- 1Key Laboratory of Soybean Biology of Chinese Education Ministry, Soybean Research Institute, Northeast Agricultural University, Harbin, China
- 2Jiamusi Branch of Heilongjiang Academy of Agricultural Sciences, Jiamusi, China
- 3Key Laboratory of Soybean Cultivation of Ministry of Agriculture China, Soybean Research Institute, Heilongjiang Academy of Agricultural Sciences, Harbin, China
- 4Heihe Branch of Heilongjiang Academy of Agricultural Sciences, Heihe, China
Phytophthora root and stem rot caused by the oomycete pathogen Phytophthora sojae is a destructive disease of soybean worldwide. Plant dirigent proteins (DIR) are proposed to have roles in biosynthesis of either lignan or lignin-like molecules, and are important for defense responses, secondary metabolism, and pathogen resistance. In the present work, a novel DIR gene expressed sequence tag is identified as up-regulated in the highly resistant soybean cultivar ‘Suinong 10’ inoculated with P. sojae. The full length cDNA is isolated using rapid amplification of cDNA ends, and designated GmDIR22 (GenBank accession no. HQ_993047). The full length GmDIR22 is 789 bp and contains a 567 bp open reading frame encoding a polypeptide of 188 amino acids. The sequence analysis indicated that GmDIR22 contains a conserved dirigent domain at amino acid residues 43–187. The quantitative real-time reverse transcription PCR demonstrated that soybean GmDIR22 mRNA is expressed most highly in stems, followed by roots and leaves. The treatments with stresses demonstrated that GmDIR22 is significantly induced by P. sojae and gibberellic acid (GA3), and also responds to salicylic acid, methyl jasmonic acid, and abscisic acid. The GmDIR22 is targeted to the cytomembrane when transiently expressed in Arabidopsis protoplasts. Moreover, The GmDIR22 recombinant protein purified from Escherichia coli could effectively direct E-coniferyl alcohol coupling into lignan (+)-pinoresinol. Accordingly, the overexpression of GmDIR22 in transgenic soybean increased total lignan accumulation. Moreover, the lignan extracts from GmDIR22 transgenic plants effectively inhibits P. sojae hyphal growth. Furthermore, the transgenic overexpression of GmDIR22 in the susceptible soybean cultivar ‘Dongnong 50’ enhances its resistance to P. sojae. Collectively, these data suggested that the primary role of GmDIR22 is probably involved in the regulation of lignan biosynthesis, and which contributes to resistance to P. sojae.
Introduction
Dirigent proteins (DIR), whose name is derived from the Latin word, dirigere (to align or guide), were first identified in Forsythia suspensa (Davin et al., 1997). The dirigent gene family has several members in numerous plant species, including lichens, ferns, gymnosperms, and angiosperms (Ralph et al., 2006, 2007; Wu et al., 2009). Many dirigent genes are inducible by different types of abiotic and biotic stress factors, including wounding (Ralph et al., 2007), dehydration, low temperature, abscisic acid (ABA) (Wu et al., 2009), H2O2, NaCl, and polyethylene glycol (Magalhaes et al., 2010). The apparent upregulation of dirigent genes in response to attacks by fungi (Zhu et al., 2007; Reboledo et al., 2015) and insects (Ralph et al., 2007) are of particular interest. The predominant roles of the dirigent gene family are in defense responses, secondary metabolism, and fiber biosynthesis (Davin and Lewis, 2003; Ralph et al., 2007; Berim et al., 2008). Moreover, they have important roles in plant secondary metabolism, including contribution to lignan and lignin formation (Liu et al., 2008; Pickel et al., 2010; Kim H.J. et al., 2012; Dalisay et al., 2015; Effenberger et al., 2015). In addition, DIR have been implicated in certain developmentally controlled lignification processes, where they contribute to the formation of the casparian strip in the developing Arabidopsis root (Hosmani et al., 2013).
Groundbreaking in vitro biochemical analysis by Davin et al. (1997) demonstrated that a Forsythia intermedia dirigent protein (FiDIR) could stereoselectively couple two coniferyl alcohol molecules to produce a (+)-pinoresinol in the presence of an oxidase or electron oxidant. The same specificity has been reported for DIRs from Thuja plicata (TpDIR5, 8) (Kim et al., 2002), Schisandra chinensis (ScDIR) (Kim K.W. et al., 2012), Pisum sativum (PsDRR206) (Seneviratne et al., 2015), and Linum usitatissimum (LuDIR1) (Dalisay et al., 2015). Coniferyl alcohol is the direct precursor for formation of lignan dimers. Meanwhile, coniferyl and sinapyl alcohol, together with other monolignol p-coumaryl alcohols, are also polymerized into the macromolecule lignin, certain biosynthetic pathways overlap in production of lignans and lignin (Davin and Lewis, 2003). These two major classes of plant metabolic products are apparently unique to vascular land plants, and together they constitute approximately 20–30% of their organic carbon (Lewis and Yamamoto, 1990; Lewis and Davin, 1998, 1999; Lewis et al., 1999). Lignans are structurally defined as 8, 8′-coupled phenylpropanoid dimers (Halls et al., 2004; Davin and Lewis, 2005), their primary formation can be under either constitutive or inducible control, and their composition and amounts vary markedly among plant species (Lewis and Davin, 1999). Lignans are present in different tissues and organs of vascular plants (roots, stems, leaves, flowers, seeds, etc.) (Ayres and Loike, 1990), and have potent plant defense properties, including antiviral activities (Chang et al., 1995), antioxidant (Lundberg et al., 1944; Osawa et al., 1985), antifeedant (Matsui and Munakata, 1975), and antimicrobial (Nakatani et al., 1988; Pauletti et al., 2000; Harmatha and Dinan, 2003; Saleem et al., 2005), which are increasingly considered of practical importance (MacRae and Towers, 1984; Lewis and Davin, 1999). Lignin is the main component of plant cell walls, and has an important role in development. Accumulation of lignin and callose in the cell walls enhances plants mechanical strength and prevents pathogen spread between cells (Tiburzy and Reisener, 1990). The lignin formed in response to pathogens or elicitors is significantly different from the type that occurs in undamaged plant structures; therefore, it has been speculated that the regulation of lignin synthesis in defense responses and development may be controlled by different pathways (Lange et al., 1995; Pomar et al., 2002).
The dirigent phenomenon was discovered by Davin et al. (1997), who studied the coupling of two coniferyl alcohol radicals generated from coniferyl alcohol by single-electron transfer (e.g., by laccases or peroxidases) as the first step in the biosynthesis of lignans (Davin and Lewis, 2000). Although DIR were discovered more than a decade ago, the biological functions of many DIR proteins remain ambiguous, and direct experimental evidence substantiating the contributions of DIR to lignin biosynthesis is rare (Önnerud et al., 2002; Davin et al., 2008; Ma and Liu, 2015).
Phytophthora root and stem rot of soybean (PRR), caused by the oomycete pathogen Phytophthora sojae, is a destructive soil-borne disease of soybean all around the world (Wrather et al., 1997; Tyler, 2007). In our previous study, a cDNA library was constructed from the leaf tissues of the highly resistant soybean ‘Suinong 10,’ using suppression subtractive hybridization, and the mRNAs encoding expressed sequence tags (ESTs) showed increased expression during P. sojae infection (Xu et al., 2012). In this study, a full-length cDNA corresponding to an up-regulated EST encoding a conservative dirigent domain was isolated from soybean ‘Suinong 10’ using rapid amplification of cDNA ends (RACE). The full-length cDNA belonged to a new member of the soybean DIR family and was designated GmDIR22 (GenBank accession no. HQ_993047). The expression patterns of the GmDIR22 gene were characterized in response to abiotic and biotic stresses, and its expression levels were determined in different soybean tissues. GmDIR22 protein expressed in Escherichia coli was purified, and its function was investigated. Furthermore, the content of lignan and lignin in transgenic plants overexpressing GmDIR22 was established and the antimicrobial activity of lignan extracts against P. sojae hyphal growth was evaluated. Moreover, the transgenic soybean plants over-expressing GmDIR22 gene were produced, and their antimicrobial properties were also investigated.
Materials and Methods
Plant Materials and Growth Conditions
The resistant soybean cultivar ‘Suinong 10’ against the predominant race of P. sojae (race 1) in Heilongjiang, China (Zhang et al., 2010), was used for gene isolation and the seedlings at the first-node stage (V1; Fehr et al., 1971) were used for various treatments. P. sojae race 1, PSR01, isolated from infected soybean plants in Heilongjiang (Zhang et al., 2010), was used in the experiment. ‘Suinong 10’ seeds were grown with a photoperiod of 16/8 h light/dark and maintained at 22°C and 70% relative humidity in a greenhouse. The susceptible soybean cultivar ‘Dongnong 50’ was obtained from Key Laboratory of Soybean Biology of Chinese Education Ministry, Harbin, and used for gene transformation experiments.
Isolation and Sequence Analysis of GmDIR22
Rapid amplification of cDNA ends was performed to isolate the full-length cDNA, corresponding to an EST up-regulated in soybean ‘Suinong 10’ in response to P. sojae infection, using the SMART RACE cDNA amplification Kit (Clontech, CA, United States). The gene-specific primers GSP1 for 5′- RACE and GSP2 for 3′-RACE (and) were designed to produce the antisense and sense strands, respectively. The full-length cDNA sequence of GmDIR22 was obtained by PCR amplification from ‘Suinong 10’ cDNA using the primer pair GmDIR22F and GmDIR22R (Supplementary Table S1). PCR products were ligated into the pMDTM18-T vector (Takara Biotech Inc, Dalian, China), then transformed into E. coli DH5α cells (Shanghai Biotech Inc, Shanghai, China) and sequenced (GENEWIZ, Beijing, China).
NCBI bioinformatics tools were used to analyze nucleotide and protein sequence data1. The predicted protein structure was analyzed using SMART2. Sequence alignments were performed using DNAMAN software3. A phylogenetic tree, based on the nucleotide sequences of GmDIR22 and other DIR members, was generated using MEGA 5.1 software4. The three-dimensional structure of GmDIR22 was predicted using the online program, Phyre25.
Stress Treatments
To investigate GmDIR22 gene expression patterns in response to various stress factors, ‘Suinong 10’ soybean seedlings were treated with 50 mg.L-1 gibberellic acid (GA3), 100 μM methyl jasmonic acid (MeJA), 0.5 mM salicylic acid (SA), and 50 mM ABA. The unifoliolate leaves were harvested at 0, 3, 6, 9, 12, and 24 h after the treatment, wrapped in aluminum foil, immediately frozen in liquid nitrogen, and stored at –80°C until RNA extraction and cDNA synthesis. The untreated soybean leaves were used as controls. For P. sojae treatment, soybean plants were infected with P. sojae zoospores following the method described by Ward et al. (1979). The unifoliolate leaves were harvested for RNA isolation at 0, 6, 12, 24, 48, and 72 h after the treatments, wrapped in aluminum foil, immediately frozen in liquid nitrogen and stored at –80°C.
