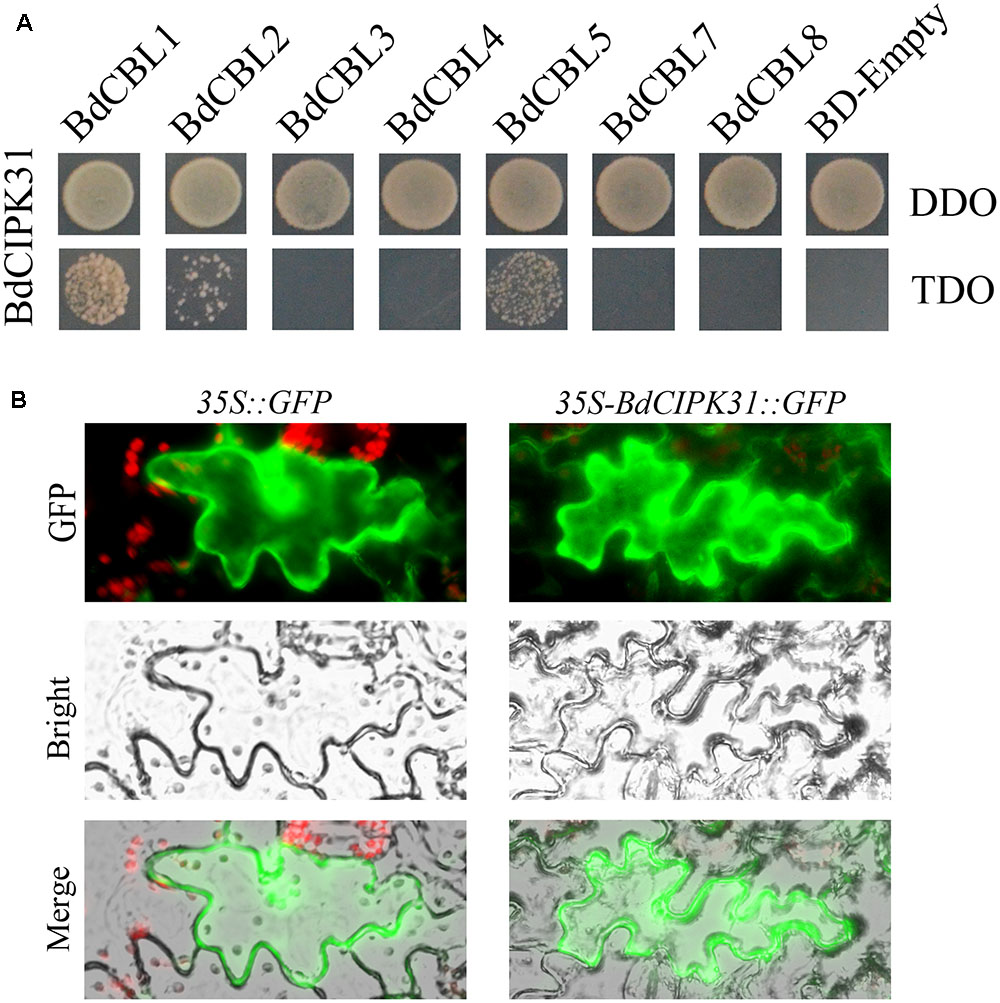
FIGURE 1. BdCIPK31-BdCBLs interaction and BdCIPK31 subcellular localization analysis. (A) Yeast-two-hybrid analysis of the interactions between BdCIPK31 and BdCBLs. The pGADT7-BdCIPK31 and pGBKT7-BdCBLs vectors were co-transformed into AH109. The transformants were screened by DDO/-Leu/-Trp and TDO/-Leu/-Trp/-His for 3 days. (B) Subcellular localization analysis of BdCIPK31::GFP. The left column shows the epidermal cells expressing the free GFP protein as controls. The right column shows the epidermal cells expressing the BdCIPK31::GFP fusion protein. Three independent biological replicates were performed and produced similar results.
To determine the subcellular localization of BdCIPK31, a recombinant vector was constructed to fuse the BdCIPK31 with fluorescent protein GFP. The constructed plasmid was transformed into tobacco leaves. The transformed epidermis was examined after 72 h. The fluorescence of BdCIPK31::GFP fusion protein was observed throughout the cell, similar to that of the green fluorescent protein used as control (Figure 1B).
BdCIPK31 Expression Responds to Abiotic Stress and Exogenous ABA
Quantitative reverse-transcription PCR was employed to examine the expression patterns of BdCIPK31. Organ-specific expression analysis showed that BdCIPK31 was expressed in all B. distachyon organs tested (Figure 2A). Young seedlings were treated with osmotic stress generator PEG6000, salt stress generator NaCl, oxidative stress inducer H2O2, and stress-related phytohormone ABA. As shown in Figures 2B–F, BdCIPK31 expression was significantly suppressed 1 h after all treatments. After 6 h of treatment, the expression levels raised again after a fall (0.5-fold in PEG, 1-fold in NaCl, 0.4-fold in H2O2, and 0.76-fold in ABA) and then downregulated after a longer period (>6 h) of exposure to all treatments. In general, the results showed that the BdCIPK31 expression was downregulated by PEG6000, NaCl, H2O2, and ABA (Supplementary Figure S10).
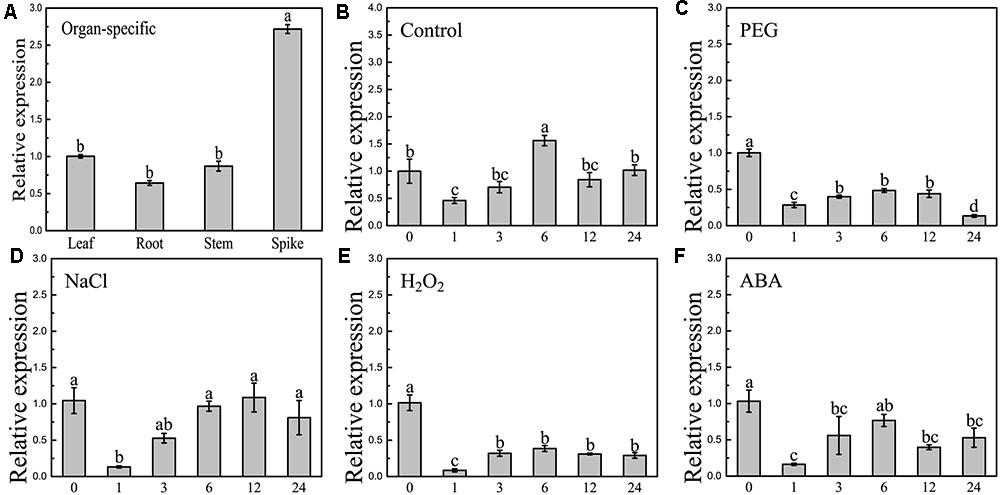
FIGURE 2. Expression patterns analyses of BdCIPK31 gene. (A) Organ-specific expressions of BdCIPK31 in leaf, root, stem, and spike of 6-week-old plant. Ten-day-old seedlings were treated by: (B) no treatment control, (C) 20% PEG6000, (D) 200 mM NaCl, (E) 100 mM H2O2, and (F) 100 μM ABA treatment, respectively. The BdCIPK31 expression in the leaves of treated seedlings was detected. Data in this figure represent the mean ± SE of three independent replicates. Different letters represent significant difference in each condition (Duncan’s test, P < 0.05).
Promoter Analysis of BdCIPK31
To investigate the transcriptional regulation mechanism of BdCIPK31, we obtained a 2 kb fragment from the 5′-flanking region of the transcriptional initiation site (ATG) of BdCIPK31 from Ensembl Plants2 and analyzed the sequence by the PlantCARE3 database. Various cis elements associated with the response to stress and hormone, such as the ABRE, were found in this region. Notably, no dehydration response element was discovered (Supplementary Table S3). These findings suggest that the transcriptional regulation of BdCIPK31 may be through ABA-dependent pathway.
The result was confirmed by introducing the promoter fragment into pBI121 to generate the ProBdCIPK31::GUS construct. The constructed vector was then transformed into tobacco (N. tabacum var. Samsun). Ten-day-old transgenic seedlings were treated with 300 mM mannitol, 200 mM NaCl, or 5 μM ABA. After 1 day treatment, the transgenic seedlings displayed lighter GUS staining than the controls (Supplementary Figure S3), and the observation was further confirmed by quantitative GUS activity assay (Supplementary Figure S4). This finding is consistent with the qRT-PCR results.
Overexpression of BdCIPK31 Enhances Drought and Salt Stress Tolerance in Tobacco
To characterize the function of BdCIPK31, transgenic tobacco overexpressing BdCIPK31 under control of CaMV 35S promoter was generated. Five independent transgenic lines (T1) were obtained (Supplementary Figure S5). The transcript level of BdCIPK31 in each T2 homozygous line was examined by semi-quantitative RT-PCR. Among these lines, OE2, OE4, and OE8, which expressed BdCIPK31 at different levels, were selected as experimental materials (Supplementary Figure S6). Transgenic tobacco plants transformed with vacant pCAMBIA1303 vectors were also generated as VCs.