Quantitative RT-PCR Analysis
To determine the abundance of the GmDIR22 transcript, qRT-PCR analysis was performed. Total RNA was extracted from ‘Suinong 10’ soybean leaves at different time points after diverse stress treatments using TRIzol reagent (Invitrogen, Shanghai, China). First-strand cDNA was synthesized from 1 μg RNA using a Moloney murine leukemia virus reverse transcriptase kit (Takara, Dalian, China), according to the manufacturer’s protocol. qRT-PCR analysis was performed using a real-time RT-PCR kit (Takara, Japan) on a CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad, United States). DNA levels were determined using SYBR Green dye. The soybean housekeeping gene, GmActin4 (GenBank accession no. AF049106), was used as an internal control (see Supplementary Table S1 for primer sequences). For tissue distribution analysis, the transcript levels of the GmEF1β gene (GenBank accession no. NM_001248778) was used as a quantitative control (see Supplementary Table S1 for primer sequences). Relative expression levels were determined using the comparative threshold method (2-ΔΔCT). Three biological replicates with three technical replicates of each qRT-PCR were performed.
Subcellular Localization of GmDIR22 Protein
To determine the subcellular localization of GmDIR22, the full-length GmDIR22 was cloned in frame at the 5′-terminus of the green fluorescent protein (GFP) coding sequence in the 35S:GFP vector using GmDIR22-GF and GmDIR22-GR (Supplementary Table S1) as the primer pair, to generate the fusion construct 35S:GmDIR22-GFP. GFP-fused GmDIR22 was transiently expressed in Arabidopsis protoplast cells, following the instructions described by Yoo et al. (2007) with minor modifications.
Expression and Purification of GmDIR22 Fusion Protein
The full-length GmDIR22 cDNA was framed into the NcoI/XhoI site of the pET29b(+) vector (Novagen, Germany), to generate pET29b(+)-GmDIR22. The recombinant fusion plasmid was transformed into E. coli BL21 (DE3) cells (TransGen Biotech, China). His-tagged GmDIR22 protein expression was induced using 0.5 mM isopropyl-β-D-thiogalactoside (IPTG) at 37°C for 6 h in LB medium containing 50 mg. mL-1 kanamycin. The analysis and purification of the recombinant protein were performed by SDS–PAGE.
Analysis of Coupling Properties
Coupling assays were performed as described by Davin et al. (1997) with minor modifications. The reaction mixtures consisted of ammonium peroxydisulfate (2 μmol.mL-1) as an oxidant, recombinant protein (1.5 nmol.mL-1), and E-coniferyl alcohol (10 μmol.mL-1) in MES-NaOH buffer (0.1 M, pH 5.0) to a total volume of 250 μL. After incubation for 3 h at 30°C, the mixture was extracted three times with ethyl acetate. The extract was evaporated to dryness under vacuum, and the residue re-dissolved in 50% methanol. High Performance Liquid Chromatography (HPLC) was used to separate the substrate and product.
Vector Construction and Soybean Transformation
The full-length coding region of GmDIR22 was PCR amplified and inserted into the NcoI/ApalI site of pCAMBIA3301 vector containing the bar gene as the selective marker with the primer pairs GmDIR22-oF and GmDIR22-oR (Supplementary Table S1). The recombinant construct 35S:GmDIR22 was introduced into Agrobacterium tumefaciens strain LBA4404 using the freeze-thaw method, as described by Holsters et al. (1978). The susceptible soybean ‘Dongnong 50’ was used for gene transformation using the Agrobacterium-mediated method described by Paz et al. (2004). Transgenic soybean plants (T4) were identified by PCR amplification, and developed to T5 transgenic soybean plants for further analysis.
Assays of Pathogen Responses of Transgenic Soybean Plants
The response assays of GmDIR22-transformed plants to P. sojae, the living cotyledons and detached leaves of T5 transgenic soybean plants were treated with pathogen inoculum following the methods described by Dou et al. (2003) with minor modifications. Non-transformed plants treated in the same way served as controls. Disease symptoms on each cotyledon were photographed after inoculation using a Nikon D7000 camera. The relative biomass of P. sojae in infected cotyledons was assessed after 72 h based on the transcript levels of the P. sojae TEF1 gene (GenBank accession no. EU079791) using soybean GmEF1β as a reference gene, determined according to the method described by Chacón et al. (2010) (see Supplementary Table S1 for TEF1 and GmEF1β primer sequences). The pathogen response assays were performed on three biological replicates with their respective three technical replicates.
Analyses of the Lignan and Lignin Content of Transgenic Plants
Samples from air-dried stems were subjected to quantitative analysis for ligninreferred to the Klason method by Kirk and Obst (1988). The cell wall residue (% CWR) was expressed relative to lignin content. Total lignan was extracted from leaf samples referred to the method by Chen et al. (2010). The extract was dried under vacuum, and the residue was re-dissolved in ethanol buffer. The chromotropic acid spectrophotometric method as described by Lu et al. (2011) was used to determine the total lignan.
Verification of the Antimicrobial Effects of Lignan Extracts
The antimicrobial activities of lignan extracts were assayed using the hyphal growth inhibition method, as described by Schlumbaum et al. (1986). P. sojae race 1 was cultured on V8 juice agar plates at 25°C for 72 h and the sterile filter-paper disks containing lignan extracts from different transgenic plants were placed around the colony. After incubation for 24 h at 25°C, the pathogen growth zones were observed. In further experiments, the inhibition of the hyphal growth of P. sojae with various concentrations of lignan were Researched, as described by Giannakopoulou et al. (2014) with minor modifications.
Results
GmDIR22 Sequence and Bioinformatic Analysis
The full-length GmDIR22 cDNA (GenBank accession no. HQ_993047) was isolated from ‘Suinong10’ soybean by RACE. The sequence analysis showed that GmDIR22 has an open reading frame of 567 bp and encodes a polypeptide of 188 amino acids (Supplementary Figure S1) with a predicted molecular mass of 20.88 kDa and a pI of 9.02. The deduced protein has a central 145 amino acid dirigent domain at amino acid residues 43–187 (Supplementary Figure S1). The predicted three-dimensional model of the GmDIR22 protein consists of eight β-strands, forming an eight-stranded antiparallel β-barrel (Supplementary Figure S2), similar to the previously reported (+)-pinoresinol-forming DIR from pea, DDR206 (Kim et al., 2015). A phylogenetic tree was constructed using the neighbor-joining algorithm based on the nucleotide sequences of GmDIR22 that contained other members of the DIR family, indicating that these neighboring genes might display similar functions (Figure 1). A total of 101 dir or dir-like nucleotide sequences from Glycine max (1), Arabidopsis thaliana (19), Oryza sativa (28), Picea glauca (26), Thuja plicata (9), and an additional 18 dir genes identified from a variety of species, including pea, cotton, corn, and sesame, etc. (see Supplementary Table S2) were used to construct the phylogenetic tree. In the phylogenetic tree, GmDIR22 is close to five AtDIR members (belonging to dir-b/d subfamily) and shares 55.15, 54.70, 53.66, 52.95, and 47.97 % nucleotide identity with AtDIR20, AtDIR7, AtDIR19, AtDIR3, and AtDIR8, respectively (Figure 1). We then performed sequence comparison between GmDIR22 and the five AtDIR nucleotides to analyze the homology among them. The alignments of the nucleotide sequences show five well-conserved motifs (Supplementary Figure S3).
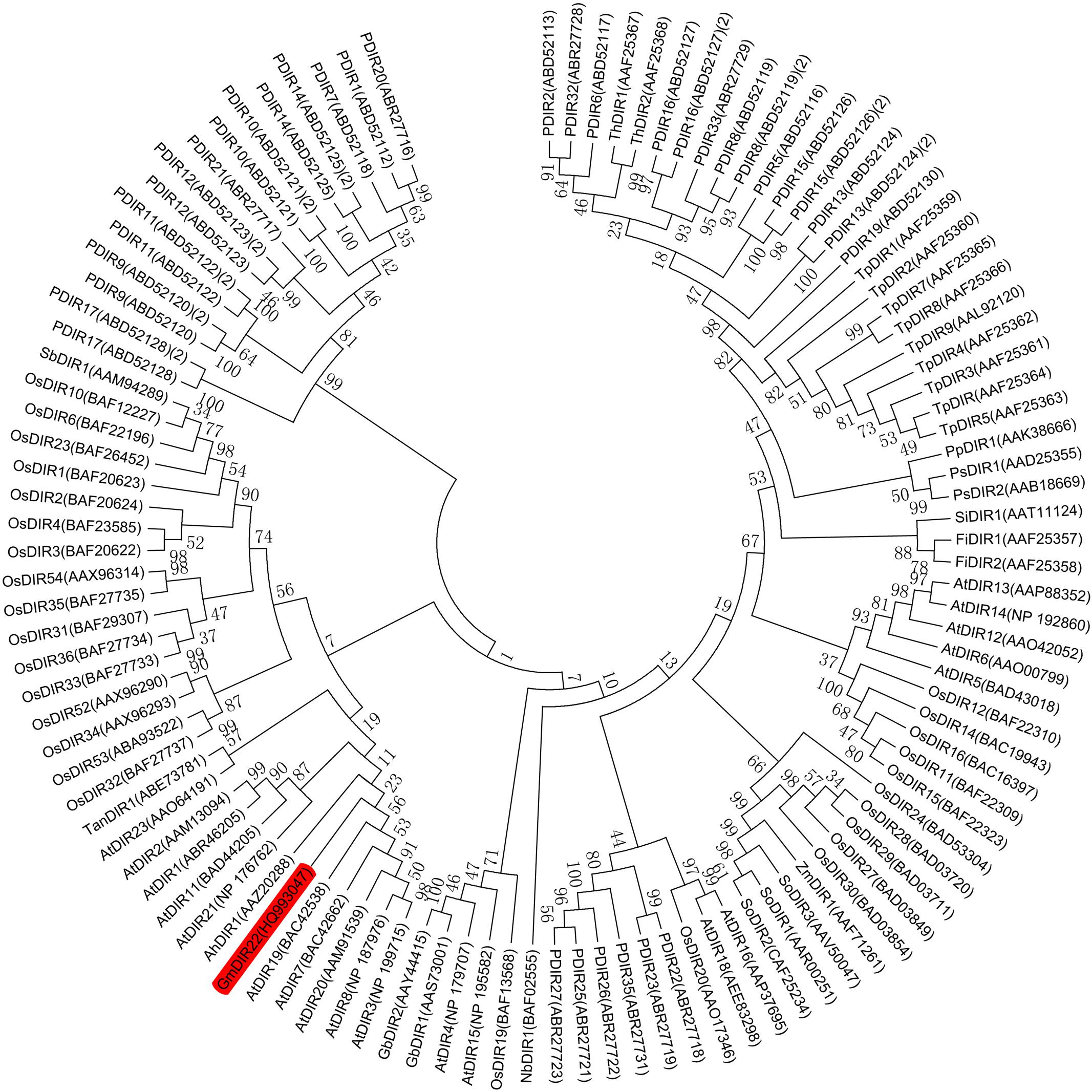
FIGURE 1. Phylogenetic tree constructed using soybean GmDIR22 nucleotide and DIR or DIR-like nucleotide sequences from various plant species. The nucleotide sequences of 101 dirigent genes were analyzed by neighbor-joining algorithm using Mega 5.1. The stability of internal nodes was assessed by bootstrap analysis with 1000 replicates. DIR nomenclature is as follows: Ah, Arachis hypogaea; At, Arabidopsis thaliana; Fi, Forsythia intermedia; Gb, Gossypium barbadense; Gm, Glycine max; Nb, Nicotiana benthamiana; Os, Oryza sativa; P, Picea glauca, Picea sitchensis, or P. glauca x engelmannii; Pp, Podophyllum peltatum; Ps, Pisum sativum; Sb, Sorghum bicolor; Si, Sesamum indicum; So, Saccharum officinarum; Ta, Tamarix androssowii; Th, Tsuga heterophylla; Tp, Thuja plicata; and Zm, Zea mays.