The responses of transgenic young seedlings to drought and salt were initially examined. The young seedlings were grown on 1/2 MS media supplied with 150/300 mM mannitol or 150/200 mM NaCl after seed germination. The seedlings of all lines displayed similar root length on the medium without treatments after 14 days (Figure 3A). However, OE seedlings exhibited longer root lengths than the controls under osmotic and salt stress (Figures 3B–I).
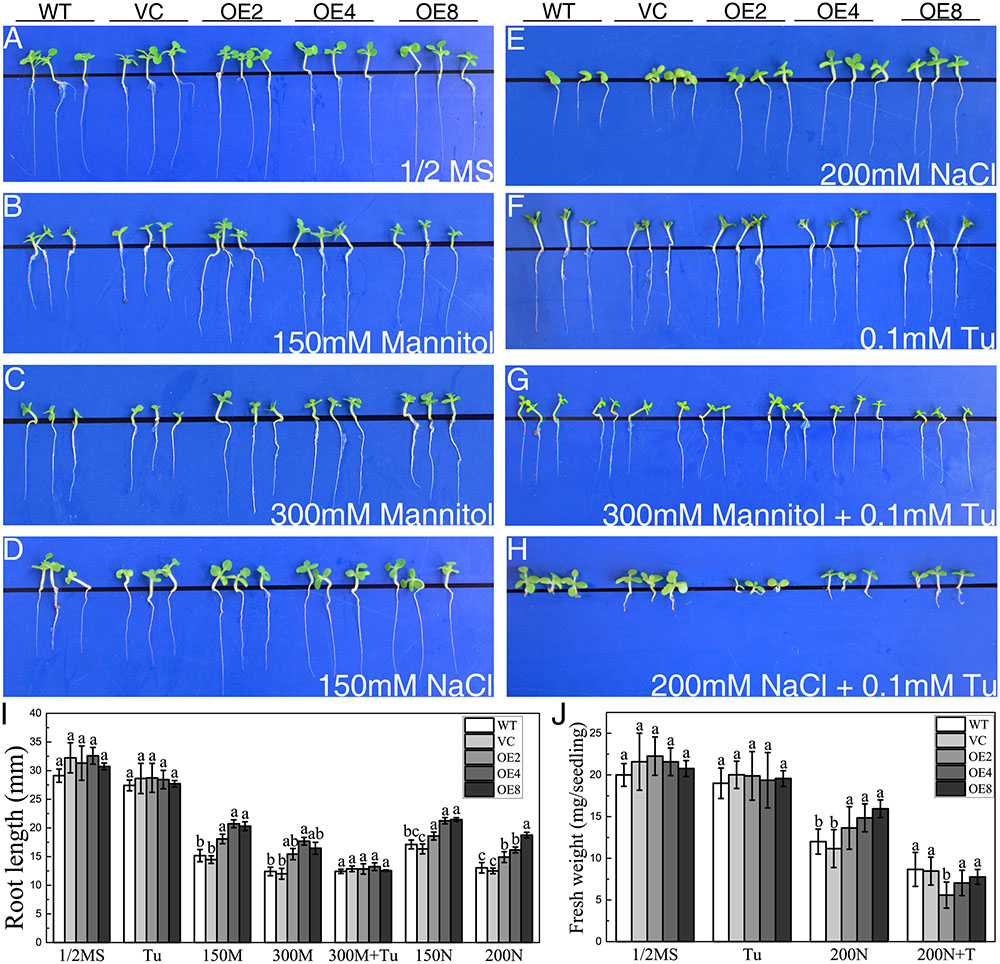
FIGURE 3. Analysis of root elongation and fresh weight in control and OE plants exposed to osmotic and salt stress. Seedlings grown on (A) 1/2 MS medium, (B,C) 1/2 MS medium containing (B) 150/(C) 300 mM mannitol, (D,E) 1/2 MS medium containing (D) 150/(E) 200 mM NaCl, (F) 1/2 MS medium containing 0.1 mM Tu, (G) 1/2 MS medium containing 300 mM mannitol and 0.1 mM Tu, and (H) 1/2 MS medium with 200 mM NaCl and 0.1 mM Tu for 14 days. (I) Statistical analysis of root length. (J) Statistical analysis of fresh weight. Data in (I,J) represent the mean ± SE from three independent replicates. Different letters represent significant difference in each condition (Duncan’s test, P < 0.05).
To elucidate whether endogenous ABA participates in the mechanism of the stress-tolerance phenotype, Tu was employed as a ABA biosynthesis inhibitor. WT, VC, and OE seedlings exhibited similar growth status on the medium containing 0.1 mM Tu (Figure 3F). However, the stress-tolerance phenotype of OE lines disappeared under 300 mM mannitol treatment and even reversed under 200 mM NaCl treatment by supplying with 0.1 mM Tu (Figures 3G,H). The results suggested that the stress tolerance regulation of BdCIPK31 was dependent on endogenous ABA.
BdCIPK31 also functioned in stress tolerance during vegetative growth. Drought stress was given to 4-week-old plants. Both controls and OE lines shriveled after a 35 days exposure to drought. However, the control lines were more severely wilted, whereas the leaves of the OE lines remained expanded (Figure 4B). After re-watering for 7 days, more than 50% of the OE plants survived compared with only 14.8% of the WT plants. Meanwhile, 13.2% of VC plants were recovered from drought (Figures 4C,F).
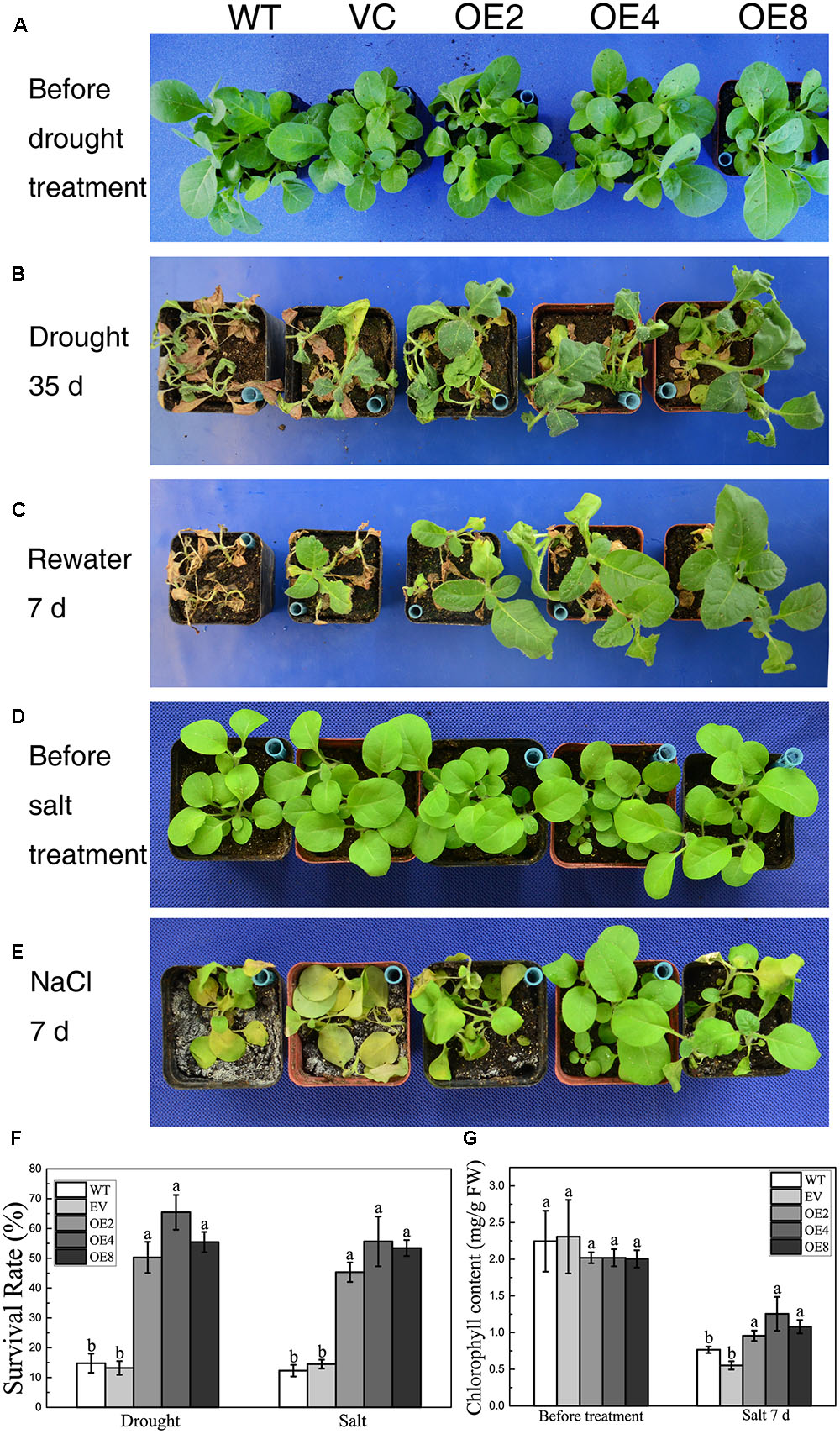
FIGURE 4. Analysis of the drought and salt tolerance in control plants and transgenic plants overexpressing BdCIPK31. (A,D) Four-week-old tobacco plants grown in normal condition. (B) Plants were subjected to drought for 35 days. (C) Plants in (B) were re-watered for 7 days. (E) Plants were subjected to high salinity for 7 days. (F) Statistical analysis of survival rates. (G) Chlorophyll content analysis of plants subjected to salt treatment. Data in (F,G) represent the means ± SE from three independent replicates. Different letters represent significant difference in each condition (Duncan’s test, P < 0.05).