Changes of GmDIR22 Transcript Levels in Response to Stress Treatments
To determine the expression pattern of GmDIR22 in different soybean cultivars, qRT-PCR was performed to examine the transcript levels of GmDIR22 (Supplementary Table S3). The expression of GmDIR22 was higher in resistant cultivars (‘Suinong 10,’ ‘Williams 82,’ ‘Hefeng 34,’ ‘Nenfeng 15,’ and ‘Kangxian 1’) than that in susceptible cultivars (‘Hefeng 25,’ ‘Heinong 37,’ ‘Dongnong 50,’ ‘Kendou 18,’ and ‘Hefeng 35’) (Figure 2). The responsiveness of GmDIR22 mRNA transcript levels to biotic and abiotic stresses in ‘Suinong10’ soybean plants was evaluated by qRT-PCR (Supplementary Tables S5, S6, S7, S8, S9). The investigation of tissue-specific transcript abundance in ‘Suinong 10’ soybean (Supplementary Table S4) demonstrated that GmDIR22 is constitutively and highly expressed in the stems, followed by the roots and leaves (Figure 3A). After P. sojae treatment, the levels of GmDIR22 mRNA rapidly increases, reaching a maximum level at 48 h after treatment, followed by a decline (Figure 3B). GmDIR22 expression is also induced with the treatments of GA3, SA, MeJA, and ABA. The levels of GmDIR22 mRNA increases rapidly in response to GA3 treatment, reaching maximum levels at 9 h, followed by a rapid decline. With SA, MeJA, and ABA treatment, GmDIR22 transcripts accumulates and reaches a maximum level at 6, 6, and 9 h, respectively, but the expression of GmDIR22 is relatively lower than that under P. sojae and GA3 stresses (Figure 3C).
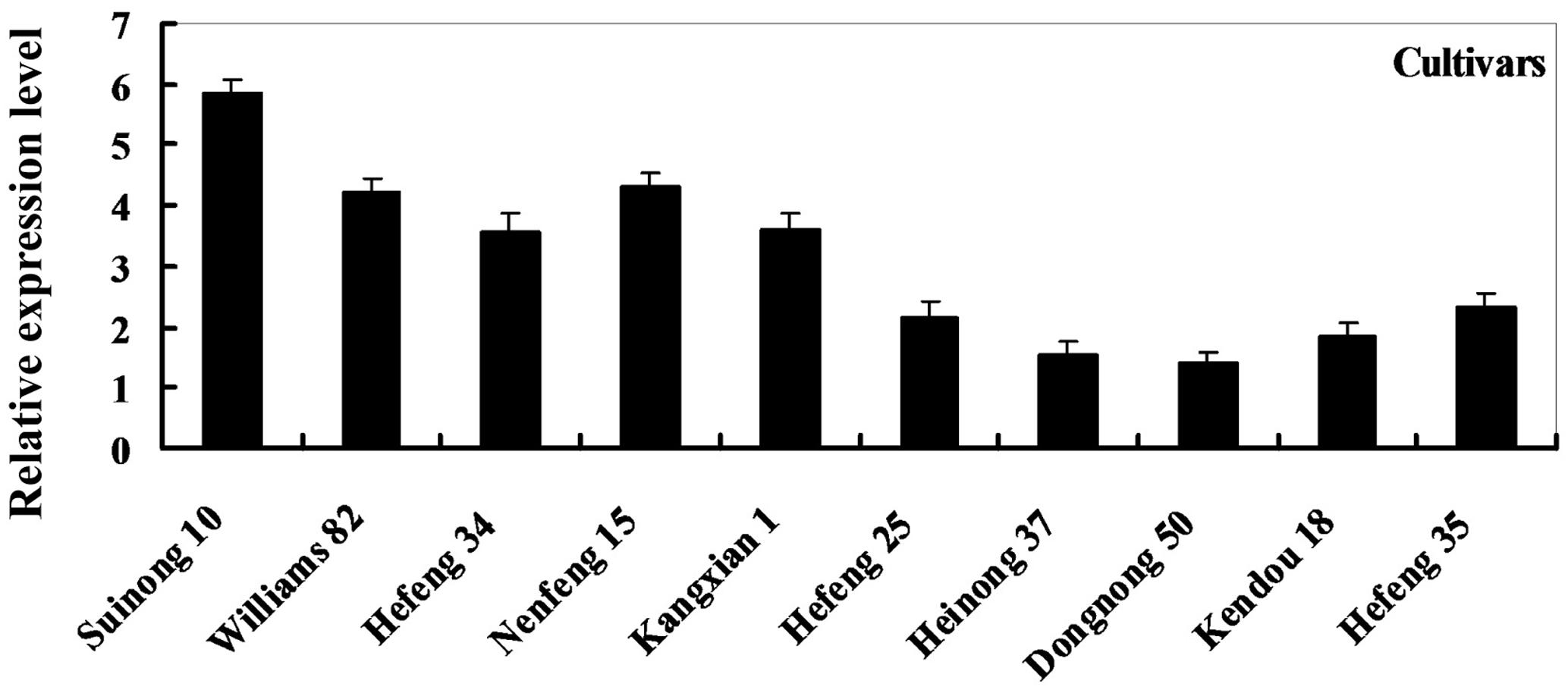
FIGURE 2. Relative expression levels of GmDIR22 in resistant and susceptible soybean cultivars, at 48 h after P. sojae infection. The soybean cultivars ‘Suinong 10,’ ‘Williams 82,’ ‘Hefeng 34,’ ‘Nenfeng 15,’ and ‘Kangxian 1,’ which were resistant to P. sojae race 1. The soybean cultivars ‘Hefeng 25,’ ‘Heinong 37,’ ‘Dongnong 50,’ ‘Kendou 18,’ and ‘Hefeng 35’ which were susceptible to P. sojae race 1. Experiments were performed using three biological replicates with three technical replicates each. Bars indicated standard errors of the mean.
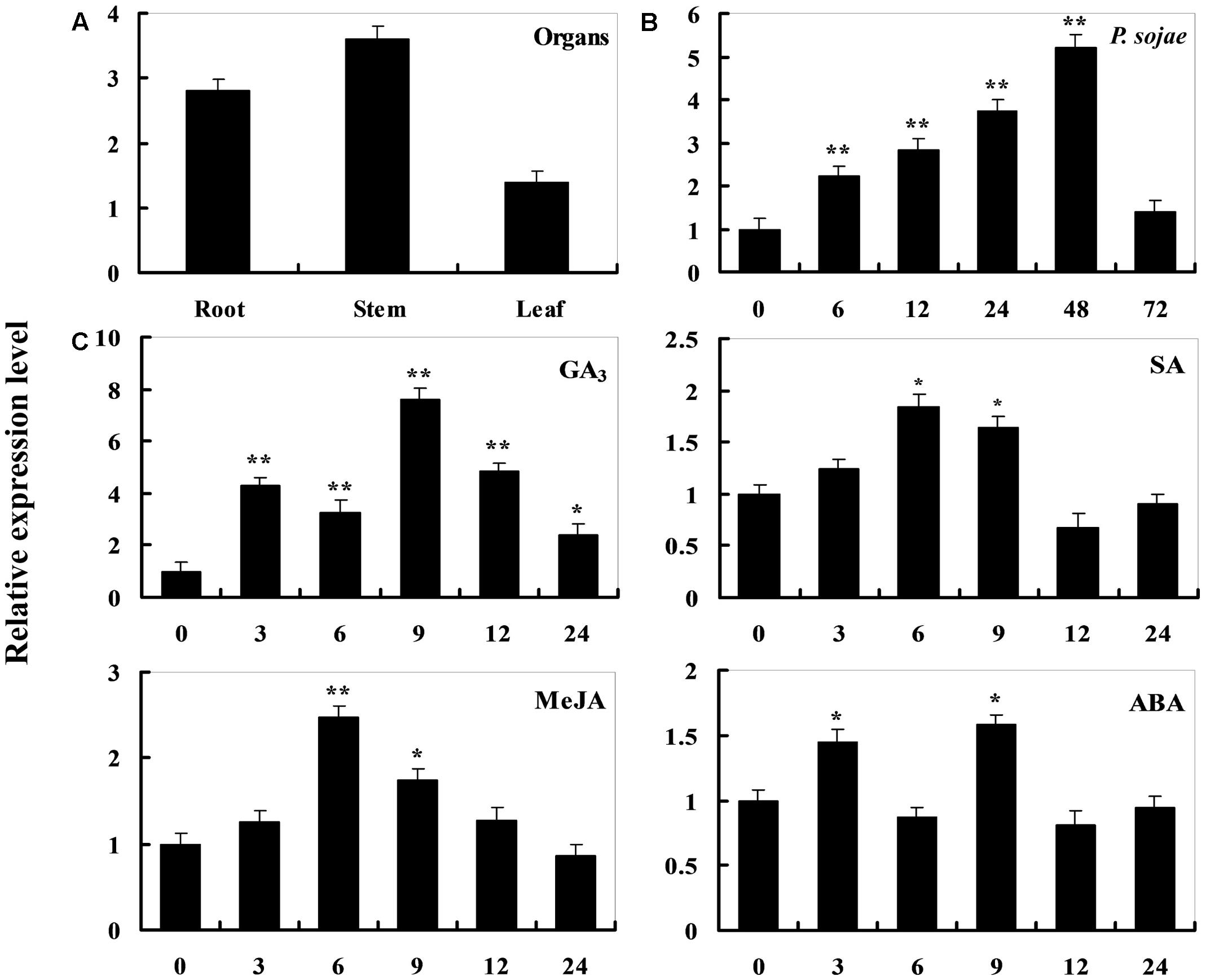
FIGURE 3. Analysis of GmDIR22 expression patterns by qRT-PCR. (A) Roots, stems, and leaves were separated from 14-day-old seedlings. The amplification of the soybean EF1 (GmEF1) gene was used as an internal control for data normalization. (B) GmDIR22 expression in soybean leaves infected with P. sojae. Soybean leaves were harvested at 0, 6, 12, 24, 36, 48, and 72 h after infection. The relative GmDIR22 transcript levels were quantified and compared between infected and mock-treated control plants at the same time point. (C) GmDIR22 expression in soybean leaves in response to exogenous hormone: SA (0.5 mM), MeJA (100 μM), ABA (50 mM), and GA3 (50 mg.L-1) treatment for 0, 3, 6, 9, 12, and 24 h. The soybean Actin (GmActin4) gene was used as an internal control for data normalization. Experiments were performed using three biological replicates with three technical replicates each, and statistically analyzed using Student’s t-tests (∗P < 0.05, ∗∗P < 0.01). Bars indicated standard errors of the mean.
Subcellular Localization of the GmDIR22 Protein
The subcellular localization of the GmDIR22 protein was analyzed by expressing a gene encoding a GmDIR22-GFP fusion protein under the control of the 35S promoter in Arabidopsis protoplasts. As shown in Figure 4, the observation by confocal microscopy demonstrated that GFP fluorescence is dispersed throughout the entire cells expressing the control 35S-GFP plasmid. In contrast, the GmDIR22-GFP fusion protein is localized exclusively to the membrane of the Arabidopsis cells, indicating that GmDIR22 is a membrane-localized protein.
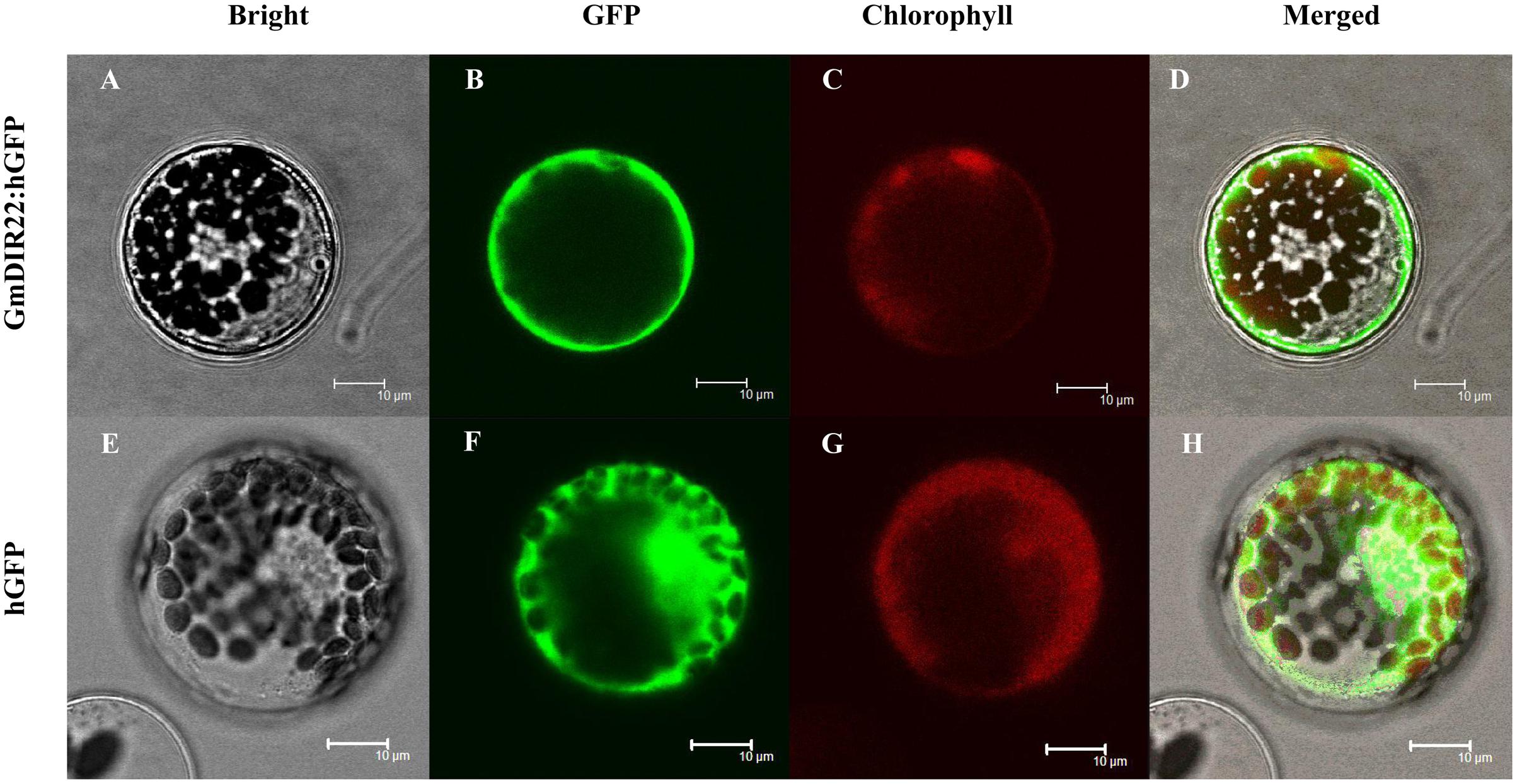
FIGURE 4. Analysis of the subcellular localization of GmDIR22-GFP protein in Arabidopsis protoplasts. GmDIR22-GFP fusion protein expression was driven by the cauliflower mosaic virus 35S promoter and transiently expressed in Arabidopsis protoplasts. Bright-field images (A,E), GFP fluorescence (green) only (B,F), chlorophyll autofluorescence (red) only (C,G), cytoplasmic marker fluorescence localization, and combined images (D,H) are shown. Bars = 10 μm.
The Recombinant GmDIR22 Protein Directs Pinoresinol Formation
The expression of recombinant GmDIR22 protein is remarkably enhanced by adding 0.5 mM IPTG at 37°C and increases from 1 to 6 h (Figure 5A). The maximum expression of the protein is achieved at 4 h. The recombinant protein was purified using His-Bind Kits (EMD Millipore, Billerica, MA, USA), and the molecular weight of the purified protein is determined as approximately 23 kDa by SDS–PAGE (Figure 5A). Furthermore, the dirigent ability of recombinant GmDIR22 to couple monolignols was investigated. As shown in Figures 5B,C, GmDIR22 could direct conversion of E-coniferyl alcohol into lignan (+)-pinoresinol, in the presence of ammonium peroxydisulfate as oxidant. The result is similar to that reported for FiDIR (Davin et al., 1997).
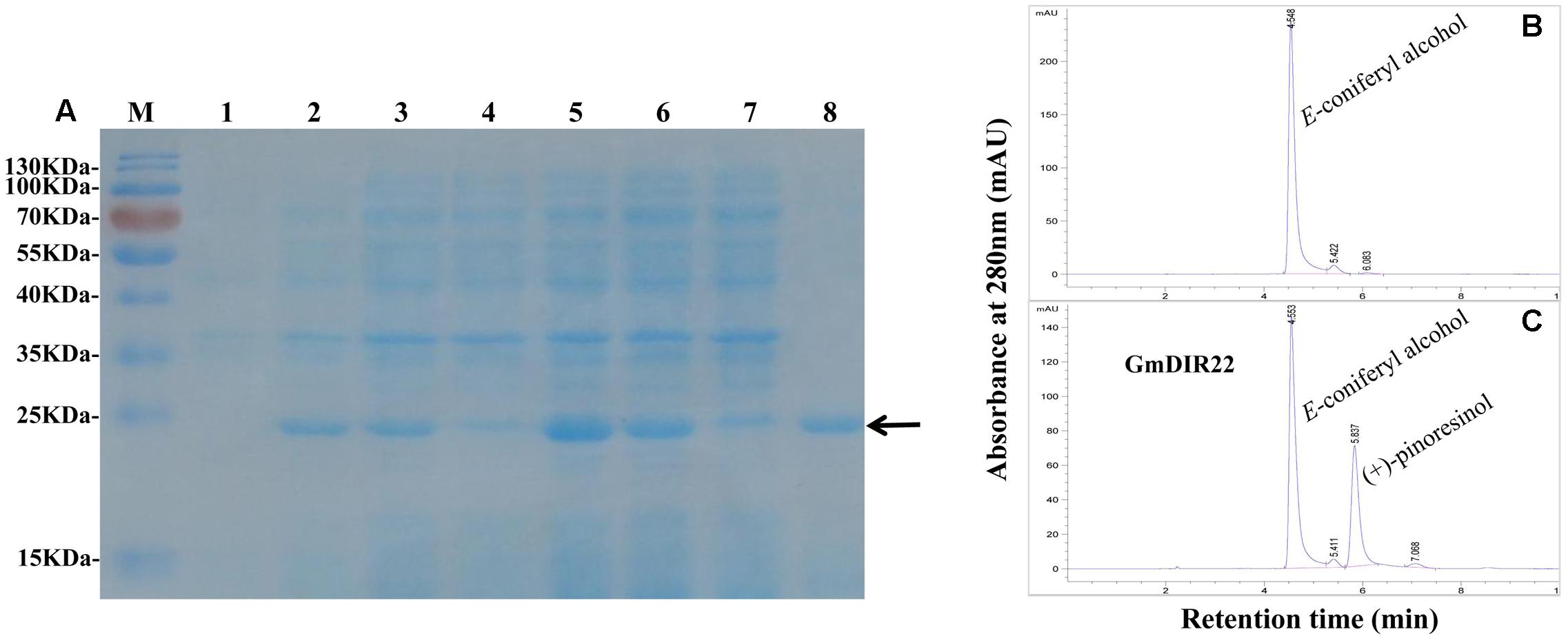
FIGURE 5. Analysis of GmDIR22 dirigent activity by HPLC. (A) Expression of recombinant GmDIR22 protein in E. coli BL21 (DE3) was induced using 0.5 mM IPTG at 37°C for 1–6 h and purified from the soluble fraction of induced cells using resin with affinity for the His-Tag. Lane 1 pET-29b(+) vector induced by IPTG for 3 h, Lane 2–7 pET29b(+)-GmDIR22 induced by IPTG for 1, 2, 3, 4, 5, and 6 h, Lane 8 purified soluble GmDIR22 protein with Nickel-CL agarose affinity chromatography. (B) HPLC chromatogram at 280 nm absorption of E-coniferyl alcohol at retention time 4.5 min. (C) HPLC chromatogram at 280 nm absorption of reaction mixture of E-coniferyl alcohol with purified soluble GmDIR22 protein, the product of (+)-pinoresinol at retention time 5.8 min.