In salt treatment, most of the control plants displayed languish and chlorosis after a 7-day-exposure to salt, whereas the OE plants remained thriving and green (Figures 4D,E). Less than 15% of the control seedlings survived. By contrast, the survival rates of the OE lines were 45% (OE2), 55% (OE4), and 53% (OE8), respectively (Figure 4F). Moreover, chlorophyll content analysis showed that the OE plants contained more chlorophyll than the control after salt treatment (Figure 4G). These results suggest that BdCIPK31 improved drought and high-salinity tolerance in transgenic tobacco.
Overexpression of BdCIPK31 Confers Hypersensitivity to Exogenous ABA in Tobacco but Does Not Affect ABA Biosynthesis under Drought and Salt Stress
The downregulation of BdCIPK31 in response to exogenous ABA implied that BdCIPK31 might participate in ABA signaling. Seedlings were grown on the 1/2 MS supplied with 2 or 5 μM ABA for 21 days. All seedlings exhibited growth inhibition under ABA treatment. However, the OE lines showed shorter root lengths than those of the control lines (Figures 5A–D). The result indicates that BdCIPK31 overexpression increases plant sensitivity to ABA.
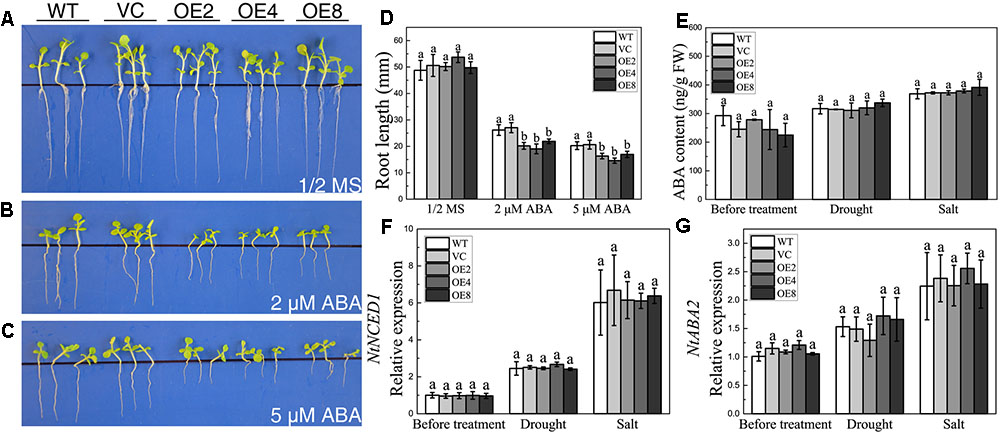
FIGURE 5. Analyses of the plant sensitivity exogenous ABA, ABA content, and ABA synthesis-related gene expression and under drought and salt treatments. Seedlings grown on (A) 1/2 MS medium, (B,C) 1/2 MS medium with (B) 2 μM or (C) 5 μM ABA for 21 days. (D) Root length analysis of control and OE seedlings. (E) Endogenous ABA level of control and OE plants before and after treatments. (F,G) Expression levels of (F) NtNCED1 and (G) NtABA2 in control and OE plants before and after treatments. Data in (D–G) represent the means ± SE from three independent replicates. Different letters represent significant difference in each condition (Duncan’s test, P < 0.05).
To investigate whether BdCIPK31 affected ABA biosynthesis, endogenous ABA levels were examined under drought or salt condition. Interestingly, the endogenous ABA contents in the OE seedlings were similar to those in the controls after treatment (Figure 5E). The key enzymes in ABA biosynthesis were also examined at the transcript level. The transcripts of NtNCED1 and NtABA2 in the OE seedlings were similar to those in the controls under both drought and salt stress (Figures 5F,G). These findings suggest that BdCIPK31 has no effect on ABA biosynthesis, and BdCIPK31 may function in the downstream of ABA.
Overexpression of BdCIPK31 Confers Better Water Status and Decreased Stomatal Aperture in Response to Drought Stress
Enhanced drought tolerance implied the differences in water status between the OE and control plants. Fully expanded leaves were obtained in normal growth condition, and the FW changes of the leaves were measured regularly in 24 h. The water loss rates were calculated, and the OE lines displayed significantly lower water loss rates than the controls (Figure 6A). RWCs were also measured. After 14 days of drought treatment, the RWC of the control lines was 20% lower than that of controls (Figure 6B).
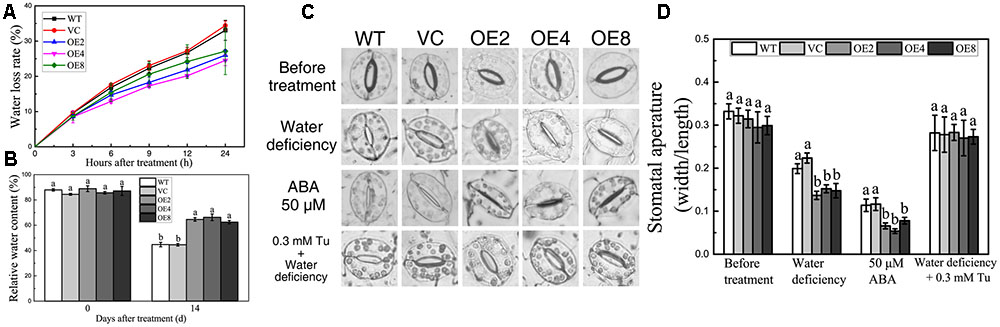
FIGURE 6. Analyses of RWC, water loss, and stomatal aperture. (A) Water loss rate of control and OE leaves in 24 h. (B) RWC of control and OE plants after drought treatment. Data in (A,B) represent the means ± SE from three independent replicates. (C) Stomatal closure under water deficiency, ABA, and Tu treatments. (D) Statistical analysis of the stomatal width:length ratio. Data in (D) represent the means ± SE from three independent replicates. Different letters represent significant difference in each condition (Duncan’s test, P < 0.05).
Transpiration through stomata is the main way of water loss under water-deficient conditions (Li et al., 2013). Thus, we investigated whether BdCIPK31 affects stomatal closure under drought stress. The stomatal width:length ratio was adopted as the index of stomatal closure. The leaves of all the lines were floated in buffer solution under light to cause the stomata to fully open prior to treatment application (Figure 6C). After exposing to low-humidity environment for 2 h, the OE lines displayed lower stomatal width:length ratios (Figures 6C,D).
Since stomatal closure is mediated by ABA, we performed an investigation on whether BdCIPK31 functioned in ABA-mediated stomatal closure. Leaves were incubated in 25 or 50 μM ABA solution for 4 h, and the OE lines displayed smaller stomatal aperture than the controls (Figures 6C,D). However, when the leaves were pretreated by Tu, the humidity-induced stomatal closure was inhibited, no significant differences in stomatal aperture between control and transgenic lines were observed (Figures 6C,D). These findings suggest that BdCIPK31 participates in ABA-mediated stomatal closure.
Overexpression of BdCIPK31 Affects Plant Ion Homeostasis under Salt Stress
High salinity can disrupt intracellular ion homeostasis. To determine whether BdCIPK31 influences ion accumulation under high-salinity conditions, the Na+ and K+ contents in shoots and roots were assessed. The Na+ levels were similar in all seedlings without treatment (Figure 7A). By contrast, the K+ contents were higher in OE lines (Figure 7B). After salt treatment, Na+ content increased, whereas K+ content decreased, in all plants. However, the OE seedlings contained more Na+ than the control in both shoots and roots (Figure 7A). Notably, the K+ level in the shoots of the OE seedlings was equal to that in the controls after salt treatment, whereas the roots of the OE plants accumulated more K+ than the control did (Figure 7B). These findings indicate that BdCIPK31 is involved in intracellular Na+/K+ homeostasis under salt stress. Further analysis of the salt-shock-induced K+ efflux in OE plants was performed. Before treatment, the WT seedlings showed a slightly higher K+ efflux compared with OE seedlings. After addition of 150 mM NaCl, the OE seedlings displayed significantly lower K+ efflux than the WT (Figure 7C). This result supports the notion that BdCIPK31 functions in preventing K+ loss from root under salt stress.