Overexpression of GmDIR22 Can Enhance Soybean Resistance to P. sojae
To investigate whether overexpression of GmDIR22 could enhance resistance to P. sojae in transgenic plants, the T5 transgenic soybean plants were selected by qRT-PCR (Supplementary Table S11) to assay the pathogen response (Figure 6A). The living cotyledons and detached leaves were treated with a P. sojae inoculum. As shown in Figure 6B, the cotyledons of the non-transgenic soybean plants exhibits clearer and larger water-soaked lesions compared with those of the transgenic plants after 72 h of incubation with P. sojae, and the lesion areas are significantly smaller in the transgenic lines than those of non-transgenic soybean plants 72 h after inoculation (P < 0.01) (Figure 6C). After 96 h incubation with P. sojae, the similar results are observed in the detached leaves (Figure 6E), and the lesion areas in transgenic soybean lines were significantly smaller than those in non-transgenic plants (P < 0.01) (Figure 6D). The biomass of P. sojae, based on the transcript levels of the P. sojae TEF1 gene (Supplementary Table S10), was significantly lower in transgenic GmDIR22-overexpressing plants than that in non-transgenics (P < 0.01) (Figure 6F). These data indicated that the over-expression of GmDIR22 in soybean plants in a certain extent improves their resistance to P. sojae.
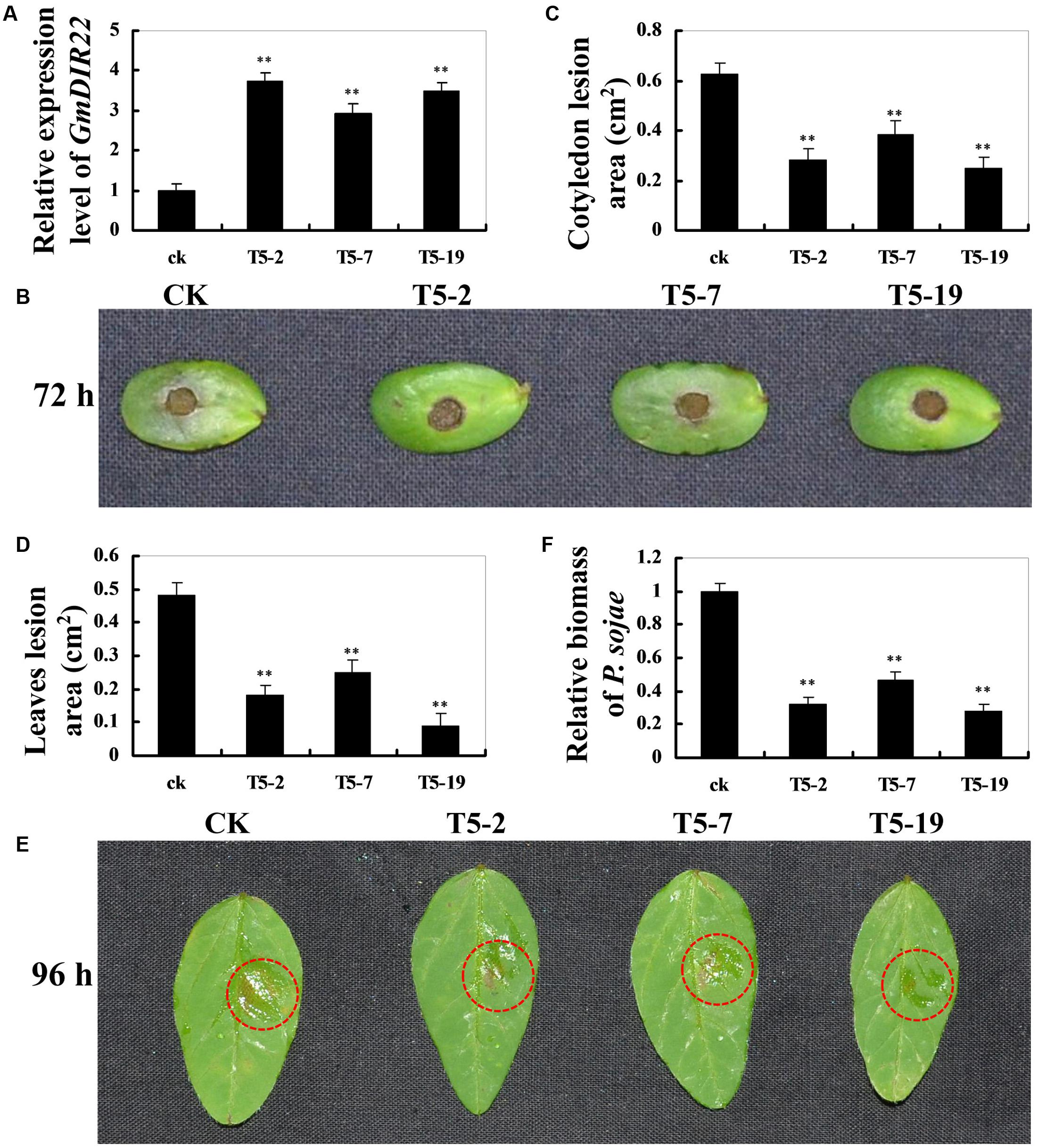
FIGURE 6. Resistance analysis of GmDIR22 transgenic soybean plants to P. sojae infection. (A) The qRT-PCR results demonstrating the relative abundance of GmDIR22 in transgenic soybean plants. (B,E) The disease symptoms on the living cotyledons and detached leaves of transgenic lines treated with P. sojae inoculum at 72 h and 96 h, respectively. (C) The relative lesion area of transgenic soybean cotyledons infected with P. sojae after 72 h of inoculation. (D) The relative lesion area of transgenic soybean leaves infection with P. sojae after 96 h of inoculation. (F) qRT-PCR analysis of P. sojae relative biomass based on the transcript level of the P. sojae TEF1 gene in infected living cotyledons after 72 h of inoculation. Experiments were performed using three biological replicates with three technical replicates each, and statistically analyzed using Student’s t-tests (∗P < 0.05, ∗∗P < 0.01). Bars indicated standard errors of the mean.
Overexpression of GmDIR22 Leads to Increased Lignan Accumulation
The dirigent phenomenon was discovered by Davin et al. (1997), and then Davin and Lewis (2000) studied the coupling of two E-coniferyl alcohol molecules to produce a (+)-pinoresinol as the first step in the biosynthesis of lignans. Since coniferyl and sinapyl alcohols, together with other monolignol p-coumaryl alcohols, are also polymerized into the macromolecule, lignin, there is some overlap in the biosynthetic pathways involved in lignans and lignin production (Davin and Lewis, 2003). Thus, the changes in the DIR expression levels may cause alterations in the lignan and lignin content of soybean plants. As shown in Figure 7A, the relative lignan content of transgenic plants overexpressing GmDIR22 is significantly higher than that of non-transgenic control plants, indicating an important role for GmDIR22 in the synthesis of lignan in soybean. In contrast, the levels of acid-insoluble lignin remain almost unchanged in transgenic plants compared with those of controls (Figure 7B).
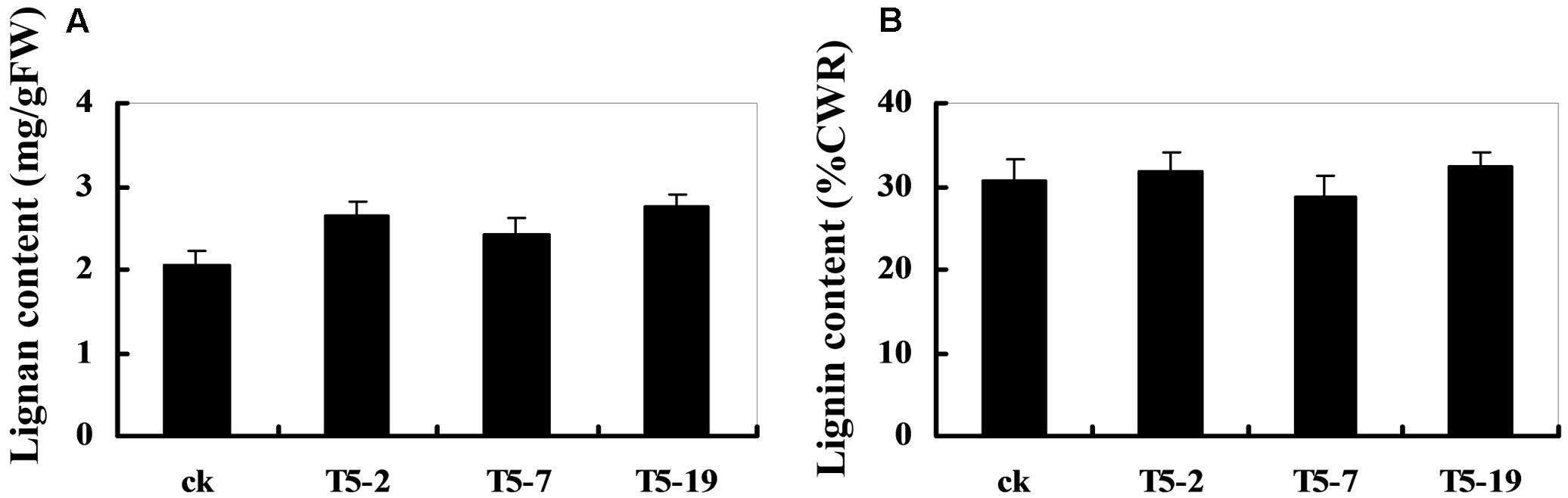
FIGURE 7. Lignan and lignin content in GmDIR22 transgenic lines. (A) Lignan content. (B) Lignin content. Experiments were performed using three biological replicates with three technical replicates each. Bars indicated standard errors of the mean.