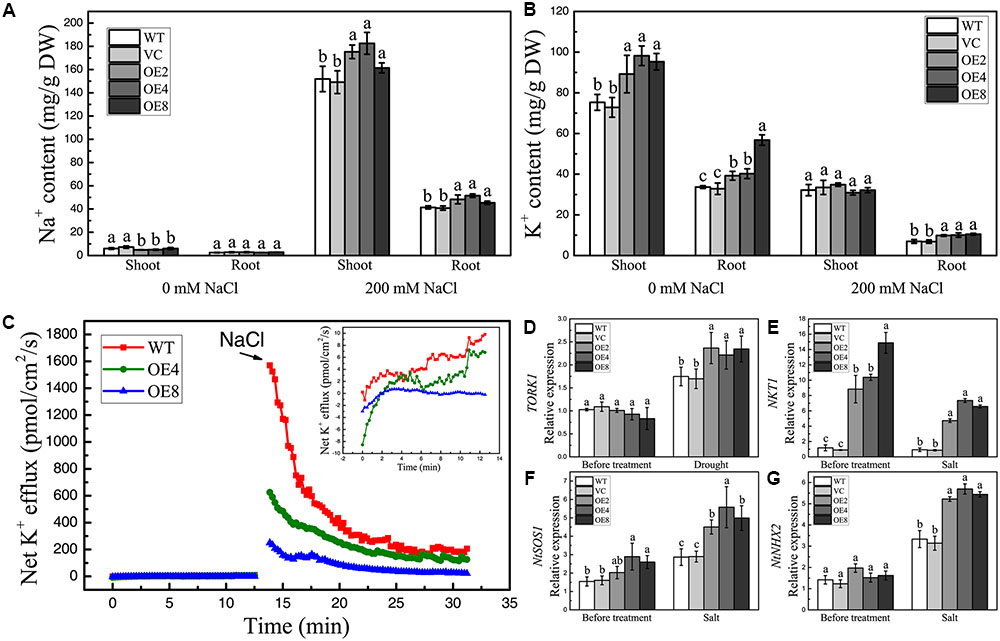
FIGURE 7. Analyses of ion content, K+ efflux in seedlings under salt condition, and expression analyses of ion-transporter genes in plants. (A) Na+ and (B) K+ levels in shoots and roots of control and OE seedlings under salt stress. (C) Net K+ efflux in root tips of WT and OE seedlings at the addition of 150 mM NaCl. The insert shows the K+ efflux before NaCl addition. (D) Expression levels of TORK1 in the leaves of control and OE plants under drought treatment. (E–G) Expression levels of (E) NKT1, (F) NtSOS1, and (G) NtNHX2 in the control and OE young seedlings under salt stress. Data in (A,B,D–G) represent the means ± SE from three independent replicates. Different letters represent significant difference in each condition (Duncan’s test, P < 0.05).
Overexpression of BdCIPK31 Modulates the Expression of Ion-Transporter Genes under Drought and Salt Stress
Ion channels and transporters play important roles in stomatal movement and intracellular ion homeostasis. To clarify whether BdCIPK31 affects ion-transporter gene expression, we measured the transcripts of tobacco K+ influx channel genes NKT1, NKT2, NTKC1, K+ efflux channel TORK1, Na+/H+ antiporters NtSOS1, and Na+, K+/H+ antiporter NtNHX2.
The outward K+ channels play essential roles in stomatal closure. TORK1 is the homolog of Arabidopsis guard cell K+ efflux channel GORK in N. tabacum (Eisenach et al., 2014). The transcripts of TORK1 in the leaves of OE lines showed at least a 1.5-fold increase compared with that in the controls under drought conditions (Figure 7D). This finding implies a stronger K+ efflux in the guard cells of OE plants.
Selective uptake of K+ and extrusion or compartmentalization of Na+ are main strategies for plants to cope with ion stress caused by high salinity. In Arabidopsis, K+-channel proteins AKT1 and AtKC1 functions in K+ uptake (Hirsch et al., 1998; Reintanz et al., 2002; Pilot et al., 2003), where AKT2 participates in long-distance K+ transport (Lacombe et al., 2000). The expression levels of their homologs NKT1, NKT2, and NTKC1 in tobacco were selected and examined under salt treatment together with the NtSOS1 and NtNHX2. Results showed that NKT1 exhibited significantly higher transcript levels in transgenic seedlings than in controls under normal and high-salinity conditions (Figure 7E). The transcripts of NKT2 and NtKC1 in OE seedlings were equal to those in the controls before and after treatment (data not shown). Moreover, the OE lines showed higher NtSOS1 and NtNHX2 expression levels than the controls only under NaCl treatment (Figures 7F,G). These findings demonstrate that BdCIPK31 affects the expressions of several ion channel and transporter genes under high-salinity stress.
Overexpression of BdCIPK31 Enhances the Plant’s Scavenging Ability for Reactive Oxygen Species (ROS) and Maintains Plasma Membrane Stability under Drought and Salt Stress
Both drought and salt stress can cause oxidative and osmotic damages to plants. To explore whether BdCIPK31 functions in resistance to these damages, we detected the malondialdehyde (MDA) content, the IL, and the H2O2 content. The results showed that the IL and MDA levels were lower in OE seedlings (Supplementary Figures S7A,B), and the OE plants accumulated less H2O2 than the controls (Supplementary Figure S7C), suggesting that the OE seedlings suffered milder damage than the control under drought and salt stress.
Reactive oxygen species are essential signal-transduction molecules as well as toxic by-products of stress metabolism (Huang et al., 2012). Antioxidative enzymes, such as CAT, POD, SOD, and GST, and antioxidants, such as anthocyanin and ascorbic acid, function in preventing damage from over-produced ROS (Julkowska and Testerink, 2015).
The activities of CAT, POD, SOD, and GST were examined, and the results showed that the activities of all these antioxidative enzymes were higher in the OE seedlings than that in the controls (Supplementary Figure S8). Meanwhile, the OE plants accumulated more ascorbic acid and anthocyanin than the controls after treatment (Supplementary Figure S8). Moreover, the expression levels of NtCAT1, NtPOX2, NtSOD, NtGST, ascorbate peroxidase gene NtAPX, and flavonoid biosynthetic gene NtDFR were also higher in OE plants. By contrast, the ROS-producer genes NtRbohD and NtRbohF showed lower transcript levels in OE plants (Supplementary Figure S8).
Osmolyte accumulation is an important way to balance cell osmotic pressure. The proline and soluble sugar contents were examined. The OE seedlings accumulated more proline and soluble sugar under treatments (Supplementary Figure S9).
The results of the increased osmolyte accumulation in OE plants were also confirmed at transcriptional level. The expression levels of NtP5CS1, which encodes a rate-limiting enzyme of proline biosynthesis and a tobacco sucrose synthase gene NtSUS1 were detected, together with polyamine biosynthesis genes NtADC1 and NtSAMDC1. All these genes exhibited higher transcript levels in OE seedlings under drought or salt stress (Supplementary Figure S9). The above results contribute to explain the enhancement of the stress tolerance in the transgenic tobacco.
Overexpression of BdCIPK31 Enhances Plant Tolerance to Oxidative Stress
Overexpression of BdCIPK31 confers enhanced ROS-scavenging ability in transgenic tobacco, which implies that BdCIPK31 may participate in plant response to oxidative stress. Leaf disks were incubated in 20 or 50 μM methyl viologen (MV) for 72 h. All leaf disks exhibited bleaching, and the controls displayed more severe results than those of the OE leaves (Figure 8A). Furthermore, when seedlings were grown on 1/2 MS media with 2 or 5 μM MV for 14 days, the OE lines disclosed a better phenotype than that of the controls (Figure 8B). These observed results were further confirmed by chlorophyll content and FW assay results (Figures 8C,D). All the results suggest that BdCIPK31 enhanced plant tolerance to oxidative stress.
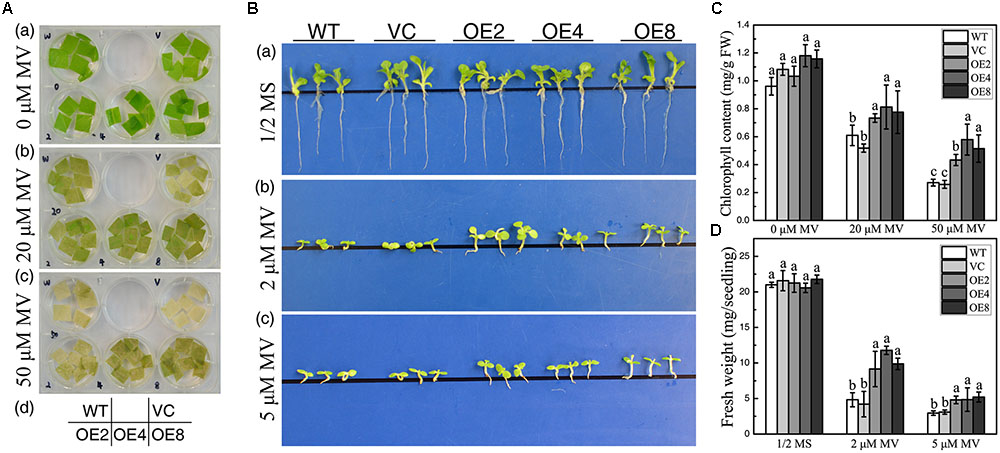
FIGURE 8. Analyses of oxidative tolerance control and OE plants. (A) Leaf disks obtained from control and OE seedlings treated with methyl viologen (MV). (B) Seedlings grown on 1/2 MS containing 0, 2, or 5 μM MV. (C) Chlorophyll content in leaves treated with 20 or 50 μM MV. (D) Average fresh weight of seedlings treated by 2 or 5 μM MV. Data in (C,D) represent the means ± SE from three independent replicates. Different letters represent significant difference in each condition (Duncan’s test, P < 0.05).
BdCIPK31 Modulates the Expressions of Some Stress-Related Genes in Transgenic Tobacco
The transcripts of some stress-related genes in OE and control plants were also examined under drought or salt treatment. The selected genes included stress-responsive transcription factor genes, such as NtABF1, NtABF2, NtDREB3, and NtRD26, and stress-defense genes, such as NtERD10C, NtERD10D, NtLEA5, and TobLTP1. The transcripts of all these tested genes were higher in the OE plants under stress conditions (Figures 9A–H). Notably, the transcripts of NtRD26, NtDREB3, NtLEA5, and TobLTP1 were evidently higher in the OE plants than in the controls before and after treatments (Figures 9B–D,G,H). These findings suggest that BdCIPK31 can elevate the expression levels of stress signal transduction related genes together with stress-defense genes under drought and salt conditions.
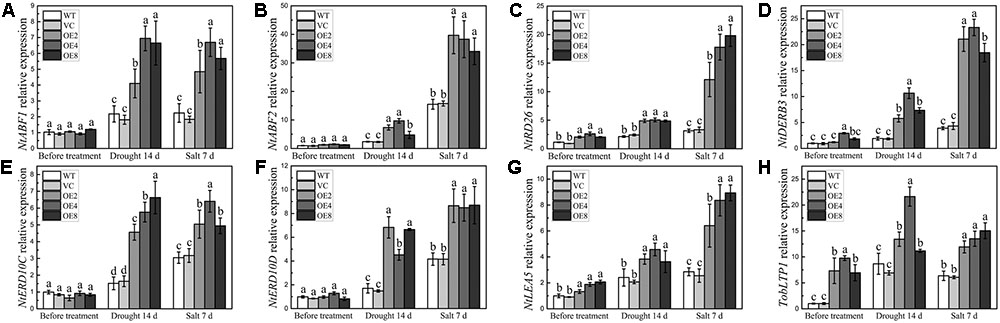
FIGURE 9. Expression analyses of stress-related genes in plants under drought and salt treatments. Expression levels of (A) NtABF1, (B) NtABF2, (C) NtRD26, (D) NtDREB3, (E) NtERD10C, (F) NtERD10D, (G) NtLEA5, and (H) TobLTP1 were examined by qRT-PCR. Data represent the means ± SE from three independent replicates. Different letters represent significant difference in each condition (Duncan’s test, P < 0.05).
Discussion
BdCIPK31 Is a Positive Regulator in Plant Response to Drought, High Salinity, and ABA
Previous studies on CIPKs revealed that numerous CIPK genes regulate plant responses to abiotic stress. In our previous study, TaCIPK14 and TaCIPK29 were found to confer single or multiple stress tolerance in transgenic tobacco (Deng et al., 2013a,b). In the present work, overexpression of BdCIPK31, a stress-responsive CIPK gene, conferred improved drought and salt tolerance in transgenic tobacco (Figures 3, 4). These findings demonstrate that BdCIPK31 has a positive function in plant responses to drought and high salinity.
Abscisic acid is a pivotal phytohormone in plant stress responses (Finkelstein et al., 2002), which plays a crucial role in the coordination of plant growth and stress tolerance (Raghavendra et al., 2010). It was previously demonstrated that CIPKs participate in stress response through the ABA signaling pathway (Chen et al., 2012; Wang et al., 2016; Zhou et al., 2016). In the present study, BdCIPK31 increased plant sensitivity to ABA (Figure 5), and accelerated ABA-mediated stomatal closure (Figures 6C,D). Moreover, when ABA biosynthesis inhibitor Tu was applied together with osmotic and salt treatments (Yan et al., 2014), the phenotype of enhanced tolerance in OE lines disappeared (Figures 3G,H). Furthermore, BdCIPK31 exerted no effect on ABA biosynthesis (Figures 5E–G), indicating that the alternation of ABA content is not the key reason for stress tolerance in BdCIPK31-overexpressing tobacco. The above results demonstrate that BdCIPK31 serves as a positive regulator in ABA signaling pathway. Moreover, the improved stress tolerance in transgenic tobacco is dependent on endogenous ABA.
Our results indicated that the BdCIPK31 was a positive regulator in stress and ABA response, while BdCIPK31 was downregulated by abiotic stress and ABA, these were unexpected. However, similar discrepancies are not rare in studies of CIPK and other gene families. OsCIPK31 and OsCIPK03 were the homologs of BdCIPK31 in rice. The stress-repressed OsCIPK31 gene functioned positively in stress response, whereas the stress-induced OsCIPK03 gene conferred stress sensitivity to transgenic rice (Piao et al., 2010; Rao et al., 2011). Similar contradictions were also reported in other gene families of different species. ONAC095, a NAC transcription factor gene in rice, was induced by drought and ABA, repressed by low temperature. However, overexpressing ONAC095 in rice showed no effect on drought tolerance, while the dominant chimeric repressor-mediated suppression of ONAC095 enhanced drought tolerance and ABA sensitivity (Huang et al., 2016).
BdCIPK31 Improves Plant Tolerance to Water Stress
Under water-deficient conditions, plants need to maintain favorable water status. Transpiration from stomata is the major means to eliminate water (Li et al., 2013), and ABA-mediated stomatal movement is a crucial process in preventing water loss (Leung and Giraudat, 1998; Pitzschke and Hirt, 2009). ABA-mediated stomatal movement is associated with the K+ flux in guard cells (Hosy et al., 2003; Kim et al., 2010). CBL-CIPK networks were reported to function in the stomatal movements. AtCIPK23, together with AtCBL1 and AtCBL9, activates the K+ channel AKT1 and affects stomatal closure (Li et al., 2006; Xu et al., 2006; Cheong et al., 2007; Lee et al., 2007). In this study, overexpression of BdCIPK31 improved the sensitivity of stomatal closure to drought treatment, and ABA is confirmed to be involved in this process (Figures 6C,D). This finding confirmed that BdCIPK31 could promote stomatal closure via the ABA signaling pathway. Thus, the OE seedlings displayed lower water loss rate and higher RWC than the control plants under drought treatment (Figures 6A,B). This result indicates that the OE plants possessed a better water status. Moreover, the expression level of the guard cell K+ efflux channel gene TORK1 elevated more obviously in the leaves of the OE plants under drought treatment, and the outflow of K+ from guard cells can cause stomatal closing (Ache et al., 2000; Hosy et al., 2003; Osakabe et al., 2013). Hence, the above findings indicate that BdCIPK31 plays a positive role in stomatal closure.
BdCIPK31 Improves Plant Tolerance to Ionic Stress
High salinity causes ionic stress to plants. High Na+ concentration in growth medium disrupts plant ion uptake and leads to an abnormal Na+ accumulation, whereas the K+ accumulation decreases (Bartels and Sunkar, 2005). Excess Na+ in the cytoplasm is toxic, whereas potassium is essential for many biochemical and physiological processes. The chief mechanisms maintaining intracellular K+/Na+ homeostasis include restricting Na+ uptake by selective ion uptake, and reducing cytoplasmic Na+ content by Na+ extrusion and/or intracellular Na+ compartmentalization (Lv et al., 2012). These mechanisms must be implemented by regulating ion channels and transporters. Earlier studies revealed that CBL-CIPK networks function in regulating ion transport under salt stress. The classical Arabidopsis SOS pathway functions in extruding Na+ (Liu and Zhu, 1997, 1998; Qiu et al., 2002). Transgenic tobacco overexpressing TaCIPK14 and TaCIPK29 can maintain higher K+/Na+ ratios under salt treatment (Deng et al., 2013a,b). Furthermore, the AtCIPK23 can activate the K+ influx channel AKT1 in the root, leading to an influx of K+ (Li et al., 2006; Cheong et al., 2007; Lee et al., 2007). Therefore, we tried to clarify whether BdCIPK31 functions in intracellular ion homeostasis under salt stress. Interestingly, transgenic plants overexpressing BdCIPK31 retained significantly more Na+ than the control, despite the former’s salt-tolerant phenotype (Figure 7A). Similar phenomena have also been reported previously. The atcbl10 mutant shows hypersensitivity to salt stress, but the Na+ content of this mutant is significantly lower than that in the control. Further investigations indicate that AtCIPK24 interacts with AtCBL10, and the complex is involved in intracellular Na+ compartmentalization (Kim et al., 2007). Transgenic tomatos overexpressing SlCIPK24 display more tolerance to salt, and the enhanced tolerance is associated with higher Na+ level in aerial parts. SlCIPK24 induces and upregulates SlSOS1, endosomal K+, Na+/H+ antiporter LeNHX2, and vacuolar Na+/H+ antiporter LeNHX4, indicating that SlCIPK24 functions in Na+ partitioning and compartmentalization under salt stress (Huertas et al., 2012). In our work, BdCIPK31 overexpression upregulated the transcripts of NtSOS1 and NtNHX2 under salt stress (Figures 7F,G). This finding is consistent with the higher sodium content in BdCIPK31-overexpressing plants relative to the control, implying that BdCIPK31 may be involved in Na+ compartmentalization in endosomes or vacuoles under salt stress. Moreover, BdCIPK31 also functions in preventing K+ loss. Transgenic plants overexpressing BdCIPK31 contained more K+ in whole seedlings under normal conditions (Figure 7B). Under salt treatment, although the OE seedlings retained more Na+ than the control (Figure 7A), OE seedlings still retained more K+ in roots, whereas the shoot K+ accumulation was similar between OE and control seedlings (Figure 7B). This result could be explained by the expression analyses of the K+ channel genes. Overexpression of BdCIPK31 constitutively elevated the expression of NKT1 (Figure 7E), and the NKT1 may function in root K+ uptake (Sano et al., 2007). However, BdCIPK31 does not affect the expression of NKT2 that might be involved in long-distance K+ transport (Lacombe et al., 2000; Sano et al., 2007). Thus, the above results imply that BdCIPK31 might participate in K+ uptake in the root, but does not affect K+ transport from roots to shoots. Further investigation revealed that the OE plants exhibited lower net K+ efflux in roots than the WT (Figure 7C). The root plasma membrane is depolarized under salt treatment, thus the channel-mediated K+ uptake are inhibited in such condition (Shabala et al., 2016). Therefore, our finding implies that BdCIPK31 also functions in preventing K+ loss.