Verification of Antimicrobial Effects of Lignan Extracts
To examine the antimicrobial effects of lignan extracts on P. sojae race 1, the filter-paper disks containing lignan extracts from different transgenic plants were placed around a P. sojae colony. After incubation for 72 h, the results showed that the lignan extracts from transgenic plant lines T5-2 and T5-19 inhibit P. sojae hyphal growth, leading to 1 and 2 mm inhibition zones, respectively, indicating that they exhibit an enhanced antimicrobial effect compared to those from non-transgenic plants (Figure 8A). To further examine the effect of lignan extracts on P. sojae, we conducted inhibition assays using an equal amount hyphal plugs of 1-week-old of P. sojae race 1, which were placed in sterilized plates with rich medium, supplemented with varying concentrations of lignans. Hyphal growth was assessed by visual inspection after 7 days, the results confirmed that the antibacterial effect gradually increased with increasing of the concentration of lignans (Figure 8B). These results demonstrated that lignans from soybean possess antimicrobial activity against P. sojae.
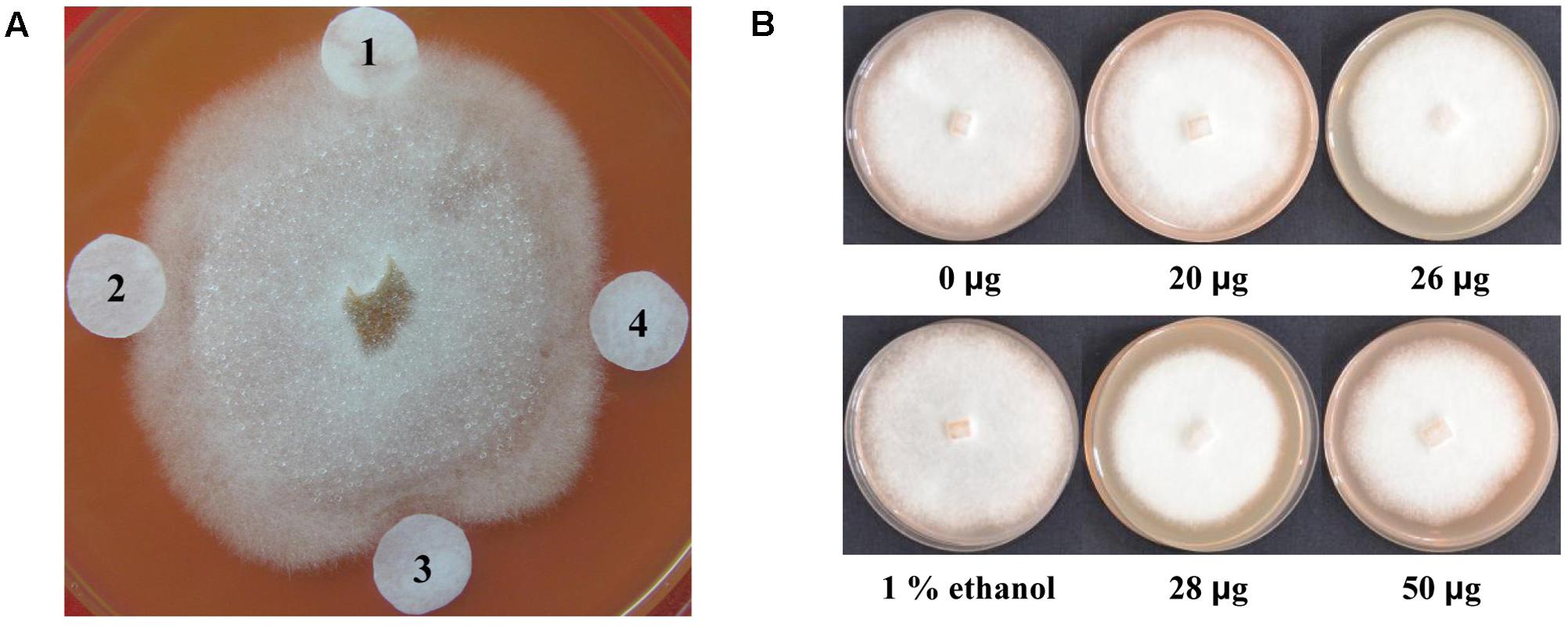
FIGURE 8. Antimicrobial activity of lignan extracts. (A) Inhibition of P. sojae race 1 growth by lignan extracts. (1) Ethanol buffer; (2) lignan extracts (20 μg) from a non-transgenic control plant; (3) lignan extracts (28 μg) from the T5-19 transgenic plant line; (4) lignan extracts (26 μg) from the T5-2 transgenic plant line. (B) Growth inhibition assay of P. sojae race 1 after 7 days of exposure of hyphal plugs to varying concentrations of lignan extracts. The experiment was performed three times.
Discussion
Dirigent proteins were first identified in Forsythia (Davin et al., 1997) and are considered to be responsible for an important enzymatic reaction in the production of lignan and lignin, which evolved during the adaptation of aquatic plants to terrestrial environments (Kim et al., 2002). In addition, this gene family has primary roles in defense responses, secondary metabolism, and fiber biosynthesis (Davin and Lewis, 2000; Burlat et al., 2001; Ralph et al., 2007). However, little is known about dirigent and dirigent-like proteins and their biological functions in soybean, especially the likely effect on P. sojae resistance in soybean. In this study, a novel dirigent gene, GmDIR22, that plays a positive role in soybean during infection with P. sojae. Here, we isolated and characterized the GmDIR22 gene and our findings contributed to improved understanding of the function of dirigent genes in defense responses to P. sojae and secondary metabolism.
Dirigent proteins are members of a large family, initially subdivided into five subfamilies (DIR-a, DIR-b, DIR-c, DIR-d, and DIR-e) (Ralph et al., 2006). With the increasing numbers of DIR proteins, the DIR-b and DIR-d subfamilies are combined together with the appearance of the DIR-f and DIR-g subfamilies (Ralph et al., 2007). The phylogenetic analysis demonstrated that GmDIR22 is close to five AtDIR members that had been confirmed belonging to DIR-b/d subfamily (Ralph et al., 2007), indicating that GmDIR22 might display similar functions with DIR-b/d. Many members of which are indicated to play a role in plants responses to biotic and abiotic stresses, including H2O2, NaCl, and PEG (Guo et al., 2012), SA, GA, and MeJA (Damaj et al., 2010). SA and JA are signaling molecules associated with pathogens, insect and mechanical damage (wound) resistance. The apparent upregulation of many DIR-b/d genes in response to attacks by fungi (Williams et al., 2002; Zhu et al., 2007) and insects (Ralph et al., 2007) are of particular interest. Moreover, they have important roles in plant secondary metabolism, especially contribute to lignin formation (Zhu et al., 2007; Ma and Liu, 2015). The expression pattern determined that the transcript levels of GmDIR22 was higher in resistant cultivars than that in susceptible cultivars with P. sojae infection. The result indicated that GmDIR22 may contribute to naturally occurring resistance to the pathogen. Moreover, GmDIR22 mRNA transcripts are strongly induced by GA3 stress; therefore, we speculate that GmDIR22 may have an important role in soybean plant resistance to P. sojae, primarily through GA3 signaling, which is an important signal transduction component involved in activation of plant defense responses against pathogen attack (Mellersh et al., 2002). Future studies are required to determine how this process may be regulated by GA3 signaling.
Dirigent proteins proteins are involved in plant defenses against pathogens and are proposed to mediate the free radical coupling of monolignol plant phenols to yield the cell wall polymers, lignans and/or lignin-like compounds (Espiñeira et al., 2011; Alvarez et al., 2016). In this study, the soybean plants overexpressing the GmDIR22 gene exhibit increased lignan accumulation, compared with controls. In contrast, the levels of acid-insoluble lignin remain almost unchanged in transgenic plants relative to those in controls. We propose two possible explanations for this result: first, GmDIR22 protein may direct the formation of lignan precursors, and is mainly, if not completely, involved in lignan biosynthesis; second, in the defense against pathogen invasion, the GmDIR22 dirigent protein may induce production of a lignin-like defense barrier, consisting of suberized or lignified metabolites associated with specialized phloem parenchyma cells or resin ducts, rather than being directly involved in the production of lignin. This is conceivable when spatial separation of lignan and lignin synthesis systems is considered, and the former occurs in the cytoplasm, while the latter takes place in the cell wall compartment (Davin et al., 2008; Bonawitz and Chapple, 2010). The subcellular localization of the GmDIR22 protein was investigated, and the results indicated that GmDIR22 is a membrane protein, suggesting that the expression of this protein in the membrane would probably be conducive when receiving stimulus signals and generate corresponding responses.
Lignans are phenylpropanoid dimers, synthesized via the phenylpropanoid pathway (Davin and Lewis, 2003). The precursor of lignan is pinoresinol, which is derived from E-coniferyl alcohol through the action of DIR (Burlat et al., 2001), and there is overlap between the biosynthetic pathways producing lignin and lignans (Davin and Lewis, 2003). They occur naturally in a number of plant families and are thought to have important physiological and ecological roles in interactions with insects and pathogens, due to their antifeedant activities and antibacterial (Wang et al., 1999; Harmatha and Dinan, 2003; Schroeder et al., 2006). Lignans have been studied extensively and are reported to possess a number of biological activities, including anti-inflammatory, antimicrobial, antioxidant, and anti-estrogenic properties, which may reduce the risk of cardiovascular diseases, as well as certain types of cancer (Adlercreutz and Mazur, 1997; Raffaelli et al., 2002; Arts and Hollman, 2005; Saleem et al., 2005).
In this study, lignan extracts from GmDIR22 transgenic plants exhibit a certain effect inhibition on P. sojae hyphal growth. Furthermore, we determined that the recombinant GmDIR22 protein could effectively direct E-coniferyl alcohol coupling into (+)-pinoresinol, suggesting that overexpression of soybean GmDIR22 is likely to promote the synthesis of lignans, leading to enhanced resistance to P. sojae in soybean to a certain degree. The results of analysis of lignan extracts on the inhibition of P. sojae hyphal growth and the living cotyledons and detached leaves of transgenic soybean plants enhancing resistance to P. sojae also support this hypothesis.
Authors Contributions
Conceived and designed the experiments: PX and SZ. Performed the experiments: NL, MZ, and TL. Analyzed the data: NL, LD, QC, JW, LW, XC, CZ, and WL. Contributed reagents/materials/analysis tools: SZ and PX.