BdCIPK31 Improves Plant Tolerance to Oxidative Stress
Exposure to drought or high salinity results in some common reactions in plants (Bartels and Sunkar, 2005), such as oxidative and osmotic stress. Both drought and salt stress leads to ROS overproduction, which acts as important stress signal-transduction molecules. However, excess ROS brings cell toxicity, leading to enzyme activity disruption, membrane injury, and cell death (Gouiaa et al., 2012). Thus, plants have developed an efficient antioxidative system, which includes ROS-scavenging enzymes and antioxidants, to regulate ROS content to appropriate levels (Ruiz-Lozano et al., 2012). CIPKs affect the antioxidant system in both physiologic and transcript levels have been reported previously (Yang et al., 2008; Piao et al., 2010; Chen et al., 2012; Deng et al., 2013a,b; Drerup et al., 2013; He et al., 2013). We demonstrated that overexpression of BdCIPK31 significantly improved the ROS-scavenging system in both physiological and transcriptional level, whereas the transcripts of ROS-producer genes were downregulated (Supplementary Figure S8). As a result, the OE plants exhibited lower H2O2, MDA, and IL levels relative to the control (Supplementary Figure S7). Such finding indicates a decreased ROS accumulation and milder membrane damage in BdCIPK31-overexpressing plants. The above results indicate that BdCIPK31 functions in enhancing the plant ROS-scavenging system. Further study showed that BdCIPK31 overexpression conferred enhanced tolerance to oxidative stress (Figure 8), confirming that BdCIPK31 participated in the antioxidation process.
BdCIPK31 Improves Plant Tolerance to Osmotic Stress
Both drought and high salinity cause cell dehydration, which leads to osmotic stress (Bartels and Sunkar, 2005). Plants synthesize additional osmolytes to maintain turgor to cope with osmotic stress. In this study, the BdCIPK31-overexpressing tobacco exhibited higher proline and soluble sugar levels under stress together with higher transcripts of the NtP5CS1 and NtSUS1 genes (Supplementary Figures S9A–D). The transcripts of the genes on polyamine biosynthesis, NtADC1 and NtSAMDC1, were also obviously elevated in the OE plants (Supplementary Figures S9E,F). These results indicate that BdCIPK31 could enhance ROS-scavenging system to alleviate cell damage and enhance osmolyte synthesis to balance cellular osmotic pressure.
BdCIPK31 Modulates the Expression of Stress-Related Genes
The expression of stress-related genes are modulated through ABA-dependent and ABA-independent pathways (Yamaguchi-Shinozaki and Shinozaki, 2006). The ABFs are the major transcription factors in the ABA-dependent pathway, whereas DREBs have key functions in ABA-independent pathway (Yoshida et al., 2014). In this study, the transcripts of NtABF1, NtABF2, and NtRD26, which encodes a NAC transcription factor in the ABA-dependent pathway (Fujita et al., 2004), were significantly higher in the BdCIPK31-overexpressing tobacco under drought and salt stress (Figures 9A–C). Moreover, NtDREB3 also presented higher transcripts in OE seedlings under the same conditions (Figure 9D). The transcripts of the stress-inducible genes, including NtERD10C, NtERD10D, NtLEA5, and TobLTP1, were higher in the BdCIPK31-overexpressing plants under stress (Figures 9E–H). The NtERD10C and NtERD10D genes encode group 2 LEA proteins, and the NtLEA5 gene encodes a group 5 LEA protein. Both NtERD10C/D and NtLEA5 function in binding water, stabilizing enzyme and macromolecular structures, and then preventing cells from damages caused by abiotic stress (Becker et al., 1996; Hundertmark and Hincha, 2008; Liu et al., 2009). TobLTP1 encodes a lipid-transfer protein, which participates in abiotic stress responses (Trevino and O’Connell, 1998; Zhang et al., 2005). The stress-inducible genes are recognized as targets of ABF and DREB genes (Shinozaki et al., 2003; Fujita et al., 2005); thus, these findings demonstrated that BdCIPK31 modulates the transcripts of stress-responsive genes through both ABA-dependent and ABA-independent pathways.
Conclusion
In conclusion, BdCIPK31 confers enhanced drought and salt stress tolerance and ABA hypersensitivity in plants. Overexpressing BdCIPK31 functions in stomatal closure, ion homeostasis, ROS scavenging, osmolyte biosynthesis, and transcriptional regulation of stress-related genes. Overall, BdCIPK31 functions in plant responses to drought and salt stress as a positive regulator in ABA-mediated Ca2+ signaling pathway.
Author Contributions
QL performed all the experiments, analyzed the data and wrote the manuscript. QW generated the transgenic tobacco plants and helped to perform other experiments. RW, YZ, FZ, YH, SZ, and JF helped to perform the physiological experiments and promoter analysis. GH and GY conceived the study, designed experiments and wrote the manuscript.
Funding
This work was supported by National Genetically Modified New Varieties of Major Projects of China (2016ZX08010004-004), and the National Natural Science Foundation of China (No.31570261).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Analytical and Testing Center of Wuhan University for providing atomic absorption spectroscopy assay. We also thank Mr. Yuanzhi Wang for the non-injuring ion flux tests.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01184/full#supplementary-material
Abbreviations
ABA, abscisic acid; ABF, ABRE-binding factor; ABRE, ABA-responsive element; CaMV, Cauliflower mosaic virus; CAT, catalase; CBL, calcineurin B-like protein; CDPK, Ca-dependent protein kinase; CIPK, CBL-interacting protein kinase; DREB, DRE-/CRT-binding protein; GST, glutathione S-transferase; IL, ion leakage; MDA, malondialdehyde; MS, Murashige and Skoog; POD, peroxidase; qRT-PCR, quantitative reverse-transcription PCR; ROS, reactive oxygen species; RWC, relative water content; SOD, superoxide dismutase; SOS, salt overly sensitive; Tu, tungstate sodium; VC, vector control; WT, wild type.
Footnotes
- ^http://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^http://plants.ensembl.org/index.html
- ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
References
Ache, P., Becker, D., Ivashikina, N., Dietrich, P., Roelfsema, M. R. G., and Hedrich, R. (2000). GORK, a delayed outward rectifier expressed in guard cells of Arabidopsis thaliana, is a K+-selective, K+-sensing ion channel. FEBS Lett. 486, 93–98. doi: 10.1016/s0014-5793(00)02248-1
Asano, T., Tanaka, N., Yang, G., Hayashi, N., and Komatsu, S. (2005). Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol. 46, 356–366. doi: 10.1093/pcp/pci035
Bartels, D., and Sunkar, R. (2005). Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24, 23–58. doi: 10.1080/07352680590910410
Batistic, O., and Kudla, J. (2004). Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 219, 915–924. doi: 10.1007/s00425-004-1333-3
Becker, W., Heukelbach, J., Kentrup, H., and Joost, H. G. (1996). Molecular cloning and characterization of a novel mammalian protein kinase harboring a homology domain that defines a subfamily of serine/threonine kinases. Eur. J. Biochem. 235, 736–743.