Funding
This work was supported by NSFC Projects (31171577, 31671719), Outstanding Talents and Innovative Team of Agricultural Scientific Research, Young and Middle-aged scientific and Technological innovation leader (MOST), Special Fund for Agro-scientific Research in the Public Interest (201303018), Natural Science Foundation of Heilongjiang Province (JC201308, C2015010), Changjiang Scholar Candidates Program for Provincial Universities in Heilongjiang (2013CJHB003).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01185/full#supplementary-material
FIGURE S1 | Nucleotide and amino acid sequence of GmDIR22.
FIGURE S2 | The predicted three-dimensional structure of GmDIR22.
FIGURE S3 | Alignment of the nucleotide sequences of GmDIR22 and the nearby 5 AtDIR.
Footnotes
- ^ http://www.ncbi.nlm.nih.gov/blast
- ^ http://smart.embl-heidelberg.de/
- ^ http://www.lynnon.com/
- ^ http://www.megasoftware.net
- ^ http://www.sbg.bio.ic.ac.uk/phyre2
References
Adlercreutz, H., and Mazur, W. (1997). Phyto-oestrogens and western diseases. Ann. Med. 29, 95–120. doi: 10.3109/07853899709113696
Alvarez, A., Montesano, M., Schmelz, E., and León, I. P. D. (2016). Activation of shikimate, phenylpropanoid, oxylipins, and auxin pathways in pectobacterium carotovorum elicitors-treated Moss. Front. Plant Sci. 7:328. doi: 10.3389/fpls.2016.00328
Arts, I. C. W., and Hollman, P. C. H. (2005). Polyphenols and disease risk in epidemiologic studies. Am. J. Clin. Nutr. 81, 317–325.
Ayres, D. C., and Loike, J. D. (1990). Lignans: Chemical, Biological and Clinical Properties. Cambridge: Cambridge University Press.
Berim, A., Ebel, R., Schneider, B., and Petersen, M. (2008). UDP-glucose: (6-methoxy) podophyllotoxin 7-O-glucosyltransferase from suspension cultures of Linum nodiflorum. Phytochemistry 69, 374–381. doi: 10.1016/j.phytochem.2007.07.030
Bonawitz, N. D., and Chapple, C. (2010). The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu. Rev. Genet. 44, 337–363. doi: 10.1146/annurev-genet-102209-163508
Burlat, V., Kwon, M., Davin, L. B., and Lewis, N. G. (2001). Dirigent proteins and dirigent sites in lignifying tissues. Phytochemistry 57, 883–897. doi: 10.1016/S0031-9422(01)00117-0
Chacón, O., González, M., López, Y., Portieles, R., Pujol, M., González, E., et al. (2010). Over-expression of a protein kinase gene enhances the defense of tobacco against Rhizoctonia solani. Gene 452, 54–62. doi: 10.1016/j.gene.2009.11.011
Chang, C. W., Lin, M. T., Lee, S. S., Chen Liu, K. C. S., Hsu, F. L., and Lin, J. Y. (1995). Differential inhibition of reverse transcriptase and cellular DNA polymerase-α activities by lignans isolated from the Chinese herbs, Phyllanthus myrtifolius Moon, and tannins from Lonicera japonica Thunb and Castanopsis hystrix. Antiviral Res. 27, 367–374. doi: 10.1016/0166-3542(95)00020-M
Chen, Y., Shi, Z. W., and Huang, X. M. (2010). Ultrasonic assisted extraction of total lignans and its content analysis in soybeans. Soybean Sci. 29, 168–170.
Dalisay, D. S., Kim, K. W., Lee, C., Yang, H., Rübel, O., Bowen, B. P., et al. (2015). Dirigent protein-mediated lignan and cyanogenic glucoside formation in flax seed: integrated omics and MALDI mass spectrometry imaging. J. Nat. Prod. 78, 1231–1242. doi: 10.1021/acs.jnatprod.5b00023
Damaj, M. B., Kumpatla, S. P., Emani, C., Beremand, P. D., Avutu, S., Reddy, A. S., et al. (2010). Sugarcane DIRIGENT and O-methyltransferase promoters confer stem-regulated gene expression in diverse monocots. Planta 231, 1439–1458. doi: 10.1007/s00425-010-1138-5
Davin, L. B., Jourdes, M., Patten, A. M., Kim, K. W., Vassao, D. G., and Lewis, N. G. (2008). Dissection of lignin macromolecular configuration and assembly: comparison to related biochemical processes in allyl/propenyl phenol and lignan biosynthesis. Nat. Prod. Rep. 25, 1015–1090. doi: 10.1039/b510386j
Davin, L. B., and Lewis, N. G. (2000). Dirigent proteins and dirigent sites explain the mystery of specificity of radical precursor coupling in lignan and lignin biosynthesis. Plant Physiol. 123, 453–462. doi: 10.1104/pp.123.2.453
Davin, L. B., and Lewis, N. G. (2003). An historical perspective on lignan biosynthesis: monolignol, allylphenol and hydroxycinnamic acid coupling and downstream metabolism. Phytochem. Rev. 2, 257–288. doi: 10.1023/b:phyt.0000046175.83729.b5
Davin, L. B., and Lewis, N. G. (2005). Dirigent phenoxy radical coupling: advances and challenges. Curr. Opin. Biotechnol. 16, 398–406. doi: 10.1016/j.copbio.2005.06.010
Davin, L. B., Wang, H. B., Crowell, A. L., Bedgar, D. L., Martin, D. M., Sarkanen, S., et al. (1997). Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science 275, 362–366. doi: 10.1126/science.275.5298.362
Dou, D. L., Wang, B. S., Zhu, S. W., Tang, Y. X., Wang, Z. X., Sun, J. S., et al. (2003). Transgenic tobacco with NDR1 gene improved its resistance to two fungal disease. Sci. Agric. Sin. 36, 1120–1124.
Effenberger, I., Zhang, B., Li, L., Wang, Q., Liu, Y., Klaiber, I., et al. (2015). Dirigent proteins from cotton (Gossypium sp.) for the atropselective synthesis of gossypol. Angew. Chem. Int. Ed. Engl. 54, 14660–14663. doi: 10.1002/anie.201507543
Espiñeira, J. M., Novo, U. E., Gómez Ros, L. V., Carrión, J. S., Merino, F., Ros Barceló, A., et al. (2011). Distribution of lignin monomers and the evolution of lignification among lower plants. Plant Biol. 13, 59–68. doi: 10.1111/j.1438-8677.2010.00345.x
Fehr, W. R., Caviness, C. E., Burmood, D. T., and Pennington, J. (1971). Stage of development descriptions for soybeans, Glycine max (L.) Merrill. Crop Sci. 11, 929–931. doi: 10.2135/cropsci1971.0011183X001100060051x
Giannakopoulou, A., Schornack, S., Bozkurt, T. O., Haart, D., Ro, D. K., Faraldos, J. A., et al. (2014). Variation in capsidiol sensitivity between Phytophthora infestans and Phytophthora capsici is consistent with their host range. PLoS ONE 9:e107462. doi: 10.1371/journal.pone.0107462
Guo, J. L., Xu, L. P., Fang, J. P., Su, W. C., Fu, H. Y., Que, Y. X., et al. (2012). A novel dirigent protein gene with highly stem-specific expression from sugarcane, response to drought, salt and oxidative stresses. Plant Cell Rep. 31, 1801–1812. doi: 10.1007/s00299-012-1293-1
Halls, S. C., Davin, L. B., Kramer, D. M., and Lewis, N. G. (2004). Kinetic study of coniferyl alcohol radical binding to the (+)-pinoresinol forming dirigent protein. Biochemistry 43, 2587–2595. doi: 10.1021/bi035959o
Harmatha, J., and Dinan, L. (2003). Biological activities of lignans and stilbenoids associated with plant-insect chemical interaction. Phytochem. Rev. 2, 321–330. doi: 10.1023/B:PHYT.0000045494.98645.a3
Holsters, M., Waele, D., Depicke, R. A., Messens, E., Montagu, M., and Schell, J. (1978). Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 163, 181–187. doi: 10.1007/BF00267408
Hosmani, P. S., Kamiya, T., Danku, J., Naseer, S., Geldner, N., Guerinot, M. L., et al. (2013). Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc. Natl. Acad. Sci. U.S.A. 110, 14498–14503. doi: 10.1073/pnas.1308412110
Kim, H. J., Lim, J. S., Kim, W. K., and Kim, J. S. (2012). Soybean glyceollins: biological effects and relevance to human health. Proc. Nutr. Soc. 71, 166–174. doi: 10.1017/S0029665111003272
Kim, K. W., Moinuddin, S. G. A., Atwell, K. M., Costa, M. A., Davin, L. B., and Lewis, N. G. (2012). Opposite stereoselectivities of dirigent proteins in Arabidopsis and Schizandra species. J. Biol. Chem. 287, 33957–33972. doi: 10.1074/jbc.M112.387423
Kim, K. W., Smith, C. A., Daily, M. D., Cort, J. R., Davin, L. B., and Lewis, N. G. (2015). Trimeric structure of (+)-pinoresinol-forming dirigent protein at 1.95 Å resolution with three isolated active sites. J. Biol. Chem. 290, 1308–1318. doi: 10.1074/jbc.M114.611780
Kim, M. K., Jeon, J. H., Fujita, M., Davin, L. B., and Lewis, N. G. (2002). The western red cedar (Thuja plicata) 8-8’ DIRIGENT family displays diverse expression patterns and conserved monolignol coupling specificity. Plant Mol. Biol. 49, 199–214. doi: 10.1023/A:1014940930703
Kirk, T. K., and Obst, J. R. (1988). Lignin determination. Methods Enzymol. 161, 87–101. doi: 10.1016/0076-6879(88)61014-7
Lange, B. M., Lapierre, C., and Sandermann, H. Jr. (1995). Elicitor-induced spruce stress lignin (structural similarity to early developmental lignins). Plant Physiol. 108, 1277–1287. doi: 10.1104/pp.108.3.1277
Lewis, N. G., and Davin, L. B. (1998). The biochemical control of monolignol coupling and structure during lignan and lignin biosynthesis. ACS Symp. Ser. 22, 334–361. doi: 10.1021/bk-1998-0697.ch022
Lewis, N. G., and Davin, L. B. (1999). Lignans: biosynthesis and function. Nat. Prod. Chem. 1, 639–712.
Lewis, N. G., Davin, L. B., and Sarkanen, S. (1999). The nature and function of lignins. Nat. Prod. Chem. 3, 618–745. doi: 10.1016/B978-0-08-091283-7.00085-0
Lewis, N. G., and Yamamoto, E. (1990). Lignin: occurrence, biosynthesis and biodegradation. Plant Mol. Biol. 41, 455–496. doi: 10.1146/annurev.pp.41.060190.002323
Liu, J., Stipanovic, R. D., Bell, A. A., Puckhaber, L. S., and Magill, C. W. (2008). Stereoselective coupling of hemigossypol to form (+)-gossypol in moco cotton is mediated by a dirigent protein. Phytochemistry 69, 3038–3042. doi: 10.1016/j.phytochem.2008.06.007
Lu, X., Li, J., Jiang, X. J., Zhang, K. F., and Duan, X. Q. (2011). Content determination of lignans in Kadsura coccinea (Lem.) A. C. Smith. J. Anhui. Agri. Sci. 39, 12152–12153.
Lundberg, W. O., Halvorson, H. O., and Burr, G. O. (1944). The antioxidant properties of nordihydroguaiaretic acid. Oil Soap 21, 33–35. doi: 10.1007/BF02593156
Ma, Q. H., and Liu, Y. C. (2015). TaDIR13, a dirigent protein from wheat, promotes lignan biosynthesis and enhances pathogen resistance. Plant Mol. Biol. Rep. 33, 143–152. doi: 10.1007/s11105-014-0737-x
MacRae, W. D., and Towers, G. H. N. (1984). Biological activities of lignans. Phytochemistry 23, 1207–1220. doi: 10.1016/S0031-9422(00)80428-8
Magalhaes, J. C. M. S., Valencise Bonine, C. A. V., Dornelas, M. C., and Mazzafera, P. (2010). Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 52, 360–376. doi: 10.1111/j.1744-7909.2010.00892.x
Matsui, K., and Munakata, K. (1975). The structure of piperenone, a new insect antifeedant substance from Piper futokadzura. Tetrahedron Lett. 24, 1905–1908. doi: 10.1016/S0040-4039(00)72318-5
Mellersh, D. G., Foulds, I. V., Higgins, V. J., and Heath, M. C. (2002). H2O2 plays different roles in determining penetration failure in three diverse plant-fungal interactions. Plant J. 29, 257–268. doi: 10.1046/j.0960-7412.2001.01215.x
Nakatani, N., Ikeda, K., Kikuzaki, H., Kido, M., and Yamaguchi, Y. (1988). Diaryldimethylbutane lignans from Myristica argentea and their antimicrobial action against Streptococcus mutans. Phytochemistry 27, 3127–3129. doi: 10.1016/ochem.2008.06.007
Önnerud, H., Zhang, L., Gellerstedt, G., and Henriksson, G. (2002). Polymerization of monolignols by redox shuttle-mediated enzymatic oxidation: a new model in lignin biosynthesis I. Plant Cell 14, 1953–1962. doi: 10.1105/tpc.001487
Osawa, T., Nagata, M., Namiki, M., and Fukuda, Y. (1985). Sesamolinol, a novel antioxidant isolated from sesame seeds. Agric. Biol. Chem. 49, 3351–3352. doi: 10.1080/00021369.1985.10867272
Pauletti, P. M., Araujo, A. R., Young, M. C. M., Giesbrecht, A. M., and Bolzani, V. (2000). Nor-Lignans from the leaves of Styrax ferrugineus (Styracaceae) with antibacterial and antifungal activity. Phytochemistry 55, 597–601. doi: 10.1016/S0031-9422(00)00225-9
Paz, M. M., Shou, H., Guo, Z., Zhang, Z., Banerjee, A. K., and Wang, K. (2004). Assessment of conditions affecting Agrobacterium-mediated soybean transformation using the cotyledonary node explants. Euphytica 136, 167–179. doi: 10.1016/S0168-7972(00)80013-4
Pickel, B., Constantin, M. A., Pfannstiel, J., Conrad, J., Beifuss, U., and Schaller, A. (2010). An enantiocomplementary dirigent protein for the enantioselective laccase-catalyzed oxidative coupling of phenols. Angew. Chem. Int. Ed. Engl. 49, 202–204. doi: 10.1002/anie.200904622
Pomar, F., Merino, F., and Barceló, A. R. (2002). O-4-Linked coniferyl and sinapyl aldehydes in lignifying cell walls are the main targets of the Wiesner (phloroglucinol-HCl) reaction. Protoplasma 220, 17–28. doi: 10.1007/s00709-002-0030-y
Raffaelli, B., Hoikkala, A., Leppälä, E., and Wähälä, K. (2002). Enterolignans. J. Chromatogr. B. 777, 29–43. doi: 10.1016/S1570-0232(02)00092-2
Ralph, S., Park, J. Y., Bohlmann, J., and Mansfield, S. D. (2006). Dirigent proteins in conifer defense: gene discovery, phylogeny, and differential wound-and insect-induced expression of a family of DIR and DIR-like genes in spruce (Picea spp). Plant Mol. Biol. 60, 21–40. doi: 10.1007/s11103-005-2226-y
Ralph, S. G., Jancsik, S., and Bohlmann, J. (2007). Dirigent proteins in conifer defense II: extended gene discovery, phylogeny, and constitutive and stress-induced gene expression in spruce (Picea spp). Phytochemistry 68, 1975–1991. doi: 10.1016/j.phytochem.2007.04.042
Reboledo, G., Del Campo, R., Alvarez, A., Montesano, M., Mara, H., and Ponce de León, I. (2015). Physcomitrella patens activates defense responses against the pathogen Colletotrichum gloeosporioides. Int. J. Mol. Sci. 16, 22280–22298. doi: 10.3390/ijms160922280
Saleem, M., Kim, H. J., Ali, M. S., and Lee, Y. S. (2005). An update on bioactive plant lignans. Nat. Prod. Rep. 22, 696–716. doi: 10.1039/b514045p
Schlumbaum, A., Mauch, F., Vogeli, U., and Boller, T. (1986). Plant chitinases are potent inhibitors of fungal growth. Nature 324, 365–367. doi: 10.1038/324365a0
Schroeder, F. C., Del Campo, M. L., Grant, J. B., Weibel, D. B., Smedley, S. R., Bolton, K. L., et al. (2006). Pinoresinol: a lignol of plant origin serving for defense in a caterpillar. Proc. Natl. Acad. Sci. U.S.A. 103, 15497–15501. doi: 10.1073/pnas.0605921103
Seneviratne, H. K., Dalisay, D. S., Kim, K. W., Moinuddin, S. G., Yang, H., Hartshorn, C. M., et al. (2015). Non-host disease resistance response in pea (Pisum sativum) pods: biochemical function of DRR206 and phytoalexin pathway localization. Phytochemistry 113, 140–148. doi: 10.1016/j.phytochem.2014.10.013
Tiburzy, R., and Reisener, H. J. (1990). Resistance of wheat to Puccinia graminis f. sp. tritici: association of the hypersensitive reaction with the cellular accumulation of lignin-like material and callose. Physiol. Mol. Plant Pathol. 36, 109–120. doi: 10.1016/0885-5765(90)90100-C
Tyler, B. M. (2007). Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 8, 1–8. doi: 10.1111/j.1364-3703.2006.00373.x
Wang, Y., Nowak, G., Culley, D., Hadwiger, L. A., and Fristensky, B. (1999). Constitutive expression of pea defense gene DRR206 confers resistance to blackleg (Leptosphaeria maculans) disease in transgenic canola (Brassica napus). Mol. Plant Microbe Interact 12, 410–418.
Ward, E. W. B., Lazarovits, G., Unwin, C. H., and Buzzell, R. I. (1979). Hypocotyl reactions and glyceollin in soybeans inoculated with zoospores of Phytophthora megaspuma var. sojae. Phytopathology 69, 951–955. doi: 10.1094/Phyto-69-951
Williams, C. E., Collier, C. C., Nemacheck, J. A., Liang, C., and Cambron, S. E. (2002). A lectin-like wheat gene responds systemically to attempted feeding by avirulent first-instar Hessian fly larvae. J. Chem. Ecol. 28, 1411–1428. doi: 10.1023/A:1016200619766
Wrather, J. A., Anderson, T. R., Arsyad, D. M., Tan, Y., Ploper, L. D., Porta-Puglia, A., et al. (1997). Soybean disease loss estimates for the top 10 soybean producing countries in 1998. Plant Dis. 81, 107–110. doi: 10.1094/PDIS.1997.81.1.107
Wu, R. H., Wang, L. L., Wang, Z., Shang, H. H., Liu, X., Zhu, Y., et al. (2009). Cloning and expression analysis of a dirigent protein gene from the resurrection plant Boea hygrometrica. Prog. Nat. Sci. 19, 347–352. doi: 10.3724/sp.j.1259.2012.00044
Xu, P. F., Chen, W. Y., Lv, H. Y., Fan, S. J., Wang, X., Jiang, L. Y., et al. (2012). Differentially expressed genes of soybean during infection by Phytophthora sojae. J. Integr. Agric. 11, 368–377.
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, S. Z., Xu, P. F., Wu, J. J., Allen, X., Zhang, J. X., Li, W. B., et al. (2010). Races of Phytophthora sojae and their virulences on commonly grown soybean varieties in Heilongjiang, China. Plant Dis. 94, 87–91. doi: 10.1094/PDIS-94-1-0087
Keywords: Glycine max, dirigent protein, lignan, Phytophthora sojae, gene expression
Citation: Li N, Zhao M, Liu T, Dong L, Cheng Q, Wu J, Wang L, Chen X, Zhang C, Lu W, Xu P and Zhang S (2017) A Novel Soybean Dirigent Gene GmDIR22 Contributes to Promotion of Lignan Biosynthesis and Enhances Resistance to Phytophthora sojae. Front. Plant Sci. 8:1185. doi: 10.3389/fpls.2017.01185
Received: 22 April 2017; Accepted: 21 June 2017;
Published: 04 July 2017.
Edited by:
Stanislav Kopriva, University of Cologne, GermanyReviewed by:
Roger W. Innes, Indiana University Bloomington, United StatesMark Gijzen, Agriculture and Agri-Food Canada, Canada
Copyright © 2017 Li, Zhao, Liu, Dong, Cheng, Wu, Wang, Chen, Zhang, Lu, Xu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pengfei Xu, xupengfei@neau.edu.cn Shuzhen Zhang, zhangshuzhen@neau.edu.cn
†These authors have contributed equally to this work.
 Ninghui Li1,2†
Ninghui Li1,2† Pengfei Xu
Pengfei Xu Shuzhen Zhang
Shuzhen Zhang