Chen, L., Ren, F., Zhou, L., Wang, Q. Q., Zhong, H., and Li, X. B. (2012). The Brassica napus calcineurin B-Like 1/CBL-interacting protein kinase 6 (CBL1/CIPK6) component is involved in the plant response to abiotic stress and ABA signalling. J. Exp. Bot. 63, 6211–6222. doi: 10.1093/jxb/ers273
Chen, X., Gu, Z., Xin, D., Hao, L., Liu, C., Huang, J., et al. (2011). Identification and characterization of putative CIPK genes in maize. J. Genet. Genomics 38, 77–87. doi: 10.1016/j.jcg.2011.01.005
Cheong, Y. H., Pandey, G. K., Grant, J. J., Batistic, O., Li, L., Kim, B.-G., et al. (2007). Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 52, 223–239. doi: 10.1111/j.1365-313X.2007.03236.x
Cui, L.-G., Shan, J.-X., Shi, M., Gao, J.-P., and Lin, H.-X. (2014). The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 80, 1108–1117. doi: 10.1111/tpj.12712
D’Angelo, C., Weinl, S., Batistic, O., Pandey, G. K., Cheong, Y. H., Schultke, S., et al. (2006). Alternative complex formation of the Ca2+-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. Plant J. 48, 857–872. doi: 10.1111/j.1365-313X.2006.02921.x
Deng, X., Hu, W., Wei, S., Zhou, S., Zhang, F., Han, J., et al. (2013a). TaCIPK29, a CBL-interacting protein kinase gene from wheat, confers salt stress tolerance in transgenic tobacco. PLoS ONE 8:e69881. doi: 10.1371/journal.pone.0069881
Deng, X., Zhou, S., Hu, W., Feng, J., Zhang, F., Chen, L., et al. (2013b). Ectopic expression of wheat TaCIPK14, encoding a calcineurin B-like protein-interacting protein kinase, confers salinity and cold tolerance in tobacco. Physiol. Plant. 149, 367–377. doi: 10.1111/ppl.12046
Drerup, M. M., Schlucking, K., Hashimoto, K., Manishankar, P., Steinhorst, L., Kuchitsu, K., et al. (2013). The Calcineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the Arabidopsis NADPH oxidase RBOHF. Mol. Plant 6, 559–569. doi: 10.1093/mp/sst009
Eisenach, C., Papanatsiou, M., Hillert, E. K., and Blatt, M. R. (2014). Clustering of the K+ channel GORK of Arabidopsis parallels its gating by extracellular K+. Plant J. 78, 203–214. doi: 10.1111/tpj.12471
Finkelstein, R. R., Gampala, S. S. L., and Rock, C. D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14, S15–S45. doi: 10.1105/tpc.010441
Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., et al. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876. doi: 10.1111/j.1365-313X.2004.02171.x
Fujita, Y., Fujita, M., Satoh, R., Maruyama, K., Parvez, M. M., Seki, M., et al. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17, 3470–3488. doi: 10.1105/tpc.105.035659
Gouiaa, S., Khoudi, H., Leidi, E. O., Pardo, J. M., and Masmoudi, K. (2012). Expression of wheat Na+/H+ antiporter TNHXS1 and H+- pyrophosphatase TVP1 genes in tobacco from a bicistronic transcriptional unit improves salt tolerance. Plant Mol. Biol. 79, 137–155. doi: 10.1007/s11103-012-9901-6
He, L., Yang, X., Wang, L., Zhu, L., Zhou, T., Deng, J., et al. (2013). Molecular cloning and functional characterization of a novel cotton CBL-interacting protein kinase gene (GhCIPK6) reveals its involvement in multiple abiotic stress tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 435, 209–215. doi: 10.1016/j.bbrc.2013.04.080
Heath, R. L., and Packer, L. (1968). Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125, 189–198. doi: 10.1016/0003-9861(68)90654-1
Held, K., Pascaud, F., Eckert, C., Gajdanowicz, P., Hashimoto, K., Corratge-Faillie, C., et al. (2011). Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res. 21, 1116–1130. doi: 10.1038/cr.2011.50
Hirsch, R. E., Lewis, B. D., Spalding, E. P., and Sussman, M. R. (1998). A role for the AKT1 potassium channel in plant nutrition. Science 280, 918–921. doi: 10.1126/science.280.5365.918
Horsch, R. B., Fry, J. E., Hoffmann, N. L., Eichholtz, D., Rogers, S. C., and Fraley, R. T. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1231.
Hosy, E., Vavasseur, A., Mouline, K., Dreyer, I., Gaymard, F., Poree, F., et al. (2003). The Arabidopsis outward K+ channel GORK is involved in regulation of stomatal movements and plant transpiration. Proc. Natl. Acad. Sci. U.S.A. 100, 5549–5554. doi: 10.1073/pnas.0733970100
Hu, W., Huang, C., Deng, X., Zhou, S., Chen, L., Li, Y., et al. (2013). TaASR1, a transcription factor gene in wheat, confers drought stress tolerance in transgenic tobacco. Plant Cell Environ. 36, 1449–1464. doi: 10.1111/pce.12074
Hu, Y., Chen, L. G., Wang, H. P., Zhang, L. P., Wang, F., and Yu, D. Q. (2013). Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 74, 730–745. doi: 10.1111/tpj.12159
Huang, G. T., Ma, S. L., Bai, L. P., Zhang, L., Ma, H., Jia, P., et al. (2012). Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 39, 969–987. doi: 10.1007/s11033-011-0823-1
Huang, L., Hong, Y., Zhang, H., Li, D., and Song, F. (2016). Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol. 16:203. doi: 10.1186/s12870-016-0897-y
Huertas, R., Olias, R., Eljakaoui, Z., Galvez, F. J., Li, J., De Morales, P. A., et al. (2012). Overexpression of SlSOS2 (SlCIPK24) confers salt tolerance to transgenic tomato. Plant Cell Environ. 35, 1467–1482. doi: 10.1111/j.1365-3040.2012.02504.x
Hundertmark, M., and Hincha, D. K. (2008). LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9:118. doi: 10.1186/1471-2164-9-118
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Jin, X., Sun, T., Wang, X. T., Su, P. P., Ma, J. F., He, G. Y., et al. (2016). Wheat CBL-interacting protein kinase 25 negatively regulates salt tolerance in transgenic wheat. Sci. Rep. 6:28884. doi: 10.1038/srep28884
Julkowska, M. M., and Testerink, C. (2015). Tuning plant signaling and growth to survive salt. Trends Plant Sci. 20, 586–594. doi: 10.1016/j.tplants.2015.06.008
Kim, B. G., Waadt, R., Cheong, Y. H., Pandey, G. K., Dominguez-Solis, J. R., Schultke, S., et al. (2007). The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J. 52, 473–484. doi: 10.1111/j.1365-313X.2007.03249.x
Kim, K. N., Cheong, Y. H., Grant, J. J., Pandey, G. K., and Luan, S. (2003). CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15, 411–423. doi: 10.1105/tpc.006858
Kim, T. H., Boehmer, M., Hu, H., Nishimura, N., and Schroeder, J. I. (2010). Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61, 561–591. doi: 10.1146/annurev-arplant-042809-112226
Kolukisaoglu, U., Weinl, S., Blazevic, D., Batistic, O., and Kudla, J. (2004). Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 134, 43–58. doi: 10.1104/pp.103.033068
Kong, X., Pan, J., Zhang, M., Xing, X., Zhou, Y., Liu, Y., et al. (2011). ZmMKK4, a novel group C mitogen-activated protein kinase kinase in maize (Zea mays), confers salt and cold tolerance in transgenic Arabidopsis. Plant Cell Environ. 34, 1291–1303. doi: 10.1111/j.1365-3040.2011.02329.x
Kudla, J., Xu, Q., Harter, K., Gruissem, W., and Luan, S. (1999). Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc. Natl. Acad. Sci. U.S.A. 96, 4718–4723. doi: 10.1073/pnas.96.8.4718
Lacombe, B., Pilot, G., Michard, E., Gaymard, F., Sentenac, H., and Thibaud, J. B. (2000). A shaker-like K+ channel with weak rectification is expressed in both source and sink phloem tissues of Arabidopsis. Plant Cell 12, 837–851. doi: 10.1105/tpc.12.6.837
Lee, S. C., Lan, W.-Z., Kim, B.-G., Li, L., Cheong, Y. H., Pandey, G. K., et al. (2007). A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc. Natl. Acad. Sci. U.S.A. 104, 15959–15964. doi: 10.1073/pnas.0707912104
Leung, J., and Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 199–222. doi: 10.1146/annurev.arplant.49.1.199
Li, J., Besseau, S., Toronen, P., Sipari, N., Kollist, H., Holm, L., et al. (2013). Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 200, 457–472. doi: 10.1111/nph.12378
Li, L., Kim, B.-G., Cheong, Y. H., Pandey, G. K., and Luan, S. (2006). A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 12625–12630. doi: 10.1073/pnas.0605129103
Liu, J., and Zhu, J. K. (1997). An Arabidopsis mutant that requires increased calcium for potassium nutrition and salt tolerance. Proc. Natl. Acad. Sci. U.S.A. 94, 14960–14964. doi: 10.1073/pnas.94.26.14960
Liu, J., and Zhu, J. K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280, 1943–1945. doi: 10.1126/science.280.5371.1943
Liu, X., Wang, Z., Wang, L., Wu, R., Phillips, J., and Deng, X. (2009). LEA 4 group genes from the resurrection plant Boea hygrometrica confer dehydration tolerance in transgenic tobacco. Plant Sci. 176, 90–98. doi: 10.1016/j.plantsci.2008.09.012
Luan, S. (2009). The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 14, 37–42. doi: 10.1016/j.tplants.2008.10.005
Lv, S., Jiang, P., Chen, X., Fan, P., Wang, X., and Li, Y. (2012). Multiple compartmentalization of sodium conferred salt tolerance in Salicornia europaea. Plant Physiol. Biochem. 51, 47–52. doi: 10.1016/j.plaphy.2011.10.015
Osakabe, Y., Arinaga, N., Umezawa, T., Katsura, S., Nagamachi, K., Tanaka, H., et al. (2013). Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 25, 609–624. doi: 10.1105/tpc.112.105700
Pandey, G. K., Kanwar, P., Singh, A., Steinhorst, L., Pandey, A., Yadav, A. K., et al. (2015). Calcineurin B-like protein-interacting protein kinase CIPK21 regulates osmotic and salt stress responses in Arabidopsis. Plant Physiol. 169, 780–792. doi: 10.1104/pp.15.00623
Piao, H. L., Xuan, Y. H., Park, S. H., Je, B. I., Park, S. J., Park, S. H., et al. (2010). OsCIPK31, a CBL-interacting protein kinase is involved in germination and seedling growth under abiotic stress conditions in rice plants. Mol. Cells 30, 19–27. doi: 10.1007/s10059-010-0084-1
Pilot, G., Gaymard, F., Mouline, K., Cherel, I., and Sentenac, H. (2003). Regulated expression of Arabidopsis Shaker K+ channel genes involved in K+ uptake and distribution in the plant. Plant Mol. Biol. 51, 773–787. doi: 10.1023/a:1022597102282
Pitzschke, A., and Hirt, H. (2009). Disentangling the complexity of mitogen-activated protein kinases and reactive oxygen species signaling. Plant Physiol. 149, 606–615. doi: 10.1104/pp.108.131557
Qiu, Q. S., Guo, Y., Dietrich, M. A., Schumaker, K. S., and Zhu, J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U.S.A. 99, 8436–8441. doi: 10.1073/pnas.122224699
Raghavendra, A. S., Gonugunta, V. K., Christmann, A., and Grill, E. (2010). ABA perception and signalling. Trends Plant Sci. 15, 395–401. doi: 10.1016/j.tplants.2010.04.006
Rao, X.-L., Zhang, X.-H., Li, R.-J., Shi, H.-T., and Lu, Y.-T. (2011). A calcium sensor-interacting protein kinase negatively regulates salt stress tolerance in rice (Oryza sativa). Funct. Plant Biol. 38, 441–450. doi: 10.1071/fp10205
Reintanz, B., Szyroki, A., Ivashikina, N., Ache, P., Godde, M., Becker, D., et al. (2002). AtKC1, a silent Arabidopsis potassium channel alpha-subunit modulates root hair K+ influx. Proc. Natl. Acad. Sci. U.S.A. 99, 4079–4084. doi: 10.1073/pnas.052677799
Ruiz-Lozano, J. M., Porcel, R., Azcon, C., and Aroca, R. (2012). Regulation by arbuscular mycorrhizae of the integrated physiological response to salinity in plants: new challenges in physiological and molecular studies. J. Exp. Bot. 63, 4033–4044. doi: 10.1093/jxb/ers126
Sano, T., Becker, D., Ivashikina, N., Wegner, L. H., Zimmermann, U., Roelfsema, M. R., et al. (2007). Plant cells must pass a K+ threshold to re-enter the cell cycle. Plant J. 50, 401–413. doi: 10.1111/j.1365-313X.2007.03071.x
Shabala, S., Bose, J., Fuglsang, A. T., and Pottosin, I. (2016). On a quest for stress tolerance genes: membrane transporters in sensing and adapting to hostile soils. J. Exp. Bot. 67, 1015–1031. doi: 10.1093/jxb/erv465
Shinozaki, K., Yamaguchi-Shinozaki, K., and Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417. doi: 10.1016/S1369-5266(03)00092-X
Sparkes, I. A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Trevino, M. B., and O’Connell, M. A. (1998). Three drought-responsive members of the nonspecific lipid-transfer protein gene family in Lycopersicon pennellii show different developmental patterns of expression. Plant Physiol. 116, 1461–1468. doi: 10.1104/pp.116.4.1461
Tripathi, V., Parasuraman, B., Laxmi, A., and Chattopadhyay, D. (2009). CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. Plant J. 58, 778–790. doi: 10.1111/j.1365-313X.2009.03812.x
Vogel, J. P., Garvin, D. F., Mockler, T. C., Schmutz, J., Rokhsar, D., Bevan, M. W., et al. (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768. doi: 10.1038/nature08747
Wang, L., Hu, W., Sun, J., Liang, X., Yang, X., Wei, S., et al. (2015). Genome-wide analysis of SnRK gene family in Brachypodium distachyon and functional characterization of BdSnRK2.9. Plant Sci. 237, 33–45. doi: 10.1016/j.plantsci.2015.05.008
Wang, R. K., Li, L. L., Cao, Z. H., Zhao, Q., Li, M., Zhang, L. Y., et al. (2012). Molecular cloning and functional characterization of a novel apple MdCIPK6L gene reveals its involvement in multiple abiotic stress tolerance in transgenic plants. Plant Mol. Biol. 79, 123–135. doi: 10.1007/s11103-012-9899-9
Wang, Y., Sun, T., Li, T. T., Wang, M., Yang, G. X., and He, G. Y. (2016). A CBL-interacting protein kinase TaCIPK2 confers drought tolerance in transgenic tobacco plants through regulating the stomatal movement. PLoS ONE 11:e0167962. doi: 10.1371/journal.pone.0167962
Xiang, Y., Huang, Y., and Xiong, L. (2007). Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 144, 1416–1428. doi: 10.1104/pp.107.101295
Xu, J., Li, H.-D., Chen, L.-Q., Wang, Y., Liu, L.-L., He, L., et al. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125, 1347–1360. doi: 10.1016/j.cell.2006.06.011
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803.
Yan, H., Jia, H., Chen, X., Hao, L., An, H., and Guo, X. (2014). The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 55, 2060–2076. doi: 10.1093/pcp/pcu133
Yang, Q., Chen, Z.-Z., Zhou, X.-F., Yin, H.-B., Li, X., Xin, X.-F., et al. (2009). Overexpression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol. Plant 2, 22–31. doi: 10.1093/mp/ssn058
Yang, W., Kong, Z., Omo-Ikerodah, E., Xu, W., Li, Q., and Xue, Y. (2008). Calcineurin B-like interacting protein kinase OsCIPK23 functions in pollination and drought stress responses in rice (Oryza sativa L.). J. Genet. Genomics 35, 531–543, S1–S2. doi: 10.1016/S1673-8527(08)60073-9
Yoshida, T., Mogami, J., and Yamaguchi-Shinozaki, K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. doi: 10.1016/j.pbi.2014.07.009
Yu, Q., An, L., and Li, W. (2014). The CBL-CIPK network mediates different signaling pathways in plants. Plant Cell Rep. 33, 203–214. doi: 10.1007/s00299-013-1507-1
Zhang, H., Yang, B., Liu, W. Z., Li, H., Wang, L., Wang, B., et al. (2014). Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.). BMC Plant Biol. 14:8. doi: 10.1186/1471-2229-14-8.
Zhang, X., Zhang, Z. J., Chen, J., Chen, Q., Wang, X. C., and Huang, R. F. (2005). Expressing TERF1 in tobacco enhances drought tolerance and abscisic acid sensitivity during seedling development. Planta 222, 494–501. doi: 10.1007/s00425-005-1564-y
Zhou, L., Lan, W., Chen, B., Fang, W., and Luan, S. (2015). A calcium sensor-regulated protein kinase, CALCINEURIN B-LIKE PROTEIN-INTERACTING PROTEIN KINASE19, is required for pollen tube growth and polarity. Plant Physiol. 167, 1351–1360. doi: 10.1104/pp.114.256065
Keywords: ABA, Brachypodium distachyon, CIPK, drought stress, Nicotiana tabacum, salt stress
Citation: Luo Q, Wei Q, Wang R, Zhang Y, Zhang F, He Y, Zhou S, Feng J, Yang G and He G (2017) BdCIPK31, a Calcineurin B-Like Protein-Interacting Protein Kinase, Regulates Plant Response to Drought and Salt Stress. Front. Plant Sci. 8:1184. doi: 10.3389/fpls.2017.01184
Received: 29 April 2017; Accepted: 21 June 2017;
Published: 07 July 2017.
Edited by:
Junhua Peng, Center for Life Sci&Tech of China National Seed Group Co. Ltd., ChinaReviewed by:
Hong Luo, Clemson University, United StatesLi Huang, Montana State University, United States
Chang-Jie Jiang, National Institute of Agrobiological Sciences, Japan
Copyright © 2017 Luo, Wei, Wang, Zhang, Zhang, He, Zhou, Feng, Yang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangxiao Yang, ygx@hust.edu.cn Guangyuan He, hegy@hust.edu.cn
†These authors have contributed equally to this work.
 Qingchen Luo1†
Qingchen Luo1† Shiyi Zhou
Shiyi Zhou Guangxiao Yang
Guangxiao Yang Guangyuan He
Guangyuan He